Abstract
Protein–carbohydrate interactions play crucial roles in biology. Understanding and modifying these interactions is of major interest for fighting many diseases. We took a synthetic biology approach and incorporated noncanonical amino acids into a bacterial lectin to modulate its interactions with carbohydrates. We focused on tryptophan, which is prevalent in carbohydrate binding sites. The exchange of the tryptophan residues with analogs fluorinated at different positions resulted in three distinctly fluorinated variants of the lectin from Ralstonia solanacearum. We observed differences in stability and affinity toward fucosylated glycans and rationalized them by X-ray and modeling studies. While fluorination decreased the aromaticity of the indole ring and, therefore, the strength of carbohydrate–aromatic interactions, additional weak hydrogen bonds were formed between fluorine and the ligand hydroxyl groups. Our approach opens new possibilities to engineer carbohydrate receptors.
Glycans and glycoconjugates are ubiquitously found on all living cells. Protein receptors, such as lectins, bind to such glycans and decipher their structural code, which results in biological and physiological actions.1 Modifying and engineering lectins to interfere with glycan interactions is in its infancy.2 Novel synthetic biology tools are now available to explore glycobiology. Noncanonical amino acids (ncAAs) find application in diverse scientific fields, such as protein engineering,3,4 proteomics,5,6 and material sciences7,8 (for recent reviews, see refs (9−12)), which is attributed to their chemical or structural versatility. However, only little emphasis has been put on the use of ncAAs to improve or alter receptor–ligand interactions. Fluorinated amino acids were studied for their effects on protein–protein interactions (reviewed in ref (13)). For example, incorporation of 5-fluorotryptophan into the cyclic peptide inhibitor compstatin resulted in a 4-fold increased affinity toward complement factor 3.14 The effect of protein fluorination on carbohydrate binding has not been much investigated. Luck and Falke15 explored structural changes in the d-galactose chemosensory receptor induced by sugar binding. They observed a 2.2–3-fold reduced affinity for the sugar with a variant containing 5-fluorotryptophan in its ligand binding site compared to the unlabeled receptor. Similarly, fluorination of phenylalanine and tyrosine of the small antibiotic peptide AcAMP2 lowered its affinity for chitin trisaccharide.16
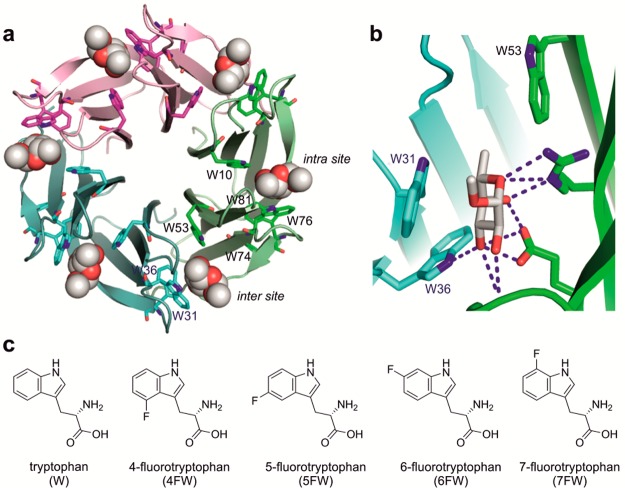
Ralstonia solanacearum lectin (RSL) is a carbohydrate binding protein with unusually strong affinity for fucose and specificity for fucosylated oligosaccharides from plants and animals.17 RSL is a homotrimer that forms a six-bladed β-propeller fold. It presents six carbohydrate binding sites, which are either intra- or intermonomeric (Figure 1a), and nonetheless are structurally very similar.17 Each RSL monomer contains seven Trp residues, six of which are directly involved in carbohydrate binding (Supporting Information, Table S1). The analysis of the RSL structure combined with modeling revealed the importance of three Trp residues present in the binding site. One makes hydrophobic contact with the sugar methyl group; another is involved in hydrogen bonding (Figure 1b). The third is responsible for a stacking interaction with the fucose residue and defines the specificity for this sugar.18,19 Such carbohydrate–aromatic interactions are frequent20,21 with a 9-fold prevalence of Trp in the carbohydrate binding site.21 Recently, Trp was also demonstrated to have a crucial role in inducing a large conformational change of a fucosylated trisaccharide in the RSL binding site.22
Figure 1.
(a) Structure of RSL (pdb 2BT9). The three monomers are colored in magenta, green, and cyan; the bound αMeFuc is represented as spheres. (b) The intermonomeric binding site with three important Trp residues: W31, W36, and W53 (structurally equivalent to W76, W81, and W10 in the intramonomeric site). (c) Structures of l-tryptophan and the fluorinated l-analogs used in this study.
RSL appears as an excellent model protein to study the effect of tryptophan fluorination on carbohydrate recognition. Here, we incorporated four fluorinated Trp analogs (Figure 1c) into RSL to examine the effect on the protein stability and affinity toward mono- and oligosaccharides. We report the first crystal structures of fluorinated lectins and elucidate the molecular details of their interaction with ligands. After appropriate parametrization, molecular modeling gave access to the dynamics of the modified proteins and demonstrated the predictive power of modeling ncAA incorporation.
Results and Discussion
Lectin Production and Physicochemical Characterization
We employed the supplementation of a tryptophan auxotrophic Escherichia coli strain with four tryptophan analogs (FWs, Figure 1c) for their residue-specific incorporation23 into RSL. Since the tryptophan analogs are not commercially available, we fed our cells with indole analogs to convert these to tryptophan analogs24−27in situ by the host tryptophan synthase (reviewed by Phillips28). The procedure resulted in titers of purified synthetic variants in the range of 35–60 mg lyophilized protein per liter of cell culture, compared to 100 mg L–1 cell culture for the unmodified parent protein. Interestingly, the incorporation of 6FW drove RSL into insolubility, while all other fluorinated RSL variants remained soluble (Figure S1). Efficient labeling of RSL with FWs was confirmed by mass spectrometry (Figure S2). The majority of the proteins (85% of RSL[4FW], 86% of RSL[5FW] and 84% of RSL[7FW]) showed quantitative replacement of Trp by the analogs, and only in a minor fraction (14–16%) the exchange was incomplete (Table S2). By assuming a random distribution of the unlabeled residues over the seven positions, one would thus expect a labeling efficiency of 97% (RSL[7FW]) to 98% (RSL[4FW] and RSL[5FW]) per site. To test the individual sites for labeling with the fluorinated Trp analogs, we subjected the fluorinated variants and the wild-type RSL proteins to different enzymatic digests to ensure nearly complete sequence coverage. Nano-LC-tandem mass spectrometry analysis showed that four Trp residues and their respective fluoro-analogs could be relatively quantified (W10, W31, W53, W74) because they were covered in well ionizing peptides with a single Trp in their sequence (Figure S3). Only minimal residual unlabeled Trp was found in the labeled protein analogs at these four positions (<3% of wild-type), which matches the proportion expected by a random distribution.
We analyzed the physico- and biochemical properties of the synthetic RSL variants. Several reports in the literature indicate that the incorporation of fluorinated ncAAs results in increased protein stability.29−31 We compared the thermostability of the variants to the parent protein using differential scanning fluorimetry (DSF) and differential scanning calorimetry (DSC). Indeed, in the presence of d-mannose, the 5FW-containing variant showed a slightly increased melting temperature in comparison to the parent protein (Tm = 90 °C vsTm = 88 °C; Figure S4a). In contrast, the incorporation of 4- and 7FW clearly decreased the melting temperatures to 79 and 72 °C, respectively. The same effects were observed by DSC measurements in the presence of the high-affinity ligand methyl α-l-fucopyranoside (αMeFuc)17 (Figure S4b). Since the interaction of the RSL variants with the high-affinity ligand exceeded the range of DSF, we used DSC for the analysis. The results exemplify the remarkably pronounced effect of the position of the fluorine atom in the indole ring on protein stability. As shown in Figure 1, Trp residues are involved in interblade contacts, and alteration of their structure may indeed affect the folding (6FW) or modify the protein’s stability.
Affinity for Different Ligands
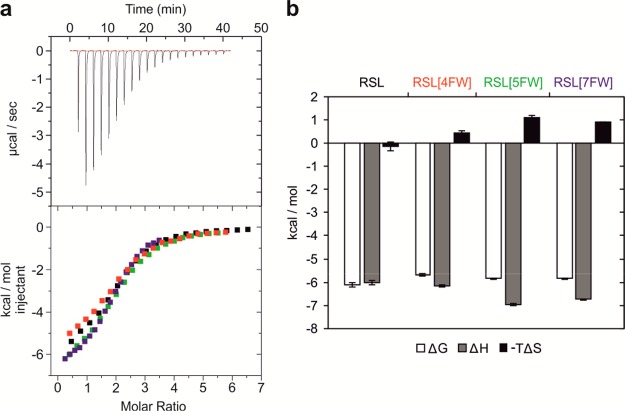
Given the prominent position of the fluorine in the ligand binding site of the synthetic RSL variants, we hypothesized that the variants could show subtle affinity differences for different sugar ligands. Therefore, we analyzed the affinity toward αMeFuc and two tetrasaccharides, blood group H type 2 antigen (HType2, Fucα1–2Galβ1–4GlcNAcβ1–3Gal) and lewis X (LeX, Galβ1–4[Fucα1–3]GlcNAcβ1–3Gal) by isothermal titration calorimetry (ITC, Figure S5, Table S3). A dissociation constant, Kd, of 1.21 ± 0.04 μM was measured for RSL interacting with αMeFuc, which is in agreement with previous reports.17 RSL[5FW] and RSL[7FW] behaved similarly, with Kd values of 0.889 ± 0.004 and 1.11 ± 0.02 μM, respectively, while RSL[4FW] displayed a slightly lower affinity (Kd = 1.73 ± 0.41 μM). We only observed minor differences in affinity toward HType2. They varied from 4.35 ± 0.35 μM for RSL[7FW] to 8.18 ± 0.99 μM for RSL[4FW]. RSL[4FW] displayed a lower affinity toward LeX (Kd = 67.8 ± 6.8 μM) than RSL (Kd = 32.5 ± 4.9 μM) and the other two variants (RSL[4FW], Kd = 52.4 ± 2.6 μM; RSL[7FW], Kd = 52.1 ± 0.8 μM). While RSL bound LeX with no entropy cost, both RSL[5FW] and RSL[7FW] displayed a stronger enthalpy contribution and some entropy cost for binding (ΔH, −TΔS; Figure 2). LeX was therefore selected for the structural study to determine the molecular basis of the thermodynamic behavior.
Figure 2.
ITC experiment of RSL and fluorinated RSL binding to the tetrasaccharide LeX. (a) Thermogram (top) of the injection of LeX aliquots to RSL in solution and corresponding integrated peaks (bottom) for RSL and the variants. (b) The thermodynamic contributions were derived from curve fitting. Entropy cost, −TΔS; enthalpy contribution, ΔH; free energy of binding, ΔG.
Structural Analysis
The three fluoro-RSL proteins were cocrystallized with the LeX tetrasaccharide using the vapor diffusion method and yielded diamond-shaped crystals diffracting to high resolution (1.15 to 1.35 Å). All structures belonged to the F23 space group with two independent monomers in the asymmetric unit (Table S4, PDB ID 5O7W, 5O7V, and 5O7U). The presence of the 3-fold axis resulted in the generation of two trimers (A and B), which are similar to the RSL native structure (2BT9) with RMSD lower than 0.2 Å (Figure 3a).
Figure 3.
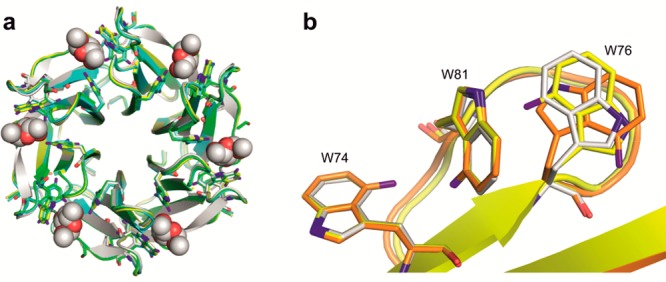
(a) Superimposition of the crystal structure of the RSL/αMeFuc complex (white; pdb 2BT9) with the structures of RSL[4FW] (yellow; pdb 5O7W), RSL[5FW] (cyan; pdb 5O7V), and RSL[7FW] (green; pdb 5O7U). (b) Superimposition of loop 76–82 from wt-RSL (white), and chain A (yellow) and chain B (orange) of RSL[4FW]. Fluorine atoms are colored in purple.
The electron density of the Trp residues in the binding sites clearly indicated the presence of the fluorine at each expected position (Figures S6–S8). The effect of fluorination on the conformation of the Trp side chains is very limited, except for Trp76 in the 4FW variant. In chain A of the RSL[4FW] structure (Figure 3b, yellow), the indole ring of Trp76 is slightly pushed back, while it undergoes a 180° rotation around the Cβ–Cγ bond in chain B (Figure 3b, orange), orienting the fluorine to the opposite side. Clearly, fluorination at this position results in a steric repulsion with the neighboring Trp81, which induces the rearrangement.
We clearly observed the electron density of the carbohydrate ligand in all structures (Figure S6–S8). Similar to previous observations in the RSL/LeX complex,22 the LeX adopted open conformations (Table S5) that were very different from those observed in solution.32 The conformational behavior of LeX in RSL was comprehensively described in previous work,32 and we focus here on the contacts between the fluorinated Trp residues and the oligosaccharide ligand. For the sake of clarity, the description of structures and models below will be centered on the intermonomeric site that does not present significant variations as the intramonomeric site does. Due to the orientation of the indole ring of the Trp residues (Figure 4a), we observed no direct contacts between the fluorine atoms and the ligand in the complex with RSL[4FW]. The fluorine on Trp31 of RSL[5FW] had limited contact (∼4 Å) to the C3 of fucose. Nevertheless, LeX made several contacts within RSL[7FW] (Figure 4b). The fluorine atom of Trp36 was close to the O3 and O4 hydroxyls of fucose with an F···O distance of 3.3 and 3.1 Å, respectively. The fluorine of Trp31 was in close proximity to O2 of galactose and to O4 of GlcNAc (3.0 and 3.2 Å). Such distances are indicative of favorable contacts, and indeed, fluorine atoms are considered as weak acceptors of hydrogen bonds.33,34 Fluorination is expected to induce a large modification of the π electron density on the indole ring,14 so the effect on the affinity may be more complex than the observed weak hydrogen bonds.
Figure 4.
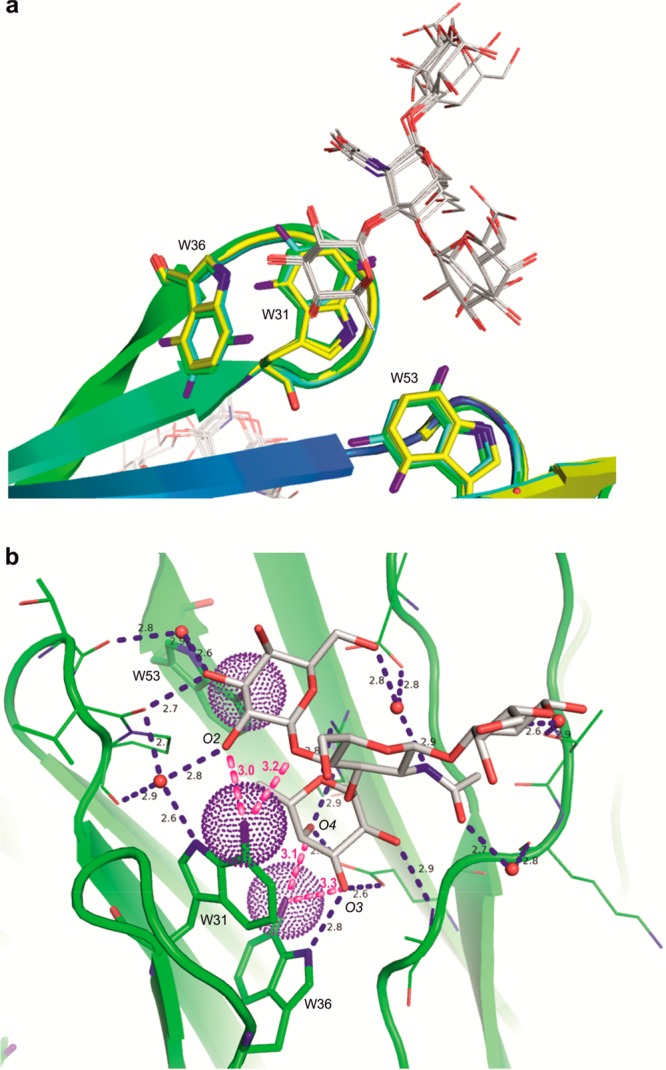
(a) Superimposition of the six intermonomeric binding sites (chains A and B of the three RSL variants) complexed with the LeX tetrasaccharide. Color coding as in Figure 3a. (b) Close-up of the intermonomeric binding site of RSL[7FW] (chain B). The network of hydrogen bonds is displayed as dashed blue lines, and contacts with the fluorine atoms are in magenta.
Modeling of FW–Ligand Interactions
We performed molecular modeling to dissect the interaction between residues Trp31 and Trp36 that are involved in stacking with fucose and in hydrogen bonding, respectively. Parameterization was necessary to include F-Trp in the model. Atomic charges (Table S6) and other parameters were obtained by quantum chemical calculations and were included in Amber Tools to prepare the topology files of the different complexes. The monosaccharide αMeFuc was selected as the ligand to avoid the superimposition of the interaction analysis with conformational effects. Each RSL variant complexed with αMeFuc was submitted to a molecular dynamics simulation of 500 ns that was used for calculating the interaction energy and its decomposition per residue.35
All modeled complexes were very stable, and the average structures of the binding sites were similar to the X-ray structures (Figure S9). The calculated energies of interaction (Table 1) predicted that the parent protein and RSL[7FW] would have a stronger affinity for αMeFuc than the other variants. This is partially in agreement with experimental ITC data, where RSL[4FW] showed the lowest affinity for αMeFuc (Figure S5).
Table 1. Interaction Energies and Average Distances Measured with Molecular Dynamics Simulationsa.
| MD simulation data | RSL-wt | RSL[4FW] | RSL[5FW] | RSL[7FW] |
|---|---|---|---|---|
| ΔG(TOT)calcb | –11.2 | –10.2 | –10.2 | –11.2 |
| (ΔGexp)c | (−8.1) | (−7.9) | (−8.3) | (−8.1) |
| ΔG(Trp31)calc | –5.0 | –4.4 | –4.7 | –4.4 |
| ΔG(Trp36)calc | –3.5 | –3.6 | –3.6 | –3.8 |
| d(H5···Trp31.CZ2)d | 2.84 ± 0.25 | 2.94 ± 0.32 | 2.85 ± 0.27 | 2.93 ± 0.29 |
| d(O3···Trp36.NHE1)d | 1.95 ± 0.14 | 1.95 ± 0.14 | 1.95 ± 0.15 | 1.92 ± 0.14 |
| d(HO4···Glu73.OE2)d | 1.80 ± 0.19 | 1.78 ± 0.17 | 1.78 ± 0.18 | 1.94 ± 0.32 |
All energies are in kcal mol–1.
Calculated binding free energies.
Data measured by ITC in this work.
Distances in Å; time-averaged values and standard deviations were computed over all binding sites of each trimer.
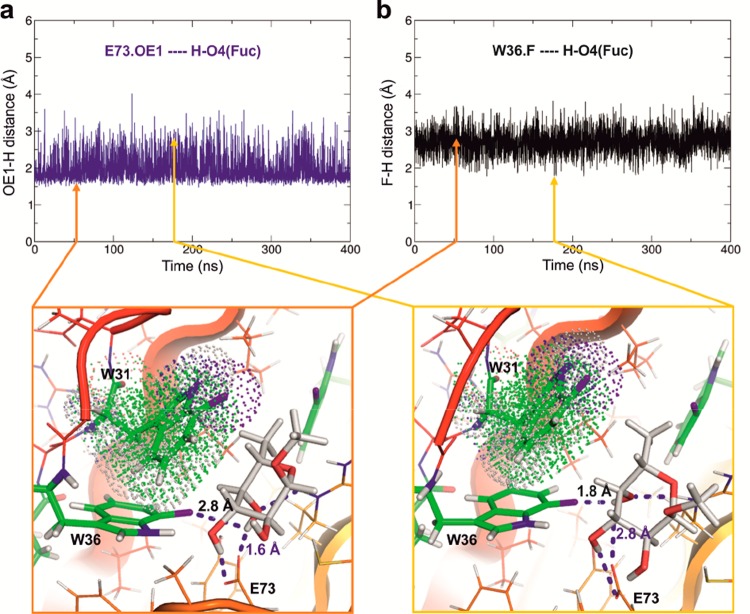
A detailed analysis of the influence of the fluorination on the two Trp residues closer to fucose, that is, Trp36 and Trp31, demonstrated opposite effects: The binding energy of Trp31 that stacks on fucose is weakened by fluorination. The alteration of the indole charge distribution by fluorine deactivates the aromaticity as previously described for a fluorinated derivative of phenylalanine.16 In contrast, the energy of interaction of Trp36 that establishes an NH···O hydrogen bond with the O3 of fucose is not decreased in the variants compared to the parent RSL (Table 1). In the case of RSL[7FW], the energy of interaction is even increased, which is in agreement with the formation of a fluorine-based hydrogen bond. A closer analysis of the molecular dynamics trajectories indicates that this variant, which shows direct contact between the fluorine atom and the ligand, presents a very unique behavior of fucose in the binding site (Figure 5). While the average structure is similar to the one observed by X-ray, rapid fluctuation in the orientation of fucose creates a sporadic strong hydrogen bond between fluorine and HO4 (distance F···H shorter than 2 Å), which competes with the hydrogen bond between HO4 and Glu73.OE1 that is observed in all crystal structures. The rapid change of orientation of fucose is also correlated with a partial loss of stacking with Trp31. The average contacts of fucose in the binding site are not significantly affected (Table 1), but the stability of the complex is lower as indicated by larger deviation in hydrogen bond distances in the RSL[7FW] variant.
Figure 5.
Instability of the hydrogen bond network involving the Fuc-HO4 hydroxyl group in RSL[7FW]. History of distances for competing hydrogen bonds between HO4 and either Trp36.F or Glu73.OE1, and snapshots demonstrating the occurrence of different orientations of fucose in the binding site, which results in alternative hydrogen bond networks. Arrows indicate the position of the two snapshots in the trajectories.
Differences in Specificity
Since the Trp residues can interact with other oligosaccharides in addition to the fucose residue, we assessed the specificity of the synthetic lectin variants for >300 carbohydrates via a previously described36 glycan microarray. RSL and all variants demonstrated rather similar binding patterns (Figure S10). They bound to all fucosylated oligosaccharides, as described previously for RSL17 and the structurally related BambL.37 Nevertheless, a more thorough comparison of the binding pattern to a subset of fucosylated oligosaccharides, that is, the histo-blood group epitopes, revealed some variations as a function of the position of the fluorine atom (Figure S11). While RSL bound more strongly to oligosaccharides of blood group A and B than to blood group O(H) and Lewis, this tendency was reversed for the fluorinated variants. Particularly RSL[7W] showed weaker binding to blood group B trisaccharide than to other glycans (Figure 6a, B-tri).
Figure 6.
(a) Comparison of the blood group oligosaccharide-binding specificity of RSL and RSL[7FW]. Full names and chemical structures of the glycans are shown in Figure S11. (b) Docking of blood group B trisaccharide into the intermonomeric site of RSL[7FW] reveals nonfavorable short contacts between the fluorine atoms at Trp53 and Trp31 and the CH groups at positions C3 and C4 of galactose.
The relative change in specificity can be rationalized by manually docking the blood group B trisaccharide into the RSL binding site. We used the structure of the blood group B trisaccharide, which had been docked into the related BambL,37 and superimposed it on the glycan binding site of RSL[7FW]. Evidently, the galactose residue of blood group B trisaccharide comes in close contact to the fluorine atoms of both Trp53 and Trp31 resulting in local steric conflicts (Figure 6b).
Conclusions
We present here the first structural characterization of lectins containing fluorinated tryptophan residues and their interaction with carbohydrate ligands. The global fluorination of the Trp residues moderately affected the glycan affinity, and it changed the specificity for blood group glycans. The structural analysis and the free energy decomposition by molecular dynamics calculations pointed out that fluorination may induce different effects. The stacking effect is partially lost, due to the deactivation of the aromaticity. Based on our experiments and calculations, the hydrogen bond property of the NH group of the indole ring does not seem to be affected by the presence of the neighboring fluorine. Of interest for the future design of synthetic proteins with noncanonical amino acids is the possibility to create novel contacts, based on the weak hydrogen bond acceptor character of fluorine and the strong capacity of sugars to be involved in such contacts. Either the introduction of additional hydrogen bonds via the fluorine or the weakening of the stacking interaction with the ligand might be exploited. Here, we used a lectin in which several Trp residues participate in glycan binding and their global exchange for fluorinated derivatives leads to effects that interfere with each other. The site-specific incorporation of Trp analogs could be beneficial for a more surgical lectin–carbohydrate interaction analysis. Yet at the moment, this approach is limited to a small set of Trp analogs excluding fluorinated derivatives.38 The approach described here could be used in the future for incorporation of fluorinated Trp analogs into lectins or other carbohydrate-binding proteins that involve a single Trp residue in the glycan binding. This could be a promising compromise until the advent of more versatile site-specific incorporation systems for these analogs. Precise insertion of fluorination in engineered proteins and the exploration of other hydrogen bond accepting groups represent a promising strategy for the conception of novel receptors for glycans.
Methods
Protein Expression and Purification
The incorporation of tryptophan analogs into the Ralstonia solanacearum lectin (RSL) was performed in E. coli BWEC47, a tryptophan auxotrophic derivative of strain BL21-Gold (DE3) (Agilent Technologies, Palo Alto, CA), genotype E. coli B F–ompT hsdS(rB– mB–) dcm+ Tetrgal λ (DE3) endA Hte ΔtrpC. The construction of this strain will be described elsewhere.
The amino acid sequence of RSL (PDB 2BS5) was back-translated into a DNA sequence codon optimized for the expression in E. coli and ordered as a gBlock (Integrated DNA Technologies, Coralville, IA). The rsl gene was amplified using primers pBP654 and pBP643 and cloned into the EcoRI/HindIII (Thermo Fisher Scientific, Waltham, MA) digested pQE80L vector (Qiagen, Hilden, Germany) via Gibson assembly39 resulting in plasmid pQE80L-RSL. The DNA, primer, and amino acid sequences can be found in Table S1.
The tryptophan auxotrophic E. coli BWEC47 harboring the pQE80L-RSL plasmid was grown in M9 medium (47.76 mM Na2HPO4, 22.04 mM KH2PO4, 8.56 mM NaCl, 18.69 mM NH4Cl, 22 mM α-d-glucose, 1 mM MgSO4, 0.1 mM CaCl2, 8.63 μM FeSO4, 3.55 μM MnSO4, 2.49 μM AlCl3, 1.84 μM CoCl2, 0.42 μM ZnSO4, 0.5 μM Na2MoO4, 0.35 μM CuCl2, 0.49 μM H3BO3) supplemented with 1% (w/v) casamino acids (BD Biosciences, San Jose, CA), 47 μM l-tryptophan, and 100 μg mL–1 ampicillin at 37 °C and 120 rpm to allow for growth until tryptophan depletion occurred at a cell density of D600 ≈ 3. Subsequently, the cells were incubated for an additional hour to ensure complete depletion of the canonical amino acid, before indole (Sigma-Aldrich, St. Louis, MO) or an indole-analog (4-fluoroindole, Tokyo Chemical Industry Europe, Zwijndrecht, Belgium; 5-, 6-, and 7-fluoroindole, Molekula, Newcastle Upon Tyne, U.K.) was added to a final concentration of 1 mM and protein expression was induced by the addition of 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG, Sigma-Aldrich). After expression for 18 h at 30 °C and 120 rpm, the cells were harvested by centrifugation (20 min at 8000g and 4 °C). The pellet was resuspended in lysis buffer (20 mM Tris/Cl, 1 M NaCl, pH 7.4), and the cells were disintegrated by sonication. Cell-free extract was prepared by centrifugation (50 min at 20 000g and 4 °C) and loaded onto a mannose-agarose column (Mannose Separapore 4B, BioWorld, Dublin, OH). Unbound protein was removed by washing with washing buffer (20 mM Tris/Cl, 100 mM NaCl, pH 7.4), and RSL or the synthetic variants were eluted with washing buffer containing 100 mM d(+)-mannose (Carl Roth GmbH, Karlsruhe, Germany). The proteins were dialyzed against ddH2O for 5 days at 4 °C, changing water twice a day, lyophilized, and stored at 4 °C until use.
Intact Protein LC-MS Analysis of RSL Species
Protein solutions were diluted to 10 ng μL–1 with water containing 5% ACN and 0.1% FA. Possible protein variations were separated by nano-HPLC (Dionex Ultimate 3000) equipped with a Pepswift precolumn (monolithic, 5 × 0.2 mm2) and a ProSwift RP-4H column (monolithic, 100 μm × 25 cm) (all Thermo Fisher Scientific, Vienna, Austria). One microliter of protein sample was injected and concentrated on the enrichment column for 2 min at a flow rate of 5 μL min–1 with 0.1% formic acid as isocratic solvent. Separation was carried out on the nanocolumn at a flow rate of 1 μL min–1 at 37 °C using the following gradient: solvent A is 0.1% formic acid in water and solvent B acetonitrile containing 0.1% formic acid: 0–2 min, 5% B; 2–17 min, 5–60% B; 17–20 min, 60% B; 20–20.1 min, 60–5% B; 20.1–29 min, 5% B. The maXis II ETD mass spectrometer (Bruker, Bremen, Germany) was operated with the captive spray source in positive mode with following settings: mass range, 300–3000 m/z; 1 Hz; source voltage, 1.6 kV; dry gas flow, 3 L min–1; 180 °C. The protein mass spectra were deconvoluted by the data analysis software, using the MaxEnt2 algorithm. The following main parameters were applied: charge carrier, H+; m/z range, min. 800 to max. 2000; minimum instrument resolving power was set to 50 000. For peak detection SNAP algorithm with the following parameters was used: quality factor threshold 0.9, S/N threshold 2, and maximum charge state of 12.
Amino Acid Sequence Analysis by LC-MS/MS of RSL Species
Twenty micrograms of RSL protein species was reduced with tris(2-carboxyethyl)phosphine (TCEP, final 10 mM) and alkylated with chloroacetamide (final 40 mM) for 10 min by shaking at 550 rpm at 95 °C. Proteins were digested separately by adding 0.2 μg of Promega modified trypsin and shaking overnight at 550 rpm at 37 °C or adding 0.2 μg of chymotrypsin (Promega, Madison, WI) according to the manufacturer’s instructions. The resulting peptide solutions were acidified by adding formic acid to a final concentration of 0.1%. Fifty nanograms of the digests was injected and concentrated on the enrichment column (C18, 5 μm, 100 Å, 5 × 0.3 mm2) for 2 min using 0.1% formic acid as isocratic solvent at 5 μL min–1 flow rate. The column was then switched in the nanoflow circuit, and the sample was loaded on the Acclaim PepMap RSLC nanocolumn (C18, 2 μm, 100 Å, 500 × 0.075 mm2) at a flow rate of 250 nL min–1 at 60 °C and separated using the following gradient: solvent A, water, 0.1% formic acid; solvent B, acetonitrile, 0.1% formic acid; 0–2 min, 4% B; 2–90 min, 4–25% B; 90–95 min, 25–95% B, 96–110 min, 95% B; 110–110.1 min, 4% B; 110.1–125 min, 4% B. The sample was ionized in the nanospray source equipped with stainless steel emitters (Thermo Fisher Scientific) and analyzed in a Thermo Orbitrap velos pro mass spectrometer in positive ion mode by alternating full scan MS (m/z 300 to 2000, 60000 resolution) in the ICR cell and MS/MS by CID of the 20 most intense peaks in the ion trap with dynamic exclusion enabled. The LC-MS/MS data were analyzed by searching a homemade database containing all common contaminants with Proteome Discoverer 1.4 (Thermo Fisher Scientific) and Mascot 2.4.1 (MatrixScience, London, UK). Carbamidomethylation on cysteine was entered as fixed and oxidation on methionine and fluorination on tryptophan as variable modification. Detailed search criteria were used as follows: trypsin or chymotrypsin; maximum missed cleavage sites, 2; search mode, MS/MS ion search with decoy database search included; precursor mass tolerance, ±10 ppm; product mass tolerance, ±0.7 Da; label free quantitation of precursor ions was performed with Proteome Discoverer 1.4. Peptide precursor ion intensities were normalized on total RSL protein intensity and compared for each peptide over the different protein species.
Differential Scanning Fluorimetry (DSF)
DSF was performed with a 7500 Real Time PCR System (Applied Biosystems PerkinElmer Corp., Foster City, CA). Protein samples of 3.5–11.0 μM were measured in PBS (9.55 mM Na2HPO4, 136.89 mM NaCl, 2.68 mM KCl, 1.47 mM KH2PO4) with 8× Sypro Orange (Sigma-Aldrich) in the presence (8 μM) or absence of d-(+)-mannose (Carl Roth). Melting temperatures of triplicate measurements performed on three different days were analyzed with the Protein Thermal Shift software v1.3 (Applied Biosystems). SigmaPlot (Systat software, San Jose, CA) was used for the statistical analysis of the significance of the generated melting temperatures and for box plot preparation.
Differential Scanning Calorimetry (DSC)
DSC was performed with a VP-DSC (MicroCal, Inc., Northampton, MA). Protein samples were prepared in 20 mM Tris/Cl, 100 mM NaCl, 1 mM αMeFuc, pH 7.5, with concentrations of 1.5 mg mL–1 for RSL and RSL[5FW] and 1.0 mg mL–1 for RSL[4FW] and RSL[7FW]. Measurements were performed at 30 psi pressure and a scan rate of 1 °C min–1. Data was analyzed with the MicroCal Origin software (VP-DSC version).
Glycan Microarray Analysis
The lectins were biotinylated using the EZ-Link Sulfo-NHS-LC-Biotinylation Kit (Thermo Fisher), according to the manufacturers recommendations. The glycan microarrays contained 317 carbohydrate ligands and were printed as described previously.36 The microarray slides were blocked with blocking buffer (50 mM ethanolamide in 50 mM borate buffer, pH 8.0) for one hour and subsequently thoroughly rinsed with ddH2O. The biotinylated protein samples were diluted in PLI-P buffer (0.5 M NaCl, 3 mM KCl, 1.5 mM KH2PO4, 6.5 mM Na2HPO4, 1% (w/v) BSA, 1% (v/v) Triton-X-100, pH 7.4) to a protein concentration of 1 μg mL–1 and incubated on the microarray slides in a humidified chamber at RT with gentle agitation for one hour. After incubation, the protein sample was removed and the slides were washed with PBS buffer (10 mM Na2HPO4, 10 mM NaH2PO4, 138 mM NaCl, 2.7 mM KCl, pH 7.4). The microarray slides were then incubated with cyanine-3 labeled streptavidin (Thermo Fisher) diluted 1:500 in PLI-P buffer for 1 h, as described above. Subsequently, the slides were washed with PBS buffer, rinsed with ddH2O, and stored in the dark until scanning. Fluorescent measurements were performed using the ScanArray 4000 Microarray Analysis System (PerkinElmer, Waltham, MA). Fluorescence intensities were quantified using the ScanArray Express Microarray Analysis System 4.0 (PerkinElmer) and the data was further analyzed with Microsoft Excel (Microsoft Corporation, Redmond, WA).
Isothermal Titration Calorimetry (ITC)
ITC experiments were performed on a MicroCal iTC200 microcalorimeter (Malvern Instruments Ltd., Worcestershire, UK). The experiments were carried out at 25 °C. αMeFuc (Tokyo Chemical Industry Europe), HType2, and LeX tetrasacharides (ELICITYL, Crolles, France), as well as the lyophilized proteins, were dissolved in the same buffer (20 mM Tris/Cl, 100 mM NaCl, pH 7.5). Protein concentrations in the microcalorimeter cell (204 μL) varied from 0.0476 to 0.0554 mM and between 0.242 and 0.266 mM for measurements with αMeFuc/HType2 and LeX, respectively. Twenty injections with 1 μL of sugar solutions (1 or 1.5 mM of αMeFuc and HType2 or 5 mM of LeX) were performed at intervals of 120 s with stirring at 750 rpm. The obtained data was integrated and fitted to a theoretical titration curve with Origin software (OriginLab Corporation, Northampton, MA) applying the one-set-of-sites model. The protein concentration was determined using NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). An extinction coefficient of 43.68 × 104 M–1 cm–1 was used. For the variants, the concentration was adjusted in the software to obtain the binding site stoichiometry of n = 2, that is, one inter- and intramonomeric binding site per monomer as confirmed by the crystal structures.
Crystallization and Structure Determination
The three fluorinated RSL samples were dissolved in 20 mM Hepes 7.5 and 150 mM NaCl to 8 mg mL–1 and cocrystallized with 1 mM LeX tetrasaccharide (Ellicityl, Crolles, France). Crystallization screening was performed using the hanging drop vapor diffusion at 19 °C. Diamond shaped crystals were obtained in a few days from solution 1–8 and 1–9 of the Morpheus screen or 1–30 from the Midas screen (Molecular Dimension Ltd., Newmarket, UK). Single crystals were directly mounted in a cryoloop and flash frozen in liquid nitrogen. RSL[7FW] diffraction data were collected at 100 K at the European Synchrotron Radiation Facility (Grenoble, France) on BM30A-FIP using an ADSC Q315r detector, while those for RSL[4FW] and RSL[5FW] were collected at Synchrotron Soleil (Saint Aubin, France) on Proxima 1 using a Pilatus 6 M detector. The data were processed using XDS,40 and their quality statistics are summarized in Table S4. All further computing was performed using the CCP4 suite unless otherwise stated.41 The RSL[7FW] and RSL[5FW] structures were solved by molecular replacement (MR) using PHASER and the coordinates of chain A from PDB-ID 2BT9 to search for two copies in the asymmetric unit.42 After initial rebuilding with ARP/WARP,43 the model was refined with restrained maximum likelihood refinement using REFMAC 5.8,44 iterated with manual rebuilding in Coot.45 The RSL[4FW] structure was also solved by MR using PHASER, but the coordinates of the protein chain A of RSL[5FW] were used as the search model. No rebuilding with ARP/WARP was required. Anisotropic refinement was introduced at the end of the refinement of all structures. Five percent of the observations were set aside for cross-validation analysis, and hydrogen atoms were added in their riding positions and used for geometry and structure-factor calculations. Incorporation of the ligand was performed after inspection of 2Fo – DFc weighted maps. The 7FW ligand library was constructed with Acedrg.46 Water molecules, introduced first with ARP/WARP and then automatically using Coot, were inspected manually. The model was validated with the wwPDB Validation server (http://wwpdb-validation.wwpdb.org), and the glycans were checked with Privateer.47 The coordinates were deposited in the Protein Data Bank under codes 5O7U, 5O7V, and 5O7W, for the structures of RSL[7FW], RSL[5FW], and RSL[4FW], respectively.
We clearly observed the electron density of the carbohydrate ligand in both binding sites of all structures with the exception of chain A of RSL[4FW], where a glycerol molecule was present in the intramonomeric site. The electron density in the intermonomeric site was of higher quality than in the intramonomeric site in all structures, and the tetrasaccharide was distinctly defined. In contrast, some parts were conformationally disordered in the intramonomeric sites.
Molecular Modeling
The crystal structure of wild-type RSL complexed with αMeFuc was used as a starting reference to build all models (PDB ID 2BT9).17 The fluorinated tryptophans were parametrized for compatibility with the AMBER-ff14SB force field,48 using available standard parameters for fluorine for bonding and van der Waals parameters. Fluorination is expected to induce a large modification of the π electron density on the indole ring.14 Therefore we performed quantum chemistry calculations in order to obtain representative atomic charges for each fluorinated tryptophan. RESP charges49 were calculated following the recommended setup of the PyRED server50 (http://upjv.q4md-forcefieldtools.org/REDServer-Development/). The atomic charges and modified parameters were included in Amber Tools to prepare the topology files of the different systems. Molecular dynamics simulations were carried out using the pmemd module of the AMBER12 code (http://ambermd.org). The AMBER-ff14SB48 and GLYCAM06-j51 force fields were used for protein and saccharides, respectively, together with the TIP3 water model.52 The protonation states of titratable residues were assigned using the H++ web server (http://biophysics.cs.vt.edu/H++). Standard protonation states were observed for each RSL variant. Short-range van der Waals interactions were switched to zero at a cutoff distance of 8 Å. The long-range electrostatic interactions were computed by means of the particle mesh Ewald (PME) method53 using a real-space cutoff of 8 Å. The SHAKE algorithm was applied to constrain bonds involving hydrogen atoms, and thus an integration time step of 2 fs could be used. The temperature was maintained at 300 K using the Langevin thermostat with a collision frequency of 2 ps–1. A length of 500 ns of NpT simulation was performed for each RSL variant. The MD trajectories were postprocessed to compute free energies of binding using the MMPBSA.py program.35 The intramonomeric binding site of chain A complexed with α-l-Me-fucoside was used to make the calculations. The free energy of binding was computed for each RSL variant using the Poisson–Boltzmann surface area (PBSA) framework combined with normal modes analysis. Due to computational cost of the Poisson–Boltzmann method, the energy decomposition per residue was performed from generalized Born surface area (GBSA) calculations.35
Acknowledgments
We are grateful to K. Lohner and A. Zenz for excellent assistance with DSC measurements and analysis and to P. Megusar and L. Gajdos for crystallization trials. We are grateful to Synchrotron SOLEIL (Saint Aubin, France) and ESRF (Grenoble, France) for access and technical support with beamlines PROXIMA1 and BM30A-FIP, respectively. MD calculations were partially performed on the CECIC platform of ICMG.
Glossary
Abbreviations
- RSL
Ralstonia solanacearum lectin
- 4FW
4-fluorotryptophan
- 5FW
5-fluorotryptophan
- 6FW
6-fluorotryptophan
- 7FW
7-fluorotryptophan
- αMeFuc
α-l-methyl fucopyranoside
- HType2
blood group H type 2 antigen
- LeX
lewis X
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.8b00377.
Sequences of RSL and primers used, quantification of FW incorporation into RSL, affinity constants and thermodynamics parameters, data collection and refinement statistics for fluorinated RSL co-crystallized with LeX tetrasaccharide, conformation of the LeX tetrasaccharide in each binding site, atomic charges used for Amber parameterization of the fluorinated tryptophan residues, expression and purification of fluorotryptophan-containing RSL variants, mass analysis of the fluorinated RSL variants, quantification of the incorporation of FW at individual positions, melting temperatures of RSL and fluorinated variants, ITC data of the interaction of RSL and fluorinated variants with αMeFuc and the two tetrasaccharides, electron density in the binding sites of the variant lectins complexed with the tetrasaccharide LeX, superimposition of X-ray crystal structure and averaged one from MD calculations in the intermonomeric binding site of fucose in RSL[7FW], image analysis of glycan microarrays with RSL and the FW-containing variants, and interaction of the RSL and variants with the eight selected oligosaccharides (PDF)
The authors acknowledge support by the Austrian Science Fund (FWF) project number I 1708-B22 in the framework of the EU ERASynBio project SynGlycTis as well as by the FWF project number W901 (DK Molecular Enzymology). This work has been supported by the Austrian BMWFW, BMVIT, SFG, Standortagentur Tirol, Government of Lower Austria, and ZIT through the Austrian FFG-COMET-Funding Program (grant number 282482) and the French ANR for Glyco@Alps (ANR-15-IDEX-02) and Labex ARCANE (ANR-11-LABX-003).
The authors declare no competing financial interest.
Supplementary Material
References
- Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., and Etzler M. E. (2009) Essentials of Glycobiology, 2nd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Arnaud J.; Audfray A.; Imberty A. (2013) Binding sugars: from natural lectins to synthetic receptors and engineered neolectins. Chem. Soc. Rev. 42, 4798–4813. 10.1039/c2cs35435g. [DOI] [PubMed] [Google Scholar]
- Yoo T. H.; Link A. J.; Tirrell D. A. (2007) Evolution of a fluorinated green fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 104, 13887–13890. 10.1073/pnas.0701904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Bae J.; Rubini M.; Jung G.; Wiegand G.; Seifert M. H. J.; Azim M. K.; Kim J.-S.; Zumbusch A.; Holak T. A.; Moroder L.; Huber R.; Budisa N. (2003) Expansion of the genetic code enables design of a novel “gold” class of green fluorescent proteins. J. Mol. Biol. 328, 1071–1081. 10.1016/S0022-2836(03)00364-4. [DOI] [PubMed] [Google Scholar]
- Dieterich D. C.; Link A. J.; Graumann J.; Tirrell D. A.; Schuman E. M. (2006) Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. U. S. A. 103, 9482–9487. 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty K. E.; Tirrell D. A. (2008) Two-color labeling of temporally defined protein populations in mammalian cells. Bioorg. Med. Chem. Lett. 18, 5995–5999. 10.1016/j.bmcl.2008.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K.; Sugawara A.; Tirrell D. A. (2009) Generation of surface-bound multicomponent protein gradients. ChemBioChem 10, 2617–2619. 10.1002/cbic.200900542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasuhn D. E. Jr.; Singh P.; Strable E.; Brown S.; Manchester M.; Finn M. G. (2008) Plasma clearance of bacteriophage Qβ particles as a function of surface charge. J. Am. Chem. Soc. 130, 1328–1334. 10.1021/ja075937f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K.; Chin J. W. (2014) Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem. Rev. 114, 4764–4806. 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- Dumas A.; Lercher L.; Spicer C. D.; Davis B. G. (2015) Designing logical codon reassignment – Expanding the chemistry in biology. Chem. Sci. 6, 50–69. 10.1039/C4SC01534G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar Y.; Nadarajan S. P.; Hyeon Yoo T.; Lee C.-S.; Yun H. (2015) Incorporating unnatural amino acids to engineer biocatalysts for industrial bioprocess applications. Biotechnol. J. 10, 1862–1876. 10.1002/biot.201500153. [DOI] [PubMed] [Google Scholar]
- Ravikumar Y.; Nadarajan S. P.; Hyeon Yoo T.; Lee C.-s.; Yun H. (2015) Unnatural amino acid mutagenesis-based enzyme engineering. Trends Biotechnol. 33, 462–470. 10.1016/j.tibtech.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Salwiczek M.; Nyakatura E. K.; Gerling U. I. M.; Ye S.; Koksch B. (2012) Fluorinated amino acids: compatibility with native protein structures and effects on protein–protein interactions. Chem. Soc. Rev. 41, 2135–2171. 10.1039/C1CS15241F. [DOI] [PubMed] [Google Scholar]
- Katragadda M.; Magotti P.; Sfyroera G.; Lambris J. D. (2006) Hydrophobic effect and hydrogen bonds account for the improved activity of a complement inhibitor, compstatin. J. Med. Chem. 49, 4616–4622. 10.1021/jm0603419. [DOI] [PubMed] [Google Scholar]
- Luck L. A.; Falke J. J. (1991) 19F NMR studies of the D-galactose chemosensory receptor. 1. Sugar binding yields a global structural change. Biochemistry 30, 4248–4256. 10.1021/bi00231a021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez M. I.; Andreu C.; Vidal P.; Aboitiz N.; Freire F.; Groves P.; Asensio J. L.; Asensio G.; Muraki M.; Canada F. J.; Jimenez-Barbero J. (2005) On the importance of carbohydrate-aromatic interactions for the molecular recognition of oligosaccharides by proteins: NMR studies of the structure and binding affinity of AcAMP2-like peptides with non-natural naphthyl and fluoroaromatic residues. Chem. - Eur. J. 11, 7060–7074. 10.1002/chem.200500367. [DOI] [PubMed] [Google Scholar]
- Kostlanova N.; Mitchell E. P.; Lortat-Jacob H.; Oscarson S.; Lahmann M.; Gilboa-Garber N.; Chambat G.; Wimmerova M.; Imberty A. (2005) The fucose-binding lectin from Ralstonia solanacearum: A new type of β-propeller architecture formed by oligomerization and interacting with fucoside, fucosyllactose, and plant xyloglucan,. J. Biol. Chem. 280, 27839–27849. 10.1074/jbc.M505184200. [DOI] [PubMed] [Google Scholar]
- Mishra S. K.; Adam J.; Wimmerova M.; Koca J. (2012) In silico mutagenesis and docking study of Ralstonia solanacearum RSL lectin: performance of docking software to predict saccharide binding. J. Chem. Inf. Model. 52, 1250–1261. 10.1021/ci200529n. [DOI] [PubMed] [Google Scholar]
- Wimmerova M.; Kozmon S.; Necasova I.; Mishra S. K.; Komarek J.; Koca J. (2012) Stacking interactions between carbohydrate and protein quantified by combination of theoretical and experimental methods. PLoS One 7, e46032 10.1371/journal.pone.0046032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asensio J. L.; Arda A.; Canada F. J.; Jimenez-Barbero J. (2013) Carbohydrate-aromatic interactions. Acc. Chem. Res. 46, 946–954. 10.1021/ar300024d. [DOI] [PubMed] [Google Scholar]
- Hudson K. L.; Bartlett G. J.; Diehl R. C.; Agirre J.; Gallagher T.; Kiessling L. L.; Woolfson D. N. (2015) Carbohydrate–aromatic interactions in proteins. J. Am. Chem. Soc. 137, 15152–15160. 10.1021/jacs.5b08424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topin J.; Lelimousin M.; Arnaud J.; Audfray A.; Perez S.; Varrot A.; Imberty A. (2016) The hidden conformation of Lewis x, a human histo-blood group antigen, is a determinant for recognition by pathogen lectins,. ACS Chem. Biol. 11, 2011–2020. 10.1021/acschembio.6b00333. [DOI] [PubMed] [Google Scholar]
- Wiltschi B. (2012) Expressed protein modifications: Making synthetic proteins. Methods Mol. Biol. 813, 211–225. 10.1007/978-1-61779-412-4_13. [DOI] [PubMed] [Google Scholar]
- Lepthien S.; Hoesl M. G.; Merkel L.; Budisa N. (2008) Azatryptophans endow proteins with intrinsic blue fluorescence. Proc. Natl. Acad. Sci. U. S. A. 105, 16095–16100. 10.1073/pnas.0802804105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann R. D.; Adams M. D.; White O.; Clayton R. A.; Kirkness E. F.; Kerlavage A. R.; Bult C. J.; Tomb J. F.; Dougherty B. A.; Merrick J. M.; et al. (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512. 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Sloan M. J.; Phillips R. S. (1992) Enzymatic synthesis of aza-L-tryptophans: The preparation of 5- and 6-aza-L-tryptophan,. Bioorg. Med. Chem. Lett. 2, 1053–1056. 10.1016/S0960-894X(00)80617-4. [DOI] [Google Scholar]
- Lee M. S.; Phillips R. S. (1992) Enzymatic synthesis of chloro-L-tryptophans. Bioorg. Med. Chem. Lett. 2, 1563–1564. 10.1016/S0960-894X(00)80430-8. [DOI] [Google Scholar]
- Phillips R. S. (2004) Synthetic applications of tryptophan synthase. Tetrahedron: Asymmetry 15, 2787–2792. 10.1016/j.tetasy.2004.06.054. [DOI] [Google Scholar]
- Holmgren S. K.; Taylor K. M.; Bretscher L. E.; Raines R. T. (1998) Code for collagen’s stability deciphered. Nature 392, 666–667. 10.1038/33573. [DOI] [PubMed] [Google Scholar]
- Montclare J. K.; Son S.; Clark G. A.; Kumar K.; Tirrell D. A. (2009) Biosynthesis and stability of coiled-coil peptides containing (2S,4R)-5,5,5-trifluoroleucine and (2S,4S)-5,5,5-trifluoroleucine. ChemBioChem 10, 84–86. 10.1002/cbic.200800164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoulders M. D.; Satyshur K. A.; Forest K. T.; Raines R. T. (2010) Stereoelectronic and steric effects in side chains preorganize a protein main chain. Proc. Natl. Acad. Sci. U. S. A. 107, 559–564. 10.1073/pnas.0909592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aeschbacher T.; Zierke M.; Smiesko M.; Collot M.; Mallet J. M.; Ernst B.; Allain F. H.; Schubert M. (2017) A secondary structure element present in a wide range of fucosylated glycoepitopes. Chem. - Eur. J. 23, 11598–11610. 10.1002/chem.201701866. [DOI] [PubMed] [Google Scholar]
- Dunitz J. D.; Taylor R. (1997) Organic Fluorine Hardly Ever Accepts Hydrogen Bonds. Chem. - Eur. J. 3, 89–98. 10.1002/chem.19970030115. [DOI] [Google Scholar]
- Samsonov S. A.; Salwiczek M.; Anders G.; Koksch B.; Pisabarro M. T. (2009) Fluorine in protein environments: a QM and MD study. J. Phys. Chem. B 113, 16400–16408. 10.1021/jp906402b. [DOI] [PubMed] [Google Scholar]
- Miller B. R. 3rd; McGee T. D. Jr.; Swails J. M.; Homeyer N.; Gohlke H.; Roitberg A. E. (2012) MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 8, 3314–3321. 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- Frederiksen R. F.; Yoshimura Y.; Storgaard B. G.; Paspaliari D. K.; Petersen B. O.; Chen K.; Larsen T.; Duus J. O.; Ingmer H.; Bovin N. V.; Westerlind U.; Blixt O.; Palcic M. M.; Leisner J. J. (2015) A diverse range of bacterial and eukaryotic chitinases hydrolyzes the LacNAc (Galbeta1–4GlcNAc) and LacdiNAc (GalNAcbeta1–4GlcNAc) motifs found on vertebrate and insect cells,. J. Biol. Chem. 290, 5354–5366. 10.1074/jbc.M114.607291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audfray A.; Claudinon J.; Abounit S.; Ruvoën-Clouet N.; Larson G.; Smith D. F.; Wimmerová M.; Le Pendu J.; Römer W.; Varrot A.; Imberty A. (2012) The fucose-binding lectin from opportunistic pathogen Burkholderia ambifaria binds to both plant and human oligosaccharidic epitopes,. J. Biol. Chem. 287, 4335–4347. 10.1074/jbc.M111.314831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A.; Xiao H.; Yang P.-Y.; Soundararajan G.; Schultz P. G. (2013) A tryptophanyl-tRNA synthetase/tRNA pair for unnatural amino acid mutagenesis in E. coli. Angew. Chem., Int. Ed. 52, 5106–5109. 10.1002/anie.201301094. [DOI] [PubMed] [Google Scholar]
- Gibson D. G.; Young L.; Chuang R.-Y.; Venter J. C.; Hutchison C. A.; Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (2010) Xds. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M. D.; Ballard C. C.; Cowtan K. D.; Dodson E. J.; Emsley P.; Evans P. R.; Keegan R. M.; Krissinel E. B.; Leslie A. G.; McCoy A.; McNicholas S. J.; Murshudov G. N.; Pannu N. S.; Potterton E. A.; Powell H. R.; Read R. J.; Vagin A.; Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr., Sect. D: Biol. Crystallogr. 67, 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer G.; Cohen S. X.; Lamzin V. S.; Perrakis A. (2008) Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179. 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N.; Skubak P.; Lebedev A. A.; Pannu N. S.; Steiner R. A.; Nicholls R. A.; Winn M. D.; Long F.; Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr., Sect. D: Biol. Crystallogr. 67, 355–367. 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. (2010) Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F.; Nicholls R. A.; Emsley P.; Gražulis S.; Merkys A.; Vaitkus A.; Murshudov G. N. (2017) AceDRG: a stereochemical description generator for ligands. Acta Crystallogr., Sect. D: Biol. Crystallogr. 73, 112–122. 10.1107/S2059798317000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirre J.; Iglesias-Fernandez J.; Rovira C.; Davies G. J.; Wilson K. S.; Cowtan K. D. (2015) Privateer: software for the conformational validation of carbohydrate structures. Nat. Struct. Mol. Biol. 22, 833–834. 10.1038/nsmb.3115. [DOI] [PubMed] [Google Scholar]
- Maier J. A.; Martinez C.; Kasavajhala K.; Wickstrom L.; Hauser K. E.; Simmerling C. (2015) ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713. 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly C. I.; Cieplak P.; Cornell W.; Kollman P. A. (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model,. J. Phys. Chem. 97, 10269–10280. 10.1021/j100142a004. [DOI] [Google Scholar]
- Vanquelef E.; Simon S.; Marquant G.; Garcia E.; Klimerak G.; Delepine J. C.; Cieplak P.; Dupradeau F. Y. (2011) R.E.D. Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 39, W511–W517. 10.1093/nar/gkr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner K. N.; Yongye A. B.; Tschampel S. M.; Gonzalez-Outeirino J.; Daniels C. R.; Foley B. L.; Woods R. J. (2008) GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 29, 622–655. 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Darden T.; York D.; Pedersen L. (1993) Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.