Abstract
p53 is an important tumour suppressor gene, with loss of p53 contributing to the development of most human cancers. However, the activation of p53 in response to stress signals underpins a role for p53 in diverse aspects of health and disease. Activities of p53 that regulate metabolism can play a role in maintaining homeostasis and protecting cells from damage – so preventing disease development. By contrast, either loss or over-activation of p53 can contribute to numerous metabolic pathologies, including aging, obesity and diabetes.
1. Introduction
The p53 protein has been studied for almost 40 years in the context of tumour development and suppression. Initially identified through its interaction with viral oncoproteins, p53 has been found to be mutated in over half of all human cancers across many organ types, including breast, lung and colon [1]. Inheritance of one mutant p53 allele gives rise to Li Fraumeni syndrome, in which affected individuals show early onset of a variety of malignancies [2]. Furthermore, a plethora of mouse studies have shown that loss or perturbation of p53 enhances tumour development in virtually any cancer model tested [3]. Initially, the ability of p53 to prevent tumour development was ascribed largely to the ability of p53 to provoke responses that inhibit cell growth – such as cell death, senescence or differentiation. p53 is induced by signals that might be associated with oncogenic transformation (Figure 1), leading to a simple model in which p53 becomes activated by oncogenic stress to eliminate nascent tumour cells, but lies dormant under normal growth conditions – the latter contention reinforced by the observation that p53 null mice can develop ostensibly normally. However, accumulating evidence shows that p53 biology is far more complex. Other activities of p53, including support of DNA repair and genomic integrity, control of metabolism or the ability to influence the microenvironment, are also likely to play important roles in the modulation of cancer development [4]. The cell elimination functions of p53 can have ramifications beyond tumour suppression, contributing to pathologies such as metabolic syndrome, neurodegenerative diseases, ischemia and even aging [5]. Furthermore, as seen with other arbiters of cell fate, p53 can also support cell survival under some stress conditions, with loss of this activity affecting a host of outcomes at the cell and organismal level. Many excellent reviews have been written on various p53 functions [4, [6], [7], [8]], and here we will attempt to focus on some of the physiological consequences of the metabolic activities of p53 in normal and disease processes.
Fig. 1.
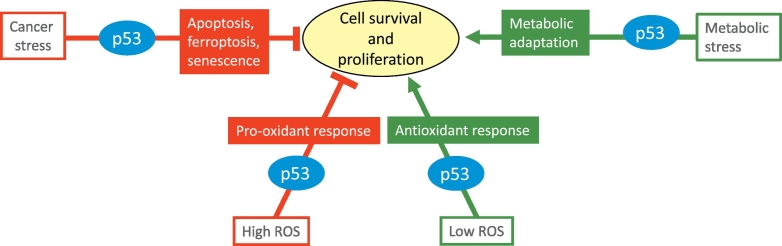
p53 regulates cell survival and proliferation in vitro. p53 can repress or support cell proliferation and survival depending on the cellular signal and context. Cancer associated stress signals including high ROS levels, DNA damage, oncogene activation, telomere shortening and hypoxia activate a p53 response that frequently promotes cell death and cell elimination. Metabolic stress signals including transient oxidative stress, nutrient starvation and growth factor deprivation activate a p53 response that more commonly promotes cell survival and proliferation.
2. Regulation of p53 function
It is clear that excessive p53 activity is highly deleterious to cell survival, and under normal growth conditions p53 function is held firmly in check. In part, this is achieved by controlling p53 protein levels through continuous ubiquitination and degradation – largely mediated by the E3 ubiquitin ligase MDM2 [9]. Exposure of cells to various stress signals – such as oxidative stress, oncogene activation, telomere shortening and hypoxia -can block the action of MDM2 and so lead to the stabilization of p53. Mediators of this pathway to p53 activation include stress induced kinases - such as ATM, ATR, Chk1 and Chk2 - which phosphorylate p53 to modulate binding to MDM2, and the deployment of small proteins such as Arf or ribosomal protein L11 (RPL11), which bind and inactivate MDM2 [10]. By contrast, phosphorylation of MDM2 by Akt serves to enhance p53 degradation in response to growth stimulatory signals [11]. Numerous other post-transcriptional modifications of p53 – including acetylation and methylation - contribute to the regulation of p53 function and may also serve to fine tune the activity of p53 to determine the ultimate outcome of the response [12]. Stabilization and activation of p53 is not limited to conditions of oncogenic stress, however. Most clearly, acute DNA damage induces a strong p53 response that can lead to cell death in several tissues and loss of p53 can protect from radiation sickness that results from the death of intestinal cells [13]. Elegant studies have shown that the acute response to DNA damage is not required for the subsequent limitation of cancer development, suggesting that the role of p53 as a tumour suppressor is subtler than simply eliminating all damaged cells [14]. Indeed, the p53 response to genomic damage may be more important for the elimination of damaged germ cells and maintaining germline integrity [15]. Other forms of stress can also activate p53, including a battery of signals that have been clustered under the term “metabolic stress”. These include oxidative stress and nutrient, growth factor or oxygen deprivation, and can be transmitted through established metabolic sensors like mTOR, AMPK and eIF2a as well as bespoke pathways specifically induced by each type of stress [16]. In some circumstances, the outcome of exposure to metabolic stress is the canonical cell elimination response to p53. However, p53 can also promote cell survival functions that become important when the stress signal is transient, and cells are able to revert back to normal growth. Under these conditions loss of p53 tracks with loss of cell adaptability and viability, resulting in decreased overall organismal fitness. Depending on the nature and persistence of the stress signal, p53 can help or hinder various normal and pathological responses.
3. Activities of p53 – in vitro
p53 is as a transcription factor that functions as a sequence specific DNA binding protein with the potential to regulate the expression of thousands of target genes [6, 17]. While the binding of p53 to response elements in DNA does not appear to be regulated [18], the transcriptional response to p53 can be influenced by numerous factors, including modifications of p53, the presence or absence of co-factors and the chromatin architecture [19]. Consequently, the outcome of p53 activation can differ greatly depending on the nature of the activating signal and the cellular context in which the p53 response is induced. Transcriptional regulation by p53 – of both coding genes and miRNAs – is responsible for many of these responses, but p53 can also play a role in regulation of the translation of some mRNAs. Furthermore, various transcriptionally independent activities of p53 resulting from direct interactions of p53 with other proteins have also been described.
3.1. p53 in cell death and senescence
One of the best-understood functions of p53 is its ability to induce apoptosis, in large part through the transcriptional activation of pro-apoptotic Bcl-2 family proteins like Noxa and Puma [20]. p53 can also interact directly with some of these proteins in the cytoplasm or at the mitochondrial outer membrane and so modulates their activity in response to different stimuli [21]. Intriguingly, cytoplasmic p53 not only regulates Bcl2 family proteins, but can also be regulated by them. A recent study demonstrated the limitation of apoptosis in glioblastoma cells following glucose depletion was a consequence of Bcl-xL sequestration of cytoplasmic p53 [22]. Modifications of p53 can also enhance its apoptotic functions. For example, Pin-1 catalyses the cis-trans interconversion of p53 proline 47 to enhance p53 dependent Bax activation [23], while serine 392 phosphorylation is necessary for p53 to relocate to the mitochondria where it binds to Bax and Bak to trigger apoptosis [24]. p53 can also prime cells to undergo apoptosis in response to extracellular adenosine - which accumulates under conditions of metabolic stress - by activating the receptor Adora2B [25]. In addition to apoptosis, p53 promotes various non-apoptotic forms of cell death, including ferroptosis. This iron-dependent form of cell death is the result of increased ROS and lipid peroxidation and can be inhibited by the anti-oxidant functions of glutathione [26]. As glutathione synthesis requires cysteine and glutamate, alterations in the metabolism or availability of these amino acids can regulate this form of death. p53 impacts ferroptosis through several mechanisms. By regulating the cystine/glutamate antiporter SLC7A11 and glutaminase 2 – one of the enzymes that produce glutamate from glutamine - p53 limits cystine uptake and glutamate production respectively, so sensitising cells to ROS induced ferroptosis [27, 28]. p53-dependent induction of SAT1 – a protein that functions in the polyamine pathway and promotes lipid peroxidation – also contributes the ferroptotic response [29]. The ability to promote apoptosis and ferroptosis are separable functions of p53 and, strikingly, the ability to promote ferroptosis – not apoptosis - tracks with tumour suppressor activity in various mouse model systems [28, 30]. However, an additional complication has been introduced by studies showing that p53 can limit ferroptosis – either by preventing the relocalisation of dipeptidyl-peptidase-4 (DPP4) from the nucleus to the plasma membrane [31] or as a result of a p21-mediated cell cycle arrest, which helps cells to conserve glutathione [32]. Other cell eliminating activities of p53 include the ability to induce autophagy and senescence. Autophagy generally functions as a cell survival mechanism, but several studies have identified the autophagic response to p53 activation as important for p53-mediated cell death [33, 34]. A p53/Bnip axis was also shown to be important in promoting autophagy and cell death in ventricular myocytes [35]. Several p53-dependent target genes – including p21 and PAI1 – can promote senescence and so inhibit cell growth [36]. This response may contribute to tumour suppression, although senescence itself can be both tumour inhibiting and promoting in an in vivo setting.
3.2. p53 in cell survival and adaptation to stress
While each of the cell elimination functions of p53 described above is likely to contribute to tumour suppression, there is a growing realisation that p53 not only responds to damage (in which case the response is often cell elimination) but can also help to prevent damage and maintain homeostasis under normal and conditions of non-genotoxic stress (Fig. 1). These activities can be tumour suppressive, but the protective functions of p53 can also play a role in limiting other pathologies and may - paradoxically - contribute to malignant progression.
3.2.1. DNA damage
While some cells respond to DNA damage by activating p53-dependent apoptosis, non-lethal DNA damage can also induce a reversible p53-dependent cell cycle arrest (principally though the activation of p21), allowing for DNA repair [37, 38]. p53 also activates many DNA damage repair genes and helps to maintain genome stability by regulating centrosome duplication [39, 40] and replication fidelity [[41], [42], [43]]. The loss of p53 can therefore be associated with increased levels of genomic perturbations that might cause oncogenic progression, as well as leading to a tolerance of the stress signals that accompany malignant progression [5].
3.2.2. Metabolic stress
A general ability of p53 to help cells adapt and survive under conditions of nutrient depletion has recently been described, with functions for p53 in supporting cells under glucose [44], glutamine [45] and serine starvation [46]. The transient activation of p21 plays a role in many of these protective responses – for example in the case of serine starvation helping to channel de novo synthesised serine into glutathione production rather than nucleotide synthesis [46] or by delaying ferroptosis in response to cystine starvation [32]. Activation of catabolic pathways such as autophagy and fatty acid oxidation by p53 may also help to sustain cells temporarily in the absence of exogenous nutrients. Of note, however, the interaction between autophagy and p53 in the control of cancer development is complex and discussed in more detail elsewhere [47].
Of course, long term starvation leads to cell death, to which p53 can also contribute. For example, p53-dependent transcriptional repression of PHGDH – the rate limiting enzyme in de novo serine synthesis – can prevent cell survival when exogenous serine is removed [48]. What controls whether p53 supports survival or hastens death under these conditions is not well understood. However, p53 seems to play a key role in responding to starvation – mobilising alternative nutrient sources and catabolic pathways to maintain cell survival while also playing a role in the elimination of cells that cannot recover.
3.2.3. Regulation of metabolism
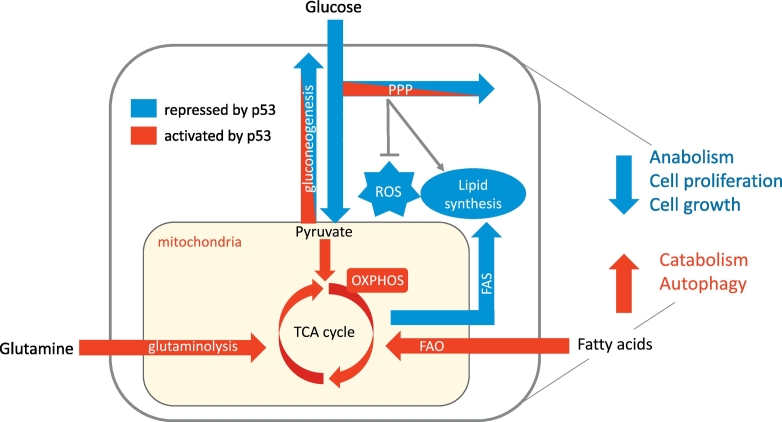
Beyond the overt stress responses are activities of p53 that reflect functions contributing to normal metabolic homeostasis (Fig. 2). Activities of p53 that help to maintain mitochondrial health, increase mitochondrial respiration and lower glycolysis have been described [49] and the full gamut of p53-regulated metabolic target genes is discussed in detail in many recent reviews [5, 16, 50, 51] Briefly, p53 represses the expression of several glucose transporters including GLUT1 and GLUT4 and indirectly controls GLUT3 expression through repression of IKK [52]. p53 can also limit glycolytic flow by activating TIGAR and RRAD or by regulating PGAM and Hexokinase 2. A recent study also showed that p53 transcriptionally down regulates PFKFB4, an enzyme that controls the levels of fructose-2,6-bisphosphate, an important allosteric regulator of glycolysis [53]. On the other hand, p53 promotes the flux from pyruvate and glutamate into the mitochondria for use in the TCA cycle by regulating enzymes like PDK2, PARK2 and GLS2 [[54], [55], [56], [57]], while also controlling lactate levels in tumour cells through repression of MCT1 [58]. The regulation of SCO2 and AIF1 by p53 can promote efficient mitochondrial respiration while p53 also plays a role in regulating mitochondrial DNA copy number and mitochondrial mass [59]. Overall, p53 seems to favour mitochondrial respiration over glycolysis (thus opposing the “Warburg effect” of increased aerobic glycolysis exhibited by many cancer cells). The pentose phosphate pathways (PPPs), alternative branches of glycolysis that provides ribose for nucleotide synthesis and NADPH for reductive biosynthesis and antioxidant control, can be supported by p53 through activation of TIGAR, which encourages glycolytic intermediates to flow into the oxidative PPP or by the activation of AKT and SP-1 which promote PPP gene expression [[60], [61], [62]]. However, p53 can also suppress the PPP by repressing the expression of PFKFB4 [53] or by directly binding and inhibiting G6PD, a key enzyme in the oxidative arm of the PPP [63].
Fig. 2.
Regulation of energy metabolism by p53. Under conditions of metabolic stress, p53 induces catabolic pathways and autophagy to maintain energy production and cell survival while inhibiting energy and nutrient consuming anabolic pathways, cell growth and proliferation. p53 inhibits glycolysis and promotes mitochondrial respiration, while also limiting lipid biosynthesis and promoting lipolysis. Note that while oxidative phosphorylation is a major source of mitochondrial ROS, maintenance of mitochondrial health by p53 is likely to limit oxidative stress through this source. In addition to PPP activation, p53 controls ROS through numerous other mechanisms (not shown). Pathways depicted in blue are repressed by p53 while pathways depicted in red are activated by p53. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The role of p53 in the regulation of gluconeogenesis is less clear. p53 has been reported to both increase [64] and decrease [65] the expression of gluconeogenic enzymes. Gluconeogenesis is supported by p53 through the induction of Pank1, which catalyses the rate limiting step in CoA production [66], while the activation of SIRT6 by p53 deacetylates and deactivates FOXO1, a positive regulator of PCK1 and G6PC [67]. Gluconeogenesis by the liver is an important survival response to glucose starvation, but the impact of regulating gluconeogenesis in cancers and the effect of p53 on such a response has not yet been established. Another key pathway influenced by p53 is lipid metabolism, where p53 promotes the switch from the anabolism to the catabolism of lipids [68]. Various genes that are directly involved in fatty acid oxidation (FAO) can be regulated by p53, including three carnitine acyltransferases CROT, CPTA1 and CPT1C [69, 70]. p53 has also been shown to upregulate LPIN1 during fasting, resulting in increased FAO [71]. Conversely, p53 inhibits fatty acid synthesis (FAS), in part by repressing the expression of genes directly involved in this process, such as FASN and ACL, or by downregulating the expression of SREBP1c, a transcription factor that plays a key role in driving expression of FAS genes [69, 72]. Additionally, as mentioned above, p53 can inhibit the oxidative PPP - the main source of NADPH for fatty acid synthesis – leading to decreased lipogenesis [63].
Finally, p53 can more indirectly integrate with other regulators of the response to metabolic stress. p53 is activated by AMPK during energetic stress through serine 15 phosphorylation leading to a transient cell-cycle arrest [44]. During genotoxic stress however, p53 can activate AMPK via sestrin 1 and 2 which inhibit mTORC leading to inhibition of cell growth and proliferation [73]. p53 also transcriptionally activates several other targets that negatively regulates mTOR including PTEN, Plk2 and TSC1/2 [16, 74]. These p53 responses function to limit cell growth and so – in concert with the inhibition of cell cycle progression and proliferation - allows for the preservation of resources under nutrient limiting conditions.
3.2.4. Regulation of ROS
Central to many of the protective responses elicited by p53 is the ability to limit or control ROS. p53 directly activates the expression of numerous pro- and antioxidant pathways that are discussed in much more detail elsewhere [75]. An increase in ROS is driven by p53-dependent activation of genes like proline-oxidase [76] and PIG3 [77], inhibition of the antioxidant defence factors like SOD2 [78] and limitation of NADPH production by repressing the PPP [53, 63] and ME1 and 2 [79]. Downregulation of cystine uptake can also limit glutathione synthesis, enhancing the vulnerability of the cell to death through ferroptosis [27]. However, under conditions where p53 contributes to cell survival, such as serine starvation, induction of p53 responses that limit ROS become important [46]. This function of p53 can be mediated by repression of pro-oxidant genes like NOS2 and COX2, or the activation of anti-oxidant defence genes such as the Sestrins, ALDH4, GPX1 and SOD2 [80], while other p53 target genes support NADPH production through the PPP [81] or activate Nrf2 antioxidant activity [82]. The ability of p53 to maintain mitochondrial health, as discussed above, will also limit ROS production.
4. Activities of p53 – in vivo
The concept that p53 exerts protective functions that allow cells to adapt to metabolic stress and limit ROS, but can also drive cell elimination pathways accompanied by increased oxidative stress (Fig. 2), has been developed largely through extensive analysis of the functions of p53 in vitro. These activities of p53 are implicated in mechanisms of tumour suppression and promotion, but have also prompted a reassessment of the cancer independent functions of p53. The challenge now is to understand how these multiple p53 activities contribute to health and disease in vivo.
The identification of metabolic functions for p53 has implications for mechanisms of tumour suppression and promotion but has also prompted a reassessment of cancer-independent functions of p53.
4.1. p53 and cancer
Loss of p53 function Is strongly associated with cancer development across a wide range of organs, as discussed in numerous excellent recent reviews [6, 83, 84]. Many of the activities of p53 outlined above could play a role in tumour suppression, by preventing or repairing potentially oncogenic damage or by eliminating cells that have embarked on oncogenic progression. The ability of p53 to regulate transcription is important for its role in tumour protection, and animals bearing p53 mutants with severely reduced transcriptional activity are prone to tumour development [85]. However, deletion of the key p53 target genes that mediate cell cycle arrest and apoptosis generated the surprising conclusion that neither of these functions is necessary for tumour suppression [86, 87]. The generation of mice expressing p53 mutants with selectively compromised transcriptional activity further supported this observation, as discussed elsewhere [4]. Examples include mice carrying p53 mutated in the first transcriptional activation domain, which are defective in the induction of key p53 target genes that drive cell cycle arrest and apoptosis, but nevertheless retain tumour suppressive protection [85]. Similarly, mice expressing p53 mutated in lysine residues that are targets of acetylation show impaired apoptotic and cell cycle arrest functions, yet also retain the ability to limit cancer development [88]. These studies raise the interesting question of which p53 functions are responsible for tumour suppression? The answer seems to be that numerous p53 responses can help to limit malignant progression, with the importance of each likely to vary depending on oncogenic signal and organ under stress. Limitation of stem cell function and maintenance of genome integrity, as well as the elimination of cells through various pathways including ferroptosis and senescence, probably all contribute to p53's tumour suppressor activities [89]. The regulation of metabolic pathways described above may also play a role here, and it is interesting to note that several of the tumour suppressive p53 mutants that have lost apoptotic and cell cycle arrest functions retain the ability to control metabolic targets [88, 90].
While these studies are consistent with a role for metabolic regulation by p53 in tumour suppression, the ability of p53 to protect cells from DNA damage or induce a cell cycle arrest/cell survival response is thought to underscore the puzzling observation that wild type p53 expressing cancers can respond more poorly to genotoxic therapy than tumours with mutated p53 [91, 92]. Indeed, retention of wild type p53 may even support tumour development in some contexts, potentially reflecting the cell protection functions of p53 described above. Consistent with this line of thought, it is clear that some of the individual components of the p53 response can be strongly associated with cancer progression. For example, Tigar – the product of a p53 responsive gene that can limit oxidative stress [81] – can support tumour development [93]. Consistently, p53-independent overexpression of Tigar is seen in many cancers. The link between reactive oxygen species (ROS) and cancer is complex, but there is evidence that while increased ROS can promote cancer initiation, limitation of excess ROS production is necessary for cancer progression [94]. The antioxidant functions of p53 may therefore help suppress tumour development while supporting tumour progression. Similarly, although the ability of p53 to promote autophagy has been shown to suppress the transformation of cells in culture [34], more generally autophagy is linked with tumour promotion [47]. Taken together, a theme emerges that while the complete p53 response generally functions to limit cancer development, individual activities of p53 that assist cell survival could also be advantageous to cancer progression, especially if these activities can be uncoupled from the cell elimination functions of p53.
The observations that some functions of p53 might be helpful to cancers also prompts a reassessment of the activities of tumour associated p53 mutations. While alterations in the p53 gene occur in well over half of all cancers, many of these are subtle missense mutations, giving rise to the expression of a p53 protein altered at only a single amino acid residue [95]. These mutant p53 proteins often accumulate to high levels in the cancer cells and generally have lost the canonical cell cycle arrest and cell death functions (i.e. the cell elimination functions) of p53. However, several studies have shown that these mutants also acquire new activities (so-called gain of function), which are not shown by the wild type p53 and contribute to cancer cell survival and metastasis [96]. Intriguingly, several of these novel functions relate to the ability of mutant p53s to regulate metabolism and are discussed in more detail in several recent papers and reviews [[97], [98], [99]]. By contrast to the gain of novel function, mutant p53s may also contribute to cancer development by selective retention of a subset of wild type p53 activities. The ability of some mutant p53s to support cancer cells under nutrient starvation [100] raises the intriguing possibility that mutant p53 showing loss of cell elimination functions while maintaining cell survival functions are most effective in promoting tumour development. Tumours harbour many types of p53 mutation, and closer analyses of these may help to deconvolute the multiple activities of wild type p53.
4.2. p53 and obesity
In the modern world, overeating has become a significant health problem. By 2010, excess weight had overtaken lack of food in reducing years of healthy living, with WHO estimates that almost 2 billion adults and 380 million children are now overweight or obese, triple the number seen in 1975 (WHO Fact Sheet, 2018). The metabolic stress associated with obesity is linked to the development of cardiovascular disease, diabetes and cancer - diseases that cause huge burdens of morbidity and mortality worldwide. Not surprisingly, there has been an increasing interest in understanding the role – if any - for p53 in the response to overeating or excess weight.
In mice, overeating and obesity leads to the activation of p53 in adipose tissue, liver, muscle and other organs [72, [101], [102], [103]] – as does starvation [104] - consistent with a potential role for p53 in responding to nutrient imbalance. However, assessing the function of p53 in regulating long term, chronic metabolic disease can be challenging. Systemic deletion of p53 may compromise studies due to the rapid development of cancer, so models in which p53 is more subtly modulated have been developed. Humans express a common p53 polymorphism at codon 72 producing an R72 or P72 variant of the protein. While both variants retain some tumour suppressor function, R72 has a greater apoptotic activity than P72, which is more adept at inducing p21 and a cell cycle arrest [[105], [106], [107], [108]]. However, while these activities translate into enhanced transformation suppression by R72 p53 in cell culture, studies in human populations failed to uncover a strong association with cancer risk [109, 110]. Unfortunately, this polymorphism is not found in mice, necessitating the use of mice with humanised p53 to study these variants. As in humans, these mice did not show an obvious difference in tumour development [105]. However, some interesting differences in metabolism are now being identified using this model. Mice carrying the R72 polymorphism showed more fat accumulation on a high fat diet than their P72 p53 expressing counterparts [111]. The R72 mice also displayed increased insulin resistance and glucose tolerance, adipose tissue inflammation and non-alcoholic fatty liver disease (NAFLD), all signs of metabolic disease that were functionally linked with an increased expression of inflammatory and cholesterol pathway regulating genes in the R72 mice. The relevance of these observations is strongly enhanced by population studies showing that the R72 p53 variant is associated with increased body weight and susceptibility to type 2 diabetes in humans [[112], [113], [114], [115]]. Intriguingly, under conditions of nutrient deprivation it is the R72 version of p53 that more effectively promotes growth arrest (as oppose to apoptosis) and so helps survival under starvation conditions [116]. The ability to promote fat accumulation when food is plenty and support survival when food is scarce may explain why the R72 p53 variant becomes more common in harsher northern latitudes [117].
Mice carrying specific MDM2 mutations have also been used to assess the effect of defects in the p53 response. Substitution of amino acid 305 in MDM2 (C305F mutant) prevents the binding of MDM2 to RPL11, so limiting the ability of RPL11 to activate p53 [118]. While DNA damaging signals do not require this RPL11 axis to induce p53, oncogenic and metabolic signals that induce ribosomal stress depend on this pathway [119]. Mice carrying C305F MDM2 are resistant to high fat diet induced obesity, which was associated with increased energy expenditure and enhanced expression of genes required for energy consumption in the adipose tissue of these mice. Importantly, p53 levels in the adipose tissue of these mice were reduced in response to HFD, with mice expressing only one p53 allele showing a similar phenotype [120]. Consistently, mice treated with Pifithrin alpha, an inhibitor of p53, showed reduced weight gain on a high fat diet [121]. p53 null mice were also shown to be resistant to weight gain induced by Ghrelin, a hormone that stimulates feeding and weight gain [122]. Furthermore, p53 is induced in endothelial cells in response to a high calorie diet, in this model exacerbating fat gain and insulin resistance [123]. Loss of p53 function in endothelial cells provoked mitochondrial biosynthesis in muscle, increased glucose uptake and increased energy expenditure - a response also seen p53 null mice [124] and in the mutant MDM2 mice [120]. However, other studies have shown p53 can support mitochondrial metabolism and exercise capacity [125, 126].
The studies linking p53 to weight gain are countered somewhat by other work showing that whole body loss of p53 leads to an exacerbation of weight gain in response to a high fat diet [127, 128]. In the latter study, this response was linked to elevated testosterone due to the loss of p53-induced aromatase expression and – importantly – was limited to male mice.
Clearly the metabolic role of p53 in vivo is complex and signal dependent. p53 can be induced in response to metabolic stress, and while under some conditions the expression of p53 may help to maintain homeostasis, sustained p53 activity is linked to the development of obesity and other markers of metabolic syndrome. Importantly, cell extrinsic activities of p53 (which have been predominantly studied in the context of cancer) may allow for the activation of p53 in some organs to systemically affect whole body metabolism. The outcome to p53 induction will therefore vary depending on which tissue is subject to stress and activation of a p53 response.
4.3. p53 in the pancreas and diabetes
Numerous studies have shown that the activation of p53 is associated with cell death in pancreatic beta cells. Free fatty acids such as palmitate can induce p53 and promote death in pancreatic islets [129, 130], while apoptosis induced by diabetes-associated hyperglycolysis is rescued by loss of p53 [131]. miR-200, which is induced in the islets of diabetic mice, promotes the activation of cell death through p53, and loss of p53 partially rescued diabetic phenotypes in susceptible mice [132]. Mice expressing an N-terminally truncated form of p53 (deltaNp53) – which is thought to hyperactivate p53 - show early onset diabetes associated with a severe decrease in beta cell mass [133]. Induction of p53 through deletion of ARF-BP1, a regulator of p53 stability, induced beta cell loss and diabetes in mice that was rescued by deletion of p53 [134]. While several studies suggest that the pathologies associated with p53 activation reflects enhanced activation of cell death, mice carrying a mutant p53 that has no apoptotic activity but retains cell cycle arrest function were also found to have a greater susceptibility to diabetes [135], suggesting a role for p53-induced senescence. More direct control of metabolic pathways by p53 has also been reported in mice carrying beta-cell specific depletion of MDM2. Loss of MDM2 resulted in the activation of p53, which inhibited pyruvate carboxylase activity and so resulted in mitochondrial dysfunction and, importantly, impaired glucose stimulated insulin secretion and glucose tolerance [136]. In a genetic model of obesity and diabetes, high levels of p53 were detected in the islets and deletion of p53 rescued free fatty acid induced lipotoxicity without affecting beta cell mass.
While p53 induction is generally detrimental to islet cells, some individual facets of the p53 response can help to protect these cells. The overexpression of p21 (one of the key mediators of p53-induced cell cycle arrest) inhibits beta cell proliferation, but also promotes beta cell survival and limits the onset of diet-induced diabetes [137, 138]. Consistent with these observations, mice deficient for p21 show accelerated diet induced diabetes. Intriguingly, treatment of diabetic mice with Nutlin – an activator of p53 – improved glucose homeostasis, while Nutlin was also shown to protect mice from STZ (a beta cell toxin) induced diabetes [139]. These studies attributed the protective effect of Nutlin to the p53-dependent activation of p21 and illustrate a recurring conundrum – that activation of p53 can either kill or protect cells and tissues depending on criteria that are - as yet - not well understood. In another model of STZ-induced pancreatic damage, loss of p53 protected from the development of diabetes. In this case, cytosolic p53 was found to interact with Parkin and prevent mitophagy, leading to mitochondrial damage and defects in insulin secretion [140]. It would therefore seem that in similar models, p53-dependent activation of p21 protects cells from toxin-induced damage while cytosolic p53 promotes damage.
4.4. p53 in adipocytes
Mice ectopically expressing the agouti peptide (Ay mice) overeat and develop obesity and diabetes that is associated with an induction of oxidative damage in the fat tissue, leading to senescence and the induction of pro-inflammatory cytokines. This increase in senescent adipocytes is also seen in human obese diabetic patients [103]. As seen in other obese mice [72], p53 accumulation and activation is evident in the adipose tissue from the Ay mice [103] and seems to be responsible for the adverse pathology, as deletion of p53 diminished the senescent and inflammatory response, and substantially improved the insulin resistance in these mice. Other studies have also linked loss of p53 with an improvement in insulin resistance linked to heart failure [141]. Consistent with these results, an important target of p53 in adipocytes is semaphorin 3E, which induces an inflammatory response and insulin resistance in mice [142]. More recent studies have confirmed the ability of p53 to drive chemokine production and an inflammatory response in adipocytes [143]. The senescence response to p53 activation is associated with increased p21 expression and p21 has also been implicated in adipocyte differentiation. While p21 expression appears to be helpful in limiting diabetes (as described above), retention of p21 is associated with increased susceptibility to high fat diet induced weight gain [144]. Taken together, it seems that excessive caloric intake and other chronic metabolic stress signals activate adipocyte p53, which contributes to the induction of an inflammatory response, further promoting weight gain and the development of insulin resistance.
More detailed analyses of adipocytes suggest a complex contribution of p53 in these cells. One study demonstrated that p53 can suppress white adipocyte differentiation and promote brown adipocytes differentiation, with whole body loss of p53 exacerbating fat gain in overfed mice [127]. Consistently, p53 null male mice on a HFD were shown to have increased levels of epididymal WAT [128]. By contrast, p53 null mice kept under thermoneutral conditions were resistant to weight gain on a high fat diet and showed increased expression of UCP1 (a marker of browning) in white adipose tissue [145]. A third report confirmed that whole body loss of p53 leads to reduced weight gain on HFD, which was associated with an increase in brown adipose tissue (BAT) activity [124]. However, while BAT-specific deletion of p53 did not dramatically affect weight gain in this study, activation of p53 in BAT reduced weigh gain in overfed mice, due to an increased thermogenic activity [124].
4.5. p53 in the liver
p53 is induced in the liver in response to metabolic stress and p53 expression correlates with various forms of liver damage and diseases such as steatosis, fibrosis, alcohol exposure, inflammation and HCV infection [101, 121, [146], [147], [148], [149]]. While obesity is linked to increased liver p53, this activation of p53 is also seen in a mouse model of SREBP1-driven fatty liver, which occurs in the absence of systemic obesity [101]. These observations suggest that the signal for p53 induction may be lipotoxic stress and excessive lipid accumulation. Consistently, starvation – which promotes an acute (or transient) increase in hepatic lipid accumulation [150] - also induces an increase in liver p53 levels [104]. This response is mediated through the activation of AMPK, a well-established transducer of metabolic stress signalling. Therefore, consistent with a general role in stress responses, liver p53 is mobilised in both starvation and nutrient excess conditions.
While the stress-induced induction of liver p53 is clear, the role of p53 and consequences of loss of p53 are complex - exacerbating or protecting from disease development depending on the context. Activation of p53 (through deletion of MDM2) can lead to fibrosis [151] while loss of p53 limited liver damage in genetically obese mice [101] or in models of steatohepatitis [152]. On the other hand, other studies suggest a protective role for p53 in limiting liver fat accumulation and hepatic steatosis [63, 128]. Acute viral-mediated deletion of p53 specifically in the liver (rather than the whole animal) also led to increased hepatic steatosis [104, 153], while deletion of p53 in hepatic stellate cells increased fibrosis in the context of toxin induced chronic liver damage [154]. Importantly, the latter study showed that activation of p53 promoted senescence and the release of factors from stellate cells that induced tumour inhibiting macrophages. In this case, therefore, the tumour suppressive function of p53 was a reflection of the p53 response in stromal cells, rather than the tumour cells themselves.
The protective effect of liver p53 may reflect the control of lipid metabolism described above, favouring the catabolic use of lipids through FAO while also limiting the production of lipids through FAS. Consistent with this proposal, depletion of liver p53 (either directly or through the action of miR-21) resulted in increased lipid accumulation [104, 155]. Mice unable to fully activate p53 due to the expression of a mutant form of MDM2 also showed increased liver fat under normal feeding conditions and hepatosteatosis following starvation [156]. However, these mice showed reduced accumulation of liver fat on a high fat diet [120] – an interesting paradox suggesting that the role of p53 in controlling hepatic lipid metabolism may depend on the extent of p53 activation or the nature of the nutrient stress. It is worth remembering that this model impedes p53 activation in all tissues. So, while the steatosis and lipid accumulation in the livers of starved mice may reflect defects in lipid metabolism in the liver itself, the decreased lipid accumulation in the overfed mice may reflect altered responses in cells such as adipocytes, that indirectly impact liver fat accumulation.
The effect of loss of liver p53 is not limited to fat accumulation but can also modulate gluconeogenesis and hepatic glucose production in response to starvation. The ability of p53 to down-regulate the expression of gluconeogenic genes correlates with the improved pyruvate tolerance of p53 null mice [67]. However, others have found that mice with liver specific downregulation of p53 show defects in glycogen storage and amino acid catabolism and become hypoglycaemic upon food withdrawal [104]. Activation of a p53 response was also shown to support glucose tolerance and insulin sensitivity [157, 158]. One consequence of loss of p53 in the liver is an increase in expression of the p53 related protein TAp63, which is also induced by a high fat diet [153]. TAp63 induction increases IKK activation, ER stress and consequent steatosis. However, loss of p63 in the whole animal dramatically perturbed lipid metabolism, resulting in obesity and liver damage [159, 160] – a response at least in part due to the loss of Coiled-Coil Domain Containing 3 protein (ccdc3) expression, a secretory protein activated by p63 [159]. Finally, mice with mutations in p53 also show increased liver steatosis and fibrosis in a model of liver inflammation linked to an increased infiltration of inflammatory cells [161]. On balance, most of the studies to date suggest that p53 plays a general protective role when expressed in the liver, maintaining homeostasis and limiting the development of fatty liver disease. Of note, however, systemic p53 induction in other tissues (such as adipose tissue) can overwhelm this response and indirectly lead to liver damage. Finally, and not surprisingly, persistent induction of p53 (potentially resulting from irreparable liver damage) may also contribute to the induction of cell death and consequent development of more severe disease.
Given the role of p53 in the control of liver metabolism, several studies have investigated the potential utility of drugs that modulate p53 for the treatment of liver disease. Consistent with a role for p53 in promoting steatosis, mice treated with Pifithrin alpha (a p53 inhibitor) showed improved liver function on a high fat diet [121]. By contrast, and consistent with a protective function of p53, activation of p53 using low dose doxorubicin protected mice from diet-induced steatosis [162].
4.6. p53 and aging
The strong tumour suppressive function of p53, and the relatively mild impact of p53 deficiency on normal development, gave an initial impression that p53 was exclusively a guardian against disease. Subsequent work showing a role for p53 in promoting numerous adverse pathologies soon corrected this view, and the darker side of p53 is further revealed during aging [163]. While strong systemic activation of p53 is acutely deleterious to survival [164], more subtle but persistent increases in p53 activity reduced cancer development but lead to early aging [165, 166]. The consequences of constitutive p53 activity have been further supported by a family carrying a mutation in MDM2 that weakens its ability to control p53, and that display progeria or early aging syndrome [167]. Importantly, however, a model in which p53 was more readily activated than normal but still held under control in unstressed conditions allowed for enhanced cancer protection without compromising longevity [168].
How subtly but persistently elevated levels of p53 promote aging is not fully understood, and may reflect activities of different p53 isoforms [169]. Functions of p53 that deplete stem cell populations or induce senescence may each play a role here [36, [170], [171], [172]]. Intriguingly, in some contexts mice with enhanced but normally regulated p53 show delayed aging [173] and the maintenance (rather than loss) of stem cell pools [174]. It is tempting to speculate that some of the protective and survival functions of p53, such the limitation of DNA damage and oxidative stress, could play a role in both preventing cancer and promoting healthy aging. Some insight into which functions of p53 are important here may be gained from the study of the R72P polymorphism mentioned above, where the P72 variant tracks with increased lifespan [175, 176].
5. Conclusion
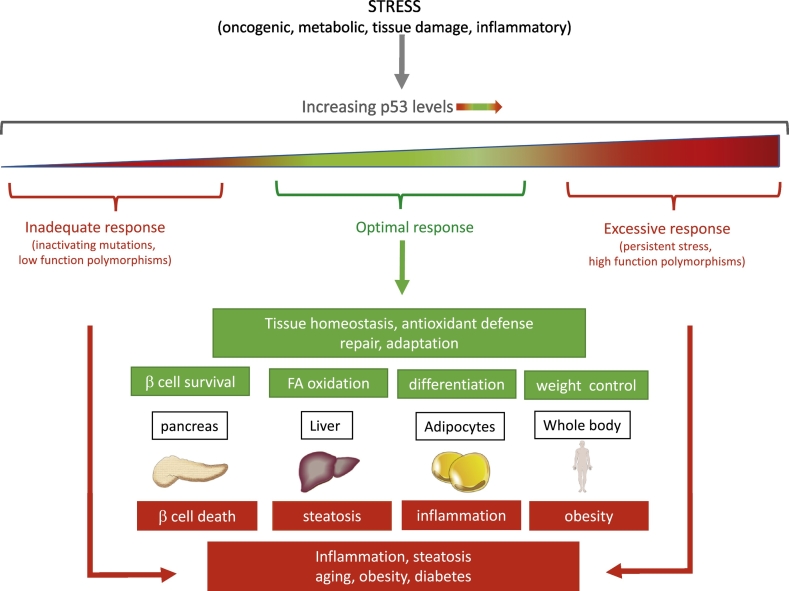
An enormous volume of in vitro data has shown that p53 can play many roles in response to metabolic stress and activities that can be broadly grouped into cell elimination and cell survival functions (Fig. 2). Not surprisingly, therefore, analysis of p53 functions in vivo has revealed a similar - if not more - complex pattern of responses (Fig. 3). p53 activation in individual organs or body-wide, promotes ameliorating or protective responses – most notably under conditions where the stress signal is transient or can be resolved. In several models that have been examined, this can be linked to the activation of a reversible cell cycle arrest through the induction of p21 and the activity of various p53-induced antioxidant defences. However, under acute or persistent stress conditions p53 turns to the induction of cell death or elimination, leading to multiple pathologies that include diabetes (as discussed here), reperfusion injury and neurodegenerative disease. An emerging model suggests that optimal p53 function is important to maintain numerous aspects of health, while too little or too much p53 can have profoundly deleterious consequences to the organism (Fig. 3). Evolutionarily, it's clear that the homeostatic and tumour suppressor activities of p53 are too important to lose, and trump the collateral damage associated with p53-induced cell death. But understanding the various responses to p53 in different circumstances will allow us to use modulation of p53 for therapeutic gain much more effectively.
Fig. 3.
p53 regulates stress response and homeostasis in vivo. Numerous stress signals activate p53. An optimal p53 response (green) is required to maintain whole body and tissue homeostasis. However, insufficient or excessive p53 (red) has deleterious effects and results in cell death, lipid accumulation, inflammation and compromised tissue functionality. As a consequence, failure to maintain optimal p53 levels can lead to numerous adverse pathologies, beyond cancer. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Transparency document
Transparency document.
Acknowledgements
We would like to thank Joe Brock for help with the illustrations. This work was funded by Cancer Research UK grant C596/A26855 and supported by the Francis Crick Institute which receives its core funding from Cancer Research UK, the UK Medical Research Council and the Wellcome Trust.
Footnotes
This article is part of a Special Issue entitled: Cancer Metabolism edited by Dr. Chi Van Dang.
The Transparency document associated with this article can be found, in online version.
References
- 1.Hainaut P., Pfeifer G.P. Somatic TP53 mutations in the era of genome sequencing. Cold Spring Harb. Perspect. Med. 2016;6 doi: 10.1101/cshperspect.a026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guha T., Malkin D. Inherited TP53 mutations and the Li-Fraumeni syndrome. Cold Spring Harb. Perspect. Med. 2017;7 doi: 10.1101/cshperspect.a026187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donehower L.A., Lozano G. 20 years studying p53 functions in genetically engineered mice. Nat. Rev. Cancer. 2009;9:831–841. doi: 10.1038/nrc2731. [DOI] [PubMed] [Google Scholar]
- 4.Kaiser A.M., Attardi L.D. Deconstructing networks of p53-mediated tumor suppression in vivo. Cell Death Differ. 2018;25:93–103. doi: 10.1038/cdd.2017.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruiswijk F., Labuschagne C.F., Vousden K.H. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 6.Kastenhuber E.R., Lowe S.W. Putting p53 in context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haupt S., Haupt Y. P53 at the start of the 21st century: lessons from elephants. F1000Res. 2017;6:2041. doi: 10.12688/f1000research.12682.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aylon Y., Oren M. The paradox of p53: what, how, and why? Cold Spring Harb. Perspect. Med. 2016;6 doi: 10.1101/cshperspect.a026328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hock A.K., Vousden K.H. The role of ubiquitin modification in the regulation of p53. Biochim. Biophys. Acta. 2014;1843:137–149. doi: 10.1016/j.bbamcr.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 10.Zhou X., Cao B., Lu H. Negative auto-regulators trap p53 in their web. J. Mol. Cell Biol. 2017;9:62–68. doi: 10.1093/jmcb/mjx001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayo L.D., Donner D.B. The PTEN, Mdm2, p53 tumor suppressor-oncoprotein network. Trends Biochem. Sci. 2002;27:462–467. doi: 10.1016/s0968-0004(02)02166-7. [DOI] [PubMed] [Google Scholar]
- 12.Gu B., Zhu W.G. Surf the post-translational modification network of p53 regulation. Int. J. Biol. Sci. 2012;8:672–684. doi: 10.7150/ijbs.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merritt A.J., Potten C.S., Kemp C.J., Hickman J.A., Balmain A., Lane D.P., Hall P.A. The role of p53 in spontaneous and radiation-induced apoptosis in the gastrointestinal tract of normal and p53-deficient mice. Cancer Res. 1994;54:614–617. [PubMed] [Google Scholar]
- 14.Christophorou M.A., Ringshausen I., Finch A.J., Swigart L.B., Evan G.I. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–217. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- 15.Gebel J., Tuppi M., Krauskopf K., Coutandin D., Pitzius S., Kehrloesser S., Osterburg C., Dotsch V. Control mechanisms in germ cells mediated by p53 family proteins. J. Cell Sci. 2017;130:2663–2671. doi: 10.1242/jcs.204859. [DOI] [PubMed] [Google Scholar]
- 16.Humpton T.J., Vousden K.H. Regulation of cellular metabolism and hypoxia by p53. Cold Spring Harb. Perspect. Med. 2016;6 doi: 10.1101/cshperspect.a026146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36:3943–3956. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hafner A., Stewart-Ornstein J., Purvis J.E., Forrester W.C., Bulyk M.L., Lahav G. p53 pulses lead to distinct patterns of gene expression albeit similar DNA-binding dynamics. Nat. Struct. Mol. Biol. 2017;24:840–847. doi: 10.1038/nsmb.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan K.D., Galbraith M.D., Andrysik Z., Espinosa J.M. Mechanisms of transcriptional regulation by p53. Cell Death Differ. 2018;25:133–143. doi: 10.1038/cdd.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villunger A., Michalak E.M., Coultas L., Mullauer F., Bock G., Ausserlechner M.J., Adams J.M., Strasser A. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science (New York, N.Y.) 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 21.Speidel D. Transcription-independent p53 apoptosis: an alternative route to death. Trends Cell Biol. 2010;20:14–24. doi: 10.1016/j.tcb.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Mai W.X., Gosa L., Daniels V.W., Ta L., Tsang J.E., Higgins B., Gilmore W.B., Bayley N.A., Harati M.D., Lee J.T., Yong W.H., Kornblum H.I., Bensinger S.J., Mischel P.S., Rao P.N., Clark P.M., Cloughesy T.F., Letai A., Nathanson D.A. Cytoplasmic p53 couples oncogene-driven glucose metabolism to apoptosis and is a therapeutic target in glioblastoma. Nat. Med. 2017;23:1342–1351. doi: 10.1038/nm.4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Follis A.V., Llambi F., Merritt P., Chipuk J.E., Green D.R., Kriwacki R.W. Pin1-induced proline isomerization in cytosolic p53 mediates BAX activation and apoptosis. Mol. Cell. 2015;59:677–684. doi: 10.1016/j.molcel.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castrogiovanni C., Waterschoot B., De Backer O., Dumont P. Serine 392 phosphorylation modulates p53 mitochondrial translocation and transcription-independent apoptosis. Cell Death Differ. 2018;25:190–203. doi: 10.1038/cdd.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long J.S., Crighton D., O'Prey J., Mackay G., Zheng L., Palmer T.M., Gottlieb E., Ryan K.M. Extracellular adenosine sensing-a metabolic cell death priming mechanism downstream of p53. Mol. Cell. 2013;50:394–406. doi: 10.1016/j.molcel.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 26.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S., Morrison B., III, Stockwell B.R. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang L., Kon N., Li T., Wang S.J., Su T., Hibshoosh H., Baer R., Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jennis M., Kung C.P., Basu S., Budina-Kolomets A., Leu J.I., Khaku S., Scott J.P., Cai K.Q., Campbell M.R., Porter D.K., Wang X., Bell D.A., Li X., Garlick D.S., Liu Q., Hollstein M., George D.L., Murphy M.E. An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes Dev. 2016;30:918–930. doi: 10.1101/gad.275891.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ou Y., Wang S.J., Li D., Chu B., Gu W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E6806–E6812. doi: 10.1073/pnas.1607152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S.J., Li D., Ou Y., Jiang L., Chen Y., Zhao Y., Gu W., Acetylation Is. Crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep. 2016;17:366–373. doi: 10.1016/j.celrep.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie Y., Zhu S., Song X., Sun X., Fan Y., Liu J., Zhong M., Yuan H., Zhang L., Billiar T.R., Lotze M.T., Zeh H.J., III, Kang R., Kroemer G., Tang D. The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 2017;20:1692–1704. doi: 10.1016/j.celrep.2017.07.055. [DOI] [PubMed] [Google Scholar]
- 32.Tarangelo A., Magtanong L., Bieging-Rolett K.T., Li Y., Ye J., Attardi L.D., Dixon S.J. p53 suppresses metabolic stress-induced ferroptosis in cancer cells. Cell Rep. 2018;22:569–575. doi: 10.1016/j.celrep.2017.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crighton D., Wilkinson S., O'Prey J., Syed N., Harrison P.R., Gasco M., Garrone O., Crook T., Ryan K.M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;14:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 34.Kenzelmann Broz D., Spano Mello S., Bieging K.T., Jiang D., Dusek R.L., Brady C.A., Sidow A., Attardi L.D. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013;27:1016–1031. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang E.Y., Gang H., Aviv Y., Dhingra R., Margulets V., Kirshenbaum L.A. p53 mediates autophagy and cell death by a mechanism contingent on Bnip3. Hypertension. 2013;62:70–77. doi: 10.1161/HYPERTENSIONAHA.113.01028. [DOI] [PubMed] [Google Scholar]
- 36.Rufini A., Tucci P., Celardo I., Melino G. Senescence and aging: the critical roles of p53. Oncogene. 2013;32:5129–5143. doi: 10.1038/onc.2012.640. [DOI] [PubMed] [Google Scholar]
- 37.Deng C., Zhang P., Harper J.W., Elledge S.J., Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 38.Brugarolas J., Chandrasekaran C., Gordon J.I., Beach D., Jacks T., Hannon G.J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–556. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 39.Fukasawa K., Choi T., Kuriyama R., Rulong S., Vande Woude G.F. Abnormal centrosome amplification in the absence of p53. Science (New York, N.Y.) 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 40.Vitre B.D., Cleveland D.W. Centrosomes, chromosome instability (CIN) and aneuploidy. Curr. Opin. Cell Biol. 2012;24:809–815. doi: 10.1016/j.ceb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeo C.Q.X., Alexander I., Lin Z., Lim S., Aning O.A., Kumar R., Sangthongpitag K., Pendharkar V., Ho V.H.B., Cheok C.F. p53 maintains genomic stability by preventing interference between transcription and replication. Cell Rep. 2016;15:132–146. doi: 10.1016/j.celrep.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 42.Klusmann I., Rodewald S., Muller L., Friedrich M., Wienken M., Li Y., Schulz-Heddergott R., Dobbelstein M. p53 activity results in DNA replication fork processivity. Cell Rep. 2016;17:1845–1857. doi: 10.1016/j.celrep.2016.10.036. [DOI] [PubMed] [Google Scholar]
- 43.Kumari S.R., Mendoza-Alvarez H., Alvarez-Gonzales R. Functional interactions of p53 with poly (ADP-ribose) polymerase (PARP) during apoptosis following DNA damage: covalent poly (ADP-ribosyl)ation of p53 by exogenous PARP and noncovalent binding of p53 to the Mr 85,000 proteolytic fragment. Cancer Res. 1998;58:5075–5078. [PubMed] [Google Scholar]
- 44.Jones R.G., Plas D.R., Kubek S., Buzzai M., Mu J., Xu Y., Birnbaum M.J., Thompson C.B. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 45.Reid M.A., Wang W.I., Rosales K.R., Welliver M.X., Pan M., Kong M. The B55alpha subunit of PP2A drives a p53-dependent metabolic adaptation to glutamine deprivation. Mol. Cell. 2013;50:200–211. doi: 10.1016/j.molcel.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 46.Maddocks O.D., Berkers C.R., Mason S.M., Zheng L., Blyth K., Gottlieb E., Vousden K.H. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White E. Autophagy and p53. Cold Spring Harb. Perspect. Med. 2016;6:a026120. doi: 10.1101/cshperspect.a026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ou Y., Wang S.J., Jiang L., Zheng B., Gu W. p53 protein-mediated regulation of phosphoglycerate dehydrogenase (PHGDH) is crucial for the apoptotic response upon serine starvation. J. Biol. Chem. 2015;290:457–466. doi: 10.1074/jbc.M114.616359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lago C.U., Sung H.J., Ma W., Wang P.Y., Hwang P.M. p53, aerobic metabolism, and cancer. Antioxid. Redox Signal. 2011;15:1739–1748. doi: 10.1089/ars.2010.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berkers C.R., Maddocks O.D., Cheung E.C., Mor I., Vousden K.H. Metabolic regulation by p53 family members. Cell Metab. 2013;18:617–633. doi: 10.1016/j.cmet.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Floter J., Kaymak I., Schulze A. Regulation of metabolic activity by p53. Meta. 2017;7 doi: 10.3390/metabo7020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawauchi K., Araki K., Tobiume K., Tanaka N. p53 regulates glucose metabolism though an IKK-NF-kB pathway and inhibits cell transformation. Nat. Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 53.Ros S., Floter J., Kaymak I., Da Costa C., Houddane A., Dubuis S., Griffiths B., Mitter R., Walz S., Blake S., Behrens A., Brindle K.M., Zamboni N., Rider M.H., Schulze A. 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 is essential for p53-null cancer cells. Oncogene. 2017;36:3287–3299. doi: 10.1038/onc.2016.477. [DOI] [PubMed] [Google Scholar]
- 54.Hu A., Zhang C., Wu R., Sun Y., Levine A.J., Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki S., Tanaka T., Poyurovsky M.V., Nagano H., Mayama T., Ohkubo S., Lokshin M., Hosokawa H., Nakayama T., Suzuki Y., Sugano S., Sato E., Nagao T., Yokote K., Tatsuno I., Prives C. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Contractor T., Harris C.R. p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer Res. 2012;72:560–567. doi: 10.1158/0008-5472.CAN-11-1215. [DOI] [PubMed] [Google Scholar]
- 57.Zhang C., Lin M., Wu R., Wang X., Yang B., Levine A.J., Hu W., Feng Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boidot R., Vegran F., Meulle A., Le Breton A., Dessy C., Sonveaux P., Lizard-Nacol S., Feron O. Regulation of monocarboxylate transporter MCT1 expression by p53 mediates inward and outward lactate fluxes in tumors. Cancer Res. 2012;72:939–948. doi: 10.1158/0008-5472.CAN-11-2474. [DOI] [PubMed] [Google Scholar]
- 59.Itahana Y., Itahana K. Emerging roles of p53 family members in glucose metabolism. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19030776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee P., Vousden K.H., Cheung E.C. TIGAR, TIGAR, burning bright. Cancer Metab. 2014;2:1. doi: 10.1186/2049-3002-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li M., Sun M., Cao L., Gu J.H., Ge J., Chen J., Han R., Qin Y.Y., Zhou Z.P., Ding Y., Qin Z.H. A TIGAR-regulated metabolic pathway is critical for protection of brain ischemia. J. Neurosci. 2014;34:7458–7471. doi: 10.1523/JNEUROSCI.4655-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duan L., Perez R.E., Chen L., Blatter L.A., Maki C.G. p53 promotes AKT and SP1-dependent metabolism through the pentose phosphate pathway that inhibits apoptosis in response to Nutlin-3a. J. Mol. Cell Biol. 2017 doi: 10.1093/jmcb/mjx051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang P., Du W., Wang X., Mancuso A., Gao X., Wu M., Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goldstein I., Yizhak K., Madar S., Goldfinger N., Ruppin E., Rotter V. p53 promotes the expression of gluconeogenesis-related genes and enhances hepatic glucose production. Cancer Metab. 2013;1:9. doi: 10.1186/2049-3002-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harami-Papp H., Pongor L.S., Munkacsy G., Horvath G., Nagy A.M., Ambrus A., Hauser P., Szabo A., Tretter L., Gyorffy B. TP53 mutation hits energy metabolism and increases glycolysis in breast cancer. Oncotarget. 2016;7:67183–67195. doi: 10.18632/oncotarget.11594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang S.J., Yu G., Jiang L., Li T., Lin Q., Tang Y., Gu W. p53-Dependent regulation of metabolic function through transcriptional activation of pantothenate kinase-1 gene. Cell Cycle. 2013;12:753–761. doi: 10.4161/cc.23597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang P., Tu B., Wang H., Cao Z., Tang M., Zhang C., Gu B., Li Z., Wang L., Yang Y., Zhao Y., Wang H., Luo J., Deng C.X., Gao B., Roeder R.G., Zhu W.G. Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proc. Natl. Acad. Sci. U. S. A. 2014;111:10684–10689. doi: 10.1073/pnas.1411026111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goldstein I., Rotter V. Regulation of lipid metabolism by p53- fighting two villains with one sword. Trends Endocrinol. Metab. 2012;23:567–575. doi: 10.1016/j.tem.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 69.Goldstein I., Ezra O., Rivlin N., Molchadsky A., Madar S., Goldfinger N., Rotter V. p53, a novel regulator of lipid metabolism pathways. J. Hepatol. 2012;56:656–662. doi: 10.1016/j.jhep.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 70.Zaugg K., Yao Y., Reilly P.T., Kannan K., Kiarash R., Mason J., Huang P., Sawyer S.K., Fuerth B., Faubert B., Kalliomaki T., Elia A., Luo X., Nadeem V., Bungard D., Yalavarthi S., Growney J.D., Wakeham A., Moolani Y., Silvester J., Ten A.Y., Bakker W., Tsuchihara K., Berger S.L., Hill R.P., Jones R.G., Tsao M., Robinson M.O., Thompson C.B., Pan G., Mak T.W. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25:1041–1051. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Assaily W., Rubinger D.A., Wheaton K., Lin Y., Ma W., Xuan W., Brown-Endres L., Tsuchihara K., Mak T.W., Benchimol S. ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol. Cell. 2011;44:491–501. doi: 10.1016/j.molcel.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 72.Yahagi N., Shimano H., Matsuzaka T., Najima Y., Sekiya M., Nakagawa Y., Ide T., Tomita S., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., Gotoda T., Nagai R., Kimura S., Ishibashi S., Osuga J., Yamada N. p53 activation in adipocytes of obese mice. J. Biol. Chem. 2003;278:25395–25400. doi: 10.1074/jbc.M302364200. [DOI] [PubMed] [Google Scholar]
- 73.Budanov A.V., Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hasty P., Sharp Z.D., Curiel T.J., Campisi J. mTORC1 and p53: clash of the gods? Cell Cycle. 2013;12:20–25. doi: 10.4161/cc.22912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu D., Xu Y. p53, oxidative stress, and aging. Antioxid. Redox Signal. 2011;15:1669–1678. doi: 10.1089/ars.2010.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rivera A., Maxwell S.A. The p53-induced gene-6 (proline oxidase) mediates apoptosis through a calcineurin-dependent pathway. J. Biol. Chem. 2005;280:29346–29354. doi: 10.1074/jbc.M504852200. [DOI] [PubMed] [Google Scholar]
- 77.Polyak K., Xia Y., Zweier J.L., Kinzler K.W., Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 78.Zhao Y., Chaiswing L., Velez J.M., Batinic-Haberle I., Colburn N.H., Oberley T.D., St Clair D.K. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer Res. 2005;65:3745–3750. doi: 10.1158/0008-5472.CAN-04-3835. [DOI] [PubMed] [Google Scholar]
- 79.Jiang P., Du W., Mancuso A., Wellen K.E., Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493:689–693. doi: 10.1038/nature11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Budanov A.V. The role of tumor suppressor p53 in the antioxidant defense and metabolism. Subcell. Biochem. 2014;85:337–358. doi: 10.1007/978-94-017-9211-0_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bensaad K., Tsuruta A., Selak M.A., Vidal M.N., Nakano K., Bartrons R., Gottlieb E., Vousden K.H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 82.Chen W., Sun Z., Wang X.J., Jiang T., Huang Z., Fang D., Zhang D.D. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol. Cell. 2009;34:663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mello S.S., Attardi L.D. Deciphering p53 signaling in tumor suppression. Curr. Opin. Cell Biol. 2017;51:65–72. doi: 10.1016/j.ceb.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sabapathy K., Lane D.P. Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat. Rev. Clin. Oncol. 2018;15:13–30. doi: 10.1038/nrclinonc.2017.151. [DOI] [PubMed] [Google Scholar]
- 85.Brady C.A., Jiang D., Mello S.S., Johnson T.M., Jarvis L.A., Kozak M.M., Kenzelmann Broz D., Basak S., Park E.J., McLaughlin M.E., Karnezis A.N., Attardi L.D. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aubrey B.J., Kelly G.L., Janic A., Herold M.J., Strasser A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018;25:104–113. doi: 10.1038/cdd.2017.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Valente L.J., Gray D.H., Michalak E.M., Pinon-Hofbauer J., Egle A., Scott C.L., Janic A., Strasser A. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep. 2013;3:1339–1345. doi: 10.1016/j.celrep.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 88.Li T., Kon N., Jiang L., Tan M., Ludwig T., Zhao Y., Baer R., Gu W. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mello S.S., Attardi L.D. Deciphering p53 signaling in tumor suppression. Curr. Opin. Cell Biol. 2018;51:65–72. doi: 10.1016/j.ceb.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jiang D., LaGory E.L., Kenzelmann Broz D., Bieging K.T., Brady C.A., Link N., Abrams J.M., Giaccia A.J., Attardi L.D. Analysis of p53 transactivation domain mutants reveals Acad11 as a metabolic target important for p53 pro-survival function. Cell Rep. 2015;10:1096–1109. doi: 10.1016/j.celrep.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bertheau P., Espie M., Turpin E., Lehmann J., Plassa L.-F., Varna M., Janin A., de The H. TP53 status and response to chemotherapy in breast cancer. Pathobiology. 2008;75:132–139. doi: 10.1159/000123851. [DOI] [PubMed] [Google Scholar]
- 92.Jackson J.G., Pant V., Li Q., Chang L.L., Quintas-Cardama A., Garza D., Tavana O., Yang P., Manshouri T., Li Y., El-Naggar A.K., Lozano G. p53-mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer Cell. 2012;21:793–806. doi: 10.1016/j.ccr.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheung E.C., Athineos D., Lee P., Ridgway R.A., Lambie W., Nixon C., Strathdee D., Blyth K., Sansom O.J., Vousden K.H. TIGAR is required for efficient intestinal regeneration and tumorigenesis. Dev. Cell. 2013;25:463–477. doi: 10.1016/j.devcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gorrini C., Harris I.S., Mak T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 95.Petitjean A., Mathe E., Kato S., Ishioka C., Tavtigian S.V., Hainaut P., Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 96.Muller P.A., Vousden K.H. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu J., Zhang C., Hu W., Feng Z. Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Lett. 2015;356:197–203. doi: 10.1016/j.canlet.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Basu S., Gnanapradeepan K., Barnoud T., Kung C.P., Tavecchio M., Scott J., Watters A., Chen Q., Kossenkov A.V., Murphy M.E. Mutant p53 controls tumor metabolism and metastasis by regulating PGC-1alpha. Genes Dev. 2018;32:230–243. doi: 10.1101/gad.309062.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gnanapradeepan K., Basu S., Barnoud T., Budina-Kolomets A., Kung C.P., Murphy M.E. The p53 tumor suppressor in the control of metabolism and ferroptosis. Front. Endocrinol. 2018;9:124. doi: 10.3389/fendo.2018.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tran T.Q., Lowman X.H., Reid M.A., Mendez-Dorantes C., Pan M., Yang Y., Kong M. Tumor-associated mutant p53 promotes cancer cell survival upon glutamine deprivation through p21 induction. Oncogene. 2017;36:1991–2001. doi: 10.1038/onc.2016.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yahagi N., Shimano H., Matsuzaka T., Sekiya M., Najima Y., Okazaki S., Okazaki H., Tamura Y., Iizuka Y., Inoue N., Nakagawa Y., Takeuchi Y., Ohashi K., Harada K., Gotoda T., Nagai R., Kadowaki T., Ishibashi S., Osuga J., Yamada N. p53 involvement in the pathogenesis of fatty liver disease. J. Biol. Chem. 2004;279:20571–20575. doi: 10.1074/jbc.M400884200. [DOI] [PubMed] [Google Scholar]
- 102.Homayounfar R., Jeddi-Tehrani M., Cheraghpour M., Ghorbani A., Zand H. Relationship of p53 accumulation in peripheral tissues of high-fat diet-induced obese rats with decrease in metabolic and oncogenic signaling of insulin. Gen. Comp. Endocrinol. 2015;214:134–139. doi: 10.1016/j.ygcen.2014.06.029. [DOI] [PubMed] [Google Scholar]
- 103.Minamino T., Orimo M., Shimizu I., Kunieda T., Yokoyama M., Ito T., Nojima A., Nabetani A., Oike Y., Matsubara H., Ishikawa F., Komuro I. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat. Med. 2009;15:1082–1087. doi: 10.1038/nm.2014. [DOI] [PubMed] [Google Scholar]
- 104.Prokesch A., Graef F.A., Madl T., Kahlhofer J., Heidenreich S., Schumann A., Moyschewitz E., Pristoynik P., Blaschitz A., Knauer M., Muenzner M., Bogner-Strauss J.G., Dohr G., Schulz T.J., Schupp M. Liver p53 is stabilized upon starvation and required for amino acid catabolism and gluconeogenesis. FASEB J. 2017;31:732–742. doi: 10.1096/fj.201600845R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhu F., Dolle M.E., Berton T.R., Kuiper R.V., Capps C., Espejo A., McArthur M.J., Bedford M.T., van Steeg H., de Vries A., Johnson D.G. Mouse models for the p53 R72P polymorphism mimic human phenotypes. Cancer Res. 2010;70:5851–5859. doi: 10.1158/0008-5472.CAN-09-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dumont P., Leu J.I., Della Pietra A.C., George D.L., Murphy M.P. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 107.Bonafe M., Salvioli S., Barbi C., Mishto M., Trapassi C., Gemelli C., Storci G., Olivieri F., Monti D., Franceschi C. p53 codon 72 genotype affects apoptosis by cytosine arabinoside in blood leukocytes. Biochem. Biophys. Res. Commun. 2002;299:539–541. doi: 10.1016/s0006-291x(02)02691-8. [DOI] [PubMed] [Google Scholar]
- 108.Pim D., Banks L. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int. J. Cancer. 2004;108:196–199. doi: 10.1002/ijc.11548. [DOI] [PubMed] [Google Scholar]
- 109.Murphy M.E. Polymorphic variants in the p53 pathway. Cell Death Differ. 2006;13:916–920. doi: 10.1038/sj.cdd.4401907. [DOI] [PubMed] [Google Scholar]
- 110.Dahabreh I.J., Linardou H., Bouzika P., Varvarigou V., Murray S. TP53 Arg72Pro polymorphism and colorectal cancer risk: a systematic review and meta-analysis. Cancer Epidemiol. Biomark. Prev. 2010;19:1840–1847. doi: 10.1158/1055-9965.EPI-10-0156. [DOI] [PubMed] [Google Scholar]
- 111.Kung C.P., Leu J.I., Basu S., Khaku S., Anokye-Danso F., Liu Q., George D.L., Ahima R.S., Murphy M.E. The P72R polymorphism of p53 predisposes to obesity and metabolic dysfunction. Cell Rep. 2016;14:2413–2425. doi: 10.1016/j.celrep.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bonfigli A.R., Sirolla C., Testa R., Cucchi M., Spazzafumo L., Salvioli S., Ceriello A., Olivieri F., Festa R., Procopio A.D., Brandoni G., Boemi M., Marra M., Franceschi C. The p53 codon 72 (Arg72Pro) polymorphism is associated with the degree of insulin resistance in type 2 diabetic subjects: a cross-sectional study. Acta Diabetol. 2013;50:429–436. doi: 10.1007/s00592-012-0450-x. [DOI] [PubMed] [Google Scholar]
- 113.Gloria-Bottini F., Banci M., Saccucci P., Magrini A., Bottini E. Is there a role of p53 codon 72 polymorphism in the susceptibility to type 2 diabetes in overweight subjects? A study in patients with cardiovascular diseases. Diabetes Res. Clin. Pract. 2011;91:e64–e67. doi: 10.1016/j.diabres.2010.11.031. [DOI] [PubMed] [Google Scholar]
- 114.Gaulton K.J., Willer C.J., Li Y., Scott L.J., Conneely K.N., Jackson A.U., Duren W.L., Chines P.S., Narisu N., Bonnycastle L.L., Luo J., Tong M., Sprau A.G., Pugh E.W., Doheny K.F., Valle T.T., Abecasis G.R., Tuomilehto J., Bergman R.N., Collins F.S., Boehnke M., Mohlke K.L. Comprehensive association study of type 2 diabetes and related quantitative traits with 222 candidate genes. Diabetes. 2008;57:3136–3144. doi: 10.2337/db07-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Burgdorf K.S., Grarup N., Justesen J.M., Harder M.N., Witte D.R., Jorgensen T., Sandbaek A., Lauritzen T., Madsbad S., Hansen T., Consortium D., Pedersen O. Studies of the association of Arg72Pro of tumor suppressor protein p53 with type 2 diabetes in a combined analysis of 55,521 Europeans. PLoS One. 2011;6 doi: 10.1371/journal.pone.0015813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kung C.P., Liu Q., Murphy M.E. The codon 72 polymorphism of p53 influences cell fate following nutrient deprivation. Cancer Biol. Ther. 2017;18:484–491. doi: 10.1080/15384047.2017.1323595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Beckman G., Birgander R., Sjalander A., Saha N., Holmberg P.A., Kivela A., Beckman L. Is p53 polymorphism maintained by natural selection? Hum. Hered. 1994;44:266–270. doi: 10.1159/000154228. [DOI] [PubMed] [Google Scholar]
- 118.Lindstrom M.S., Jin A., Deisenroth C., White Wolf G., Zhang Y. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol. Cell. Biol. 2007;27:1056–1068. doi: 10.1128/MCB.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Macias E., Jin A., Deisenroth C., Bhat K., Mao H., Lindstrom M.S., Zhang Y. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell. 2010;18:231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu S., Kim T.H., Franklin D.A., Zhang Y. Protection against high-fat-diet-induced obesity in MDM2(C305F) mice due to reduced p53 activity and enhanced energy expenditure. Cell Rep. 2017;18:1005–1018. doi: 10.1016/j.celrep.2016.12.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Derdak Z., Villegas K.A., Harb R., Wu A.M., Sousa A., Wands J.R. Inhibition of p53 attenuates steatosis and liver injury in a mouse model of non-alcoholic fatty liver disease. J. Hepatol. 2013;58:785–791. doi: 10.1016/j.jhep.2012.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Porteiro B., Diaz-Ruiz A., Martinez G., Senra A., Vidal A., Serrano M., Gualillo O., Lopez M., Malagon M.M., Dieguez C., Nogueiras R. Ghrelin requires p53 to stimulate lipid storage in fat and liver. Endocrinology. 2013;154:3671–3679. doi: 10.1210/en.2013-1176. [DOI] [PubMed] [Google Scholar]
- 123.Yokoyama M., Okada S., Nakagomi A., Moriya J., Shimizu I., Nojima A., Yoshida Y., Ichimiya H., Kamimura N., Kobayashi Y., Ohta S., Fruttiger M., Lozano G., Minamino T. Inhibition of endothelial p53 improves metabolic abnormalities related to dietary obesity. Cell Rep. 2014;7:1691–1703. doi: 10.1016/j.celrep.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 124.Al-Massadi O., Porteiro B., Kuhlow D., Kohler M., Gonzalez-Rellan M.J., Garcia-Lavandeira M., Diaz-Rodriguez E., Quinones M., Senra A., Alvarez C.V., Lopez M., Dieguez C., Schulz T.J., Nogueiras R. Pharmacological and genetic manipulation of p53 in brown fat at adult but not embryonic stages regulates thermogenesis and body weight in male mice. Endocrinology. 2016;157:2735–2749. doi: 10.1210/en.2016-1209. [DOI] [PubMed] [Google Scholar]
- 125.Matoba S., Kang J.G., Patino W.D., Wragg A., Boehm M., Gavrilova O., Hurley P.J., Bunz F., Hwang P.M. p53 regulates mitochondrial respiration. Science (New York, N.Y.) 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 126.Park J.Y., Wang P.Y., Matsumoto T., Sung H.J., Ma W., Choi J.W., Anderson S.A., Leary S.C., Balaban R.S., Kang J.G., Hwang P.M. p53 improves aerobic exercise capacity and augments skeletal muscle mitochondrial DNA content. Circ. Res. 2009;105:705–712. doi: 10.1161/CIRCRESAHA.109.205310. (711 p following 712) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Molchadsky A., Ezra O., Amendola P.G., Krantz D., Kogan-Sakin I., Buganim Y., Rivlin N., Goldfinger N., Folgiero V., Falcioni R., Sarig R., Rotter V. p53 is required for brown adipogenic differentiation and has a protective role against diet-induced obesity. Cell Death Differ. 2013;20:774–783. doi: 10.1038/cdd.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang X., Zhao X., Gao X., Mei Y., Wu M. A new role of p53 in regulating lipid metabolism. J. Mol. Cell Biol. 2013;5:147–150. doi: 10.1093/jmcb/mjs064. [DOI] [PubMed] [Google Scholar]
- 129.Wrede C.E., Dickson L.M., Lingohr M.K., Briaud I., Rhodes C.J. Protein kinase B/Akt prevents fatty acid-induced apoptosis in pancreatic beta-cells (INS-1) J. Biol. Chem. 2002;277:49676–49684. doi: 10.1074/jbc.M208756200. [DOI] [PubMed] [Google Scholar]
- 130.Lovis P., Roggli E., Laybutt D.R., Gattesco S., Yang J.Y., Widmann C., Abderrahmani A., Regazzi R. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes. 2008;57:2728–2736. doi: 10.2337/db07-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tornovsky-Babeay S., Dadon D., Ziv O., Tzipilevich E., Kadosh T., Schyr-Ben Haroush R., Hija A., Stolovich-Rain M., Furth-Lavi J., Granot Z., Porat S., Philipson L.H., Herold K.C., Bhatti T.R., Stanley C., Ashcroft F.M., In'T Veld P., Saada A., Magnuson M.A., Glaser B., Dor Y. Type 2 diabetes and congenital hyperinsulinism cause DNA double-strand breaks and p53 activity in beta cells. Cell Metab. 2014;19:109–121. doi: 10.1016/j.cmet.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 132.Belgardt B.F., Ahmed K., Spranger M., Latreille M., Denzler R., Kondratiuk N., von Meyenn F., Villena F.N., Herrmanns K., Bosco D., Kerr-Conte J., Pattou F., Rulicke T., Stoffel M. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat. Med. 2015;21:619–627. doi: 10.1038/nm.3862. [DOI] [PubMed] [Google Scholar]
- 133.Hinault C., Kawamori D., Liew C.W., Maier B., Hu J., Keller S.R., Mirmira R.G., Scrable H., Kulkarni R.N. Delta40 Isoform of p53 controls beta-cell proliferation and glucose homeostasis in mice. Diabetes. 2011;60:1210–1222. doi: 10.2337/db09-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kon N., Zhong J., Qiang L., Accili D., Gu W. Inactivation of arf-bp1 induces p53 activation and diabetic phenotypes in mice. J. Biol. Chem. 2012;287:5102–5111. doi: 10.1074/jbc.M111.322867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tavana O., Puebla-Osorio N., Sang M., Zhu C. Absence of p53-dependent apoptosis combined with nonhomologous end-joining deficiency leads to a severe diabetic phenotype in mice. Diabetes. 2010;59:135–142. doi: 10.2337/db09-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li X., Cheng K.K., Liu Z., Yang J.K., Wang B., Jiang X., Zhou Y., Hallenborg P., Hoo R.L., Lam K.S., Ikeda Y., Gao X., Xu A. The MDM2-p53-pyruvate carboxylase signalling axis couples mitochondrial metabolism to glucose-stimulated insulin secretion in pancreatic beta-cells. Nat. Commun. 2016;7:11740. doi: 10.1038/ncomms11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mihailidou C., Chatzistamou I., Papavassiliou A.G., Kiaris H. Regulation of P21 during diabetes-associated stress of the endoplasmic reticulum. Endocr. Relat. Cancer. 2015;22:217–228. doi: 10.1530/ERC-15-0018. [DOI] [PubMed] [Google Scholar]
- 138.Yang J., Zhang W., Jiang W., Sun X., Han Y., Ding M., Shi Y., Deng H. P21cip-overexpression in the mouse beta cells leads to the improved recovery from streptozotocin-induced diabetes. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Secchiero P., Toffoli B., Melloni E., Agnoletto C., Monasta L., Zauli G. The MDM2 inhibitor Nutlin-3 attenuates streptozotocin-induced diabetes mellitus and increases serum level of IL-12p40. Acta Diabetol. 2013;50:899–906. doi: 10.1007/s00592-013-0476-8. [DOI] [PubMed] [Google Scholar]
- 140.Hoshino A., Ariyoshi M., Okawa Y., Kaimoto S., Uchihashi M., Fukai K., Iwai-Kanai E., Ikeda K., Ueyama T., Ogata T., Matoba S. Inhibition of p53 preserves Parkin-mediated mitophagy and pancreatic beta-cell function in diabetes. Proc. Natl. Acad. Sci. U. S. A. 2014;111:3116–3121. doi: 10.1073/pnas.1318951111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shimizu I., Yoshida Y., Katsuno T., Tateno K., Okada S., Moriya J., Yokoyama M., Nojima A., Ito T., Zechner R., Komuro I., Kobayashi Y., Minamino T. p53-induced adipose tissue inflammation is critically involved in the development of insulin resistance in heart failure. Cell Metab. 2012;15:51–64. doi: 10.1016/j.cmet.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 142.Shimizu I., Yoshida Y., Moriya J., Nojima A., Uemura A., Kobayashi Y., Minamino T. Semaphorin3E-induced inflammation contributes to insulin resistance in dietary obesity. Cell Metab. 2013;18:491–504. doi: 10.1016/j.cmet.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 143.Vergoni B., Cornejo P.J., Gilleron J., Djedaini M., Ceppo F., Jacquel A., Bouget G., Ginet C., Gonzalez T., Maillet J., Dhennin V., Verbanck M., Auberger P., Froguel P., Tanti J.F., Cormont M. DNA damage and the activation of the p53 pathway mediate alterations in metabolic and secretory functions of adipocytes. Diabetes. 2016;65:3062–3074. doi: 10.2337/db16-0014. [DOI] [PubMed] [Google Scholar]
- 144.Inoue N., Yahagi N., Yamamoto T., Ishikawa M., Watanabe K., Matsuzaka T., Nakagawa Y., Takeuchi Y., Kobayashi K., Takahashi A., Suzuki H., Hasty A.H., Toyoshima H., Yamada N., Shimano H. Cyclin-dependent kinase inhibitor, p21WAF1/CIP1, is involved in adipocyte differentiation and hypertrophy, linking to obesity, and insulin resistance. J. Biol. Chem. 2008;283:21220–21229. doi: 10.1074/jbc.M801824200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hallenborg P., Fjaere E., Liaset B., Petersen R.K., Murano I., Sonne S.B., Falkerslev M., Winther S., Jensen B.A., Ma T., Hansen J.B., Cinti S., Blagoev B., Madsen L., Kristiansen K. p53 regulates expression of uncoupling protein 1 through binding and repression of PPARgamma coactivator-1alpha. Am. J. Physiol. Endocrinol. Metab. 2016;310:E116–E128. doi: 10.1152/ajpendo.00119.2015. [DOI] [PubMed] [Google Scholar]
- 146.Lieber C.S., Leo M.A., Wang X., Decarli L.M. Alcohol alters hepatic FoxO1, p53, and mitochondrial SIRT5 deacetylation function. Biochem. Biophys. Res. Commun. 2008;373:246–252. doi: 10.1016/j.bbrc.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 147.Otsuka M., Kato N., Lan K., Yoshida H., Kato J., Goto T., Shiratori Y., Omata M. Hepatitis C virus core protein enhances p53 function through augmentation of DNA binding affinity and transcriptional ability. J. Biol. Chem. 2000;275:34122–34130. doi: 10.1074/jbc.M000578200. [DOI] [PubMed] [Google Scholar]
- 148.Akyol G., Dursun A., Poyraz A., Uluoglu O., Ataoglu O., Edaly N., Memis L. P53 and proliferating cell nuclear antigen (PCNA) expression in non-tumoral liver diseases. Pathol. Int. 1999;49:214–221. doi: 10.1046/j.1440-1827.1999.00849.x. [DOI] [PubMed] [Google Scholar]
- 149.Attallah A.M., Shiha G.E., Ismail H., Mansy S.E., El-Sherbiny R., El-Dosoky I. Expression of p53 protein in liver and sera of patients with liver fibrosis, liver cirrhosis or hepatocellular carcinoma associated with chronic HCV infection. Clin. Biochem. 2009;42:455–461. doi: 10.1016/j.clinbiochem.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 150.Lin X., Yue P., Chen Z., Schonfeld G. Hepatic triglyceride contents are genetically determined in mice: results of a strain survey. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;288:G1179–G1189. doi: 10.1152/ajpgi.00411.2004. [DOI] [PubMed] [Google Scholar]
- 151.Kodama T., Takehara T., Hikita H., Shimizu S., Shigekawa M., Tsunematsu H., Li W., Miyagi T., Hosui A., Tatsumi T., Ishida H., Kanto T., Hiramatsu N., Kubota S., Takigawa M., Tomimaru Y., Tomokuni A., Nagano H., Doki Y., Mori M., Hayashi N. Increases in p53 expression induce CTGF synthesis by mouse and human hepatocytes and result in liver fibrosis in mice. J. Clin. Invest. 2011;121:3343–3356. doi: 10.1172/JCI44957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tomita K., Teratani T., Suzuki T., Oshikawa T., Yokoyama H., Shimamura K., Nishiyama K., Mataki N., Irie R., Minamino T., Okada Y., Kurihara C., Ebinuma H., Saito H., Shimizu I., Yoshida Y., Hokari R., Sugiyama K., Hatsuse K., Yamamoto J., Kanai T., Miura S., Hibi T. p53/p66Shc-mediated signaling contributes to the progression of non-alcoholic steatohepatitis in humans and mice. J. Hepatol. 2012;57:837–843. doi: 10.1016/j.jhep.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 153.Porteiro B., Fondevila M.F., Delgado T.C., Iglesias C., Imbernon M., Iruzubieta P., Crespo J., Zabala-Letona A., Ferno J., Gonzalez-Teran B., Matesanz N., Hernandez-Cosido L., Marcos M., Tovar S., Vidal A., Sanchez-Ceinos J., Malagon M.M., Pombo C., Zalvide J., Carracedo A., Buque X., Dieguez C., Sabio G., Lopez M., Aspichueta P., Martinez-Chantar M.L., Nogueiras R. Hepatic p63 regulates steatosis via IKKbeta/ER stress. Nat. Commun. 2017;8:15111. doi: 10.1038/ncomms15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lujambio A., Akkari L., Simon J., Grace D., Tschaharganeh D.F., Bolden J.E., Zhao Z., Thapar V., Joyce J.A., Krizhanovsky V., Lowe S.W. Non-cell-autonomous tumor suppression by p53. Cell. 2013;153:449–460. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wu H., Ng R., Chen X., Steer C.J., Song G. MicroRNA-21 is a potential link between non-alcoholic fatty liver disease and hepatocellular carcinoma via modulation of the HBP1-p53-Srebp1c pathway. Gut. 2016;65:1850–1860. doi: 10.1136/gutjnl-2014-308430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu Y., He Y., Jin A., Tikunov A.P., Zhou L., Tollini L.A., Leslie P., Kim T.H., Li L.O., Coleman R.A., Gu Z., Chen Y.Q., Macdonald J.M., Graves L.M., Zhang Y. Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2414–E2422. doi: 10.1073/pnas.1315605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Armata H.L., Golebiowski D., Jung D.Y., Ko H.J., Kim J.K., Sluss H.K. Requirement of the ATM/p53 tumor suppressor pathway for glucose homeostasis. Mol. Cell. Biol. 2010;30:5787–5794. doi: 10.1128/MCB.00347-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang X., Duan W., Lee W.N., Zhang Y., Xiang F., Liu Q., Go V.L., Xiao G.G. Overexpression of p53 improves blood glucose control in an insulin resistant diabetic mouse model. Pancreas. 2016;45:1010–1017. doi: 10.1097/MPA.0000000000000637. [DOI] [PubMed] [Google Scholar]
- 159.Liao W., Liu H., Zhang Y., Jung J.H., Chen J., Su X., Kim Y.C., Flores E.R., Wang S.M., Czarny-Ratajczak M., Li W., Zeng S.X., Lu H. Ccdc3: a new P63 target involved in regulation of liver lipid metabolism. Sci. Rep. 2017;7:9020. doi: 10.1038/s41598-017-09228-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Su X., Gi Y.J., Chakravarti D., Chan I.L., Zhang A., Xia X., Tsai K.Y., Flores E.R. TAp63 is a master transcriptional regulator of lipid and glucose metabolism. Cell Metab. 2012;16:511–525. doi: 10.1016/j.cmet.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dibra D., Xia X., Mitra A., Cutrera J.J., Lozano G., Li S. Mutant p53 in concert with an interleukin-27 receptor alpha deficiency causes spontaneous liver inflammation, fibrosis, and steatosis in mice. Hepatology. 2016;63:1000–1012. doi: 10.1002/hep.28379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Porteiro B., Fondevila M.F., Buque X., Gonzalez-Rellan M.J., Fernandez U., Mora A., Beiroa D., Senra A., Gallego R., Ferno J., Lopez M., Sabio G., Dieguez C., Aspichueta P., Nogueiras R. Pharmacological stimulation of p53 with low-dose doxorubicin ameliorates diet-induced nonalcoholic steatosis and steatohepatitis. Mol. Metab. 2018;8:132–143. doi: 10.1016/j.molmet.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wu D., Prives C. Relevance of the p53-MDM2 axis to aging. Cell Death Differ. 2018;25:169–179. doi: 10.1038/cdd.2017.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ringshausen I., O'Shea C.C., Finch A.J., Swigart L.B., Evan G.I. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell. 2006;10:501–514. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 165.Tyner S.D., Venkatachalam S., Choi J., Ghebranious N., Igelmann H., Lu X., Soron G., Cooper B., Brayton C., Park S.H., Thompson T., Karsenty G., Bradley A., Donehower L.A. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 166.Maier B., Gluba W., Bernier B., Turner T., Mohammad K., Guise T., Sutherland A., Thorner M., Scable H. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lessel D., Wu D., Trujillo C., Ramezani T., Lessel I., Alwasiyah M.K., Saha B., Hisama F.M., Rading K., Goebel I., Schutz P., Speit G., Hogel J., Thiele H., Nurnberg G., Nurnberg P., Hammerschmidt M., Zhu Y., Tong D.R., Katz C., Martin G.M., Oshima J., Prives C., Kubisch C. Dysfunction of the MDM2/p53 axis is linked to premature aging. J. Clin. Invest. 2017;127:3598–3608. doi: 10.1172/JCI92171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Garcia-Cao I., Garcia-Cao M., Martin-Caballero J., Criado L.M., Klatt P., Flores J.M., Weill J.C., Blasco M.A., Serrano M. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21:6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.von Muhlinen N., Horikawa I., Alam F., Isogaya K., Lissa D., Vojtesek B., Lane D.P., Harris C.C. p53 isoforms regulate premature aging in human cells. Oncogene. 2018;37:2379–2393. doi: 10.1038/s41388-017-0101-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dumble M., Moore L., Chambers S.M., Geiger H., Van Zant G., Goodell M.A., Donehower L.A. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood. 2007;109:1736–1742. doi: 10.1182/blood-2006-03-010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 172.Zhao T., Xu Y. p53 and stem cells: new developments and new concerns. Trends Cell Biol. 2010;20:170–175. doi: 10.1016/j.tcb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 173.Matheu A., Maraver A., Klatt P., Flores I., Garcia-Cao I., Borras C., Flores J.M., Vina J., Blasco M.A., Serrano M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 174.Carrasco-Garcia E., Arrizabalaga O., Serrano M., Lovell-Badge R., Matheu A. Increased gene dosage of Ink4/Arf and p53 delays age-associated central nervous system functional decline. Aging Cell. 2015;14:710–714. doi: 10.1111/acel.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Orsted D.D., Bojesen S.E., Tybjaerg-Hansen A., Nordestgaard B.G. Tumor suppressor p53 Arg72Pro polymorphism and longevity, cancer survival, and risk of cancer in the general population. J. Exp. Med. 2007;204:1295–1301. doi: 10.1084/jem.20062476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.van Heemst D., Mooijaart S.P., Beekman M., Schreuder J., de Craen A.J., Brandt B.W., Slagboom P.E., Westendorp R.G. Variation in the human TP53 gene affects old age survival and cancer mortality. Exp. Gerontol. 2005;40:11–15. doi: 10.1016/j.exger.2004.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.