Abstract
Patients with rheumatoid arthritis (RA) suffer from pain, which is associated with inflammation, peripheral and central pain processing, and joint structure damage. The aim of the present study was to investigate a key microRNA (miR) and its target genes that are involved in the pain responses of RA, and to clarify the mechanism of pain regulation. Collagen-induced arthritis (CIA) was induced in DBA/1 and C57BL/6 mice. The paw swelling, mechanical withdrawal threshold (MWT), thermal withdrawal latency (TWL), and expression levels of tumor necrosis factor (TNF)-α and prostaglandin (PG)E2 in the sera were investigated. Decreased MWT and TWL, and increased TNF-α and PGE2, in the CIA model group were observed in DBA/1 and C57BL/6 mice. DBA/1 mice exhibited greater hyperalgesia and higher levels of inflammatory mediators. miR-143-3p expression in the blood and the dorsal root ganglion (DRG) were detected, and low miR-143-3p expression was demonstrated in the blood and DRG tissue of CIA mice. The target genes of miR-143 were predicted and analyzed. A total of 1,305 genes were predicted and 55 pain-associated genes were obtained. Prostaglandin-endoperoxide synthase 2 (Ptgs2), MAS related GPR family member E (Mrgpre), prostaglandin D2 receptor and Tnf were selected as target genes of miR-143. DRG cells were cultured and transfected with miR-143-3p inhibitor or mimic. The expression of Mrgpre, Ptgs2 and Tnf was significantly inhibited following miR-143-3p mimic transfection, while the expression of Mrgpre, Ptgs2 and Tnf was increased following inhibitor transfection. Additionally, the expression of pain-associated genes in the DRG of mice was investigated and the expression of Ptgs2, Mrgpre and Tnf in the DRG of CIA mice was also significantly upregulated. These results revealed that CIA mice exhibited marked hyperalgesia and high levels of inflammatory pain mediators. Low expression of miR-143-3p negatively regulated the pain-associated target genes, including Mrgpre, Ptgs2 and Tnf, thereby affecting chronic inflammatory pain and neuropathic pain in RA.
Keywords: rheumatoid arthritis, microRNA-143, pain, inflammatory mediator, MAS related GPR family member E
Introduction
Rheumatoid arthritis (RA) is a common inflammatory autoimmune disease and the associated inflammation has been extensively studied. RA patients suffer from pain (1–3), which affects their daily lives (4). RA pain is frequently characterized by neuropathic pain. The dorsal root ganglion (DRG) is a place where primary sensory neurons accumulate in the pain conduction pathway (5), which may selectively perceive noxious stimulation and potential tissue damage. The sensory neuron sensitization of the DRG is one of the principal mechanisms underlying pain responses that are affected by the interaction of inflammatory cells with neurons in the DRG and the central nervous system. In addition, the release of inflammatory mediators (e.g., various prostaglandins, leukotrienes and interleukins) serves a crucial role in mediating the chronic pain of RA. The inflammatory mediators lead to peripheral sensitization of sensory nerves, and alterations in nerve growth and gene expression. Therefore, RA pain originates from joint inflammation and pain signal transduction processes, with complex regulatory mechanisms. It is, therefore, of great interest to identify whether there is a modulator that simultaneously influences the release of inflammatory pain mediators and pain conduction.
Previously, microRNAs (miRs) have been associated with certain pathological mechanisms for chronic inflammatory pain and neuropathic pain (6,7). The expression of miRs is significantly altered in different nerve tissues of previous pain models. In addition, selective accumulation or deletion of miRs in local synapses can regulate gene expression, protein synthesis and inflammatory pain-associated transcription factors (8), being closely associated with the occurrence and maintenance of pain. The authors' previous studies have also confirmed abnormalities in miR expression and alterations in miR-143 expression altered in RA patients and animal models (9,10). Certain reports also demonstrated that miR-143 had an association with inflammatory pain responses. For example, the expression of hsa-miR-143-3p is reduced in patients with fibromyalgia (11); miR-143 degrades cyclooxygenase (COX)-2 mRNA and regulates pain mediator production in pancreatic cancer (12). Tam et al (13) demonstrated that the expression levels of miR-143 were significantly lower in DRGs ipsilateral to complete Freund's adjuvant (CFA) injection. Therefore, the present study aimed to investigate the role of miR-143 in the complex mechanism of RA pain and to identify key pain targets for regulating pain responses.
In the present study, collagen-induced arthritis (CIA) was first compared by observing pain responses and detecting the levels of inflammatory pain mediators in the DBA/1 and C57BL/6 mice. miR-143 expression in the peripheral blood and DRG of CIA mice was further investigated, and the target genes that regulated RA pain responses were clarified. The results may provide novel insights into the development of strategies to prevent or alleviate RA pain.
Materials and methods
Animals
A total of 18 DBA/1 and 12 C57BL/6 male mice aged 6–8 weeks (16–20 g) were provided by the Laboratory Animal Center of the Academy of Military Medical Sciences (Beijing, China). The experiment was performed in the Experimental Animal Center of Nanjing University of Chinese Medicine (Jiangsu, China). Mice were housed in a temperature- (22±3°C) and humidity-controlled (40–70%) animal room under a 12-h light/dark cycle, with free access to food and water. All experimental protocols performed in this study were approved by the Ethics Review Committee for Animal Experimentation of Nanjing University of Chinese Medicine and were in accordance with the Declaration of the National Institutes of Health Guide for Care and Use of Laboratory Animals (14).
Establishment of the DBA/1 or C57BL/6 mouse model of CIA (15)
Bovine type II collagen (CII; Chondrex, Inc., Redmond, WA, USA) was dissolved in 0.01 mol/l acetic acid into a 2 g/l solution and shaken overnight at 4°C. The solution was mixed with CFA (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at a ratio of 1:1 and emulsified. A total of 12 DBA/1 or C57BL/6 mice were separately divided into two groups (n=6). In the model group, a mixed emulsion (100 µl) of CII and CFA was intradermally injected at the tail root of mice for primary immunization, and this day was recorded as d1. On d21, CII was mixed evenly with incomplete Freund's adjuvant (Sigma-Aldrich; Merck KGaA) at a ratio of 1:1, 100 µl of which was injected at the tail root to boost immunization. The mice in the control group were intradermally injected with normal saline at the tail root on d1 and d21, respectively.
Detection of inflammatory responses and pain mediator release in DBA/1 or C57BL/6 mice with CIA
The paw thickness of DBA/1 mice was measured on d21, d28, d35, d41 and d49, and the mice were sacrificed by anesthesia on d49. The paw thickness of C57BL/6 mice was measured on d5, d12, d19, d26, d33, d40, d47 and d54, and the mice were sacrificed by anesthesia on d54. The sera of all mice were collected and the levels of tumor necrosis factor (TNF)-α and prostaglandin (PG)E2 were measured using mouse TNF-α (cat. no. YY02868B) and PGE2 ELISA kits (cat. no. YY02798B; Shanghai Yuanye Biotechnology Co., Ltd., Shanghai, China).
Detection of pain responses in DBA/1 or C57BL/6 mice with CIA
Pain responses, including the mechanical withdrawal threshold (MWT) and thermal withdrawal latency (TWL) were measured on d20, d24, d30, d36, d42 and d48 in DBA/1 mice, and on d5, d12, d19, d26, d33, d40, d47 and d54 in C57BL/6 mice. The tests were performed as follows. The experimental environment was kept quiet. The mice were placed in a plexiglass box with metal mesh bottom. Following >30 min of adaptation, the experiment was begun when the exploration activities had stopped. Von Frey filaments (0.008–4.0 g) were used to stimulate the mouse paw vertically into a slight S shape. Obvious paw withdrawal, lifting or licking was regarded as a positive response, and no response was negative. MWT was measured and calculated using the ‘up and down’ method described by Chaplan et al (16).
Under the same conditions described above, the mouse paw was irradiated with a radiation source using a 37370 thermal plantar analgesia instrument and the avoidance time, from the initiation of irradiation to leg lifting and paw withdrawal, was recorded. Each mouse was tested three times with a 5-min interval, and the average was recorded as TWL.
Detection of miR-143-3p expression in the peripheral blood and DRG using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
A total of 6 DBA/1 mice were equally divided into a control group and a CIA group, and the CIA model was established with the method described above. Peripheral blood and DRG tissues were obtained from the control and CIA mice on d49, from which the RNA was extracted with ice-cold TRIzol® (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The RNA concentration and purity were spectrophotometrically determined at 260 and 280 nm. cDNA was synthesized using a Transcriptor First-Strand cDNA Synthesis kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). The reverse transcription reaction was performed under the following conditions: 16°C for 30 min, 42°C for 40 min and 85°C for 5 min. The target miR of each sample and the internal reference (mmu-miR-425-5p) were detected using an Applied Biosystems detection instrument (Thermo Fisher Scientific, Inc.). mmu-miR-425-5p was used for normalization. The SYBR® Green qPCR Master Mix (Arraystar Inc, Rockville, MD, USA) was used and the PCR was performed using the following cycling conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and 60°C for 60 sec. The data were analyzed using the 2−ΔΔCq method (17). A total of three replicate wells were set up and the results were averaged. The primers were as follows: mmu-miR-143-3p gene specific primer (GSP), 5′-GGGATGAGATGAAGCACT-3′ and reverse (R), 5′-TGCGTGTCGTGGAGTC-3′; mmu-miR-425-5p GSP, 5′-GGGGAATGACACGATCACTC-3′ and R, 5′-GTGCGTGTCGTGGAGTCG-3′.
Bioinformatics analysis of pain-associated target genes of miR-143
The target genes of miR-143 were predicted using bioinformatics databases, including miRanda (http://www.mirbase.org), miRDB v. 4.0 (http://mirdb.org/miRDB), TargetScan v. 6.2 (http://www.targetscan.org), miRWalk v. 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2) and RNA22 v. 2 (https://cm.jefferson.edu/rna22/Interactive). The target genes of the three intersects were selected to minimize the false positive rate. All the target genes were analyzed by Cytoscape software v. 3.5.0 (http://www.cytoscape.org/) to obtain the microRNA-gene regulatory network. Using the gene database from the Pain Genetics Lab (http://www.psych.mcgill.ca/labs/mogillab/paingenetics/lab/) of McGill University (Montreal, Canada), the predicted target genes of miR-143 were analyzed comprehensively to determine potential pain-associated genes.
Gene ontology (GO) analysis and pathway analysis of miR-143 target genes
GO functional annotation enrichment analysis of the predicted miR-143 target genes was conducted using the GO database (http://www.geneontology.org), including cell components and molecular function. Using the Kyoto Encyclopedia of Gene and Genomes (KEGG) database (http://www.genome.jp/kegg) in the DAVID database, biological pathway enrichment analysis was performed on the predicted target genes.
Isolation, culture and transfection of DRG cells
In an ice bath, the spinal canal of a normal mouse was cut open under a dissecting microscope to remove the spinal cord and other nerve fibers to isolate the DRG. Subsequently, 1 ml mixed digestive enzyme was added, digestion was performed in a 37°C water bath for 25–30 min and the resulting tissue was pipetted repeatedly into a single cell suspension. The supernatant was discarded following centrifugation (4°C, 12,000 × g, 15 min) and 1 ml DH10 medium (WISENT Inc., Quebec, QC, Canada) and 1% nerve growth factor (NGF; R&D Systems, Inc., Minneapolis, MN, USA) were added to the precipitate. The cell suspension was pipetted into a single cell suspension and grown in a pre-coated petri dish. When the density of DRG cells reached 50–60% confluence, the cell culture medium was replaced prior to transfection.
The mmu-miR-143-3p sequence (Shanghai GenePharma Co., Ltd., Shanghai, China) was dissolved into a 10 µM stock solution, aliquoted and stored at-20°C. When the density of DRG cells reached 50–60%, the culture medium was refreshed prior to transfection and 2 ml antibiotic-free DH10 medium was added into each dish. A total of 3 µl miR mimic, inhibitor or miR negative control (NC) (Shanghai GenePharma Co., Ltd.) was mixed gently with 150 µl Opti-MEM reduced serum medium (Gibco; Thermo Fisher Scientific, Inc.). A total of 9 µl Lipofectamine® RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.) was diluted with 150 µl Opti-MEM reduced serum medium. Diluted Lipofectamine® RNAiMAX was mixed gently with diluted miR. miR-143 mimic NC-FAM (carboxyfluorescein): Sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. miR-143 mimic: Sense, 5′-UGAGAUGAAGCACUGUAGCUC-3′ and antisense, 5′-GCUACAGUGCUUCAUCUCAUU-3′. miR-143 inhibitor: Sense, 5′-GAGCUACAGUGCUUCAUCUCA-3′. miR-143 inhibitor NC: Sense, 5′-CAGUACUUUUGUGUAGUACAA-3′. Following incubation at room temperature for 5 min, the mixture was added into the dish containing the DRG cells and culture medium, mixed by gentle agitation and cultured in a CO2 incubator at 37°C. The cells were observed using fluorescence microscopy at 6, 12, 24, 48 and 72 h following transfection to determine the suitable transfection conditions.
Detection of the expression of miR-143-3p target genes in transfected DRG cells and DRG tissues of CIA mice
DRG cells were cultured and transfected with the method described above. miR-143-3p mimic, inhibitor and miR NCs were transfected into DRG cells for 48 h. RNA was extracted from DRG cells and the expression of mmu-miR-143, MAS related GPR family member E (Mrgpre), prostaglandin endoperoxide synthase 2 (Ptgs2, also known as COX-2), prostaglandin D2 receptor (Ptgdr) and Tnf were detected using qPCR.
A total of 6 DBA/1 mice were divided into a control group and a CIA group. The CIA model was established by the method described above. DRG tissues were obtained from the control and CIA mice on d49, from which RNA was extracted. The expression of Mrgpre, Ptgs2 and Tnf were detected using RT-qPCR, as described above.
The primers were as follows: Mrgpre forward (F), 5′-CAGAACCACCCGTAGGCAAAT-3′ and R, 5′-TTCCAGGTCACCGTCCATCAC-3′; Ptgs2 F, 5′-CAGATGACTGCCCAACTCCCA-3′ and R, 5′-GTGAACCCAGGTCCTCGCTTA-3′; Ptgdr F, 5′-AACACCGTCTCACTGTAGGCTT-3′ and R, 5′-CTGGTTTCCCAACTCATTTCTC-3′; Tnf F, 5′-GAGTCCGGGCAGGTCTACTTT-3′ and R, 5′-CAGGTCACTGTCCCAGCATCT-3′; U6 F, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and R, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; and GAPDH F, 5′-CACTGAGCAAGAGAGGCCCTAT-3′ and R, 5′-GCAGCGAACTTTATTGATGGTATT-3′.
Data processing and statistics
The experiments were repeated three times. Experimental data are expressed as the means ± standard deviation. T-test was used for the comparison of two groups and analysis of variance followed by Tukey's test was used for the comparison of multiple groups. Data processing was performed using the SPSS v. 13.0 statistical software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Comparison of inflammatory pain responses and pain mediator release in DBA/1 or C57BL/6 mice with CIA
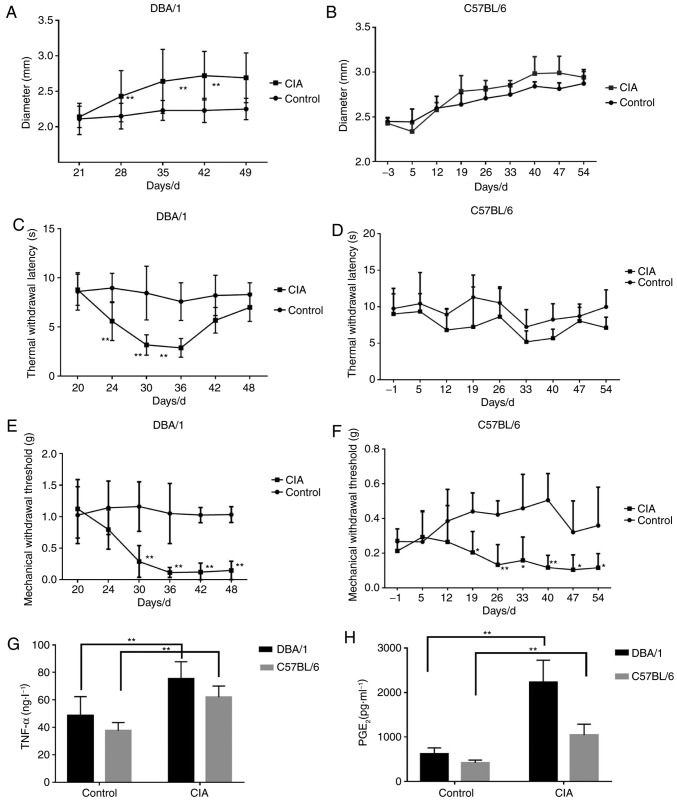
DBA/1 and C57BL/6 mouse models of CIA were constructed, and the alterations of CIA-associated indices were investigated. The forelimb and hindlimb became red and swollen from d24 (DBA/1 mice) and d33 (C57BL/6 mice), and their paws and ankle joints were also notably affected on d28 (DBA/1 mice) and d40 (C57BL/6 mice). The paw thickness markedly increased with the progression of arthritis in the CIA group compared with the control group (P<0.01; Fig. 1A and B).
Figure 1.
Inflammatory reactions and pain responses in DBA/1 or C57BL/6 mice with CIA. Paw thickness was measured between d21 and d49. TWL and MWT were detected between d20 and d48. The levels of TNF-α and PGE2 in the sera were determined by ELISA on d49. (A) Footpad diameter of DBA/1 mice. (B) Footpad diameter of C57BL/6 mice. (C) TWL of DBA/1 mice. (D) TWL of C57BL/6 mice. (E) MWT of DBA/1 mice. (F) MWT of C57BL/6 mice. (G) TNF-α levels in the sera. (H) PGE2 levels in the sera. *P<0.05, **P<0.01 vs. respective control. TWL, thermal withdrawal latency; MWT, mechanical withdrawal threshold; TNF, tumor necrosis factor; PGE2, prostaglandin E2; CIA, collagen-induced arthritis.
MWT and TWL, which indicate pain responses, were also detected. For DBA/1 mice, the TWL of the CIA model group decreased significantly from d24 (P<0.01; Fig. 1C) and gradually recovered from d42, and MWT dropped significantly from d30 (P<0.01; Fig. 1E). For C57BL/6 mice, TWL was slightly lower compared with the control group (P>0.05; Fig. 1D) and the MWT of the CIA model group significantly decreased (P<0.05; Fig. 1F).
Additionally, the levels of inflammatory pain mediators, including TNF-α and PGE2, significantly increased in the CIA model group compared with the control group for the two strains of mice (P<0.01; Fig. 1G and H).
The results indicated that DBA/1 mice underwent more marked hyperalgesia and had increased levels of inflammatory mediators compared with the C57BL/6 mice, thus they were selected for the following experiments.
miR-143-3p expression in the peripheral blood and DRG
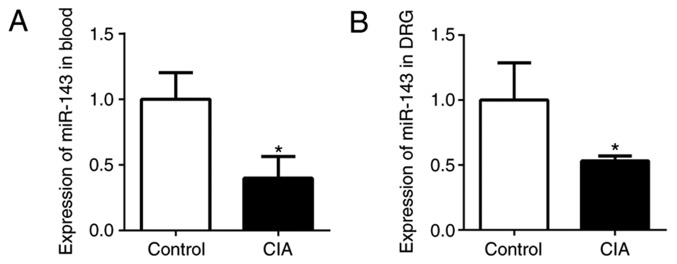
In the authors' previous studies, miRs in CIA mice and normal mice were screened, revealing that miR-143-3p was expressed at a low level (9,10). In the present study, miR-143-3p expression in the peripheral blood and DRG was detected by RT-qPCR. The relative expression of miR-143-3p in the peripheral blood of CIA mice was significantly lower compared with the normal control mice (P<0.05; Fig. 2A). In addition, miR-143-3p expression in the DRG tissue of the CIA mice was also significantly downregulated (P<0.05; Fig. 2B). These results further confirmed that low miR-143-3p expression was an important feature of the CIA mouse model.
Figure 2.
miR-143-3p expression measured by reverse transcription quantitative polymerase chain reaction in DBA/1 mice. miR-143-3p expression in (A) the peripheral blood and (B) DRG of mice.*P<0.05 vs. control. CIA, collagen-induced arthritis; miR, microRNA; DRG, dorsal root ganglion.
Prediction of pain-associated target genes of miR-143
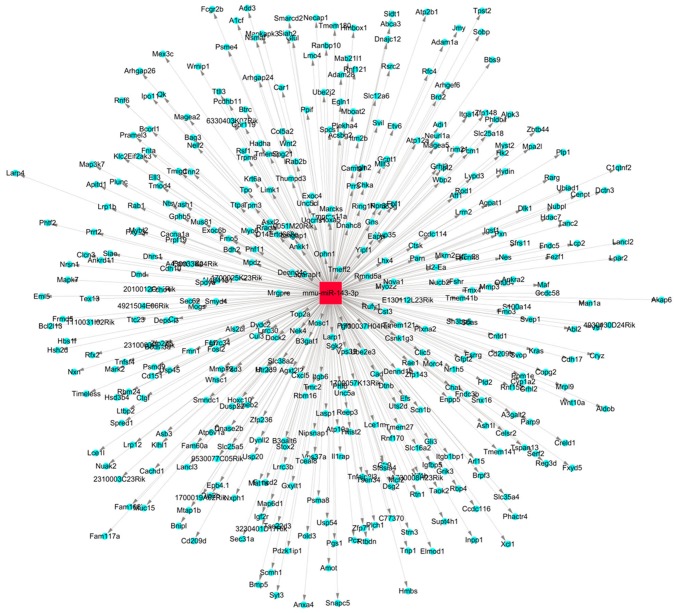
The target genes of miR-143 were predicted and analyzed. The miR-target gene regulatory network is exhibited in Fig. 3. A total of 1,305 genes, including mitogen-activated protein kinase (MAPK)7, MAPK3, COX2, matrix metalloproteinase and TNF, were predicted to be the target genes of miR-143. Using the gene database of the Pain Genetics Lab, 55 pain-associated genes were obtained as potential target genes of miR-143 (Table I).
Figure 3.
miR-143 target gene regulatory network. The red square represents the miR, the blue circles represent the target genes and the lines with grey arrowheads indicate their interactions. miR, microRNA.
Table I.
Potential pain-associated target genes of miR-143.
| Gene | Protein name | Protein acronym |
|---|---|---|
| Accn1 | Amiloride-sensitive cation channel 1, neuronal (degenerin) | ASIC2 |
| Adcy1 | Adenylatecyclase 1 | AC1 |
| Adra1d | Adrenergic receptor, α1d | a1D-AR |
| Adra2c | Adrenergic receptor, α2c | a2C-AR |
| Adrbk2 | Adrenergic receptor kinase, β2 | GRK3 |
| Agtr2 | Angiotensin II receptor, type 2 | AT2R |
| Bace1 | β-site APP cleaving enzyme 1 | – |
| Cacnb3 | Calcium channel, voltage-dependent, β3 subunit | b3 |
| Calb1 | Calbindin1 | Calb-1 |
| Ccr5 | Chemokine (C-C motif) receptor 5 | CCR5 |
| Cd274 | CD274 antigen | B7-H1 |
| Cd40 | CD40 antigen | CD40 |
| Chrna4 | Cholinergic receptor, nicotinic, α polypeptide 4 | Acra4 |
| Clock | Circadian locomoter output cycles kaput | CLOCK |
| Cnr1 | Cannabinoid receptor 1 (brain) | CB1 |
| Cxcr3 | Chemokine (C-C-C motif) receptor 3 | CXCR3 |
| Dicer1 | Dcr-1 homolog (Drosophila) | – |
| Dlg2 | Discs, large homolog 2 (Drosophila) | – |
| Drd1a | Dopamine receptor D1A | DRD1 |
| Foxn1 | Forkhead box N1 | nu |
| Gabbr1 | γ-aminobutyric acid (GABA-B) receptor, 1 | GABA-B(1) |
| Gad2 | Glutamic acid decarboxylase 2 | GAD65 |
| Gja1 | Gap junction protein, α1 | Cx43 |
| Gria1 | Glutamate receptor, ionotropic, AMPA1 (α1) | GluR-1 |
| Ifngr1 | Interferon γ receptor 1 | IFNgR |
| Ikbke | Inhibitor of κB kinase ε | IKKepsilon |
| Il1rap | Interleukin 1 receptor accessory protein | IL-1RAcP |
| Kcna1 | Potassium voltage-gated channel, shaker-related subfamily, member 1 | Kv1.1 |
| L1cam | L1 cell adhesion molecule | L1 |
| Mgll | Monoglyceride lipase | MAGL |
| Mmp24 | Matrix metalloproteinase 24 | MT5-MMP |
| Mrgpre | MAS related GPR, member E | MrgE |
| Nf1 | Neurofibromatosis 1 | Nf-1 |
| Ngfr | Nerve growth factor receptor (TNFR superfamily, member 16) | p75 |
| Nlgn2 | Neuroligin 2 | NL2 |
| Nptx1 | Neuronal pentraxin 1 | NP1 |
| Npy | Neuropeptide Y | NPY |
| Nts | Neurotensin | NT |
| Ntsr1 | Neurotensin receptor 1 | NT1R |
| Pcsk2 | Proprotein convertase subtilisin/kexin type 2 | PC2 |
| Pik3cg | Phosphoinositide-3-kinase, catalytic, γ polypeptide | PI3Kg |
| Plp1 | Proteolipid protein (myelin) 1 | Plp |
| Ppp1r9b | Protein phosphatase 1, regulatory subunit 9B | – |
| Ptgdr | Prostaglandin D receptor | DP |
| Ptgfr | Prostaglandin F receptor | FP |
| Ptgs2 | Prostaglandin-endoperoxide synthase 2 | COX-2 |
| Rabggta | Rab geranylgeranyl transferase, a subunit | gm |
| Slc12a6 | Solute carrier family 12, member 6 | KCC3 |
| Slc6a2 | Solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 | NET |
| Slc6a4 | Solute carrier family 6, member 4 | 5-HTT |
| Stx1a | Syntaxin 1A (brain) | HPC-1 |
| Tacr1 | Tachykinin receptor 1 | NK1R |
| Tnf | Tumor necrosis factor | TNF-α |
| Vip | Vasoactive intestinal polypeptide | VIP |
CD, cluster of differentiation; miR, microRNA.
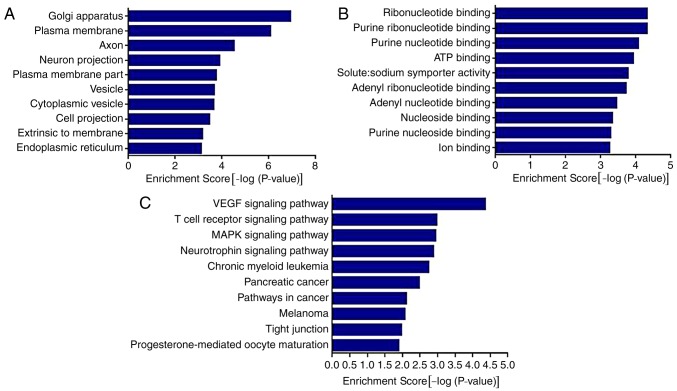
The results of the GO analysis demonstrated that the genes were associated with ‘Golgi apparatus’, ‘plasma membrane’, ‘axon’, ‘neuron projection’, ‘vesicle’, ‘cytoplasmic vesicle’ and ‘endoplasmic reticulum’. The results also demonstrated that the genes were associated with various molecular functions (Fig. 4A), including ‘ribonucleotide binding’, ‘purine ribonucleotide binding’, ‘ATP binding’, ‘solute: sodium symporter activity’ and ‘ion binding’.
Figure 4.
GO and KEGG analysis of miR-143 target pain-associated genes. (A) GO analysis of cell components. (B) GO analysis of molecular functions. (C) KEGG analysis of miR-143. GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; miR, microRNA; VEGF, vascular endothelial growth factor; MAPK, mitogen-activated protein kinase; ATP, adenosine triphosphate.
The results of the KEGG analysis demonstrated that miR-143 was involved in the regulation of multiple pathways and its target genes were significantly enriched in ‘VEGF signaling pathway’, ‘T cell receptor signaling pathway’, ‘MAPK signaling pathway’, ‘neurotrophin signaling pathway’, ‘pathways in cancer’ and other signaling pathways (P<0.01).
Following this, Ptgs2, Mrgpre, Ptgdr and Tnf were selected as associated target genes for further verification, based on the results of the GO and KEGG analysis and the factors in RA inflammatory pain.
Effects of miR-143 on pain-associated target genes
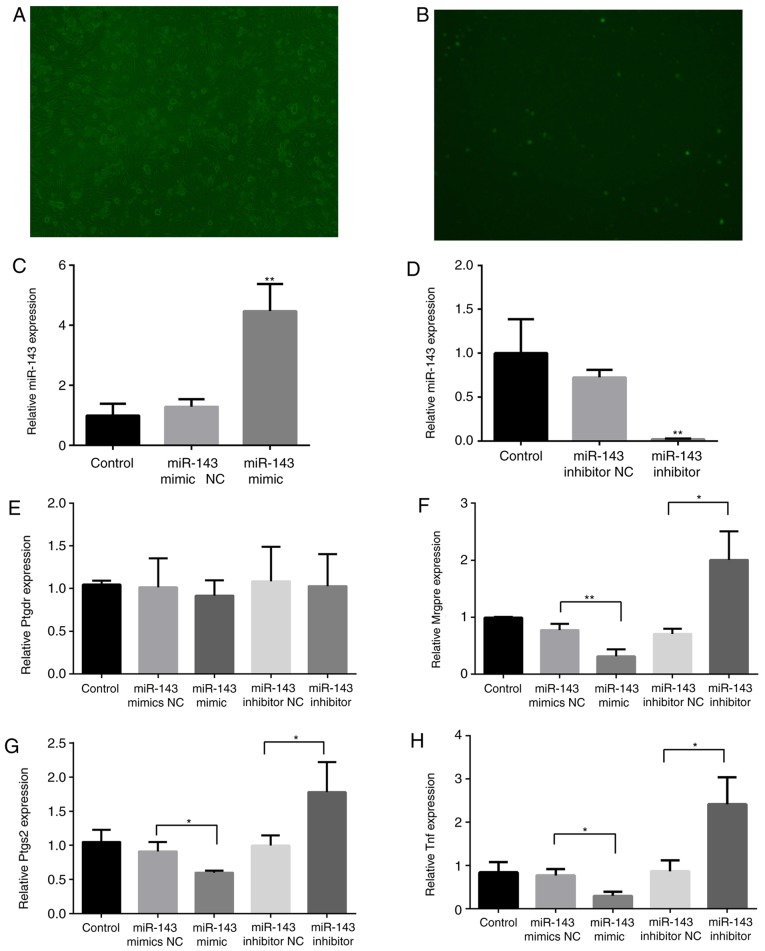
To determine the association between miR-143 and target genes, DRG cells were cultured and transfected with Lipofectamine RNAiMAX transfection reagent and the miR-143-3p mimic NC-FAM. The transfected cells were observed using microscopy at 6, 12, 24, 48 and 72 h. As demonstrated in Fig. 5A and B, DRG cells were successfully transfected, and the transfection efficiency peaked (~70%) at 48 h. miR-143-3p expression significantly decreased following miR-143 inhibitor transfection for 48 h (P<0.01), whereas it increased 3.5-fold following miR-143 mimic transfection (P<0.01; Fig. 5C and D), verifying the effects of inhibitor or mimic transfection.
Figure 5.
Effects of miR-143 on pain-associated target genes. Mouse DRG cells were transfected with Lipofectamine RNAiMAX transfection reagent and the miR-143-3p mimic NC-carboxyfluorescein. (A) Bright field (magnification, ×100). (B) Fluorescence microscopy (magnification, ×100). RNA was extracted from the DRG cells 48 h following transfection with an miR-143-3p inhibitor or mimic. (C) miR-143-3p expression following transfection with the mimic. (D) miR-143-3p expression following transfection with the inhibitor. (E) Ptgdr expression, (F) Mrgpre expression, (G) Ptgs2 expression and (H) Tnf expression. *P<0.05, **P<0.01 vs. NC group. miR, microRNA; TNF, tumor necrosis factor; DRG, dorsal root ganglion; Mrgpre, MAS related GPR family member E; Ptgdr, prostaglandin D2 receptor; Ptgs2, prostaglandin-endoperoxide synthase 2; NC, negative control.
Subsequently, the expression of Mrgpre, Ptgs2, Ptgdr and Tnf in DRG cells was detected. As illustrated in Fig. 5E-H, the expression of Mrgpre, Ptgs2 and Tnf was significantly inhibited (P<0.05) following miR-143-3p mimic transfection, although it was significantly promoted following miR-143-3p inhibitor transfection (P<0.05). The mimic and inhibitor transfections did not markedly affect Ptgdr expression.
Expression of pain-associated genes in the DRG of CIA mice
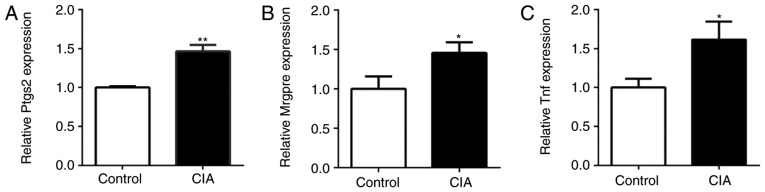
The expression of Ptgs2, Mrgpre and Tnf in the DRG of CIA and control mice was also detected by RT-qPCR. Compared with the control mice, the expression of Ptgs2, Mrgpre and Tnf in the DRG of CIA mice was significantly upregulated (P<0.05; Fig. 6). The expression of miR-143-3p was negatively associated with the expression of Mrgpre, Ptgs2 and Tnf in CIA mice, being consistent with the results of the in vitro experiments.
Figure 6.
Expression of pain-associated genes in the dorsal root ganglia of CIA mice. (A) Ptgs2 expression, (B) Mrgpre expression and (C) Tnf expression. *P<0.05, **P<0.01 vs. control. Mrgpre, MAS related GPR family member E; Ptgs2, prostaglandin-endoperoxide synthase 2; TNF, tumor necrosis factor; CIA, collagen-induced arthritis.
Discussion
The pain of RA patients is associated with multiple complex mechanisms, including inflammation, peripheral and central pain processing, and joint structure damage (18). Therefore, it may be useful for effective treatment to investigate the mechanisms of pain. In this study, the role of miR-143 in RA pain was investigated and the mechanism by which miR-143 may regulate the pain of CIA is described for the first time, to the best of the authors' knowledge.
The CIA model has a similar pathological mechanism to that of human RA (19) and it is the most widely used mouse model for RA (20). CII is the principal constituent collagen form in articular cartilage, and immunoreactivity to CII maybe identified in certain patients with RA. Immunization with CII leads to the development of severe polyarticular arthritis mediated by immune mechanisms. The autoimmune response involves CII-specific T cells and B cells and their products, proinflammatory cytokines and anti-CII antibodies (21). Thus, the CIA model may mimic the responses of RA pain more accurately compared with other pain models. Considering that different strains of mice may have a number of different symptoms, two strains of mice were tested, and it was demonstrated that they were subjected to apparent CIA. MWT and TWL are the common indices that reflect pain sensitivity to mechanical and thermal stimuli. In the present study, the MWT and TWL values of DBA/1 and C57BL/6 CIA mice decreased, demonstrating that the CIA animal models exhibited pain behaviors. Furthermore, DBA/1 CIA mice underwent increased hyperalgesia compared with the C57BL/6 mice. Also, the levels of PGE2 and TNF-α increased in the CIA mice. The results of the present study also confirmed the increase in PGE2 and TNF-α levels in CIA mice. As a well-known mediator of inflammation and pain, PGE2 serves a key role in the occurrence and maintenance of RA pain and hyperalgesia (22). TNF-α, an essential inflammatory factor for RA (23), is able to induce the release of bradykinin, promote the release of substance P from nerve terminal receptors and accelerate the synthesis of PGE2 through COX activation (24,25). Therefore, increasing inflammatory mediators may be one of the mechanisms underlying the pain response. DBA/1 CIA mice exhibited increased levels of inflammatory mediators compared with C57BL/6 mice. Accordingly, DBA/1 mice were selected to construct the CIA model for further investigation.
Low-level expression of miR-143 in the peripheral blood of patients with RA and CIA mice has been previously demonstrated (9). In addition, miR-143 has been associated with inflammatory pain responses (11,12). In the present study, RT-qPCR demonstrated that miR-143 was expressed at a low level in the peripheral blood of CIA mice. Considering the role of the DRG in pain conduction, miR-143 expression was also measured, and the results demonstrated that miR-143 may be an important modulator of the responses to CIA pain.
Bioinformatics analysis also revealed that certain potential target genes of miR-143 were pain-associated and others were associated with the production of inflammatory mediators. As suggested by GO and KEGG analysis, the cytokines may be synthesized in the Golgi apparatus and may be transported in a soluble form via the endoplasmic reticulum. Alternatively, they may be present in a membrane-bound form or processed into an intracellular form and transported in the cell. Given that Ptgs2, Ptgdr and Tnf have been closely associated with the release of inflammatory pain mediators in RA, they may be the miR-143-3p target genes for pain regulation. In addition, pain conduction is also worthy of attention according to the results of the GO analysis. For example, target-gene associated axons, the output channels of the neurons, are the principal signal transmission channels in the nervous system. The analysis of the molecular functions also indicated that miR-143 may regulate gene expression by influencing nucleotide binding. It may also regulate the selective transport of sodium ions, which maintain the cell excitability and signal transduction, and affect peripheral and central nervous system sensitization. miR-143 may also influence adenosine triphosphate (ATP) release and its combination with the ATP receptor in the primary afferent, and subsequently modulate the excitement of the DRG nociceptive neurons. Among the potential genes, Mrgpre is selectively highly expressed in the DRG and small neurons of the trigeminal ganglion, which are closely associated with pain conduction, being involved in the onset of chronic pain and hyperalgesia (26). Therefore, Mrgpre may be the target gene of miR-143-3p.
To further clarify the target genes of miR-143-3p in CIA pain responses, mouse DRG cells were cultured and transfected with an miR-143-3p inhibitor or mimic. The expression of certain potential genes in the transfected cells was compared, and it was demonstrated that the expression of Mrgpre, Ptgs2 and Tnf was negatively associated with miR-143-3p, although Ptgdr expression was not notably affected. Therefore, Mrgpre, Ptgs2 and Tnf may be the target genes of miR-143-3p. The results of the in vivo experiment were similar. In detail, the expression of Mrgpre, Ptgs2 and Tnf increased in the DRG of CIA mice. Low miR-143-3p expression in RA may induce upregulation of Mrgpre in nervous tissues, therefore affecting the onset of hyperalgesia and pain conduction. In addition, Ptgs2 and Tnf expression were also upregulated. Ptgs2 in DRG cells may facilitate the synthesis of PGE2 (27), activate the corresponding receptors in the DRG and a variety of cellular signaling pathways, augment the sensitivity of nociceptors to pain stimulation, and finally cause hyperalgesia and allodynia (18,28). Tnf mRNA may further regulate the release of TNF-α and subsequently activate COX to enhance the regulation of PGEs (23,25). Pham et al (12) also reported that miR-143 degraded COX-2 mRNA and regulated PGE production. Therefore, miR-143-3p may modulate the levels of inflammatory algogenic substances (PGE2 and TNF-α) by regulating its target genes. Similarly, the increasing levels of PGE2 and TNF-α in CIA mice concurred with the above analysis. Taken together, miR-143-3p regulated pain transduction and the release of inflammatory pain mediators, thereby regulating the complex mechanisms of RA pain.
In conclusion, CIA mice were subjected to hyperalgesia and the release of inflammatory pain mediators. Low expression of miR-143-3p negatively regulated pain-associated target genes, including Mrgpre, Ptgs2 and Tnf, thereby affecting the chronic inflammatory pain and neuropathic pain of RA. Seeking drugs that regulate miR-143 may be useful for the treatment of RA pain.
Acknowledgements
The authors would like to express their sincere gratitude to Professor Zongxiang Tang for his constructive comments and useful suggestions as well as his technical assistance for this project.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 81673937, 81573869 and 81573929) and the National Natural Science Foundation for the Youth of Jiangsu Province (grant no. BK20161043).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LLZ and XPZ conceived the project and designed the experiments. LLZ, YMZ, FYQ, CCY and DPY performed the experiments. LLZ and YMZ analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All experimental protocols performed in this study were approved by the Ethics Review Committee for Animal Experimentation of Nanjing University of Chinese Medicine and were in accordance with the Declaration of the National Institutes of Health Guide for Care and Use of Laboratory Animals (14).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Heiberg T, Kvien TK. Preferences for improved health examined in 1,024 patients with rheumatoid arthritis: pain has highest priority. Arthritis Rheum. 2002;47:391–397. doi: 10.1002/art.10515. [DOI] [PubMed] [Google Scholar]
- 2.Walsh DA, Mcwilliams DF. Pain in rheumatoid arthritis. Curr Pain Headache Rep. 2012;16:509–517. doi: 10.1007/s11916-012-0303-x. [DOI] [PubMed] [Google Scholar]
- 3.Mcwilliams DF, Zhang W, Mansell JS, Kiely PD, Young A, Walsh DA. Predictors of change in bodily pain in early rheumatoid arthritis: An inception cohort study. Arthrit Care Res (Hoboken) 2012;64:1505–1513. doi: 10.1002/acr.21723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leffler AS, Kosek E, Lerndal T, Nordmark B, Hansson P. Somatosensory perception and function of diffuse noxious inhibitory controls (DNIC) in patients suffering from rheumatoid arthritis. Eur J Pain. 2002;6:161–176. doi: 10.1053/eujp.2001.0313. [DOI] [PubMed] [Google Scholar]
- 5.Gold MS, Gebhart GF. Nociceptor sensitization in pain pathogenesis. Nat Med. 2010;16:1248–1257. doi: 10.1038/nm.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai G, Ambalavanar R, Wei D, Dessem D. Downregulation of selective microRNAs in trigeminal ganglion neurons following inflammatory muscle pain. Mol Pain. 2007;3:15. doi: 10.1186/1744-8069-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aldrich BT, Frakes EP, Kasuya J, Hammond DL, Kitamoto T. Changes in expression of sensoryorgan-specific microRNAs in rat dorsal root ganglia in association with mechanical hypersensitivity induced by spinal nerve ligation. Neuroscience. 2009;164:711–723. doi: 10.1016/j.neuroscience.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Lee MC, Momin A, Cendan CM, Shepherd ST, Baker MD, Asante C, Bee L, Bethry A, Perkins JR, et al. Small RNAs control sodium channel expression, nociceptor excitability, and pain thresholds. J Neurosci. 2010;30:10860–10871. doi: 10.1523/JNEUROSCI.1980-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu YM, Zhou LL, Peng XW, Geng S, Zhou XP. Study of Qingluo Tongbi Compound treating rheumatoid arthritis based on miRNA network. Chin J Immunol. 2016;32:495–499. (In Chinese) [Google Scholar]
- 10.Yuan CC, Geng SA, Zhu YM, et al. Comparision of abnormal expression of miRNAsin peripheral blood of rheumatoid arthritis patients and osteoclasts in rat and analysis of MiRNAs. Chin J Immunol. 2017;33:715–720. (In Chinese) [Google Scholar]
- 11.Cerdá-olmedo G, Mena-durán AV, Monsalve V, Oltra E. Identification of a MicroRNA Signature for the Diagnosis of Fibromyalgia. PLoS One. 2015;10:e0121903. doi: 10.1371/journal.pone.0121903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pham H, Rodriguez CE, Donald GW, Hertzer KM, Jung XS, Chang HH, Moro A, Reber HA, Hines OJ, Eibl G. miR-143 decreases COX-2 mRNA stability and expression in pancreatic cancer cells. Biochem Biophys Res Commun. 2013;439:6–11. doi: 10.1016/j.bbrc.2013.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tam Tam S, Bastian I, Zhou XF, Vander Hoek M, Michael MZ, Gibbins IL, Haberberger RV. MicroRNA-143 expression in dorsal root ganglion neurons. Cell Tissue Res. 2011;346:163–173. doi: 10.1007/s00441-011-1263-x. [DOI] [PubMed] [Google Scholar]
- 14.National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals: Guide for the Care and Use of Laboratory Animals. 8th edition. Vol. 327. The National Academies Press; Washington, DC: 2011. pp. 963–965. Publication no. 85-23 (rev.) [PubMed] [Google Scholar]
- 15.Brand DD, Kang AH, Rosloniec EF. The mouse model of collagen-induced arthritis. Methods Mol Med. 2004;102:295–312. doi: 10.1385/1-59259-805-6:295. [DOI] [PubMed] [Google Scholar]
- 16.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Meth. 1994;53:55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Schweinhardt P, Kalk N, Wartolowska K, Chessell I, Wordsworth P, Tracey I. Corrigendum to Investigation into the neural correlates of emotional augmentation of clinical pain. Neuroimage. 2011;56:384. doi: 10.1016/j.neuroimage.2011.02.047. [DOI] [PubMed] [Google Scholar]
- 19.Billiau A, Matthys P. Collagen-induced arthritis and related animal models: How much of their pathogenesis is auto-immune, how much is auto-inflammatory. Cytokine Growth F R. 2011;22:339–344. doi: 10.1016/j.cytogfr.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Luross JA, Williams NA. The genetic and immunopathological processes underlying collagen-induced arthritis. Immunology. 2001;103:407–416. doi: 10.1046/j.1365-2567.2001.01267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cook AD, Rowley MJ, Stockman A, Muirden KD, Mackay IR. Specificity of antibodies to type II collagen in early rheumatoid arthritis. J Rheumatol. 1994;21:1186–1191. [PubMed] [Google Scholar]
- 22.Walsh DA, Mcwilliams DF. Mechanisms, impact and management of pain in rheumatoid arthritis. Nat Rev Rheumatol. 2014;10:581–592. doi: 10.1038/nrrheum.2014.64. [DOI] [PubMed] [Google Scholar]
- 23.Junger H, Sorkin LS. Nociceptive and inflammatory effects of subcutaneous TNFalpha. Pain. 2000;85:145–151. doi: 10.1016/S0304-3959(99)00262-6. [DOI] [PubMed] [Google Scholar]
- 24.Weinblatt ME, Kremer JM, Bankhurst AD, Bulpitt KJ, Fleischmann RM, Fox RI, Jackson CG, Lange M, Burge DJ. A trial of etanercept, a recombinant tumor necrosis factor receptor: Fc fusion protein, in patients with rheumatoid arthritis receiving methotrexate. N Engl J Med. 1999;340:253–259. doi: 10.1056/NEJM199901283400401. [DOI] [PubMed] [Google Scholar]
- 25.Boettger MK, Weber K, Grossmann D, Gajda M, Bauer R, Bär KJ, Schulz S, Voss A, Geis C, Bräuer R, Schaible HG. Spinal tumor necrosis factor α neutralization reduces peripheral inflammation and hyperalgesia and suppresses autonomic responses in experimental arthritis: A role for spinal tumor necrosis factor α during induction and maintenance of peripheral inflamma. Arthritis Rheum. 2010;62:1308–1318. doi: 10.1002/art.27380. [DOI] [PubMed] [Google Scholar]
- 26.Cox PJ, Pitcher T, Trim SA, Bell CH, Qin W, Kinloch RA. The effect of deletion of the orphan G- protein coupled receptor (GPCR) gene MrgE on pain-like behaviours in mice. Mol Pain. 2008;4:2. doi: 10.1186/1744-8069-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julius D, Basbaum AI. Molecular Mechanisms of nociception. Nature. 2001;413:203–210. doi: 10.1038/35093019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.