Abstract
Helicobacter pylori (H. pylori) is an organism that is widespread in the human population and is sometimes responsible for some of the most common chronic clinical disorders of the upper gastrointestinal tract in humans, such as chronic-active gastritis, duodenal and gastric ulcer disease, low-grade B-cell mucosa associated lymphoid tissue lymphoma of the stomach, and gastric adenocarcinoma, which is the third leading cause of cancer death worldwide. The routes of infection have not yet been firmly established, and different routes of transmission have been suggested, although the most commonly accepted hypothesis is that infection takes place through the faecal-oral route and that contaminated water and foods might play an important role in transmission of the microorganism to humans. Furthermore, several authors have considered H. pylori to be a foodborne pathogen because of some of its microbiological and epidemiological characteristics. H. pylori has been detected in drinking water, seawater, vegetables and foods of animal origin. H. pylori survives in complex foodstuffs such as milk, vegetables and ready-to-eat foods. This review article presents an overview of the present knowledge on the microbiological aspects in terms of phenotypic characteristics and growth requirements of H. pylori, focusing on the potential role that foodstuffs and water may play in the transmission of the pathogen to humans and the methods successfully used for the detection of this microorganism in foodstuffs and water.
Keywords: Helicobacter pylori, Viable but nonculturable state, Foodborne pathogen, Food, Water, Animal reservoirs, Culture methods, Molecular methods
Core tip: To date, the transmission routes and reservoirs of Helicobacter pylori (H. pylori) are topics of debate. Epidemiological evidence and the occurrence of H. pylori in foods of animal origin, vegetables and water corroborate the hypothesis advanced by numerous authors that H. pylori may be a foodborne pathogen. The present review is focused on the evidence supporting the role of foods and water in the transmission of H. pylori to humans and on the methods for detecting the pathogen in foodstuffs and water.
INTRODUCTION
The first isolation of Helicobacter pylori (H. pylori) in 1982 by Marshall and Warren[1,2] marked a turning point in understanding gastrointestinal microbial ecology and disease[3]. Following the initial scepticism regarding the aetiologic importance of this organism, it is now recognized that infections with H. pylori are linked to some of the most common chronic clinical disorders of the upper gastrointestinal tract in humans[4]. In fact, H. pylori has been acknowledged as a major cause of chronic-active gastritis and is associated with duodenal and gastric ulcer disease, low-grade B-cell mucosa-associated lymphoid tissue lymphoma of the stomach (MALToma)[5], and gastric adenocarcinoma, which is the third leading cause of cancer death worldwide[4,6,7]. Furthermore, H. pylori has been linked to a variety of extra-gastric disorders, including coronary heart disease, dermatological disorders such as rosacea and idiopathic urticaria, autoimmune thyroid disease, thrombocytopenic purpura, and iron deficiency anaemia[8].
Human infection by H. pylori is a great public health hazard because H. pylori colonizes the gastric mucosa of approximately half of the world’s population[9-12]. The infection is usually acquired in infancy and early childhood, and it is long lasting, often remaining for the entire lifespan[13]. The prevalence of H. pylori shows large geographical variation, with infection rates much higher in developing countries (in some areas > 85%) than in Europe and North America (approximately 30%-40%)[14,15]. In various developing countries, more than 80% of the population is H. pylori positive, even at young ages[16]. The prevalence of H. pylori in industrialized countries generally remains under 40% and is considerably lower in children and adolescents than in adults and elderly people. A comparison of prevalence rates by age suggests that the acquisition of H. pylori is decreasing in recent cohorts, and this finding is most apparent in developed countries and may be linked to improvements in hygiene practices. Furthermore, it has been estimated that between two and 20 percent of people infected with H. pylori develop peptic ulcer disease[17].
Although H. pylori can cause severe illnesses with a high rate of morbidity and mortality, the complex interactions between this microbe and humans, particularly its transmission pathways to humans and reservoirs, are largely unknown, although multiple routes of transmission have been suggested[3,18-20]. The current literature suggests that the transmission of H. pylori occurs from person to person via the oral-oral, faecal-oral, and gastric-oral routes and that the infection dose for humans is low[6,21]. H. pylori may be a sex-transmitted pathogen[22,23] and may lead to fibrocystic breast changes[24]. The oral cavity can be primarily colonized by H. pylori, and this can be linked to later gastric infection[25]. Faecal-oral transmission has more important implications than oral-oral transmission because H. pylori may occur in food and water supplies subsequent to faecal contamination[26]. Furthermore, several authors have considered H. pylori to be a foodborne pathogen because of its microbiological and epidemiological characteristics[6,10,27-31]. Information on the distribution of H. pylori in water, vegetables and foods of animal origin is critical in determining its potential transmission in foods.
This review article presents a brief overview of the present knowledge on the microbiological characteristics of H. pylori in terms of its phenotypic characteristics and growth requirements, focusing on the potential role that foodstuffs and water may play in the transmission of this pathogen to humans and the methods for isolating and detecting this microorganism in foodstuffs and water.
LITERATURE SEARCH
A PubMed search was conducted using the following keywords and phrases: “Helicobacter pylori, Helicobacter pylori and food, Helicobacter pylori and milk, Helicobacter pylori and water, VBNC, survival of Helicobacter pylori”. In addition, we performed a manual review of the reference lists of the primary and review articles to ensure identification of all relevant articles.
MICROBIOLOGICAL CHARACTERISTICS
Phenotypic characteristics
H. pylori was originally thought to be a species belonging to the genus Campylobacter and was first named Campylobacter pyloridis, which was later corrected to Campylobacter pylori (C. pylori)[32]. Because subsequent 16S rRNA sequence analysis showed that the distance between the species belonging to the genus Campylobacter and C. pylori was sufficient to exclude C. pylori from this genus[33], it was renamed Helicobacter pylori[34], the first member of the new genus Helicobacter.
H. pylori organisms are spiral or curved bacilli ranging from 0.3 to 1.0 μm in width and 1.5 to 10.0 μm in length; they are gram-negative and assume a rod-like shape when cultured on solid medium[34]. Furthermore, after prolonged in vitro culture and under adverse environmental conditions, such as an insufficient supply of nutrients, desiccation, lack of protection against oxygen, and exposure to antimicrobial agents, H. pylori can survive entering the viable but non-culturable (VBNC) state, changing its rod-like shape to a coccoid shape[35-37].
When this morphological change occurs, H. pylori is unable to grow on agar plates using conventional cultivation methods[38-40].
Bacteria in the VBNC state maintain their metabolic activity, pathogenicity and ability to return to active regrowth conditions[41,42]. For H. pylori, the ability to return to active regrowth conditions has not yet been proven. Nevertheless, the aptitude of H. pylori to overcome stressed conditions is very significant for public health[29], even if the role of VBNC in the transmission of H. pylori, especially by food and water, is still controversial.
H. pylori is motile and usually possesses four to six unipolar-sheathed flagella, which may be an adaptation to survive in gastric juices[3,43].
Growth requirements
Since the discovery of H. pylori, bacterial culture has been used as a routine diagnostic test and is considered the gold standard. H. pylori culture is recommended for performing antibiotic susceptibility testing if primary resistance to clarithromycin is higher than 20% or after failure of second-line treatment[44]. Despite the long use of bacterial culture, to date, there are no defined media for the selective culture of H. pylori because of its fastidious nature with particular growth requirements of atmosphere, nutrient-rich media, high humidity (98%), and long incubation time (5-7 d)[3,44].
H. pylori is a capnophilic organism that requires an atmosphere with a high level of CO2 (from 5% to 10%). It has been considered a microaerophile, but the concentration of O2 required for its growth is still a topic of discussion[44].
H. pylori requires a complex culture substrate (solid or liquid) with some forms of supplementation, such as whole sheep or horse blood, haemoglobin, serum, coal, yeast, or yolk emulsion[45,46], which may serve as nutritional substrates. These supplements also detoxify the medium and protect the microorganism[7].
Furthermore, if isolation is attempted from samples with basic microbial flora, it is necessary to make the media selective through supplementation of several antibiotics[47].
Growth in liquid media is enhanced by agitation, which allows gas dispersion and incubation in a CO2-rich atmosphere[31,47].
H. pylori grows within a temperature range of 30 °C to 37 °C, with optimum growth at 37 °C, but is not able to grow at 25 °C[43]. At 42 °C, growth is variable[29].
Similar to C. jejuni, H. pylori survives longer at 4 °C than at room temperature, and it grows within a pH range of 4.5 to 7.3, with optimum at pH 5.5. H. pylori grows well in the presence of 0.5% and 1% NaCl but not of 2% NaCl. The minimum water activity (aw) for growth is between 0.96 and 0.98. These data suggest that this microorganism is most likely not able to grow in many types of food[47].
H. pylori is catalase and oxidase positive; it is also characterized by strong urease activity and is negative for hippurate and nitrate reduction, characteristics that discriminate it from species belonging to the genus Campylobacter[29].
EVIDENCE SUPPORTING THE ROLE OF FOODS IN TRANSMISSION OF H. PYLORI TO HUMANS
Since 1997, when the transmission of H. pylori through water and foods was hypothesized for the first time, several studies have evaluated the survival and the presence of this microorganism in different foodstuffs (Tables 1 and 2).
Table 1.
Studies evaluating the survival of Helicobacter pylori in artificially contaminated foods
| Year | Food | Method | Observations | Ref. |
| 1998 | Sterilized milk | Bacterial count on chocolate agar | 10 d at 4 °C | Fan et al[54] |
| 3 d at 25 °C | ||||
| 2001 | Pasteurized milk water tofu, tofu, yogurt, lettuce and chicken | Bacterial count on tryptic soy agar, non-selective Wilkins-Chalgren Anaerobe blood agar and selective Wilkins-Chalgren Anaerobe blood agar | from 5 to 7 d in pasteurized milk, tofu and water tofu at 4 °C | Poms et al[26] |
| for up to 2 d in lettuce and raw chicken at 4 °C | ||||
| for up to 1 d in yogurt at 4 °C | ||||
| 2000 | Ground beef packaged in vacuum and air | Bacterial count on H. pylori special peptone agar | 6 d in ground beef packaged in air at 4 °C | Stevenson et al[55] |
| 3-6 d in ground beef packaged in vacuum at 4 °C | ||||
| 3 d in ground beef packaged in air and in vacuum at -18 °C | ||||
| 2002 | Ground beef, sterile milk, and apple and orange juices | Bacterial count on brain heart infusion agar and horse serum | 7 d in ground beef at 4 °C | Jiang et al[56] |
| 11 d in irradiated ground beef at 4 °C | ||||
| 6 d in sterile milk at 4 °C | ||||
| 1 d in apple and orange juice at 4 °C and 25 °C | ||||
| 2004 | Lettuce and carrots | Bacterial count on Helicobacter special peptone agar and Columbia blood agar | 3 d in lettuce at 8 °C | Gomes et al[57] |
| 5 d in sterilized carrot at 8 °C | ||||
| 3 d in sanitized carrot at 8 °C | ||||
| 2007 | Sterile milk and pasteurized milk | Bacterial count on Wilkins-Chalgren anaerobe agar | 12 d in sterile milk at 4 °C | Quaglia et al[53] |
| 9 d in pasteurized milk at 4 °C | ||||
| 2010 | Spinach | Bacterial count on brucella blood agar, Wilkins-Chalgren anaerobe blood agar | 6 d at 8 °C | Buck et al[58] |
| 2011 | Traditional Turkish fermented sausage (sucuk) | Bacterial count on Wilkins-Chalgren anaerobe blood agar | 7 d | Guner et al[59] |
| 2017 | Spring onion, cabbage, lettuce and spinach | Bacterial count on non-selective Blood base agar with 5% horse blood | 3 d in spring onion, lettuce and spinach | Ng et al[60] |
| 4 d in cabbage stored at 4 °C |
Table 2.
Studies evaluating the occurrence of Helicobacter pylori in foods
| Year | Food | Method | Number of samples n | Observations | Ref. |
| 2001 | Raw sheep milk | Culture and PCR | 63 raw sheep milk | 60% PCR positive samples | Dore et al[65] |
| 2.6% culture positive samples | |||||
| 2002 | Raw and pasteurized cow milk | Semi nested PCR, culture method and electron microscopy | 18 raw cow milk | Raw milk: 72.2% semi-nested PCR positive samples; 1 culture positive sample | Fujimura et al[66] |
| 20 pasteurized milk | Pasteurized milk: 55% semi-nested PCR positive samples | ||||
| 2002 | Raw sheep milk | Culture | 440 raw sheep milk | 0% positive samples | Turutoglu et al[74] |
| 2008 | Raw goat, sheep, and cow milks | Nested-PCR | 160 raw goat milk | 25.6% positive goat milk | Quaglia et al[71] |
| 130 raw sheep milk | 33% positive sheep milk | ||||
| 110 raw cow milk | 50% positive cow milk | ||||
| 2008 | Raw chicken and ready- to- eat raw tuna | Multiplex PCR | 11 raw chicken | 36% positive raw chicken | Meng et al[77] |
| 18 ready-to-eat raw tuna | 44% positive ready-to-eat raw tuna | ||||
| 2011 | Raw cow milk | FISH | 20 | 20% positive samples | Angelidis et al[67] |
| 2012 | Raw cow, sheep, goat, buffalo and camel milks | PCR | 75 raw cow milk | 16.00% positive cow milk | Rahimi et al[68] |
| 58 raw sheep milk | 13.79% positive sheep milk | ||||
| 42 raw goat milk | 4.76% positive goat milk | ||||
| 20 raw buffalo milk | 13.33% positive camel milk | ||||
| 15 raw camel milk | 20.00% positive buffalo milk | ||||
| 2014 | Milk and traditional dairy products | Culture and PCR | 120 raw cow milk | 16.6% positive cow milk | Mousavi et al[69] |
| 100 raw goat milk | 28% positive goat milk | ||||
| 100 raw sheep milk | 35% positive sheep milk | ||||
| 80 raw buffalo milk | 15% positive buffalo milk | ||||
| 60 raw camel milk | 13.3% positive camel milk | ||||
| 60 raw donkey milk | 0% positive donkey milk | ||||
| 100 cheese | 30% positive cheese | ||||
| 100 butter | 15% positive cream | ||||
| 100 cream | 5% positive butter | ||||
| 100 ice cream | 27% positive ice cream | ||||
| 2014 | Vegetables and salad | Culture and PCR | 60 salad | 16.6% positive salad | Atapoor et al[81] |
| 40 basil | 12.5% positive basil | ||||
| 40 radish | 7.5% positive radish | ||||
| 40 leek | 20% positive leek | ||||
| 80 spinach | 6.25% positive spinach | ||||
| 80 lettuce | 13.75% positive lettuce | ||||
| 120 parsley | 6.6% positive parsley | ||||
| 2014 | Washed and unwashed vegetables | Culture and PCR | 430 washed and unwashed vegetable | 13.72% positive vegetables and salads | Yahaghi et al[80] |
| 2015 | Raw cow, sheep, goat, buffalo and camel milks | PCR | 75 raw cow milk | 16.00% positive cow milk | Talaei et al[72] |
| 58 raw sheep milk | 13.79% positive sheep milk | ||||
| 42 raw goat milk | 4.76% positive goat milk | ||||
| 20 raw buffalo milk | 13.33% positive camel milk | ||||
| 15 raw camel milk | 20.00% positive buffalo milk | ||||
| 2015 | Raw cow milk | Culture and nested PCR | 50 raw cow milk | 22% positive cow milk | Osman et al[73] |
| 2015 | Raw cow milk | Culture and nested PCR | 163 raw cow milk | 0% positive cow milk | Bianchini et al[75] |
| 2016 | Raw cow, sheep, goat, buffalo and camel milks and meats | Culture and PCR | 420 raw milk | 21.90% positive raw milk | Saedi et al[70] |
| 400 raw meat | 26.25% positive meat | ||||
| 2016 | Ready-to-eat food | Culture and PCR | 550 ready-to-eat food | 13.45% positive ready-to-eat food | Hemmatinezhad et al[78] |
| 2016 | Ready-to-eat food and minced meat | Culture and PCR | 60 ready-to-eat fish | 15% positive ready-to-eat fish | Ghorbani et al[79] |
| 60 ham | 8.33% positive ham | ||||
| 40 chicken sandwich | 5% positive chicken sandwich | ||||
| 40 vegetable sandwich | 45% positive vegetable sandwich | ||||
| 50 meat sandwich | 20% positive meat sandwich | ||||
| 50 minced meat | 32% positive minced meat | ||||
| 2017 | Hamburger and minced meat | Culture and nested PCR | 80 hamburger | 1.42% positive hamburger | Gilani et al[76] |
| 70 minced-meat | 12.5% positive minced-meat |
One of the most important topics supporting this thesis was the phylogenetic proximity of H. pylori to C. jejuni, which led to the hypothesis that the transmission pathways described for the latter could also be applied to H. pylori[27]. Several investigators have also considered H. pylori a foodborne pathogen based on some of its epidemiological characteristics[27,28,30] such as the high prevalence of infection within closed family groups and among individuals living in institutions[48]. These aspects suggest that in addition to direct transmission, this bacterium may be transmitted indirectly through a common source, such as through consumption of the “same foods at the same table”[26]. The finding that the prevalence of H. pylori infection is greater in geographical areas in which the hygienic conditions of life are poor also supports this hypothesis[30,49].
Additional indirect evidence of the transmission of H. pylori to humans through foods of animal origin has been provided by epidemiological studies on the presence of antibodies in slaughterhouse workers and in veterinary workers. The incidence rates in these workers were positive and were greater than those in workers who had no direct contact with carcasses[50,51] (Figure 1).
Figure 1.
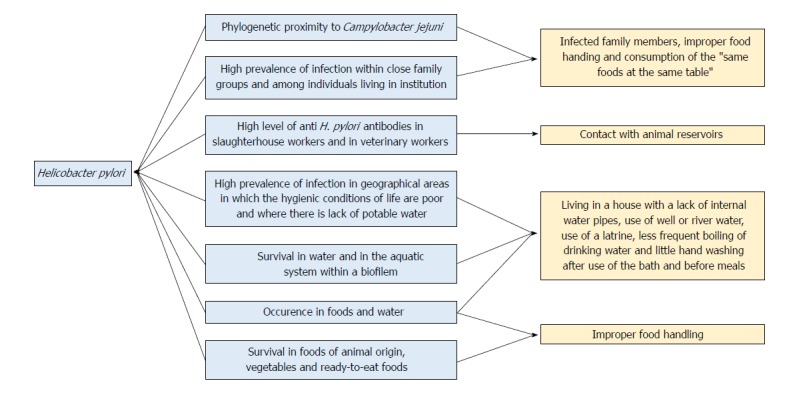
Evidence supporting the role of foods and water in the transmission of Helicobacter pylori to humans. In light blue is the epidemiological evidence supporting the hypothesis that H. pylori is a foodborne and/or a waterborne pathogen; in orange are the risks factors for H. pylori infection. H. pylori: Helicobacter pylori.
Foods presenting intrinsic factors, such as aw higher than 0.97 and pH ranging from 4.9 to 6.0, could theoretically provide conditions for H. pylori survival[30,52].
Therefore, data on survival ability may be more important than concerns about the growth of the microorganism in foods when determining the role of foods in H. pylori transmission to humans[53].
Survival of H. pylori in foodstuffs
Several studies have demonstrated the survival of H. pylori in water, milk, ready-to-eat foods, vegetables, pasteurized apple and orange juices, ground beef and dry fermented sausages[26,53-60] (Table 1).
H. pylori is able to survive in artificially contaminated milk stored at 4 °C for several days (from 5 to 9 d in pasteurized milk and from approximately 6 to 12 d in sterile milk)[26,53,54,56]. These findings corroborate the hypothesis that post-processed contaminated milk may play a more effective role than other foods in the transmission of H. pylori infection due to the intrinsic characteristics of this organism[53]. It is well known that H. pylori’s ability to survive in an acidic pH environment is urea dependent[3], and because urea is present in milk[61], the urea-dependent acid resistance of H. pylori may account for the long-term survival of H. pylori in this foodstuff[47]. Moreover, the microorganism is able to survive in milk for longer than the best-before date on an open milk package, making milk a possible source of transmission of this microorganism to humans. In fact, although the H. pylori load contaminating milk under natural conditions is unknown (although it is presumably lower than that used in vitro), the infection dose for humans is low; thus, even a small number of H. pylori cells surviving in foods may represent a potential health hazard for consumers[53].
Other studies have been conducted on the survival of microorganisms in other more complex foodstuffs. H. pylori survives for approximately 7 d in ground beef at 4 °C, up to 3 d at -18 °C[55,56] and for only 2 d in prepacked boneless, skinless chicken thighs[26]. Vacuum packaging has no impact on survival time. However, if the high level of background bacteria present in the ground beef is eliminated, survival time increases to an undetectable level (< 10 cfu/g) within 11 d[55]. The fate of H. pylori during the fermentation process of a traditional Turkish fermented sausage (sucuk) was investigated. The results of this study showed that the microorganism could survive and grow during the fermentation process of sucuk (22 °C for 7 d). A possible explanation is that some fermentation products, such as protein degradation compounds and CO2, might have been used by this pathogen and that indigenous bacteria might have created a microenvironment suitable for H. pylori growth[59]. In contrast, H. pylori is not able to survive in yogurt[26] or pasteurized fruit juice[56] because its growth is hampered by the acidic pH and organic acids from lactic acid bacteria growth[62,63].
Survival time in vegetables is shorter: 3 d in sanitized lettuce and carrot stored at 8 °C, 4 d in sterilized carrot and 5 d in carrot packaged in a modified atmosphere[57]. A possible explanation could be the not-robust nature of this bacterium that on the surface of vegetables is exposed to oxygen and desiccation as opposed to what happens in liquid food and the presence of a high load of natural bacterial flora. Moreover, H. pylori is able to survive in contaminated vegetables despite the abovementioned adverse conditions, as it is able to form biofilms[60]. However, a study on the survival of H. pylori in artificially contaminated spinach showed that this bacterium is able to survive for up to 6 d in VBNC forms that are still viable and can maintain its virulence factors despite its lack of cultivability[58].
Occurrence of H. pylori in foodstuffs
Based on these findings, several studies have attempted to prove the occurrence of H. pylori in foodstuffs (Table 2). To the best of our knowledge, the first report about the presence of H. pylori in sheep milk was prompted by the observation that Sardinian shepherds with direct animal contact had a higher prevalence of infection than did their same-household siblings[64]. H. pylori was isolated in 1 out of 38 PCR-positive raw sheep milk samples and in one out of 6 PCR-positive sheep gastric tissue samples[65].
After these findings, H. pylori has rarely been isolated from raw milk samples[66-69]. Bacteriological isolation of H. pylori occurred in one sample of raw cow milk out of 13 PCR-positive samples during a survey conducted in Japan[66]. It was also isolated in 4 samples of raw cow milk out of 20 samples analysed in Greece[67] and in Iran in 2 samples of raw sheep milk out of 11 PCR-positive samples and in 1 raw buffalo milk out of 15 PCR-positive samples[68]. Afterward, Mousavi et al[69] and Saedi et al[70] reported a higher prevalence of H. pylori in raw cow, sheep, goat, buffalo and camel milks in Iran than that previously mentioned.
Furthermore, only a few studies have been carried out on the occurrence of H. pylori in dairy products other than milk. For example, in the survey of Mousavi et al[69], 30% of Iranian traditional cheese, 15% of cream, 5% of butter and 27% of ice cream samples all made from unpasteurized milk were positive for H. pylori.
Compared to the few bacteriological isolations, the prevalence of H. pylori DNA is higher depending on the sensitivity of the method employed and the target gene[65,66,68,71-73]. Conversely, these findings were not confirmed in the studies conducted by Jiang and Doyle[56], Turutoglu et al[74] and Bianchini et al[75], which failed to detect H. pylori in cow and sheep raw milk in the United States, Turkey and Italy, respectively, by PCR and bacteriological analysis.
The attempts to culture H. pylori from the majority of PCR-positive samples may have been unsuccessful for several reasons: the low number of contaminating bacteria in milk samples, the presence of VBNC forms that are not detectable by conventional microbiological culture-based protocols, and the relatively long period of storage before analysis, which could have affected the vitality of the few H. pylori cells present in the contaminated samples[37,67,71].
Studies on the detection of H. pylori in food products other than milk are quite rare. H. pylori was isolated in 25%, 37%, 22%, 28% and 14% of cow, sheep, goat, buffalo and camel meat samples[70] and in 1.42% and 12.5% of hamburger and minced beef samples, respectively[76]. H. pylori DNA was detected in 36% and 44% of raw chicken and ready-to-eat raw tuna meat samples, respectively[77]. Furthermore, Hemmatinezhad et al[78] analysed 550 samples of ready-to-eat foods, detecting H. pylori in 74% of samples; olive salad (36%), restaurant salad (30%), fruit salad (28%) and soup (22%) were the most commonly contaminated. Additionally, Ghorbani et al[79] recovered H. pylori in 60 out of 300 ready-to-eat food samples (20%), including ready-to-eat fish (15%), ham (8.33%), chicken sandwiches (5%), vegetable sandwiches (18%), meat sandwiches (10%), and minced meat (32%).
Few reports have addressed the occurrence of H. pylori in vegetables. In two surveys conducted in Iran, many of the vegetables analysed were positive for the presence of this microorganism: 13.72%[80] and 9.56%[81] of the vegetables and traditional salads analysed.
The high prevalence of H. pylori in ready-to-eat foods, meats, milks and vegetables could be due to post-processing contamination. In fact, the high prevalence of H. pylori in healthy human carriers[11] suggests that foodstuff contamination due to poor hygiene management during milking, chilling and storage and during the handling, preparation and packaging of ready-to-eat foods may occur. Furthermore, H. pylori strains isolated from foods showed genotypes of vacA alleles similar to those in isolates from human clinical samples, endorsing the hypothesis that foods can be the source of H. pylori transmission to humans[76,78-80].
However, the existence of animal reservoirs of the microorganism cannot be excluded[65,66]. In addition, the histopathology of lesions by H. pylori in humans differs from that of many other gastric helicobacters, causing mild or absent inflammatory responses in their natural hosts[82]. These data suggest that H. pylori may have not originally evolved as a human pathogen but was likely introduced into the human population from a mammalian reservoir sometime in the distant past[65]. This hypothesis is further supported by evidence of H. pylori in the gastric mucosa of calves, pigs and horses[83] and its isolation from sheep gastric tissue and milk[65,84]. Furthermore, in the studies of Papiez et al[85] and Dore et al[65], H. pylori prevalence was higher in shepherds with direct animal contact than in controls without contact with sheep. Considering the 100% positive 13C-urea breath test in sheep, it may be reasonable to assume that these animal species may act as reservoirs and spreaders of H. pylori[85,86]. However, further epidemiological and experimental studies are needed to corroborate these few data.
EVIDENCE SUPPORTING THE ROLE OF WATER IN THE TRANSMISSION OF H. PYLORI TO HUMANS
In the last Joint Monitoring Report (JMP) of 2017, “Progress on Drinking Water, Sanitation and Hygiene” by WHO and UNICEF[87], the first global assessment of safe drinking water and sanitation services was reported. In 2015, approximately 2.1 billion people did not manage water safely, and among them, 844 million did not even have basic drinking water services, spending more than 30 min per trip to collect water from external sources, and some of them still drank untreated water from surface water sources such as streams and lakes. Globally, at least 2 billion people use a stool-contaminated source of drinking water. Contaminated water can transmit diseases such as diarrhoea, cholera, dysentery, typhoid and polio[87].
It has been estimated that H. pylori colonizes more than half of the world’s population, and contaminated water is mentioned as one of major causes[60,88-91].
Bellack et al[92] developed a conceptual model of the role of water in H. pylori transmission. The hypothesis is that both humans and animals are long-term hosts and that water is a relatively short-term reservoir. H. pylori may survive in water for a period before it is ingested as drinking water, accidentally during bathing, or through other pathways involving food. The infected person will spread H. pylori through faeces; through direct faecal-oral transmission, an infected person can infect another person or contaminate water bodies through direct contamination with faeces or indirectly with wastewater that comes into contact with the water used to drink. Animal contamination of water reserves may occur by defecating directly in surface waters or by faeces penetrating groundwater. The type of soil and heavy rain events can play an important role because they can facilitate the penetrability of manure containing bacteria in groundwater[92].
Several epidemiological studies have been conducted on the transmission of H. pylori through water, and several risk factors have been highlighted, such as living in a house with a lack of internal water pipes, the use of well or river water, the use of a latrine, less frequent boiling of drinking water and little hand washing after use of the bath and before meals[93-96] (Figure 1).
In 1991, a survey carried out on 407 Peruvian Lima children aged 2 mo to 12 years from families with different socioeconomic statuses showed an overall H. pylori prevalence of 48%. The children underwent the 13C-urea breath test, and the results showed a higher incidence among the children of low-income families than among those of high-income families (56% vs 32%). An important risk factor was the water supply; incidence increased three-fold when the water sources were outside the home compared to those whose homes had internal water sources. Furthermore, the municipal water supply seemed to be an important source of infection among Lima children from families of both low and high socioeconomic status because children from high-income families whose homes had municipal water were 12 times more likely to be infected than were those from high-income families whose water supplies came from community wells[97].
The poor basic hygiene conditions and the lack of potable water have been reported as the cause of H. pylori infection in an epidemiological study of a population in a rural area of the state of Mato Grosso in Brazil. The survey was conducted on 40 children and adolescents and 164 adults. H. pylori antibodies were detected in 31 (77.5%) children and adolescents and in 139 (84.7%) adults. The most important identified risk factor is using untreated water that could be contaminated by wastewater due to the lack of a sewage system[98].
Nurgalieva et al[49] conducted a similar study on 233 adults and 55 children in Kazakhstan. The overall prevalence of H. pylori was 86% among adults and 64% among children. The prevalence of H. pylori infection was inversely correlated with the index of clean water (CWI) (boil water before consumption, frequency of recovery and reuse of water and frequency of bath and shower). Infection was significantly lower among those with a high CWI (56%) than those with a moderate (79%) or low (95%) CWI. Moreover, the prevalence of H. pylori was inversely related to socioeconomic status. Those living in a family in which the levels of education and study were low had a higher rate of H. pylori infection (90%) than did those from a higher socioeconomic group (69%)[49].
In another epidemiological survey conducted in Germany on 3347 children from cities and rural areas, 179 children (119 from cities and 60 from rural areas) were infected by H. pylori. Among the children from rural areas, positivity significantly increased with the consumption of water from non-municipal sources[99].
Fujimura et al[100] studied the presence of H. pylori in 4 Japanese rivers and in 224 children who lived near one river using the stool antigen test for H. pylori prevalence.
The results of this study showed that H. pylori DNA was frequently present in river water from the middle and downstream reaches in which the human biosphere is embedded. The author concluded that river water in the natural environment could be a risk factor for H. pylori transmission.
Occurrence of H. pylori in water
Despite the several epidemiological studies that support the hypothesis that H. pylori is a waterborne pathogen, the real role of water in the spread of the pathogen remains a topic of discussion. As with foodstuffs, the fastidious nature of the bacterium and the difficulties in isolating it from environmental sources do not provide unequivocal evidence about the role of water as a source of transmission of this microorganism.
Culture methods, immunological methods and molecular methods have been employed to detect H. pylori in the aquatic environment.
Several studies on the occurrence of H. pylori in sewage and drinking water samples have been carried out worldwide using molecular methods. In many of these surveys, it was not possible to isolate the bacterium using culture methods (Table 3).
Table 3.
Studies evaluating the occurrence and survival of Helicobacter pylori in water
| Year | Water and study type | Method | Observations | Ref. |
| 1993 | Survival of H. pylori in artificially contaminated sterile river water | Culture | Culture up to 48 h | Shahamat et al[118] |
| Autoradiography | ||||
| 1996 | Occurrence of H. pylori in 48 water samples: 30 from municipal water system, 14 from community taps, 4 from brick tanks or plastic barrels of different households | IMS and PCR | 50% PCR positive samples | Hulten et al[101] |
| 1997 | Study on H. pylori resistance to chlorination | Culture | H. pylori were readily inactivated by free chlorine | Johnson et al[117] |
| 1999 | Occurrence of H. pylori in water from rivers and ponds | IMS and nested PCR | H. pylori-specific DNA was detected in samples | Sasaki et al[103] |
| 1999 | Occurrence of H. pylori in water from delivery truck and two lakes | Nested PCR and Southern blot hybridization | PCR positive samples from truck | McKeown et al[105] |
| PCR positive samples from two lakes | ||||
| 2001 | Occurrence of H. pylori in 10 seawater samples, 10 river water samples, 10 tap water samples, 6 well water samples | IMS, real-time PCR and nested PCR | 2 PCR positive samples of well water | Horiuchi et al[127] |
| 2001 | Occurrence of H. pylori in 139 ground water samples | PCR and Southern blot hybridization | 69% positive samples | Mazari-Hiriart et al[104] |
| 2002 | Occurrence of H. pylori in raw municipal wastewater | IMS, culture and PCR | 23 out of 37 isolated strains were confirmed to be H. pylori | Lu et al[106] |
| 11 out of 23 strains of H. pylori demonstrated vacA gene heterogeneity | ||||
| 2002 | Study on the susceptibility of H. pylori to chlorine, monochloramine, and ozone compared to that of Escherichia coli | Culture | H. pylori was more resistant than E. coli to chlorine and ozone but not monochloramine | Baker et al[116] |
| 2004 | Occurrence of H. pylori in water and biofilms: 11 samples from domestic proprieties, 7 samples from educational properties and from hydrants, and samples from reservoirs and water meters of 3 water utilities | Culture, IMS and PCR | All cultures were negative 26% PCR positive sample with the highest frequency in biofilm | Watson et al[120] |
| 2004 | Occurrence of H. pylori in seawater | Nested-PCR | H. pylori DNA only detected in fractionated water samples containing zooplanktonic organisms | Cellini et al[130] |
| 2005 | Occurrence of H. pylori in seawater | Filtration (200 mm, filter), culture and PCR | H. pylori was only isolated from fractionated water samples containing large zooplanktonic organisms | Cellini et al[131] |
| 2005 | Occurrence of H. pylori in 36 seawater samples | Culture and PCR | 30 positive samples | Carbone et al[132] |
| 2006 | Study on the ability of H. pylori to adhere on different water-exposed abiotic surfaces | Scanning electron microscope | H. pylori was able to adhere to all substrates tested | Azevedo et al[123] |
| 2007 | Study on the ability of H. pylori to adhere to stainless steel 304 in different environmental conditions | Epifluorescence microscopy | H. pylori was able to adhere to stainless steel 304 | Azevedo et al[124] |
| 2007 | Study on the resistance of H. pylori to chlorination | Culture, FISH, PCR and RT-PCR | Culture until 5 min | Moreno et al[115] |
| FISH viable cells until 3 h | ||||
| PCR samples positive after 24 h | ||||
| RT-PCR positive after 24 h | ||||
| 2007 | Survival of H. pylori in spiked bottled mineral water (drinking water) | Culture epifluorescence microscopy and PCR | Culture until 5 d | Queralt et al[114] |
| Cell viability until 14 d | ||||
| 2007 | Survival of H. pylori in spiked chlorinated filtered water (drinking water) | Culture, FISH and PCR | Culture until 5 min | Monero-Mesonero et al[115] |
| FISH viable cells until 3 h | ||||
| PCR positive after 24 h | ||||
| RT-PCR after 24 h | ||||
| 2009 | Occurrence of H. pylori in 75 drinking and environmental water samples and 21 natural water biofilms samples | Real-time PCR | 0% positive samples | Janzon et al [129] |
| 2010 | Occurrence of H. pylori in 198 drinking water samples | Culture | 10 out of 469 isolated strains were confirmed H. pylori | Al-Sulami et al[109] |
| 2011 | Occurrence of H. pylori in 137 seawater samples | PCR | 21% of the samples were positive for H. pylori | Twing et al[134] |
| 2013 | Occurrence of H. pylori in 50 tap water samples, 35 dental units’ water samples, and 40 bottled mineral water samples | Culture and PCR | 2 positive tap water samples | Bahrami et al[110] |
| 2 positive water from dental unit samples | ||||
| 1 positive water coolers sample | ||||
| 2013 | Occurrence of H. pylori in 31 seawater samples | Culture and PCR | 4 positive samples | Holman et al[135] |
| 2016 | Occurrence of H. pylori in 450 bottled mineral water samples | Culture and PCR | 8 positive samples | Ranjbar et al[111] |
| 2018 | Occurrence of H. pylori in 241 drinking water samples | PCR | 49 positive samples | Boehnke et al[102] |
H. pylori: Helicobacter pylori.
Based on the findings that there is an association between water sources and the prevalence of H. pylori infection in Peruvian children[97], H. pylori DNA was detected in drinking water samples from different locations near Lima (Peru) in two different surveys using molecular methods. These results provided evidence of the presence of H. pylori DNA in drinking water in Peru and were consistent with conclusions from a previous epidemiological study of the same population[101,102]. In addition, other studies have highlighted the presence of H. pylori DNA in samples of tap water, well water[103], aquatic systems located in Mexico City[104], trucks for water transport and lake water[105], further supporting the hypothesis of the transmissibility of H. pylori through water.
Despite the high incidence of H. pylori DNA in water, only a few studies have reported bacteriological isolation of this microorganism. Bacteriological isolation of H. pylori occurred in the study of Lu et al[106], who analysed untreated municipal wastewater samples using a series of steps beginning with immunomagnetic separation and cell culture.
In a survey carried out in Iraq, out of 198 samples of treated municipal drinking water, 10 strains of H. pylori were isolated and identified. The low concentration of chlorine in the water samples and the ability to form biofilms in water pipes[107,108] were the reasons that H. pylori was isolated[109].
H. pylori was also isolated in tap water, dental unit water, and bottled mineral water in Iran. Out of 200 water samples collected in Iran, 5 cultures were positive. Two out of 50 tap water samples (4%), 2 out of 35 dental unit water samples (5.8%), and 1 out of 40 samples (2.5%) from water coolers in public places were found to be contaminated with H. pylori[110]. Ranjbar et al[111] examined 450 bottled mineral water samples and confirmed the presence of H. pylori in bottled mineral water.
Survival studies in water samples showed that H. pylori could be cultured from 48 h up to 20 d in autoclaved distilled water. An increase in survival occurs at low temperatures; in fact, high temperature causes loss of culturability[89,91,112-118]. Shahamat et al[118] used an autoradiographic method to detect the metabolic activity of H. pylori VBNC in water. Four H. pylori strains were studied using 72-h cultures in water and incubated with [3H] thymidine for 24-72 h. After being exposed to the Kodak NTB2 emulsion for 3-28 d, the organism was vital and culturable under these conditions for up to 48 h and, in some cases, 20 to 30 d (Table 3).
One factor in support of H. pylori infection being waterborne or related to poor health practices is the association, which some authors claim, of H. pylori with free-living amoebae (FLA), such as Acanthamoeba, Naegleria, Vermamoeba or Balamuthia, which are ubiquitous protozoa commonly found in water[119-122].
Several studies have also shown that H. pylori can be present as a biofilm on the pipes of the drinking water system with the ability to adhere to different hydraulic materials, such as copper and stainless steel[121,123,124]. As a result, H. pylori is likely to survive in an aquatic system within a biofilm rather than in the planktonic state[39,125].
H. pylori cells were able to survive for short periods in chlorinated drinking water in the VBNC form, which would allow them to reach final consumption points and, at the same time, enable them to be undetectable by culture methods[115].
Moreover, in biofilms, the resistance of H. pylori to chlorine increases significantly[120,126]. Therefore, it is possible that if the organism enters a distribution system, it may survive disinfection treatment within the biofilm matrix[115]. This characteristic may be based on the lack of isolation in some surveys of H. pylori. In fact, in water samples treated with suitable potabilization systems, the lack of isolation can be caused by the formation of the biofilm or by the conversion of H. pylori in the form of VBNC[127-129].
Some important clues have emerged from Italian research on the presence of bacteria in seawater. Work by Cellini et al[130,131] and subsequent investigations[132] suggest that a significant reservoir for the microorganism is seawater, in which H. pylori can occur both in a free-living form and in association with plankton. Plankton-related H. pylori cells were detected in both summer and winter months depending on the flowering of Copepods and Cladocerans. The authors supposed that zooplankton organisms represent a sort of protected niche for survival of the microorganism. The finding of H. pylori attached to planktonic organisms is particularly interesting for the role of the latter in the seafood chain and its subsequent potential role in the spread of H. pylori infection[130-132]. More generally, the presence of H. pylori in seawater could also be a health hazard for swimmers and others using those waters for work or pleasure[133].
Moreover, H. pylori DNA has been isolated in 21% of samples from freshwater, estuarine and beach sites in Delaware (United States)[134] and in seawater sampled from 31 locations in Georgia, Trinidad and Puerto Rico[135]. In both reports, no correlation between the occurrence of H. pylori and faecal indicator bacteria was found, suggesting that standard water quality tests are ineffective in predicting the presence of this pathogen in natural waters, confirming the potential risk for H. pylori presence in marine waters.
METHODS FOR THE DETECTION OF H. PYLORI IN FOODS AND WATER
The isolation of H. pylori from food samples, particularly when they present high loads of accompanying microflora, is demanding and time consuming because it requires the use of selective media with numerous antibiotics, microaerophilic conditions and long incubation periods (7 d)[55,71]. The detection of H. pylori in food samples and water by means of conventional microbiological techniques generally employed for clinical specimens, which are unable to detect the VBNC, may yield false negative results and thus underestimate the presence of the bacterium in food; furthermore, the presence of H. pylori in VBNC state in food and water represents a potential microbiological risk for consumers, especially as a source of virulence factors[37,136,137].
Several solid and liquid culture media for the selective isolation of H. pylori from foods have been tested. The culture media most suitable for H. pylori growth often contain defibrinated horse or sheep blood acting as a reducing agent[29]. Furthermore, to achieve replication of H. pylori in broth culture, agitation is mandatory to provide good dispersion of gases throughout the liquid[31].
Brain heart infusion broth (BHIB) with growth supplement and selective agents has been evaluated[26,56]. BHIB with horse serum supplemented with porcine stomach mucin (0.3%), ferrous sulphate and sodium pyruvate (5%) or urea, along with the adjustment of the pH to 5.5 or 4.5, enhances the survival and possibly enables the growth of H. pylori in enrichment medium with fresh ground beef. In particular, pH 5.5 greatly enhances the growth and detectability of H. pylori in foods and should be considered an important factor for the detection of H. pylori in enrichment culture[56]. Stevenson et al[55] compared the growth of H. pylori in several liquid and solid media. None of the media tested presented an outstanding performance; only H. pylori special peptone agar offered the advantage of allowing the formation of the largest H. pylori colonies, and it was suitable for recovering H. pylori from environmental samples likely to be contaminated with large numbers of competing microorganisms[31,55].
Poms and Tatini[26] evaluated the efficacy for the in vitro isolation of H. pylori from foods of two solid media containing tryptic soy agar and Wilkins-Chalgren anaerobe agar supplemented with 5% defibrinated horse blood. The latter, to which several antibiotics (30 mg/L colistinmethanesulphonate, 100 mg/L cycloheximide, 30 mg/L nalidixic acid, 30 mg/L trimethoprim, and 10 mg/L vancomycin) were added, was found to be highly selective for the recovery of H. pylori from foods, but it lacked sufficient sensitivity to detect very low numbers of the bacterium.
Many authors have successfully used Wilkins-Chalgren anaerobe-agar or the broth developed by Poms et al[26] for the isolation of H. pylori in foods, both supplemented as described above[70,76,78-80].
However, there are still no standardized isolation protocols that are able to isolate the few H. pylori cells present in samples rich in microbial flora such as food. Furthermore, the pathogen is able to enter a VBNC state that remains metabolically active but fails to develop into colonies when cultured on routine media[40]. Consequently, molecular assays have been conducted to detect H. pylori DNA in water and foodstuffs.
Immuno-separation (IMS) followed by PCR has been successfully used by several investigators[138-140]. The advantage of using this protocol is that it offers excellent specificity using the IMS able to concentrate the pathogen from foods, followed by high sensitivity of the molecular methods[29]. Nevertheless, it appears expensive, exacting and time consuming. Autoradiography and ATP bioluminescence have been successfully used for the detection of H. pylori from water, human stools and pure cultures but have never been tested on food[29,118]. In addition, the ATP bioluminescence assay does not allow for distinguishing among ATP from different cell sources when applied to a complex system such as a foodstuff[141].
A multiplex touchdown PCR (MT-PCR) method for the identification (16S rRNA gene) and genotyping (vacA- s1/m1, s1/m2, and s2/m2- and cagA genes) of H. pylori directly from artificially contaminated sheep milk was developed[142]. The characterization of the genes encoding virulence factors provides important information with respect to the sanitary assessment of food items because of the greater pathogenicity of certain H. pylori genotypes. Hence, for public health purposes, the evaluation of a food containing H. pylori will thus have to include the genotyping of isolates. This rapid, sensitive (15 cfu/mL) and specific molecular method presents the advantage of detecting and genotyping H. pylori from microbiologically complex foodstuffs in a single step[142]. A nested PCR approach has been employed for the detection of the H. pylori glmM gene from seawater and sheep, goat and cow milks[71,130,141]. The sensitivity of the nested PCR technique was 3 cfu/mL in all types of milk and 62 cfu/mL in seawater samples, and therefore, compared to the previously described MT-PCR, it was more sensitive for the detection of H. pylori from foods (3 cfu/mL vs 15 cfu/mL) with the same specificity[141]. Osman et al[73] employed nested PCR in 50 cow milk samples, and 22% were positive for the presence of the H. pylori glmM gene.
H. pylori, as with many other bacteria, is able to form biofilms within which it can survive due to the different protection mechanisms that the biofilm offers. Quantitative real-time PCR was developed for the detection of H. pylori in drinking water biofilms of different ages. The target gene was the ureA subunit of the H. pylori urea gene, which showed high specificity and sensitivity[143].
As is well known, the main limit of PCR assays is their inability to distinguish live organisms from dead organisms. PCR techniques can, however, be used to screen water and foodstuffs, thus making it necessary to use conventional isolation methods only on those samples that test positive by PCR.
In a study by Buck et al[58], mRNA of known virulence factor (vacA) was detected in VBNC H. pylori cells using RT-PCR. This method exploits the unstable nature of bacterial mRNA to infer pathogenic viability when H. pylori becomes non-cultivable[144]. The half-life of mRNA of E. coli and Vibrio vulnificus cells is approximately 3-8 min and less than 60 min, respectively[145,146]. Thus, mRNA can be an excellent indicator of viability when H. pylori occurs in the VBNC state. Moreover, detection of transcripts from the vacA virulence gene may deduce continued virulence activity of H. pylori when present in the VBNC state[58]. This molecular technique offers significant promise for the detection of microorganisms in water and foodstuff and is a valid alternative to culture methods.
The fluorescence in situ hybridization (FISH) assay with the rRNA-direct molecular method has been applied for the specific detection of H. pylori in river and wastewater samples[147] and in raw bulk tank bovine milk[67] and for the assessment of its survival in chlorinated drinking water[115,148]. The authors concluded that FISH was a rapid method for the direct detection and specific identification of viable bacteria in food[67].
CONCLUSION
Several studies report the presence and survival of H. pylori in foods and water, especially in milk and in ready-to-eat products, suggesting that they can be sources of infection for humans.
Although many of the findings reported in the literature are based on indirect evidence of H. pylori in food and water through molecular methods and there are only in a few cases on the bacteriological isolation of this microorganism, the possibility that food and water can be routes of transmission among others cannot be ruled out.
Most of the bacteriological isolations of the pathogen in foods and water have been obtained in work conducted in Iran; a possible explanation could be the greater prevalence of the disease in this geographical area than in other areas. Moreover, the discrepancy in the prevalence of H. pylori in the different surveys could be related to the type and number of samples tested, sampling method, experimental methodology and climate differences in the regions from which the samples were collected.
However, to confirm a definite foodborne and waterborne role of H. pylori transmission, more surveys are needed on the presence of H. pylori in other foods of animal origin, particularly in seafood, and on the survival ability of this microorganism in dry fermented sausages and dairy products. Further investigations on the possible role of humans and animals as reservoirs of the microorganism are also required to clarify the faecal-oral route of transmission and the method of food and water contamination. Finally, the development of molecular biology methods and, above all, bacteriological isolation methods of H. pylori from water and food would add provide data that could confirm or deny the role of H. pylori as a foodborne and waterborne pathogen.
Footnotes
Manuscript source: Invited Manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Italy
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B, B
Grade C (Good): C, C, C
Grade D (Fair): D
Grade E (Poor): 0
Conflict-of-interest statement: No potential conflicts of interest. No financial support.
Peer-review started: May 4, 2018
First decision: May 17, 2018
Article in press: June 27, 2018
P- Reviewer: Hoff DA, El-Shabrawi MH, Chiba T, Chmiela M, Balaban YH, Reyes VE, Ding SZ, Chen XZ S- Editor: Gong ZM L- Editor: A E- Editor: Huang Y
Contributor Information
Nicoletta C Quaglia, Department of Emergency and Organ Transplantation, Section of Veterinary Clinic and Animal Production, University of Bari “Aldo Moro”, Valenzano 70010, Italy. nicolettacristiana.quaglia@uniba.it.
Angela Dambrosio, Department of Emergency and Organ Transplantation, Section of Veterinary Clinic and Animal Production, University of Bari “Aldo Moro”, Valenzano 70010, Italy.
References
- 1.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 2.Marshall BJ. History of the discovery of Campylobacter pylori. In: Blaser MJ, editor. Campylobacter pylori in gastritis and peptic ulcer disease. New York: Igaku Shoin Publishers; 1989. pp. 7–24. [Google Scholar]
- 3.Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kao CY, Sheu BS, Wu JJ. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed J. 2016;39:14–23. doi: 10.1016/j.bj.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leja M, Axon A, Brenner H. Epidemiology of Helicobacter pylori infection. Helicobacter. 2016;21 Suppl 1:3–7. doi: 10.1111/hel.12332. [DOI] [PubMed] [Google Scholar]
- 6.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatakeyama M, Brzozowski T. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2006;11 Suppl 1:14–20. doi: 10.1111/j.1478-405X.2006.00424.x. [DOI] [PubMed] [Google Scholar]
- 8.Kuo CH, Chen YH, Goh KL, Chang LL. Helicobacter pylori and Systemic Disease. Gastroenterol Res Pract. 2014;2014:358494. doi: 10.1155/2014/358494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres J, Pérez-Pérez G, Goodman KJ, Atherton JC, Gold BD, Harris PR, la Garza AM, Guarner J, Muñoz O. A comprehensive review of the natural history of Helicobacter pylori infection in children. Arch Med Res. 2000;31:431–469. doi: 10.1016/s0188-4409(00)00099-0. [DOI] [PubMed] [Google Scholar]
- 10.Frenck RW Jr, Clemens J. Helicobacter in the developing world. Microbes Infect. 2003;5:705–713. doi: 10.1016/s1286-4579(03)00112-6. [DOI] [PubMed] [Google Scholar]
- 11.Olivares D, Gisbert JP. Factors involved in the pathogenesis of Helicobacter pylori infection. Rev Esp Enferm Dig. 2006;98:374–386. doi: 10.4321/s1130-01082006000500008. [DOI] [PubMed] [Google Scholar]
- 12.Axon A. How to influence health providers. Helicobacter. 2007;12 Suppl 2:80–84. doi: 10.1111/j.1523-5378.2007.00569.x. [DOI] [PubMed] [Google Scholar]
- 13.Singh M, Prasad KN, Yachha SK, Saxena A, Krishnani N. Helicobacter pylori infection in children: prevalence, diagnosis and treatment outcome. Trans R Soc Trop Med Hyg. 2006;100:227–233. doi: 10.1016/j.trstmh.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Ozaydin N, Turkyilmaz SA, Cali S. Prevalence and risk factors of Helicobacter pylori in Turkey: a nationally-representative, cross-sectional, screening with the ¹³C-Urea breath test. BMC Public Health. 2013;13:1215. doi: 10.1186/1471-2458-13-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khedmat H, Karbasi-Afshar R, Agah S, Taheri S. Helicobacter pylori Infection in the general population: A Middle Eastern perspective. Caspian J Intern Med. 2013;4:745–753. [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Perez GI, Rothenbacher D, Brenner H. Epidemiology of Helicobacter pylori infection. Helicobacter. 2004;9 Suppl 1:1–6. doi: 10.1111/j.1083-4389.2004.00248.x. [DOI] [PubMed] [Google Scholar]
- 17.Breckan RK, Paulssen EJ, Asfeldt AM, Kvamme JM, Straume B, Florholmen J. The All-Age Prevalence of Helicobacter pylori Infection and Potential Transmission Routes. A Population-Based Study. Helicobacter. 2016;21:586–595. doi: 10.1111/hel.12316. [DOI] [PubMed] [Google Scholar]
- 18.Goodman KJ, Correa P. The transmission of Helicobacter pylori. A critical review of the evidence. Int J Epidemiol. 1995;24:875–887. doi: 10.1093/ije/24.5.875. [DOI] [PubMed] [Google Scholar]
- 19.Brown LM. Helicobacter pylori: epidemiology and routes of transmission. Epidemiol Rev. 2000;22:283–297. doi: 10.1093/oxfordjournals.epirev.a018040. [DOI] [PubMed] [Google Scholar]
- 20.Allaker RP, Young KA, Hardie JM, Domizio P, Meadows NJ. Prevalence of helicobacter pylori at oral and gastrointestinal sites in children: evidence for possible oral-to-oral transmission. J Med Microbiol. 2002;51:312–317. doi: 10.1099/0022-1317-51-4-312. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell H, Katelaris P. Epidemiology, clinical impacts and current clinical management of Helicobacter pylori infection. Med J Aust. 2016;204:376–380. doi: 10.5694/mja16.00104. [DOI] [PubMed] [Google Scholar]
- 22.Singh V, Trikha B, Vaiphei K, Nain CK, Thennarasu K, Singh K. Helicobacter pylori: evidence for spouse-to-spouse transmission. J Gastroenterol Hepatol. 1999;14:519–522. doi: 10.1046/j.1440-1746.1999.01908.x. [DOI] [PubMed] [Google Scholar]
- 23.Schütze K, Hentschel E, Dragosics B, Hirschl AM. Helicobacter pylori reinfection with identical organisms: transmission by the patients’ spouses. Gut. 1995;36:831–833. doi: 10.1136/gut.36.6.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kast RE. Some fibrocystic breast change may be caused by sexually transmitted H. pylori during oral nipple contact: supporting literature and case report of resolution after gut H. pylori eradication treatment. Med Hypotheses. 2007;68:1041–1046. doi: 10.1016/j.mehy.2006.09.050. [DOI] [PubMed] [Google Scholar]
- 25.Yee JKC. Are the view of Helicobacter pylori colonized in the oral cavity an illusion? Exp Mol Med. 2017;49:e397. doi: 10.1038/emm.2017.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poms RE, Tatini SR. Survival of Helicobacter pylori in ready-to-eat foods at 4 degrees C. Int J Food Microbiol. 2001;63:281–286. doi: 10.1016/s0168-1605(00)00441-4. [DOI] [PubMed] [Google Scholar]
- 27.Wesley IV. Helicobacter and Arcobacter: Potential human foodborne pathogens? Trends Food Sci Technol. 1997;8:293–299. [Google Scholar]
- 28.Meng J, Doyle MP. Emerging issues in microbiological food safety. Annu Rev Nutr. 1997;17:255–275. doi: 10.1146/annurev.nutr.17.1.255. [DOI] [PubMed] [Google Scholar]
- 29.Velázquez M, Feirtag JM. Helicobacter pylori: characteristics, pathogenicity, detection methods and mode of transmission implicating foods and water. Int J Food Microbiol. 1999;53:95–104. doi: 10.1016/s0168-1605(99)00160-9. [DOI] [PubMed] [Google Scholar]
- 30.van Duynhoven YT, de Jonge R. Transmission of Helicobacter pylori: a role for food? Bull World Health Organ. 2001;79:455–460. [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes BC, De Martinis ECP. The significance of Helicobacter pylori in water, food and environmental samples. Food Control. 2004;15:397–403. [Google Scholar]
- 32.Solnick JV, Schauer DB. Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin Microbiol Rev. 2001;14:59–97. doi: 10.1128/CMR.14.1.59-97.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romaniuk PJ, Zoltowska B, Trust TJ, Lane DJ, Olsen GJ, Pace NR, Stahl DA. Campylobacter pylori, the spiral bacterium associated with human gastritis, is not a true Campylobacter sp. J Bacteriol. 1987;169:2137–2141. doi: 10.1128/jb.169.5.2137-2141.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodwin CS, Armstrong JA. Microbiological aspects of Helicobacter pylori (Campylobacter pylori) Eur J Clin Microbiol Infect Dis. 1990;9:1–13. doi: 10.1007/BF01969526. [DOI] [PubMed] [Google Scholar]
- 35.Pinto D, Santos MA, Chambel L. Thirty years of viable but nonculturable state research: unsolved molecular mechanisms. Crit Rev Microbiol. 2015;41:61–76. doi: 10.3109/1040841X.2013.794127. [DOI] [PubMed] [Google Scholar]
- 36.Percival SL, Yates MV, Williams DW, Chalmers RM, Gray NF. Helicobacter pylori. In: Microbiology of waterborne diseases., editor. Microbiological aspects and risks. 2nd ed. London: Elsevier; 2014. pp. 119–153. ISBN: 978-0-12-415846-7. [Google Scholar]
- 37.Cellini L. Helicobacter pylori: a chameleon-like approach to life. World J Gastroenterol. 2014;20:5575–5582. doi: 10.3748/wjg.v20.i19.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cellini L, Robuffo I, Di Campli E, Di Bartolomeo S, Taraborelli T, Dainelli B. Recovery of Helicobacter pylori ATCC43504 from a viable but not culturable state: regrowth or resuscitation? APMIS. 1998;106:571–579. [PubMed] [Google Scholar]
- 39.Andersen LP, Rasmussen L. Helicobacter pylori-coccoid forms and biofilm formation. FEMS Immunol Med Microbiol. 2009;56:112–115. doi: 10.1111/j.1574-695X.2009.00556.x. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Mendis N, Trigui H, Oliver JD, Faucher SP. The importance of the viable but non-culturable state in human bacterial pathogens. Front Microbiol. 2014;5:258. doi: 10.3389/fmicb.2014.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lleò MM, Bonato B, Tafi MC, Signoretto C, Boaretti M, Canepari P. Resuscitation rate in different enterococcal species in the viable but non-culturable state. J Appl Microbiol. 2001;91:1095–1102. doi: 10.1046/j.1365-2672.2001.01476.x. [DOI] [PubMed] [Google Scholar]
- 42.Senoh M, Ghosh-Banerjee J, Ramamurthy T, Colwell RR, Miyoshi S, Nair GB, Takeda Y. Conversion of viable but nonculturable enteric bacteria to culturable by co-culture with eukaryotic cells. Microbiol Immunol. 2012;56:342–345. doi: 10.1111/j.1348-0421.2012.00440.x. [DOI] [PubMed] [Google Scholar]
- 43.Versalovic J, Fox JG. Helicobacter. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH. Manual of Clinical Microbiology 7h edition; 1999. pp. 727–736. [Google Scholar]
- 44.Lopes AI, Vale FF, Oleastro M. Helicobacter pylori infection - recent developments in diagnosis. World J Gastroenterol. 2014;20:9299–9313. doi: 10.3748/wjg.v20.i28.9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hachem CY, Clarridge JE, Evans DG, Graham DY. Comparison of agar based media for primary isolation of Helicobacter pylori. J Clin Pathol. 1995;48:714–716. doi: 10.1136/jcp.48.8.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henriksen TH, Brorson O, Schöyen R, Thoresen T, Setegn D, Madebo T. Rapid growth of Helicobacter pylori. Eur J Clin Microbiol Infect Dis. 1995;14:1008–1011. doi: 10.1007/BF01691385. [DOI] [PubMed] [Google Scholar]
- 47.Jiang X, Doyle MP. Effect of environmental and substrate factors on survival and growth of Helicobacter pylori. J Food Prot. 1998;61:929–933. doi: 10.4315/0362-028x-61.8.929. [DOI] [PubMed] [Google Scholar]
- 48.Roma-Giannikou E, Karameris A, Balatsos B, Panayiotou J, Manika Z, Van-Vliet C, Rokkas T, Skandalis N, Kattamis C. Intrafamilial spread of Helicobacter pylori (H. pylori): A genetic analysis. European Helicobacter Study Group, Abstract from Athens Workshop 2002. XV International Workshop, September 11-14. [Google Scholar]
- 49.Nurgalieva ZZ, Malaty HM, Graham DY, Almuchambetova R, Machmudova A, Kapsultanova D, Osato MS, Hollinger FB, Zhangabylov A. Helicobacter pylori infection in Kazakhstan: effect of water source and household hygiene. Am J Trop Med Hyg. 2002;67:201–206. doi: 10.4269/ajtmh.2002.67.201. [DOI] [PubMed] [Google Scholar]
- 50.Vaira D, D’Anastasio C, Holton J, Dowsett J, Londei M, Salmon P, Gandolfi L. Is Campylobacter pylori a zoonosis? Lancet. 1988;2:1149. doi: 10.1016/s0140-6736(88)90575-2. [DOI] [PubMed] [Google Scholar]
- 51.Husson MO, Vincent P, Grabiaud MH, Furon D, Leclerc H. Anti-Helicobacter pylori IgG levels in abattoir workers. Gastroenterol Clin Biol. 1991;15:723–726. [PubMed] [Google Scholar]
- 52.Beuchat LR. Ecological factors influencing survival and growth of human pathogens on raw fruits and vegetables. Microbes Infect. 2002;4:413–423. doi: 10.1016/s1286-4579(02)01555-1. [DOI] [PubMed] [Google Scholar]
- 53.Quaglia NC, Dambrosio A, Normanno G, Parisi A, Firinu A, Lorusso V, Celano GV. Survival of Helicobacter pylori in artificially contaminated ultrahigh temperature and pasteurized milk. Food Microbiol. 2007;24:296–300. doi: 10.1016/j.fm.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 54.Fan XG, Chua A, Li TG, Zeng QS. Survival of Helicobacter pylori in milk and tap water. J Gastroenterol Hepatol. 1998;13:1096–1098. doi: 10.1111/j.1440-1746.1998.tb00582.x. [DOI] [PubMed] [Google Scholar]
- 55.Stevenson TH, Bauer N, Lucia LM, Acuff GR. Attempts to isolate Helicobacter from cattle and survival of Helicobacter pylori in beef products. J Food Prot. 2000;63:174–178. doi: 10.4315/0362-028x-63.2.174. [DOI] [PubMed] [Google Scholar]
- 56.Jiang X, Doyle MP. Optimizing enrichment culture conditions for detecting Helicobacter pylori in foods. J Food Prot. 2002;65:1949–1954. doi: 10.4315/0362-028x-65.12.1949. [DOI] [PubMed] [Google Scholar]
- 57.Gomes BC, De Martinis ECP. Fate of Helicobacter pylori artificially inoculated in lettuce and carrot samples. Braz J Microbiol. 2004;35:145–150. [Google Scholar]
- 58.Buck A, Oliver JD. Survival of spinach-associated Helicobacter pylori in the viable but nonculturable state. Food Control. 2010;21:1150–1154. [Google Scholar]
- 59.Guner A, Kav K, Tekinsen KK, Dogruer Y, Telli N. Survival of Helicobacter pylori in Turkish fermented sucuk and heat-treated sucuk during production. J Food Prot. 2011;74:2055–2061. doi: 10.4315/0362-028X.JFP-11-019. [DOI] [PubMed] [Google Scholar]
- 60.Ng CG, Loke MF, Goh KL, Vadivelu J, Ho B. Biofilm formation enhances Helicobacter pylori survivability in vegetables. Food Microbiol. 2017;62:68–76. doi: 10.1016/j.fm.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 61.Verma N, Singh M. A disposable microbial based biosensor for quality control in milk. Biosens Bioelectron. 2003;18:1219–1224. doi: 10.1016/s0956-5663(03)00085-x. [DOI] [PubMed] [Google Scholar]
- 62.West AP, Millar MR, Tompkins DS. Effect of physical environment on survival of Helicobacter pylori. J Clin Pathol. 1992;45:228–231. doi: 10.1136/jcp.45.3.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Midolo PD, Lambert JR, Hull R, Luo F, Grayson ML. In vitro inhibition of Helicobacter pylori NCTC 11637 by organic acids and lactic acid bacteria. J Appl Bacteriol. 1995;79:475–479. doi: 10.1111/j.1365-2672.1995.tb03164.x. [DOI] [PubMed] [Google Scholar]
- 64.Dore MP, Bilotta M, Vaira D, Manca A, Massarelli G, Leandro G, Atzei A, Pisanu G, Graham DY, Realdi G. High prevalence of Helicobacter pylori infection in shepherds. Dig Dis Sci. 1999;44:1161–1164. doi: 10.1023/a:1026676223825. [DOI] [PubMed] [Google Scholar]
- 65.Dore MP, Sepulveda AR, El-Zimaity H, Yamaoka Y, Osato MS, Mototsugu K, Nieddu AM, Realdi G, Graham DY. Isolation of Helicobacter pylori from sheep-implications for transmission to humans. Am J Gastroenterol. 2001;96:1396–1401. doi: 10.1111/j.1572-0241.2001.03772.x. [DOI] [PubMed] [Google Scholar]
- 66.Fujimura S, Kawamura T, Kato S, Tateno H, Watanabe A. Detection of Helicobacter pylori in cow’s milk. Lett Appl Microbiol. 2002;35:504–507. doi: 10.1046/j.1472-765x.2002.01229.x. [DOI] [PubMed] [Google Scholar]
- 67.Angelidis AS, Tirodimos I, Bobos M, Kalamaki MS, Papageorgiou DK, Arvanitidou M. Detection of Helicobacter pylori in raw bovine milk by fluorescence in situ hybridization (FISH) Int J Food Microbiol. 2011;151:252–256. doi: 10.1016/j.ijfoodmicro.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 68.Rahimi E, Kheirabadi EK. Detection of Helicobacter pylori in bovine, buffalo, camel, ovine, and caprine milk in Iran. Foodborne Pathog Dis. 2012;9:453–456. doi: 10.1089/fpd.2011.1060. [DOI] [PubMed] [Google Scholar]
- 69.Mousavi S, Dehkordi FS, Rahimi E. Virulence factors and antibiotic resistance of Helicobacter pylori isolated from raw milk and unpasteurized dairy products in Iran. J Venom Anim Toxins Incl Trop Dis. 2014;20:51. doi: 10.1186/1678-9199-20-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saeidi E, Sheikhshahrokh A. vacA Genotype Status of Helicobacter pylori Isolated from Foods with Animal Origin. Biomed Res Int. 2016;2016:8701067. doi: 10.1155/2016/8701067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quaglia NC, Dambrosio A, Normanno G, Parisi A, Patrono R, Ranieri G, Rella A, Celano GV. High occurrence of Helicobacter pylori in raw goat, sheep and cow milk inferred by glmM gene: a risk of food-borne infection? Int J Food Microbiol. 2008;124:43–47. doi: 10.1016/j.ijfoodmicro.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 72.Talaei R, Souod N, Momtaz H, Dabiri H. Milk of livestock as a possible transmission route of Helicobacter pylori infection. Gastroenterol Hepatol Bed Bench. 2015;8:S30–S36. [PMC free article] [PubMed] [Google Scholar]
- 73.Osman EY, El-Eragi AM, Musa AM, El-Magboul SB, A/Rahman MB, Abdo AE. Detection of Helicobacter pylori glmM gene in bovine milk using Nested polymerase chain reaction. Vet World. 2015;8:913–917. doi: 10.14202/vetworld.2015.913-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Turutoglu H, Mudul S. Investigation of Helicobacter pylori in raw sheep milk samples. J Vet Med B Infect Dis Vet Public Health. 2002;49:308–309. doi: 10.1046/j.1439-0450.2002.00557.x. [DOI] [PubMed] [Google Scholar]
- 75.Bianchini V, Recordati C, Borella L, Gualdi V, Scanziani E, Selvatico E, Luini M. Helicobacteraceae in Bulk Tank Milk of Dairy Herds from Northern Italy. Biomed Res Int. 2015;2015:639521. doi: 10.1155/2015/639521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gilani A, Razavilar V, Rokni N, Rahimi E. VacA and cagA genotypes status and antimicrobial resistance properties of Helicobacter pylori strains isolated from meat products in Isfahan province, Iran. Iran J Vet Res. 2017;18:97–102. [PMC free article] [PubMed] [Google Scholar]
- 77.Meng X, Zhang H, Law J, Tsang R, Tsang T. Detection of Helicobacter pylori from food sources by a novel multiplex PCR assay. J Food Saf. 2008;28:609–619. [Google Scholar]
- 78.Hemmatinezhad B, Momtaz H, Rahimi E. VacA, cagA, iceA and oipA genotypes status and antimicrobial resistance properties of Helicobacter pylori isolated from various types of ready to eat foods. Ann Clin Microbiol Antimicrob. 2016;15:2. doi: 10.1186/s12941-015-0115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ghorbani F, Gheisari E, Dehkordi FS. Genotyping of vacA alleles of Helicobacter pylori strains recovered from some Iranian food items. Trop J Pharm Res. 2016;15:1631–1636. [Google Scholar]
- 80.Yahaghi E, Khamesipour F, Mashayekhi F, Safarpoor Dehkordi F, Sakhaei MH, Masoudimanesh M, Khameneie MK. Helicobacter pylori in vegetables and salads: genotyping and antimicrobial resistance properties. Biomed Res Int. 2014;2014:757941. doi: 10.1155/2014/757941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Atapoor S, Safarpoor Dehkordi F, Rahimi E. Detection of Helicobacter pylori in Various Types of Vegetables and Salads. Jundishapur J Microbiol. 2014;7:e10013. doi: 10.5812/jjm.10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thomson MA, Storey P, Greer R, Cleghorn GJ. Canine-human transmission of Gastrospirillum hominis. Lancet. 1994;343:1605–1607. doi: 10.1016/s0140-6736(94)93060-0. [DOI] [PubMed] [Google Scholar]
- 83.Dimola S, Caruso ML. Helicobacter pylori in animals affecting the human habitat through the food chain. Anticancer Res. 1999;19:3889–3894. [PubMed] [Google Scholar]
- 84.Quaglia NC, Dambrosio A, Normanno G, Alberti F, Rella A, Tamborrino C, Celano GV. Detection of Helicobacter pylori in gastric mucosa of sheep: preliminary results. Rivista dell’Associazione Italiana Veterinari Igienisti. 2009;3:45–48. [Google Scholar]
- 85.Papiez D, Konturek PC, Bielanski W, Plonka M, Dobrzanska M, Kaminska A, Szczyrk U, Bochenek A, Wierzchos E. Prevalence of Helicobacter pylori infection in Polish shepherds and their families. Dig Liver Dis. 2003;35:10–15. doi: 10.1016/s1590-8658(02)00004-x. [DOI] [PubMed] [Google Scholar]
- 86.Mladenova-Hristova I, Grekova O, Patel A. Zoonotic potential of Helicobacter spp. J Microbiol Immunol Infect. 2017;50:265–269. doi: 10.1016/j.jmii.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 87.World Health Organization (WHO) and the United Nations Children’s Fund (UNICEF). Progress on drinking water, sanitation and hygiene: 2017 update and Sustainable Development Goal baselines. The Joint Monitoring Programme (JMP) report. 2018. Available from: https://www.unicef.org/publications/index_96611.html.
- 88.Lambert JR, Lin SK, Aranda-Michel J. Helicobacter pylori. Scand J Gastroenterol Suppl. 1995;208:33–46. doi: 10.3109/00365529509107760. [DOI] [PubMed] [Google Scholar]
- 89.Vu C, Ng YY. Prevalence of Helicobacter pylori in peptic ulcer disease in a Singapore hospital. Singapore Med J. 2000;41:478–481. [PubMed] [Google Scholar]
- 90.Adams BL, Bates TC, Oliver JD. Survival of Helicobacter pylori in a natural freshwater environment. Appl Environ Microbiol. 2003;69:7462–7466. doi: 10.1128/AEM.69.12.7462-7466.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aziz RK, Khalifa MM, Sharaf RR. Contaminated water as a source of Helicobacter pylori infection: A review. J Adv Res. 2015;6:539–547. doi: 10.1016/j.jare.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bellack NR, Koehoorn MW, MacNab YC, Morshed MG. A conceptual model of water’s role as a reservoir in Helicobacter pylori transmission: a review of the evidence. Epidemiol Infect. 2006;134:439–449. doi: 10.1017/S0950268806006005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee YY, Ismail AW, Mustaffa N, Musa KI, Majid NA, Choo KE, Mahendra Raj S, Derakhshan MH, Malaty HM, Graham DY. Sociocultural and dietary practices among Malay subjects in the north-eastern region of Peninsular Malaysia: a region of low prevalence of Helicobacter pylori infection. Helicobacter. 2012;17:54–61. doi: 10.1111/j.1523-5378.2011.00917.x. [DOI] [PubMed] [Google Scholar]
- 94.Porras C, Nodora J, Sexton R, Ferreccio C, Jimenez S, Dominguez RL, Cook P, Anderson G, Morgan DR, Baker LH, et al. Epidemiology of Helicobacter pylori infection in six Latin American countries (SWOG Trial S0701) Cancer Causes Control. 2013;24:209–215. doi: 10.1007/s10552-012-0117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Syam AF, Miftahussurur M, Makmun D, Nusi IA, Zain LH, Zulkhairi, Akil F, Uswan WB, Simanjuntak D, Uchida T, et al. Risk Factors and Prevalence of Helicobacter pylori in Five Largest Islands of Indonesia: A Preliminary Study. PLoS One. 2015;10:e0140186. doi: 10.1371/journal.pone.0140186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Awuku YA, Simpong DL, Alhassan IK, Tuoyire DA, Afaa T, Adu P. Prevalence of helicobacter pylori infection among children living in a rural setting in Sub-Saharan Africa. BMC Public Health. 2017;17:360. doi: 10.1186/s12889-017-4274-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klein PD, Graham DY, Gaillour A, Opekun AR, Smith EO. Water source as risk factor for Helicobacter pylori infection in Peruvian children. Gastrointestinal Physiology Working Group. Lancet. 1991;337:1503–1506. doi: 10.1016/0140-6736(91)93196-g. [DOI] [PubMed] [Google Scholar]
- 98.Souto FJ, Fontes CJ, Rocha GA, de Oliveira AM, Mendes EM, Queiroz DM. Prevalence of Helicobacter pylori Infection in a Rural Area of the State of Mato Grosso, Brazil. Memorias do Instituto Oswaldo Cruz. 1998;93:171–174. doi: 10.1590/s0074-02761998000200006. [DOI] [PubMed] [Google Scholar]
- 99.Herbarth O, Krumbiegel P, Fritz GJ, Richter M, Schlink U, Müller DM, Richter T. Helicobacter pylori prevalences and risk factors among school beginners in a German urban center and its rural county. Environ Health Perspect. 2001;109:573–577. doi: 10.1289/ehp.01109573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fujimura S, Kato S, Kawamura T. Helicobacter pylori in Japanese river water and its prevalence in Japanese children. Lett Appl Microbiol. 2004;38:517–521. doi: 10.1111/j.1472-765X.2004.01529.x. [DOI] [PubMed] [Google Scholar]
- 101.Hulten K, Han SW, Enroth H, Klein PD, Opekun AR, Gilman RH, Evans DG, Engstrand L, Graham DY, El-Zaatari FA. Helicobacter pylori in the drinking water in Peru. Gastroenterology. 1996;110:1031–1035. doi: 10.1053/gast.1996.v110.pm8612990. [DOI] [PubMed] [Google Scholar]
- 102.Boehnke KF, Brewster RK, Sánchez BN, Valdivieso M, Bussalleu A, Guevara M, Saenz CG, Alva SO, Gil E, Xi C. An assessment of drinking water contamination with Helicobacter pylori in Lima, Peru. Helicobacter. 2018;23:e12462. doi: 10.1111/hel.12462. [DOI] [PubMed] [Google Scholar]
- 103.Sasaki K, Tajiri Y, Sata M, Fujii Y, Matsubara F, Zhao M, Shimizu S, Toyonaga A, Tanikawa K. Helicobacter pylori in the natural environment. Scand J Infect Dis. 1999;31:275–279. doi: 10.1080/00365549950163572. [DOI] [PubMed] [Google Scholar]
- 104.Mazari-Hiriart M, López-Vidal Y, Calva JJ. Helicobacter pylori in water systems for human use in Mexico City. Water Sci Technol. 2001;43:93–98. [PubMed] [Google Scholar]
- 105.McKeown I, Orr P, Macdonald S, Kabani A, Brown R, Coghlan G, Dawood M, Embil J, Sargent M, Smart G, et al. Helicobacter pylori in the Canadian arctic: seroprevalence and detection in community water samples. Am J Gastroenterol. 1999;94:1823–1829. doi: 10.1111/j.1572-0241.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- 106.Lu Y, Redlinger TE, Avitia R, Galindo A, Goodman K. Isolation and genotyping of Helicobacter pylori from untreated municipal wastewater. Appl Environ Microbiol. 2002;68:1436–1439. doi: 10.1128/AEM.68.3.1436-1439.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Al-Taee MR. Assessment of water quality due to microbial growth in drinking water distribution systems in Basrah city. Marina Mesopotamica. 2001;16:37–46. [Google Scholar]
- 108.Momba MNB, Makala N. Comparing the effect of various pipe materials on biofilm formation in chlorinated and combined chlorine-chloraminated water systems. Water SA. 2003;30:175–182. [Google Scholar]
- 109.Al-Sulami AA, Al-Taee AM, Juma’a MG. Isolation and identification of Helicobacter pylori from drinking water in Basra governorate, Iraq. East Mediterr Health J. 2010;16:920–925. [PubMed] [Google Scholar]
- 110.Bahrami AR, Rahimi E, Ghasemian Safaei H. Detection of Helicobacter pylori in city water, dental units’ water, and bottled mineral water in Isfahan, Iran. ScientificWorldJournal. 2013;2013:280510. doi: 10.1155/2013/280510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ranjbar R, Khamesipour F, Jonaidi-Jafari N, Rahimi E. Helicobacter pylori in bottled mineral water: genotyping and antimicrobial resistance properties. BMC Microbiol. 2016;16:40. doi: 10.1186/s12866-016-0647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vale FF, Vítor JM. Transmission pathway of Helicobacter pylori: does food play a role in rural and urban areas? Int J Food Microbiol. 2010;138:1–12. doi: 10.1016/j.ijfoodmicro.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 113.Azevedo NF, Almeida C, Cerqueira L, Dias S, Keevil CW, Vieira MJ. Coccoid form of Helicobacter pylori as a morphological manifestation of cell adaptation to the environment. Appl Environ Microbiol. 2007;73:3423–3427. doi: 10.1128/AEM.00047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Queralt N, Araujo R. Analysis of the survival of H. pylori within a laboratory-based aquatic model system using molecular and classical techniques. Microb Ecol. 2007;54:771–777. doi: 10.1007/s00248-007-9242-1. [DOI] [PubMed] [Google Scholar]
- 115.Moreno Y, Piqueres P, Alonso JL, Jiménez A, González A, Ferrús MA. Survival and viability of Helicobacter pylori after inoculation into chlorinated drinking water. Water Res. 2007;41:3490–3496. doi: 10.1016/j.watres.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 116.Baker KH, Hegarty JP, Redmond B, Reed NA, Herson DS. Effect of oxidizing disinfectants (chlorine, monochloramine, and ozone) on Helicobacter pylori. Appl Environ Microbiol. 2002;68:981–984. doi: 10.1128/AEM.68.2.981-984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johnson CH, Rice EW, Reasoner DJ. Inactivation of Helicobacter pylori by chlorination. Appl Environ Microbiol. 1997;63:4969–4970. doi: 10.1128/aem.63.12.4969-4970.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shahamat M, Mai U, Paszko-Kolva C, Kessel M, Colwell RR. Use of autoradiography to assess viability of Helicobacter pylori in water. Appl Environ Microbiol. 1993;59:1231–1235. doi: 10.1128/aem.59.4.1231-1235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Winiecka-Krusnell J, Wreiber K, von Euler A, Engstrand L, Linder E. Free-living amoebae promote growth and survival of Helicobacter pylori. Scand J Infect Dis. 2002;34:253–256. doi: 10.1080/00365540110080052. [DOI] [PubMed] [Google Scholar]
- 120.Watson CL, Owen RJ, Said B, Lai S, Lee JV, Surman-Lee S, Nichols G. Detection of Helicobacter pylori by PCR but not culture in water and biofilm samples from drinking water distribution systems in England. J Appl Microbiol. 2004;97:690–698. doi: 10.1111/j.1365-2672.2004.02360.x. [DOI] [PubMed] [Google Scholar]
- 121.Percival SL, Thomas JG. Transmission of Helicobacter pylori and the role of water and biofilms. J Water Health. 2009;7:469–477. doi: 10.2166/wh.2009.070. [DOI] [PubMed] [Google Scholar]
- 122.Moreno-Mesonero L, Moreno Y, Alonso JL, Ferrús MA. Detection of viable Helicobacter pylori inside free-living amoebae in wastewater and drinking water samples from Eastern Spain. Environ Microbiol. 2017;19:4103–4112. doi: 10.1111/1462-2920.13856. [DOI] [PubMed] [Google Scholar]
- 123.Azevedo NF, Pacheco AP, Keevil CW, Vieira MJ. Adhesion of water stressed Helicobacter pylori to abiotic surfaces. J Appl Microbiol. 2006;101:718–724. doi: 10.1111/j.1365-2672.2006.03029.x. [DOI] [PubMed] [Google Scholar]
- 124.Azevedo NF, Pinto AR, Reis NM, Vieira MJ, Keevil CW. Shear stress, temperature, and inoculation concentration influence the adhesion of water-stressed Helicobacter pylori to stainless steel 304 and polypropylene. Appl Environ Microbiol. 2006;72:2936–2941. doi: 10.1128/AEM.72.4.2936-2941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 126.Yu FP, McFeters GA. Physiological responses of bacteria in biofilms to disinfection. Appl Environ Microbiol. 1994;60:2462–2466. doi: 10.1128/aem.60.7.2462-2466.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Horiuchi T, Ohkusa T, Watanabe M, Kobayashi D, Miwa H, Eishi Y. Helicobacter pylori DNA in drinking water in Japan. Microbiol Immunol. 2001;45:515–519. doi: 10.1111/j.1348-0421.2001.tb02652.x. [DOI] [PubMed] [Google Scholar]
- 128.Böckelmann U, Dörries HH, Ayuso-Gabella MN, Salgot de Marçay M, Tandoi V, Levantesi C, Masciopinto C, Van Houtte E, Szewzyk U, Wintgens T, et al. Quantitative PCR monitoring of antibiotic resistance genes and bacterial pathogens in three European artificial groundwater recharge systems. Appl Environ Microbiol. 2009;75:154–163. doi: 10.1128/AEM.01649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Janzon A, Sjöling A, Lothigius A, Ahmed D, Qadri F, Svennerholm AM. Failure to detect Helicobacter pylori DNA in drinking and environmental water in Dhaka, Bangladesh, using highly sensitive real-time PCR assays. Appl Environ Microbiol. 2009;75:3039–3044. doi: 10.1128/AEM.02779-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cellini L, Del Vecchio A, Di Candia M, Di Campli E, Favaro M, Donelli G. Detection of free and plankton-associated Helicobacter pylori in seawater. J Appl Microbiol. 2004;97:285–292. doi: 10.1111/j.1365-2672.2004.02307.x. [DOI] [PubMed] [Google Scholar]
- 131.Cellini L, Grand R, Prenna M, Pasquantonio M, Pane L. Detection of Helicobacter pylori associated with zooplankton. Aquat Microb Ecol. 2005;40:115–120. [Google Scholar]
- 132.Carbone M, Maugeri TL, Gugliandolo C, La Camera E, Biondo C, Fera MT. Occurrence of Helicobacter pylori DNA in the coastal environment of southern Italy (Straits of Messina) J Appl Microbiol. 2005;98:768–774. doi: 10.1111/j.1365-2672.2004.02517.x. [DOI] [PubMed] [Google Scholar]
- 133.Dixon B. Helicobacter from seas? ASM news. 2005;71:4–5. [Google Scholar]
- 134.Twing KI, Kirchman DL, Campbell BJ. Temporal study of Helicobacter pylori presence in coastal freshwater, estuary and marine waters. Water Res. 2011;45:1897–1905. doi: 10.1016/j.watres.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 135.Holman CB, Bachoon DS, Otero E, Ramsubhag A. Detection of Helicobacter pylori in the coastal waters of Georgia, Puerto Rico and Trinidad. Mar Pollut Bull. 2014;79:354–358. doi: 10.1016/j.marpolbul.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 136.Bode G, Mauch F, Malfertheiner P. The coccoid forms of Helicobacter pylori. Criteria for their viability. Epidemiol Infect. 1993;111:483–490. doi: 10.1017/s0950268800057216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cao J, Li ZQ, Borch K, Petersson F, Mårdh S. Detection of spiral and coccoid forms of Helicobacter pylori using a murine monoclonal antibody. Clin Chim Acta. 1997;267:183–196. doi: 10.1016/s0009-8981(97)00134-4. [DOI] [PubMed] [Google Scholar]
- 138.Cohen AE. A PCR method for the detection of Helicobacter pylori in food products: US Food and Drug Administration (FDA) Study. Proceedings of the 16th Food Microbiology Symposium Current concepts in foodborne pathogens and rapid and automated methods in food microbiology; 1996 University of Wisconsin, River Falls [Google Scholar]
- 139.Enroth H, Engstrand L. Immunomagnetic separation and PCR for detection of Helicobacter pylori in water and stool specimens. J Clin Microbiol. 1995;33:2162–2165. doi: 10.1128/jcm.33.8.2162-2165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hultén K, Enroth H, Nyström T, Engstrand L. Presence of Helicobacter species DNA in Swedish water. J Appl Microbiol. 1998;85:282–286. doi: 10.1046/j.1365-2672.1998.00500.x. [DOI] [PubMed] [Google Scholar]
- 141.Quaglia NC, Dambrosio A, Normanno G, Celano GV. Evaluation of a Nested-PCR assay based on the phosphoglucosamine mutase gene (glmM) for the detection of Helicobacter pylori from raw milk. Food Control. 2009;20:119–123. [Google Scholar]
- 142.Quaglia NC, Normanno G, Dambrosio A, Celano GV, Parisi A, Firinu A, Buonavoglia C. Multiplex-touchdown PCR assay for the detection and genotyping of Helicobacter pylori from artificially contaminated sheep milk. J Food Prot. 2005;68:2136–2139. doi: 10.4315/0362-028x-68.10.2136. [DOI] [PubMed] [Google Scholar]
- 143.Linke S, Lenz J, Gemein S, Exner M, Gebel J. Detection of Helicobacter pylori in biofilms by real-time PCR. Int J Hyg Environ Health. 2010;213:176–182. doi: 10.1016/j.ijheh.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 144.Lahtinen SJ, Ahokoski H, Reinikainen JP, Gueimonde M, Nurmi J, Ouwehand AC, Salminen SJ. Degradation of 16S rRNA and attributes of viability of viable but nonculturable probiotic bacteria. Lett Appl Microbiol. 2008;46:693–698. doi: 10.1111/j.1472-765X.2008.02374.x. [DOI] [PubMed] [Google Scholar]
- 145.Sheridan GE, Masters CI, Shallcross JA, MacKey BM. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl Environ Microbiol. 1998;64:1313–1318. doi: 10.1128/aem.64.4.1313-1318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Smith B, Oliver JD. In situ and in vitro gene expression by Vibrio vulnificus during entry into, persistence within, and resuscitation from the viable but nonculturable state. Appl Environ Microbiol. 2006;72:1445–1451. doi: 10.1128/AEM.72.2.1445-1451.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Moreno Y, Ferrús MA, Alonso JL, Jiménez A, Hernández J. Use of fluorescent in situ hybridization to evidence the presence of Helicobacter pylori in water. Water Res. 2003;37:2251–2256. doi: 10.1016/S0043-1354(02)00624-3. [DOI] [PubMed] [Google Scholar]
- 148.Piqueres P, Moreno Y, Alonso JL, Ferrús MA. A combination of direct viable count and fluorescent in situ hybridization for estimating Helicobacter pylori cell viability. Res Microbiol. 2006;157:345–349. doi: 10.1016/j.resmic.2005.09.003. [DOI] [PubMed] [Google Scholar]


