Abstract
Humoral immunity involves multiple checkpoints that occur in B cell development, maturation, and activation. The pre–B-cell receptor (pre-BCR) is expressed following the productive recombination of the immunoglobulin heavy-chain gene, and sSignalsing through the pre-BCR are required for the differentiation of pre–B cells into immature B cells. However, the molecular mechanisms controlling the pre-BCR expression and signaling strength remain undefined. Herein, we probed the role of the endoplasmic reticulum–associated, stress-activated E3 ubiquitin ligase HMG-CoA reductase degradation 1 (Hrd1) in B cell differentiation. Using mice with a specific Hrd1 deletion in pro–B cells and subsequent B cell developmental stages, we showed that the E3 ubiquitin ligase Hrd1 governs a critical checkpoint during B cell development. We observed that Hrd1 is required for degradation of the pre-BCR complex during the early stage of B cell development. As a consequence, loss of Hrd1 in the B cell lineage resulted in increased pre-BCR expression levels and a developmental defect in the transition from large to small pre–B cells. This defect, in turn, resulted in reduced fewer mature B cells in bone marrow and peripheral lymphoid organs. Our results revealed a novel critical role of Hrd1 in controlling a critical checkpoint in B cell–mediated immunity and suggest that Hrd1 may functioning as an E3 ubiquitin ligase of the pre-BCR complex.
Keywords: lymphocyte, unfolded protein response (UPR), ubiquitin ligase, ubiquitin, protein turnover, pre-BCR
Introduction
B cell development and activation involves several checkpoints, which are important to orchestrate and balance survival and apoptotic signals and control the quality and size of the B cell compartment (1, 2). The earliest steps in B cell development occur in the bone marrow, where assembly of the pre–B cell receptor (pre-BCR)4 followed by the mature BCR occurs in distinct stages. The μ heavy chain (μHC) is first expressed in pro–B cells, and then pairs with the invariant VpreB and λ5 components of the surrogate light chain to form the pre-BCR complex and mark the transition from pro–B cells to pre–B cells (3, 4). Large pre–B cells undergo rapid proliferation before transitioning into resting small pre–B cells, where rearrangement of light chains and assembly of the mature BCR occurs (5, 6). Once the mature BCR is expressed, B lymphocytes then progress into the immature B cell stage, where further selection checkpoints occur before the immature B cells transition to the periphery and mature into circulating B cells (2, 7). Interrupting any stage of pre-BCR assembly, down-regulation, and subsequent BCR assembly leads to a developmental block and ultimately apoptosis of developing B cells (5, 6, 8). Unlike mature BCR, which is expressed primarily on the cell surface, pre-BCR is primarily retained in the endoplasmic reticulum (ER) (9). It has been shown in cell lines that pre-BCR and BCR can be shuttled from the ER to the endolysosomal compartment for degradation (10, 11). However, it is unclear whether degradation of the pre-BCR or BCR within the ER has a physiological role in B cell development.
Hrd1 is one of the ER-associated E3 ubiquitin ligases involved in ER stress-associated protein degradation (ERAD), which is situated in the ER along with Sel1, Derlin1, Herp, and other proteins as part of an ERAD complex (12, 13). During ERAD, misfolded proteins within the ER bind to chaperones such as BiP, and are retro-translocated into the cytosol to be ubiquitinated by the cytosolic RING domain of Hrd1 (12). Although Hrd1 is required for the clearance of misfolded proteins during ERAD, emerging evidence suggests that Hrd1 can also regulate cellular functions by controlling the availability of specific proteins, such as Nrf2, Blimp1, and PGC-1β (14–16). In addition, the ER master chaperone gp96 and heat shock proteins play important roles in both T and B cell development and functions (17–30). Therefore, in addition to the protein folding and misfolded protein responses, the ER stress pathway govern a variety of critical biological functions. In this paper, we report that Hrd1 plays a crucial role in the B cell developmental checkpoint by down-regulating pre-BCR in large pre–B cells to support further differentiation into mature B cells, which defines novel Hrd1 functions in B cell biology independent of ER stress response.
Results
Loss of Hrd1 in CD19+ cells leads to accumulation of large pre–B cells and reduces viability of later B cell development stages
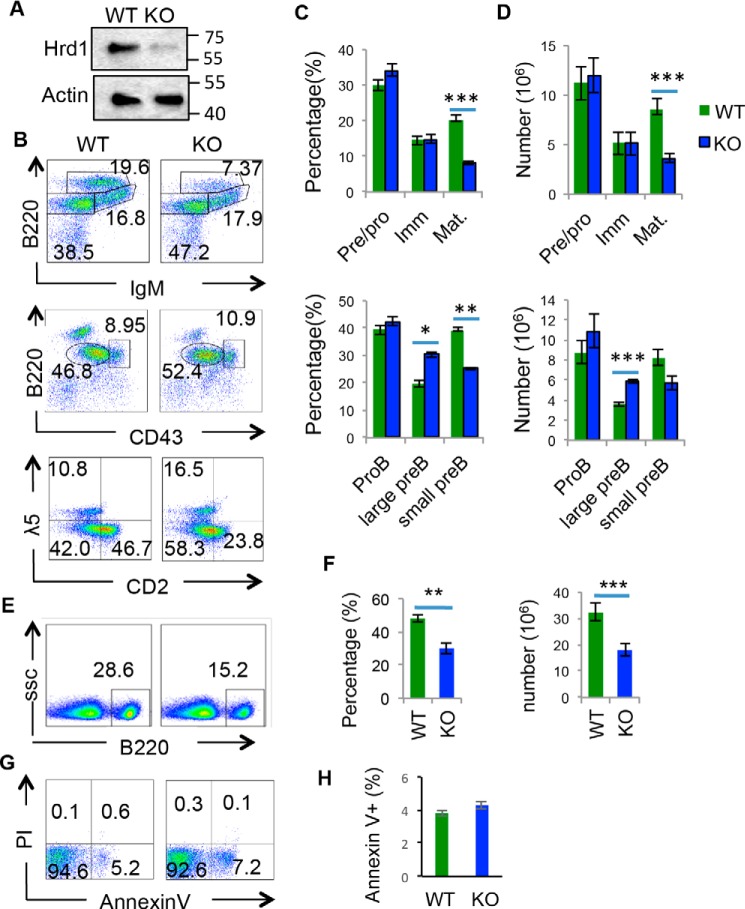
To investigate the function of Hrd1 in B cells, mice bearing loxP-flanked Hrd1 allele (Hrd1f/f) were crossed with mice expressing CD19 promoter-driven Cre recombinase (CD19-Cre) to generate mice with a conditional deletion of Hrd1 in pro–B cells and subsequent B cell developmental stages. Hrd1 protein was deleted in B220+ bone marrow cells of Hrd1 KO mice as confirmed by Western blotting (Fig. 1A). Flow cytometry analysis revealed that deletion of Hrd1 resulted in a significantly reduced number of B220HIIgM+ mature B cells in the bone marrow (Fig. 1, B–D). Consequently, peripheral B220+ cells were reduced in the spleen (Fig. 1, E and F). These results indicate that loss of Hrd1 in CD19-expressing cells resulted in loss of mature B cells and mature B cells in the periphery lymphoid organ.
Figure 1.
Deletion of Hrd1 leads to a developmental block at the large pre–B cell stage and reduced viability of immature and mature B cells. A, B220+ cells were isolated from bone marrow of WT and Hrd1 KO (KO) mice and immunoblotted for Hrd1 protein. β-Actin protein served as loading control. B, bone marrow isolated from WT and Hrd1 KO (KO) mice was analyzed for B220LOIgM− pro–B and pre–B cells, B220LOIgM+ immature B cells, and B220HIIgM+ mature B cells (top panel). B220LOIgM− populations were further analyzed for B220+CD43HI pro–B and B220+CD43LO pre–B cells (middle panel). B220LOIgM− cells were also analyzed for CD2− icλ5− pro–B, CD2− icλ5+ large pre–B, and CD2+ icλ5− small pre–B cells (bottom panel). C, percentages of B cell development stages by B220 and IgM gating (top panel) and icλ5 and CD2 gating (bottom panel). D, absolute number of B cell stages as analyzed in B. E, splenic B cell populations in WT and KO mice were analyzed by B220 expression. F, percentages (left) and absolute number (right) of B220+ cells in spleen. G, bone marrow B cells from WT and KO mice were cultured in media for 4 h and the apoptosis of B220lowIgM− pre/pro–B cells were analyzed by Annexin-V and propidium iodide staining. H, quantification of apoptosis as analyzed in G. Error bars represent S.D. n = 10. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To determine whether the reduction in mature B cells was due to a developmental defect in Hrd1 KO mice, we compared earlier stages of B cell development in WT and Hrd1 KO mice. We identified an accumulation of B220LowIgM−CD43− pre–B cells (Fig. 1B, middle panel), and further analysis revealed that the increase in pre–B cells was specific to B220LowIgM−CD43−λ5+ CD2− large pre–B cells, with B220LowIgM−CD43−λ5−CD2+ small pre–B cells significantly reduced (Fig. 1, B–D, bottom panels), indicating a developmental block in the large pre–B to resting small pre–B cell transition. Further analysis indicated that the apoptotic rates for B220Low pro–B, pre–B cells in WT and Hrd1 KO mice were comparable as detected by Annexin V and propidium iodide staining (Fig. 1, G and H), suggesting that deletion of Hrd1 resulted in a developmental block in the large pre–B to small pre–B transition.
Hrd1 regulate B cell development independent of UPR mediators IRE1α and CHOP
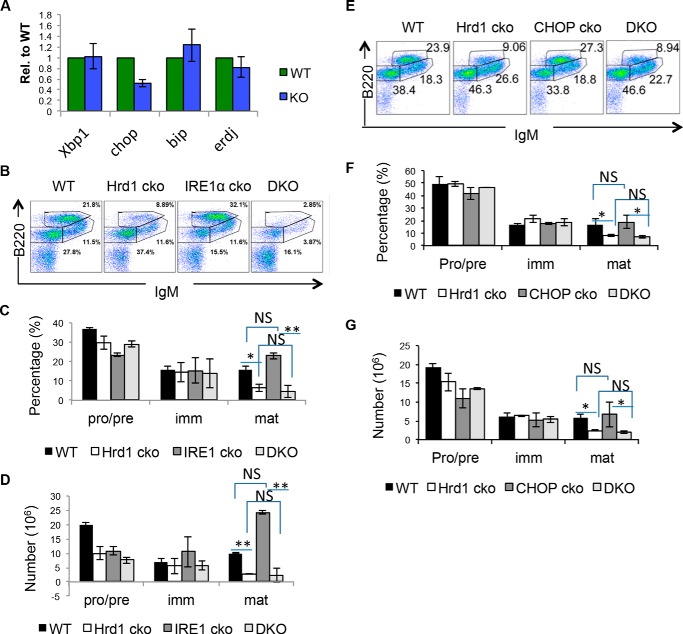
As Hrd1 is part of the ER–associated degradation pathway, it is possible that the loss of Hrd1 clearance of the misfolded proteins in Hrd1 KO mice could increase ER stress and trigger the unfolded protein response (UPR) in Hrd1 KO bone marrow. However, analysis of ER stress-induced genes Xbp1, Chop, Bip, and Erdj3 in WT and Hrd1 KO bone marrow revealed that the UPR was not activated (Fig. 2A), suggesting that loss of Hrd1 functions did not result in ER stress response that potentially blocks B cell development. To investigate whether Hrd1-mediated ubiquitination of pre-BCR is not confounded with the ERAD and UPR pathways, we crossed B cell-specific Hrd1 KO mice to mice with a conditional deletion of IRE1α (Ire1αf/f) or Chop (Chopf/f) (31). IRE1α is a key sensor of UPR and an E3 substrate of Hrd1 (32, 33). Although IRE1α KO mice had similar B220HIIgM+ mature B cells compared with WT, mice with deletions of both Hrd1 and IRE1α had similar numbers of B220LOIgM−, B220LOIgM+, and B220HIIgM+ cells as Hrd1 KO mice (Fig. 2, B–D), indicating that the requirement of Hrd1 for B cell development is independent of IRE1α function. In addition, CHOP is an ER stress-associated pro-apoptotic factor. However, deletion of CHOP was not able to rescue the reduction of B220HIIgM+ mature B cells in Hrd1 KO mice (Fig. 2, E–G), again supporting the conclusion that Hrd1-mediated ubiquitination of pre-BCR is independent of the ERAD and UPR pathways.
Figure 2.
B cell development defect in Hrd1 KO mice is independent of ER stress-associated proteins IRE1α and CHOP. A, qPCR of B220+ BM cells measuring ER stress-associated genes Xbp1, Chop, Bip, and Erdj3. B, bone marrow from WT, Hrd1 KO, Ire1α KO, and double KO (DKO) littermates were analyzed for B220LOIgM+ pro–B and pre–B cells, B220LOIgM+ immature B cells, and B220HIIgM+ mature B cells. The percentages (C) and total number (D) of B cell development stages as analyzed in B are shown. E, WT, Hrd1 KO, Chop KO, and DKO littermates were analyzed for B cell development as in B. The percentages (F) and total number (G) of B cell development stages as measured in E are shown. Error bars represent S.D. n = 5. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Loss of Hrd1 in the B cell lineage leads to accumulation of pre-BCR in large pre–B cells
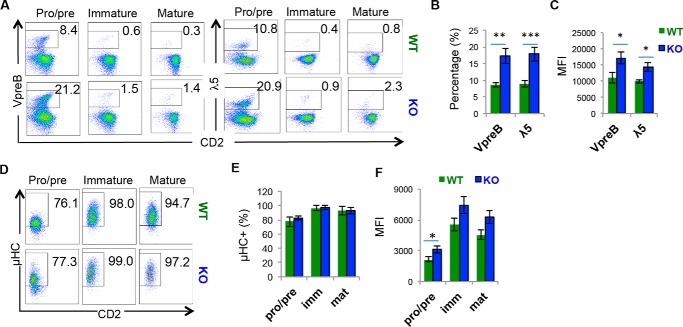
We noticed that both the percentage and fluorescence identity of λ5 expression was increased in the Hrd1-null pre–B cells (Fig. 1B, bottom panels). Because λ5 is a component of the pre-BCR complex, these results imply a possibility that Hrd1 regulates T cell development through controlling the pre-BCR complex expression. Indeed, the number of pre–B cells expressing the pre-BCR components VpreB and λ5 was significantly increased in Hrd1 KO mice (Fig. 3, A and B). In addition, an increased mean fluorescence intensity (MFI) of VpreB and λ5 expression in Hrd1 KO pre–B cells indicated that pre-BCR components are increased on a per cell basis (Fig. 3, A and C). Similarly, μ heavy chain (μHC) expression was increased, both in terms of percentage and MFI at the B220LOIgM− pro/pre–B cell stage (Fig. 3, D–F). In contrast, μHC was not up-regulated in B220LOIgM+ immature B cells and B220HIIgM+ mature B cells (Fig. 3, D–F), where μHC is complexed with light chains to form mature BCR. These results indicate that Hrd1 regulates B cell development through controlling the pre-BCR expression levels.
Figure 3.
Pre-BCR is up-regulated in Hrd1 KO pre–B cells. A, VpreB (left) and λ5 (right) expression in WT and Hrd1 KO (KO) B220LOIgM− BM cells. B, percentage of VpreB and λ5 expression B220LOIgM− cells as analyzed in A. C, MFI of VpreB and λ5 as analyzed in A. D, μHC chain expression in B220LOIgM− pro–B and pre–B cells, B220LOIgM+ immature B cells, and B220HIIgM+ mature B cells. E, percentage of μHC+ cells as analyzed in D. F, MFI of μHC in μHC+ cells as measured in E. Error bars represent S.D. n = 7. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Hrd1 promotes ubiquitination of the pre-BCR complex
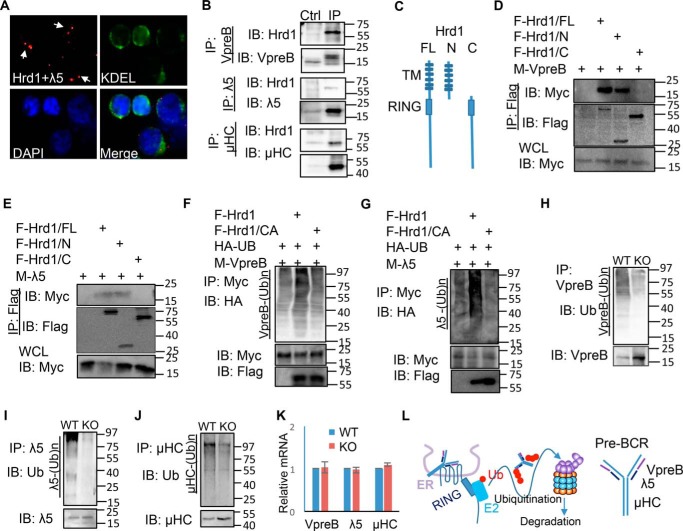
Unlike the mature BCR, <5% of the pre-BCR is expressed on the cell surface, with the majority retained in the ER (9). We therefore determined whether components of the pre-BCR co-localize with the ER-resident E3 ubiquitin ligase Hrd1. We performed a proximity ligation assay on the pre–B cell line 70Z/3, and found that the pre-BCR component λ5 co-localized with Hrd1 within the ER, as indicated by detection of proteins with the ER-retention sequence Lys-Asp-Glu-Leu (KDEL; Fig. 4A). Moreover, co-immunoprecipitation experiments using cell lysates of the pre–B cell line 70Z/3 revealed that Hrd1 interacts with pre-BCR components μHC, VpreB, and λ5 (Fig. 4B). To determine the regions of Hrd1 that mediate its interaction with the pre-BCR complex, Hrd1 truncation mutants were generated (Fig. 4C). When FLAG-tagged truncation mutants of Hrd1 were co-transfected with either Myc-tagged VpreB or Myc-tagged λ5, co-immunoprecipitation experiments revealed that the luminal N-terminal region of Hrd1 is required and sufficient for its interaction with VpreB (Fig. 4D) and λ5 (Fig. 4E), providing supporting evidence that Hrd1 interacts with pre–BCR within the ER lumen. Because Hrd1 is an E3 ubiquitin ligase, we hypothesized that pre-BCR is a substrate of Hrd1. Indeed, co-transfection of VpreB with Hrd1, but not with the E3 ligase catalytic inactive mutant Hrd1/CA, significantly increased VpreB ubiquitination when compared with VpreB co-transfected with empty vector (Fig. 4F). Similarly, Hrd1, but not its CA mutant, promoted λ5 ubiquitination (Fig. 4G). Conversely, VpreB, λ5, and μHC immunoprecipitated from the BM cell lysate of Hrd1 KO mice had reduced ubiquitination compared with WT (Fig. 4, H–J). In contrast, the mRNA expression levels of VpreB, λ5, and μHC were not altered by Hrd1 deficiency (Fig. 4K). Thus, our data indicate that Hrd1 recruits the pre-BCR complex through its N terminus in the ER to catalyze pre-BCR ubiquitination for degradation (Fig. 4L), and the loss of Hrd1 leads to the accumulation of the pre-BCR in the ER of pre–B cells.
Figure 4.
Hrd1 promotes pre-BCR ubiquitination. A, proximity ligation assay of Hrd1 and λ5 in the 70z/3 pre–B cell line. KDEL served as marker for ER proteins. B, 70Z/3 cell lysate was immunoprecipitated (IP) with anti-VpreB (top), anti-λ5 (middle), and anti-μHC (bottom) antibodies, and interaction with Hrd1 was analyzed by immunoblotting (IB). IgG antibody served as negative control (Ctrl). C–E, FLAG-tagged Hrd1 and truncated mutants (C) were co-transfected with Myc-tagged VpreB (D) or λ5 (E) and immunoprecipitated with anti-FLAG antibody. Hrd1 interaction with VpreB (D) and λ5 (E) was detected by immunoblotting with anti-Myc. F and G, Myc-tagged VpreB (F) or λ5 (G) was either co-transfected with FLAG-tagged Hrd1 or its CA mutant and HA-tagged ubiquitin. VpreB (F) or λ5 (G) in the lysates of transfected cells were then immunoprecipitated and ubiquitination was detected by anti-HA antibody. H–J, BM cell lysate from WT and KO mice was immunoprecipitated with anti-VpreB VpreB (H), anti-λ5 (I), or anti-mHC (J) antibodies and ubiquitination was detected by immunoblotting with anti-Ub. K, the mRNA expression levels of VpreB, λ5, and μHC in WT and Hrd1 KO bone marrow cells were determined by real-time RT-PCR. L, a proposed model for Hrd1-mediated pre-BCR complex ubiquitination.
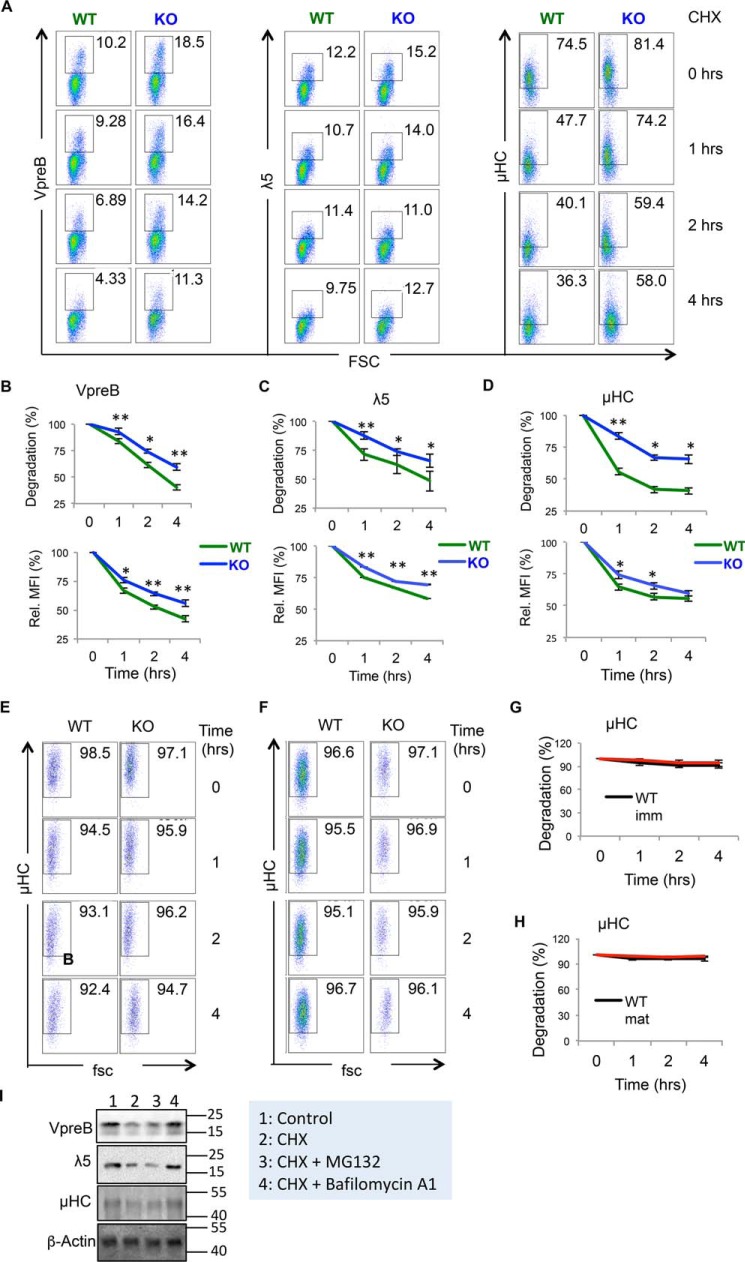
Loss of Hrd1 in pre–B cells leads to increased pre-BCR stability
The increased MFI of the pre-BCR complex in pre–B cells suggested that loss of Hrd1-mediated ubiquitination of pre-BCR could stabilize the pre-BCR in Hrd1 KO pre–B cells. To test this, we treated WT and Hrd1 KO BM cells with cyclohexamide (CHX) to halt de novo synthesis of pre-BCR, and detected expression levels of the pre-BCR over time (Fig. 5A). Prior to CHX treatment, Hrd1 KO had more VpreB and λ5-expressing B220LOIgM− cells (Fig. 5A), and based on MFI, had higher expression of VpreB, λ5, and μHC per cell compared with WT (Fig. 5A). Interestingly, upon CHX treatment, VpreB, λ5, and μHC expression decreased to ∼50% of baseline levels in WT BM B220LOIgM− cells, but all 3 components of the pre-BCR were stabilized in Hrd1 KO BM B220LOIgM− cells (Fig. 5, A–D). Therefore, our results collectively indicate that Hrd1 regulates the pre-BCR expression levels through ubiquitination-mediated protein degradation. We speculated whether Hrd1 also targets the BCR complex for degradation in immature and mature B cells. However, when treated with CHX, μHC levels decreased at similar rates in B220LOIgM+ immature B cells (Fig. 5, E and G) and B220HIIgM+ mature B cells (Fig. 5, F and H) from WT and Hrd1 KO mice, indicating that whereas the pre-BCR is stabilized in Hrd1 KO pre–B cells, μHC expression, as a component of the mature BCR, is unaffected by deletion of Hrd1. Treatment of pre–B cells with the lysosomal inhibitor bafilomycin A1, largely protected pre-BCR from degradation. In contrast, the proteasomal inhibitor MG132 failed to protect pre-BCR from degradation (Fig. 5I). Therefore, Hrd1-mediated pre-BCR degradation appears to be through a lysosome but not proteasome pathway.
Figure 5.
Pre-BCR is stabilized in the absence of Hrd1. A, WT and Hrd1 KO (KO) BM cells were treated with CHX (100 μg/ml) for 0, 1, 2, and 4 h and B220LOIgM− cells were analyzed for VpreB (left), λ5 (middle), and μHC (right) protein levels by intracellular staining. B–D, reduction in percentage (top) and MFI (bottom) of VpreB (B), λ5 (C), and μHC (D) upon CHX treatment. Percentage and MFI were normalized to expression levels at 0 h. E and G, μHC expression in B220LOIgM+ immature B cells (E) and reduction in the percentage of μHC+ cells (top) and MFI (bottom) of μHC in immature B cells (G). F and H, μHC expression in B220HIIgM+ mature B cells (H) and reduction in percentage of μHC+ cells (top) and MFI (bottom) of μHC in mature B cells (H). Error bars represent S.D. n = 8. *, p < 0.05; **, p < 0.01. I, B220LOIgM− cells from WT mice were treated with CHX or further with MG132 (25 μm) or bafilomycin A1 (100 nm) for 4 h. The protein levels of pre-BCR complex were determined by Western blotting using β-Actin as a control.
Loss of Hrd1 resulted in the developmental defect of mature B cells
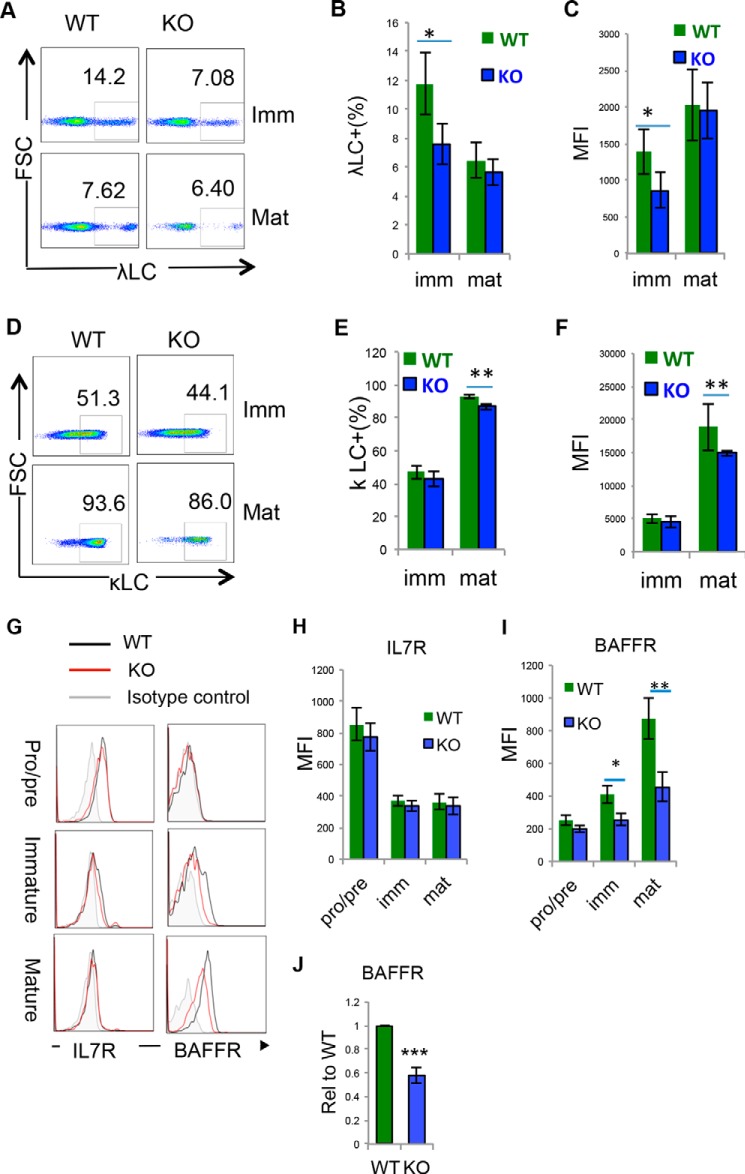
Signals through the pre-BCR are required for initiating diverse processes in pre-B cells, including proliferation and recombination of the light chain gene, which eventually lead to the differentiation of pre–B cells to immature B cells. To explore the functional consequences of Hrd1-mediated degradation of pre-BCR complexes, we analyzed the expression levels of κ and λ light chains (κLC and λLC) during B cell development. As indicated in Fig. 6, expression of κLC and λLC was significantly lower in Hrd1 KO immature and mature B cell populations compared with WT, suggesting that whereas pre-BCR is up-regulated in Hrd1 KO pre–B cells, mature light chains are down-regulated in subsequent B cell developmental stages of Hrd1 KO mice (Fig. 6, A–F). Large pre–B cells proliferate readily due to IL-7R and autonomous pre-BCR signaling. As pre–B cells differentiate into immature and mature B cells, IL-7R and pre-BCR signaling is down-regulated, and survival is maintained by tonic BCR and B cell-activating factor (BAFF) receptor (BAFFR) signaling (6, 10). However, IL-7R receptor expression was similar between WT and Hrd1 KO mice across all B cell populations including pro/pre–B, immature, and mature B cells (Fig. 6, G and H), excluding the possibility that Hrd1 regulates B cell development through modulating the IL-7R expression.
Figure 6.
Loss of Hrd1 resulted in the developmental defect of mature B cells. WT and Hrd1 KO (KO) BM cells were isolated and analyzed by flow cytometry. The B220+IgM− pro/pre–B, B220LoIgM+ immature and B220HIIgM+ mature B cells were gated for the analysis. A–F, κLC and λLC expression were analyzed. Representative flow cytometry images for the expression of κLC (A) and λLC (D) in B220LOIgM+ immature B cells and B220HIIgM− mature B cells are shown. Percentages (B) and MFI (C) of κLC expression as analyzed in A. Percentages (E) and MFI (F) of λLC as analyzed in D. G–J, the expression of IL-7R and BAFF receptor (BAFFR) were analyzed. Representative images are shown in G. The MFI of IL-7R (H) and BAFFR (I) from 7 pairs of mice are indicated. The expression levels of BAFFR mRNA in IgM+B220+ cells were determined by real-time RT-PCR (J). Error bars represent S.D. n = 8. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
It has been reported that pre-BCR signaling up-regulates BAFFR expression, and reduced pre-BCR signaling leads to loss of BAFFR-mediated survival signals. In conjunction with reduced κ and λ light chains in Hrd1 KO IgM+, we unexpectedly found that BAFFR was even down-regulated in Hrd1 KO immature and mature B cells (Fig. 6, G and I). Hrd1 deficiency impairs BAFFR expression at the mRNA expression levels, because BAFFR mRNA were significantly reduced in Hrd1 KO B220+IgM+ BM cells (Fig. 6J). The data taken together suggests that pre-BCR up-regulation impairs the transition from large to small pre–B cells, a developmental defect leads to reduced expression of κLC and λLC in immature B cells. As a consequence, the expression of co-receptor BAFFR was down-regulated, and finally impairs the development of subsequent B cell stages in Hrd1 KO mice.
Discussion
Our studies identified a novel role of Hrd1 in controlling a critical checkpoint in B cell immunity by functioning as an E3 ubiquitin ligase of the pre-BCR complex. This conclusion is supported by the following observations. First, loss of Hrd1 expression during the early stage of B cell development resulted in reduced mature B cells and in mature B cells in the peripheral lymphoid organs. Second, Hrd1 functions is required for the transition from large to small B cell development. Third, Hrd1 regulates B cell development independent of ER stress response. Fourth, Hrd1 functions as an E3 ubiquitin ligase for the pre-BCR complex and promotes pre-BCR degradation, and last, Hrd1 gene deletion resulted in reduced BAFFR expression and viability of subsequent immature and mature B cell stages. Therefore, Hrd1 is critical for maintenance of the B cell compartment by promoting B cell development in the bone marrow.
It has been reported that forced expression of the pre-BCR throughout B cell development does not promote proliferation of pre–B cells, but rather results in reduced immature and mature B cell populations (8). Similarly, genetic deletion of Sel1l, a component in the Hrd1–ERAD complex, impairs B cell development due to the altered pre-BCR expression (34). The reduced degradation of the pre-BCR in Hrd1 KO mice led to reduced viability of subsequent immature and mature B cell populations, resulting in a reduced mature B cell compartment. In addition, whereas μ heavy chain stability was comparable between WT and Hrd1 KO immature and mature B cells, other components of the mature BCR, i.e. κ and λ and Igα expression, were reduced in Hrd1 KO immature and mature B cells, suggesting that impaired degradation of pre-BCR results in reduced expression of mature BCR, which in turn is crucial for tonic B cell survival signaling. In support of our hypothesis, BAFFR mRNA and expression was reduced in mature Hrd1 KO B cells. BAFFR expression has been shown to be directly correlated and regulated by mature BCR signaling strength (35). Based on our findings, it is likely that the reduced mature B cell number in Hrd1 KO bone marrow is due to defective pre-BCR down-regulation in the pre–B cell population, which results in reduced mature BCR expression and signaling that is required for mature B cell survival. In fact, we have recently discovered that Hrd1 regulates activation-induced B cell death through targeting the death receptor Fas (36). Therefore, Hrd1 appears to be critical to regulate B cell functions at multiple distinct stages.
As pre-BCR and mature BCR share components and are highly similar in structure, it is interesting that loss of Hrd1 led to stabilization of pre-BCR, but had no significant effect on mature BCR. One possibility is that whereas mature BCR is transported to the cell surface, the majority of pre-BCR is retained in the ER, thus increasing the probability of interaction between pre-BCR and Hrd1 (9, 10). In addition, although Hrd1 was found to interact with its retrotranslocated substrates through its C-terminal RING domain within the cytoplasm (12, 13), we observed that Hrd1 interacts with VpreB and λ5 through its luminal N-terminal domain. This novel interaction mechanism suggests that Hrd1 might recognize pre-BCR through its distinct glycosylation status or through recognition of chaperone partners (38–40). In addition, unlike mature BCR, which is stably expressed in immature and mature B cells, pre-BCR is down-regulated at the large pre–B stage before further developing into resting small pre–B cells (10, 41). It is possible that upstream signals are required for Hrd1 degradation of the μ-heavy chain, which might terminate in later B cell stages. Inconsistent to our observation, Ji et al. recently reported that deletion of Sel1L, an adaptor protein that forms a complex with Hrd1 to mediate ER stress-induced protein degradation, results in the B cell developmental blockade due to the elevated pre-BCR expression (34).
During ERAD, misfolded proteins are retrotranslocated into the cytosol from the ER, where they are ubiquitinated by the cytosolic RING domain of Hrd1 (12, 13). It has also been reported that Hrd1 degrades truncated forms of μ-heavy chain (42). It is possible that the observed degradation of pre-BCR by Hrd1 is predominantly due to the accumulation of misfolded forms of pre-BCR in the ER. Despite this significant accumulation in pre-BCR, Hrd1 deletion does not appear to result in the systemic accumulation of misfolded proteins and activation of UPR in B cells, as we did not detect an increase in UPR targets such as IRE1α, CHOP, BiP, or Xbp1 in Hrd1 KO B cells. More importantly, deletion of UPR targets IRE1α or CHOP could not rescue the B cell development blockade in Hrd1 KO mice, thus further supporting our hypothesis that Hrd1 is required for selective degradation of pre-BCR in addition to its function in ERAD and regulating ER stress. In summary, our studies identified a novel role of Hrd1 in controlling the transition from large to small pre-B cells during B cell development by functioning as an E3 ubiquitin ligase of pre-BCR complex.
Materials and methods
Animals
Hrd1-floxed mice were used as described previously (15). CD19-Cre transgenic mice in the C57BL/6 genetic background were purchased from The Jackson Laboratory (Bar Harbor, ME). B cell-specific Hrd1-null mice were generated by breeding Hrd1-floxed mice with CD19-Cre transgenic mice. The IRE1α-floxed mice were generated as previously described and had been backcrossed with C57BL/6J mice for more than 10 generations to maintain the C57BL strain background. Chop-floxed mice were used as reported (31). Double knockout mice were generated by mating CD19-Cre Hrd1-floxed mice with Ire1α- or Chop-floxed mice. All mice used in this study were maintained and used at the Northwestern University Mouse Facility under pathogen-free conditions according to institutional guidelines and animal study proposals approved by the Institutional Animal Care and Use Committee.
Cell lines and reagents
Mature B cell line A20 (TIB-208) and pre–B cell line 70Z/3 (TIB-158) were purchased from ATCC and maintained in RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin and 0.05 mm 2-mercaptoethanol. Human embryonic kidney 293 cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% FBS, 1% penicillin/streptomycin. The proteasome inhibitor MG132 and the lysosome inhibitor bafilomycin A1 were purchased from Sigma.
Plasmids and antibodies
Hrd1 and the ubiquitin expression plasmids were constructed as before (32). The truncation mutants of Hrd1, Vpreb, and λ5 expression plasmids were generated by PCR and subcloned into pCMV-FLAG (Sigma) or pCMV-Myc vectors (Invitrogen). Antibodies used for immunoblotting, co-immunoprecipitation, and immunofluorescence staining were from Santa Cruz Biotechnology (Santa Cruz, CA), anti-epitope tags (FLAG, HA, and Myc), and anti-λ5 (M-60); Abcam (Cambridge, MA), anti-Hrd1 (EP7459) and anti-KDEL (MAC256); Calbiochem (Pasadena, CA), anti-Tubulin (DM1A); Sigma, anti-Hrd1 (HRD1–5); and Invitrogen, Alexa Fluor® 488 anti-rabbit IgG. Fluorescence-labeled antibodies used for flow cytometry were purchased from eBioscience (San Diego, CA), anti-CD16/32, anti-CD43 (eBioR2/60), anti-CD79a (HM47), anti-IgM (II/41), anti-mouse CD268 (BAFF receptor; eBio7H22–E16); Biolegend (San Diego, CA), anti-CD45R (B220; RA3-6B2), anti-IgM (RMM-1), anti-CD2 (RM2-5), anti-CD21/CD35 (7E9), anti-CD23 (B3B4), anti-CD5 (53-7.3), anti-Ig light chain κ (RMK-45), anti-Ig light chain λ (RML-42), and anti-CD179a (VpreB) (R3); BD Biosciences, anti-CD138 (281-2), anti-CD127 (SB199), anti-CD1179b (λ5; LM34), and anti-CD95 (Jo2); and Novus (Littleton, CO), anti-BCMA/TNFRSF17 (Vicky-2).
Flow cytometry and immunofluorescence staining
BM cells were obtained by flushing femur and tibia with PBS containing 3% FBS as reported (43). Single-cell suspensions from bone marrow and spleen were blocked with antibody against Fc receptor and then stained with the appropriate fluorophore-conjugated antibodies. For intracellular staining, cells were fixed and permeabilized using the CytoFix/Perm Kit (eBioscience) per the manufacturer's instructions, followed by staining with the appropriate fluorophore-conjugated antibodies. Cells were collected in an Accuri C6 Flow Cytometer or FACS Canto (BD Biosciences). The proximity ligation assay was performed using Duolink® In Situ Red Starter Kit Mouse/Rabbit purchased from Sigma (DUO92101). Cells were cytospun onto slides, fixed with 4% formaldehyde at room temperature for 15 min. After washing three times with PBS, cells were permeabilized with 0.1% Triton X-100 at room temperature for 5 min, and then incubated with primary antibodies diluted with 1–4% BSA for 1 h at room temperature. After washing three times with PBS, cells were incubated with secondary antibodies for 1 h at room temperature. Cells were processed and visualized with the Nikon Eclipse Ti fluorescence microscope at original magnification ×40 after staining with 4′,6-diamidino-2-phenylindole.
Cell transfection, immunoblotting, and co-immunoprecipitation assay
Human embryonic kidney 293 cells in 60-mm or 12-well dishes were transfected with appropriate plasmids using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. 24 or 48 h after transfection, cells were lysed in 1× Nonidet P-40 lysis buffer, which was freshly added with protease inhibitor mixture. After adding pre-washed protein A or G magnetic beads to the cell lysates, antibodies (1 μg) were added and the lysates were rotated in a rotating wheel at 4 °C for 2 h followed by the addition of 30 μl of protein G-Sepharose beads (GE Healthcare) for an additional 2 h at 4 °C or overnight. Immunoprecipitates were washed four times with Nonidet P-40 lysis buffer, and then were boiled at 95 °C in 20 μl of 2× Laemmli's buffer. Samples were analyzed on an 8–12% SDS-PAGE gel and electrotransferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA). After blocking with 4% milk dissolved in TBST, membranes were probed with the appropriate primary antibodies. After washing twice with TBST, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies. After washing twice with TBST, membranes were treated with enhanced chemiluminescence detection system (ECL; GE Healthcare) and exposed. If necessary, membranes were stripped with stripping buffer (Bio-Rad), washed, blocked, and then reprobed with other antibodies as reported (44, 45).
Real-time quantitative PCR
Total RNA was extracted from cells with TRIzol reagent per the manufacturer's instructions (Ambion, Grand Island, NY). First-strand cDNA was synthesized using a qScript cDNA Synthesis kit (Quanta Biosciences, Gaithersburg, MD). Real-time qPCR was performed using iQ5 and the SYBR Green Detection system (Bio-Rad Laboratories). Data were normalized to the expression of β-actin in each sample as reported (37). Primers are shown in Table S1.
Statistical analysis
All experiments used the Student's t test for statistical analysis. Data were presented as mean ± S.D. p values less than 0.05 were considered statistically significant.
Author contributions
Y. Y., S. K., B. G., and Yusi Zhang data curation; Y. Y. and S. K. formal analysis; Y. Y., J. M.-C., and Yusi Zhang validation; Y. Y., S. K., Yana Zhang, and Yusi Zhang investigation; Y. Y. visualization; Y. Y., S. K., and J. M.-C. methodology; B. G. and D. F. project administration; D. D. Z., E. T., and K. Z. resources; D. D. Z., J. S., K. Z., J. Z., and D. F. writing-review and editing; B. Z., J. S., E. T., K. Z., J. Z., and D. F. conceptualization; B. Z. and D. F. writing-original draft.
Supplementary Material
Acknowledgments
We thank Dr. Ira Tabas, MD and Ph.D., Richard J. Stock, Professor and Vice-Chairman of Research (Department of Medicine, Columbia University), for the CHOP-floxed mice. We thank Fang lab members for critical reading of the manuscript and constructive suggestions during our research.
This work was supported by National Institutes of Health R01 Grants AI079056, AI108634, and AR006634 (to D. F.) and National Natural Science Foundation of China (NSFC) Grants 81600784 (to Y. Z.) 31270939, 81471526, and 81771667, Training Program of the Major Research Plan in regional immunology of the National Natural Science Foundation of China Grant 91442110,; Natural Science Foundation of Jiangsu Province Grant BK20170349, a postdoctoral foundation of China grant (to Y. Y.), Social development project of Jiangsu Province Grant BE2016676 (to Y. Y.), and the Jiangsu Key Laboratory of Infection and Immunity, Institutes of Biology and Medical Sciences of Soochow University. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1.
- pre-BCR
- pre-B cell receptor
- ER
- endoplasmic reticulum
- ERAD
- ER stress-associated protein degradation
- UPR
- unfolded protein response
- MFI
- mean fluorescence intensity
- CHX
- cyclohexamide
- μHC
- μ heavy chain
- LC
- light chain
- BAFFR
- B-cell activating factor receptor
- FBS
- fetal bovine serum
- HA
- hemagglutinin
- qPCR
- quantitative PCR
- Hrd1
- 3-hydroxy-3-methylglutaryl-CoA reductase degradation 1.
References
- 1. Kraus M., Alimzhanov M. B., Rajewsky N., and Rajewsky K. (2004) Survival of resting mature B lymphocytes depends on BCR signaling via the Igα/β heterodimer. Cell 117, 787–800 10.1016/j.cell.2004.05.014 [DOI] [PubMed] [Google Scholar]
- 2. Levine M. H., Haberman A. M., Sant'Angelo D. B., Hannum L. G., Cancro M. P., Janeway C. A. Jr., and Shlomchik M. J. (2000) A B-cell receptor-specific selection step governs immature to mature B-cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 97, 2743–2748 10.1073/pnas.050552997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rickert R. C. (2013) New insights into pre-BCR and BCR signalling with relevance to B cell malignancies. Nat. Rev. Immunol. 13, 578–591 10.1038/nri3487 [DOI] [PubMed] [Google Scholar]
- 4. Nishimoto N., Kubagawa H., Ohno T., Gartland G. L., Stankovic A. K., and Cooper M. D. (1991) Normal pre-B cells express a receptor complex of μ heavy chains and surrogate light-chain proteins. Proc. Natl. Acad. Sci. U.S.A. 88, 6284–6288 10.1073/pnas.88.14.6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Geier J. K., and Schlissel M. S. (2006) Pre-BCR signals and the control of Ig gene rearrangements. Semin. Immunol. 18, 31–39 10.1016/j.smim.2005.11.001 [DOI] [PubMed] [Google Scholar]
- 6. Herzog S., Reth M., and Jumaa H. (2009) Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat. Rev. Immunol. 9, 195–205 10.1038/nri2491 [DOI] [PubMed] [Google Scholar]
- 7. Khan W. N. (2009) B cell receptor and BAFF receptor signaling regulation of B cell homeostasis. J. Immunol. 183, 3561–3567 10.4049/jimmunol.0800933 [DOI] [PubMed] [Google Scholar]
- 8. van Loo P. F., Dingjan G. M., Maas A., and Hendriks R. W. (2007) Surrogate-light-chain silencing is not critical for the limitation of pre-B cell expansion but is for the termination of constitutive signaling. Immunity 27, 468–480 10.1016/j.immuni.2007.07.018 [DOI] [PubMed] [Google Scholar]
- 9. Brouns G. S., de Vries E., Neefjes J. J., and Borst J. (1996) Assembled pre-B cell receptor complexes are retained in the endoplasmic reticulum by a mechanism that is not selective for the pseudo-light chain. J. Biol. Chem. 271, 19272–19278 10.1074/jbc.271.32.19272 [DOI] [PubMed] [Google Scholar]
- 10. Kawano Y., Ouchida R., Wang J.-Y., Yoshikawa S., Yamamoto M., Kitamura D., and Karasuyama H. (2012) A novel mechanism for the autonomous termination of pre-B cell receptor expression via induction of lysosome-associated protein transmembrane 5. Mol. Cell. Biol. 32, 4462–4471 10.1128/MCB.00531-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ouchida R., Kurosaki T., and Wang J. Y. (2010) A role for lysosomal-associated protein transmembrane 5 in the negative regulation of surface B cell receptor levels and B cell activation. J. Immunol. (Baltimore, Md.: 1950) 185, 294–301 10.4049/jimmunol.1000371 [DOI] [PubMed] [Google Scholar]
- 12. Okuda-Shimizu Y., and Hendershot L. M. (2007) Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol. Cell 28, 544–554 10.1016/j.molcel.2007.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Christianson J. C., Olzmann J. A., Shaler T. A., Sowa M. E., Bennett E. J., Richter C. M., Tyler R. E., Greenblatt E. J., Harper J. W., and Kopito R. R. (2012) Defining human ERAD networks through an integrative mapping strategy. Nat. Cell. Biol. 14, 93–105 10.1038/ncb2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fujita H., Yagishita N., Aratani S., Saito-Fujita T., Morota S., Yamano Y., Hansson M. J., Inazu M., Kokuba H., Sudo K., Sato E., Kawahara K., Nakajima F., Hasegawa D., Higuchi I., et al. (2015) The E3 ligase synoviolin controls body weight and mitochondrial biogenesis through negative regulation of PGC-1β. EMBO J. 34, 1042–1055 10.15252/embj.201489897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang H., Qiu Q., Gao B., Kong S., Lin Z., and Fang D. (2014) Hrd1-mediated BLIMP-1 ubiquitination promotes dendritic cell MHCII expression for CD4 T cell priming during inflammation. J. Exp. Med. 211, 2467–2479 10.1084/jem.20140283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu T., Zhao F., Gao B., Tan C., Yagishita N., Nakajima T., Wong P. K., Chapman E., Fang D., and Zhang D. D. (2014) Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 28, 708–722 10.1101/gad.238246.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng H., Dai J., Stoilova D., and Li Z. (2001) Cell surface targeting of heat shock protein gp96 induces dendritic cell maturation and antitumor immunity. J. Immunol. 167, 6731–6735 10.4049/jimmunol.167.12.6731 [DOI] [PubMed] [Google Scholar]
- 18. Zhang Y., Wu B. X., Metelli A., Thaxton J. E., Hong F., Rachidi S., Ansa-Addo E., Sun S., Vasu C., Yang Y., Liu B., and Li Z. (2015) GP96 is a GARP chaperone and controls regulatory T cell functions. J. Clin. Investig. 125, 859–869 10.1172/JCI79014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Y., Liu B., Dai J., Srivastava P. K., Zammit D. J., Lefrançois L., and Li Z. (2007) Heat shock protein gp96 is a master chaperone for Toll-like receptors and is important in the innate function of macrophages. Immunity 26, 215–226 10.1016/j.immuni.2006.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang Y., and Li Z. (2005) Roles of heat shock protein gp96 in the ER quality control: redundant or unique function? Mol. Cells 20, 173–182 [PubMed] [Google Scholar]
- 21. Wu S., Hong F., Gewirth D., Guo B., Liu B., and Li Z. (2012) The molecular chaperone gp96/GRP94 interacts with Toll-like receptors and integrins via its C-terminal hydrophobic domain. J. Biol. Chem. 287, 6735–6742 10.1074/jbc.M111.309526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu S., Dole K., Hong F., Noman A. S., Issacs J., Liu B., and Li Z. (2012) Chaperone gp96-independent inhibition of endotoxin response by chaperone-based peptide inhibitors. J. Biol. Chem. 287, 19896–19903 10.1074/jbc.M112.343848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thaxton J. E., Liu B., Zheng P., Liu Y., and Li Z. (2014) Deletion of CD24 impairs development of heat shock protein gp96-driven autoimmune disease through expansion of myeloid-derived suppressor cells. J. Immunol. 192, 5679–5686 10.4049/jimmunol.1302755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Staron M., Yang Y., Liu B., Li J., Shen Y., Zúñuniga-Pflücker J. C., Aguila H. L., Goldschneider I., and Li Z. (2010) gp96, an endoplasmic reticulum master chaperone for integrins and Toll-like receptors, selectively regulates early T and B lymphopoiesis. Blood 115, 2380–2390 10.1182/blood-2009-07-233031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Staron M., Wu S., Hong F., Stojanovic A., Du X., Bona R., Liu B., and Li Z. (2011) Heat-shock protein gp96/grp94 is an essential chaperone for the platelet glycoprotein Ib-IX-V complex. Blood 117, 7136–7144 10.1182/blood-2011-01-330464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morales C., Wu S., Yang Y., Hao B., and Li Z. (2009) Drosophila glycoprotein 93 is an ortholog of mammalian heat shock protein gp96 (grp94, HSP90b1, HSPC4) and retains disulfide bond-independent chaperone function for TLRs and integrins. J. Immunol. 183, 5121–5128 10.4049/jimmunol.0900811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu B., Yang Y., Dai J., Medzhitov R., Freudenberg M. A., Zhang P. L., and Li Z. (2006) TLR4 up-regulation at protein or gene level is pathogenic for lupus-like autoimmune disease. J. Immunol. 177, 6880–6888 10.4049/jimmunol.177.10.6880 [DOI] [PubMed] [Google Scholar]
- 28. Liu B., and Li Z. (2008) Endoplasmic reticulum HSP90b1 (gp96, grp94) optimizes B-cell function via chaperoning integrin and TLR but not immunoglobulin. Blood 112, 1223–1230 10.1182/blood-2008-03-143107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu B., Dai J., Zheng H., Stoilova D., Sun S., and Li Z. (2003) Cell surface expression of an endoplasmic reticulum resident heat shock protein gp96 triggers MyD88-dependent systemic autoimmune diseases. Proc. Natl. Acad. Sci. U.S.A. 100, 15824–15829 10.1073/pnas.2635458100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Z., and Srivastava P. K. (1993) Tumor rejection antigen gp96/grp94 is an ATPase: implications for protein folding and antigen presentation. EMBO J. 12, 3143–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou A. X., Wang X., Lin C. S., Han J., Yong J., Nadolski M. J., Borén J., Kaufman R. J., and Tabas I. (2015) C/EBP-homologous protein (CHOP) in vascular smooth muscle cells regulates their proliferation in aortic explants and atherosclerotic lesions. Circ. Res. 116, 1736–1743 10.1161/CIRCRESAHA.116.305602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gao B., Lee S. M., Chen A., Zhang J., Zhang D. D., Kannan K., Ortmann R. A., and Fang D. (2008) Synoviolin promotes IRE1 ubiquitination and degradation in synovial fibroblasts from mice with collagen-induced arthritis. EMBO Rep 9, 480–485 10.1038/embor.2008.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun S., Shi G., Sha H., Ji Y., Han X., Shu X., Ma H., Inoue T., Gao B., Kim H., Bu P., Guber R. D., Shen X., Lee A. H., et al. (2015) IRE1α is an endogenous substrate of endoplasmic-reticulum-associated degradation. Nat. Cell Biol. 17, 1546–1555 10.1038/ncb3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ji Y., Kim H., Yang L., Sha H., Roman C. A., Long Q., and Qi L. (2016) The Sel1L-Hrd1 endoplasmic reticulum-associated degradation complex manages a key checkpoint in B cell development. Cell Rep. 16, 2630–2640 10.1016/j.celrep.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mackay F., Figgett W. A., Saulep D., Lepage M., and Hibbs M. L. (2010) B-cell stage and context-dependent requirements for survival signals from BAFF and the B-cell receptor. Immunol. Rev. 237, 205–225 10.1111/j.1600-065X.2010.00944.x [DOI] [PubMed] [Google Scholar]
- 36. Kong S., Yang Y., Xu Y., Wang Y., Zhang Y., Melo-Cardenas J., Xu X., Gao B., Thorp E. B., Zhang D. D., Zhang B., Song J., Zhang K., Zhang J., Zhang J., et al. (2016) Endoplasmic reticulum-resident E3 ubiquitin ligase Hrd1 controls B-cell immunity through degradation of the death receptor CD95/Fas. Proc. Natl. Acad. Sci. U.S.A. 113, 10394–10399 10.1073/pnas.1606742113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee S. M., Gao B., and Fang D. (2008) FoxP3 maintains Treg unresponsiveness by selectively inhibiting the promoter DNA-binding activity of AP-1. Blood 111, 3599–3606 10.1182/blood-2007-09-115014 [DOI] [PubMed] [Google Scholar]
- 38. Lee Y. K., Brewer J. W., Hellman R., and Hendershot L. M. (1999) BiP and immunoglobulin light chain cooperate to control the folding of heavy chain and ensure the fidelity of immunoglobulin assembly. Mol. Biol. Cell 10, 2209–2219 10.1091/mbc.10.7.2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Keren Z., Diamant E., Ostrovsky O., Bengal E., and Melamed D. (2004) Modification of ligand-independent B cell receptor tonic signals activates receptor editing in immature B lymphocytes. J. Biol. Chem. 279, 13418–13424 10.1074/jbc.M311970200 [DOI] [PubMed] [Google Scholar]
- 40. Ubelhart R., Bach M. P., Eschbach C., Wossning T., Reth M., and Jumaa H. (2010) N-Linked glycosylation selectively regulates autonomous precursor BCR function. Nat. Immunol. 11, 759–765 10.1038/ni.1903 [DOI] [PubMed] [Google Scholar]
- 41. Schebesta M., Pfeffer P. L., and Busslinger M. (2002) Control of pre-BCR signaling by Pax5-dependent activation of the BLNK gene. Immunity 17, 473–485 10.1016/S1074-7613(02)00418-1 [DOI] [PubMed] [Google Scholar]
- 42. Cattaneo M., Otsu M., Fagioli C., Martino S., Lotti L. V., Sitia R., and Biunno I. (2008) SEL1L and HRD1 are involved in the degradation of unassembled secretory Ig-μ chains. J. Cell. Physiol. 215, 794–802 10.1002/jcp.21364 [DOI] [PubMed] [Google Scholar]
- 43. Zhang J., Lee S. M., Shannon S., Gao B., Chen W., Chen A., Divekar R., McBurney M. W., Braley-Mullen H., Zaghouani H., and Fang D. (2009) The type III histone deacetylase Sirt1 is essential for maintenance of T cell tolerance in mice. J. Clin. Invest. 119, 3048–3058 10.1172/JCI38902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lin Z., Yang H., Kong Q., Li J., Lee S. M., Gao B., Dong H., Wei J., Song J., Zhang D. D., and Fang D. (2012) USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell 46, 484–494 10.1016/j.molcel.2012.03.024 [DOI] [PubMed] [Google Scholar]
- 45. Lin Z., Yang H., Tan C., Li J., Liu Z., Quan Q., Kong S., Ye J., Gao B., and Fang D. (2013) USP10 antagonizes c-Myc transcriptional activation through SIRT6 stabilization to suppress tumor formation. Cell Rep. 5, 1639–1649 10.1016/j.celrep.2013.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.