Abstract
Endometriosis is a common gynecological disease, affecting 6–10% of women of reproductive age. The precise mechanisms underlying the development of endometriosis remain unclear. In the present study, a bioinformatics approach was applied to systematically identify the pathways and genes involved in the development of endometriosis and to discover potential biomarkers. The gene expression profiles of GSE6364, a microarray dataset of endometrial biopsies obtained from women with or without endometriosis, was downloaded from the Gene Expression Omnibus DataSets database that stores original submitter-supplied records (series, samples and platforms), as well as curated datasets. Differentially expressed gene (DEG) analysis was performed with GEO2R. DAVID was used to analyze the gene ontology enrichment of the DEGs. Gene Set Enrichment Analysis (GSEA) was conducted using the GSEA v3.0 software. Protein-protein interactions (PPI) were evaluated with the Search Tool for the Retrieval of Interacting Genes, and PPI network visualization was performed with Cytoscape. In addition, Cell Counting kit-8 and Transwell assays were performed on human endometrial stromal cells (HESCs). A total of 172 DEGs were extracted. Inflammatory response genes were significantly upregulated in the endometriosis tissues and C-X-C motif chemokine receptor 2 (CXCR2), was one of the most up-regulated genes according to DEG analysis. Cell-based experiments confirmed that CXCR2 promoted the proliferation, migration and invasion of HESCs. In conclusion, a bioinformatics approach combined with in vitro experiments in the present study revealed that CXCR2 may be associated with the development of endometriosis and has potential as a biomarker for the diagnosis of endometriosis.
Keywords: bioinformatics approach, endometriosis, differentially expressed genes, pathways, C-X-C motif chemokine receptor 2
Introduction
Endometriosis is a common gynecological disease affecting 6–10% of women of reproductive age (1). It is characterized by the presence of endometrial glands and/or stroma outside of the uterine cavity, including the ovaries, ligaments of the uterus, fallopian tubes, cervical vaginal area, peritoneum, umbilicus and urinary tract (2,3). The primary symptoms of endometriosis are pelvic pain and infertility, although other symptoms, including ovarian masses, dysmenorrhea, dyspareunia, irregular uterine bleeding and dysuria, are also common (1,4).
Various hypotheses have been postulated to explain the pathogenesis of endometriosis, including stem cell-, metaplasia- and implantation-based theories (3,5,6). However, the implantation theory is the most accepted one, which proposes that ectopic endometrioid lesions develop via the implantation of endometrial glands and/or stroma that are retrogradely transported into the pelvic cavity through menstrual blood. That is, the pathogenesis of endometriosis involves the adhesion, proliferation and invasion of endometrial cells and angiogenesis (7). Cytokines, angiogenic factors and adhesion promotion factors, including interleukin (IL)-6, IL-8, tumor growth factor-β and vascular endothelial growth factor, contribute to cell attachment, proliferation and invasion, and neovascularization (8,9). It is also considered that various factors, including inflammatory, genetic, epigenetic, hormonal, immune, anatomic and lifestyle factors, are associated with the etiology of endometriosis (6,10,11). However, despite the various hypotheses, the exact pathogenesis of endometriosis remains unclear.
Endometriosis is a chronic disease with a high risk of recurrence and histological analysis is the only definitive method of diagnosis (12,13). Hormone therapy, medication and surgery can only alleviate the symptoms (14). As efficient diagnostic methods and treatments are lacking, patients with endometriosis typically experience severe symptoms and high medical expenses, which considerably influence their quality of life (12,13,15). Thus, it is important to explore the pathological mechanisms and identify more efficient biomarkers for the diagnosis and drug treatment of endometriosis.
In the present study, a bioinformatics approach was applied to systematically identify the pathways involved in endometriosis. Gene expression profiling analysis is a useful method to distinguish differentially expressed genes (DEGs) in a particular condition (16,17). Based on Sampson's implantation theory that ectopic endometrial tissues contribute to the pathogenesis of endometriosis (18), the gene expression profiles from endometrial biopsies from women with or without endometriosis were investigated using the Gene Expression Omnibus (GEO) database. Through analysis of the biological functions and pathways of the identified genes, the present study aimed to identify critical pathways and genes contributing to endometriosis, and potential biomarkers for its diagnosis, prognosis and therapy.
Materials and methods
Bioinformatics approach
Microarray data
The GSE6364 dataset was downloaded from the GEO DataSets database (www.ncbi.nlm.nih.gov/gds). GSE6364 contained the processed and normalized gene expression profiles of endometrial biopsies from women with normal endometrial pathology and no history of endometriosis, and from women with laparoscopy-proven moderate-to-severe stage endometriosis from an Affymetrix GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array; Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA) (19). In the present study, proliferative phases (day 8–14 in regular menstruation) samples were selected, including six samples (GSM150190-150195) from patients with endometriosis and five normal samples (GSM150196-150201).
Identification of DEGs
DEG analysis of the dataset was performed using GEO2R (www.ncbi.nlm.nih.gov/geo/geo2r/), which performed comparisons using the GEO query and limma R packages of the Bioconductor project (20). P-values of the DEGs were determined using Student's t-test. An adjusted P<0.05 and a fold-change in expression ≥2 were set as thresholds to identify statistically significant DEGs. Hierarchical clustering analysis was applied to categorize the data, and a heatmap was produced with HemI 1.0 software (21).
Gene ontology (GO) and pathway enrichment analyses of DEGs
GO analysis is commonly used for gene annotation, including the molecular function (MF), biological process (BP) and cellular component (CC) categories (22). The DAVID online tool (david.ncifcrf.gov/) was applied to analyze the functional level, including GO enrichment of the DEGs (23). Gene Set Enrichment Analysis (GSEA) was conducted using the software GSEA v3.0 (www.broadinstitute.org/gsea/) (24,25).
Construction of a protein-protein interaction (PPI) network
The Search Tool for the Retrieval of Interacting Genes (STRING version 10.5; string-db.org/) database is an online tool used for the evaluation of the PPIs (26). To identify interactions among the DEGs, STRING was applied to map the DEGs. Experimentally validated interactions were included, whereas single nodes without interactions were excluded. PPI networks visualization was achieved with the Cytoscape software (version 3.5.1) (27).
Validation of key genes
Ethics statement
Endometrial tissue in the present study was collected from patients from September to November 2017 in Renmin Hospital of Wuhan University (Renmin, China) and they provided written informed consent. The present study was approved by the Ethics Committee of Renmin Hospital of Wuhan University in September 2017.
Cell isolation and culture
Primary endometrial stromal cells (HESCs) were isolated as described previously (28). Patients, ranging from 24–45-years old, had regular menstrual cycles, and were documented not to be pregnant at the time of surgery were enrolled in the present study; however, patients using any form of hormonal treatment within 3 months of biopsy were excluded. Briefly, endometrial tissue from 8 patients with endometriosis was finely minced and the cells dispersed by incubation in Hank's balanced salt solution (25 mmol/ml) containing collagenase (1 mg/ml, 15 U/mg), 1% penicillin/streptomycin and deoxyribonuclease (0.1 mg/ml, 1,500 U/mg) at 37°C for 60 min in a water bath. Following filtration through a 40-µm cell strainer (Falcon®; Corning Incorporated, Corning, NY, USA), the cells were seeded in 75 cm2 Falcon tissue culture flasks (BD Biosciences, San Jose, CA, USA) and suspended in Ham's F12:Dulbecco's modified eagle medium (1:1) containing 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences, Logan, UT, USA) and antibiotics (100 IU/ml penicillin, 100 µg/ml streptomycin and 0.25 µg/ml amphotericin B) in a 37°C incubator with 5% CO2. The purity of HESCs were determined by vimentin immunohistochemical staining. Briefly, following 10% formalin fixation in room temperature for 30 min, cells were immersed in 3% hydrogen peroxide solution for 10 min at room temperature. Then, incubation with an anti-vimentin primary antibody (1:400; ab8978) at 37°C for 1 h was conducted and followed by a second antibody goat anti-mouse Alexa Fluor® 488 IgG (1:3,000; ab150117; both Abcam, Cambridge, MA, USA) at 37°C for 20 min. Subsequently, staining with diaminobenzidine (GK6007, Gene Tech Co., Ltd., Shanghai, China) according to manufacturer recommendations and then counterstained with hematoxylin at room temperature for 10 min, followed by analysis with an inverted light microscope (magnification, ×400, Nikon Corporation, Tokyo, Japan). Cultured HESCs were used for further analysis following 3–5 passages.
Cell treatments
Overexpression of C-X-C motif chemokine receptor 2 (CXCR2) was achieved using a pLKO lentiviral vector targeting CXCR2 constructed by Thermo Fisher Scientific, Inc. Lentivirus stocks were obtained using the ViraPower™ Lentiviral Packing Mix and 293FT cell line according to the manufacturer's protocol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). HESCs at 50% confluency were cultured in a 1:1 dilution of virus:media with 5 µg/ml polybrene for 24 h at 37°C, followed by selection of stable cell lines. The knockdown of CXCR2 in HESCs was achieved using small interfering RNA (siRNA). Cells were transfected with CXCR2-siRNA at a final concentration of 50 nM using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Cells transfected with a final concentration of 50 nM scramble sequences were the control group. Sequences of the CXCR2 primers and CXCR2-siRNAs used are demonstrated in Table I. Following 24 h, cells were collected for the subsequent analyses.
Table I.
siRNA sequences and primers.
| Name | Sense or forward sequences (5′-3′) | Anti-sense or reverse sequences (5′-3′) |
|---|---|---|
| CXCR2 siRNA | AGCGACCCAGUCAGGAUUUTT | AAAUCCUGACUGGGUCGCUTT |
| CXCR2 siRNA scramble | AGCAGCUCAAUGCGUCAGUTT | ACUGACGCAUUGAGCUGCUTT |
| CXCR2 primers | AGCTGAGAATATGCAGCCGTT | CATAGCAGGCTGGGCTAACA |
| GAPDH primers | TTGATGGCAACAATCTCCAC | CGTCCCGTAGACAAAATGGT |
CXCR2, C-X-C motif chemokine receptor 2; siRNA, small interfering RNA.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted with TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The PrimeScript™ RT Reagent kit was purchased from Takara Biotechnology Co., Ltd. (Dalian, China). Details of the RNA isolation, cDNA conversion and RT-qPCR assays have been described previously (29). To examine the mRNA expression levels of CXCR2, RT-qPCR was conducted in 96-well reaction plates using the ABI Step One Plus™ real-time PCR System (Thermo Fisher Scientific, Inc.). Each well contained 1 µl cDNA template, 0.2 µl each primer, 3.6 µl diethyl pyrocarbonate-H2O and 5 µl SYBR Green dye. CXCR2 and GAPDH primers were designed and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). The qPCR cycling conditions were as follows: Pre-denaturation at 95°C for 30 sec, followed by 40 cycles of denaturation at 95°C for 5 sec, annealing at 60°C for 10 sec and extension at 72°C for 30 sec. GAPDH was used to normalize the relative mRNA expression of CXCR2. The 2−ΔΔCq method was applied to calculate the relative expression level of the target amplicon (30).
Cell proliferation and Transwell assays
For the cell proliferation assay, transfected HESCs were suspended in Ham's F12/DMEM for 3 days in 96-well plates (5×104 cells/well). A Cell Counting kit-8 (CCK-8) assay was subsequently conducted. CCK-8 (10 µl; WST-8, Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was mixed with the cells, followed by incubation at 37°C for 2 h. The absorbance at 450 nm was measured using an ELISA reader (Tecan Group, Ltd., Mannedorf, Switzerland) to detect cell viability. For the cell migration and invasion assays, Transwell permeable supports (Corning Incorporated) and Matrigel (BD Biosciences) were used. Transfected cells were cultured in 200 µl in Ham's F12:Dulbecco's modified eagle medium prior to being transferred onto the upper chambers of 24-well plates (5×105 cells/well) with or without a Matrigel coating. A total of 800 µl medium with 10% FBS was added to the lower chamber. Following 24 h incubation, cells in the lower chamber were fixed with absolute methanol and stained with 0.1% crystal violet solution (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 10 min at room temperature. Cell counting was performed at medium magnification (×100) under an inverted light microscope from thee randomly selected fields. For each condition, three independent experiments were conducted. Image analysis was performed using Image J software version 1.49 (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
Statistical analysis was performed using the Stata 13.0 software (StataCorp LP, College Station, TX, USA). One-way analysis of variance and the Scheffe post hoc test were applied. Data are presented as the mean ± standard error. P<0.05 was considered to indicate a statistically significant difference.
Results
Overview of the GEO microarray data and identification of DEGs
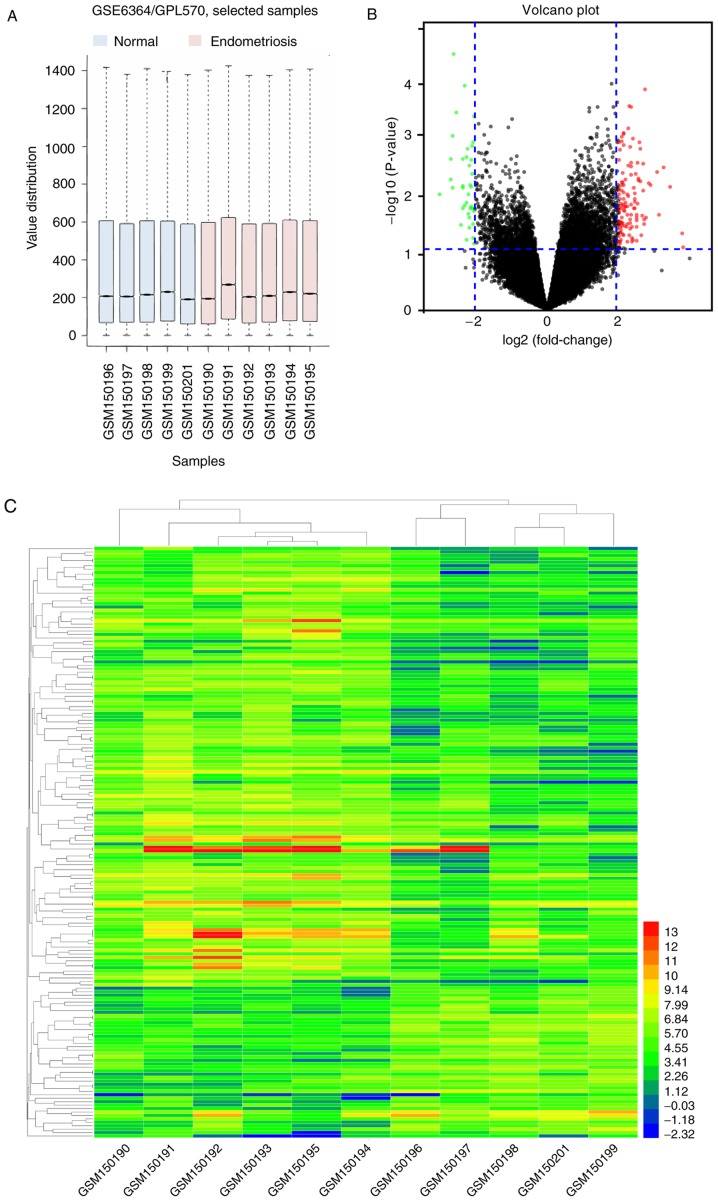
A box plot graph of the value distribution in the GSE6364 dataset was generated to assess whether the distributions of values across samples were median-centered and of conformity. The results indicated that the data of GSE6364 were generally normalized and cross-comparable (Fig. 1A). An adjusted P<0.05 and a fold-change ≥2 were set to filter for statistically significant DEGs. A total of 172 genes were identified, as demonstrated in the volcano plot (Fig. 1B) and a hierarchical cluster analysis was performed on the DEGs, as indicated in the heat map (Fig. 1C).
Figure 1.
Overview of GSE6364 and identification of common DEGs. (A) Box plot graph of value distribution of GSE6364. (B) Cutoff values and volcano plot displaying the DEGs. Red indicates upregulated genes and green indicates downregulated genes. (C) Hierarchical clustering and heat map of the DEGs in GSE6364. GSM150190-150195: Patients with endometriosis; GSM150196-150201: Normal samples. C-X-C motif chemokine receptor 2; DEG, differentially expression genes; GSEA, Gene Set Enrichment Analysis.
Pathway enrichment analysis
The results of the GO analysis of the DEGs indicated that ‘inflammatory response’, ‘innate immune response’ and ‘chemokine production’ were the most significant terms in the BP category. ‘Extracellular space’, ‘extracellular exosome’ and ‘membrane’ were the most significant in CC. The significant MF terms were ‘Toll-like receptor 4 binding’, ‘arachidonic acid binding’, ‘signaling pattern recognition receptor activity’ and ‘CXCR chemokine receptor binding’ (Table II).
Table II.
GO analysis of the differentially expressed genes (P<0.05).
| A, GOTERM_BP_DIRECT | |||
| Term | Count | P-value | Fold enrichment |
|---|---|---|---|
| GO:0006954 inflammatory response | 9 | 3.70×10−4 | 5.05 |
| GO:0045087 innate immune response | 9 | 8.48×10−4 | 4.45 |
| GO:0002523 leukocyte migration involved in inflammatory response | 3 | 1.14×10−3 | 57.97 |
| GO:0030593 neutrophil chemotaxis | 4 | 3.58×10−3 | 12.88 |
| GO:0042742 defense response to bacterium | 5 | 4.64×10−3 | 7.33 |
| GO:0002227 innate immune response in mucosa | 3 | 5.96×10−3 | 25.51 |
| GO:0006910 phagocytosis, recognition | 3 | 7.45×10−3 | 22.77 |
| GO:0070488 neutrophil aggregation | 2 | 9.27×10−3 | 212.56 |
| GO:0016337 single organismal cell-cell adhesion | 4 | 1.16×10−2 | 8.42 |
| GO:0032602 chemokine production | 2 | 1.39×10−2 | 141.70 |
| GO:0019731 antibacterial humoral response | 3 | 1.78×10−2 | 14.49 |
| GO:0032119 sequestering of zinc ion | 2 | 1.85×10−2 | 106.28 |
| GO:0002793 positive regulation of peptide secretion | 2 | 1.85×10−2 | 106.28 |
| GO:0032870 cellular response to hormone stimulus | 3 | 1.85×10−2 | 14.17 |
| GO:0007267 cell-cell signaling | 5 | 3.05×10−2 | 4.18 |
| GO:0006260 DNA replication | 4 | 3.55×10−2 | 5.49 |
| GO:0002221 pattern recognition receptor signaling pathway | 2 | 3.66×10−2 | 53.14 |
| GO:0032496 response to lipopolysaccharide | 4 | 4.09×10−2 | 5.18 |
| GO:0002544 chronic inflammatory response | 2 | 4.10×10−2 | 47.23 |
| GO:0070098 chemokine-mediated signaling pathway | 3 | 4.31×10−2 | 8.98 |
| GO:0006955 immune response | 6 | 4.61×10−2 | 3.03 |
| B, GOTERM_CC_DIRECT | |||
| Term | Count | P-value | Fold enrichment |
| GO:0005615 extracellular space | 20 | 1.30×10−5 | 3.11 |
| GO:0005576 extracellular region | 20 | 1.48×10−4 | 2.60 |
| GO:0070062 extracellular exosome | 23 | 9.88×10−3 | 1.71 |
| GO:0005886 plasma membrane | 29 | 2.18×10−2 | 1.47 |
| GO:0016020 membrane | 18 | 2.68×10−2 | 1.71 |
| GO:0009986 cell surface | 7 | 4.30×10−2 | 2.71 |
| C, GOTERM_MF_DIRECT | |||
| Term | Count | P-value | Fold enrichment |
| GO:0035662 Toll-like receptor 4 binding | 2 | 1.86×10−2 | 105.51 |
| GO:0050544 arachidonic acid binding | 2 | 2.32×10−2 | 84.41 |
| GO:0008329 signaling pattern recognition receptor activity | 2 | 3.23×10−2 | 60.29 |
| GO:0045236 CXCR chemokine receptor binding | 2 | 4.13×10−2 | 46.89 |
GO, gene ontology; BP, biological processes; CC, cellular components; MF, molecular function; CXCR, C-X-C motif chemokine receptor.
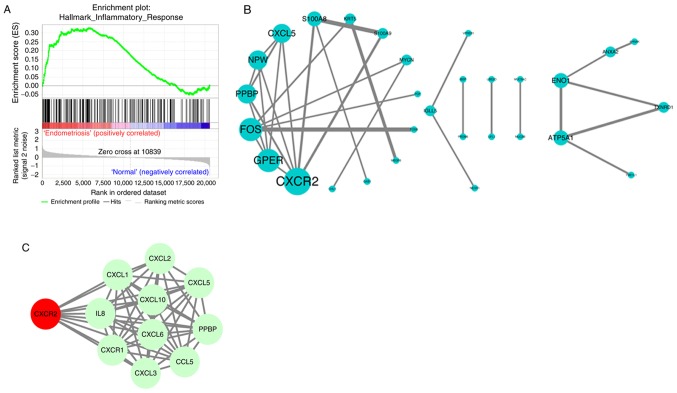
Key pathways and genes identification
To further identify the key pathways, GSEA was conducted on the GSE6364 dataset. ‘INFLAMMATORY RESPONSE’ was identified, in accordance with the results of GO analysis. The enrichment score for this signaling pathway was positively correlated to endometriosis samples, while the enrichment score was lower in normal samples (Fig. 2A). Subsequently, the PPI among the DEGs were evaluated by STRING and PPI network visualization was performed using Cytoscape. CXCR2, G protein-coupled estrogen receptor 1, Fos proto-oncogene, C-X-C motif chemokine ligand 5 and others were preserved according to the inclusion criterion (Fig. 2B). As the most significant DEG, CXCR2 was uploaded into STRING to search its potential PPI with possible proteins. All the interacting proteins were involved in the inflammatory response, which was consistent with the other findings (Fig. 2C). Therefore, CXCR2 was considered the most important identified gene.
Figure 2.
GSEA and PPI prediction. (A) Key pathways identification in GSE6364 dataset via GSEA software. (B) PPI network of the differentially expression genes from STRING. (C) PPI prediction of CXCR2 with other proteins. GSEA, Gene Set Enrichment Analysis; PPI, protein-protein interaction; CXCR2, C-X-C motif chemokine receptor 2.
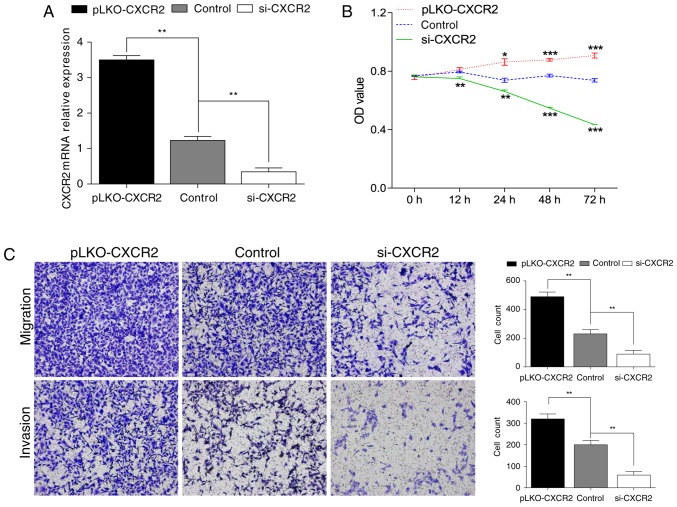
Validation of the function of CXCR2 in HESCs
To validate the critical role of CXCR2 in the development of endometriosis, HESCs were used. Overexpression and knockdown of CXCR2 in cells were achieved via transfection with pLKO lentiviral vectors and siRNA, respectively. Cells with scramble sequences were set as the control group. RT-qPCR was applied to confirm the efficacy of transfection (Fig. 3A). In the analysis of cell proliferation by CCK-8 assay, the overexpression of CXCR2 clearly increased the proliferation capacity of HESCs, while the capacity markedly decreased following the knockdown of CXCR2 (Fig. 3B). For the analysis of cell migration and invasion, Transwell assays were performed. As demonstrated in Fig. 3C, the migration and invasion capacities of HESCs were significantly promoted in the pLKO- CXCR2 group, but clearly suppressed in the si-CXCR2 group, compared with the control.
Figure 3.
CXCR2 promotes proliferation, migration and invasion of HESCs. (A) Confirmation of the efficacy of transfection by RT-qPCR. (B) Transfected HESCs were cultured for 3 days and CCK-8 assays were conducted to examine the proliferation capacity. (C) Migration and invasion capacities of transfected HESCs were measured by Transwell migration/invasion assays following 24 h in culture. Magnification, ×100. Data are demonstrated as the mean ± standards error from 3 independent wells. *P<0.05, **P<0.01, ***P<0.001 vs. control. HESCs, human endometrial stromal cells; CXCR2, C-X-C motif chemokine receptor 2; si, small interference; OD, optical density.
Discussion
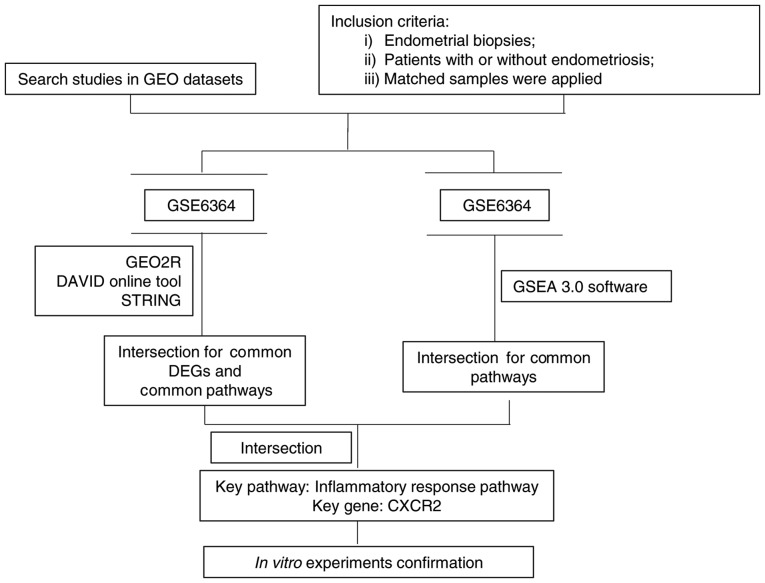
Endometriosis is characterized by the morphological and biological properties of the endometrium, including the proliferation, invasiveness, and attachment to the extracellular matrix, which are greater in ectopic endometrial cells (3), and results in a severe decline in the quality of life and psychological well-being, as well as it is a heavy economic burden, for those affected (10,31). Although various potential causative factors have been investigated, the exact mechanisms underlying the aberrant implantation of ectopic endometrial cells remain unclear. In the present study, a bioinformatics approach was applied to reveal the possible dysregulated pathways and genes in endometriosis, specifically through gene expression analysis (Fig. 4). The gene expression profiles of endometrial biopsies from women with or without endometriosis (GSE6364) were selected. A total of 172 DEGs were extracted. Gene expression profiling analysis through functional annotation and gene set enrichment analysis as well as a PPI network, demonstrated that genes associated with the inflammatory response were upregulated, and CXCR2 was one of the most important genes. Furthermore, the in vitro experiments confirmed that CXCR2 promoted proliferation, migration and invasion of HESCs, suggesting that CXCR2 may serve a critical role in the development of endometriosis.
Figure 4.
Summary of bioinformatics procedure. Flowchart demonstrating the identification and selection of microarray data from GEO, the data analysis procedure and relevant tools. GEO, Gene Expression Omnibus; GSEA, Gene Set Enrichment Analysis; DEG, differentially expression genes; CXCR2, C-X-C motif chemokine receptor 2.
According to Sampson's implantation theory that ectopic endometrial tissues contribute to the pathogenesis of endometriosis (18), the present study compared the gene expression profiles of endometriosis patients with that of normal subjects. The present results demonstrated that inflammatory response genes were significantly upregulated. Previous studies have indicated that inflammation serves a key role in the pathogenesis of endometriosis (32,33). Alterations in inflammatory factors may promote fibrotic adhesion contributing to the growth and spread of endometrial tissue (34). It has been reported that the peritoneal fluid of patients with endometriosis exhibits an increased level of activated macrophages and pro-inflammatory cytokines compared with that in normal individuals, and that the capacities for proliferation, invasion and adhesion to the extracellular matrix are greater in ectopic endometrial cells than in eutopic cells (3,35). The dynamic interactions between cytokines may serve a pivotal role in the formation of a microenvironment that favors the ectopic implantation of endometrial tissues and the progression of endometriosis (36). The presence and role of cytokines, such as IL-8, have been extensively investigated (37). IL-8 contributes to reproductive pathological processes, such as endometriosis, through affecting the proliferation and invasion of endometrial stroma cells (ESCs) in the ectopic endometrial tissue (36,38–40). CXCR2, a membrane receptor for IL-8, is a member of the G-protein-coupled receptor family, and is involved in influencing intracellular calcium concentration, the release of granular enzymes and chemotaxis in response to IL-8 (36). Through the high-affinity binding of IL8 to CXCR2, signals are transduced via a G protein-activated second messenger system to promote the proliferation and invasion of ESCs. The present results also demonstrated that CXCR2 promoted the proliferation of HESCs. This may have been the result of CXCR2 high affinity for growth regulated protein-α, which has previously been demonstrated to affect the proliferative capacity of endometriosis cells (41,42). However, the contribution of decreased proliferation to reductions in migration and invasion cannot be excluded as migration/invasion of cells is generally affected by the number of cells, which may pose as a limitation of the present study. In addition, the expression of CXCR2 is very weak in normal endometrium and detection of CXCR2 in endometrial tissues through immunohistochemistry demonstrate that the expression is higher in women with endometriosis than in those with normal endometrium (36,42), which is consistent with the present DEG analysis.
The diagnosis of endometriosis may be achieved by detecting certain factors circulating in the serum or in the peritoneal fluid of patients. For example, the expression of soluble class-I and class-II molecules has been demonstrated to be significantly reduced by >30% in patients with endometriosis (43,44). The production of cytokines, which are thought to regulate the processes involved in the progression and development of endometriosis, largely depends on the immune system status of each patient with endometriosis, detection of a combination of these factors may allow endometriosis diagnosis more rapidly than the surgical route. IL-8 is increased in the serum of patients with endometriosis (45). Thus, CXCR2 could serve as a novel biomarker for the diagnosis of endometriosis; however, further studies are required to demonstrate this.
To conclude, the application of bioinformatic methods to reveal important biomarkers in endometriosis was of importance of the present study. Overexpression of CXCR2 promotes the proliferation, migration and invasion of endometrial cells, which in situ may lead to the presence of endometrial glands and/or stroma outside of the uterine cavity. CXCR2 may be associated with the development of endometriosis via the inflammatory response and it has potential as a biomarker for the diagnosis of endometriosis.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Independent Scientific Research Program of Wuhan University (grant no. 2042018kf0170).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
AT conducted the experiments and wrote the manuscript. RL conducted software-based analysis and modified the English language of the manuscript. HL and ML collected the patient data. PR made substantial contributions to the design of the present study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Endometrial tissue in the present study was collected from patients from September to November 2017 and written informed consent was provided. The present study was approved by the Ethics Committee of Renmin Hospital of Wuhan University (Renmin, China) in September 2017.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Eskenazi B, Warner ML. Epidemiology of endometriosis. Obstet Gynecol Clin North Am. 1997;24:235–258. doi: 10.1016/S0889-8545(05)70302-8. [DOI] [PubMed] [Google Scholar]
- 2.Vetvicka V, Lagana AS, Salmeri FM, Triolo O, Palmara VI, Vitale SG, Sofo V, Králíčková M. Regulation of apoptotic pathways during endometriosis: From the molecular basis to the future perspectives. Arch Gynecol Obstet. 2016;294:897–904. doi: 10.1007/s00404-016-4195-6. [DOI] [PubMed] [Google Scholar]
- 3.Laganà AS, Vitale SG, Salmeri FM, Triolo O, Frangež Ban H, Vrtačnik-Bokal E, Stojanovska L, Apostolopoulos V, Granese R, Sofo V. Unus pro omnibus, omnes pro uno: A novel, evidence-based, unifying theory for the pathogenesis of endometriosis. Med Hypotheses. 2017;103:10–20. doi: 10.1016/j.mehy.2017.03.032. [DOI] [PubMed] [Google Scholar]
- 4.Louis Buck GM, Hediger ML, Peterson CM, Croughan M, Sundaram R, Stanford J, Chen Z, Fujimoto VY, Varner MW, Trumble A, et al. Incidence of endometriosis by study population and diagnostic method: The ENDO study. Fertil Steril. 2011;96:360–365. doi: 10.1016/j.fertnstert.2011.05.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sourial S, Tempest N, Hapangama DK. Theories on the pathogenesis of endometriosis. Int J Reprod Med. 2014;2014:179515. doi: 10.1155/2014/179515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laganà AS, Salmeri FM, Vitale SG, Triolo O, Götte M. Stem cell trafficking during endometriosis: May epigenetics play a pivotal role? Reprod Sci. 2018;25:978–979. doi: 10.1177/1933719116687661. [DOI] [PubMed] [Google Scholar]
- 7.Gupta S, Agarwal A, Krajcir N, Alvarez JG. Role of oxidative stress in endometriosis. Reprod Biomed Online. 2006;13:126–134. doi: 10.1016/S1472-6483(10)62026-3. [DOI] [PubMed] [Google Scholar]
- 8.Gazvani R, Templeton A. Peritoneal environment, cytokines and angiogenesis in the pathophysiology of endometriosis. Reproduction. 2002;123:217–226. doi: 10.1530/rep.0.1230217. [DOI] [PubMed] [Google Scholar]
- 9.Van Langendonckt A, Casanas-Roux F, Donnez J. Oxidative stress and peritoneal endometriosis. Fertil Steril. 2002;77:861–870. doi: 10.1016/S0015-0282(02)02959-X. [DOI] [PubMed] [Google Scholar]
- 10.Arablou T, Kolahdouz-Mohammadi R. Curcumin and endometriosis: Review on potential roles and molecular mechanisms. Biomed Pharmacother. 2018;97:91–97. doi: 10.1016/j.biopha.2017.10.119. [DOI] [PubMed] [Google Scholar]
- 11.Mohammadi Kolahdouz R, Arablou T. Resveratrol and endometriosis: In vitro and animal studies and underlying mechanisms (Review) Biomed Pharmacother. 2017;91:220–228. doi: 10.1016/j.biopha.2017.04.078. [DOI] [PubMed] [Google Scholar]
- 12.Laganà AS, La Rosa VL, Rapisarda AMC, Valenti G, Sapia F, Chiofalo B, Rossetti D, Frangež Ban H, Bokal Vrtačnik E, Vitale SG. Anxiety and depression in patients with endometriosis: Impact and management challenges. Int J Womens Health. 2017;9:323–330. doi: 10.2147/IJWH.S119729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laganà AS, La Rosa V, Petrosino B, Vitale SG. Comment on ‘Risk of developing major depression and anxiety disorders among women with endometriosis: A longitudinal follow-up study’. J Affect Disord. 2017;208:672–673. doi: 10.1016/j.jad.2016.07.016. [DOI] [PubMed] [Google Scholar]
- 14.Laganà AS, Vitale SG, Granese R, Palmara V, Frangež Ban H, Vrtačnik-Bokal E, Chiofalo B, Triolo O. Clinical dynamics of Dienogest for the treatment of endometriosis: From bench to bedside. Expert Opin Drug Metab Toxicol. 2017;13:593–596. doi: 10.1080/17425255.2017.1297421. [DOI] [PubMed] [Google Scholar]
- 15.Simoens S, Dunselman G, Dirksen C, Hummelshoj L, Bokor A, Brandes I, Brodszky V, Canis M, Colombo GL, DeLeire T, et al. The burden of endometriosis: Costs and quality of life of women with endometriosis and treated in referral centres. Hum Reprod. 2012;27:1292–1299. doi: 10.1093/humrep/des073. [DOI] [PubMed] [Google Scholar]
- 16.Luo Y, Wu Y, Peng Y, Liu X, Bie J, Li S. Systematic analysis to identify a key role of CDK1 in mediating gene interaction networks in cervical cancer development. Ir J Med Sci. 2016;185:231–239. doi: 10.1007/s11845-015-1283-8. [DOI] [PubMed] [Google Scholar]
- 17.Fang C, Huang Y, Pei Y, Zhang HH, Chen X, Guo H, Li S, Ji X, Hu J. Genome-wide gene expression profiling reveals that CD274 is up-regulated new-onset type 1 diabetes mellitus. Acta Diabetol. 2017;54:757–767. doi: 10.1007/s00592-017-1005-y. [DOI] [PubMed] [Google Scholar]
- 18.Benagiano G, Brosens I, Lippi D. The history of endometriosis. Gynecol Obstet Invest. 2014;78:1–9. doi: 10.1159/000358919. [DOI] [PubMed] [Google Scholar]
- 19.Burney RO, Talbi S, Hamilton AE, Vo KC, Nyegaard M, Nezhat CR, Lessey BA, Giudice LC. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology. 2007;148:3814–3826. doi: 10.1210/en.2006-1692. [DOI] [PubMed] [Google Scholar]
- 20.Sean D, Meltzer PS. GEOquery: A bridge between the gene expression omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 21.Deng W, Wang Y, Liu Z, Cheng H, Xue Y. HemI: A toolkit for illustrating heatmaps. PLoS One. 2014;9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 26.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho S, Mutlu L, Zhou Y, Taylor HS. Aromatase inhibitor regulates let-7 expression and let-7f-induced cell migration in endometrial cells from women with endometriosis. Fertil Steril. 2016;106:673–680. doi: 10.1016/j.fertnstert.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Stefano V, Wang B, Parobchak N, Roche N, Rosen T. RelB/p52-mediated NF-κB signaling alters histone acetylation to increase the abundance of corticotropin-releasing hormone in human placenta. Sci Signal. 2015;8:ra85. doi: 10.1126/scisignal.aaa9806. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Simoens S, Hummelshoj L, D'Hooghe T. Endometriosis: Cost estimates and methodological perspective. Hum Reprod Update. 2007;13:395–404. doi: 10.1093/humupd/dmm010. [DOI] [PubMed] [Google Scholar]
- 32.Mu F, Harris HR, Rich-Edwards JW, Hankinson SE, Rimm EB, Spiegelman D, Missmer SA. A prospective study of inflammatory markers and risk of endometriosis. Am J Epidemiol. 2018;187:515–522. doi: 10.1093/aje/kwx272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bulun SE. Endometriosis. N Engl J Med. 2009;360:268–279. doi: 10.1056/NEJMra0804690. [DOI] [PubMed] [Google Scholar]
- 34.Allen C, Hopewell S, Prentice A. Non-steroidal anti-inflammatory drugs for pain in women with endometriosis. Cochrane Database Syst Rev: CD004753. 2005 doi: 10.1002/14651858.CD004753.pub2. [DOI] [PubMed] [Google Scholar]
- 35.Burney RO, Giudice LC. Pathogenesis and pathophysiology of endometriosis. Fertil Steril. 2012;98:511–519. doi: 10.1016/j.fertnstert.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulukus M, Ulukus EC, Seval Y, Zheng W, Arici A. Expression of interleukin-8 receptors in endometriosis. Hum Reprod. 2005;20:794–801. doi: 10.1093/humrep/deh675. [DOI] [PubMed] [Google Scholar]
- 37.Nishida M, Nasu K, Narahara H. Role of chemokines in the pathogenesis of endometriosis. Front Biosci (Schol Ed) 2011;3:1196–1204. doi: 10.2741/220. [DOI] [PubMed] [Google Scholar]
- 38.Arici A, Seli E, Zeyneloglu HB, Senturk LM, Oral E, Olive DL. Interleukin-8 induces proliferation of endometrial stromal cells: A potential autocrine growth factor. J Clin Endocrinol Metab. 1998;83:1201–1205. doi: 10.1210/jcem.83.4.4743. [DOI] [PubMed] [Google Scholar]
- 39.Garcia-Velasco JA, Arici A. Interleukin-8 expression in endometrial stromal cells is regulated by integrin-dependent cell adhesion. Mol Hum Reprod. 1999;5:1135–1140. doi: 10.1093/molehr/5.12.1135. [DOI] [PubMed] [Google Scholar]
- 40.Iwabe T, Harada T, Tsudo T, Tanikawa M, Onohara Y, Terakawa N. Pathogenetic significance of increased levels of interleukin-8 in the peritoneal fluid of patients with endometriosis. Fertil Steril. 1998;69:924–930. doi: 10.1016/S0015-0282(98)00049-1. [DOI] [PubMed] [Google Scholar]
- 41.Premack BA, Schall TJ. Chemokine receptors: Gateways to inflammation and infection. Nat Med. 1996;2:1174–1178. doi: 10.1038/nm1196-1174. [DOI] [PubMed] [Google Scholar]
- 42.Ulukus M, Ulukus EC, Seval Y, Cinar O, Zheng W, Arici A. Expression of interleukin-8 receptors in patients with adenomyosis. Fertil Steril. 2006;85:714–720. doi: 10.1016/j.fertnstert.2005.08.053. [DOI] [PubMed] [Google Scholar]
- 43.Antiñolo G, Fernández RM, Noval JA, Molini JL, Borrego S. Analysis of the involvement of CCR5-Delta32 and CCR2-V64I variants in the development of endometriosis. Mol Hum Reprod. 2004;10:155–157. doi: 10.1093/molehr/gah026. [DOI] [PubMed] [Google Scholar]
- 44.Wieser F, Dogan S, Klingel K, Diedrich K, Taylor RN, Hornung D. Expression and regulation of CCR1 in peritoneal macrophages from women with and without endometriosis. Fertil Steril. 2005;83:1878–1881. doi: 10.1016/j.fertnstert.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 45.Chapron C, Borghese B, Streuli I, de Ziegler D. Markers of adult endometriosis detectable in adolescence. J Pediatr Adolesc Gynecol. 2011;24(5 Suppl):S7–S12. doi: 10.1016/j.jpag.2011.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.