Abstract
Exosomal micro (mi)RNAs have been suggested to have important roles in abdominal obesity, and to be associated with metabolic alterations via posttranscriptional regulation of target genes. However, exosomal miRNA profiles in subcutaneous adipose tissue (SAT) and visceral adipose tissue (VAT) have rarely been investigated. In the present study, microarray data were obtained from the Gene Expression Omnibus database with the following accession numbers: GSE68885 (exosomal miRNAs in SAT obtained from seven patients with obesity and five lean patients), GSE50574 (exosomal miRNAs in VAT obtained from seven patients with obesity and five lean patients) and GSE29718 [mRNAs in SAT (obtained from seven patients with obesity and eight lean patients) and VAT (obtained from three patients with obesity and two lean patients)]. Differentially expressed (DE)-miRNAs and differentially expressed genes (DEGs) were identified using the Linear Models for Microarray Data method, and mRNA targets of DE-miRNAs were predicted using the miRWalk2.0 database. Potential functions of DE-miRNA target genes were determined using the Database for Annotation, Visualization and Integrated Discovery. As a result, 10 exosomal DE-miRNAs were identified in SAT between patients with obesity and lean patients, while 58 DE-miRNAs were identified in VAT between patients with obesity and lean patients. miRNA (miR)-4517 was revealed to be a downregulated exosomal miRNA between SAT and VAT, while the other DE-miRNAs were SAT-(e.g. hsa-miR-3156-5p and hsa-miR-4460) or VAT-(e.g. hsa-miR-582-5p, hsa-miR-566 and miR-548) specific. Following overlapping with the target genes of DE-miRNAs, only one DEG [cluster of differentiation 86 (CD86)] was identified in SAT samples, whereas 25 DEGs (e.g. fibroblast growth factor 2 (FGF2), FOS like 2, AP-1 transcription factor subunit (FOSL2); and adenosine monophosphate deaminase 3 (AMPD3)] were identified in VAT samples. CD86 was revealed to be regulated by hsa-miR-3156-5p; whereas FGF2, FOSL2 and AMPD3 were revealed to be regulated by hsa-miR-582-5p, hsa-miR-566 and miR-548, respectively. Functional enrichment analysis demonstrated that these target genes may be associated with inflammation. In conclusion, exosomal miRNAs may represent underlying therapeutic targets for the treatment of abdominal obesity and metabolic disorders via regulation of inflammatory genes.
Keywords: obesity, adipose tissue, metabolic disorders, exosomes, microRNAs, inflammatory
Introduction
Due to the improvement of living standards, greater work stress and reduced exercise intensity, abdominal obesity has been considered to represent a major worldwide public health problem in the 21st century, with an estimated age-adjusted prevalence of ~40.0% worldwide (1,2). Abdominal obesity is associated with an increased risk of the development of numerous metabolic disorders, including diabetes, hypertension, dyslipidemia, hepatic steatosis and cardiovascular diseases, which may result in disability and sudden mortality (3,4). Therefore, it is important to investigate the mechanisms underlying the initiation of abdominal obesity, in addition to associated metabolic alterations, in order to develop novel therapeutic strategies.
Abdominal obesity is characterized by the expansion of adipose tissue (AT) mass via enlargement of existing adipocytes (hypertrophy) and increased numbers of novel adipocytes (hyperplasia) (5). Hypertrophy results in a limited blood supply to each adipocyte, which triggers tissue hypoxia and subsequent stimulation of numerous inflammatory responses, including proliferation and infiltration of M1 macrophages, in addition to the release of numerous pro-inflammatory cytokines, including interleukin (IL)-6, tumor necrosis factor (TNF)-α and IL-1β (6,7). Such inflammatory cytokines may further contribute to adipogenesis (8) and metabolic diseases (9,10). Thus, targeted anti-inflammatory therapies have been suggested to be beneficial for the treatment of obesity and the prevention of associated metabolic disorders (11–13). However, current treatment strategies have been revealed to exhibit limited therapeutic benefits (11–13), suggesting that there are further mechanisms associated with AT inflammation in obesity.
Previous studies have demonstrated the regulatory roles of micro (mi)RNAs and small non-coding RNAs (19–22 nucleotides in length) in inflammatory obesity (14,15). miRNAs function by inhibiting gene expression by binding to complementary sequences in the 3′untranslated region (UTR) of target genes, and may be present intracellularly or secreted extracellularly within nanoparticles, namely exosomes. Compared with endogenous miRNAs, adipocyte exosomes have attracted more attention in recent years due to their function as important mediators of intercellular communication and their ability to transfer phenotypic traits from mature and inflammatory adipocytes into surrounding cells of the same type (e.g. preadipocytes or non-inflammatory adipocytes) or other cells (e.g. liver or muscle cells) (16), thus facilitating novel adipogenesis and the development of metabolic diseases. For example, exosomal miRNA (miR)-450a-5p in AT has been demonstrated to enhance adipogenesis via suppression of Wnt family member 1 inducible signaling pathway protein 2 expression by targeting its 3′UTR (17). Furthermore, miR-155 has been revealed to be overexpressed in obese adipocyte-derived exosomes (16). In addition, it has been demonstrated that miR-155 knockout animals are insulin sensitive and exhibit a greater glucose tolerance compared with control animals (16). Furthermore, a previous study suggested that miR-155 interferes with insulin signaling and insulin-induced glucose uptake in adipocytes via suppression of suppressor of cytokine signaling 1 protein expression levels and subsequent suppression of signal transducer and activator of transcription 6 signaling, which promotes M1 macrophage polarization (18). A previous study also uses miRNA arrays to investigate exosomal miRNA profiles in visceral adipose tissues (VAT) obtained from patients with obesity, and reveal that miR-23b and miR-4429 may be important in the development of obesity by affecting transforming growth factor (TGF)-β signaling pathways (19). However, studies investigating exosomal miRNAs in adipocytes are rare, and, to the best of our knowledge, there have been no clinical studies investigating the use of exosomal miRNAs in adipocytes.
Abdominal ATs associated with complications arising from obesity include subcutaneous adipose tissue (SAT) and VAT (20). Despite VAT having been previously studied, increasing studies have suggested that SAT accumulation may represent a potential predictor for insulin resistance and metabolic syndrome (21–23). Therefore, understanding of which exosomal miRNAs exert important roles in SAT and how these are performed may further the understanding of the pathogenesis associated with abdominal obesity. The aim of the present study was to analyze exosomal miRNA profiles in SAT obtained from patients with obesity and from lean patients, and to preliminarily determine associated functions via target prediction. Furthermore, the microarray data of exosomal miRNAs in VAT were further analyzed using different analytical methods (Student's t-tests vs. three-way analysis of covariance) and loose threshold values [|log fold change (FC)|>0.5 vs. FC≥|1.2| (19)], which were subsequently compared with SAT to determine shared and specific exosomal miRNAs, which may reveal further potential targets for the treatment of patients with obesity and associated metabolic disorders.
Materials and methods
Microarray data
miRNA microarray data were collected from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) under accession nos. GSE68885 and GSE50574 (19), in which the exosomal miRNAs profiles from SAT and VAT obtained from seven patients with obesity and five lean patients (n=12) were investigated. Patients with obesity were recruited from adolescent bariatric surgery programs, and lean subjects were obtained from patients undergoing unrelated abdominal procedures in the Children's National Medical Center (Washington, DC, USA) (19). None of the included patients had been administered any noteworthy medication prior to surgery. SAT was excised from the anterior abdominal wall incision site and VAT was excised from the omentum (19).
mRNA microarray datasets were also obtained from the GEO database with the accession no. GSE29718 (24), which analyzed the mRNA profiles from 15 SAT samples (seven patients with obesity and eight lean patients) and five VAT samples (three patients with obesity and two lean patients). All samples were collected from patients undergoing an elective procedure in the Garvan Institute of Medical Research (Darlinghurst, New South Wales, Australia) wherein abdominal fat could be obtained (24).
Data normalization and identification of differentially expressed (DE)-miRNAs and differentially expressed genes (DEGs)
The series matrix files of the aforementioned datasets and the annotated symbols were downloaded from the Affymetrix Multispecies miRNA-3 Array platform, GPL16384 (19); or [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version] platform, GPL6244 (24) (both Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA; http://www.ncbi.nlm.nih.gov/geo). If numerous probes corresponded to the same gene, the mean value was considered to represent the expression value of this gene. Missing values of probes were imputed using the nearest neighbor averaging method in the impute package (25) in R (v1.0; http://bioconductor.org/packages/release/bioc/html/impute.html) (26) with default parameters. Subsequently, all expression values were logarithmically transformed (base 2) and quantile-normalized using the Bioconductor preprocessCore package (v1.28.0; http://www.bioconductor.org/packages/release/bioc/html/preprocessCore.html) (27).
DE-miRNAs and DEGs between patients with obesity and lean patients were identified using the Linear Models for Microarray Data method (v2.16.4; http://bioconductor.org/packages/release/bioc/html/limma.html) (28) using the Bioconductor R package. P-values were calculated using an unpaired Student's t-test. P<0.05 and |logFC|>0.5 were considered to indicate statistically significant differences regarding DE-miRNA and DEG analyses. In order to determine the intersection of target genes and DEGs, the threshold values of DEGs in SATs were set at P<0.05 and |logFC|>0.45. Heat maps of DE-miRNAs and DEGs were generated using the heatmap R package (v1.0.8; cran.r-project.org/web/packages/pheatmap/index.html) based on Euclidean clustering distances (29). Furthermore, shared and specific SAT and VAT DE-miRNAs were analyzed using a Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Target gene prediction of DE-miRNAs
mRNA targets of DE-miRNAs were predicted using the miRWalk database (v2.0; http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) (30), which is a comprehensive archive providing a collection of predicted and experimentally verified miRNA-target interactions using 12 prediction programs [DIANA-microT (v4.0), DIANA-microT-CDS (v5.0), miRanda-rel (v1.0), mirBridge (v1.0), miRDB (v4.0), miRmap (v1.0), miRNAMap (v1.0), PicTar (v2.0), PITA (v6.0), RNA22 (v2.0), RNAhybrid (v2.1) and Targetscan (v6.2)]. The DE-miRNA-target interactions were visualized using Cytoscape software (v2.8; www.cytoscape.org/) (31).
Functional enrichment analysis
In order to investigate the functions associated with the identified target genes at the functional level, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) online tool (v6.8; http://david.abcc.ncifcrf.gov) (32). P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of exosomal DE-miRNAs between patients with obesity and lean patients
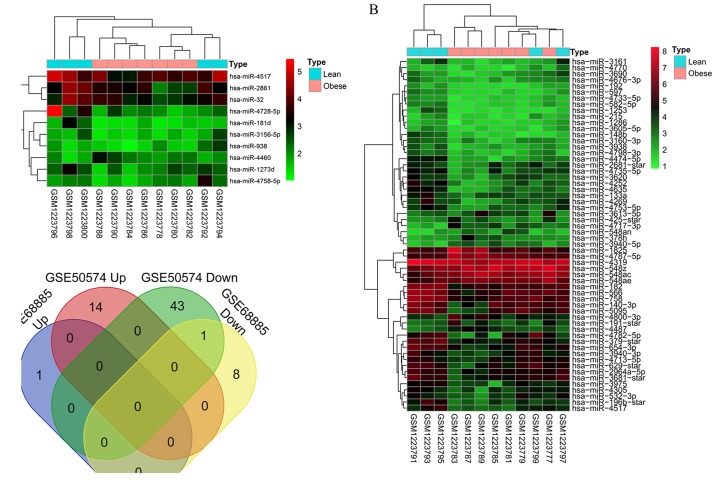
According to the threshold values of P<0.05 and |logFC|>0.5, 10 exosome-derived DE-miRNAs were identified in SATs between patients with obesity and lean patients, including nine downregulated DE-miRNAs (hsa-miR-1273d, hsa-miR-181d, hsa-miR-2861, hsa-miR-3156-5p, hsa-miR-32, hsa-miR-4517, hsa-miR-4728-5p, hsa-miR-4758-5p and hsa-miR-938) and one upregulated DE-miRNA (hsa-miR-4460; Table I); whereas, 58 exosomal DE-miRNAs were identified in VATs, including 44 downregulated DE-miRNAs (e.g. hsa-miR-4517, hsa-miR-532-3p and hsa-miR-582-5p) and 14 upregulated DE-miRNAs (e.g. hsa-miR-4487; Table I). Heatmaps generated by the pheatmap package in R suggested that the identified DE-miRNAs in SAT (Fig. 1A) and VAT (Fig. 1B) were distinguishable between patients with obesity and lean patients. Venn diagram analysis revealed that there was only one shared exosomal DE-miRNA (hsa-miR-4517) in SAT and VAT; whereas, the other nine DE-mRNAs in SAT and 57 DE-miRNAs in VAT were tissue-specific (Fig. 1C).
Table I.
Differentially expressed microRNAs of subcutaneous and visceral adipose tissues between obese and lean subjects.
| A, Subcutaneous adipose tissues | ||
|---|---|---|
| miRNA | logFC | P-value |
| hsa-miR-1273d | −0.597 | 0.032 |
| hsa-miR-181d | −0.662 | 0.023 |
| hsa-miR-2861 | −0.732 | 0.032 |
| hsa-miR-3156-5p | −0.593 | 0.011 |
| hsa-miR-32 | −0.593 | 0.048 |
| hsa-miR-4517 | −0.696 | 0.039 |
| hsa-miR-4728-5p | −1.156 | 0.038 |
| hsa-miR-4758-5p | −0.636 | 0.029 |
| hsa-miR-938 | −0.560 | 0.028 |
| hsa-miR-4460 | 0.556 | 0.018 |
| B, Visceral adipose tissues | ||
| miRNA | logFC | P-value |
| hsa-miR-4733-5p | −0.514 | 0.022 |
| hsa-miR-1286 | −0.514 | 0.019 |
| hsa-miR-4319 | −0.528 | 0.007 |
| hsa-miR-3605-5p | −0.531 | 0.036 |
| hsa-miR-4676-3p | −0.536 | 0.043 |
| hsa-miR-192 | −0.572 | 0.010 |
| hsa-miR-4770 | −0.644 | 0.023 |
| hsa-miR-4713-5p | −0.662 | 0.037 |
| hsa-miR-3160-3p | −0.689 | 0.038 |
| hsa-miR-215 | −0.723 | 0.004 |
| hsa-miR-582-5p | −0.734 | 0.012 |
| hsa-miR-3938 | −0.741 | 0.024 |
| hsa-miR-133a | −0.750 | 0.032 |
| hsa-miR-532-3p | −0.754 | 0.050 |
| hsa-miR-4798-3p | −0.761 | 0.034 |
| hsa-miR-3690 | −0.769 | 0.046 |
| hsa-miR-597 | −0.800 | 0.003 |
| hsa-miR-4305 | −0.865 | 0.033 |
| hsa-miR-2964a-5p | −0.900 | 0.009 |
| hsa-miR-379-star | −0.909 | 0.020 |
| hsa-miR-566 | −0.911 | 0.036 |
| hsa-miR-3940-3p | −0.922 | 0.020 |
| hsa-miR-3620 | −0.942 | 0.025 |
| hsa-miR-196b-star | −0.974 | 0.032 |
| hsa-miR-3975 | −0.986 | 0.023 |
| hsa-miR-1253 | −0.987 | 0.010 |
| hsa-miR-2681-star | −1.048 | 0.011 |
| hsa-miR-182 | −1.048 | 0.003 |
| hsa-miR-4517 | −1.048 | 0.008 |
| hsa-miR-4753-5p | −1.061 | 0.009 |
| hsa-miR-140-3p | −1.097 | 0.047 |
| hsa-miR-629-star | −1.110 | 0.017 |
| hsa-miR-3681-star | −1.110 | 0.005 |
| hsa-miR-4735-5p | −1.114 | 0.022 |
| hsa-miR-148b | −1.131 | 0.008 |
| hsa-miR-4269 | −1.147 | 0.019 |
| hsa-miR-4252 | −1.165 | 0.025 |
| hsa-miR-3161 | −1.172 | 0.008 |
| hsa-miR-654-3p | −1.192 | 0.002 |
| hsa-miR-758 | −1.207 | <0.001 |
| hsa-miR-4635 | −1.258 | 0.013 |
| hsa-miR-5095 | −1.360 | 0.023 |
| hsa-miR-4782-5p | −1.443 | 0.041 |
| hsa-miR-4474-5p | −1.513 | 0.002 |
| hsa-miR-548an | 1.014 | 0.006 |
| hsa-miR-4717-3p | 1.004 | 0.028 |
| hsa-miR-4487 | 1.004 | 0.001 |
| hsa-miR-3613-5p | 0.995 | 0.033 |
| hsa-miR-1825 | 0.942 | 0.042 |
| hsa-miR-191-star | 0.848 | 0.041 |
| hsa-miR-4800-3p | 0.815 | 0.022 |
| hsa-miR-378h | 0.812 | 0.037 |
| hsa-miR-425-star | 0.802 | 0.037 |
| hsa-miR-4787-5p | 0.771 | 0.014 |
| hsa-miR-3940-5p | 0.768 | 0.033 |
| hsa-miR-548ac | 0.695 | 0.040 |
| hsa-miR-548ae | 0.664 | 0.017 |
| hsa-miR-548z | 0.624 | 0.048 |
FC, fold change; miR/miRNA, microRNA.
Figure 1.
Differentially expressed exosomal miRNAs were identified between the patients with obesity and the lean patients. (A) Heat map of DE-miRNAs in the GSE68885 dataset, which included subcutaneous adipose tissue samples obtained from patients with obesity and lean patients (red indicates high expression, green indicates low expression). (B) Heat map of DE-miRNAs in the GSE50574 dataset, which included visceral adipose tissue samples obtained from patients with obesity and lean patients (red indicates high expression, green indicates low expression). (C) The Venn diagram revealed the intersection of DE-miRNAs between subcutaneous and visceral adipose tissues. DE, differentially expressed; miRNA, microRNA.
Target genes for DE-miRNAs
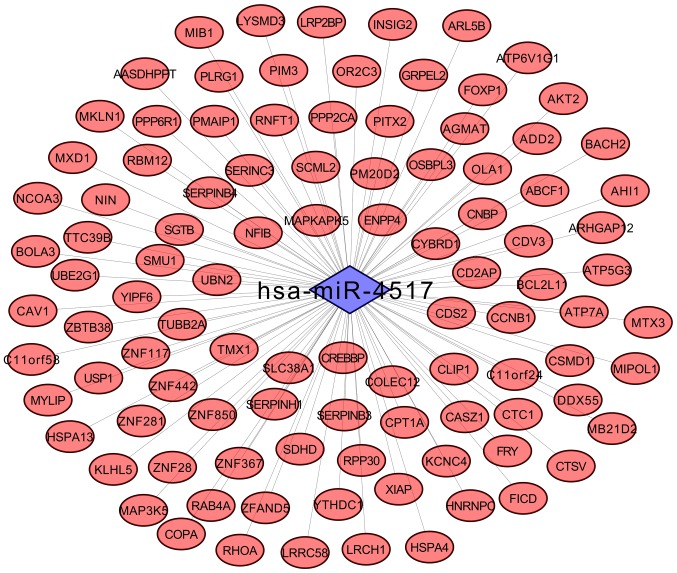
To investigate the potential involvement of the aforementioned DE-miRNAs in the pathogenesis of diseases associated with obesity, 1,622 and 3,384 potential targets were respectively identified to be associated with the 10 and 58 DE-miRNAs in SAT and VAT, respectively, using the miRWalk2 database. Among them, the shared hsa-miR-4517 between SAT and VAT was revealed to regulate 101 target genes, including ras homolog family member A (RhoA; Fig. 2).
Figure 2.
Regulatory network of shared exosomal hsa-microRNA-4517 between subcutaneous and visceral adipose tissues and associated target genes.
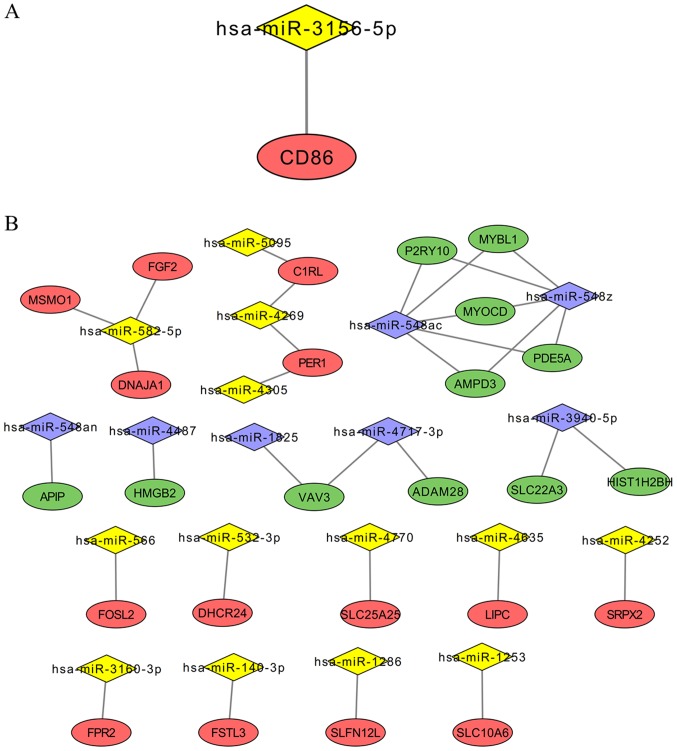
To investigate whether the expression levels of DE-miRNA target genes were differential in patients with obesity and lean patients, mRNA expression profiles in SAT and VAT obtained from patients with obesity and lean patients were also determined. The results revealed that 168 (including 61 upregulated and 107 downregulated) and 487 (including 143 upregulated and 344 downregulated) DEGs were identified in SAT and VAT when comparing patients with obesity to lean patients, respectively (Table II). Following overlapping with the target genes of 10 DE-miRNAs, only one DEG (CD86) with converse expression to the miRNAs remained for SAT. This upregulated DEG may be regulated by downregulated hsa-miR-3156-5p (Fig. 3A). Regarding the VAT samples, 25 DEGs (including 14 upregulated and 11 downregulated) were revealed to be overlapping with the target genes of 58 DE-miRNAs (Table II; Fig. 3B). Upregulated genes in patients with obesity were regulated by downregulated hsa-miR-1253, hsa-miR-1286, hsa-miR-140-3p, hsa-miR-3160-3p, hsa-miR-4252, hsa-miR-4269, hsa-miR-4305, hsa-miR-4635, hsa-miR-4770, hsa-miR-5095, hsa-miR-532-3p, hsa-miR-566 and hsa-miR-582-5p; whereas the downregulated genes in patients with obesity were regulated by upregulated hsa-miR-1825, hsa-miR-3940-5p, hsa-miR-4487, hsa-miR-4717-3p, hsa-miR-548ac, hsa-miR-548z and hsa-miR-548an (Fig. 3B).
Table II.
Differentially expressed genes of subcutaneous and visceral adipose tissues between obese and lean subjects.
| A, Subcutaneous adipose tissues | ||
|---|---|---|
| Gene | logFC | P-value |
| IFNA10 | 0.644 | 0.001 |
| TIGAR | 0.790 | <0.001 |
| FAM72A | 0.501 | 0.002 |
| FCGR2A | 0.640 | 0.003 |
| TM4SF19-TCTEX1D2 | 1.195 | 0.003 |
| CLEC12A | 0.747 | 0.004 |
| NCEH1 | 0.569 | 0.005 |
| AIF1 | 0.519 | 0.008 |
| CD86 | 0.495 | 0.022 |
| FAM105A | 0.406 | 0.027 |
| HP | 0.960 | 0.008 |
| EPDR1 | 0.530 | 0.008 |
| NIPSNAP3B | −0.734 | 0.001 |
| ABHD5 | −0.833 | 0.002 |
| MYOCD | −0.554 | 0.002 |
| PFKFB3 | −0.620 | 0.004 |
| AGTR1 | −0.700 | 0.005 |
| ZBTB16 | −0.894 | 0.006 |
| CYP4B1 | −0.815 | 0.007 |
| TSC22D3 | −0.543 | 0.007 |
| CALCRL | −0.674 | 0.008 |
| ACACB | −0.532 | 0.008 |
| SLC27A2 | −1.470 | 0.010 |
| RDH10 | −0.680 | 0.010 |
| B, Visceral adipose tissues | ||
| Gene | logFC | P-value |
| SLFN12L | 1.039 | 0.003 |
| FOSL2 | 0.835 | 0.005 |
| SRPX2 | 1.216 | 0.008 |
| DHCR24 | 1.411 | 0.012 |
| LIPC | 0.526 | 0.013 |
| PER1 | 0.803 | 0.017 |
| SLC10A6 | 0.566 | 0.018 |
| FPR2 | 1.020 | 0.025 |
| FSTL3 | 0.540 | 0.025 |
| C1RL | 0.518 | 0.026 |
| FGF2 | 0.575 | 0.027 |
| DNAJA1 | 0.576 | 0.031 |
| MSMO1 | 0.705 | 0.039 |
| SLC25A25 | 0.966 | 0.048 |
| MYOCD | −0.845 | 0.004 |
| AMPD3 | −0.663 | 0.010 |
| PDE5A | −0.544 | 0.012 |
| VAV3 | −0.723 | 0.021 |
| P2RY10 | −1.259 | 0.022 |
| MYBL1 | −0.611 | 0.022 |
| APIP | −0.508 | 0.023 |
| HMGB2 | −0.573 | 0.024 |
| HIST1H2BH | −0.544 | 0.038 |
| ADAM28 | −1.283 | 0.043 |
Figure 3.
Important genes associated with DE-miRNAs. (A) The regulatory association between the important DE-miRNA and its associated target gene in subcutaneous adipose tissues. (B) Regulatory associations between important DE-miRNAs and associated target genes in visceral adipose tissues. Diamonds represent miRNAs (yellow represents downregulated miRNAs; purple represents upregulated miRNAs); ovals represent DE target genes (red represents upregulated DE target genes; green represents downregulated DE target genes). DE, differentially expressed; miRNA, microRNA.
Functional enrichment analysis
Target genes of DE-miRNAs were subjected to functional enrichment analysis using DAVID software. As a result, the GO enrichment results demonstrated that the target genes of DE-miRNAs in SAT were involved in ‘positive regulation of transcription, DNA-templated’ (e.g. CD86) and ‘toll-like receptor 3 signaling pathway’ (e.g. CD86; Table III). A total of 28 KEGG pathways were enriched for the target genes of DE-miRNAs in exosomes, including ‘endocytosis’, ‘neurotrophin signaling pathway’, ‘TGF-β signaling pathway’ (e.g. RhoA) and ‘thyroid hormone signaling pathway’ (e.g. mediator complex subunit 13; Table IV).
Table III.
Gene Ontology biological process term enrichment results for target genes of differentially expressed microRNAs in subcutaneous and visceral adipose tissues between obese and lean subjects.
| A, SAT | ||||
|---|---|---|---|---|
| Accession no. | GO terms | P-value | Count | Genes |
| GO:0006351 | Transcription, DNA-templated | <0.001 | 236 | ITGB3BP, PRR13, ATP1B4, XRCC6, ZNF781, ZNF250, ZNF253, CRY2, MED28, MED29 |
| GO:0006355 | Regulation of transcription, DNA-templated | <0.001 | 184 | RALY, ITGB3BP, ZNF584, THRA, PRR13, ATP1B4, ZNF781, CNOT2, ZNF250, ZNF253 |
| GO:0050821 | Protein stabilization | <0.001 | 27 | HSP90AB1, ATP1B3, SOX4, HSPA1B, CALR, PTEN, SUMO1, APOA1, MORC3, CREBL2 |
| GO:0071456 | Cellular response to hypoxia | <0.001 | 20 | ICAM1, ACAA2, CPEB2, TP53, PMAIP1, PTEN, KCNMB1, KCNK3, SUV39H2, SLC29A1 |
| GO:0043161 | Proteasome-mediated ubiquitin-dependent protein catabolic process | <0.001 | 33 | TRIM13, RNF187, RLIM, CD2AP, AMER1, UBXN2A, C18ORF25, UBXN2B, BTBD2, PSMD3 |
| GO:0016567 | Protein ubiquitination | <0.001 | 50 | BACH2, MYLIP, RLIM, ZNRF3, KLHL5, G2E3, ZYG11A, KLHL26, RNF103, KLHL21 |
| GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | <0.001 | 15 | RBL2, TP53, CNOT2, SOX4, AURKA, ATM, CNOT4, CCNB1, CDKN1A, EP300 |
| GO:0097193 | Intrinsic apoptotic signaling pathway | <0.001 | 10 | CDKN1A, CUL5, HRAS, DDX3X, SGPP1, BBC3, CYCS, TP53, APAF1, PMAIP1 |
| GO:0051301 | Cell division | <0.001 | 48 | ITGB3BP, HAUS3, CLTA, SEPT2, MPLKIP, TSG101, BORA, ARF6, AURKA, NR3C1 |
| GO:0045893 | Positive regulation of transcription, DNA-templated | <0.001 | 65 | E2F3, PTGES2, RSF1, FGF7, GPBP1, XRCC6, RNF187, ZKSCAN3, ZXDA, CD86 |
| GO:0034138 | Toll-like receptor 3 signaling pathway | 0.024 | 4 | HAVCR2, CD86, TLR3, COLEC12 |
| B, VAT | ||||
| GO:0045944 | Positive regulation of transcription from RNA polymerase II promoter | 0.001 | 7 | HMGB2, FOSL2, MYOCD, FSTL3, PER1, MYBL1, FGF2 |
| GO:0007283 | Spermatogenesis | 0.013 | 4 | ADAM28, HMGB2, DNAJA1, FSTL3 |
| GO:0045893 | Positive regulation of transcription, DNA-templated | 0.029 | 4 | HMGB2, MYOCD, MYBL1, FGF2 |
| GO:0043388 | Positive regulation of DNA binding | 0.036 | 2 | HMGB2, MYOCD |
| GO:0043552 | Positive regulation of phosphatidylinositol 3-kinase activity | 0.040 | 2 | VAV3, FGF2 |
| GO:0046034 | ATP metabolic process | 0.041 | 2 | SLC25A25, AMPD3 |
| GO:0050918 | Positive chemotaxis | 0.045 | 2 | HMGB2, FGF2 |
| GO:0006695 | Cholesterol biosynthetic process | 0.049 | 2 | MSMO1, DHCR24 |
The top 10 genes are presented for each GO term if the overall count of enriched genes was >10. GO, Gene Ontology; SAT, subcutaneous adipose tissues; VAT, visceral adipose tissues.
Table IV.
KEGG pathways for target genes of differentially expressed microRNAs in subcutaneous and visceral adipose tissues between obese and lean subjects.
| A, SAT | ||||
|---|---|---|---|---|
| KEGG entry | KEGG pathways | P-value | Count | Genes |
| hsa04144 | Endocytosis | <0.001 | 46 | RAB7A, CLTA, CAV1, PARD3, HRAS, LDLR, TSG101, KIAA0196, ASAP1, RhoA |
| hsa04115 | p53 signaling pathway | <0.001 | 18 | CYCS, TP53, IGF1, PMAIP1, CCNG1, SESN2, PTEN, ATM, SESN3, CCNB1 |
| hsa04722 | Neurotrophin signaling pathway | <0.001 | 22 | HRAS, TP53, FASLG, PRKCD, IRAK3, NRAS, MAP3K5, RPS6KA3, CRKL, RHOA |
| hsa04151 | PI3K-Akt signaling pathway | 0.001 | 45 | HSP90AB1, HRAS, FGF7, PHLPP2, MCL1, OSMR, STK11, ITGA11, FASLG, BCL2L11 |
| hsa04919 | Thyroid hormone signaling pathway | 0.004 | 19 | HRAS, KAT2B, THRA, ATP1B3, ATP1B4, CREBBP, TP53, ITGB3, MED13, NRAS |
| hsa04068 | FoxO signaling pathway | 0.011 | 20 | HRAS, RBL2, SGK3, STK11, TGFBR1, CREBBP, IGF1, FASLG, STK4, BCL2L11 |
| hsa04066 | HIF-1 signaling pathway | 0.025 | 15 | CREBBP, MKNK2, IGF1, CDKN1A, EIF4EBP1, EP300, CDKN1B, TFRC, BCL2, PLCG2 |
| hsa04141 | Protein processing in endoplasmic reticulum | 0.031 | 22 | HSP90AB1, MAN1A2, UBE2G1, PDIA6, DNAJB12, HSPA1B, CALR, LMAN1, EDEM1, CANX |
| hsa04152 | AMPK signaling pathway | 0.036 | 17 | SREBF1, STK11, HMGCR, SCD, IGF1, CREB5, ADIPOQ, CPT1A, EIF4EBP1, TSC1 |
| hsa04350 | TGF-β signaling pathway | 0.037 | 13 | SMAD9, SMAD7, TGFBR1, SMAD6, CREBBP, BMPR2, EP300, PPP2CA, RHOA, ID4 |
| hsa04010 | MAPK signaling pathway | 0.037 | 30 | HRAS, FGF7, MAPKAPK5, DUSP10, CACNB1, MKNK2, FASLG, HSPA1B, MAP3K5, MAP3K3 |
| hsa04145 | Phagosome | 0.039 | 20 | MBL2, RAB7A, DYNC1LI2, RAB7B, C3, TUBB2A, HLA-A, COLEC12, ITGB3, ATP6V1G1 |
| B, VAT | ||||
| KEGG pathways | P-value | Count | Genes | |
| hsa00100 | Steroid biosynthesis | 0.040 | 2 | MSMO1, DHCR24 |
The top 10 genes are presented for each KEGG pathway if the overall count of enriched genes was >10. KEGG, Kyoto Encyclopedia of Genes and Genomes; SAT, subcutaneous adipose tissues; VAT, visceral adipose tissues.
Furthermore, the 25 overlapping DEGs regulated by DE-miRNAs in VAT were also subjected to functional enrichment analysis using DAVID software. The results revealed that ‘positive regulation of transcription from RNA polymerase II promoter’ [Fos-like antigen 2 (FOSL2)], ‘positive chemotaxis’ [fibroblast growth factor 2 (FGF2)] and ‘ATP metabolic process’ [adenosine monophosphate deaminase 3 (AMPD3)] were enriched in VAT (Table III).
Discussion
To the best of our knowledge, the present study is the first to have investigated the exosomal miRNA profiles in SAT obtained from patients with obesity and lean patients, the results of which identified 10 DE-miRNAs, including nine downregulated DE-miRNAs (hsa-miR-1273d, hsa-miR-181d, hsa-miR-2861, hsa-miR-3156-5p, hsa-miR-32, hsa-miR-4517, hsa-miR-4728-5p, hsa-miR-4758-5p and hsa-miR-938) and one upregulated DE-miRNA (hsa-miR-4460). The majority of DE-miRNAs identified in SAT were not identified in VAT, further revealing the difference between visceral and subcutaneous adipocytes in abdominal obesity, which was consistent with previous studies (33,34). Only one exosomal miRNA, hsa-miR-4517, was demonstrated to be shared between SAT and VAT. This miRNA was also identified by Ferrante et al (19), thus demonstrating its importance in obesity. Notably, hsa-miR-4517 was predicted to inhibit the expression of RhoA in the present study. Rho-kinase functions as an important regulator of inflammation, proliferation and fibrosis via activation of the mitogen-activated protein kinase (MAPK)/extracellular signa-regulated kinase, nuclear factor (NF)-κB subunit and p38MAPK pathways (35–37). Thus, RhoA has been previously reported to be activated in inflammatory obesity and metabolic syndrome. The use of RhoA inhibitors may ameliorate obesity and disorders associated with obesity (35–37). In the present study, it was hypothesized that hsa-miR-4517 may function as an inhibitor of RhoA and that exosomes may be responsible for the transfer of hsa-miR-4517 from mature adipocytes to preadipocytes or distant liver tissue cells, where hsa-miR-4517 may subsequently inhibit RhoA to maintain lean body mass and prevent distant injury; however, downregulation of exosomal hsa-miR-4517 has been previously demonstrated to lead to the development of obesity and liver diseases (16,17). The present hypothesis has been indirectly demonstrated by a recent study, which revealed that miR-4517 mimics ameliorated hepatic steatosis induced by free fatty acids (38). The results of the present study demonstrated that RhoA mRNA expression levels were not significantly different between obese and lean patients in SAT and VAT, which may be attributable to the use of small sample sizes. Therefore, further investigation is required to confirm the aforementioned hypothesized mechanisms regarding the association between exosomal miR-4517 and obesity.
Among the other nine SAT-specific exosomal DE-miRNAs, hsa-miR-3156-5p was demonstrated to negatively regulate CD86 and subsequently to be involved in the inflammatory toll-like receptor (TLR) 3 signaling pathway. Thus, aberrant expression of hsa-miR-3156-5p may represent an important factor associated with obesity. Recent studies have predominantly focused on the association between hsa-miR-3156-5p and cancer (39,40), which has a similar inflammatory mechanism to obesity. However, the association between CD86 and obesity has been extensively studied. For example, Sindhu et al (41) revealed that CD86 protein expression was significantly enhanced in adipose tissue samples obtained from patients with obesity. Furthermore, CD86 has been demonstrated to be positively correlated with the levels of pro-inflammatory cytokines in adipose tissues, including IL-18, IL-18R and TLR8 (42,43). Vitamin D supplementation, which decreases the expression of CD86 in the spleen, has been proposed as a treatment option to alleviate high-fat diet-induced obesity (44). Therefore, it was hypothesized that the downregulation of exosomal hsa-miR-3156-5p may result in the development of obesity and liver diseases due to its inability to inhibit inflammation. In the present study, hsa-miR-4460 was the only miRNA identified to be upregulated in the exosomes of SAT in patients with obesity compared with lean patients. Overexpression of hsa-miR-4460 was predicted to inhibit the expression of mediator complex subunit 13 (MED13), which was also revealed to be downregulated in SAT obtained from patients with obesity. A previous study demonstrated that cardiac overexpression of MED13 confers a lean phenotype on mice via increased lipid uptake, β-oxidation and mitochondrial content in white adipose tissues and liver tissues (45). Thus, downregulation of MED13 by hsa-miR-4460 may contribute to the development of obesity via the suppression of metabolism. Treatment with exosomes possessing anti-miR-4460 may represent a therapeutic strategy for the treatment of patients suffering from obesity and associated complications.
As opposed to the results generated by SAT analysis, further investigation revealed that exosomal hsa-miR-582-5p, hsa-miR-566 and miR-548 may be important in VAT-associated diseases by respectively upregulating FGF2, FOSL2 and AMPD3, all of which are associated with inflammation. The results of the present study were in agreement with numerous previous studies. For example, Shao et al (46) demonstrated that ectopic expression of FGF2 in mouse joints enhanced the levels of IL-17-induced inflammatory cytokines and the production of chemokines in the tissue, which resulted in exacerbated symptoms of autoimmune arthritis. Drosos et al (47) revealed that the upregulation of hypoxia-inducible factor-1α and FOSL2 in perivascular adipose tissues may enhance leptin gene transcription, which further induces vascularization and inflammation, ultimately contributing to increased atherosclerotic plaque burden in the coronary arteries. In addition, Li et al (48) demonstrated that a genetic deficiency of AMPD3 resulted in markedly enhanced infiltration of neutrophils in the lungs, which further increased reperfusion-induced lung injury. Thus, modification of exosomes via transfection with hsa-miR-582-5p, hsa-miR-566 or anti-miR-548 may represent novel therapeutic approaches for the treatment of patients with obesity and associated diseases.
There were a number of limitations to the present study. Firstly, only exosomal miRNAs and mRNA expression profile data were downloaded from the GEO database. Future studies using patient data are required to confirm the results of the present study. Secondly, the sample size was small, which may have resulted in a number of unexpected statistical deviations regarding the analyses performed to investigate the roles of miRNAs and mRNAs. Furthermore, despite the results of the present study having preliminarily speculated negative associations between numerous miRNAs and their target genes, in vitro and in vivo experiments are required to confirm these results (16–19). In addition, the results of the present study suggested that exosomal miRNAs in mature SAT and VAT may affect peripheral or distant liver tissue cells, which subsequently leads to the development of obesity and obesity-associated disorders; however, this proposed mechanism requires further experimental validation (17).
The present study preliminarily investigated the profiles of exosomal miRNAs in SAT and VAT obtained from patients with obesity, in addition to the mechanisms underlying obesity. The results of the present study revealed that hsa-miR-4517 was a shared downregulated exosomal miRNA between SAT and VAT, which upregulated its target gene, RhoA. The results demonstrated that downregulated hsa-miR-3156-5p and upregulated hsa-miR-4460 may represent important and specific exosomal miRNAs in SAT via regulation of CD86 and MED13, respectively; whereas, hsa-miR-582-5p, hsa-miR-566 and miR-548 were revealed to be important in VAT via regulation of FGF2, FOSL2 and AMPD3, respectively. All of the aforementioned target genes were revealed to be associated with inflammation, and thus such target genes and their respective miRNAs may represent novel therapeutic targets for the treatment of obesity and associated metabolic alterations.
Acknowledgements
Not applicable.
Funding
The present study was supported by Natural Science Foundation of Inner Mongolia (grant no. 2016MS0367) and Chinese Postdoctoral Science Foundation (grant no. 2014M561213).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the National Center for Biotechnology information database repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE68885; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50574; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29718).
Authors' contributions
ZW, ZY and HY participated in the design of this study. ZW and ZY performed the bioinformatics analyses. ZW, ZY and XW contributed to the acquisition and interpretation of data. ZW and ZY were involved in drafting the manuscript. XW and HY participated in the manuscript revision. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Du P, Wang HJ, Zhang B, Qi SF, Mi YJ, Liu DW, Tian QB. Prevalence of abdominal obesity among Chinese adults in 2011. J Epidemiol. 2017;27:282–286. doi: 10.1016/j.je.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranceta-Bartrina J, Pérez-Rodrigo C, Alberdi-Aresti G, Ramos-Carrera N, Lázaro-Masedo S. Prevalence of general obesity and abdominal obesity in the spanish adult population (Aged 25–64 Years) 2014–2015: The ENPE study. Rev Esp Cardiol (Engl Ed) 2016;69:579–587. doi: 10.1016/j.recesp.2016.02.010. (In English, Spanish) [DOI] [PubMed] [Google Scholar]
- 3.Wong E, Woodward M, Stevenson C, Backholer K, Sarink D, Peeters A. Prevalence of disability in Australian elderly: Impact of trends in obesity and diabetes. Prev Med. 2016;82:105–110. doi: 10.1016/j.ypmed.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Barbieri M, Désesquelles A, Egidi V, Demuru E, Frova L, Meslé F, Pappagallo M. Obesity-related mortality in France, Italy, and the United States: A comparison using multiple cause-of-death analysis. Int J Public Health. 2017;62:623–629. doi: 10.1007/s00038-017-0978-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jo J, Gavrilova O, Pack S, Jou W, Mullen S, Sumner AE, Cushman SW, Periwal V. Hypertrophy and/or hyperplasia: Dynamics of adipose tissue growth. PLoS Comput Biol. 2009;5:e1000324. doi: 10.1371/journal.pcbi.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee YS, Kim JW, Osborne O, Oh DY, Sasik R, Schenk S, Chen A, Chung H, Murphy A, Watkins SM, et al. Increased adipocyte O2 consumption triggers HIF-1α, causing inflammation and insulin resistance in obesity. Cell. 2014;157:1339–1352. doi: 10.1016/j.cell.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujisaka S, Usui I, Ikutani M, Aminuddin A, Takikawa A, Tsuneyama K, Mahmood A, Goda N, Nagai Y, Takatsu K. Adipose tissue hypoxia induces inflammatory M1 polarity of macrophages in an HIF-1α-dependent and HIF-1α-independent manner in obese mice. Diabetologia. 2013;56:1403–1412. doi: 10.1007/s00125-013-2885-1. [DOI] [PubMed] [Google Scholar]
- 8.Shin E, Shin S, Kong H, Lee S, Do SG, Jo TH, Park YI, Lee CK, Hwang IK, Kim K. Dietary aloe reduces adipogenesis via the activation of AMPK and suppresses obesity-related inflammation in obese mice. Immune Netw. 2011;11:107–113. doi: 10.4110/in.2011.11.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saltiel AR, Olefsky JM. Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest. 2017;127:1–4. doi: 10.1172/JCI92035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esser N, Legrand-Poels S, Piette J, Scheen AJ, Paquot N. Inflammation as a link between obesity, metabolic syndrome and type 2 diabetes. Diabetes Res Clin Pract. 2014;105:141–150. doi: 10.1016/j.diabres.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Lo J, Bernstein LE, Canavan B, Torriani M, Jackson MB, Ahima RS, Grinspoon SK. Effects of TNF-alpha neutralization on adipocytokines and skeletal muscle adiposity in the metabolic syndrome. Am J Physiol Endocrinol Metab. 2007;293:E102–E109. doi: 10.1152/ajpendo.00089.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Owyang AM, Maedler K, Gross L, Yin J, Esposito L, Shu L, Jadhav J, Domsgen E, Bergemann J, Lee S. XOMA 052, an anti-IL-1{beta} monoclonal antibody, improves glucose control and {beta}-cell function in the diet-induced obesity mouse model. Endocrinology. 2010;151:2515–2527. doi: 10.1210/en.2009-1124. [DOI] [PubMed] [Google Scholar]
- 13.Rissanen A, Howard CP, Botha J, Thuren T. Global Investigators: Effect of anti-IL-1β antibody (canakinumab) on insulin secretion rates in impaired glucose tolerance or type 2 diabetes: Results of a randomized, placebo-controlled trial. Diabetes Obes Metab. 2012;14:1088–1096. doi: 10.1111/j.1463-1326.2012.01637.x. [DOI] [PubMed] [Google Scholar]
- 14.Arner P, Kulyté A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol. 2015;11:276–288. doi: 10.1038/nrendo.2015.25. [DOI] [PubMed] [Google Scholar]
- 15.Yao F, Yu Y, Feng L, Li J, Zhang M, Lan X, Yan X, Liu Y, Guan F, Zhang M, Chen L. Adipogenic miR-27a in adipose tissue upregulates macrophage activation via inhibiting PPARγ of insulin resistance induced by high-fat diet-associated obesity. Exp Cell Res. 2017;355:105–112. doi: 10.1016/j.yexcr.2017.03.060. [DOI] [PubMed] [Google Scholar]
- 16.Ying W, Riopel M, Bandyopadhyay G, Dong Y, Birmingham A, Seo JB, Ofrecio JM, Wollam J, Hernandez-Carretero A, Fu W, et al. Adipose tissue Macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cells. 2017;171(372–384):e12. doi: 10.1016/j.cell.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Yu M, Dai M, Chen C, Tang Q, Jing W, Wang H, Tian W. miR-450a-5p within rat adipose tissue exosome-like vesicles promotes adipogenic differentiation by targeting WISP2. J Cell Sci. 2017;130:1158–1168. doi: 10.1242/jcs.197764. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Mei H, Chang X, Chen F, Zhu Y, Han X. Adipocyte-derived microvesicles from obese mice induce M1 macrophage phenotype through secreted miR-155. J Mol Cell Biol. 2016;8:505–517. doi: 10.1093/jmcb/mjw040. [DOI] [PubMed] [Google Scholar]
- 19.Ferrante SC, Nadler EP, Pillai DK, Hubal MJ, Wang Z, Wang JM, Gordish-Dressman H, Koeck E, Sevilla S, Wiles AA, Freishtat RJ. Adipocyte-derived exosomal miRNAs: A novel mechanism for obesity-related disease. Pediatr Res. 2015;77:447–454. doi: 10.1038/pr.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neeland IJ, Ayers CR, Rohatgi AK, Turer AT, Berry JD, Das SR, Vega GL, Khera A, McGuire DK, Grundy SM, de Lemos JA. Associations of visceral and abdominal subcutaneous adipose tissue with markers of cardiac and metabolic risk in obese adults. Obesity. 2013;21:E439–E447. doi: 10.1002/oby.20135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niijima K, Shimoda Y, Saito T, Yamada E, Niijima Y, Okada S, Yamada M. Subcutaneous abdominal adipose tissue is associated with an index of insulin sensitivity/resistance. Adipocyte. 2016;5:375–377. doi: 10.1080/21623945.2016.1240138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goel K, Misra A, Vikram NK, Poddar P, Gupta N. Subcutaneous abdominal adipose tissue is associated with the metabolic syndrome in Asian Indians independent of intra-abdominal and total body fat. Heart. 2010;96:579–583. doi: 10.1136/hrt.2009.183236. [DOI] [PubMed] [Google Scholar]
- 23.Marinou K, Hodson L, Vasan SK, Fielding BA, Banerjee R, Brismar K, Koutsilieris M, Clark A, Neville MJ, Karpe F. Structural and functional properties of deep abdominal subcutaneous adipose tissue explain its association with insulin resistance and cardiovascular risk in men. Diabetes Care. 2014;37:821–829. doi: 10.2337/dc13-1353. [DOI] [PubMed] [Google Scholar]
- 24.Tam CS, Heilbronn LK, Henegar C, Wong M, Cowell CT, Cowley MJ, Kaplan W, Clément K, Baur LA. An early inflammatory gene profile in visceral adipose tissue in children. Int J Pediatr Obes. 2011;6:e360–e363. doi: 10.3109/17477166.2011.575152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Wen Y, Lu Q, Fu WJ. An imputation approach for oligonucleotide microarrays. PLoS One. 2013;8:e58677. doi: 10.1371/journal.pone.0058677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The R Foundation for Statistical Computing. Vienna, Austria: 2011. R Core Team: R: a language and environment for statistical computing. R Foundation for Statistical Computing. [Google Scholar]
- 27.Bolstad B. PreprocessCore: A collection of pre-processing functions. R package. 2013 version 1. [Google Scholar]
- 28.Smyth GK. Limma: Linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York, NY: 2005. pp. 397–420. [DOI] [Google Scholar]
- 29.Kolde R. (2015) pheatmap: Pretty Heatmaps. R package. 2015 version 1.0. 8. [Google Scholar]
- 30.Dweep H, Gretz N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 31.Kohl M, Wiese S, Warscheid B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 32.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Vohl MC, Sladek R, Robitaille J, Gurd S, Marceau P, Richard D, Hudson TJ, Tchernof A. A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obes Res. 2004;12:1217–1222. doi: 10.1038/oby.2004.153. [DOI] [PubMed] [Google Scholar]
- 34.Romao JM, Jin W, He M, Mcallister T, Guan LL. Altered MicroRNA expression in bovine subcutaneous and visceral adipose tissues from cattle under different diet. PLoS One. 2012;7:e40605. doi: 10.1371/journal.pone.0040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tapia PC. RhoA, Rho kinase, JAK2, and STAT3 may be the intracellular determinants of longevity implicated in the progeric influence of obesity: Insulin, IGF-1, and leptin may all conspire to promote stem cell exhaustion. Med Hypotheses. 2006;66:570–576. doi: 10.1016/j.mehy.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 36.Noda K, Nakajima S, Godo S, Saito H, Ikeda S, Shimizu T, Enkhjargal B, Fukumoto Y, Tsukita S, Yamada T, et al. Rho-kinase inhibition ameliorates metabolic disorders through activation of AMPK pathway in mice. PLoS One. 2014;9:e110446. doi: 10.1371/journal.pone.0110446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hara Y, Wakino S, Tanabe Y, Saito M, Tokuyama H, Washida N, Tatematsu S, Yoshioka K, Homma K, Hasegawa K, et al. Rho and Rho-kinase activity in adipocytes contributes to a vicious cycle in obesity that may involve mechanical stretch. Sci Signal. 2011;4:ra3. doi: 10.1126/scisignal.2001227. [DOI] [PubMed] [Google Scholar]
- 38.Wu YL, Zhu YB, Huang RD, Peng XE, Lin X. Multiple MicroRNAs ameliorate hepatocyte steatosis and injury by suppressing FABP1 expression. Cell Physiol Biochem. 2017;44:2243–2255. doi: 10.1159/000486058. [DOI] [PubMed] [Google Scholar]
- 39.Xia YF, Pei GH, Wang N, Che YC, Yu FS, Yin FF, Liu HX, Luo B, Wang YK. miR-3156-3p is downregulated in HPV-positive cervical cancer and performs as a tumor-suppressive miRNA. Virol J. 2017;14:20. doi: 10.1186/s12985-017-0695-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moratin J, Hartmann S, Brands R, Brisam M, Mutzbauer G, Scholz C, Seher A, Müller-Richter U, Kübler AC, Linz C. Evaluation of miRNA-expression and clinical tumour parameters in oral squamous cell carcinoma (OSCC) J Craniomaxillofac Surg. 2016;44:876–881. doi: 10.1016/j.jcms.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 41.Sindhu S, Thomas R, Shihab PK, Al-Shawaf E, Hasan A, Alghanim M, Behbehani K, Ahmad R. Changes in the adipose tissue expression of CD86 costimulatory ligand and CD163 scavenger receptor in obesity and type-2 diabetes: Implication for metabolic disease. J Glycomics Lipidomics. 2015;5:134. doi: 10.4172/2153-0637.1000135. [DOI] [Google Scholar]
- 42.Ahmad R, Thomas R, Kochumon S, Sindhu S. Increased adipose tissue expression of IL-18R and its ligand IL-18 associates with inflammation and insulin resistance in obesity. Immun Inflamm Dis. 2017;5:318–335. doi: 10.1002/iid3.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmad R, Kochumon S, Thomas R, Atizado V, Sindhu S. Increased adipose tissue expression of TLR8 in obese individuals with or without type-2 diabetes: Significance in metabolic inflammation. J Inflamm. 2016;13:38. doi: 10.1186/s12950-016-0147-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomaa AM, El-Aziz EA. Vitamin D reduces high-fat diet induced weight gain and C-reactive protein, increases interleukin-10, and reduces CD86 and caspase-3. Pathophysiology. 2017;24:31–37. doi: 10.1016/j.pathophys.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Baskin KK, Grueter CE, Kusminski CM, Holland WL, Bookout AL, Satapati S, Kong YM, Burgess SC, Malloy CR, Scherer PE, et al. MED13-dependent signaling from the heart confers leanness by enhancing metabolism inadipose tissue and liver. EMBO Mol Med. 2014;6:1610–1621. doi: 10.15252/emmm.201404218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shao X, Chen S, Yang D, Cao M, Yao Y, Wu Z, Li N, Shen N, Li X, Song X, Qian Y. FGF2 cooperates with IL-17 to promote autoimmune inflammation. Sci Rep. 2017;7:7024. doi: 10.1038/s41598-017-07597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drosos I, Chalikias G, Pavlaki M, Kareli D, Epitropou G, Bougioukas G, Mikroulis D, Konstantinou F, Giatromanolaki A, Ritis K, et al. Differences between perivascular adipose tissue surrounding the heart and the internal mammary artery: Possible role for the leptin-inflammation-fibrosis-hypoxia axis. Clin Res Cardiol. 2016;105:887–900. doi: 10.1007/s00392-016-0996-7. [DOI] [PubMed] [Google Scholar]
- 48.Li P, Ogino K, Hoshikawa Y, Morisaki H, Toyama K, Morisaki T, Morikawa K, Ninomiya H, Yoshida A, Hashimoto K, et al. AMP deaminase 3 plays a critical role in remote reperfusion lung injury. Biochem Biophys Res Commun. 2013;434:131–136. doi: 10.1016/j.bbrc.2013.03.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the National Center for Biotechnology information database repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE68885; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50574; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29718).