Abstract
N-(2-{[2-(1H-Indol-2-ylcarbonyl)hydrazinyl](oxo)acetyl}phenyl)acetamides (5a–h) and N-[2-(2-{[2-(acetylamino)phenyl](oxo)acetyl}hydrazinyl)-2-oxoethyl]-1H-indole-2-carboxamides (5i–l) were synthesized and characterized with different analytical tools. N-Acetylisatines 4a–d were subjected to ring opening at their C2 carbons with the aid of different indole-bearing hydrazides 3a,b and 7 to afford the respective glyoxylamides 5a–l. The antimicrobial activity of the target compounds 5a–l was assessed with the aid of Diameter of the Inhibition Zone (DIZ) and Minimum Inhibitory Concentration (MIC) assays against a panel of Gram-positive and Gram-negative bacteria and certain fungal strains. The antimicrobial screening revealed that Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, and Candida albicans are the most sensitive microorganisms towards the synthesized compounds 5a–l. In addition, compounds 5c and 5h emerged as the most active congeners towards Staphylococcus aureus and Candida albicans, respectively. Molecular docking studies revealed the possible binding mode of compounds 5c and 5h to their target proteins.
Keywords: indole, N-Acetylisatins, ring opening, antimicrobial, glyoxylamides
1. Introduction
Indole is a hetero-aromatic bicyclic ring system and indoles represent an important class in drug discovery and development process [1]. Indole-bearing compounds are commonly identified and isolated from natural resources and are widely used as precursors in fine organic synthesis to develop new pharmacological lead pharmaceuticals across a broad range of therapeutic areas [2,3,4]. Biological activities exhibited by different indole derivatives include anti-inflammatory [5], anticancer [6], antihypertensive [7], antiviral [8], antibacterial [9] and antifungal activities [10].
In the same vein, 5-methoxyindole constitutes the backbone of the natural hormone melatonin, which helps sleep regulation and wake cycles through manipulation of three different melatoninergic receptors (M1–M3) [11,12]. Melatonin exhibits various therapeutic applications like anti-inflammatory, antioxidant [13], and antitumor activities [14]. In addition, 5-methoxyindole fragment was incorporated in a number of bioactive melatoninergic ligands [15,16].
2,3-Dioxindole (isatin) is another heterocyclic aromatic nucleus that was identified as an endogenous compound in humans and other mammals [17]. Isatin has a broad synthetic utility owing to its incorporation of reactive function groups which have been functionalized to prepare diverse bioactive molecules such as anticancers [18], anticonvulsants [19], and antimicrobials [20]. Moreover, N-acetylisatins are privileged structures in drug discovery and development processes due to their facile utility to prepare the corresponding glyoxylamides via attacking their C2-carbonyl functionality with different nucleophilic amines. The presence of two carbonyl groups with two different spatial orientations in glyoxylamides significantly enhances their H-bonding with protein targets and hence improving their biological activities. In addition, glyoxylamide derivatives have broad applications in organic chemistry and they are incorporated in a vast of bioactive molecules [21,22,23,24].
Therefore, the aforementioned premises encouraged us to synthesize the title glyoxylamides 5a–l via opening certain N-acetylisatines 4a–d with different indole-bearing hydrazides 3a,b and 7. The antimicrobial profile of the title compounds 5a–l was in vitro evaluated against a panel of microorganisms including Gram-positive and Gram-negative bacteria as well as filamentous and non-filamentous fungi.
2. Results and Discussion
2.1. Chemistry
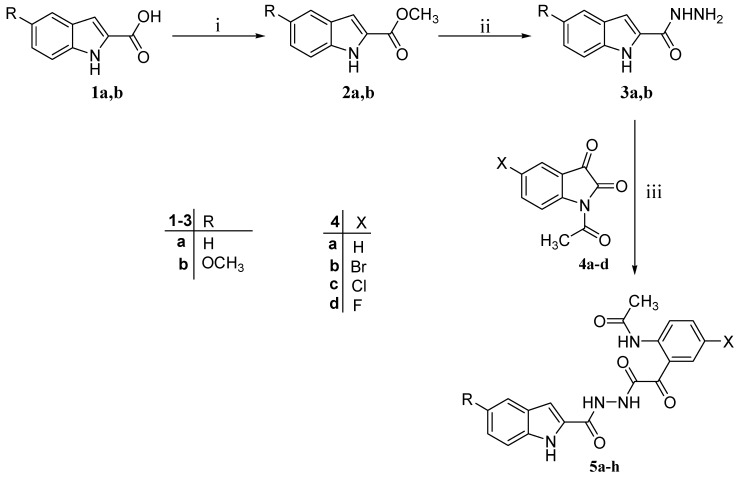
The target compounds 5a–l were successfully achieved as portrayed in Scheme 1 and Scheme 2. Thus, N-acetylisatines 4a–d were allowed to react with the appropriate hydrazide 3a,b in acetonitrile, a polar non-nucleophile solvent, to afford the respective targets 5a–h (Scheme 1). NMR (1H and 13C) as well as mass spectral data of compounds 5a–h are consistent with their proposed chemical structures. The single crystal X-ray structure of compound 5c [25], as a representative example of compounds 5a–h, confirmed doubtlessly the assigned chemical structures of 5a–h.
Scheme 1.
Synthesis of the target compounds 5a–h. Reagents and conditions: (i) Methanol, drops of H2SO4, reflux, 4 h; (ii) Methanol, H2N-NH2.H2O, reflux, 2 h; and (iii) Acetonitrile, reflux 2 h.
| Compound No. | R | X |
| 5a | H | H |
| 5b | H | Br |
| 5c | H | Cl |
| 5d | H | F |
| 5e | OCH3 | H |
| 5f | OCH3 | Br |
| 5g | OCH3 | Cl |
| 5h | OCH3 | F |
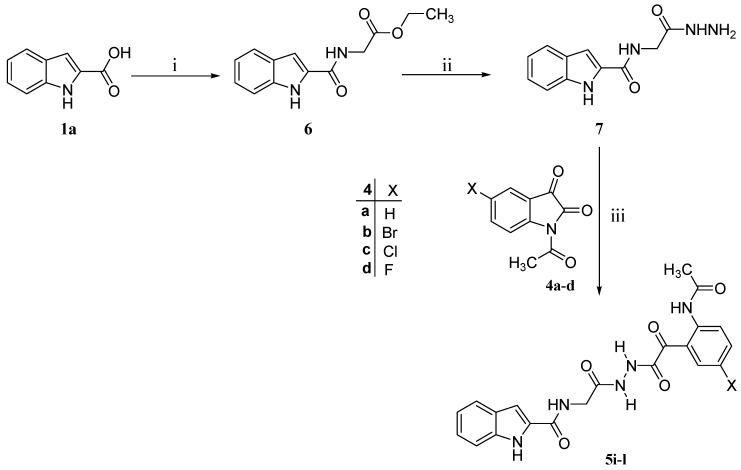
Scheme 2.
Synthesis of the target compounds 5i–l. Reagents and conditions: (i) Tetrahydrofuran, ethyl glycinate hydrochloride, carbonyldiimidazole, rt, 18 h; (ii) Methanol, H2N-NH2.H2O, reflux, 2 h; and (iii) Acetonitrile, reflux 2 h.
| Compound No. | R | X |
| 5i | H | H |
| 5j | H | Br |
| 5k | H | Cl |
| 5l | H | F |
Ethyl glycinate was coupled with the commercially available indole-2-carboxylic acid (1a) in the presence of carbonyldiimidazole to furnish the coupled product 6 (Scheme 2). Hydrazinolysis of the ester functionality of compound 6 with hydrazine hydrate yielded the respective hydrazide 7. Subsequently, N-acetylisatines 4a–d were allowed to react with hydrazide 7 to give the corresponding target compounds 5i–l (Scheme 2). The assigned chemical structures of compounds 5i-l were confirmed via their NMR (1H and 13C) and mass spectral data.
2.2. Antimicrobial Evaluation
The title glyoxylamides 5a–l were divided into two sets, the first set contains compounds 5a–h in which either indole hydrazide 3a or 5-methoxyindole hydrazide 3b was used for ring opening of N-acetylisatin derivatives 4a–d. The second set comprises compounds 5i–l in which the hydrazide 7 was used for ring opening of compounds 4a–d.
Table 1 presented the results of the preliminary antimicrobial activity of the title compounds 5a–l against certain Gram-positive and Gram-negative bacteria as well as certain fungi using Diameter of the Inhibition Zone (DIZ) assay. Compounds 5c and 5h manifested the best activity against the tested Gram-positive bacteria with DIZ values of 21 and 22 mm towards S. aureus and B. subtilis, respectively. On the other hand, compounds 5b and 5f showed the best activity against the tested Gram-negative bacteria with DIZ values of 19 and 18 mm against E. coli and Ps. Aeruginosa, respectively. C. albicans was the most sensitive fungus towards compound 5h with DIZ value of 25 mm.
Table 1.
Diameter of the Inhibition Zone (DIZ) of the title compounds 5a–l, AMP, and FLC against Gram-positive bacteria, Gram-negative bacteria, and fungi.
| Compound No. | DIZ in mm ± S.D.* | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | ||||||||||||
| Gram-Positive Bacteria | Gram-Negative Bacteria | Fungi | ||||||||||
|
B. Subtilis |
E. Fecalis |
MRSA |
S. Aureus |
E. Coli |
K. Pneumonia |
P. Vulgaris |
Ps. Aeruginosa |
S. Enteridis |
A. Niger |
C. Albicans |
P. Notatum |
|
| 5a | 9 ± 0.0 | 9 ± 1.0 | 9 ± 0.0 | 19 ± 0.0 | 13 ± 0.9 | −ve | −ve | 13 ± 0.4 | 9 ± 0.0 | 8 ± 0.0 | 11 ± 0.4 | 15 ± 0.1 |
| 5b | 15 ± 1.0 | 9 ± 0.0 | −ve | 19 ± 0.0 | 19 ± 1.1 | 11 ± 0.6 | −ve | 14 ± 0.3 | 9 ± 0.0 | 8 ± 0.0 | 10 ± 0.3 | 15 ± 0.7 |
| 5c | 15 ± 0.8 | 9 ± 0.0 | 11 ± 0.4 | 21 ± 0.7 | 13 ± 0.3 | −ve | −ve | 9 ± 0.0 | 9 ± 0.0 | 17 ± 0.4 | 8 ± 0.0 | 18 ± 0.3 |
| 5d | 16 ± 0.2 | 18 ± 1.3 | 11 ± 0.0 | 14 ± 0.3 | 14 ± 0.8 | 11 ± 0.3 | −ve | 11 ± 0.3 | 9 ± 0.0 | 8 ± 0.0 | 14 ± 0.2 | 8 ± 0.0 |
| 5e | 11 ± 0.8 | 11 ± 0.0 | −ve | −ve | 13 ± 0.44 | −ve | −ve | 9 ± 0.2 | 9 ± 0.0 | 15 ± 0.7 | 14 ± 1.1 | 9 ± 0.5 |
| 5f | 9 ± 0.0 | 11 ± 0.6 | 13 ± 1.6 | 16 ± 0.5 | 14 ± 0.0 | −ve | −ve | 18 ± 0.5 | 15 ± 0.7 | 8 ± 0.0 | 18 ± 0.0 | 15 ± 1.6 |
| 5g | 9 ± 0.4 | 9 ± 0.5 | 11 ± 0.0 | 14 ± 0.4 | 15 ± 1.0 | −ve | −ve | 9 ± 0.0 | 9 ± 0.0 | 17 ± 0.1 | 18 ± 0.0 | 14 ± 1.0 |
| 5h | 22 ± 1.6 | 14 ± 0.3 | 11 ± 0.5 | 14 ± 0.4 | 14 ± 0.3 | −ve | 11 ± 0.7 | 11 ± 0.6 | 9 ± 0.0 | 11 ± 0.1 | 25 ± 1.6 | 16 ± 0.5 |
| 5i | 13 ± 0.6 | 12 ± 0.9 | −ve | 19 ± 0.0 | 17 ± 0.6 | −ve | −ve | 14 ± 0.5 | 16 ± 0.2 | 8 ± 0.0 | 11 ± 0.2 | 18 ± 1.2 |
| 5j | 12 ± 0.6 | 11 ± 0.2 | −ve | 9 ± 0.2 | 13 ± 0.2 | −ve | 11 ± 0.2 | 9 ± 0.7 | 9 ± 0.0 | 16 ± 0.5 | 13 ± 0.3 | 14 ± 0.5 |
| 5k | 14 ± 0.4 | 11 ± 0.5 | −ve | 9 ± 0.0 | 14 ± 0.0 | −ve | −ve | 9 ± 0.6 | 9 ± 0.1 | 8 ± 0.0 | 13 ± 1.0 | 16 ± 0.4 |
| 5l | 15 ± 0.4 | 9 ± 0.8 | −ve | 9 ± 0.0 | 14 ± 0.0 | −ve | −ve | 12 ± 0.8 | 9 ± 0.0 | 8 ± 0.0 | 15 ± 0.7 | 14 ± 0.5 |
| AMP | 30 ± 0.0 | −ve | 36 ± 0.7 | −ve | 45 ± 1.0 | 32 ± 0.4 | 18 ± 0.4 | 35 ± 1.0 | 30 ± 0.5 | ND | ND | ND |
| FLC | ND | ND | ND | ND | ND | ND | ND | ND | ND | 21 ± 0.5 | 16 ± 0.8 | 15 ± 0.0 |
The results of the Minimum Inhibitory Concentration (MIC) assay for the target compounds 5a–l are presented in Table 2. S. aureus is the most sensitive Gram-positive bacteria towards the tested compounds 5a–l with MIC values of 3.9, 31.25, and 62.5 μg/mL for compounds 5c, 5d and 5b or 5i, respectively. Also, B. subtilis was sensitive to compound 5h with MIC value of 62.5 μg/mL. Regarding the tested strains of Gram-negative bacteria, compounds 5d, 5i, and 5k are the best candidates towards E. coli (compounds 5i and 5k) and compound 5d towards Ps. Aeruginosa being equipotent with an MIC value of 62.5 μg/mL. Compound 5h manifested the best antifungal profile for the whole synthesized series 5a–l as it showed MIC values of 7.8, 31.25, and 62.5 μg/mL against C. albicans, A. niger, and P. notatum, respectively.
Table 2.
Minimum Inhibitory Concentrations (MICs) of the title compounds 5a–l, AMP, and FLC against Gram-positive bacteria, Gram-negative bacteria, and fungi.
| MIC Values (μg/mL) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound No. | Strain Name | ||||||||||||
| Gram-Positive Bacteria | Gram-Negative Bacteria | Fungi | |||||||||||
|
B. Subtilis |
E. Fecalis |
MRSA |
S. Aureus |
E. Coli |
K. Pneumonia | P. Vulgaris | Ps. Aeruginosa | S. Enteridis |
A. Niger |
C. Albicans | P. Notatum | ||
| 5a | 500 | 500 | 500 | 125 | 125 | 500 | 250 | 125 | 500 | 250 | 250 | 125 | |
| 5b | 125 | 500 | 500 | 62.5 | 125 | 500 | 250 | 250 | 250 | 62.5 | 250 | 250 | |
| 5c | 250 | 500 | 500 | 3.9 | 125 | 500 | 250 | 125 | 125 | 250 | 250 | 250 | |
| 5d | 250 | 250 | >1000 | 31.25 | 125 | 500 | 250 | 62.5 | 250 | 15.6 | 62.5 | 62.5 | |
| 5e | 250 | 500 | 500 | 500 | 125 | 500 | 250 | 250 | 250 | 250 | 125 | 125 | |
| 5f | 250 | 500 | 500 | 250 | 125 | 500 | 250 | 250 | 500 | 250 | 125 | 250 | |
| 5g | 250 | 500 | 500 | 250 | 125 | 500 | 250 | 250 | 250 | 125 | 31.25 | 125 | |
| 5h | 62.5 | 125 | 500 | 250 | 250 | 500 | 250 | 250 | 500 | 31.25 | 7.8 | 62.5 | |
| 5i | 500 | 1000 | 500 | 62.5 | 62.5 | 500 | 250 | 125 | 125 | 125 | 250 | 250 | |
| 5j | 500 | 500 | 500 | 500 | 125 | 500 | 250 | 125 | 500 | 125 | 250 | 250 | |
| 5k | 500 | 500 | 500 | 500 | 62.5 | 500 | 1000 | 125 | 250 | 250 | 250 | 250 | |
| 5l | 250 | 1000 | 500 | 500 | 125 | 250 | 125 | 125 | 250 | 250 | 250 | 250 | |
| AMP | 15.6 | >1000 | <7.8 | >1000 | <7.8 | 250 | 500 | 3.9 | 1000 | ND | ND | ND | |
| FLC | ND | ND | ND | ND | ND | ND | ND | ND | ND | 15.6 | 31.25 | 250 | |
In summary, it can be deduced from the above antimicrobial screening tests that S. aureus, E. coli, Ps. Aeruginosa, and C. albicans are the most sensitive microorganisms towards the synthesized compounds 5a–l. Compound 5c bearing indole hydrazide 3a fragment and a chloro substituent in the first set 5a–h, is the best candidate against S. aureus. In the same set, compound 5h bearing 5-methoxyindole hydrazide 3b fragment and a fluoro substituent and is the most active congener towards both B. subtilis and the tested three fungal strains. On the other hand, compounds 5i and 5k, bearing indole hydrazide 7 fragment in the second set 5i–l, are the most active compounds against E. coli being equipotent, while compound 5d is the most active candidate towards Ps. Aeruginosa.
2.3. Molecular Docking
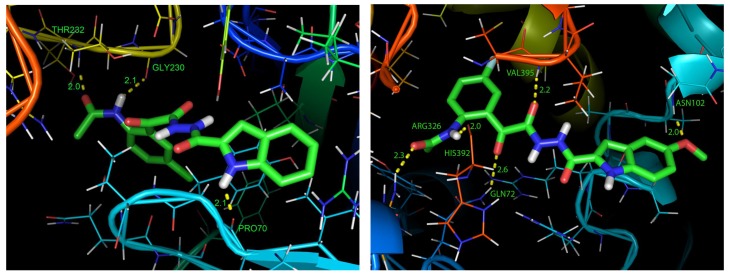
The three-dimensional (3D) structural coordinates of compounds 5c and 5h were sketched by ChemDraw Ultra 7.0.1 program [26]. The energy of compounds 5c and 5h was minimized using PRODRG online server on basis of GROMACS force field method [27]. The antibacterial (PDB ID: 4DH6) [28] and antifungal (PDB ID: 1EA1) [29] target proteins have been chosen for the docking study for compounds 5c and 5h, respectively. The 3D structural coordinates of the target proteins were downloaded from the RCSB protein data bank [30]. The target proteins manipulation has been carried out by following steps: (i) all water molecules were removed; (ii) hydrogen atoms were added to the crystal structure; (iii) Kollaman′s charges were added; and (iv) the docked inhibitors were removed from the target proteins. The protein (rigid) and ligand (flexible) docking was performed with the aid of AutoDock 4.2 [31] program interfaced with MGL Tools 1.5.6 rc3 [32] to create affinity grids centered on the active site with 90 × 90 × 90 grid size. The results of the predicted free binding energy (∆E), inhibition constant (Ki) as well as the bounded amino acid resides of the complex are given in Table 3. The docking results were evaluated by sorting the free binding energies predicted by their docking conformations. The best conformation binding energy is predicted to be –7.88 kcal/mol with Ki value of 1.67 μM for compound 5c. Its hydrogen bonding interactions were observed with THR232, GLY230, and PRO70 amino acid residues of 4DH6 target protein (Figure 1).
Table 3.
Binding energies and inhibition constants results of compounds 5c and 5h with their target proteins.
| Protein ID | Binding Energy (∆E) [kcal/mol] |
Estimated Inhibition Constant (Ki) [μM] |
Bounded Residues |
|---|---|---|---|
| 4DH6 | –7.88 | 1.67 | THR232, GLY230 and PRO70 |
| 1EA1 | –7.28 | 4.63 | ARG326, HIS392, GLN72, VAL395, and ASN102 |
Figure 1.
Hydrogen bonding projection of protein-ligand interactions and binding pose for compound 5c (left) and compound 5h (right).
On the other hand, the best conformation binding energy is predicted to be –7.28 kcal/mol with Ki value of 4.63 μM for compound 5h. Its hydrogen bonding interactions were noted with ARG326, HIS392, GLN72, VAL395, and ASN102 amino acid residues of 1EA1 target protein (Figure 1). The molecular docking investigations manifested the possible binding pose of compounds 5c and 5h inside their target bacterial and fungal proteins, respectively.
3. Experimental
3.1. General
A Gallenkamp device was used to measure melting points and they are uncorrected. Bruker NMR spectrometer (Bruker, Reinstetten, Germany) was used to record the NMR spectra of the synthesized compounds 5a–l in DMSO-d6 at 500 MHz for 1H and 125.76 MHz for 13C at the Research Center, College of Pharmacy, King Saud University, Saudi Arabia. Chemical shifts are expressed in δ-values (ppm) relative to TMS as an internal standard. Elemental analyses were carried out at Microanalysis Laboratory, Cairo University, Cairo, Egypt and the results agreed favorably with the proposed structures within ± 0.4% of the theoretical values. Agilent Quadrupole 6120 LC/MS with ESI (Electrospray ionization) source (Agilent Technologies, Palo Alto, CA, USA) was used to record mass spectra of the synthesized compounds. High-resolution mass spectrometry (HR-MS) measurements were performed on an LTQ-Orbitrap XL coupled to matrix-assisted laser desorption ionization (MALDI). Compounds 2a,b [33], 3a,b [34], 4a–d [21], and 6 [35] were prepared according to literature procedures. Ampicillin (AMP) was obtained from Sigma-Aldrich Co. (St. Louis, MO, USA) and fluconazole (FLC) was purchased from Shouguang-Fukang Pharmaceutical Ltd. (Shandong, China).
3.2. Chemistry
Methyl 1H-indole-2-carboxylate (2a): White powder; melting point (m.p.) 150–151 °C [33].
Methyl 5-methoxy-1H-indole-2-carboxylate (2b): Yellow powder; m.p. 176–177 °C [36].
1H-Indole-2-carbohydrazide (3a): Off-White powder; m.p. 251–253 °C [37].
5-Methoxy-1H-indole-2-carbohydrazide (3b): Off-White powder; m.p. 266–268 °C [38].
1-Acetyl-1H-indole-2,3-dione (4a): Yellow crystals; m.p. 141–143 °C [39].
1-Acetyl-5-bromo-1H-indole-2,3-dione (4b): Brown powder; m.p. 167–169 °C [21].
1-Acetyl-5-chloro-1H-indole-2,3-dione (4c): Light brown powder; m.p. 240–242 °C [40].
1-Acetyl-5-fluoro-1H-indole-2,3-dione (4d): Yellow powder; m.p. 147-149 °C [21].
3.2.1. General Procedure for the Synthesis of the Target Compounds 5a–h
The appropriate N-acetylisatin 4a–d (1 mmol) was added to a suspension containing the proper acid hydrazide 3a,b (1 mmol) in acetonitrile (15 mL). The reaction mixture was heated to reflux for two hours, cooled to room temperature, and filtered. The collected solid was dried and re-crystallized from ethanol to give the title compounds 5a–h.
N-(2-{[2-(1H-Indol-2-ylcarbonyl)hydrazinyl](oxo)acetyl}phenyl)acetamide (5a): Yellow powder; m.p. 249–250 °C (yield 58%); 1H-NMR (DMSO-d6): δ (ppm) 2.20 (s, 3H, CH3), 7.09 (t, J = 7.5 Hz, 1H, Ar-H), 7.24 (t, J = 7.5 Hz, 1H, Ar-H), 7.27 (d, J = 1.5 Hz, 1H, CH-3-indole), 7.31 (t, J = 7.5 Hz, 1H, Ar-H), 7.47 (d, J = 8.5 Hz, 1H, Ar-H), 7.68 (d, J = 8.0 Hz, 1H, Ar-H), 7.73 (dd, J = 1.5, 8.5 Hz, 1H, Ar-H), 8.11 (dd, J = 1.0, 8.0 Hz, 1H, Ar-H), 8.21 (d, J = 8.0 Hz, 1H, Ar-H), 10.71 (s, 1H, NH), 10.75 (s, 1H, NH), 10.90 (s, 1H, NH), 11.85 (s, 1H, NH-indole); 13C-NMR (DMSO-d6): δ (ppm) 24.9 (CH3), 104.2, 112.9, 120.5, 121.5, 121.9, 122.3, 123.6, 124.4, 127.4, 129.5, 133.5, 135.9, 137.3, 140.3 (Ar-CH and Ar-C), 158.3, 164.5, 169.6, 192.7 (4× C=O); MS m/z (ESI): 363 [M − H]−; HR-MS (MALDI) calcd for C19H16N4O4: 363.1093, found: 363.1028 (M − H).
N-(4-Bromo-2-{[2-(1H-indol-2-ylcarbonyl)hydrazinyl](oxo)acetyl}phenyl)acetamide (5b): Yellow powder; m.p. 261–263 °C (yield 56%); 1H-NMR (DMSO-d6): δ (ppm) 2.19 (s, 3H, CH3), 7.08 (t, J = 7.5 Hz, 1H, Ar-H), 7.24 (t, J = 7.5 Hz, 1H, Ar-H), 7.27 (d, J = 1.5 Hz, 1H, CH-3-indole), 7.49 (d, J = 8.5 Hz, 1H, Ar-H), 7.68 (d, J = 8.0 Hz, 1H, Ar-H), 7.89 (dd, J = 2.0, 8.5 Hz, 1H, Ar-H), 7.96 (d, J = 8.5 Hz, 1H, Ar-H), 8.08 (d, J = 2.5 Hz, 1H, Ar-H), 10.65 (s, 1H, NH), 10.68 (s, 1H, NH), 10.91 (s, 1H, NH), 11.83 (s, 1H, NH-indole); 13C-NMR (DMSO-d6): δ (ppm) 24.7 (CH3), 104.3, 112.9, 114.0, 115.4, 120.2, 122.3, 123.7, 124.4, 127.3, 129.5, 133.4, 134.6, 137.5, 138.6 (Ar-CH and Ar-C), 158.3, 160.6, 169.4, 192.6 (4× C=O); MS m/z (ESI): 441 [M − H]−, 442 [(M + 1) − H]−, 443[(M + 2) − H]−; HR-MS (MALDI) calcd for C19H15BrN4O4: 441.0198, found: 441.0206 (M − H).
N-(4-Chloro-2-{[2-(1H-indol-2-ylcarbonyl)hydrazinyl](oxo)acetyl}phenyl)acetamide (5c): Yellow powder; m.p. 259–260 °C (yield 77%); 1H-NMR (DMSO-d6): δ (ppm) 2.19 (s, 3H, CH3), 7.08 (t, J = 7.5 Hz, 1H, Ar-H), 7.24 (t, J = 7.5 Hz, 1H, Ar-H), 7.27 (d, J = 1.5 Hz, 1H, CH-3-indole), 7.48 (d, J = 8.0 Hz, 1H, Ar-H), 7.68 (d, J = 8.0 Hz, 1H, Ar-H), 7.77 (dd, J = 2.5, 8.5 Hz, 1H, Ar-H), 7.99 (d, J = 2.5 Hz, 1H, Ar-H), 7.02 (d, J = 8.5 Hz, 1H, Ar-H), 10.66 (s, 1H, NH), 10.69 (s, 1H, NH), 10.91 (s, 1H, NH), 11.83 (s, 1H, NH-indole); 13C-NMR (DMSO-d6): δ (ppm) 24.7 (CH3), 104.3, 112.9, 120.5, 122.3, 123.8, 124.4, 124.8, 127.4, 127.7, 129.5, 131.7, 134.9, 137.2, 138.2 (Ar-CH and Ar-C), 160.9, 163.4, 169.7, 190.2 (4 × C=O); MS m/z (ESI): 397 [M − H]−, 398 [(M + 1) − H]−, 399 [(M + 2) − H]−; HR-MS (MALDI) calcd for C19H15ClN4O4: 397.0704, found: 397.0738 (M − H).
N-(4-Fluoro-2-{[2-(1H-indol-2-ylcarbonyl)hydrazinyl](oxo)acetyl}phenyl)acetamide (5d): Yellow powder; m.p. 268–270 °C(yield 73%); 1H-NMR (DMSO-d6): δ (ppm) 2.17 (s, 3H, CH3), 7.08 (t, J = 7.5 Hz, 1H, Ar-H), 7.24 (t, J = 8.0 Hz, 1H, Ar-H), 7.27 (d, J = 1.5 Hz, 1H, CH-3-indole), 7.47 (d, J = 8.5 Hz, 1H, Ar-H), 7.60 (ddd, J = 2.5, 3.0, 8.5 Hz, 1H, Ar-H), 7.68 (d, J = 8.0 Hz, 1H, Ar-H), 7.84 (dd, J = 3.0, 9.5 Hz, 1H, Ar-H), 7.97–7.99 (m, 1H, Ar-H), 10.55 (s, 1H, NH), 10.69 (s, 1H, NH), 10.89 (s, 1H, NH), 11.85 (s, 1H, NH-indole); 13C-NMR (DMSO-d6): δ (ppm) 24.5 (CH3), 104.3, 112.9, 118.4, 118.5, 120.5, 122.2, 122.3, 124.4, 127.4, 129.5, 130.4, 133.2, 135.6, 137.7 (Ar-CH and Ar-C), 161.1, 163.6, 169.6, 190.1 (4× C=O); MS m/z (ESI): 381 [M − H]−; HR-MS (MALDI) calcd for C19H15FN4O4: 381.0999, found: 381.0951 (M − H).
N-{2-[{2-[(5-Methoxy-1H-indol-2yl)carbonyl]hydrazinyl}(oxo)acetyl]phenyl}acetamide (5e): Pale yellow powder; m.p. 228-230 °C (yield 52%); 1H-NMR (DMSO-d6): δ (ppm) 2.19 (s, 3H, CH3), 3.78 (s, 3H, OCH3), 6.90 (dd, J = 2.5, 8.5 Hz, 1H, Ar-H), 7.15 (d, J = 1.0 Hz, 1H, Ar-H), 7.19 (s, 1H, CH-3-indole), 7.31(t, J = 7.5 Hz, 1H, Ar-H), 7.35 (d, J = 9.0 Hz, 1H, Ar-H), 7.72 (t, J = 8.0 Hz, 1H, Ar-H), 8.11 (d, J = 8.0 Hz, 1H, Ar-H), 8.20 (d, J = 8.5 Hz, 1H, Ar-H), 10.66 (s, 1H, NH), 10.75 (s, 1H, NH), 10.89 (s, 1H, NH), 11.71 (s, 1H, NH-indole); 13C-NMR (DMSO-d6): δ (ppm) 24.9 (CH3), 55.7 (OCH3), 102.6, 103.9, 113.7, 115.8, 121.5, 121.9, 123.6, 127.8, 129.8, 132.5, 133.5, 135.9, 140.3, 154.4 (Ar-CH and Ar-C), 160.9, 164.5, 169.6, 192.7 (4× C=O); MS m/z (ESI): 393 [M − H]−; HR-MS (MALDI) calcd for C20H18N4O5: 393.1199, found: 393.1139 (M − H).
N-{4-Bromo-2-[{2-[(5-methoxy-1H-indol-2-yl)carbonyl]hydrazinyl}(oxo)acetyl]phenyl}acetamide (5f): Light brown powder; 248–250 °C (yield 65%); 1H-NMR (DMSO-d6): δ (ppm) 2.18 (s, 3H, CH3), 3.78 (s, 3H, OCH3), 6.89 (dd, J = 2.5, 9.0 Hz, 1H, Ar-H), 7.14 (d, J = 2.0 Hz, 1H, Ar-H), 7.19 (d, J = 1.5 Hz, 1H, CH-3-indole), 7.36 (d, J = 8.5 Hz, 1H, Ar-H), 7.89 (dd, J = 2.0, 8.5 Hz, 1H, Ar-H), 7.97 (d, J = 9.0 Hz, 1H, Ar-H), 8.08 (d, J = 2.5 Hz, 1H, Ar-H), 10.62 (s, 1H, NH), 10.65 (s, 1H, NH), 10.88 (s, 1H, NH), 11.68 (s, 1H, NH-indole); 13C-NMR (DMSO-d6): δ (ppm) 24.7 (CH3), 55.7 (OCH3), 102.5, 103.9, 113.7, 115.4, 115.7, 123.9, 125.0, 127.7, 129.8, 132.5, 134.5, 137.8, 138.6, 154.4 (Ar-CH and Ar-C), 160.9, 163.4, 169.7, 190.1 (4× C=O); MS m/z (ESI): 471 [M − H]−, 472 [(M + 1) − H]−, 473 [(M + 2) − H]−; HR-MS (MALDI) calcd for C20H17BrN4O5: 471.0304, found: 471.0365 (M − H).
N-{4-Chloro-2-[{2-[(5-methoxy-1H-indol-2-yl)carbonyl]hydrazinyl}(oxo)acetyl]phenyl}acetamide (5g): Yellow powder; 255–257 °C (yield 77%); 1H-NMR (DMSO-d6): δ (ppm) 2.18 (s, 3H, CH3), 3.78 (s, 3H, OCH3), 6.89 (dd, J = 2.0, 9.0 Hz, 1H, Ar-H), 7.14 (d, J = 2.5 Hz, 1H, Ar-H), 7.19 (d, J = 2.0 Hz, 1H, CH-3-indole), 7.36 (d, J = 8.5 Hz, 1H, Ar-H), 7.78 (dd, J = 2.5, 9.0 Hz, 1H, Ar-H), 7.99 (d, J = 2.5 Hz, 1H, Ar-H), 8.03 (d, J = 9.0 Hz, 1H, Ar-H), 10.63 (s, 1H, NH), 10.65 (s, 1H, NH), 10.89 (s, 1H, NH), 11.68 (s, 1H, NH-indole); 13C-NMR (DMSO-d6): δ (ppm) 24.7 (CH3), 55.7 (OCH3), 102.5, 103.9, 113.7, 115.7, 123.8, 124.7, 127.6, 127.7, 129.7, 131.7, 132.6, 134.9, 138.3, 154.4 (Ar-CH and Ar-C), 160.9, 163.5, 169.7, 190.2 (4× C=O); MS m/z (ESI): 427 [M − H]−, 428 [(M + 1) − H]−, 429 [(M + 2) − H]−; HR-MS (MALDI) calcd for C20H17ClN4O5: 427.0809, found: 427.0845 (M − H).
N-{4-Fluoro-2-[{2-[(5-methoxy-1H-indol-2-yl)carbonyl]hydrazinyl}(oxo)acetyl]phenyl}acetamide (5h): Yellow powder; 238–240 °C (yield 60%); 1H-NMR (DMSO-d6): δ (ppm) 2.17 (s, 3H, CH3), 3.78 (s, 3H, OCH3), 6.89 (dd, J = 2.5, 9.0 Hz, 1H, Ar-H), 7.14 (d, J = 2.0 Hz, 1H, Ar-H), 7.18 (d, J = 1.5 Hz, 1H, CH-3-indole), 7.35 (d, J = 9.0 Hz, 1H, Ar-H), 7.60 (ddd, J = 2.5, 3.0, 9.0 Hz, 1H, Ar-H), 7.85 (dd, J = 3.0, 9.0 Hz, 1H, Ar-H), 7.99 (dd, J = 1.0, 9.0 Hz, 1H, Ar-H), 10.55 (s, 1H, NH), 10.64 (s, 1H, NH), 10.87 (s, 1H, NH), 11.70 (s, 1H, NH-indole); 13C-NMR (DMSO-d6): δ (ppm) 24.5 (CH3), 55.8 (OCH3), 102.6, 103.9, 113.7, 115.8, 122.2, 122.4, 124.4, 124.5, 127.8, 129.7, 132.6, 135.9, 142.4, 154.4 (Ar-CH and Ar-C), 160.9, 163.7, 169.6, 190.3 (4× C=O); MS m/z (ESI): 411 [M − H]−; HR-MS (MALDI) calcd for C20H17FN4O5: 411.1105, found: 411.1148 (M − H).
Ethyl [(1H-indol-2-ylcarbonyl)amino]acetate (6): White powder; m.p. 222-224 °C [35].
3.2.2. Synthesis of N-(2-Hydrazinyl-2-oxoethyl)-1H-indole-2-carboxamide (7)
Hydrazine hydrate (50 mmol) was added to a suspension containing compound 6 (5 mmol) in methanol (15 mL). The reaction mixture was heated to reflux for three hours under stirring. The cooled reaction mixture was filtered off and dried to furnish compound 7 in 79% yield as a white powder, m.p. 227–229 °C, which was pure enough to be used for further reactions. 1H-NMR (DMSO-d6) δ (ppm): 3.89 (d, J = 6.0 Hz, 2H, CH2), 4.52 (s, 2H, NH2), 7.02–7.06 (m, 1H, Ar-H), 7.10 (s, 1H, CH-3-indole), 7.16–7.30 (m, 1H, Ar-H), 7.43–7.46 (m, 1H, Ar-H), 7.60 (d, J = 7.7 Hz, 1H, Ar-H), 9.76 (t, J = 6.0 Hz, 1H, -CH2-NH), 9.80 (s, 1H, NH), 11.63 (s, 1H, NH); 13C-NMR (DMSO-d6) δ (ppm): 41.3 (CH2), 102.3, 112.8, 120.2, 121.9, 123.6, 127.5, 130.9, 136.8 (Ar-CH and Ar-C), 161.7, 168.8 (2× C=O); MS m/z: 231 [M − H]−; HR-MS (MALDI) calcd for C11H12N4O2: 231.0882, found: 231.0808 (M − H).
3.2.3. General Procedure for the Synthesis of the Target Compounds 5i–l
Compounds 5i–l were prepared by adopting the aforementioned procedure for the synthesis for compounds 5a–h.
N-[2-(2-{[2-(Acetylamino)phenyl](oxo)acetyl}hydrazinyl)-2-oxoethyl]-1H-indole-2-carboxamide (5i): Pale yellow powder; m.p. 228–230 °C (yield 61%); 1H-NMR (DMSO-d6): δ (ppm) 2.19 (s, 3H, CH3), 4.08 (d, J = 6.0 Hz, 2H, CH2), 7.07 (t, J = 7.0 Hz, 1H, Ar-H), 7.16–7.24 (m, 1H, Ar-H), 7.27 (s, 1H, CH-3-indole), 7.29–7.32 (m, 1H, Ar-H), 7.46 (t, J = 7.5 Hz, 1H, Ar-H), 7.62–7.68 (m, 1H, Ar-H), 7.69–7.74 (m, 1H, Ar-H), 8.11 (d, J = 8.0 Hz, 1H, Ar-H), 8.20 (d, J = 8.5 Hz, 1H, Ar-H), 8.88 (t, J = 6.0 Hz, 1H, -CH2-NH), 10.70 (s, 1H, NH), 10.74 (s, 1H, NH), 11.63 (s, 1H, NH), 11.85 (s, 1H, NH); 13C-NMR (DMSO-d6): δ (ppm) 24.9 (CH3), 63.4 (CH2), 103.6, 104.2, 112.9, 120.5, 122.3, 123.6, 124.4, 127.4, 127.6, 129.5, 131.8, 135.9, 136.9, 140.2 (Ar-CH and Ar-C), 161.0, 161.9, 168.7, 169.6, 192.7 (5× C=O); MS m/z (ESI): 420 [M − H]−; HR-MS (MALDI) calcd for C21H19N5O5: 420.1307, found: 420.1353 (M − H).
N-[2-(2-{[2-(Acetylamino)-5-bromophenyl](oxo)acetyl}hydrazinyl)-2-oxoethyl]-1H-indole-2-carboxamide (5j): Green-yellow powder; m.p. 238-240 °C (yield 47%); 1H-NMR (DMSO-d6): δ (ppm) 2.13 (s, 3H, CH3), 4.07 (d, J = 6.0 Hz, 2H, CH2), 7.04–7.09 (m, 1H, Ar-H), 7.19–7.24 (m, 1H, Ar-H), 7.27 (s, 1H, CH-3-indole), 7.45–7.49 (m, 1H, Ar-H), 7.64–7.69 (m, 1H, Ar-H), 7.84–7.86 (m, 1H, Ar-H), 7.89–7.95 (m, 1H, Ar-H),7.98 (d, J = 2.0 Hz, 1H, Ar-H), 8.89 (t, J = 6.0 Hz, 1H, -CH2-NH), 10.27 (s, 1H, NH), 10.59 (s, 1H, NH), 11.63 (s, 1H, NH), 11.82 (s, 1H, NH); 13C-NMR (DMSO-d6): δ (ppm) 24.7 (CH3), 63.4 (CH2), 103.6, 112.8, 115.4, 122.1, 122.3, 123.9, 124.0, 125.2, 127.5, 129.8, 131.8, 134.5, 137.7, 138.5 (Ar-CH and Ar-C), 161.9, 162.9, 168.7, 169.7, 189.9 (5× C=O); MS m/z (ESI): 498 [M − H]−, 499 [(M + 1) − H]−, 500 [(M + 2) − H]–; HR-MS (MALDI) calcd for C21H18BrN5O5: 498.0413, found: 498.0467 (M − H).
N-[2-(2-{[2-(Acetylamino)-5-chlorophenyl](oxo)acetyl}hydrazinyl)-2-oxoethyl]-1H-indole-2-carboxamide (5k): Yellow powder; m.p. 237–239 °C (yield 72%); 1H-NMR (DMSO-d6): δ (ppm) 2.14 (s, 3H, CH3), 4.07 (d, J = 6.0 Hz, 2H, CH2), 7.04–7.09 (m, 1H, Ar-H), 7.19–7.24 (m, 1H, Ar-H), 7.27 (s, CH-3-indole), 7.45–7.49 (m, 1H, Ar-H), 7.64–7.69 (m, 1H, Ar-H), 7.73–7.78 (m, 1H, Ar-H), 7.88 (d, J = 2.0 Hz, 1H, Ar-H),7.97–8.03 (m, 1H, Ar-H), 8.89(t, J = 6.0 Hz, 1H, -CH2-NH),10.59 (s, 1H, NH), 10.66 (s, 1H, NH), 11.63 (s, 1H, NH), 11.83 (s, 1H, NH); 13C-NMR (DMSO-d6): δ (ppm) 24.6 (CH3), 63.4 (CH2), 103.6, 104.3, 112.9, 122.1, 122.3, 123.8, 124.4, 127.4, 127.7, 129.5, 131.8, 134.8, 137.3, 138.2 (Ar-CH and Ar-C), 160.9, 161.9, 168.7, 169.7, 190.0 (5× C=O); MS m/z (ESI): 454 [M − H]–, 455 [(M + 1) − H]–, 456 [(M + 2) – H]–; HR-MS (MALDI) calcd for C21H18ClN5O5: 454.0918, found: 454.0973 (M − H).
N-[2-(2-{[2-(Acetylamino)-5-fluorophenyl](oxo)acetyl}hydrazinyl)-2-oxoethyl]-1H-indole-2-carboxamide (5l): Pale yellow powder m.p. 239–241 °C (yield 58%); 1H-NMR (DMSO-d6): δ (ppm) 2.12 (s, 3H, CH3), 4.07 (d, J = 6.0 Hz, 2H, CH2), 7.04–7.09 (m, 1H, Ar-H), 7.19–7.24 (m, 1H, Ar-H), 7.27 (s, 1H, CH-3-indole), 7.45–7.48 (m, 1H, Ar-H), 7.54–7.60 (m, 1H, Ar-H), 7.64–7.69 (m, 1H, Ar-H), 7.84 (dd, J = 9.0, 3.0 Hz, 1H, Ar-H), 7.97–7.99 (m, 1H, Ar-H), 8.88 (t, J = 6.0 Hz, 1H, -CH2-NH), 10.48 (s, 1H, NH), 10.55 (s, 1H, NH), 11.62 (s, 1H, NH), 11.85 (s, 1H, NH); 13C-NMR (DMSO-d6): δ (ppm) 24.4 (CH3), 63.1 (CH2), 100.0, 103.6, 112.9, 120.5, 122.3, 123.9, 124.4, 127.4, 127.5, 129.5, 131.8, 135.6, 136.9, 140.9 (Ar-CH and Ar-C), 161.0, 161.9, 168.7, 169.5, 192.8 (5× C=O); MS m/z (ESI): 438 [M – H]–; HR-MS (MALDI) calcd for C21H18FN5O5: 438.1213, found: 438.1261 (M − H).
3.3. Antimicrobial Activity
3.3.1. Isolates
The common pathogenic microorganisms were selected: four Gram-positive isolates, namely Bacillus subtilis (B. subtilis), Enterococcus fecalis (E. fecalis), Methicillin resistant Staphylococcus aureus (MRSA), and Staphylococcus aureus (S. aureus); five Gram-negative organisms, namely Escherichia coli (E. coli), Klebsiella pneumonia (K. pneumonia), Proteus vulgaris (P. vulgaris), Pseudomonas aeruginosa (Ps. Aeruginosa), and Salmonella enteridis (S. enteridis); and three fungal isolates, namely Asperagillus niger (A. niger), Candida albicans (C. albicans), and Penicillum notatum (P. notatum). All isolates were obtained from King Khaled Hospital, Riyadh, Saudi Arabia.
3.3.2. Disk Diffusion Assay
Disk diffusion assay for the title compounds 5a–l was carried out at 1000 μg/mL concentration as previously reported [34] (Supplementary Materials: Antimicrobial Activity).
3.3.3. Determination of Minimum Inhibitory Concentrations (MICs)
The MIC values for the title compounds 5a–l and the reference compounds were determined by adopting the previously reported method [34].
4. Conclusions
Opening N-acetylisatins 4a–d with the aid of different hydrazides 3a,b and/or 7 has been successfully achieved to furnish the corresponding glyoxylamides 5a–l. The new acetamides 5a–h and carboxamides 5i–l were characterized with various spectroscopic techniques. In vitro antimicrobial potential of the title glyoxylamides 5a–l was examined using DIZ and MIC assays towards a panel of Gram-positive and Gram-negative bacteria as well as filamentous and non filamentous fungi. S. aureus, E. coli, Ps. Aeruginosa, and C. albicans are the most sensitive microorganisms towards the synthesized compounds 5a–l. Compounds 5b–d bearing indole hydrazide 3a fragment are the most active candidates towards S. aureus. Compound 5h bearing 5-methoxyindole hydrazide 3b moiety manifested the best antifungal profile against the tested three fungal strains being about three-fold more potent than fluconazole. Compounds 5i and 5k bearing indole hydrazide 7 fragment are the most active congeners against E. coli being equipotent with MIC value of 62.5 μg/mL. Molecular docking investigations predicted the possible binding pose of compounds 5c and 5h to their target proteins. It is believed that the results of the current investigation could support the development of new indole-based bioactive glyoxylamides.
Acknowledgments
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for its funding of this research through the Research Group Project No. RG-1438-083.
Supplementary Materials
Supplementary materials are available online.
Author Contributions
Maha S. Almutairi, Reem I. Al-Wabli, and Mohamed I. Attia designed, prepared, and interpreted the spectra of the title compounds. Azza S. Zakaria conducted the microbiological assays and analyzed their results. I. Hubert Joe performed molecular modeling study. Ali S. Abdelhameed carried out high resolution mass analysis. All authors discussed the contents of the manuscript.
Conflicts of Interest
The authors have declared no conflict of interest.
Footnotes
Sample Availability: Samples of the synthesized compounds are available from the corresponding author.
References
- 1.De Sa A., Fernando R., Barreiro E.J., Fraga M., Alberto C. From nature to drug discovery: The indole scaffold as a ‘privileged structure’. Mini Rev. Med. Chem. 2009;9:782–793. doi: 10.2174/138955709788452649. [DOI] [PubMed] [Google Scholar]
- 2.Joshi K.C., Chand P. Biologically active indole derivatives. Pharmazie. 1982;37:1–12. doi: 10.1002/chin.198221355. [DOI] [PubMed] [Google Scholar]
- 3.Horton D.A., Bourne G.T., Smythe M.L. The combinatorial synthesis of bicyclic privileged structures or privileged substructures. Chem. Rev. 2003;103:893–930. doi: 10.1021/cr020033s. [DOI] [PubMed] [Google Scholar]
- 4.Diss L.B., Robinson S.D., Wu Y., Fidalgo S., Yeoman M.S., Patel B.A. Age-related changes in melatonin release in the murine distal colon. ACS Chem. Neurosci. 2013;4:879–887. doi: 10.1021/cn4000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandra T., Garg N., Kumar A. Synthesis and anti-inflammatory activity of indole derivatives. Int. J. ChemTech Res. 2010;2:762–773. [Google Scholar]
- 6.Farghaly A.-R. Synthesis of some new indole derivatives containing pyrazoles with potential antitumor activity. ARKIVOC. 2010;11:177–187. [Google Scholar]
- 7.Monge A., Aldana I., Alvarez T., Losa M., Font M., Cenarruzabeitia E., Lasheras B., Frechilla D., Castiella E., Fernandez-Alvarez E. 1-Hydrazino-4-(3,5-dimethyl-1-pyrazolyl)-5H-pyridazino [4,5-b] indole. A new antihypertensive agent. Eur. J. Med. Chem. 1991;26:655–658. doi: 10.1016/0223-5234(91)90202-X. [DOI] [Google Scholar]
- 8.Zhang M.-Z., Chen Q., Yang G.-F. A review on recent developments of indole-containing antiviral agents. Eur. J. Med. Chem. 2015;89:421–441. doi: 10.1016/j.ejmech.2014.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClay K., Mehboob S., Yu J., Santarsiero B.D., Deng J., Cook J.L., Jeong H., Johnson M.E., Steffan R.J. Indole trimers with antibacterial activity against Gram-positive organisms produced using combinatorial biocatalysis. AMB Expr. 2015;5:38. doi: 10.1186/s13568-015-0125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Sawy E., Bassyouni F., Abu-Bakr S., Rady H., Abdlla M. Synthesis and biological activity of some new 1-benzyl and 1-benzoyl-3-heterocyclic indole derivatives. Acta Pharm. 2010;60:55–71. doi: 10.2478/v10007-010-0004-0. [DOI] [PubMed] [Google Scholar]
- 11.Csernus V., Mess B. Biorhythms and pineal gland. Neuroendocrinol. Lett. 2003;24:404–411. [PubMed] [Google Scholar]
- 12.Nosjean O., Ferro M., Cogé F., Beauverger P., Henlin J.-M., Lefoulon F., Fauchère J.-L., Delagrange P., Canet E., Boutin J.A. Identification of the melatonin-binding site MT3 as the quinone reductase 2. J. Biol. Chem. 2000;275:31311–31317. doi: 10.1074/jbc.M005141200. [DOI] [PubMed] [Google Scholar]
- 13.Tahan G., Gramignoli R., Marongiu F., Aktolga S., Cetinkaya A., Tahan V., Dorko K. Melatonin expresses powerful anti-inflammatory and antioxidant activities resulting in complete improvement of acetic-acid-induced colitis in rats. Dig. Dis. Sci. 2011;56:715–720. doi: 10.1007/s10620-010-1364-5. [DOI] [PubMed] [Google Scholar]
- 14.Lu J.-J., Fu L., Tang Z., Zhang C., Qin L., Wang J., Yu Z., Shi D., Xiao X., Xie F. Melatonin inhibits AP-2β/hTERT, NF-κB/COX-2 and Akt/ERK and activates caspase/Cyto C signaling to enhance the antitumor activity of berberine in lung cancer cells. Oncotarget. 2016;7:2985–3001. doi: 10.18632/oncotarget.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attia M.I., Witt-Enderby P.A., Julius J. Synthesis and pharmacological evaluation of pentacyclic 6a, 7-dihydrodiindole and 2, 3-dihydrodiindole derivatives as novel melatoninergic ligands. Bioorg. Med. Chem. 2008;16:7654–7661. doi: 10.1016/j.bmc.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 16.Attia M.I., Güclü D., Hertlein B., Julius J., Witt-Enderby P.A., Zlotos D.P. Synthesis, NMR conformational analysis and pharmacological evaluation of 7, 7a, 13, 14-tetrahydro-6 H-cyclobuta[b]pyrimido[1, 2-a: 3, 4-a′]diindole analogues as melatonin receptor ligands. Org. Biomol. Chem. 2007;5:2129–2137. doi: 10.1039/B705550A. [DOI] [PubMed] [Google Scholar]
- 17.Pandeya S.N., Smitha S., Jyoti M., Sridhar S.K. Biological activities of isatin and its derivatives. Acta Pharm. 2005;55:27–46. [PubMed] [Google Scholar]
- 18.Zou H., Zhang L., Ouyang J., Giulianotti M.A., Yu Y. Synthesis and biological evaluation of 2-indolinone derivatives as potential antitumor agents. Eur. J. Med. Chem. 2011;46:5970–5977. doi: 10.1016/j.ejmech.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Bhattacharya S.K., Chakrabarti A. Dose-related proconvulsant and anticonvulsant activity of isatin, a putative biological factor, in rats. Indian j. Exp. Biol. 1998;36:118–121. [PubMed] [Google Scholar]
- 20.Pandeya S., Sriram D., Nath G., De Clercq E. Synthesis, antibacterial, antifungal and anti-HIV evaluation of Schiff and Mannich bases of isatin derivatives with 3-amino-2-methyl mercapto quinazolin-4(3H)-one. Pharm. Acta Helv. 1999;74:11–17. doi: 10.1016/S0031-6865(99)00010-2. [DOI] [PubMed] [Google Scholar]
- 21.Aldilla V.R., Nizalapur S., Martin A., Marjo C.E., Rich A., Yee E., Suwannakot P., Black D.S., Thordarson P., Kumar N. Design, synthesis, and characterisation of glyoxylamide-based short peptides as self-assembled gels. New J. Chem. 2017;41:13462–13471. doi: 10.1039/C7NJ02248D. [DOI] [Google Scholar]
- 22.Chen J., Cunico R.F. Synthesis of α-Ketoamides from a carbamoylsilane and acid chlorides. J. Org. Chem. 2004;69:5509–5511. doi: 10.1021/jo040164o. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y.-H., Zhang Y.-H., Zhang H.-J., Liu D.-Z., Gu M., Li J.-Y., Wu F., Zhu X.-Z., Li J., Nan F.-J. Design, synthesis, and biological evaluation of isoquinoline-1,3,4-trione derivatives as potent caspase-3 inhibitors. J. Med. Chem. 2006;49:1613–1623. doi: 10.1021/jm050896o. [DOI] [PubMed] [Google Scholar]
- 24.James D.A., Koya K., Li H., Liang G., Xia Z., Ying W., Wu Y., Sun L. Indole-and indolizine-glyoxylamides displaying cytotoxicity against multidrug resistant cancer cell lines. Bioorg. Med. Chem. Lett. 2008;18:1784–1787. doi: 10.1016/j.bmcl.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 25.Al-Wabli R.I., Salman A., Shyni V., Ghabbour H.A., Joe I.H., Almutairi M.S., Maklad Y.A., Attia M.I. Synthesis, crystal structure, vibrational profiling, DFT studies and molecular docking of N-(4-chloro-2-{[2-(1H-indol-2-ylcarbonyl)hydrazinyl](oxo)acetyl}phenyl)acetamide. DMSO: A new antiproliferative agent. J. Mol. Struct. 2018;1155:457–468. doi: 10.1016/j.molstruc.2017.10.116. [DOI] [Google Scholar]
- 26.Li Z., Wan H., Shi Y., Ouyang P. Personal experience with four kinds of chemical structure drawing software: Review on ChemDraw, ChemWindow, ISIS/Draw, and ChemSketch. J. Chem. Inf. Comput. Sci. 2004;44:1886–1890. doi: 10.1021/ci049794h. [DOI] [PubMed] [Google Scholar]
- 27.SchuÈttelkopf A.W., van Aalten D.M. PRODRG: A tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 28.Kotaiah Y., Nagaraju K., Harikrishna N., Rao C.V., Yamini L., Vijjulatha M. Synthesis, docking and evaluation of antioxidant and antimicrobial activities of novel 1,2,4-triazolo[3,4-b][1,3,4] thiadiazol-6-yl)selenopheno [2,3-d]pyrimidines. Eur. J. Med. Chem. 2014;75:195–202. doi: 10.1016/j.ejmech.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Podust L.M., Poulos T.L., Waterman M.R. Crystal structure of cytochrome P450 14α-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc. Nat. Acad. Sci. USA. 2001;98:3068–3073. doi: 10.1073/pnas.061562898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose P.W., Prlić A., Altunkaya A., Bi C., Bradley A.R., Christie C.H., Costanzo L.D., Duarte J.M., Dutta S., Feng Z., et al. The RCSB protein data bank: Integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017;45:D271–D281. doi: 10.1093/nar/gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanner M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 33.Almutairi M.S., Ghabbour H.A., Attia M.I. Crystal structure of methyl 1H-indole-2-carboxylate, C10H9NO2. Z. Krist. New Cryst. Struct. 2017;232:431–432. doi: 10.1515/ncrs-2016-0303. [DOI] [Google Scholar]
- 34.Al-Wabli R.I., Zakaria A.S., Attia M.I. Synthesis, spectroscopic characterization and antimicrobial potential of certain new isatin-indole molecular hybrids. Molecules. 2017;22:1958. doi: 10.3390/molecules22111958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pigulla J., Röder E. Darstellung von Indol-2-carboxamiden und Estern von (Indol-2-ylcarbonylamino)carbonsäuren nach der Imidazolidmethode. Arch. Pharm. 1979;312:12–18. doi: 10.1002/ardp.19793120104. [DOI] [Google Scholar]
- 36.Attia M.I., Ghabbour H.A., Fun H.-K. Crystal structure of methyl 5-methoxy 1H-indole-2-carboxylate, C11H11NO3. Z. Krist. New Cryst. Struct. 2016;231:313–314. doi: 10.1515/ncrs-2015-0162. [DOI] [Google Scholar]
- 37.Boraei A.T., El Ashry E.S.H., Barakat A., Ghabbour H.A. Synthesis of new functionalized indoles based on ethyl indol-2-carboxylate. Molecules. 2016;21:333. doi: 10.3390/molecules21030333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almutairi M.S., Zakaria A.S., Ignasius P.P., Al-Wabli R.I., Joe I.H., Attia M.I. Synthesis, spectroscopic investigations, DFT studies, molecular docking and antimicrobial potential of certain new indole-isatin molecular hybrids: Experimental and theoretical approaches. J. Mol. Struct. 2018;1153:333–345. doi: 10.1016/j.molstruc.2017.10.025. [DOI] [Google Scholar]
- 39.El-Faham A., Al Marhoon Z., Abdel-Megeed A., Albericio F. OxymaPure/DIC: An efficient reagent for the synthesis of a aovel series of 4-[2-(2-acetylaminophenyl)-2-oxo-acetylamino]benzoyl amino acid ester derivatives. Molecules. 2013;18:14747–14759. doi: 10.3390/molecules181214747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azevedo L.D., Bastos M.M., Vasconcelos F.C., Hoelz L.V., Junior F.P., Dantas R.F., de Almeida A.C., de Oliveira A.P., Gomes L.C., Maia R.C. Imatinib derivatives as inhibitors of K562 cells in chronic myeloid leukemia. Med. Chem. Res. 2017;26:2929–2941. doi: 10.1007/s00044-017-1993-8. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.