Abstract.
We present a breast lesion classification methodology, based on four-dimensional (4-D) dynamic contrast-enhanced magnetic resonance images (DCE-MRI), using recurrent neural networks in combination with a pretrained convolutional neural network (CNN). The method enables to capture not only the two-dimensional image features but also the temporal enhancement patterns presented in DCE-MRI. We train a long short-term memory (LSTM) network on temporal sequences of feature vectors extracted from the dynamic MRI sequences. To capture the local changes in lesion enhancement, the feature vectors are obtained from various levels of a pretrained CNN. We compare the LSTM method’s performance to that of a CNN fine-tuned on “RGB” MRIs, formed by precontrast, first, and second postcontrast MRIs. LSTM significantly outperformed the fine-tuned CNN, resulting in and , , in the task of distinguishing benign and malignant lesions. Our method captures clinically useful information carried by the full 4-D dynamic MRI sequence and outperforms the standard fine-tuning method.
Keywords: convolutional neural networks, breast cancer, dynamic contrast-enhanced magnetic resonance imaging, four-dimensional data
1. Introduction
Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) plays a significant role in high-risk breast cancer screening, staging, and monitoring response to therapy.1–3 The imaging procedure allows for visualization of the tumor’s temporal enhancement changes, necessary for accurate tumor characterization. However, image evaluation is performed visually, by expert radiologists, leading to the possibility of human error and to long evaluation times. In this work, we propose an automated deep learning-based methodology that captures not only tumor morphological characteristics from two-dimensional (2-D) images but also the temporal enhancement changes presented in dynamic MRI sequence. The combination of the two components of DCE-MR images allows for more accurate breast cancer diagnostic decision-making.
Deep learning approaches, specifically deep convolutional neural networks (CNNs), have become state of the art in many computer vision tasks.4 CNNs consist of multiple transformation layers (e.g., convolutional, pooling, and fully connected), which extract features from pixel-level data, generating new image representations in their respective feature spaces. Image features extracted from earlier layers of CNN are more general and are related to local image structures, such as edges and shapes.5 On the other hand, later layers, such as fully connected layers, are more class-specific and responsible for representing increasingly more abstract features, hierarchically composed of lower-level features. CNNs have shown great success in standard image classification tasks6–8 and are being adapted in medical image analysis to improve accuracy and speed of image-based diagnosis and prognosis.9 However, training an accurate and generalized CNN requires large amounts of data. Due to the lack of large-scale medical image datasets, medical analyses have been frequently performed with CNNs pretrained on a natural image dataset, such as ImageNet.10–13 The typical approach to using pretrained CNN is to fine-tune the last, fully connected layers of the network for the medical classification tasks. Fine-tuning is limited in dimensionality and is performed on 2-D images, making it difficult to apply it to imaging exams that have temporal or volumetric components.
In this work, we propose a deep learning-based methodology that enables incorporation of temporal component of DCE-MRI sequences. Many breast lesion diagnostic and prognostic decisions often rely on lesion enhancement over time, as shown in DCE-MRIs. The sequential imaging yields enhancement patterns that carry clinically useful information. For example, in lesion malignancy assessment, benign lesions typically have moderate uptake and slow washout of the ejected contrast agent, whereas malignant lesions have both rapid uptake and washout. Therefore, the focus of this work is to incorporate the temporal component of the dynamic DCE-MRIs into breast lesion classification using deep learning methods.
We achieve our goal by training a recurrent neural network (RNN), specifically long short-term memory (LSTM), which exploits the temporal correlations. We train LSTM on sequences of feature vectors extracted from dynamic MRIs with a pretrained CNN.14,15 Higher-level CNN features represent information, important for class discrimination. On the other hand, lower-level CNN features possess local pattern information valuable for further differentiating within a given class.16,17 RNNs perform classifications based on sequences of input data (image feature vectors in our case) and rely on the fact that a sequence itself carries useful information for a given task. Thus, to capture the lesion enhancement changes presented in MRI images of a given DCE-MRI sequence, we form each image feature vector by concatenating features from various levels of pretrained CNN. We compare LSTM method’s performance to that of CNN fine-tuned on 2-D MRIs. The results suggest that incorporation of enhancement patterns observed over the dynamic MRI sequence into lesion classification with deep learning methods improves malignancy assessment for breast cancer.
2. Methods
2.1. Dynamic Contrast-Enhanced Magnetic Resonance Images Dataset
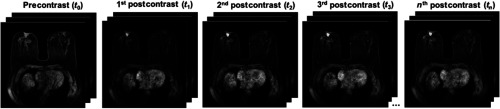
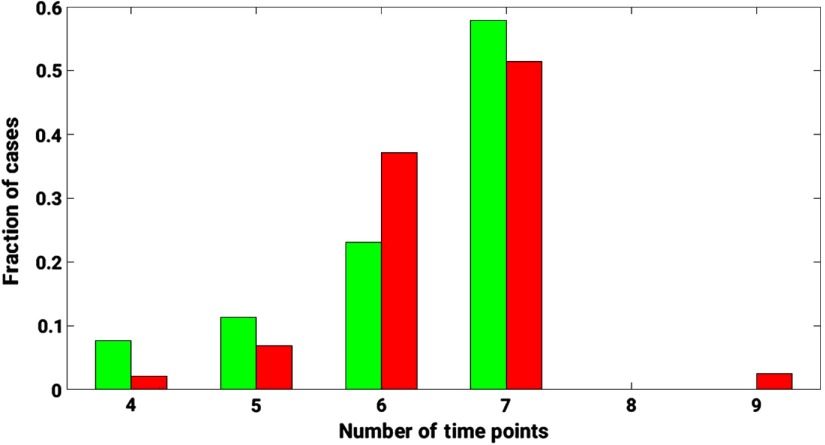
The proposed method was demonstrated for the task of discriminating malignant and benign lesions on a dataset of 703 DCE-MRI cases. The dataset was retrospectively collected under a HIPAA-Compliant Institutional Review Board protocol and annotated as benign and malignant based on pathology or radiology reports. Other clinical characteristics of the dataset are detailed in Table 1. DCE-MR images were acquired on 1.5- and 3-Tesla Philips Achieva scanners with T1-weighted spoiled gradient sequence over the period of 10 years, 2006 to 2016. Image slice thickness varied across the dataset, with 2/3 cases having slice thickness of 2 mm and 1/3 cases having slice thickness 1.5 or 1.6 mm. The image sequence included one image (precontrast) acquired prior to and multiple images (postcontrast) acquired after contrast injection (Fig. 1). A histogram of the dynamic sequence lengths for the DCE-MRIs in the database is demonstrated in Fig. 2.
Table 1.
Clinical characteristics of the UC dataset, studied for benign versus malignant lesion discrimination.
| Benign/malignant prevalence: # of cases (%) | Benign: 221 (31.4%) |
| Malignant: 482 (68.6%) | |
| Total: 703 | |
| Age: mean (STD) |
54.5 (13.2) |
| Unknown: 103 | |
| Benign tumor characteristics | |
| Tumor subtypes |
Fibroadenoma: 92 |
| Fibrocystic change: 79 | |
| Papilloma: 14 | |
| Unknown: 36 | |
| Malignant tumor characteristics | |
| Tumor subtypes | Invasive ductal carcinoma: 135 |
| Ductal carcinoma in situ: 20 | |
| Invasive ductal carcinoma + ductal carcinoma in situ: 264 | |
| Invasive lobular carcinoma: 20 | |
| Invasive lobular carcinoma mixed: 19 | |
| Unknown: 24 | |
| Estrogen receptor status: # of cases | Positive: 328 |
| Negative: 108 | |
| Unknown: 46 | |
| Progesterone receptor status: # of cases | Positive: 274 |
| Negative: 159 | |
| Unknown: 49 | |
| HER2 status: # of cases | Positive: 72 |
| Negative: 349 | |
| Equivocal: 3 | |
| Unknown: 58 | |
Fig. 1.
DSE-MRI sequence consisting of a series of 3-D images, with one 3-D image acquired prior to contrast injection and multiple 3-D images acquired after contrast injection.
Fig. 2.
A histogram of the dynamic sequence lengths for the DCE-MRIs in the database. Green bars represent the distribution of benign cases. Red bars represent the distribution of malignant cases. The length of the dynamic sequence is plotted on the -axis. The fraction of cases relative to the total number of benign/malignant cases is plotted on the -axis.
Prior to CNN fine-tuning and image feature extraction, we selected regions of interest (ROIs) around each lesion for each DCE-MRI slice and timepoint. The ROIs were selected based on lesion segmentations from the four-dimensional MRIs, performed prior to this research with the fuzzy-c means algorithm.18 These ROIs around a lesion were delineated from each transverse slice of the three-dimensional (3-D) lesion image and from each DCE timepoint (those include precontrast and multiple postcontrast timepoints ). The ROI coordinates were unchanged across DCE timepoints. The number of ROIs for an individual lesion varied based on the number of slices containing the lesion and on the dynamic sequence length.
2.2. Convolutional Neural Network Fine-tuning
In our experiments, 19-layer VGGNet, pretrained on ImageNet, was used as a base model.6,19 First, we fine-tuned the VGGNet for the task of distinguishing malignant and benign lesions and used its performance as a baseline. For fine-tuning, we utilized the convolutional base of the VGGNet and added a fully connected top. Since our dataset consisted of ROIs of various sizes, we adapted a global average-pooling layer after the last convolutional block of VGGNet.20 The average-pooling layer was followed by two fully connected layers, with dropout applied after each of the two layers. All layers, prior to and including the fourth max-pooling layer, were frozen and the rest were fine-tuned.
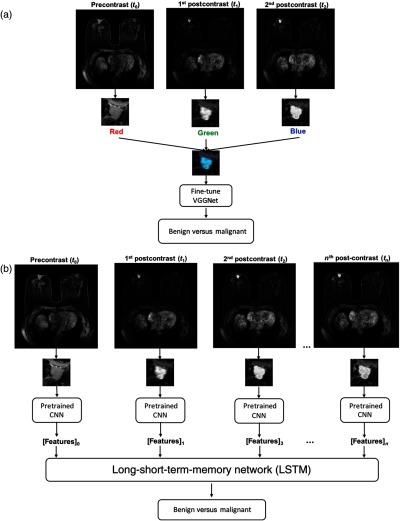
Fine-tuning was simultaneously performed on precontrast, first, and second postcontrast ROIs. VGGNet requires an image input consisting of three channels, red (R), green (G), and blue (B). Since precontrast, first, and second postcontrast ROIs are grayscale, we made use of the network’s color channels and input these ROIs into the R, G, and B channels, respectively. Thus, we fine-tuned the network on these artificially made RGB lesion ROIs [Fig. 3(a)].10
Fig. 3.
(a) Lesion classification methodologies: first, VGGNet was fine-tuned on RGB ROIs (RGB ROI is formed by ROIs at the precontrast, first, and second postcontrast DCE timepoints) and its performance was taken as a baseline and (b) image feature was extracted from various levels of VGGNet from the lesion ROIs at each DCE timepoint and utilized for LSTM network training.
2.3. Multilevel Image Features
Next, the fine-tuned VGGNet was utilized to extract feature vectors from DCE-MRIs for LSTM training and evaluation. To capture intraclass changes, i.e., contrast enhancement changes of one lesion, the feature vectors were extracted at various network depths from the five max-pooling layers of VGGNet. These features from each level were average-pooled and normalized with Euclidian distance. The pooled features were further concatenated to form a CNN feature vector for a given ROI.10
For a given slice of a 3-D MRI, the image feature vectors were extracted at each DCE timepoint. The resulting sequence of feature vectors was used as an input into LSTM network. Since the DCE-MRI sequences were of variable lengths across the dataset, the sequences were padded with zeros to the length of the longest sequence. The padded part of the sequences was not taken into account when calculating the loss of the model, described further in Sec. 2.5. Figure 3(b) demonstrates the end-to-end lesion classification pipeline with LSTM network based on DCE-MRIs.
2.4. Long Short-Term Memory Network
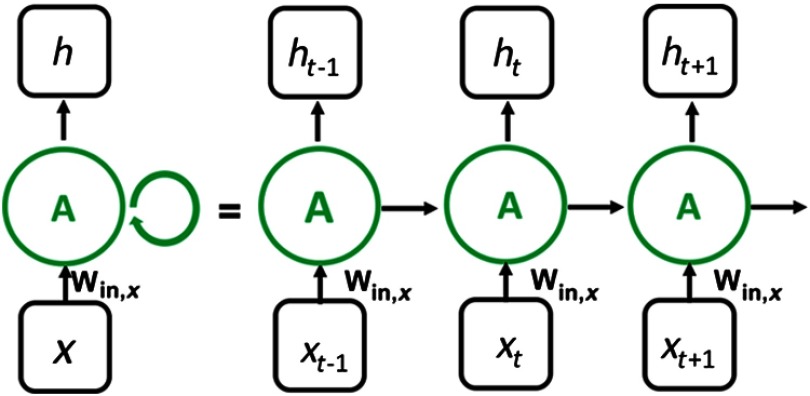
The multilevel feature vector sequences were utilized to train an LSTM network. During its training, the model captures the changes presented in a given sequences. Let , , represent a sequence of inputs, where each is an input at timestep . In our work, represents image feature vector obtained from lesion ROI at DCE timepoint . An RNN has an internal hidden state at time , , which gets updated based on the current input and its previous hidden state (Fig. 4).
Fig. 4.
General structure of an RNN. The network recurrently computes its hidden state based on its previous hidden state and the current input . The final classification output is computed based on the hidden state of the network, which depends on the previous steps.
An LSTM, a type of RNN, takes this idea further by maintaining an additional distinctive feature, a “memory cell.” Along with the hidden state , a memory cell’s state is updated as the network steps through the sequence of the inputs. This update is based on the previous step’s hidden state and the current input and is performed by mechanisms called gates, i.e., the “input gate,” the “forget gate,” and the “output gate.” Each gate has its own responsibility in information retention from and and regulates it with a sigmoid activation function
| (1) |
which takes values from 0 to 1, where is a linear combination of and .
The hidden state update involves multiple steps. First, an LSTM cell receives two inputs, the current input and previous hidden state , and transforms them into candidate value to be added to the cell state by
| (2) |
Simultaneously, the three gates, described above, monitor the flow of information into and out of the memory cell state:
-
1.The “input gate” chooses the values of the network to be updated
(3) -
2.The “forget gate” decides on which information from the past to keep and which to discard
(4) -
3.The “output gate” controls which information to let through to the hidden state update
where , , and are the weight matrices, responsible for updating the current input vectors , and the previous hidden state of the cell and , , , and represent the bias terms for the corresponding update operation.(5)
The memory cell state is updated based on the transformed input from Eq. (2) and the “input gate” and “forget gate” decisions from Eqs. (3) and (4)
| (6) |
Finally, the hidden state is updated based on memory cell state from Eq. (6) and the “output gate” decision Eq. (5)
| (7) |
2.5. Model Training and Evaluation
The prediction errors of our models were evaluated with binary cross entropy loss. As we iterate through model training, the loss function calculates the amount of penalty the algorithm receives for making a wrong prediction and is used to evaluate the algorithm’s performance. For training examples, the binary cross entropy loss function is defined as
| (8) |
where and are the true and predicted label for the case , respectively.
We utilized stochastic gradient descent as an optimizer and set batch size to 64 for VGGNet fine-tuning and LSTM training. The hyperparameters of the LSTM network were optimized using a validation set. To avoid overfitting, early stopping was used to stop network training, when validation loss started increasing.
The dataset was separated into training + validation (80%) and testing (20%) sets by lesion, with cancer prevalence among the cases being constant across the sets. All transverse slices of the MRIs containing a lesion were utilized to train the models. This totaled slices for the training set. The model was tested and evaluated only on the lesion center slices in the validation and test subsets. To avoid bias, all image slices from the same lesion were retained in either the training, validation, or testing subsets, but not shared across the three.
2.6. Evaluation Metrics
Receiver operating characteristic (ROC) analysis was applied to evaluate binary classification performance of the models in the task of distinguishing benign and malignant lesions.21 We measured their ability to discriminate the two classes using area under the ROC curve (AUC). The statistical difference in AUC values was evaluated using Delong tests.22 Furthermore, specificity, positive predictive value (PPV), and negative predictive value (NPV) were compared between the models for the same sensitivity threshold. Specificity measures the fraction of negative cases correctly identified. PPV and NPV measure the probabilities of a positive classification actually being positive and a negative classification actually being negative.
2.7. Implementation Details
The ROI extraction was performed with the MATLAB software, developed specifically for the tasks. The deep learning-based methods were implemented in Python using the Keras library with Tensorflow backend.23 Training and evaluation of the models were performed on an NVIDIA Titan X GPU.
2.8. Experiments and Results
All of the lesions in the study had undergone biopsy, resulting in sensitivity of 100% and specificity of 0% for an expert radiologist. For the testing set, Table 2 summarizes the performance metrics, specificity, PPV, and NPV, for varying the decision thresholds for the two deep learning methods studied, i.e., the fine-tuned VGGNet and the LSTM. For a sensitivity of 0.92 and below, LSTM results in higher specificity and PPV and NPV, as compared with the performance of VGGNet. These results demonstrate that the LSTM method achieves reduced number of false positives and calls a higher number of benign lesions as benign and malignant lesions as malignant. Above a sensitivity of 0.92, VGGNet shows slightly better specificity and predictive values.
Table 2.
The performance metrics for fine-tuned VGGNet and LSTM network on the DCE-MRI test subset. For a given sensitivity value, we compare specificity, PPV, and NPV for the two methods.
| Specificity | PPV | NPV | ||||
|---|---|---|---|---|---|---|
| Sensitivity | LSTM | Fine-tuned VGGNet | LSTM | Fine-tuned VGGNet | LSTM | Fine-tuned VGGNet |
| 0.80 | 0.82 | 0.73 | 0.94 | 0.91 | 0.53 | 0.51 |
| 0.82 | 0.78 | 0.71 | 0.93 | 0.91 | 0.55 | 0.53 |
| 0.84 | 0.75 | 0.68 | 0.92 | 0.90 | 0.57 | 0.54 |
| 0.86 | 0.70 | 0.64 | 0.91 | 0.90 | 0.58 | 0.57 |
| 0.88 | 0.64 | 0.61 | 0.90 | 0.89 | 0.60 | 0.59 |
| 0.90 | 0.58 | 0.56 | 0.88 | 0.88 | 0.62 | 0.61 |
| 0.92 | 0.50 | 0.51 | 0.87 | 0.87 | 0.64 | 0.64 |
| 0.94 | 0.40 | 0.45 | 0.85 | 0.86 | 0.65 | 0.68 |
| 0.96 | 0.29 | 0.37 | 0.83 | 0.84 | 0.67 | 0.72 |
| 0.98 | 0.15 | 0.26 | 0.80 | 0.82 | 0.67 | 0.78 |
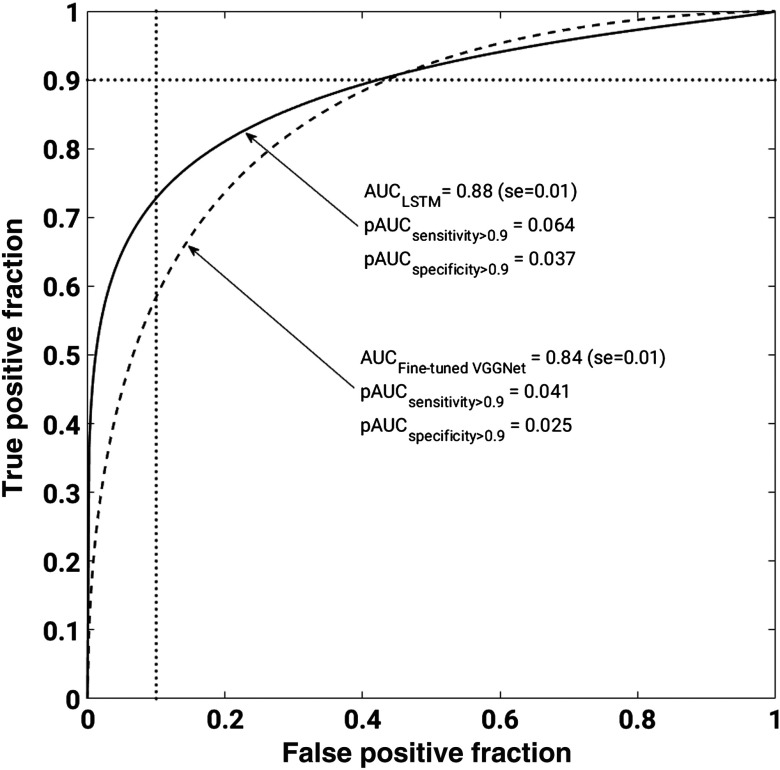
Even though both PPV and NPV are useful metrics in the performance evaluation, class prevalence directly influences them. While holding all other variables constant and increasing just the class prevalence, PPV will increase and NPV will decrease. Our work was performed on an unbalanced dataset, with 68.6% malignant and 31.4% benign lesions. Given that, we conducted ROC analysis, which yields AUC values, a metric independent of class prevalence. Figure 5 shows the ROC curves for the lesion classification performance of the two models. This figure demonstrates that LSTM significantly outperformed the fine-tuned VGGNet, resulting in (se = 0.01) and (se = 0.01), with , in the task of distinguishing benign and malignant lesions.
Fig. 5.
ROC curves corresponding to fine-tuned VGGNet and LSTM model performances in discriminating benign and malignant lesions. Solid line represents LSTM model and dashed line represents fine-tuned VGGNet. LSTM significantly outperformed the fine-tuned VGGNet, resulting in and , with , in the task of distinguishing benign and malignant lesions. We note that ROC curves cross. To better understand methods’ performances, we calculate partial AUC values for specificity (1 – false positive fraction) range from 0.9 to 1 and sensitivity (true positive fraction) range from 0.9 to 1. The vertical and horizontal dotted lines correspond specificity = 0.9 and sensitivity = 0.9, respectively.
Note that the ROC curves cross, showing some ambiguity in the performances. Thus, we calculated the partial AUCs for the sensitivity range from 1 to 0.9 and the specificity range of 1 to 0.9.24 LSTM yielded improved partial AUCs of 0.064 and 0.037 as compared to those of the fine-tuned VGGNet, 0.041 and 0.025, for sensitivity and specificity ranges, respectively.
2.9. Discussion and Conclusions
We present a breast lesion classification pipeline that captures both morphological and temporal information about a lesion presented on DCE-MRIs in the task of distinguishing malignant and benign lesions. Compared to previous works, our method enables to incorporate the entire DCE-MRI sequence into deep learning-based lesion classification. For a given lesion, we extract a sequence of image feature vectors corresponding to the DCE-MRI sequence, which is further used to train an LSTM network. The image feature vectors are obtained from five max-pooling layers of pretrained VGGNet to capture the local changes in lesion enhancement.
The LSTM method significantly outperformed the VGGNet, fine-tuned on RGB MRIs. To make sure that the results are not just due to higher-feature dimensionality, we also trained a simple two-layer, fully connected neural network on multilevel image features extracted from RGB MRIs. This method incorporates the features extracted from various levels of VGGNet, but not the full DCE-MRI sequence. The classification performance of the network resulted in AUC value of 0.82, which is lower than the performance of the fine-tuned VGGNet as well as the LSTM network.
LSTMs have achieved superior results for machine translation, language modeling, and image captioning tasks, outperforming other recurrent architectures. The main benefit of LSTMs is that they prevent vanishing or exploding gradients during error back propagation with long input sequences. Therefore, the network can retain useful classification information from the beginning of the sequence. Furthermore, LSTMs are well suited for working with sequences of various lengths as well as time lags between the sequence elements, which is characteristic of our DCE-MRI data. DCE sequences contain one image taken prior to contrast injection and 2 to 10 images taken after contrast injection. We also studied another RNN type, gated recurrent units (GRUs). GRUs have a similar, but simpler architecture than LSTMs, resulting in fewer parameters and more efficient computation.25 However, compared with LSTMs, GRUs do not control their hidden state with a memory unit. After investigating both architectures, we observed higher classification performance with the LSTM network.
The proposed method is inspired by the fact that human experts base various breast diagnostic and prognostic decisions on temporal changes in lesion enhancement observed in DCE-MRIs. Specifically, kinetic enhancement curve patterns are often visually analyzed during benign versus malignant discrimination. Benign lesions tend to demonstrate moderate uptake and slow washout of a contrast agent, while malignant lesions tend to have both rapid uptake and washout. Therefore, the sequences of image features extracted from the DCE-MRIs should be different for benign and malignant lesions. Among other clinical questions, radiologists’ evaluation of breast cancer response to therapy is also guided by temporal changes of lesion enhancement. Lesion enhancement patterns and DCE-MRI quantitative pharmokinetic parameters are used to assess breast cancer’s response to primary and neoadjuvant chemotherapies.1,26,27 These clinical questions are left for future work due to lack of availability of sufficient datasets.
Deep learning methods enable capturing of data patterns that have been previously unexploited, leading to more accurate, rapid, and accessible medical decision-making. In this work, we demonstrate a deep learning method that captures clinically useful information presented in DCE-MRI sequence for breast lesion malignancy assessment.
Acknowledgments
The authors acknowledge the other members of Giger Lab, Sasha Edwards, MS, John Papaioannou, MS, and Karen Drukker, PhD, MBA, the Department of Radiology, the University of Chicago, Chicago, Illinois. Special thanks to colleague Simas Glinskis, Physics Department, The University of Chicago, for fruitful discussions to a deep learning. This work was partially supported by the National Institutes of Health Quantitative Imaging Network under Grant No. U01CA195564 and the Chicago Metcalf program. We acknowledge the support of NVIDIA Corporation with the donation of a Titan X Pascal GPU used for this research.
Biographies
Natalia Antropova conducted this research as a PhD student in the medical physics program at the University of Chicago, working in Dr. Maryellen Giger’s Lab. Her research focuses on developing deep learning and classical methods for clinical decision-making based on medical images. Her projects involve breast cancer detection, diagnosis, and prognosis using dynamic contrast-enhanced magnetic resonance imaging.
Benjamin Huynh works on deep learning methods of medical analysis. After conducting research in Giger’s Lab at the University of Chicago, he is now pursuing his PhD in biomedical informatics at Stanford University. His research interests include computational statistics, computer vision, and nonparametric Bayesian techniques with applications of biomedical tasks.
Hui Li has been working on quantitative imaging analysis on medical images for over a decade. His research interests include breast cancer risk assessment, diagnosis, prognosis, and response to therapy, understanding the relationship between radiomics and genomics, and their future roles in precision medicine.
Maryellen L. Giger has been working, for multiple decades, on computer-aided diagnosis and quantitative image methods in cancer diagnosis and management. Her research interests include understanding the role of quantitative radiomics, computer vision, and deep learning in personalized medicine.
Disclosures
M.L.G. is a stockholder in R2 Technology/Hologic and receives royalties from Hologic, GE Medical Systems, MEDIAN Technologies, Riverain Medical, Mitsubishi, and Toshiba. She is a cofounder and stockholder in Quantitative Insights. It is the University of Chicago Conflict of Interest Policy that investigators disclose publicly actual or potential significant financial interest that would reasonably appear to be directly and significantly affected by the research activities.
References
- 1.Turkbey B., et al. , “The role of dynamic contrast enhanced MR imaging in cancer diagnosis and treatment,” Diagn. Interv. Radiol. 16(3), 186–192 (2009). 10.4261/1305-3825.DIR.2537-08.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Cancer Society, Breast Cancer Facts & Figures 2015–2016, American Cancer Society, Inc., Atlanta, Georgia: (2015). [Google Scholar]
- 3.Siegel R. L., Miller K. D., Jemal A., “Cancer statistics, 2017,” CA: A Cancer J. Clin. 67, 7–30 (2017). 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 4.Schmidhuber J., “Deep learning in neural networks: an overview,” Neural Networks 61, 85–117 (2015). 10.1016/j.neunet.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 5.Li F. F., Karpathy A., Johnson J., “CS231n: convolutional neural networks for visual recognition,” University Lecture (2015).
- 6.Simonyan K., Zisserman A., “Very deep convolutional networks for large-scale image recognition,” in Int. Conf. on Learning Representations (ICLR) (2015). [Google Scholar]
- 7.Szegedy C., et al. , “Going deeper with convolutions,” in Conf. on Computer Vision and Pattern Recognition (CVPR), 1–9 (2015). [Google Scholar]
- 8.He K., et al. , “Spatial pyramid pooling in deep convolutional networks for visual recognition,” IEEE Trans. Pattern Anal. Mach. Intell. 37, 1904–1916 (2015). 10.1109/TPAMI.2015.2389824 [DOI] [PubMed] [Google Scholar]
- 9.Litjens G., et al. , “A survey on deep learning in medical image analysis,” Med. Image Anal. 42, 60–88 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Antropova N., Huynh B. Q., Giger M. L., “A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets,” Med. Phys. 44(10), 5162–5171 (2017). 10.1002/mp.2017.44.issue-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng J.-Z., et al. , “Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans,” Sci. Rep. 6, 24454 (2016). 10.1038/srep24454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huynh B. Q., Li H., Giger M. L., “Digital mammographic tumor classification using transfer learning from deep convolutional neural networks,” J. Med. Imaging (Bellingham) 3, 034501 (2016). 10.1117/1.JMI.3.3.034501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteva A., et al. , “Dermatologist-level classification of skin cancer with deep neural networks,” Nature 542, 115–118 (2017). 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochreiter S., Schmidhuber J., “Long short-term memory,” Neural Comput. 9, 1735–1780 (1997). 10.1162/neco.1997.9.8.1735 [DOI] [PubMed] [Google Scholar]
- 15.Goodfellow I., Bengio Y., Courville A., Deep Learning, The MIT Press; (2016). [Google Scholar]
- 16.Ng J. Y.-H., Yang F., Davis L. S., “Exploiting local features from deep networks for image retrieval,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition Workshops, pp. 53–61 (2015). [Google Scholar]
- 17.Zheng L., et al. , “Good practice in CNN feature transfer,” arXiv:1604.00133 (2016).
- 18.Chen W., Giger M. L., Bick U., “A fuzzy c-means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR images,” Acad. Radiol. 13, 63–72 (2006). 10.1016/j.acra.2005.08.035 [DOI] [PubMed] [Google Scholar]
- 19.Russakovsky O., et al. , “ImageNet large scale visual recognition challenge,” Int. J. Comput. Vision 115, 211–252 (2015). 10.1007/s11263-015-0816-y [DOI] [Google Scholar]
- 20.Lin M., Chen Q., Yan S., “Network in network,” in Proc. Int. Conf. on Learning Representations (ICLR) (2014). [Google Scholar]
- 21.Parikh R., et al. , “Understanding and using sensitivity, specificity and predictive values,” Indian J. Ophthalmol. 56, 45–50 (2008). 10.4103/0301-4738.37595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLong E. R., DeLong D. M., Clarke-Pearson D. L., “Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach,” Biometrics 44, 837 (1988). 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- 23.Chollet F., “Keras. GitHub,” GitHub repository, https://github.com/fchollet/keras (2015).
- 24.Jiang Y., Metz C. E., Nishikawa R. M., “A receiver operating characteristic partial area index for highly sensitive diagnostic tests,” Radiology 201, 745–750 (1996). 10.1148/radiology.201.3.8939225 [DOI] [PubMed] [Google Scholar]
- 25.Chung J., et al. , “Empirical evaluation of gated recurrent neural networks on sequence modeling,” NIPS Deep Learning Workshop; (2014). [Google Scholar]
- 26.Pickles M. D., et al. , “Role of dynamic contrast enhanced MRI in monitoring early response of locally advanced breast cancer to neoadjuvant chemotherapy,” Breast Cancer Res. Treat. 91, 1–10 (2005). 10.1007/s10549-004-5819-2 [DOI] [PubMed] [Google Scholar]
- 27.Martincich L., et al. , “Monitoring response to primary chemotherapy in breast cancer using dynamic contrast-enhanced magnetic resonance imaging,” Breast Cancer Res. Treat. 83, 67–76 (2004). 10.1023/B:BREA.0000010700.11092.f4 [DOI] [PubMed] [Google Scholar]