Abstract
The ability of pathogens to perceive environmental conditions and modulate gene expression accordingly is a crucial feature for bacterial survival. In this respect, the heat-shock response, a universal cellular response, allows cells to adapt to hostile environmental conditions and to survive during stress. In the major human pathogen Helicobacter pylori the expression of chaperone-encoding operons is under control of two auto-regulated transcriptional repressors, HrcA and HspR, with the latter acting as the master regulator of the regulatory circuit. To further characterize the HspR regulon in H. pylori, we used global transcriptome analysis (RNA-sequencing) in combination with Chromatin Immunoprecipitation coupled with deep sequencing (ChIP-sequencing) of HspR genomic binding sites. Intriguingly, these analyses showed that HspR is involved in the regulation of different crucial cellular functions through a limited number of genomic binding sites. Moreover, we further characterized HspR-DNA interactions through hydroxyl-radical footprinting assays. This analysis in combination with a nucleotide sequence alignment of HspR binding sites, revealed a peculiar pattern of DNA protection and highlighted sequence conservation with the HAIR motif (an HspR-associated inverted repeat of Streptomyces spp.). Site-directed mutagenesis demonstrated that the HAIR motif is fundamental for HspR binding and that additional nucleotide determinants flanking the HAIR motif are required for complete binding of HspR to its operator sequence spanning over 70 bp of DNA. This finding is compatible with a model in which possibly a dimer of HspR recognizes the HAIR motif overlapping its promoter for binding and in turn cooperatively recruits two additional dimers on both sides of the HAIR motif.
Keywords: heat-shock response, HspR repressor, ChIP-seq, RNA-sequencing, transcriptome
Introduction
Helicobacter pylori represents one of the most widespread human pathogens, recognized as the principal causative agent of different gastrointestinal severe diseases such as atrophic gastritis, peptic ulcer, MALT-lymphoma and gastric adenocarcinoma (Gisbert and Calvet, 2011; Salama et al., 2013). A peculiar feature of H. pylori is its small genome (1.66 Mb), characterized by a relative low abundance of genes encoding regulators of transcription. In this respect, it has been speculated that this paucity of transcriptional regulators could reflect the adaptation of H. pylori to its very restricted niche in the mucus layer of the human stomach and could be linked to the lack of competition from other microorganisms (Marshall et al., 1998). To date, only 17 transcriptional regulators have been identified and shown to control key physiological responses of the bacterium, which are necessary to successfully colonize the gastric niche (Scarlato et al., 2001).
Helicobacter pylori transcriptional regulators appear to be arranged in different regulatory modules, transducing separate environmental inputs. Such regulatory modules are constituted by a master regulator, followed by intermediate regulators and regulatory interactions finally resulting into a coordinated output of target gene’s expressions (Danielli et al., 2010). Intriguingly, these regulatory modules do not appear as segregated, but highly interconnected and show a significant crosstalk among different signal transduction pathways. H. pylori does not possess a homologue of the Escherichia coli heat-shock sigma factor σ32. However, the heat stress response is governed by two dedicated transcriptional repressors, HrcA and HspR that negatively regulate the expression of the highly conserved class of stress-induced proteins, known as Heat-Shock Proteins (HSPs). The main function of HSPs is to assist the folding of newly synthetized polypeptides, as well as assembly, transport and degradation of cellular proteins under both normal and adverse growth condition (Roncarati and Scarlato, 2017). Besides their general role in protecting cellular proteins against different kind of stresses and in maintaining cellular homeostasis, some HSPs and chaperones are considered virulence factors and seem to be involved in specific pathogenic processes. Specifically, several lines of evidence show that the heat-shock proteins of H. pylori play also non-canonical roles and some of them seem to have undertaken additional functions during the interaction with the host (Evans et al., 1992; Huesca et al., 1996; Phadnis et al., 1996; Dunn et al., 1997; Kao et al., 2016). Because of these important functions in the cell, H. pylori has developed regulatory strategies to tightly modulate HSPs expression level in response to environmental signals.
In previous works (Spohn and Scarlato, 1999; Spohn et al., 2004) it has been demonstrated that the transcription of groES-groEL, hrcA-grpE-dnaK, and cbpA-hspR-rarA operons, encoding the major chaperones of H. pylori, is negatively regulated by HspR and/or HrcA repressors, with the former acting as the master regulator of this circuit. Indeed, HspR represses transcription of the cbpA operon alone, thereby auto-regulating its own synthesis, while it represses the expression of the other two heat-shock operons groES-groEL and hrcA-grpE-dnaK in combination with HrcA. Genes with sequence similarity to H. pylori hspR were annotated in several other bacteria, including species belonging to the Streptomyces genus. In particular, H. pylori HspR is a homologue of the repressor that controls the expression of the dnaK operon in Streptomyces coelicolor by binding three copies of the HspR consensus binding sequence called HAIR (for HspR Associated Inverted Repeat) (Bucca et al., 1995; Grandvalet et al., 1999). Also in Streptomyces albus it was demonstrated that HspR binds to an inverted repeat identical to S. coelicolor HAIR sequence, mapping in the promoter region of the protease gene clpB (Grandvalet et al., 1999). In H. pylori, DNaseI footprinting experiments with the purified protein showed that HspR binds extended DNA regions located in the promoters of the three heat-shock operons and recognizes sequences similar to the HAIR motif (Delany et al., 2002c; Roncarati et al., 2007). While in the case of cbpA promoter HspR binding occurs in close proximity to the transcription start site, on the groEL and hrcA promoters HspR binds far upstream of the core promoter region, in an atypical position for a repressor (Roncarati et al., 2007). The identification of additional genes controlled by HspR was pursued through array-based whole transcriptome analyses both in H. pylori (Roncarati et al., 2007) and in several other bacterial species (Stewart et al., 2002; Andersen et al., 2005; Schmid et al., 2005; Holmes et al., 2010). Moreover, from an in vitro selection of genomic DNA fragments bound by purified H. pylori HspR protein, two novel HspR binding sites were identified in the 3′ regions of both speA and tlpB genes coding for proteins with functions unrelated with those of chaperones (Delany et al., 2002c).
In the present study, by combining ChIP-sequencing and RNA-sequencing we investigated more in detail the direct or indirect contribution of H. pylori HspR to the heat-shock regulon. While the heat-shock regulon includes many genes with key cellular functions, HspR only binds to a limited number of genomic sites. Furthermore, by means of the high-resolution hydroxyl-radical footprinting technique, we further characterized the HspR-DNA interactions at the molecular level. The data provide a more detailed comprehension of the interaction between HspR and its target DNA sequences and, at least on its own promoter, let us to propose a cooperative DNA-binding mechanism of three HspR dimers per operator sequence.
Materials and Methods
Bacterial Strains and Growth Conditions
Helicobacter pylori strains (Table 1) were recovered from frozen glycerol stocks on Brucella broth agar plates, containing 5% fetal calf serum (FCS), in a 9% CO2-91% air atmosphere at 37°C and 95% humidity in a water jacketed incubator (Thermo Scientific). Liquid cultures were grown in Brucella Broth supplemented with 5% FCS with gentle agitation (120 rpm). E. coli strains DH5α and BL21 (DE3) (Table 1) were grown on Luria-Bertani (LB) agar plates or LB liquid broth with vigorous agitation (250 rpm); when required, ampicillin was added to the medium to achieve a final concentration of 100 μg/ml.
Table 1.
Bacterial strains and plasmids used in this study.
| Bacterial strains/ Plasmids | Description | Source/ Reference |
|---|---|---|
| Strains | ||
| H. pylori G27 wild type | Clinical isolate, wild type | Xiang et al., 1995 |
| H. pylori G27 (hspR::Km) | G27 derivative; bp 66 to 334 of the hspR coding sequence replaced by a Kanamycin (Km) cassette. | Spohn and Scarlato, 1999 |
| H. pylori G27 (vacA::Pcbpwt-lux) | G27 derivative; containing the Pcbp wild type promoter region upstream of the luxC gene in the vacA locus; Cpr | This work |
| H. pylori G27 (vacA::Pcbpwt-lux, hspR::Km) | G27 derivative; containing the Pcbp wild type promoter region upstream of the luxC gene in the vacA locus and the hspR coding sequence (bp 66 to 334) replaced by a Kanamycin (Km) cassette; Cpr, Kmr | This work |
| H. pylori G27 (vacA::PcbpM1 + 2-lux, hspR::Km) | G27 derivative; containing the Pcbp HAIR-mutant promoter region upstream of the luxC gene in the vacA locus and the hspR coding sequence (bp 66 to 334) replaced by a Kanamycin (Km) cassette; Cpr, Kmr | This work |
| H. pylori G27 (vacA::PcbpM1 + 2-lux) | G27 derivative; containing the Pcbp HAIR-mutant promoter region upstream of the luxC gene in the vacA locus; Cpr | This work |
| E. coli DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Hanahan, 1983 |
| E. coli BL21(DE3) | hsdS gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1). | Studier et al., 1990 |
| Plasmids | ||
| pGEM-T-Easy | Cloning vector, Ampr | Promega |
| pGEM-T-Easy-RBS gro | pGEM-T-Easy derivative, containing a 147 bp DNA fragment corresponding to the region from 9,452–9,543 of H. pylori G27 genome amplified by PCR with oligonucleotides RBS GroF/RBS GroR. This region corresponds to a portion of the promoter region of HPG27_RS00075 (HP0011 according to 26695 annotation). | This work |
| pGEM-T-Easy-RBS hrc | pGEM-T-Easy derivative, containing a 252 bp DNA fragment corresponding to the region from 118,944–119,035 of H. pylori G27 genome amplified by PCR with oligonucleotides RBShrcF/PhrcF. This region corresponds to a portion of the promoter region of HPG27_RS00580 (HP0111 according to 26695 annotation). | This work |
| pGEM-T-Easy-HBS cbp | pGEM-T-Easy derivative, containing a 146 bp DNA fragment corresponding to the region from 433,049 to 433,140 of H. pylori G27 genome amplified by PCR with oligonucleotides HBSCbpF/HBSCbpF. This region corresponds to a portion of the promoter region and coding sequence of HPG27_RS02130 (HP1024 according to 26695 annotation). | This work |
| pGEM-T-Easy-HBSspeA | pGEM-T-Easy derivative, containing a 135 bp DNA fragment corresponding to the region from 1,035,104–1,035,195 of H. pylori G27 genome amplified by PCR with oligonucleotides HBSspeAF /HBSspeAR. This region corresponds to a portion of the coding sequence of HPG27_RS02130 (HP0422 according to 26695 annotation). | This work |
| pGEM-T-Easy-HBScbpM1 | pGEM-T-Easy derivative, containing a 91 bp DNA fragment corresponding to the region from 432,991–433,071 of H. pylori G27 genome generated annealing oligonucleotides Mut1F/Mut1R. This region corresponds to a portion of the promoter region and coding sequence of HPG27_RS02130 (HP1024 according to 26695 annotation). | This work |
| pGEM-T-Easy-HBScbpM2 | pGEM-T-Easy derivative, containing a 146 bp DNA fragment corresponding to the region from 433,049–433,140 of H. pylori G27 genome amplified by all around PCR with oligonucleotides Mut2F/Mut2R and using as DNA template the plasmid pGEM-T-Easy-HBScbp. This region corresponds to a portion of the promoter region and coding sequence of HPG27_RS02130 (HP1024 according to 26695 annotation). | This work |
| pGEM-T-Easy-HBScbpM1 + 2 | pGEM-T-Easy derivative, containing a 146 bp DNA fragment corresponding to the region from 433,049–433,140 of H. pylori G27 genome amplified by all around PCR with oligonucleotides Mut2F/Mut2DR and using as DNA template the plasmid pGEM-T-Easy-HBScbp. This region corresponds to a portion of the promoter region and coding sequence of HPG27_RS02130 (HP1024 according to 26695 annotation). | This work |
| pGEM-T-Easy-HBScbpM3 | pGEM-T-Easy derivative, containing a 91 bp DNA fragment corresponding to the region from 432,991–433,071 of H. pylori G27 genome generated annealing oligonucleotides Mut3F/Mut3R. This region corresponds to a portion of the promoter region and coding sequence of HPG27_RS02130 (HP1024 according to 26695 annotation). | This work |
| pGEM-T-Easy-HBScbpM4 | pGEM-T-Easy derivative, containing a 91 bp DNA fragment corresponding to the region from 432,991–433,071 of H. pylori G27 genome generated annealing oligonucleotides Mut4F/Mut4R. This region corresponds to a portion of the promoter region and coding sequence of HPG27_RS02130 (HP1024 according to 26695 annotation). | This work |
| pGEM-T-Easy-HBScbpM5 | pGEM-T-Easy derivative, containing a 91 bp DNA fragment corresponding to the region from 432,991–433,071 of H. pylori G27 genome generated annealing oligonucleotides Mut5F/Mut5R. This region corresponds to a portion of the promoter region and coding sequence of HPG27_RS02130 (HP1024 according to 26695 annotation). | This work |
| pVCC | Vector carrying the luxCDABE cassette | Vannini et al., 2014 |
| pVAC::Km | Cloning Vector, Kmr | Delany et al., 2002b |
| pVAC::CAT | pVAC::Km derivative, carrying a BglII/BamHI cat cassette from pBS::cat (Vannini et al., 2012). | This work |
| pVAC-Pcbpwt-lux | pVAC-CAT derivative, containing a 146 bp DNA fragment amplified by PCR with oligonucleotides HBSCbpFEco/HBSCbpRBamHI, encompassing the Pcbp wt promoter region and a 1,000 bp DNA fragment amplified by PCR with oligonucleotides LuxF/LuxR of the luxC gene. | This work |
| pVAC-PcbpM1+2-lux | pVAC-CAT derivative, containing a 146 bp DNA fragment amplified by PCR with oligonucleotides HBSCbpFEco/HBSCbpRBamHI (using as DNA template the plasmid pGEM-T-Easy-HBScbpM1 + 2), encompassing the Pcbp M1 + 2 promoter region and a 1,000 bp DNA fragment amplified by PCR with oligonucleotides LuxF/LuxR of the luxC gene. | This work |
| pGEM3-hspR::Km | pGEM3 vector carrying the Campylobacter coli Kanamycin cassette flanked by a 1067 bp fragment comprising the cbpA gene (HPG27_RS02130) and a 716 bp fragment comprising the 5′ region of the rarA gene (HPG27_RS02120). | Spohn and Scarlato, 1999 |
| pET22b | Expression vector, allow C-terminal histidine-tag gene fusion; Ampr | Novagen |
| pET22b-HspR | pET22b derivative, containing the HspR coding sequence amplified by PCR | Spohn and Scarlato, 1999 |
RNA Isolation
Helicobacter pylori strains (Table 1) were grown with gentle agitation (120 rpm) in 30 ml of Brucella broth at 37°C until mid-exponential phase (OD = 0.7). For heat-shock treatment, the wild type (WT) culture was split into 15 ml-aliquots and one sample was subjected to heat-shock at 42°C for 30 min (heat-shock sample, HS). A volume of 10 ml cell culture was then added to 1.25 ml of ice-cold EtOH-phenol stop solution (5% acid phenol, in EtOH) to stop growth and prevent RNA degradation. Cells were pelleted, stored at −80°C, and then used to extract total RNA with TRI-reagent (Sigma-Aldrich), according to manufacturer’s protocol.
RNA-seq: Library Preparation, Sequencing and Analyses
Ribosomal RNAs were depleted starting from 1 μg of total RNA from each of the conditions analyzed by using the RiboZero Gram negative kit (Epicentre, Illumina). Strand specific RNA-seq libraries were prepared by using the ScriptSeqTM v2 RNAseq library preparation kit (Epicentre, Illumina) starting from 50 ng of previously rRNA-depleted RNA from each biological replicate and for all the conditions analyzed. Then, each library was multiplexed in equal amounts and sequenced on a GAIIX Illumina sequencer and 85 bp reads were produced. A minimum of 7 Million reads were obtained for each of the samples and for each replica. Bowtie 2 (v2.2.6) (Langmead and Salzberg, 2012) was used to align raw reads to H. pylori G27 genome selecting end-to-end mapping and specifying non-deterministic option. High quality reads were selected requiring: for uniquely mapping reads MAPQ > 30 (mapping quality) and alignment score >−15; for multi-mapping reads alignment score was set ≥−15. H. pylori G27 RefSeq annotation (GCF_000021165.1) in the version released on sept-2017 was used as the reference for gene annotation to which we manually added validated ncRNAs (Pelliciari et al., 2017; Vannini et al., 2017) (highlighted in yellow in Supplementary Table S1). We also revised the annotation of protein coding genes that, based on our sequencing data, were improperly annotated as pseudogenes in this version of the reference genome (e.g., rpoB, rpoA, hspR), indicating them as “protein-coding∗” in Supplementary Table S1. BEDTools (v2.20.1) (Quinlan and Hall, 2010) and SAMtools (v0.1.19) (Li et al., 2009) were used to verify the library preparation and sequencing performances. In particular, we measured the level of rRNA depletion, which was very efficient (less than 6% of the mapping reads) and strand specific gene coverage, considering only strand specific reads overlapping for at least 50% of their length to the annotated transcripts (see Supplementary Table S2). This analysis revealed that 99% of the transcripts were covered by at least one strand specific read and a minimum of 46 reads were counted on 90% of them. The R package DESeq2 (v1.4.5) (Love et al., 2014) was then used to normalize the counts and to identify differentially expressed genes (DEGs) showing BH adjusted p-value (padj) lower than 0.01 and log2 fold changes (log2FC) > |1| . Raw data are publicly available at Sequence Reads Archive under accession number BioProject PRJNA421261.
To evaluate functional enrichments in the DEGs lists, we retrieved COG functional classes for all the protein coding genes present in our annotation file through the NCBI CDD database (Tatusov et al., 1997). We obtained COG records for 1047 genes, 88 of them were annotated as “function unknown” or “general function prediction only” categories, so we considered a final list of 959 COG annotated genes for functional enrichment analysis. The genes classified as: (1) not coding for proteins, (2) coding for proteins but not annotated in COG or (3) annotated in COG to “function unknown” or “general function prediction only” categories were merged together into the “Unknown function” in the annotation file (see Supplementary Table S1).
Chromatin Immunoprecipitation (ChIP) With α-HspR Polyclonal Antibody
Available α–HspR polyclonal antibody from immunized mice (Vannini et al., 2016) were purified by 3 sequential precipitations with 35% saturated (NH4)2SO4 and subsequent resuspension in water. H. pylori G27 wild type and hspR mutant strains were liquid-grown to an OD600 of 0.7, crosslinked, sonicated and immunoprecipitated as previously described (Pelliciari et al., 2017; Vannini et al., 2017). Briefly, protein-DNA complexes were chemically crosslinked with 1% of formaldehyde, and then DNA was sonicated, at high power, with Bioruptor (Diagenode). HspR-DNA complexes were immunoprecipitated by incubating whole cellular extracts with the α-HspR polyclonal antibody at a 1:30 dilution and then captured with Protein-G conjugated sepharose beads. Cross-linking was reverted for 6h at 65°C. DNA was extracted once with phenol-chloroform and further extracted with chloroform. Finally, DNA was ethanol precipitated with the addition of 1% glycogen (Sigma-Aldrich) and resuspended in 50 μl of double-distilled water as previously described (Vannini et al., 2017).
ChIP-seq: Library Preparation, Sequencing and Analyses
Illumina libraries were prepared following the Illumina TruSeq ChIP-seq DNA sample preparation protocol starting from 5 ng of immunoprecipitated-DNA for each of the strains and each of the two biological replicates. Each library was sequenced on a GAIIx or MiSeq Illumina sequencer and 51 bp single stranded reads were produced. At least 2 Million of raw reads were obtained for each IP sample and biological replicate. Bowtie 2 (v2.2.6) (Langmead and Salzberg, 2012) was used to align raw reads deriving from Input (IP ΔhspR) and IP (IP wt) samples sequencing on the H. pylori G27 genome. End-to-end mapping was performed and non-deterministic option was specified. High quality reads were then selected requiring: for uniquely mapping reads MAPQ > 30 (mapping quality) and alignment score > −10 while, for multi-mapping reads, the alignment score was set ≥−10. On average, more than 98% of them mapped on the H. pylori G27 reference genome. The ChIP-seq data quality was evaluated by using ENCODE quality metrics1 and the values obtained are provided in Supplementary Table S2. To perform peak calling, the Homer (v4.7.2) (Heinz et al., 2010) algorithm was used with default parameters. Briefly, the Homer algorithm finds non-random clusters of reads by looking at the tested sample alone, then each peak is required to have: (1) 4-fold more normalized reads in the sample experiment than in the background control and a cumulative Poisson p-value of 0.0001 to assess the chance that the differences in reads counts between sample and background are statistically significant; (2) read density 4-fold greater than in the surrounding 10 kb region; (3) the ratio between the number of unique positions containing reads in the peak and the expected number of unique positions given the total number of reads in the peak lower than 2. Only the peaks identified in both biological replicates and having overlapping genomic coordinates were considered significantly reliable and included in the final peak list. The peaks having their center within −100/ + 30 bp from a transcription start site (TSS) were defined as promotorial, while the remaining peaks were divided in intragenic, when their center was mapping within a predicted coding region, or intergenic, when it was mapping outside from annotated regions (see Supplementary Table S1 and RNA-seq analysis paragraph for details). TSS were identified first by blasting 50 bp upstream of each of the transcription initiation sites reported by Sharma et al. (2010) in the HP26695 genome on the G27 genome and then by the positioning of our RNA-seq signals. Raw data are publicly available in Sequence Reads Archive under accession number BioProject PRJNA421261.
DNA Techniques
DNA manipulations were performed as described by Sambrook et al. (1989). All restriction and modification enzymes were used according to the manufacturers’ instructions (New England Biolabs). Preparations of plasmid DNA were carried out with NucleoBond Xtra Midi plasmid purification kit (Macherey-Nagel).
Overexpression and Purification of Recombinant HspR Protein
His6-tagged recombinant HspR protein was overexpressed in E. coli BL21 (DE3) cells and affinity purified as previously described (Spohn and Scarlato, 1999; Roncarati et al., 2007). The purified His-HspR protein was dialyzed against two changes of 1X footprinting buffer (10 mM Tris-HCl, pH 8.0; 50 mM NaCl; 10 mM KCl; 5 mM MgCl2; 0.1 mM DTT; 0.01% NP40) avoiding any trace of glycerol, prior to the DNA binding experiment, and stored at −80°C. Protein concentration was determined by Bradford colorimetric assay (BioRad) and purity assayed by SDS-PAGE.
Construction of DNA Probes for in vitro DNA-Binding Assays
Genomic regions of H. pylori G27 encompassing HspR binding sites on hrcA, groES, cbpA promoters and speA coding region were PCR amplified with specific primers (Table 2) and cloned into the pGEM-T-Easy plasmid (Table 1). The M1, M3, M4 and M5 Pcbp mutant probes were generated by annealing complementary oligonucleotides to form a double stranded DNA fragment with compatible overhangs required to clone it in the pGEM-T-Easy plasmid previously digested with the appropriate restriction enzymes. The M2 and M1+2 Pcbp mutant probes were generated through site-directed mutagenesis using the plasmid pGEM-T-Easy, harboring the Pcbp wild type sequence, as DNA template and primers listed in Table 2.
Table 2.
Oligonucleotides used in this study.
| Oligonucleotides | Nucleotide sequences (5’ to 3’)a | Restriction recognition site |
|---|---|---|
| RBS GroF | TCTTCAAAAAGGTTTGTTAATGACGC | None |
| RBS GroR | AGCACATTTTTAGGGATAAGTCAAGC | None |
| RBS hrc F | CGATTTTTCTTTAAAGTTTAGTCTGTATCAC | None |
| Phrc F | ATATGGATCCTACGTCAAGCAAGCGATAACTTTAC | None |
| HBS CbpF | AATTCCTTTTAATTGCACTGAAACGGG | None |
| HBS CbpR | GGTATAAACTCTTGCTCATGAATCACC | None |
| HBS speAF | CCACGAAGCCCTTGTTTTTGC | None |
| HBS speAR | CGCTAAATTCCGTAGGGTGC | None |
| Mut1 F | GATCCAAAATAGTTTTATTAGAATACTATCATAAATCAGGTACCTTAGTCAATCAAGTTTATTGATAATGTTTAGTGGTAATTGAGATTTG | None |
| Mut1 R | GTTTTATCAAAATAATCTTATGATAGTATTTAGTCCATGGAATCAGTTAGTTCAAATAACTATTACAAATCACCATTAACTCTAAACTTAA | EcoRI |
| Mut2 F | AGTCGACAGTTTATTGATAATGTTTAG | SalI |
| Mut2 R | CTAACACTAAAGATTTATGATAGTATTC | None |
| MUTD2R | CTAAGGTACCTGATTTATGATAGTATTC | KpnI |
| Mut3 F | GATCCAAAATAGTTTTATTAGAATACTATCATAAATCTTTAGTGGAGCTCAATCAAGTTTATTGATAATGTTTAGTGGTAATTGAGATTTG | None |
| Mut3 R | GTTTTATCAAAATAATCTTATGATAGTATTTAGAAATCACCTCGAGTTAGTTCAAATAACTATTACAAATCACCATTAACTCTAAACTTAA | None |
| Mut4 F | GATCCAAAATAGTTTTATTAGAATACAGGTACCAATCTTTAGTGTTAGTCAATCAAGTTTATTGATAATGTTTAGTGGTAATTGAGATTTG | None |
| Mut4 R | GTTTTATCAAAATAATCTTATGTCCATGGTTAGAAATCACAATCAGTTAGTTCAAATAACTATTACAAATCACCATTAACTCTAAACTTAA | None |
| Mut5 F | GATCCAAAATAGTTTTATTAGAATACTATCATAAATCTTTAGTGTTAGTCAATCAAGTTTTAGTCGACTGTTTAGTGGTAATTGAGATTTG | None |
| Mut5 R | GTTTTATCAAAATAATCTTATGATAGTATTTAGAAATCACAATCAGTTAGTTCAAAATCAGCTGACAAATCACCATTAACTCTAAACTTAA | None |
| LuxF | ATATGGATCCCAGGCTTGGAGGATACGTATGAC | BamHI |
| LuxR | ATATGGATCCGGCATTCGGTAATATATGCGC | BamHI |
| LuxRTF | ATCATCCGATAACGCGCTCTT | None |
| LuxRTR | ACCGCCCAATTAATCGCATC | None |
| cbpEco | ATATGAATTCAATTCCTTTTAATTGCACTGAAACGGG | EcoRI |
| cbpBam | ATATGGATCCGGTATAAACTCTTGCTCATGAATCACC | BamHI |
| cbpRTRev | TTTAGCTAGGCAATACCACCCGGA | None |
| hspRC | ATATATCTCGAGTTTTTTAAATAAAATCAGTTCATA | XhoI |
| cys-F | CGTTTTAGGGACTTTGGGAGG | None |
| vacA-R | GCTGGTTTTATGCTCTAAACTGG | None |
| ppk RT-F | CGCGCCTTTCTAAATTTCTGGGCA | None |
| ppk RT-R | CCCAAGTCAAAGGCTTGAGCGAAA | None |
aNucleotides added to reconstitute the indicated restriction recognition sites are underlined.
Hydroxyl-Radical Footprinting Assay
Probe DNA fragments obtained by digestion with the appropriate restriction enzymes were 5′ end-labeled with [γ32 P]-ATP and T4 polynucleotide kinase and gel purified. Hydroxyl-radical footprinting experiments were performed as previously described (Pelliciari et al., 2017) with some modifications. Approximately 20 fmol of labeled probes were incubated with increasing concentration of HspR protein in hydroxyl-radical footprinting buffer (10 mM Tris-HCl, pH 8.0; 50 mM NaCl; 10 mM KCl; 5 mM MgCl2; 0.1 mM DTT; 0.01% NP40) for 15 min at room temperature, including 200 ng of sonicated salmon sperm DNA as a non-specific competitor in a final volume of 30 μl. Partial digestions of the labeled probes were achieved using 2 μl each of the following solutions: 125 mM Fe (NH4)2(SO4)2 250 mM EDTA, 1% H2O2 and 100 mM DTT. After 2 min incubation, the reaction was quenched by the addition of 25 μl of OH Stop Buffer (4% glycerol; 600 mM NaOAc, pH 5.2; 100 ng/μl sonicated salmon sperm DNA). Samples were phenol/chloroform extracted, ethanol precipitated and resuspended in 12 μl of Formamide Loading Buffer. Next, samples were denatured at 100°C for 5 min, separated on a 8M urea-8.4% polyacrylamide sequencing gel in TBE buffer and autoradiographed.
Generation of H. pylori Pcbp-lux Reporter Strains
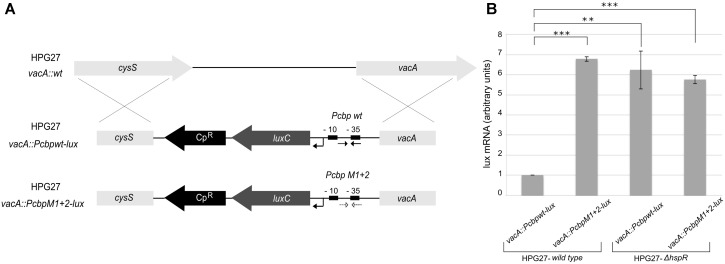
The wild type Pcbp and mutated Pcbp M1 + 2 promoter regions were PCR amplified with specific primers listed in Table 2, using as template the H. pylori G27 genomic DNA and the plasmid pGEM-T-Easy-HBScbp M1 + 2 (Table 1), respectively. Then, the generated DNA fragments were digested with appropriate restriction enzymes and cloned into the pVAC plasmid (Table 1). The luxC gene was PCR amplified from the pVCC vector (Table 1) and cloned downstream the Pcbp promoters into the pVAC vector. These plasmids were used to transform the H. pylori G27 wild type acceptor strain in the vacA locus. The chloramphenicol-selected mutant strains were expanded and the correct insertion was confirmed by PCR using oligonucleotides pair cys-F/vacA-R as primers (Table 2). The H. pylori Pcbp-lux, hspR::Km reporter strains (Table 1) were generated transforming the H. pylori vacA::Pcbp wt-lux and the H. pylori vacA::PcbpM1 + 2-lux acceptor strains (Table 1) with the plasmid pGEM3-hspR::Km (Table 1). The kanamycin-selected mutant strains were expanded and the correct insertion was confirmed by PCR using oligonucleotides pair cbpRTRev/hspRC as primers (Table 2).
qRT-PCR Analysis
Synthesis of cDNA and qRT-PCR analysis were carried out as previously described (Pelliciari et al., 2015). Briefly, for cDNA synthesis 1 μg of DNA-free RNA was incubated with 50 ng of random hexamers (Invitrogen), dNTPs mix (1 mM each), 5 U of AMV-Reverse Transcriptase (Promega), and incubated for 1 h at 37°C. For qRT-PCR analyses, 2 μl of diluted (1:10) cDNA samples were mixed with 5 μl of 2X Power Up SYBR Green master mix (ThermoFisher Scientific) and specific oligonucleotides for the genes of interest (Table 2) at 400 nM concentration in a final volume of 10 μl. qRT-PCR experiments were performed using the following cycling protocol: 95°C for 2 min, then 40 cycles consisting of a denaturation step for 5 s at 95°C followed by 30 s at 60°C (annealing and extension steps). Data were analyzed using the ΔΔCt method, using the housekeeping ppk gene, known to be constitutively expressed, as internal reference, using oligonucleotides ppk RT-F and ppk RT-R as primers (Table 2) (Muller et al., 2011; Agriesti et al., 2014).
Results
RNA-seq Analysis Identifies HspR-Dependent Heat-Shock Gene Transcripts
To define the HspR contribution to the heat-shock response, we performed a strand-specific whole transcriptome analysis of the wild type H. pylori G27 strain and of ΔhspR mutant both grown to the exponential growth phase at 37°C and of the wild type strain subjected to 30 min heat-shock at 42°C (Supplementary Table S2, Materials and Methods).
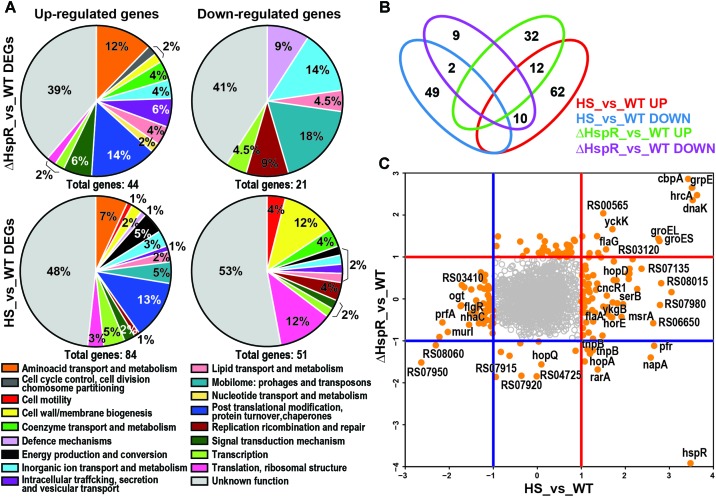
We defined the role of HspR in standard growth conditions by comparing the transcriptome of the ΔhspR mutant to that of the H. pylori wild type strain, both grown at 37°C (ΔhspR_vs_WT). This analysis showed a total of 65 deregulated genes (log2FC > |1| padj < 0.01) upon hspR gene deletion. Of these, 21 were down-regulated and 44 were up-regulated genes (Figure 1A and Supplementary Table S3, sheet B). As expected, among the up-regulated genes we found groES, groEL, grpE, dnaK, and cbpA (Spohn and Scarlato, 1999), which strongly contributed to “Post-translational modification, protein turnover, chaperones” functional enrichment among (padj = 0.01) ΔhspR de-repressed genes (Figures 1A,B), and hrcA transcriptional regulator (Figure 1B). As expected, because of the hspR genomic deletion, this gene appeared as down-regulated. The rarA gene, mapping downstream of the hspR gene, possibly due a polar effect of the deletion mutant also appeared as a down-regulated gene. Several transposase coding genes, producing the “Mobilome: prophages, transposons” category enrichment (padj = 0.00009), and several genes involved in the “Inorganic ion transport and metabolism” were also down-regulated.
FIGURE 1.
Role of HspR in heat-shock response. (A) Pie charts showing COGs functional annotation of the differentially expressed genes outlined in the ΔHspR_vs_WT and HS_vs_WT (Heat-Shock vs. Wilde Type) comparisons, respectively, subdivided into up-regulated (left) and down-regulated (right) groups. The abundance of each category is indicated as a percentage as well as the total number of genes included in each group. (B) Venn diagram showing the number of genes in common or peculiar in the four previously described gene groups, red numbers highlight commonly regulated genes. (C) log2FC plot showing for each gene HS_vs_WT value on x-axis and HspR_vs_WT value on y-axis. DEGs (log2FC > |1| and padj < 0.01 are represented as orange filled circles; empty gray circles correspond to non-differentially expressed genes. Blue and red lines indicate log2FC < −1 and log2FC > 1 thresholds, respectively, subdividing coherently and incoherently regulated genes.
Upon heat-shock at 42°C the transcriptome of the H. pylori wild type G27 strain showed a total of 134 differentially expressed genes (log2FC > 1 padj < 0.01) when compared to the wild type sample not subjected to heat-shock (HS_vs_WT). Of these genes, 83 were up-regulated and 51 were down-regulated (Supplementary Table S3, sheet A). Functional annotation and enrichment analysis (see Materials and Methods) revealed that up-regulated genes were enriched in “Post-translational modification, protein turnover, chaperones” (padj = 0.0007) and “Mobilome: prophages, transposons” class (padj = 0.008; Figure 1A). Several of the 51 down-regulated genes were also annotated to “Cell wall/membrane/envelope biogenesis” and “Translation, ribosomal structure and biogenesis” categories (Figure 1A) without statistical significance.
Comparing the transcriptome of the heat-shock response (134 deregulated genes) to that of the hspR deletion mutant (65 deregulated genes) we identified 25 genes that were deregulated in both datasets (Figures 1B,C, red numbers). Among these genes, 10 were oppositely regulated and 14 were similarly regulated: 2 down-regulated and 12 up-regulated (Figure 1B, bottom left and right quadrants of the graph and Figure 1C). The similarly up-regulated genes included all the genes of the operons already known to be directly repressed by HspR and induced by heat-shock (Spohn and Scarlato, 1999). In addition, we found genes coding for amino acid transporters yckK and yckJ, flagellar protein flaG, a polyisoprenoid-binding protein and two genes coding for hypothetical proteins, which could be new targets under HspR direct control (Figures 1B,C). The remaining eleven genes were induced by heat-shock and repressed in the hspR mutant strain, thus showing opposite behavior. Considering that HspR acts as a repressor factor, these genes are probably indirectly controlled by HspR.
Overall, this analysis highlights that HspR could be involved in the control of maximum 18% (25 out of 134) of the genes whose transcription is heat responsive and that it represses the transcription of only 14 genes.
Genome-Wide in vivo Identification of HspR Binding Sites Through ChIP-seq
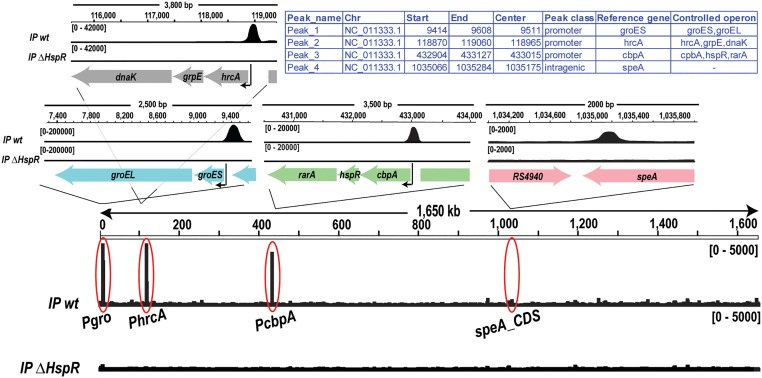
To identify the genomic regions bound in vivo by HspR, therefore, the genes directly controlled by HspR binding, we performed a Chromatin Immunoprecipitation assays followed by deep sequencing (ChIP-seq) in H. pylori G27 wild type and ΔhspR strains (Figure 2). To identify HspR bound regions (peaks), ChIP-seq signals obtained from each wild type sample (IP wt) were compared to those resulting from the pool of ΔhspR mutant samples (IP ΔhspR) (see Material and Methods for details). Only the significant peaks in both replicates were considered in the final peak list. Surprisingly, this analysis identified only four reproducible HspR binding regions (Figure 2), which were annotated with respect to the latest genome annotation of the H. pylori G27 strain (GCF_000021165.1). According to this annotation, three peaks were classified as promotorial and confirmed HspR direct control for 8 genes of the groES-groEL, hrcA-grpE-dnaK and cbpA-hspR-rarA operons. The remaining peak, mapping inside the coding region of speA gene, was classified as intragenic. Thus, these data suggest that the HspR direct regulon is very restricted and limited to the three multicistronic heat-shock operons. Moreover, through ChIP-seq analysis we confirmed in vivo also the existence of at least one intracistronic binding site, apparently not associated to transcriptional regulation.
FIGURE 2.
Genome-wide in vivo identification of the HspR binding sites. Genome-wide signals of HspR bindings in the wild type genotype (IP wt, upper track) and in hspR deletion mutant used as negative control (IP ΔHspR, lower track). The regions corresponding to the four HspR binding sites revealed by the analysis are magnified in the upper panels showing each peak in its genomic context including annotated genes (thick arrows) and the transcriptional start site (black thin arrow), when present.
HspR Binds Extended DNA Regions and Adopts a Peculiar Binding Architecture
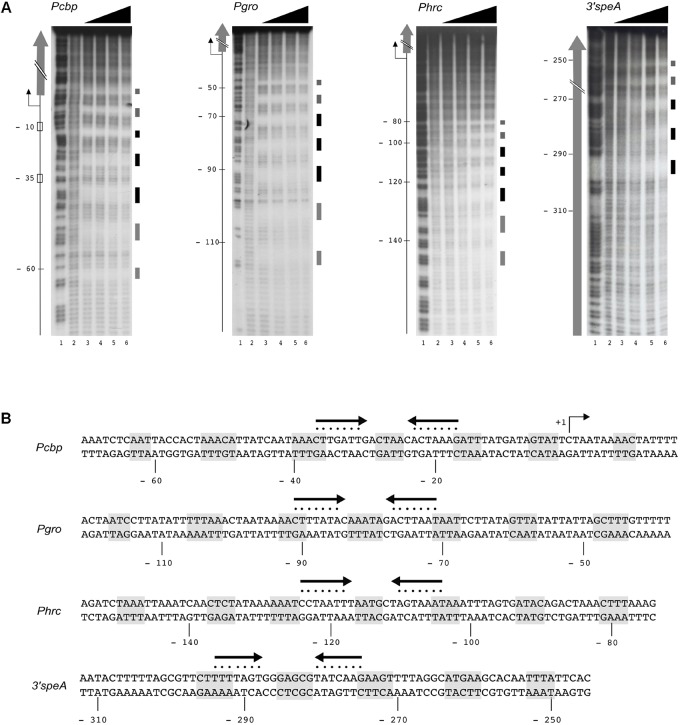
Previous DNase I footprinting assays showed that H. pylori HspR binds directly to extended DNA regions of the promoters of the three heat-shock operons (Spohn and Scarlato, 1999; Roncarati et al., 2007). In particular, binding of HspR to its operators resulted in protection of large DNA regions of 70–80 bp with six bands of enhanced DNase I sensitivity at both adjacent and internal sites. In order to further characterize HspR-DNA interactions and to get more detailed information on the HspR DNA-binding architecture, we carried out hydroxyl-radical footprinting experiments on the genomic targets found in ChIP-seq assay. Figure 3A shows the results of hydroxyl-radical footprintings performed with increasing concentrations of recombinant purified HspR on the promoter region of the three heat-shock operons (Pcbp, Pgro, Phrc) and on the 3′ region of the speA coding sequence. According to previous observations (Spohn and Scarlato, 1999; Delany et al., 2002c; Roncarati et al., 2007) the protected regions on the Pcbp, Pgro and Phrc promoters map, respectively, from position −63 to +10, from −117 to −44 and from −150 to −82 with respect to the transcriptional start site. Furthermore, the HspR binding site located in the coding region of speA gene spans from nucleotide position −298 to −250 with respect to the translational stop codon. Intriguingly, HspR binding on these targets results in a peculiar periodic pattern of short protected regions from radical digestion, which appears to be slightly different between the promotorial and intragenic binding sites. Indeed, HspR binding on Pcbp, Pgro and Phrc promoter regions results in 7 short protected DNA tracts separated by non-protected regions of 7/8 nucleotides, while the binding site located inside the speA coding sequence appears at higher protein concentrations and is characterized by 5 short protected regions instead of 7. Data obtained from DNA-binding assays are schematized in Figure 3B, which reports the nucleotide sequence of the HspR binding sites (operator) on the Pcbp, Pgro, Phrc promoters and on the speA coding region. Notably, in all the operators we identify one inverted repeat (represented by converging arrows in Figure 3B) similar to the HAIR motif proposed as a consensus sequence for the HspR protein of S. coelicolor (CTTGAGT-N7-ACTCAAG) (Grandvalet et al., 1999). It is worth noting that in the operators of the three heat-shock operons’ promoters the inverted repeat is located in a central position of the HspR binding sites, suggesting that it could play an important role in nucleating HspR binding to DNA. Therefore, the heat-shock transcriptional repressor HspR binds, with a peculiar DNA-binding pattern of short stretches of protected regions, spanning over about 70-80 bp of DNA harboring a conserved inverted repeat similar to the HAIR consensus sequence of Streptomyces spp.
FIGURE 3.
HspR binds extended DNA regions. (A) Hydroxyl-radical footprint experiments on HspR positive target genes identified by ChIP-seq analysis. From left to right, specific DNA probes for Pcbp, Pgro, Phrc, and speA fragments were radioactively end-labeled on the coding strands, and incubated with increasing amounts of purified HspR protein prior hydroxyl-radical digestion. Pcbp, Pgro, Phrc labeled probes were mixed with 0; 47.5; 95; 190; 380 nM HspR (lanes 2–6); while speA probe was mixed with 0; 60.7; 121.5; 243; 486 nM HspR (lanes 2–6). Purified DNA fragments were separated on a polyacrylamide denaturing gel along with a G + A sequence reaction ladder as reference marker (lane 1 in all the panels). On the right of each panel, black and gray boxes represent strong and weak HspR protected regions, respectively. On the left, the –10 and –35 regions and the transcriptional start site (+1, bent arrow) are indicated and the open reading frames are depicted with vertical gray arrows. The relative position of the HspR binding site on the promoter regions of Pcbp, Pgro, Phrc is reported with respect to the transcriptional start site, while those mapped on the coding sequence of speA gene are reported with respect to the translational stop codon. (B) Nucleotide sequences of HspR binding sites on the promoter regions of the three heat-shock operons (Pcbp, Pgro, Phrc) and on the 3′ coding region of the speA gene. The HspR protected regions identified in (A) are shaded in gray, while the inverted repeat sequences similar to the HAIR consensus motif are depicted as converging black arrows and each nucleotide of the motif is marked with a dot. Nucleotide positions with respect to the transcription initiation sites are reported on the non-coding strand.
The Central HAIR-Like Motif Is Essential for HspR Binding to DNA
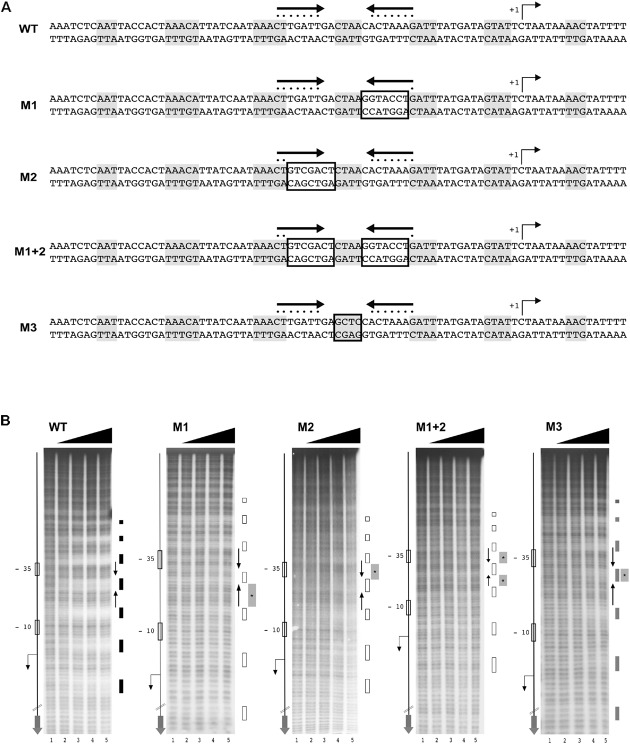
To ascertain the functional importance of the HAIR-like sequence elements in HspR binding to DNA, we decided to introduce bases substitutions in the inverted repeat and monitor their effects on HspR DNA-binding through hydroxyl-radical footprinting assays. A schematic representation of the wild type and mutant sequences of the HspR binding site on the Pcbp region is reported in Figure 4A. As shown in Figure 4B, mutation of one or both arms (M1, M2, or M1 + 2) of the HAIR-like sequence completely abolished HspR binding to the DNA in the concentration range tested. In particular, partial (M1 and M2) or total substitution (M1 + 2) of the HAIR-like sequence prevented HspR binding to both, the mutagenized sequence (central portion of the probe) and the flanking upstream and downstream regions of the HAIR-like motif. It is worth mentioning that regions protected in hydroxyl-radical footprinting experiments reflect limited accessibility of radical ions to the DNA minor groove and, for this reason, these protected regions do not necessarily represent the portions of the probe directly contacted by the HspR protein, but regions surrounding short stretches of contacted nucleotides. Considering that HspR footprint regions surround the conserved HAIR-like element (Figure 4), our data suggest that HspR could interact with the HAIR-like element in the DNA major groove narrowing the adjacent minor grooves, which results in the protection observed in vitro by hydroxyl-radical footprinting. Furthermore, mutation of the non-conserved sequence in between the two inverted repeats (probe M3 in Figure 4A) showed a barely detectable protection of HspR only upon addition of high amount of the protein to the reaction (Figure 4B, lane 6 of probe M3). This is likely due to a significant loss of protein affinity to the DNA, suggesting that, besides the HAIR-like motif, also this non-conserved DNA element is important for HspR binding. These data show for the first time that in H. pylori the HAIR-like sequence is an essential DNA element for the specific binding of HspR to the Pcbp promoter region. Likely, HspR binding to the operator is driven by a specific recognition of determinants within the HAIR-like sequence, including the non-conserved spacer between the inverted repeat.
FIGURE 4.
The HAIR-like motif is essential for HspR binding to DNA. (A) Schematic representation of wild type and four different Pcbp mutant probes, in which one (M1 and M2) or both arms (M1 + 2) of the inverted repeat and the protected region between these sequences (M3) have been mutagenized by base substitution. HspR protected regions on Pcbp are shaded in gray and the inverted repeat sequences (HAIR-like motif) is represented by converging black arrows and their nucleotides marked with dots. In each DNA probe, mutagenized nucleotides are boxed. (B) From left to right, hydroxyl-radical footprint experiments on wild type and the indicated mutants of the Pcbp promoter region. Symbols are as described in the legend to Figure 3A. Wild type (WT) and indicated mutants DNA probes were incubated with increasing amounts of purified HspR protein and submitted to hydroxyl-radical digestion (see legend to Figure 3). Black, gray, and empty boxes to the right of each panel denote strong, weak, and loss of protection by HspR, respectively. Black converging arrows to the right of each panel mark the positions of the HAIR-like inverted repeat sequences, while gray boxes with an internal asterisk indicate the mutagenized regions.
To characterize in vivo the functional significance of the central HAIR-like motif, we generated a reporter construct in which the wild type or HAIR-like mutant Pcbp promoter (PcbpM1 + 2) was fused upstream of a 5′ fragment of the luxCDABE reporter cassette and, upon integration in H. pylori chromosome, transcript level was assayed (through RT–qPCR with luxC specific primers) during exponential growth (Figure 5). When the central HAIR-like inverted repeat was mutated in the G27 wild type strain, a consistent increase in the amount of transcript from the PcbpM1 + 2 promoter with respect to the wild type Pcbp promoter was observed (Figure 5B). The about 6.5-fold increase of transcript from the PcbpM1 + 2 promoter is in line with the previously observed de-repression of the Pcbp promoter in the H. pylori hspR-mutant strain (Spohn et al., 2004). Accordingly, in a hspR null background a similar high amount of transcripts from the Pcbpwt and the PcbpM1 + 2 promoters was observed (Figure 5B). These in vivo data are consistent with the in vitro observations, indicating that the central HAIR-like motif drives specific binding of HspR to its operator on Pcbp with concomitant promoter transcriptional repression.
FIGURE 5.
In vivo H. pylori transcripts levels from the Pcbp promoter harboring WT or mutant HAIR-like sequences. (A) Schematic representation of Pcbpwt-lux and PcbpM1 + 2-lux reporter constructs obtained transforming the H. pylori G27 wild type acceptor strain by double homologous recombination in the vacA locus, and selected by chloramphenicol resistance (CpR). The wild type (Pcbpwt-lux) or HAIR-like mutant (PcbpM1 + 2-lux) Pcbp promoter is inserted upstream of a luxC reporter gene. The HAIR inverted repeat sequences are indicated by converging black arrows in the Pcbp WT promoter (Pcbpwt-lux) and by converging dotted arrows in the Pcbp HAIR-like mutant promoter (PcbpM1 + 2-lux). In each reporter construct, the –10 and –35 regions are depicted as black boxes and the transcriptional start site as a bent arrow. (B) Transcript levels of Pcbp wild type and Pcbp HAIR-like mutant promoters fused with lux reporter gene were assayed by qRT-PCR in the wild type and hspR deletion mutant strains using specific oligonucleotides for the luxC gene (LuxRTF/R). Mean values from three independent biological samples are reported in the graph, with error bars indicating standard deviation and asterisks marking statistical significance calculated by a Student’s t-test (∗∗∗p-value < 0.001; ∗∗p-value < 0.01).
Non-conserved DNA Regions, Surrounding the HAIR-Like Motif, Are Necessary for HspR to Fully Occupy Its Extended DNA Binding Site
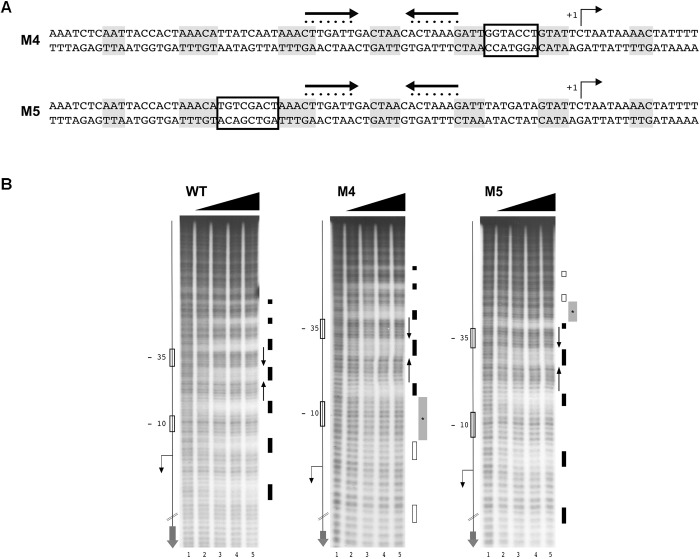
In order to understand if DNA sequences flanking the HAIR-like motif are important for HspR-DNA recognition and binding, we designed two mutant probes of the Pcbp promoter and assayed for HspR binding by hydroxyl-radical footprintings. Mutations were introduced in the non-conserved spacer region between two 4-bp protected tracts on both sides of the inverted repeat (Figure 6A, M4 and M5). Surprisingly, as shown in Figure 6B, the addition of increasing amounts of HspR to the mutant probes led to the disappearance of regions of protection (marked in light gray) only on the side of the mutated region, while it was unaffected on both the inverted repeat of the HAIR-like sequence and on the opposite side of the mutation (Figure 6B, M4 and M5). In conclusion, these data support the pivotal role of the HAIR-like motif for HspR-DNA binding in H. pylori and demonstrate that also other non-conserved DNA regions surrounding the HAIR-like motif are important elements that allow HspR to completely occupy its extended binding sites.
FIGURE 6.
Non-conserved regions surrounding the HAIR-motif are required for HspR binding. (A) Schematic representation of WT and two Pcbp mutant probes, in which the spacer regions highlighted with empty boxes were mutagenized by base substitution. Symbols are as in the legend to Figure 3. (B) From left to right, hydroxyl-radical footprint experiments on WT and mutant probes described in (A) (M4 and M5) with increasing amounts of purified HspR protein detailed in the legend to Figure 3.
Discussion
The heat-shock response is a universal mechanism of cellular protection against sudden adverse environmental growth conditions and has been observed in every bacterial species investigated (Roncarati and Scarlato, 2017). It consists of a set of well-coordinated responses and processes, mostly involving the strictly regulated expression of various heat-shock proteins (HSPs) and chaperones. In H. pylori their expression level is tightly modulated by the concerted action of two transcriptional repressors, HrcA and HspR, with this latter acting as the master regulator of this circuit (Danielli and Scarlato, 2010). In the present study, we examined the heat-shock regulon in H. pylori and discriminated between direct and indirect transcriptional response mediated by HspR. As shown by transcriptome analyses, heat-shock treatment triggered changes in the transcript levels of 135 genes. Of these, 83 genes appeared up-regulated and 51 genes appeared down regulated (Figure 1A). Accordingly, the heat-shock operons groES-groEL, hrcA-grpE-dnaK and cbpA-hspR-rarA, coding for the major chaperones and heat-shock proteins of H. pylori were clearly up-regulated. With the exception of the hspR and rarA genes, these operons, known to be directly repressed by HspR (Spohn and Scarlato, 1999; Roncarati et al., 2007), were coherently up-regulated also in the ΔhspR mutant strain. On the other hand, HspR seems to affect in a positive or negative manner the transcription of other 59 genes involved in diverse cellular processes and not strictly associated to heat-shock (Figure 1A). In contrast, ChIP-seq analysis showed only four already known in vivo HspR genomic binding sites, three of which are associated to the promoter regions of the heat-shock operons and one mapping within the coding sequence of the speA gene (Figure 2 and Spohn and Scarlato, 1999; Delany et al., 2002c; Roncarati et al., 2007). This apparent discrepancy between the RNA-seq and ChIP-seq results is in agreement with other HspR studies conducted in S. coelicolor and Mycobacterium tuberculosis (Stewart et al., 2002; Bucca et al., 2003). In these bacteria, HspR alone or in combination with other transcriptional regulators controls transcription of a limited number of genes coding for chaperones and heat-shock proteins. In H. pylori, hspR deletion affects the transcript abundance of a high number of genes not directly controlled by HspR. Likely, regulation by HspR can also be exerted in an indirect manner through a still unknown molecular mechanism. Recalling the presence of the hrcA gene among the direct targets of HspR, it is tempting to speculate that many of the genes deregulated by the hspR deletion and lacking a HspR binding site arise from altered levels of the HrcA regulator in the ΔhspR mutant. Furthermore, the possibility that up- and/or down-regulated genes in the ΔhspR mutant strain might arise from the enhanced synthesis of one or more members of the HspR regulon cannot be ruled out and remains to be elucidated.
Although several chaperones and heat-shock proteins of H. pylori are induced by a temperature upshift (Supplementary Table S3), surprisingly, the transcription of genes coding for stress related proteases seems to be unaffected by the heat challenge. Furthermore, while in some other bacteria, like for example Campylobacter jejuni and S. coelicolor, several protease-encoding genes have been shown to belong to the HspR regulon (Bucca et al., 2003; Holmes et al., 2010), we demonstrated that in H. pylori, these genes are not controlled by the HspR repressor. Considering that the amount of proteases is expected to increase in response to different stress insults encountered by the pathogen, an interesting hypothesis considers the existence of post-transcriptional or post-translational control strategies, which would provide enhanced levels of these crucial players during adverse environmental growth conditions.
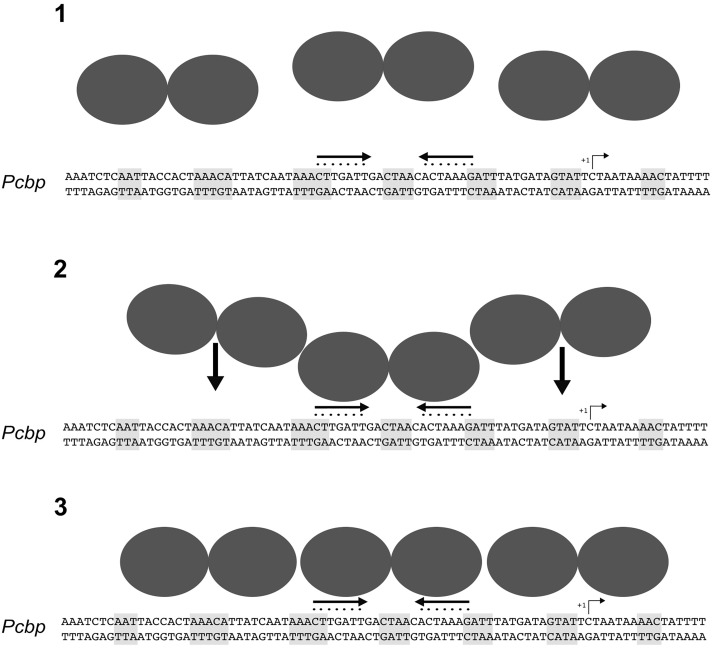
Previous DNase I footprinting assays of HspR showed protection of large DNA regions of about 70 bp upstream of the promoters controlling transcription of three heat-shock operons (Spohn and Scarlato, 1999; Roncarati et al., 2007). To deepen our understanding of the molecular mechanisms controlling HspR-DNA interactions, we set up high-resolution hydroxyl-radical footprinting assays on its target gene probes. Results showed that HspR binds extended DNA regions with a peculiar short periodic pattern, which appears to be slightly different between promoter and intragenic regions (Figure 3A). Binding of HspR to promoter regions shows 7 short protected DNA tracts spaced by non-protected regions of 7/8 nucleotides, while binding within the speA coding sequence appears at higher protein concentrations and shows only 5 short protected regions. This different binding pattern could be related to the fact that the latter binding site on the coding sequence of speA gene appears to be not associated to transcriptional regulation. In fact, RNA-seq analysis revealed no changes in the level of speA transcript in the hspR mutant strain, nor of neighboring genes. This is not surprising, as the advent of the “omics” era highlighted binding of regulatory proteins to a number of binding sites not associated to regulation, such as the H. pylori Fur repressor (Danielli et al., 2006; Vannini et al., 2017), the E. coli CRP activator, and the RNA polymerase enzyme (Grainger et al., 2005). However, all four H. pylori HspR binding sites show an inverted repeat with similarities to the HAIR consensus sequence of Streptomyces spp. (Grandvalet et al., 1999) (Figure 3B). These HAIR-like motifs map in the central position of the HspR binding sites on the three heat-shock operons’ promoters and appear to be an essential DNA element for specificity of protein binding. In fact, mutation of one or both arms of this inverted repeat completely abolished the HspR binding to the operator (Figure 4) and prevented in vivo HspR-dependent repression of Pcbp (Figure 5), demonstrating for the first time that the HAIR-like sequence is functionally important also in H. pylori. The DNA-binding mechanism of HspR on its target operators is unique among H. pylori transcriptional regulators characterized so far. Well-studied H. pylori regulatory proteins, such as HP1043, HrcA, NikR and Fur appear to recognize conserved sequence motifs as dimers and protect limited DNA regions (Roncarati et al., 2014, 2016; Pelliciari et al., 2017; Vannini et al., 2017). In the case of Fur repressor, however, it has been shown that several Fur-regulated promoters harbor multiple Fur boxes and, upon Fur binding, large regions of the promoter result occupied by this metal-dependent repressor (Delany et al., 2002a; Roncarati et al., 2016). However, data presented in this work suggest a completely different DNA-binding mechanism for HspR. HspR-controlled promoters are characterized by a single, conserved inverted repeat (HAIR-like sequence) that drives DNA specific recognition. In contrast to other H. pylori regulators, HspR DNA binding extends over a large portion of the promoter DNA, resulting in the characteristic extended protection in in vitro footprinting assays (Roncarati et al., 2007). This peculiar behavior of H. pylori HspR appears to be different from the other HspR homologs that have been characterized at the molecular level. For example, in S. coelicolor HspR binds extended DNA regions harboring three inverted repeat sequences (IR1, IR2, IR3) in the promoter region of its DNA targets (Bucca et al., 1995), while in H. pylori HspR requires only one inverted repeat on the center of the binding site. Also, no additional conserved sequences similar to the HAIR-like motif have been detected. Therefore, we suppose that the molecular mechanism through which H. pylori HspR binds to such extended DNA regions is peculiar and differs from the one adopted by the S. coelicolor HspR. Moreover, site-directed mutagenesis of the Pcbp promoter pointed out that additional non-conserved DNA regions located between (Figure 4B, M3) and flanking (Figure 6B, M4 and M5) the HAIR-like motif are important elements for DNA recognition and binding of HspR to the operator. Intriguingly, mutation of one of these elements impaired binding of HspR on the side of the mutation and showed no effects on the binding on the HAIR-like motif on the opposite side of the mutation (Figure 6). This finding is compatible with a cooperative mechanism of HspR binding. Possibly, a dimer of HspR recognizes and binds to the HAIR-like sequence and this in turn drives binding of additional dimers on both sides of the HAIR-like motif (Figure 7). Evidences that HspR could bind its DNA sequences as a dimer have been shown in previous studies, in which it was demonstrated that homologs of H. pylori HspR repressor protein could exists in a dynamic state between the dimeric and monomeric forms in solution (Parijat and Batra, 2015). Moreover, Spohn et al. (2004) provided evidences that H. pylori HspR is able to form high order oligomers. In conclusion, these results provide a more detailed comprehension of the interaction between HspR and its target DNA sequences and, at least for the Pcbp promoter, let us to propose a cooperative DNA-binding mechanism of three HspR dimers on this operator as schematized in Figure 7. In this model, an HspR dimer binds to the central DNA region harboring the HAIR-like motif that acts as a nucleation center for protein–protein interactions driving cooperative binding of HspR homodimers to the specific and still unknown determinants of the DNA sequences surrounding the HAIR-like motif. Giving the low level of sequence conservation of the HAIR-like motifs across the HspR binding sites and the imperfect nature of the inverted repeats (i.e., low conservation of the two arms of the inverted repeats), it can also be speculated that HspR DNA binding could be driven by sequence-dependent local shape variations rather than by a base readout mechanism, or by a combination of these two mechanisms. In other words, the crucial role of the HAIR-like motifs for HspR binding could be explained by taking into account local DNA shape variations imposed by a peculiar succession of purine/pyrimidine, rather than considering only the unique chemical signatures of the individual DNA bases. This behavior is common among regulators of MerR family, to which the H. pylori HspR belongs to, employing indirect (shape) readout as part of their DNA binding mechanisms. These considerations could help also to explain our data on the importance of the central region of the HAIR-like motif and of the non-conserved regions on both sides of the HAIR-like motif (Figure 4, M3 and Figure 6, M4 and M5).
FIGURE 7.
Model of the molecular mechanism for HspR-DNA binding. In solution, HspR dimerizes (1) and binds with high affinity to the DNA region containing the HAIR consensus sequence (2). This binding could act as a nucleation center driving cooperative binding of two HspR dimers on the flanking regions (3), likely recognizing still unknown sequence determinants.
Author Contributions
AD, CP, DR, and VS conceived and designed the experiments. SPE, EP, EF, TB, CP, and AV performed the experiments. SPE, EP, and SPU carried out the data analysis. DR and VS wrote the paper with contributors CP and EP. All the authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by Grants from the Italian Ministry of Education and University (2010P3S8BR_003 to VS, and 2010P3S8BR_002 to CP) and by a grant from the University of Bologna to VS. SPE and TB are the recipients of fellowships from the Ph.D. program in Cellular and Molecular Biology of the University of Bologna.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01887/full#supplementary-material
Annotation of coding genes.
Library preparation and sequencing performances.
Transcriptome of the wild type and ΔhspR strains.
References
- Agriesti F., Roncarati D., Musiani F., Del Campo C., Iurlaro M., Sparla F., et al. (2014). FeON-FeOFF: the Helicobacter pylori Fur regulator commutates iron-responsive transcription by discriminative readout of opposed DNA grooves. Nucleic Acids Res. 42 3138–3151. 10.1093/nar/gkt1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen M. T., Brondsted L., Pearson B. M., Mulholland F., Parker M., Pin C., et al. (2005). Diverse roles for HspR in Campylobacter jejuni revealed by the proteome, transcriptome and phenotypic characterization of an hspR mutant. Microbiology 151 905–915. 10.1099/mic.0.27513-0 [DOI] [PubMed] [Google Scholar]
- Bucca G., Brassington A. M. E., Hotchkiss G., Mersinias V., Smith C. P. (2003). Negative feedback regulation of dnaK, clpB and lon expression by the DnaK chaperone machine in Streptomyces coelicolor, identified by transcriptome and in vivo DnaK-depletion analysis. Mol. Microbiol. 50153–166. 10.1046/j.1365-2958.2003.03696.x [DOI] [PubMed] [Google Scholar]
- Bucca G., Ferina G., Puglia A. M., Smith C. P. (1995). The dnaK operon of Streptomyces coelicolor encodes a novel heat-shock protein which binds to the promoter region of the operon. Mol. Microbiol. 17 663–674. 10.1111/j.1365-2958.1995.mmi_17040663.x [DOI] [PubMed] [Google Scholar]
- Danielli A., Amore G., Scarlato V. (2010). Built shallow to maintain homeostasis and persistent infection: insight into the transcriptional regulatory network of the gastric human pathogen Helicobacter pylori. PLoS Pathog. 6:e1000938. 10.1371/journal.ppat.1000938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielli A., Roncarati D., Delany I., Chiarini V., Rappuoli R., Scarlato V. (2006). In vivo dissection of the Helicobacter pylori fur regulatory circuit by genome-wide location analysis. J. Bacteriol. 188 4654–4662. 10.1128/JB.00120-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielli A., Scarlato V. (2010). Regulatory circuits in Helicobacter pylori: network motifs and regulators involved in metal-dependent responses. FEMS Microbiol. Rev. 34 738–752. 10.1111/j.1574-6976.2010.00233.x [DOI] [PubMed] [Google Scholar]
- Delany I., Spohn G., Pacheco A. B. F., Ieva R., Alaimo C., Rappuoli R., et al. (2002a). Autoregulation of Helicobacter pylori Fur revealed by functional analysis of the iron-binding site. Mol. Microbiol. 46 1107–1122. 10.1046/j.1365-2958.2002.03227.x [DOI] [PubMed] [Google Scholar]
- Delany I., Spohn G., Rappuoli R., Scarlato V. (2002b). Growth phase-dependent regulation of target gene promoters for binding of the essential orphan response regulator HP1043 of Helicobacter pylori. J. Bacteriol. 184 4800–4810. 10.1128/JB.184.17.4800-4810.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delany I., Spohn G., Rappuoli R., Scarlato V. (2002c). In vitro selection of high affinity HspR-binding sites within the genome of Helicobacter pylori. Gene 283 63–69. 10.1016/S0378-1119(01)00785-5 [DOI] [PubMed] [Google Scholar]
- Dunn B. E., Vakil N. B., Schneider B. G., Miller M. M., Zitzer J. B., Peutz T., et al. (1997). Localization of Helicobacter pylori urease and heat shock protein in human gastric biopsies. Infect. Immun. 65 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D., Evans D. J., Graham D. (1992). Adherence and internalization of Helicobacter pylori by HEp-2 cells. Gastroenterology 102 1557–1567. 10.1016/0016-5085(92)91714-F [DOI] [PubMed] [Google Scholar]
- Gisbert J. P., Calvet X. (2011). Review article: common misconceptions in the management of Helicobacter pylori-associated gastric MALT-lymphoma. Aliment. Pharmacol. Ther. 34 1047–1062. 10.1111/j.1365-2036.2011.04839.x [DOI] [PubMed] [Google Scholar]
- Grainger D. C., Hurd D., Harrison M., Holdstock J., Busby S. J. W. (2005). Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. U.S.A. 102 17693–17698. 10.1073/pnas.0506687102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandvalet C., de Crecy-Lagard V., Mazodier P. (1999). The ClpB ATPase of Streptomyces albus G belongs to the HspR heat shock regulon. Mol. Microbiol. 31 521–532. 10.1046/j.1365-2958.1999.01193.x [DOI] [PubMed] [Google Scholar]
- Hanahan D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166 557–580. 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., et al. (2010). Simple combinations of lineage-determining transcription factors prime Cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes C. W., Penn C. W., Lund P. A. (2010). The hrcA and hspR regulons of Campylobacter jejuni. Microbiology 156 158–166. 10.1099/mic.0.031708-0 [DOI] [PubMed] [Google Scholar]
- Huesca M., Borgia S., Hoffman P. (1996). Acidic pH changes receptor binding specificity of Helicobacter pylori (Stress) proteins mediate sulfatide recognition in gastric colonization. Infect. Immun. 64 2643–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C., Sheu B., Wu J. (2016). ScienceDirect Helicobacter pylori infection: an overview of bacterial virulence factors and pathogenesis. Biomed. J. 39 14–23. 10.1016/j.bj.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 252078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall D. G., Dundon W. G., Beesley S. M., Smyth C. J. (1998). REVIEW Helicobacter pylori diversity - a conundrum of genetic. Microbiology 144 2925–2939. 10.1099/00221287-144-11-2925 [DOI] [PubMed] [Google Scholar]
- Muller C., Bahlawane C., Aubert S., Delay C. M., Schauer K., Michaud-Soret I., et al. (2011). Hierarchical regulation of the NikR-mediated nickel response in Helicobacter pylori. Nucleic Acids Res. 39 7564–7575. 10.1093/nar/gkr460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parijat P., Batra J. K. (2015). Role of DnaK in HspR-HAIR interaction of Mycobacterium tuberculosis. IUBMB Life 67 816–827. 10.1002/iub.1438 [DOI] [PubMed] [Google Scholar]
- Pelliciari S., Pinatel E., Vannini A., Peano C., Puccio S., De Bellis G., et al. (2017). Insight into the essential role of the Helicobacter pylori HP1043 orphan response regulator: genome-wide identification and characterization of the DNA-binding sites. Sci. Rep. 7:41063. 10.1038/srep41063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelliciari S., Vannini A., Roncarati D., Danielli A. (2015). The allosteric behavior of Fur mediates oxidative stress signal transduction in Helicobacter pylori. Front. Microbiol. 6:840. 10.3389/fmicb.2015.00840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis S. H., Parlow M. H., Levy M., Ilver D. A. G., Caulkins C. M., Connors J. B., et al. (1996). Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect. Immun. 64 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncarati D., Danielli A., Scarlato V. (2014). The HrcA repressor is the thermosensor of the heat-shock regulatory circuit in the human pathogen Helicobacter pylori. Mol. Microbiol. 92 910–920. 10.1111/mmi.12600 [DOI] [PubMed] [Google Scholar]
- Roncarati D., Danielli A., Spohn G., Delany I., Scarlato V. (2007). Transcriptional regulation of stress response and motility functions in Helicobacter pylori is mediated by HspR and HrcA. J. Bacteriol. 189 7234–7243. 10.1128/JB.00626-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncarati D., Pelliciari S., Doniselli N., Maggi S., Vannini A., Valzania L., et al. (2016). Metal-responsive promoter DNA compaction by the ferric uptake regulator. Nat. Commun. 7:12593. 10.1038/ncomms12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncarati D., Scarlato V. (2017). Regulation of heat-shock genes in bacteria: from signal sensing to gene expression output. FEMS Microbiol. Rev. 41 549–574. 10.1093/femsre/fux015 [DOI] [PubMed] [Google Scholar]
- Salama N. R., Hartung M. L., Muller A. (2013). Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat. Rev. Microbiol. 11 385–399. 10.1038/nrmicro3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Scarlato V., Delany I., Spohn G., Beier D. (2001). Regulation of transcription in Helicobacter pylori: simple systems or complex circuits? Int. J. Med. Microbiol. 291 107–117. 10.1078/1438-4221-00107 [DOI] [PubMed] [Google Scholar]
- Schmid A. K., Howell H. A., Battista J. R., Peterson S. N., Lidstrom M. E. (2005). HspR is a global negative regulator of heat shock gene expression in Deinococcus radiodurans. Mol. Microbiol. 55 1579–1590. 10.1111/j.1365-2958.2005.04494.x [DOI] [PubMed] [Google Scholar]
- Sharma C. M., Hoffmann S., Darfeuille F., Reignier J., Findeiss S., Sittka A., et al. (2010). The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464 250–255. 10.1038/nature08756 [DOI] [PubMed] [Google Scholar]
- Spohn G., Danielli A., Roncarati D., Delany I., Rappuoli R., Scarlato V. (2004). Dual control of Helicobacter pylori heat shock gene transcription by HspR and HrcA. J. Bacteriol. 186 2956–2965. 10.1128/JB.186.10.2956-2965.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spohn G., Scarlato V. (1999). The autoregulatory HspR repressor protein governs chaperone gene transcription in Helicobacter pylori. Mol. Microbiol. 34 663–674. [DOI] [PubMed] [Google Scholar]
- Stewart G. R., Wernisch L., Stabler R., Mangan J. A., Hinds J., Laing K. G., et al. (2002). Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148 3129–3138. 10.1099/00221287-148-10-3129 [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. (1990). Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185 60–89. 10.1016/0076-6879(90)85008-C [DOI] [PubMed] [Google Scholar]
- Tatusov R. L., Koonin E. V., Lipman D. J. (1997). A genomic perspective on protein families. Science 278 631–637. 10.1126/science.278.5338.631 [DOI] [PubMed] [Google Scholar]
- Vannini A., Agriesti F., Mosca F., Roncarati D., Scarlato V., Danielli A. (2012). A convenient and robust in vivo reporter system to monitor gene expression in the human pathogen Helicobacter pylori. Appl. Environ. Microbiol. 78 6524–6533. 10.1128/AEM.01252-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A., Pinatel E., Costantini P. E., Pelliciari S., Roncarati D., Puccio S., et al. (2017). Comprehensive mapping of the Helicobacter pylori NikR regulon provides new insights in bacterial nickel responses. Sci. Rep. 7:45458. 10.1038/srep45458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A., Roncarati D., Danielli A. (2016). The cag-pathogenicity island encoded CncR1 sRNA oppositely modulates Helicobacter pylori motility and adhesion to host cells. Cell. Mol. Life Sci. 73 151–168. 10.1007/s00018-016-2151-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A., Roncarati D., Spinsanti M., Scarlato V., Danielli A. (2014). In depth analysis of the Helicobacter pylori cag pathogenicity island transcriptional responses. PLoS One 9:e98416. 10.1371/journal.pone.0098416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z., Censini S., Bayeli P. F., Telford J. L., Figura N., Rappuoli R., et al. (1995). Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect. Immun. 63 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Annotation of coding genes.
Library preparation and sequencing performances.
Transcriptome of the wild type and ΔhspR strains.