Abstract
The aim of the present study was to determine the potential core genes in the pathogenesis of human thoracic aortic dissection (TAD) by analyzing microarray profiles of long non-coding (lnc)-RNAs between TAD and normal thoracic aorta (NTA). The differentially expressed lncRNA profiles of the aorta tissues between TAD patients (TAD group, n=6) and age-matched donors with aortic diseases (NTA group, n=6) were analyzed by lncRNAs microarray. Gene ontology (GO), pathway and network analyses were used to further investigate candidate lncRNAs and mRNAs. Differentially expressed lncRNAs and mRNAs were validated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). In total, the present study identified 765 lncRNAs and 619 mRNAs with differential expression between TAD and NTA (fold change >2.0, P<0.01). GO analysis demonstrated that the differentially upregulated lncRNAs are associated with cell differentiation, homeostasis, cell growth and angiogenesis. Kyoto Encyclopedia of Gene and Genomes pathway analysis demonstrated that the differentially downregulated lncRNAs are mainly associated with arrhythmogenic right ventricular cardiomyopathy, hypertrophic cardiomyopathy and dilated cardiomyopathy. To reduce the lncRNAs for further investigation and to enrich those potentially involved in TAD, a total of 16 candidate lncRNAs with a significant expression (fold change >4, P<0.01) were selected, that were associated with an annotated protein-coding gene through the GO term and scientific literatures. Then a set of significantly expressed lncRNAs [purinergic receptor P2X7 (P2RX7), hypoxia inducing factor (HIF)-1A-AS2, AX746823, RP11-69I8.3 and RP11-536K7.5) and the corresponding mRNAs (P2RX7, cyclin dependent kinase inhibitor 2B, HIF-1A, runt-related transcription factor 1, connective tissue growth factor and interleukin 2 receptor a chain] were confirmed using RT-qPCR. The present study revealed that the expression profiles of lncRNAs and mRNAs in aorta tissues from TAD were significantly altered. These results may provide important insights into the pathogenesis of TAD disease.
Keywords: long noncoding RNA, messenger RNA, microarray analysis, thoracic aortic dissection
Introduction
Thoracic aortic dissection (TAD) is a highly lethal cardiovascular disease, which is characterized by the separation of thoracic aortic medial layer along the length of the vessel (1). Typical TAD begins with the sudden initial tear in the aortic intima, and intraluminal pulsatile blood enters into the medial layer through an intimal tear, resulting in rapid aortic dilation and rupture. In spite of the improvement of diagnostic and therapeutic techniques over the years, the overall mortality of TAD remains high (2). The understanding of the pathogenesis of this serious illness may produce a better outcome in the future. Many studies have been performed to explore the pathogenesis of TAD. Most studies were mainly focused on the genetic diversity, clinical pathology, and hemodynamics. However, the potential molecular mechanism of TAD remains unclear.
Long non-coding RNAs (lncRNAs) are defined as more than 200 nt in length and found to regulate protein-coding gene expression at both the transcriptional and post-transcriptional levels. Studies showed that lncRNAs play critical roles in cardiovascular physio-pathological processes (3). For example, lncRNA Braveheart (Bvht) was associated with cardiovascular development (4). Upregulated myocardial infarction-associated transcript 1 (Mirt1) and Mirt2 can promote cardiac contractile function and decrease left ventricular remodeling (5). Myosin heavy chain-associated RNA transcript (Mhrt) can protect the heart from pathological cardiac hypertrophy (6). Although an increasing number of lncRNAs have been characterized, the role of lncRNAs in TAD has not been investigated. A recent study revealed that the overexpressed lncRNA HIF1 alpha-antisense RNA 1 (HIF1A-AS1) in the thoraco-abdominal aortic aneurysm (TAAA) promoted the proliferation and apoptosis of vascular smooth muscle cells (VSMCs), which may contribute to the pathogenesis of TAAA (7). Wang et al (8) reported that the interaction between BRG1 and HIF1A-AS1 may be involved in the pathogenesis of thoracic aortic aneurysms. Since TAA and TAD possesses similar pathological basis, these results suggested the potential role of lncRNAs in the pathogenesis of TAD. However, until now, there was no study on the expression profile of lncRNAs in TAD.
In the present study, we demonstrated expression profile of lncRNAs between TAD and normal thoracic aorta (NTA) using third-generation lncRNA microarray techniques. These results will help provide further insight into the pathogenesis of TAD.
Materials and methods
Acquisition of clinical specimens
Ascending aorta specimens were obtained from TAD patients undergoing surgical repair (TAD group, n=6; mean age, 51.4±13.4 years) at Fuwai Hospital and organ donors without aortic diseases (NTA group, n=6; mean age, 49.6±12.6 years). No significant difference in the age was found between TAD and NTA (P>0.05). All patients were confirmed to have acute Stanford type I aortic dissection within 14 days of the symptom onset before surgery. All subjects have not any history of Marfan syndrome, bicuspid aortic valve, or any other connective tissue disease. The clinical characteristics of patients and donors were in Table I. The study protocol was approved by the international review board of Beijing Yuho Rehabilitation Hospital, (Beijing, China). Written informed consent was obtained from each of the patients. Aortic media tissues were sectioned into smaller sizes and briefly stored at −80°C until RNA extraction.
Table I.
Clinical characteristics.
| Variables | NTA (n=6) | TAD (n=6) |
|---|---|---|
| Age (years) | 49.6±12.6 | 51.4±13.4 |
| Males/females | 6/0 | 6/0 |
| Hypertension | 2 (33.3%) | 3 (50.0%) |
| Atherosclerosis | 1 (16.7%) | 2 (33.3%) |
| Smoking | 2 (33.3%) | 3 (50.0%) |
| Aortic size (mm) | 31.4±9.7 | 63.4±15.2 |
Data are presented as the mean ± standard deviation, as the n number of individuals or as n (%). NTA, normal thoracic aorta; TAD, thoracic aortic dissection.
RNA isolation and lncRNA microarray analysis
Total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) following the manufacturer's instruction. For the global profiling of human lncRNAs and protein-coding transcripts, we utilized the third-generation lncRNA microarray (Arraystar v3.0; KangChen, Shanghai, China), which contains more than 30,000 capture probes, covering all lncRNAs from authoritative databases (UCSC Knowngenes, RefSeq and Ensembl) and their coding proteins. The microarray hybridization was performed based on manufacturer's instruction. Quantile normalization and subsequent data analysis was performed using GeneSpring GX 11.5.1 software (Agilent Technologies, Inc., Santa Clara, CA, USA).
Validation by reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
To confirm the microarray data, expressions of selected lncRNA and coding proteins were tested using RT-qPCR with the GoTaq qPCR Master Mix (Promega Corporation, Madison, WI, USA) on an Mx3005P real-time PCR System (Agilent Technologies, Inc.). The primers used for RT-qPCR were listed in Table II. Each RNA sample was evaluated in triplicate. Gene expression results were analyzed with the 2−∆∆Cq method and normalized to GAPDH expression.
Table II.
Primers used in the present study.
| A, lncRNA | ||
|---|---|---|
| Gene name | Forward (5′-3′) | Reverse (3′-5′) |
| CERKL | TGTAAAGTTGAATAAAGCGG | TGCCTTGGGCTGTCGTAG |
| RP11-395B7.4 | AGGACTTGGAACTATGGG | AAGGGAGAATTAAAGGCTA |
| RP11-887P2.1 | TGTGCCTGGCTGCCTTAG | TAGCCGGAGGGTTAGGGAT |
| lncP2RX7 | GGGGGTTGAATGTGAGATGA | GTTAGGGAATGTTCTGAAGG |
| CDKN2B-AS1 | AGATGCAGAGGACATGCACGG | GGCAAGGCAAGTGAGGAG |
| RP11-796E2.4 | CGCCTGTAATCCCAGCAC | TCCGGGTTCACGCCATTC |
| HIF1A-AS2 | TTCAACCCTGCTTCCTGG | TGCTACCTTCTGTCCTGG |
| HOXD-AS2 | CCTCAGTACATAAACTTCCTAA | CTTTCACCTTCTCACAGG |
| AC007254.3 | ACGACCAGGCTTACGGAG | TCATTTCCCGTGTTGGGC |
| OGFR-AS1 | ACTGCCCTTGTTAATGTCA | TGGTCGTATTGTTCTTGC |
| AX746823 | TCGATTTCGTCAAGCATT | GAGGGCAGAAAGATCACA |
| RP11-69I8.3 | GAAGTCAGAAATACTGTGGGTA | CAAATGTGTGTTCGTGGG |
| RP11-317J10.2 | CTCCAAACTGTCTCACCC | AAAACCACCGTTACCAAA |
| RP11-318K15.2 | GCTCTTACCGCTGGGTGT | CCTTGGGGAGGGAAGTTAG |
| RP11-536K7.5 | CACAGTAGAGGCGAAACC | CAGGGAATAAGTGGACAGAT |
| NPPA-AS1 | GTGAAGAAAGACTTCAT | GTAGGGAAGGCAGTAGGGTGGAG |
| B, mRNAs | ||
| Gene name | Forward (5′-3′) | Reverse (3′-5′) |
| ITGA4 | TGGACAGCTAGAATTGGT | GTAGCTCTTAGATCCGGTG |
| MUC12 | CATGTACGTCTGCTATCCAGG | CTCCTTAGTCATACCACGATG |
| SOCS2 | TATAGGTGCAGGAGATGGTTC | GCTGTTATGGGTGAAACTCTG |
| P2RX7 | GGCGTCGTGGTGAATGATG | CCTAACTTGGCGTCCTATGC |
| CDKN2B | TCAACGCTCGATTCTATTGTGG | TCAACGAGATTCTGATTGTGGG |
| BTG1 | GGGGGCGGATACTAATCCC | CACGACAAAAGCGTACTCTGT |
| HIF1A | TCTCTGACGGCTATCCAAAGAA | GCAGCAGTTCTGGGTTAGTTG |
| HOXD3 | CTTGGAGGACGGCGATTA | CCGTCACGAACTTCTTCGT |
| SLC8A1 | GCTAGAAGCCGTGACGGCT | CTGACGGTACTCTAGTGGAG |
| OGFR | TCGTCGTCTAGACATTA | CTCTAGTGGGTGACTC |
| RUNX1 | GACGGCTGTCTATCCAAA | GAAAGACGTCTCTTC |
| CTGF | AAGGTCAAGCTAGACTG | GGAGACTAAGGCAGTG |
| CA2 | ATTGCTTATGAGTGAACGCC | AGGCATAGGTAGGCAGAG |
| LYN | GGTGGTGAAGATTAATCGC | ATGGGACATTTAAGGCAATC |
| IL2RA | GGAAGTAATACAATAGA | GCTGTTAACAACGTAAACTC |
| NPPA | AGCATGTAGGAGAGTTAACA | TAGAGGGGACCTTACAAG |
| GAPDH | GCCGTCTAGACCTGACCT | AGGATTGTCGCGTGGTGG |
Gene ontology (GO) and pathway analysis
The GO analysis (www.geneontology.org) and Kyoto Encyclopedia of Gene and Genomes database (KEGG, www.genome.jp/kegg) were used to analyze the main function and pathway of differentially expressed lncRNAs and mRNAs.
LncRNA and mRNA interaction network analysis
MetaCoreTM software (Auto Expand Algorithm) was used to analyze the regulatory network of differentially expressed genes associated with lncRNAs.
Statistical analysis
All statistical data were analyzed by SPSS v20.0 software (IBM Corp., Armonk, NY, USA). Data were presented as the mean ± standard deviation of at least 3 independent experiments. The results were analyzed by analysis of variance with a Bonferroni post hoc test, or by Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
LncRNA microarray profiling
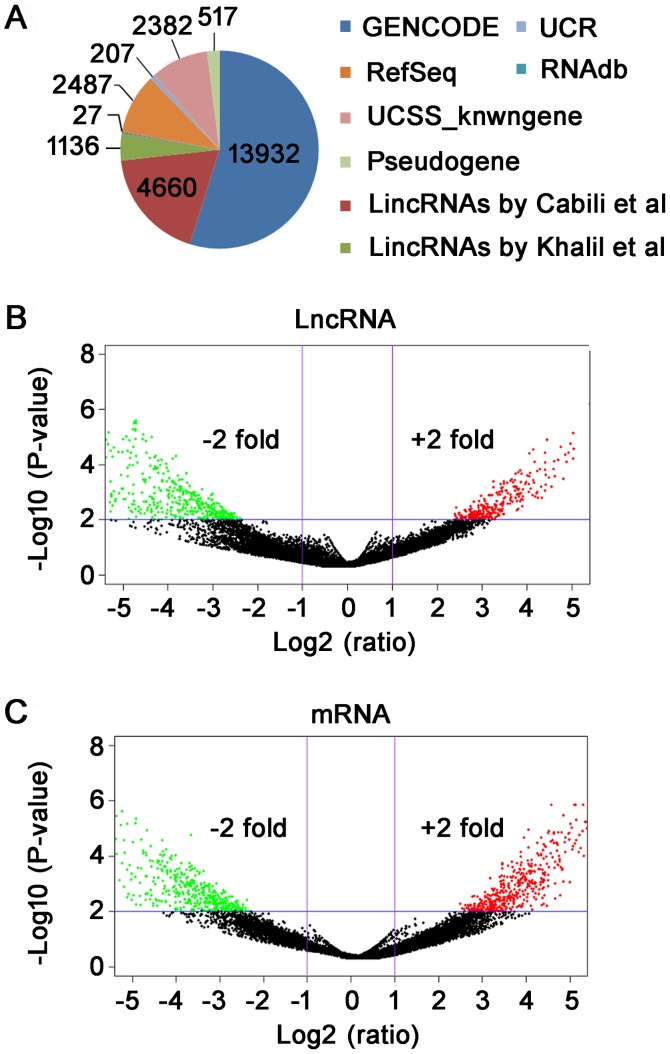
Arraystar lncRNA Microarray (v3.0), which can detect about 30,586 lncRNAs and 26,109 coding transcripts, was used for the comprehensive analysis of core genes associated with the pathogenesis of TAD. Using third-generation lncRNA microarray, we detected a total of 25,348 lncRNAs and 24,852 mRNAs. The statistical information of lncRNAs was shown in Fig. 1A. Furthermore, the differentially expressed lncRNAs and mRNAs were screened as per the fold-change and P-value. The screening criteria for differentially expressed lncRNAs and mRNAs is fold-change ≥2 and P-value <0.01. Subsequently, we have used log2 (fold-change) as the X-axis and log10 (P-value) as the Y-axis to show the distribution of lncRNAs and mRNAs in Volcano plot (Fig. 1B and C). In total, 765 differentially expressed lncRNA were identified between TAD and NTA, including 289 up-regulated and 476 down-regulated (fold-change ≥2, P<0.01). Through microarray, we found 619 differentially expressed mRNA (fold-change ≥2, P<0.01). Among mRNAs, 265 were up-regulated and 354 were down-regulated in TAD compared with NTA.
Figure 1.
Differential expression of lncRNAs between TAD and NTA by microarray analysis. (A) A pie chart presenting the relative numbers of lncRNAs from the majority of authoritative databases. Volcano plots were constructed using fold-change and P-values to present the distribution of (B) lncRNAs and (C) mRNAs, respectively. The vertical lines represent P<0.05 differences, and the horizontal lines correspond to 2.0-fold up and down. The red and green points in the plot represent the differentially expressed genes with statistical significance. TAD, thoracic aortic dissection; NTA, normal thoracic aorta, LncRNA, long non-coding RNA.
Expression signatures of dysregulated lncRNAs between TAD and NTA
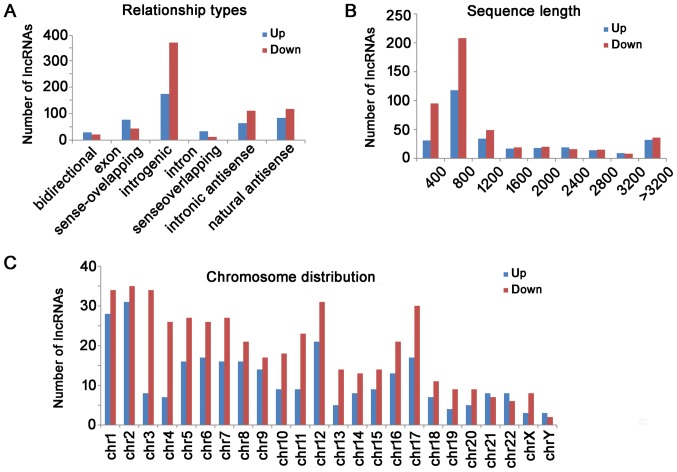
The distribution of relationship types, the sequence length and chromosomes of differentially expressed lncRNA are shown in Fig. 2. For those up-regulated lncRNAs, there were 156 intergenic, 75 natural antisense, 69 exon sense-overlapping, 57 intronic antisense, 30 intron sense-overlapping and 26 bidirectional lncRNAs. For those down-regulated lncRNAs, there were 329 intergenic, 105 natural antisense, 99 intronic antisense, 39 exon sense-overlapping, 19 bidirectional, and 11 intron sense-overlapping lncRNAs (Fig. 2A). The lncRNAs were mainly concentrated between 400 and 800 bp in length (Fig. 2B). The chromosome distribution showed the up-regulated and down-regulated lncRNAs were located at various chromosomes, respectively (Fig. 2C).
Figure 2.
Differential lncRNAs were classified according to their distribution. (A) For those upregulated lncRNAs, there were 156 intergenic, 75 natural antisense, 69 exon sense-overlapping, 57 intronic antisense, 30 intron sense-overlapping and 26 bidirectional lncRNAs. For those downregulated lncRNAs, there were 329 intergenic, 105 natural antisense, 99 intronic antisense, 39 exon sense-overlapping, 19 bidirectional and 11 intron sense-overlapping lncRNAs. (B) Length distribution of the dysregulated lncRNAs. The lncRNAs were mainly between 400 and 1200 bp in length. (C) The chromosome distribution indicated the numbers of upregulated and downregulated lncRNAs locations in various chromosomes. LncRNA, long non-coding RNA; Chr, chromosome.
GO and pathway analysis
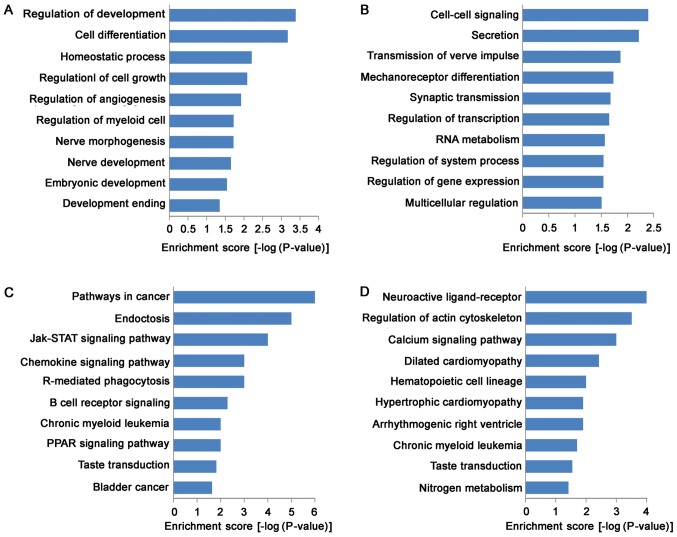
GO analysis indicated that the functions of up-regulated mRNAs were involved in a variety of biological processes, including VSMC development and vascular homeostasis, such as in cell growth (GO:0001558), cell differentiation (GO:0045597), homeostatic process (GO:0032844) and angiogenesis (GO:0045766; Fig. 3A). Meanwhile, the functions of down-regulated mRNAs were mainly involved in cell-cell signaling (GO:0007267), transcription regulation (GO:0045893), RNA metabolism (GO:0051254), and gene expression (GO:0010628; Fig. 3B). KEGG pathway analysis indicated that 31 pathways were down-regulated and 24 pathways were up-regulated. Most of up-regulated pathways were enriched in Jak-STAT signaling pathway (KEGG: hsa04630), chemokine signaling pathway (KEGG: hsa04062), PPAR (KEGG: hsa03320) signaling pathway and B cell receptor signaling pathway (KEGG: hsa04662), suggested that up-regulated pathways in TAD were closely associated with signal transduction (Fig. 3C). Moreover, down-regulated pathways were enriched in calcium signaling pathway (KEGG: hsa04020), arrhythmogenic right ventricular cardiomyopathy (ARVC) (KEGG: hsa05412), hypertrophic cardiomyopathy (HCM) (KEGG: hsa05410) and dilated cardiomyopathy (KEGG: hsa05414; Fig. 3D).
Figure 3.
GO and pathway analysis. The top 10 GO terms that were associated with the coding gene function of (A) upregulated lncRNAs and (B) downregulated lncRNAs are listed. The top 10 pathways that were associated with the coding gene of (C) upregulated lncRNAs and (D) downregulated lncRNAs are listed. GO, gene ontology; lncRNA, long non-coding RNA.
Selection of core genes in TAD
To reduce the lncRNAs for further investigation and to enrich those potentially involved in TAD, we first selected candidate lncRNAs with a significant expression (fold-change >4, P<0.01) that were associated with an annotated protein-coding gene through the GO term enrichment and scientific literatures. After functional annotation and enrichment analysis, we found: i) 8 protein-coding genes (BTG1; CA2; CDKN2B; HIF1A; IL2RA; LYN; RUNX1; HOXD3) are associated with cell differentiation; ii) 5 genes (CA2, HIF1A, IL2RA, LYN, P2RX7) are related to homeostasis; iii) 6) genes (CTGF, SOCS2, NPPA, MUC12, OGFR, BTG1) are correlated with cell growth; iv) 3 genes (BTG1, HIF1A, RUNX1) have positive regulation on angiogenesis and v) 2 genes (ITGA4, SLC8A1) are associated with cardiac disease. Finally, 16 lncRNAs associated with important genes were selected as candidates for the pathogenesis of TAD (Table III).
Table III.
Details of the 16 candidate lncRNAs in thoracic aortic dissection.
| Gene symbol | Regulation | RNA length | Chromosome | Strand | Relationship | mRNA | TAD/NTA | P-value |
|---|---|---|---|---|---|---|---|---|
| CERKL | Down | 3,030 | Chr2 | − | Natural antisense | ITGA4 | 0.04 | 0.005 |
| RP11-395B7.4 | Up | 372 | Chr7 | − | Natural antisense | MUC12 | 8.58 | 0.014 |
| RP11-887P2.1 | Up | 3,325 | Chr12 | − | Intronic antisense | SOCS2 | 7.75 | 0.017 |
| lncP2RX7 | Up | 3,604 | Chr12 | + | Exon sense-overlapping | P2RX7 | 12.48 | 0.009 |
| CDKN2B-AS1 | Up | 1,469 | Chr9 | + | Intronic antisense | CDKN2B | 10.92 | <0.001 |
| RP11-796E2.4 | Down | 1,898 | Chr12 | + | Natural antisense | BTG1 | 5.49 | 0.003 |
| HIF1A-AS2 | Up | 2,051 | Chr14 | − | Natural antisense | HIF1A | 12.67 | <0.001 |
| HOXD-AS2 | Up | 692 | Chr2 | − | Natural antisense | HOXD3 | 6.63 | 0.012 |
| AC007254.3 | Down | 344 | Chr2 | + | Intronic antisense | SLC8A1 | 0.01 | 0.008 |
| OGFR-AS1 | Down | 668 | Chr20 | − | Intronic antisense | OGFR | 11.00 | <0.001 |
| AX746823 | Up | 2,943 | Chr21 | − | Intron sense-overlapping | RUNX1 | 9.80 | 0.023 |
| RP11-69I8.3 | Up | 495 | Chr6 | + | Natural antisense | CTGF | 10.29 | 0.019 |
| RP11-317J10.2 | Up | 430 | Chr8 | − | Bidirectional | CA2 | 13.79 | <0.001 |
| RP11-318K15.2 | Up | 651 | Chr8 | + | Intron sense-overlapping | LYN | 13.34 | 0.004 |
| RP11-536K7.5 | Up | 480 | Chr10 | + | Natural antisense | IL2RA | 21.43 | 0.006 |
| NPPA-AS1 | Up | 660 | Chr1 | + | Natural antisense | NPPA | 6.51 | <0.001 |
TAD, thoracic aortic dissection; NTA, normal thoracic aorta; Chr, chromosome.
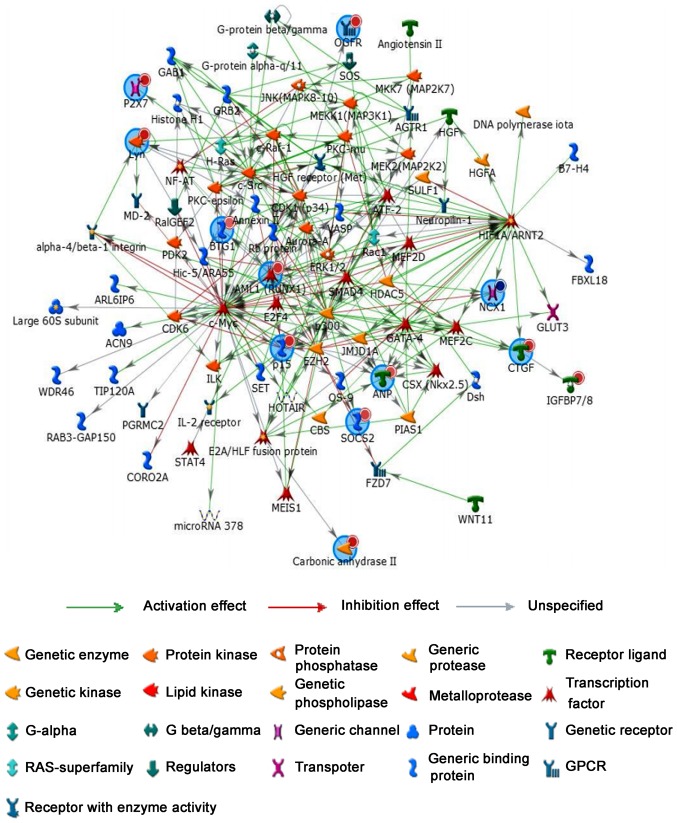
Regulatory network analysis
To further study relationship between coding genes, regulatory network analysis was performed by MetaCoreTM software to show network objects of 16 most promising lncRNA candidates (Fig. 4). Network objects that were associated with 16 candidate lncRNAs were listed Table IV. Similarly, 16 mRNA candidates (ITGA4, MUC12, SOCS2, P2RX7, CDKN2B, BTG1, HIF1A, HOXD3, SLC8A1, OGFR, RUNX1, CTGF, CA2, LYN, IL2RA and NPPA) also played an important role in the regulatory network. Additionally, more genes and signal pathways were involved in this network, suggested regulatory mechanisms of lncRNAs and mRNAs are complex in the pathogenesis of TAD.
Figure 4.
Regulatory network analysis was performed to present the network objects of 16 lncRNA candidates. LncRNA, long non-coding RNA.
Table IV.
Network objects associated with 16 lncRNA candidates.
| No. | Tag | Gene | Network object |
|---|---|---|---|
| 1 | NM_001122607 | RUNX1 (Runt-related transcription factor 1) | AML1 RUNX1 |
| 2 | NM_006172 | NPPA (Natriuretic peptide A) | ANP |
| 3 | NM_001731 | BTG1 (B-cell translocation gene 1) | BTG1 |
| 4 | NM_000067 | CA2 (Carbonic anhydrase II) | Carbonic anhydrase II |
| 5 | NM_001901 | CTGF (Connective tissue growth factor) | CTGF IGFBP7/8 |
| 6 | NM_181054 | HIF1A (Hypoxia inducible factor 1, α subunit) | HIF1A |
| 7 | NM_000417 | IL2RA (Interleukin 2 receptor α) | IL-2R α chains IL2RA |
| 8 | NM_000885 | ITGA4 (Integrin, α 4) | ITGA4 |
| 9 | NM_002350 | LYN (LYN proto-oncogene, Src family tyrosine kinase) | Lyn |
| 10 | NM_001164462 | MUC12 (Mucin 12, cell surface associated') | Mucin 12 |
| 11 | NM_001112802 | SLC8A1 (Solute carrier family 8 (sodium/calcium exchanger), member 1) | NCX1 |
| 12 | NM_007346 | OGFR (Opioid growth factor receptor) | OGFR |
| 13 | NM_078487 | CDKN2B (Cyclin-dependent kinase inhibitor 2B) | p15 |
| 14 | NM_002562 | P2RX7 (Purinergic receptor P2X, ligand gated ion channel, 7) | P2X7 |
| 15 | NM_003877 | SOCS2 (Suppressor of cytokine signaling 2) | SOCS2 |
| 16 | NM_006898 | HOXD3 (Homeobox D3) | HOXD3 |
Validation of lncRNA and mRNA candidates by RT-qPCR
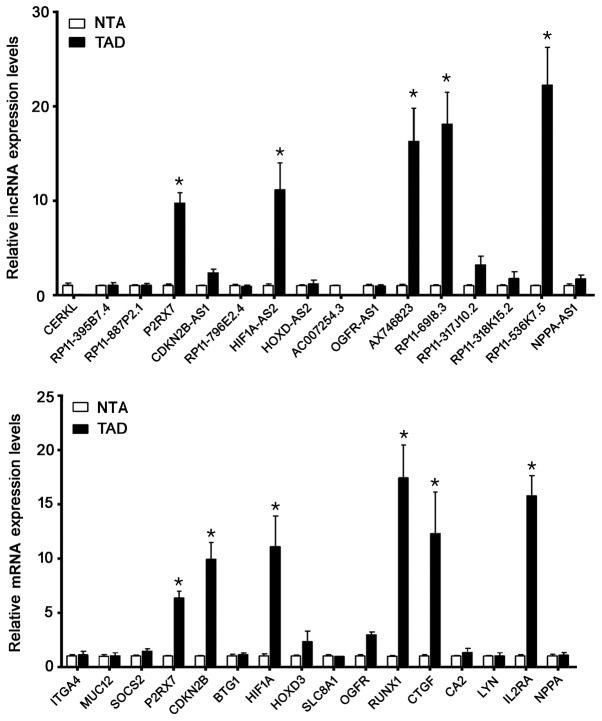
Validation of lncRNAs candidates by RT-qPCR was shown in Fig. 5A. The detection of the expression level of selected 16 lncRNAs demonstrated a good consistency with the microarray results. Among them, lncP2RX7, HIF1A-AS2, AX746823, RP11-69I8.3 and RP11-536K7.5 increased dramatically in the TAD group compared with the NTA group (P<0.01, respectively). Validation of 16 mRNAs candidates by RT-qPCR was shown in Fig. 5B. Among mRNAs, P2RX7, CDKN2B, HIF-1A, RUNX1, CTGF and IL2RA increased significantly in the TAD group compared with the NTA group (P<0.01, respectively). These genes may play critical roles in the pathogenesis of TAD.
Figure 5.
Candidate genes were validated by reverse transcription-quantitative polymerase chain reaction. Among the lncRNAs, lncP2RX7, HIF1A-AS2, AX746823, RP11-69I8.3 and RP11-536K7.5 increased markedly in the TAD group when compared with the control group. Among the mRNAs, P2RX7, CDKN2B, HIF-1A, RUNX1, CTGF and IL2RA increased markedly in the TAD group when compared with the NTA group. *P<0.05 vs. NTA. LncRNA, long non-coding RNA; P2RX7, purinergic receptor P2X7; HIF, hypoxia inducing factor; TAD, thoracic aortic dissection; CDKN2B, cyclin dependent kinase inhibitor 2B; RUNX1, runt-related transcription factor 1; CTGF, connective tissue growth factor; IL2RA, interleukin 2 receptor a chain; NTA, normal thoracic aorta.
Discussion
TAD is a life-threatening vascular disease that involves the separation of the layers within the aortic wall. Its formation, progression, and rupture cannot be reliably prevented by pharmacological therapies because the molecular mechanisms of the pathogenesis are still currently unclear (9). LncRNAs are a newly discovered class of non-coding RNAs, and they have important functions in regulating a variety of physio-pathological processes at transcriptional and post-transcriptional levels (10). Comprehensive comparison study of lncRNA expression profiles may help to identify candidate genes for the pathogenesis of TAD. In this study, the microarray that contains more than 30,000 known human lncRNAs and coding genes was utilized to detect fold-changes in the expressions of lncRNAs and mRNAs in TAD compared with those of NTA. After detailed analysis of data, we identified 765 differentially expressed lncRNAs, in which 289 were up-regulated and 476 were down-regulated. In 619 differentially expressed mRNA, 265 mRNAs were up-regulated and 354 mRNAs were down-regulated. Through bioinformatics approaches of GO, pathways and network analysis, 16 lncRNAs and their coding genes were selected as candidates for the pathogenesis of TAD. These candidates were verified by RT-qPCR. Results showed that 5 lncRNAs (lncP2RX7, HIF1A-AS2, AX746823, RP11-69I8.3 and RP11-536K7.5) and 6 mRNAs (P2RX7, CDKN2B, HIF-1A, RUNX1, CTGF and IL2RA) were significant expressed in dissected thoracic aortas, suggested they may be core genes and play critical roles in the pathogenesis of TAD.
So far, exploratory studies in the cardiovascular setting have identified several lncRNAs associated cardiovascular diseases (11). For example, lncRNA Bvht was identified as a key regulator of cardiovascular commitment from nascent mesoderm, suggested the potential implication in cardiovascular development (12). Both Mirt1 and Mirt2 lncRNAs were up-regulated in a mouse model of myocardial infarction. Increased expression of both Mirt1 and Mirt2 can promote cardiac contractile function and decrease left ventricular remodeling (5). The H19 lncRNA is a novel negative regulator of cardiomyocyte hypertrophy (13). A cardiac specific lncRNA Mhrt has been demonstrated to protect the heart from pathological cardiac hypertrophy (6). However, specific studies looking at lncRNAs in TAD are still lacking. Recent studies showed some exciting reports on lncRNAs in aortic aneurysm. The inhibition of lncRNA HIF1A-AS1 in VSMCs suppressed cell apoptosis and enhanced cell proliferation, which may participate in the pathogenesis of thoraco-abdominal aorta aneurysm (TAAA) (7,14). BRG1 expression is increased in TAA and regulates proliferation and apoptosis of VSMCs through the HIF1A-AS1 (6). These results may shed light on important functions of lncRNAs in TAD. Using third-generation lncRNA microarray, we revealed differential expression profiles of lncRNAs between TAD and NTA. The fact that some lncRNAs were differentially expressed in the developing or diseased heart provides a strong indication for their involvement in cardiac physio-pathology (15–17). Our results may provide important insights into the pathogenesis of TAD disease.
GO term and KEGG pathway analyses were utilized to gain insight into the function of differentially expressed genes (18). GO analysis indicated that the functions of dysregulated mRNAs were involved in a variety of biological processes, including VSMC development and vascular homeostasis. These both processes were closely associated with the formation and development of TAD. The aortic media is mainly composed of VSMCs, which are the source of extracellular matrix (ECM) proteins such as collagen and elastin. Medial degeneration is the main histopatholocgic characteristics of dissected thoracic aorta (18). GO results may imply the therapeutic potential of TAD by modulating lncRNAs. Moreover, KEGG pathway analysis indicated that 31 pathways were down-regulated and 24 pathways were up-regulated. Most of up-regulated pathways were enriched in Jak-STAT, chemokine, PPAR and B cell receptor signaling pathways, suggested that up-regulated pathways in TAD were closely associated with signal transduction. In addition, down-regulated pathways were mainly enriched in ARVC, HCM and dilated cardiomyopathy, indicated that these diseases may have a common pathogenic mechanism. To reduce the lncRNAs for further investigation and to find those potentially involved in TAD, we selected 16 lncRNAs as candidate genes for TAD through the significant expression (fold-change >4, P<0.01) and GO term enrichment. Regulatory network analysis demonstrated complex regulatory mechanisms of 16 mRNAs in the pathogenesis of TAD. Microarray results of lncRNAs and mRNAs was validated by RT-qPCR. Among lncRNAs, lncP2RX7, HIF1A-AS2, AX746823, RP11-69I8.3 and RP11-536K7.5 increased dramatically in the TAD group compared with the NTA group (P<0.01, respectively). With the exception of HIF1A-AS2 reported in TAAA, other lncRNAs were first found in the dissected thoracic aorta. According to protein-coding genes, lncP2RX7, AX746823, RP11-69I8.3 and RP11-536K7.5 were related to the activation of nuclear receptor (P2RX7), nuclear transcription (RUNX1), connective tissue development (CTGF) and inflammation (IL2RA). These genes may play important roles in the pathogenesis of TAD. Whether therapeutic modulation of these lncRNAs could decrease TAD development and whether this effect translates into improved survival after TAD remains to be determined.
PCR results showed that P2RX7, CDKN2B, HIF-1A, RUNX1, CTGF and IL2RA were significant expressed in dissected thoracic aortas, suggested they may be core genes and play critical roles in the pathogenesis of TAD. To date, HIF-1A has been involved in the proliferation, migration and morphological changes of VSMCs. Mechanically, hypoxia promoted the expression of HIF-1A by PI3K-AKT pathway in human aortic SMCs; HIF-1A further suppressed the expressions of AEG-1, a-SMA and SM22a, and promoted osteopontin (OPN) expression. Functionally, HIF-1A inhibited the proliferation and migration of human aortic SMCs (19). Moreover, CTGF is a matricellular protein expressed in the vascular wall, which regulates diverse cellular functions. Ungvari et al (20) showed that the expression of CTGF was increased in the abdominal aorta of ApoE−/− mice and in the adventitial region of the abdominal aorta in human AAA. CTGF is principally regulated at the level of transcription and is induced by mechanical stresses and a number of cytokines and growth factors, including TGF-β (21). Additionally, used vascular injury models, Leeper et al (22) found that CDKN2B knockout mice displayed reduced neointimal lesions and developed larger aortic aneurysms. In situ and in vitro studies suggested that these effects were attributable to increased smooth muscle cell apoptosis (23).
Besides, P2X7 receptor activation is the initial event leading to vascular dysfunction following lipopolysaccharide (LPS) treatment. Activation of P2X7 receptor amplifies LPS-induced hyporeactivity in mouse endothelium-intact aorta, which is associated with IL-1β-mediated release of nitric oxide by iNOS (24). And then, Pahl et al (25) reported that mRNA and protein expression of transcription factor RUNX1 in human abdominal aortic aneurysm. Furthermore, an integral-membrane protein, soluble IL2RA has been isolated and determined to result from extracellular proteolysis. This suggested the pathogenesis and development of human TAD may be related to inflammation of the local environment in the aorta (26). However, the molecular mechanisms of these genes are not completely understood in TAD.
lncRNAs typically show tissue-specific and vascular disease-specific patterns of expression (27,28). Give this specificity, lncRNAs may be favorable biomarkers than current coding proteins for TAD. In addtion, lncRNAs are functional molecules, thus their expressions may be a better indicator of disease states. Because of the limited sample size, the screening biomarker had some limitations, hoping to expand the sample size in the future. Future studies would also be needed to reveal whether P2RX7, CDKN2B, HIF-1A, RUNX1, CTGF and IL2RA expressions were associated to TAD and which cells expressed these proteins in TAD lesion.
This is first report about the differentially expressed lncRNA profiles between TAD and NTA. These results may provide important insights into the pathogenesis of TAD disease. Expanding our understanding about differential expression profiles of lncRNAs will assist into novel diagnostics and therapeutics, which will ultimately improve outcomes for patients with TAD.
Acknowledgements
The authors would like to thank the Department of Cardiovascular Surgery, The Army General Hospital (Beijing, China).
Funding
The present study was supported by the Project Supported by the National Natural Science Foundation of China (grant no. 81400854) and Beijing Postdoctoral Research Foundation (grant nos. 2015ZZ-50 and 2016ZZ-44).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YL and NY conceived and designed the experiments. XZ and XB conducted RNA sequencing and analysis. GQ and MZ performed the remaining experiments. HL and DL conducted data analysis. YL and NY produced the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The study protocol was approved by the international review board of Beijing Yuho Rehabilitation Hospital, (Beijing, China). Written informed consent was obtained from each of the patients.
Consent for publication
Written informed consent was obtained from each of the patients.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Clouse WD, Hallett JW, Jr, Schaff HV, Spittell PC, Rowland CM, Ilstrup DM, Melton LJ., III Acute aortic dissection: Population-based incidence compared with degenerative aortic aneurysm rupture. Mayo Clin Proc. 2004;79:176–180. doi: 10.4065/79.2.176. [DOI] [PubMed] [Google Scholar]
- 2.Leitman IM, Suzuki K, Wengrofsky AJ, Menashe E, Poplawski M, Woo KM, Geller CM, Lucido D, Bernik T, Zeifer BA, Patton B. Early recognition of acute thoracic aortic dissection and aneurysm. World J Emerg Surg. 2013;8:47. doi: 10.1186/1749-7922-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batista PJ, Chang HY. Long noncoding RNAs: Cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 6.Miner GH, Faries PL, Costa KD, Hanss BG, Marin ML. An update on the etiology of abdominal aortic aneurysms: Implications for future diagnostic testing. Expert Rev Cardiovasc Ther. 2015;13:1079–1090. doi: 10.1586/14779072.2015.1082906. [DOI] [PubMed] [Google Scholar]
- 7.He Q, Tan J, Yu B, Shi W, Liang K. Long noncoding RNA HIF1A-AS1A reduces apoptosis of vascular smooth muscle cells: Implications for the pathogenesis of thoracoabdominal aorta aneurysm. Pharmazie. 2015;70:310–315. [PubMed] [Google Scholar]
- 8.Wang S, Zhang X, Yuan Y, Tan M, Zhang L, Xue X, Yan Y, Han L, Xu Z. BRG1 expression is increased in thoracic aortic aneurysms and regulates proliferation and apoptosis of vascular smooth muscle cells through the long non-coding RNA HIF1A-AS1 in vitro. Eur J Cardiothorac Surg. 2015;47:439–446. doi: 10.1093/ejcts/ezu215. [DOI] [PubMed] [Google Scholar]
- 9.Duggirala A, Delogu F, Angelini TG, Smith T, Caputo M, Rajakaruna C, Emanueli C. Non coding RNAs in aortic aneurysmal disease. Front Genet. 2015;6:125. doi: 10.3389/fgene.2015.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ENCODE Project Consortium, corp-author. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudzinski DM, Isselbacher EM. Diagnosis and management of thoracic aortic disease. Curr Cardiol Rep. 2015;17:106. doi: 10.1007/s11886-015-0655-z. [DOI] [PubMed] [Google Scholar]
- 12.Papait R, Kunderfranco P, Stirparo GG, Latronico MV, Condorelli G. Long noncoding RNA: A new player of heart failure? J Cardiovasc Transl Res. 2013;6:876–883. doi: 10.1007/s12265-013-9488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L, An X, Li Z, Song Y, Li L, Zuo S, Liu N, Yang G, Wang H, Cheng X, et al. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc Res. 2016;111:56–65. doi: 10.1093/cvr/cvw078. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Feng G, Wang Y, Yue Y, Zhao W. Regulation of apoptosis by long non-coding RNA HIF1A-AS1 in VSMCs: Implications for TAA pathogenesis. Int J Clin Exp Pathol. 2014;7:7643–7652. [PMC free article] [PubMed] [Google Scholar]
- 15.Magistri M, Faghihi MA, St Laurent G, III, Wahlestedt C. Regulation of chromatin structure by long noncoding RNAs: Focus on natural antisense transcripts. Trends Genet. 2012;28:389–396. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung A, Trac C, Jin W, Lanting L, Akbany A, Sætrom P, Schones DE, Natarajan R. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res. 2013;113:266–278. doi: 10.1161/CIRCRESAHA.112.300849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freedman JE, Miano JM. National Heart, Lung, and Blood Institute Workshop Participants*: Challenges and opportunities in linking long noncoding RNAs to cardiovascular, lung, and blood diseases. Arterioscler Thromb Vasc Biol. 2017;37:21–25. doi: 10.1161/ATVBAHA.116.308513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu K, Fang C, Shen Y, Liu Z, Zhang M, Ma B, Pang X. Hypoxia-inducible factor 1a induces phenotype switch of human aortic vascular smooth muscle cell through PI3K/AKT/AEG-1 signaling. Oncotarget. 2017;8:33343–33352. doi: 10.18632/oncotarget.16448. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Ungvari Z, Valcarcel-Ares MN, Tarantini S, Yabluchanskiy A, Fülöp GA, Kiss T, Csiszar A. Connective tissue growth factor (CTGF) in age-related vascular pathologies. Geroscience. 2017;39:491–498. doi: 10.1007/s11357-017-9995-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sachdeva J, Mahajan A, Cheng J, Baeten JT, Lilly B, Kuivaniemi H, Hans CP. Smooth muscle cell-specific Notch1 haploinsufficiency restricts the progression of abdominal aortic aneurysm by modulating CTGF expression. PLoS One. 2017;12:e0178538. doi: 10.1371/journal.pone.0178538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leeper NJ, Raiesdana A, Kojima Y, Kundu RK, Cheng H, Maegdefessel L, Toh R, Ahn GO, Ali ZA, Anderson DR, et al. Loss of CDKN2B promotes p53-dependent smooth muscle cell apoptosis and aneurysm formation. Arterioscler Thromb Vasc Biol. 2013;33:e1–e10. doi: 10.1161/ATVBAHA.112.300399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiao CW, Tostes RC, Webb RC. P2X7 receptor activation amplifies lipopolysaccharide-induced vascular hyporeactivity via interleukin-1 beta release. J Pharmacol Exp Ther. 2008;326:864–870. doi: 10.1124/jpet.107.135350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiao CW, da Silva-Santos JE, Giachini FR, Tostes RC, Su MJ, Webb RC. P2X7 receptor activation contributes to an initial upstream mechanism of lipopolysaccharide-induced vascular dysfunction. Clin Sci (Lond) 2013;125:131–141. doi: 10.1042/CS20120479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pahl MC, Erdman R, Kuivaniemi H, Lillvis JH, Elmore JR, Tromp G. Transcriptional (ChIP-Chip) analysis of ELF1, ETS2, RUNX1 and STAT5 in human abdominal aortic aneurysm. Int J Mol Sci. 2015;16:11229–11258. doi: 10.3390/ijms160511229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Yang SH, Yao Y, Xie YQ, Yang YQ, Wang YH, Yin XY, Ma HD, Gershwin M, Lian ZX. Block of both TGF-β and IL-2 signaling impedes Neurophilin-1+ regulatory T cell and follicular regulatory T cell development. Cell Death Dis. 2016;7:e2439. doi: 10.1038/cddis.2016.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 28.Jiang X, Ning Q. The emerging roles of long noncoding RNAs in common cardiovascular diseases. Hypertens Res. 2015;38:375–379. doi: 10.1038/hr.2015.26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.