Abstract
The lytic transglycosylases (LTs) are bacterial enzymes that catalyze the non-hydrolytic cleavage of the peptidoglycan structures of the bacterial cell wall. They are not catalysts of glycan synthesis as might be surmised from their name. Notwithstanding the seemingly mundane reaction catalyzed by the LTs, their lytic reactions serve bacteria for a series of astonishingly diverse purposes. These purposes include cell-wall synthesis, remodeling, and degradation; for the detection of cell-wall-acting antibiotics; for the expression of the mechanism of cell-wall-acting antibiotics; for the insertion of secretion systems and flagellar assemblies into the cell wall; as a virulence mechanism during infection by certain Gram-negative bacteria; and in the sporulation and germination of Gram-positive spores. Significant advances in the mechanistic understanding of each of these processes have coincided with the successive discovery of new LTs structures. In this review, we provide a systematic perspective on what is known on the structure-function correlations for the LTs, while simultaneously identifying numerous opportunities for the future study of these enigmatic enzymes.
Keywords: Lytic Transglycosylase, Peptidoglycan, Muropeptide, Bacteria, Cell-Wall Recycling, Secretion System, AmpC, AmpR
Graphical abstract
Introduction
Bacteria preserve the integrity of their cell envelope during growth and cell division by the integration of an array of interwoven pathways (Pazos et al., 2017). A key component of this envelope and focus of these pathways is a peptidoglycan polymer—the cell wall—that fully surrounds the bacterium. The gross structure of the peptidoglycan is of strands of a (NAG-NAM)n (NAG: N-acetylglucosamine and NAM: N-acetylmuramic acid) polysaccharide, covalently interconnected to neighboring strands through peptide stems attached to the NAM saccharide. The intact polymer is called the murein sacculus, and the individual strands that comprise the sacculus are collectively referred to as muropeptides (Wientjes et al., 1991; Silhavy et al., 2010; Vollmer & Seligman, 2010). The peptidoglycan contributes directly to the shape of the bacterium (Young, 2010). The peptidoglycan of the monoderm Gram-positive bacterium is typically multi-layered (15–30 nm thick), while the peptidoglycan of the diderm Gram-negative bacterium is primarily mono- or bilayered (3–6 nm thick). The peptidoglycan is an exterior component of the Gram-positive cell envelope, while in the Gram-negative bacterium the peptidoglycan is located in the periplasm between the two membranes. The peptidoglycan is a structurally dynamic polymer that grows, remodels, and separates. For example, in the course of doubling the peptidoglycan in a single generation of exponential growth, the Gram-negative bacterium Escherichia coli remodels and recycles nearly half of its existing peptidoglycan (Goodell & Schwarz, 1985; Goodell, 1985; Park, 1993). As the Gram-negative bacterium ages and enters stationary growth, the extent of the cross-linking of its peptide stem increases, and the barrier function of the peptidoglycan is reinforced (Pisabarro et al., 1985). The peptidoglycan prevents the undesired passage of macromolecules into and out of the cell, while also serving as a scaffold through which critical proteins of the bacterium interact (Typas et al., 2012). A final and no less important function of peptidoglycan growth, and of the recycling of the muropeptides liberated during growth, is the provision of sensory mechanisms to detect the presence of cell-wall-targeted antibiotics (Park, 1995).
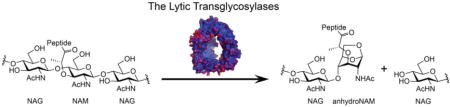
Superfamilies of the amide and glycoside hydrolases orchestrate the dynamic peptidoglycan structure to enable wall expansion during cell growth, for the splitting of the septum to permit separation of the divided cells, for the excavation of the cell wall for the insertion of protein complexes (for example, secretion systems, flagella, and pili), for the recycling of the muropeptides liberated during these processes, for endospore sporulation and germination, and for other functions that await discovery (Höltje, 1998; Koraimann, 2003; Keep et al., 2006; Baron & Coombes, 2007; Scheurwater et al., 2008; Uehara & Park, 2008; Scheurwater & Burrows, 2011; van Heijenoort, 2011; Alcorlo et al., 2017). A key superfamily of enzymes at the nexus of both cell-wall recycling and cell-wall-antibiotic detection is the lytic transglycosylases (LTs). Although their structures define them as members of the glycoside hydrolase superfamily, the reaction catalyzed by the LTs is non-hydrolytic (Höltje et al., 1975). The reactions of LTs fragment the polysaccharide of the peptidoglycan at the NAM-NAG glycosidic bond, by an intramolecular cyclization of the N-acetylmuramyl moiety to yield a 1,6-anhydro-N-acetyl-β-D-muramyl (1,6-anhydroMurNAc) product (Figure 1). This transformation is the hallmark of LT catalysis. In Gram-negative bacteria, the 1,6-anhydroMurNAc-containing muropeptides are transported from the periplasm to the cytoplasm through the transmembrane protein AmpG (Jacobs et al., 1994). These muropeptides are degraded in the cytoplasm and their constituent components are used for Lipid II biosynthesis. The Lipid II assembled in the cytoplasm is delivered to the periplasm for de novo synthesis of the peptidoglycan (Figure 2) (Barreteau et al., 2008; Bouhss et al., 2008; Vollmer & Bertsche, 2008; Butler et al., 2013; Sieger et al., 2013; Mohammadi et al., 2014; Sham et al., 2014; Meeske et al., 2015; Scheffers & Tol, 2015; Ruiz, 2016; Kuk et al., 2017; Leclercq et al., 2017). The elegantly preserved balance among peptidoglycan synthesis, remodeling, and degradation sustains the integrity of the cell wall (Höltje & Heidrich, 2001).
Figure 1.
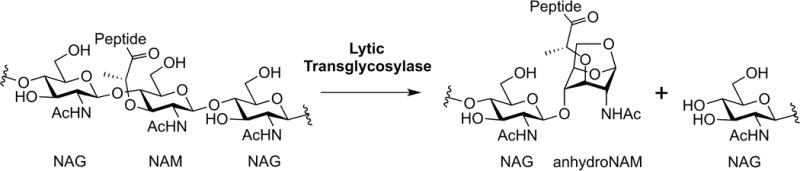
The hallmark reaction of the lytic transglycosylases.
Figure 2.
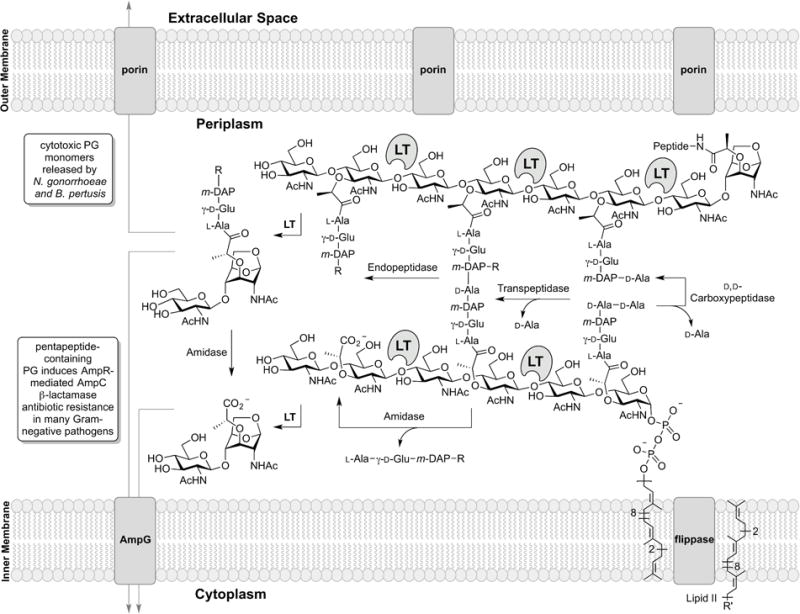
The periplasmic events (where the LTs exist) of Gram-negative cell-wall recycling are depicted. The membrane anchoring of the LTs is not represented in the figure. The LT reaction sites are shown. In the scheme R = D-Ala-D-Ala (penta), or D-Ala (tetra), or H (tri) and R’ = diphosphoryl-N-acetyl muramoyl (glucosamine)-L-Ala-γ-D-Glu-meso-DAP-D-Ala-D-Ala.
Herein, we review the LT enzymes, with particular emphasis on their structure and their roles in the Gram-negative bacteria E. coli, Pseudomonas aeruginosa, Neisseria gonorrhoeae, and Stenotrophomonas maltophilia and in the Gram-positive bacteria Clostridium difficile, Bacillus anthracis, and Bacillus cereus. This review discusses first the reaction products of LT catalysis, and progresses to the protein structural studies that organize the LT superfamily of the Gram-negative bacteria. We expand the previous six-family classification for the Gram-negative LTs (Herlihey & Clarke, 2016) and introduce two new families to accommodate the Gram-positive LTs. This latter section comprises the major topic of this review. We provide a concise mechanistic perspective, and discuss the different assay methods used to evaluate LT catalysis. Lastly, we promote the LTs as potentially important drug targets, and summarize efforts toward this objective.
The reaction products of LTs initiate both offensive and defensive mechanisms
Escherichia coli, the first organism for which LT activity was documented, recycles more than 90% of its LT reaction products (Uehara et al., 2005). Other Gram-negative organisms are less efficient and release un-recycled muropeptides to the surrounding extracellular environment (Greenway & Perkins, 1985). The iconic products of LT catalysis, the 1,6-anhydroMurNAc muropeptides, contribute to bacterial virulence in many organisms (Adin et al., 2009; Sorbara & Philpott, 2011; Boudreau et al., 2012; Bertsche et al., 2015). An example is the Gram-negative pathogen Bordatella pertussis, the notorious cause of whooping cough, that exports the “tetrapeptide” GlcNAc-1,6-anhydroMurNAc-L-Ala-γ-D-Glu-meso-DAP-D-Ala muropeptide—appropriately named tracheal cytotoxin (TCT)—from the bacterium (Rosenthal et al., 1987; Cookson et al., 1989; Luker et al., 1993; Chang et al., 2006; Kawasaki et al., 2008). This muropeptide causes both ciliostasis and extrusion of ciliated cells in infected patients. The mechanism for this cytotoxic effect is induction of interleukin-1 (IL-1), which subsequently upregulates nitric oxide (NO) production, a toxin to sensitive ciliated cells (Heiss et al., 1994). Similarly, export of structurally related muropeptides (such as 1,6-anhydroMurNAc tripeptide, lacking the terminal D-Ala of the “tetrapeptide” muropeptide) by the Gram-negative N. gonorrhoeae during pelvic infection causes an inflammation-dependent loss of the ciliated cells of the fallopian tubes (Chan et al., 2012; Chan & Dillard, 2016). These released muropeptides induce inflammatory cytokine (including IL-1β, IL-8, and TNF-α) production via NOD1 activation of the inflammasome (Philpott et al., 2014; Wheeler et al., 2014; Caruso & Núñez, 2016; Keestra-Gounder et al., 2016). Muropeptide-induced inflammatory dysfunction is associated also with infections caused by methicillin-resistant Staphylococcus aureus (Müller et al., 2015), Helicobacter pylori (Suarez et al., 2015), and the Chlamydia (Zou et al., 2016).
LT reaction products enable bacterial defense mechanisms. An efficient resistance mechanism to β-lactam antibiotics—mainstays of antibacterial chemotherapy—in many Gram-negative bacteria (including P. aeruginosa and most Enterobacteriaceae) is the induction of an enzyme, a β-lactamase, to hydrolytically deactivate the β-lactam antibiotics. β-Lactams enter the periplasm of these bacteria through outer-membrane porins to encounter their biological target, the penicillin-binding proteins (PBPs) of peptidoglycan biosynthesis. β-Lactam antibiotics obstruct cell-wall synthesis by covalent inactivation of these enzymes. The β-lactam ring (of the penicillin, cephalosporin, and carbapenem antibiotics) mimics the D-Ala-D-Ala segment of the muropeptide stem of the peptidoglycan (Lee et al., 2001; Lee et al., 2003; Pratt, 2016). PBPs act on this same muropeptide stem as their substrate. The consequence of this mimicry—the inactivation of these PBPs—results in failed crosslinking of the peptidoglycan. The resulting aberrant peptidoglycan is degraded by the LTs. Notably, the soluble LT of E. coli (Slt70) enriches the periplasmic muropeptide pool of tripeptide and tetrapeptide containing muropeptides, preventing the misincorporation of nascent peptidoglycan into the cell wall by nonspecific transpeptidases in E. coli (Cho et al., 2014). Other LTs enrich the pool of pentapeptide-containing muropeptides. These muropeptides are transported to the cytoplasm by the inner-membrane protein AmpG (Johnson et al., 2013; Fisher & Mobashery, 2014). In the cytoplasm, pentapeptide-containing muropeptides derepress (in P. aeruginosa and many Enterobacteriaceae) the AmpR transcription regulator so as to initiate β-lactamase expression (among other effects) as a key resistance mechanism (Wiedemann et al., 1998; Balcewich et al., 2010; Balasubramanian et al., 2015; Vadlamani et al., 2015; Lee et al., 2016a; Dik et al., 2017). The enabling function of LTs intimately links muropeptide recycling to antibiotic resistance (Figure 2). Antibiotic resistance is a problem of profound societal consequence and contributes to an estimated 700,000 deaths per year globally from multidrug-resistant bacterial infections (O’Neill, 2016).
Other physiological responses in eukaryotes caused by muropeptide release include somnogenic, arthritogenic, and pyrogenic activities (Krueger et al., 1984; Fleming et al., 1986; Johannsen, 1993). Furthermore, muropeptides cause appetite suppression and weight loss in rats, and are suggested to be the cause of appetite loss during bacterial illness (Biberstine et al., 1996).
A large diverse family of LTs has evolved
Bacteria depend on an array of cellular tasks performed by LTs, and encode a seemingly matching array of LTs for these purposes. Although each member is presumed to have specialized tasks, extensive study implicates functional redundancy so as to compensate for the loss of any single member. The inability to create a viable pan-LT knockout suggests that at least some of the LT functions are essential (Scheurwater & Clarke, 2008). Nonetheless (and accordingly) in many cases the primary cellular task performed by each LT is unknown. Six unique LT catalytic folds are known in Gram-negative bacteria, with each annotated as a conserved domain in the Pfam database (Figure 3). These six domains were organized previously into six distinct families (Herlihey & Clarke, 2016). LTs may be further organized by their substrate preference for either exolytic or endolytic strand cleavage. Exolytic LT activity cleaves a terminal NAG-NAM disaccharide from the end of the glycan strand. Endolytic LT activity cleaves the chains internally and hence gives products that are at least four saccharides in length (Lee et al., 2013; Lee et al., 2017). These dual characteristics provide a basis for LT organization, and in turn provide insight into their tasks within the cell. In this review, we classify the known LTs (and LT homologues) of four Gram-negative and three Gram-positive organisms. Additionally, we classify recently discovered protein homologues that are not yet proven to have bona fide LT catalytic activity.
Figure 3.

Domain architecture of Gram-negative LTs. Domains were assigned based on consensus analysis of Pfam Database and NCBI conserved domain database. Protein name and locus tag are given for E. coli K12 (ECK…), P. aeruginosa PAO1 (PA…), S. maltophilia KJ (Smlt…), and N. gonorrhoeae FA1090 (NGO…). Locus tags are not given for Enteropathogenic E. coli (EPEC) or Rhodobacter sphaeroides (RS). For the bacteriophage LTs (family 1H and 4), the name of the infected host organism (ECK, PA, Smlt, and NG) and the identities of the bacteriophages encoding these LTs are provided. The PG_binding_1 domain of Family 1H and 3B is abbreviated as PG_b1. A color version of this figure is available at www.tandfonline.com/ibmg.
Gram-negative Family 1 Lytic Transglycosylases
Overview
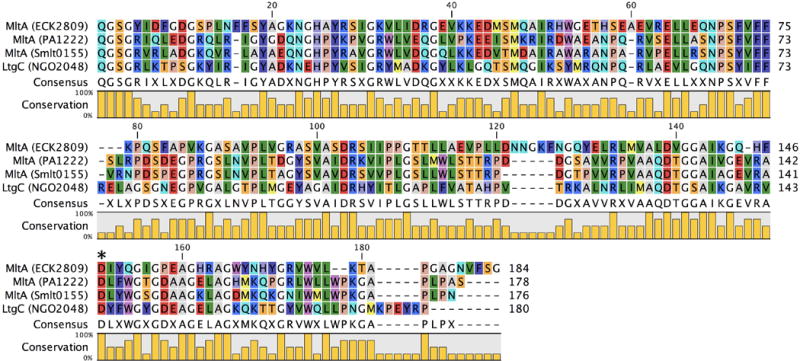
The largest family of LTs is Family 1. Family 1 previously encompassed five subfamilies in E. coli (1A, 1B, 1C, 1D, 1E) and three subfamilies in P. aeruginosa (1A, 1D, 1E) (Blackburn & Clarke, 2002). It is herein expanded to six subfamilies in E. coli (1A, 1B, 1C, 1D, 1E, 1G), four subfamilies in P. aeruginosa (1A, 1D, 1E; and as a phage 1H), two subfamilies in S. maltophilia (1A, 1D), and four subfamilies in N. gonorrhoeae (1A, 1B, 1D, 1G). The specific LTs of the subfamilies include: (1A) E. coli Slt70, P. aeruginosa Slt, S. maltophilia Slt, N. gonorrhoeae LtgA; (1B) E. coli MltC, N. gonorrhoeae LtgB; (1C) E. coli MltE; (1D) E. coli MltD, P. aeruginosa MltD, S. maltophilia MltD1 and MltD2, N. gonorrhoeae LtgE; (1E) E. coli MltF, P. aeruginosa MltF and MltF2; (1F) R. sphaeroides SltF and their respective homologues in other Gram-negative organisms (see Table 1 for complete description). Additionally the Family 1 LTs include the herein newly assigned non-chromosomally encoded EtgA (1G) of EPEC E. coli (Garcia-Gomez et al., 2011) and the LtgX (1G) residing in the gonococcal genetic island (GGI) of N. gonorrhoeae (Kohler et al., 2007). Additionally, the endolysin of bacteriophage ΦKZ GP144 (1H) is a member of Family 1. Two proteins from Mycobacterium tuberculosis, resuscitation-promoting factor B and E, give structural evidence in support of their identification as LTs, but formation of the LT reaction product has not yet been shown (Squeglia et al., 2013; Mavrici et al., 2014; Sexton et al., 2015; Ruggiero et al., 2016). Family 1 is a diverse family in both sequence (Figure 4) and function (Lee et al., 2013). Family 1 includes both primarily soluble subfamilies (1A, 1F, 1G, 1H) and lipoprotein (membrane-bound) subfamilies (1B, 1C, 1D, 1E). Each protein has a highly conserved catalytic domain, comprised primarily of α-helices (Figure 5).
Table 1.
Summary of the Gram-negative Family 1 LTs.
| Length (AA) | MW (Da) | Apo Structure | Complex Structure | |
|---|---|---|---|---|
| Family 1A | ||||
| Slt70 (ECK4384) | 645 | 73353.1 | 1QSA (van Asselt et al., 1999b) | 1QTE (1,6-Anhydromurotripeptide) (van Asselt et al., 1999b) 1SLY (Bulgecin A) (Thunnissen et al., 1995b) |
| Slt (PA3020) | 642 | 73390.4 | ||
| Slt (Smlt4007) | 661 | 73023.3 | ||
| LtgA (NGO2135) | 616 | 67726.2 | 5MPQ (Bulgecin A) (Williams et al., 2017) | |
| Family 1B | ||||
| MltC (ECK2958) | 359 | 40112.5 | 4C5F (Artola-Recolons et al., 2014) | 4CFO (Tetrasaccharide) (Artola-Recolons et al., 2014) 4CFP (Tetrasaccharide) (Artola-Recolons et al., 2014) 4CHX (Disaccharide pentapeptide) (Artola-Recolons et al., 2014) |
| LtgB (NGO1033) | 207 | 23374.9 | ||
| Family 1C | ||||
| MltE (ECK1181) | 203 | 22226.5 | 2Y8P (Artola-Recolons et al., 2011a) 3T36 (Fibriansah et al., 2012) |
4HJV (Bulgecin & Murodipeptide) (Fibriansah et al., 2012) 4HJY (Catalytic Mutant, Chitopentaose) (Fibriansah et al., 2012) 4HJZ (Catalytic Mutant, Chitopentaose) (Fibriansah et al., 2012) |
| Family 1D | ||||
| MltD (ECK0211) | 452 | 49417.4 | 1E0G (LysM domain) (Bateman & Bycroft, 2000) | |
| MltD (PA1812) | 534 | 59734.9 | ||
| MltD1 (Smlt0994) | 398 | 41874.6 | ||
| MltD2 (Smlt3434) | 537 | 57751.6 | ||
| LtgE (NGO0608) | 659 | 72159.9 | ||
| Family 1E | ||||
| MltF (ECK2556) | 518 | 58302.1 | ||
| MltF (PA3764) | 490 | 55226.5 | 4P11 5A5X (Dominguez-Gil et al., 2016) |
4OWD (Cysteine) 4OXV (Valine) 4OYV (Leucine) 4OZ9 (Isoleucine) 4P0G (Bulgecin A and Muropeptide) 5AA1 (NAG-anhNAM-pentapeptide) (Dominguez-Gil et al., 2016) 5AA2 (NAM-pentapeptide) (Dominguez-Gil et al., 2016) 5AA3 (tetrasaccharide and tetrapeptide) (Dominguez-Gil et al., 2016)5AA4 (tetrapeptide) (Dominguez-Gil et al., 2016) |
| MltF2 (PA2865) | 476 | 53310.4 | ||
| Family 1F | ||||
| SltF | ||||
| Family 1G | ||||
| EtgA (EPEC) | 152 | 17026.5 | 4XP8 (Catalytic Mutant) (Burkinshaw et al., 2015) | |
| LtgX (NG5004) | 153 | 17106.9 | ||
| Family 1H | ||||
| GP144 (Bacteriophage ΦKZ) | 260 | 28815.0 | 3BKH | 3BKV (chitotetraose, (NAG)4) (Fokine et al., 2015) |
Figure 4.
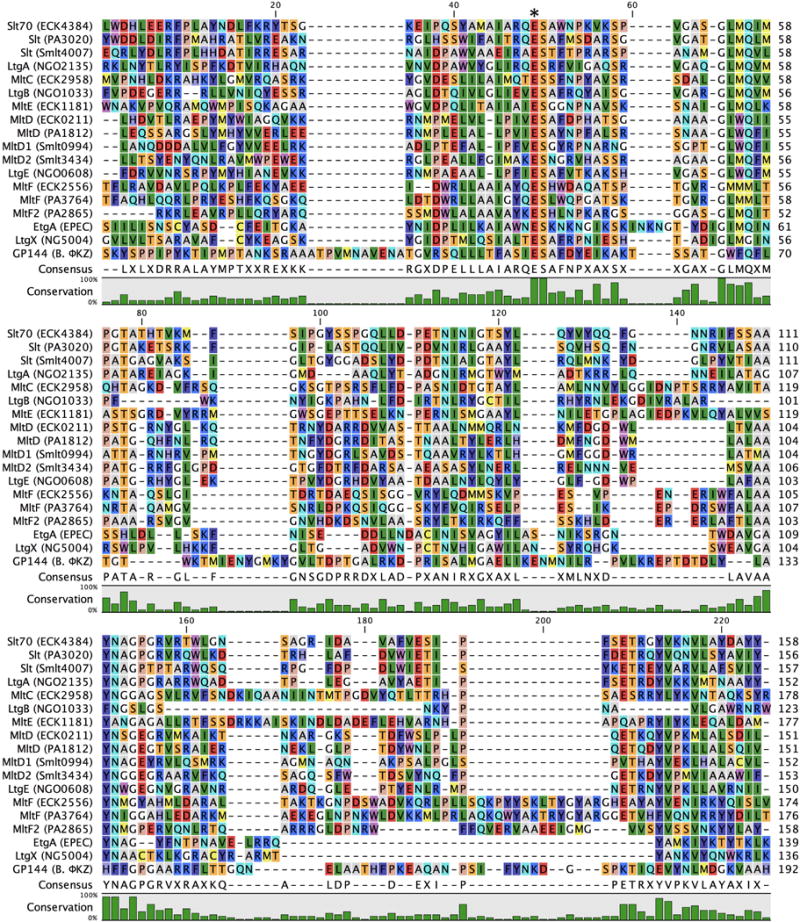
Multiple-sequence alignment displaying the LT domains of Gram-negative Family 1 LTs. In consideration of the size of the Family 1 alignment, R. sphaeroides SltF is not included as the SLT domain of the protein is considerably larger than the other Family 1 members. The asterisk denotes the position of the catalytic residue. A color version of this figure is available at www.tandfonline.com/ibmg.
Figure 5.
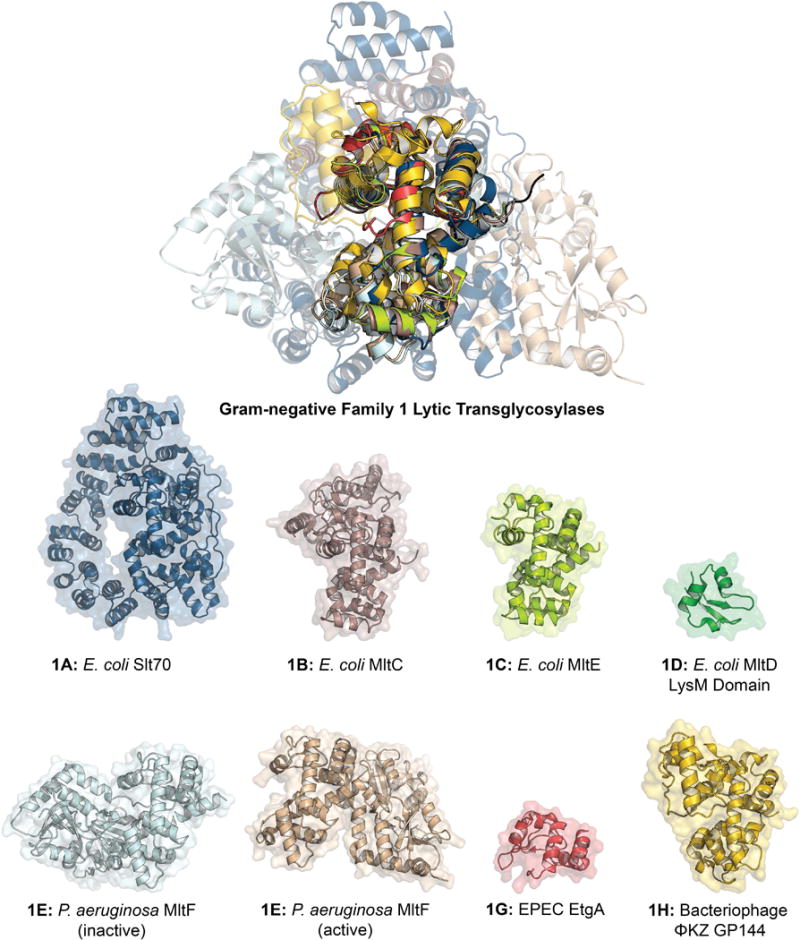
X-ray structure alignment of Family 1 LTs, displaying the conservation of the core SLT domain (Pfam: PF01464) and the diversity of the peripheral domains. The ribbon representation of each apo LT crystal structure is displayed below with a transparent surface representation. The structure of an LT domain of Family 1D has not been solved, therefore only the peripheral domain is displayed. Family 1E is displayed in both the active and inactive conformations. Refer to table 1 for PDB codes. A color version of this figure is available at www.tandfonline.com/ibmg.
Blackburn and Clarke previously defined four consensus motifs for LT assignment to Family 1. Motif I is the two residue ES, numbered as E64 and S65 in the MltE (ECK1181) LT of E. coli. This LT is chosen for representative numbering protein due to its structural simplicity, in that it lacks the additional domains of the other E. coli LTs. Motif I is invariant among Family 1 LTs (with the exception of ET in a few species encoding Family 1F). The ES dipeptide is positioned between substrate binding subsites –1 and +1 (Davies et al., 1997; Alcorlo et al., 2017). E64 is the primary catalytic residue of Family 1 LTs (Dijkstra & Thunnissen, 1994; Thunnissen et al., 1994; van Asselt et al., 1999b). The role of S65 in catalytic function has not been elucidated. Motif II (G–L–M–Q, 1A, 1B, 1C), (G–I/L–W/M–Q, 1D), (G–L/M–M–M/Q, 1E), (G–C–M/F–Q, 1F), (G–L/I–M–G/Q, 1G), and (G–W–F–Q, 1H), appearing as G79-L-M-Q82 in E. coli MltE is positioned on a loop at the +1 subsite. Motif III (A/G–Y–N, 1A), (A/R–Y–N, 1B), (Y–A–N, 1C), (A–Y–N, 1D, 1E, 1G), (A–Y–H, 1F), (A–H–F, 1H), appearing as Y146-A-N148 in E. coli MltE, is positioned on an α-helix at the –2 subsite. In all but two LT subfamilies, Motif IV is a conserved Y192 flanked by a hydrophobic residue. Residue Y192 is positioned on an α-helix at the –1 subsite adjacent to the catalytic E64, and is believed to have a catalytic role. The sequence identity at the consensus motif sites of N. gonorrhoeae LtgB is less conserved, but the sequence similarity to E. coli MltC is indisputable. The consensus motifs of Family 1 LTs are shared with other members of glycoside hydrolase Family 23 (GH23). However, the diversity of structures (Figure 5) and functions of these enzymes has at this time prevented the identification of a consensus mechanism.
Family 1A
The founding member of the LT family is the soluble lytic transglycosylase Slt70 (73 kDa; ECK4384) of E. coli that was discovered in 1975 by Höltje et al. (Höltje et al., 1975). The protein of the slt gene subsequently was cloned and purified in 1989 (Betzner & Keck, 1989; Rozeboom et al., 1990). Slt70 was initially proposed to be a cytoplasmic enzyme (Mett et al., 1980; Keck et al., 1985). Keck et al. later discovered that the N-terminal amino-acid residues comprise a signal peptide that directs the protein to the periplasm, with loss of the signal peptide (Engel et al., 1991). Slt70 is acknowledged as a representative exolytic enzyme of the LT superfamily (Kusser & Schwarz, 1980; Beachey et al., 1981; Keck et al., 1985; Betzner et al., 1990). Although Slt70 is primarily exolytic, Lee et al. demonstrated that a lower level of endolytic activity was present. Slt70 also shows strong preference for non-crosslinked muropeptides (Lee et al., 2013). A deletion mutant of the slt gene yielded E. coli without a growth defect phenotype (Roeder & Somerville, 1979; Templin et al., 1992). The structure of Slt70 (Figure 5) was solved by X-ray crystallography in 1994 by Thunnissen et al. It has a “doughnut-shaped” three-domain structure, including a dual-domained “superhelical” ring of α-helices on its N-terminus (Thunnissen et al., 1994; Thunnissen et al., 1995a). The ring is not closed. This characteristic may allow the protein to navigate, by simple rotation, around peptide stems. The third and catalytic domain is at the C-terminus. This SLT domain (Pfam: PF01464) has a characteristic lysozyme-like fold with the catalytic glutamate found as E478. Thunnissen et al. later reported Slt70 structures having bound both 1,6-anhydromurotripeptide and the disaccharide-mimetic LT inhibitor bulgecin A, shedding light on a mechanism (Thunnissen et al., 1995b; van Asselt et al., 1999b). Enzyme affinity assays revealed protein-protein interactions between E. coli Slt70 and PBPs 1b, 1c, 2, 3, 7/8 (Romeis & Höltje, 1994; von Rechenberg et al., 1996). P. aeruginosa Slt (PA3020), N. gonorrhoeae LtgA (NGO2135), and S. maltophilia Slt (Smlt4007) are homologous proteins to Slt70 of E. coli (33% (96%), 25% (78%), 27% (95%) for sequence identity (query coverage1), respectively). Unique to its family, LtgA is believed to localize to the inner leaflet of the outer membrane near the septum, due to the presence of a lipobox-processing site adjacent to the N-terminal signal peptide (Schaub et al., 2016). LtgA is the first Family 1 LT proven to cleave the glycosidic bond of a synthetic peptidoglycan tetrasaccharide (Schaub et al., 2016). A crystal structure of LtgA was recently reported by Williams and colleagues, but is not yet available in the PDB bank (Williams et al., 2017). A notable difference between the structure of LtgA and E. coli Slt70 is an overlap between the U-domain and the L-domain of the Neisseria protein that locks the protein in the “doughnut-shape”. Knockout mutants of N. gonorrhoeae LtgA displayed substantial loss of peptidoglycan monomer production. However, like the E. coli Slt70 knockout, growth of the N. gonorrhoeae LtgA-knockout is unperturbed (Cloud & Dillard, 2002). Interestingly, in the presence of primary human neutrophils, an N. gonorrhoeae strain with both ltgA and ltgD (an enzyme with a similar cellular role, as discussed later) knockouts exhibited reduced viability. This consequence was suggested to imply a protective role for the two LTs in the maintenance of the envelope, so as to limit exposure to antimicrobial proteins (Ragland et al., 2017). Nonetheless, LtgA and LtgD have essential roles in the cell-wall recycling of N. gonorrhoeae, even though they are not necessary for cell growth or division (Chan et al., 2012). Recent studies demonstrated that LtgA-produced peptidoglycan monomers are recycled at a higher rate than LtgD-produced peptidoglycan monomers (Schaub et al., 2016). Yet, the LtgA mutant was fortified compared to its wild-type counterpart, proving to be more resistant to autolysis and cell death in its stationary phase of growth (Cloud & Dillard, 2002). Similarities between LtgA and other Slt70 homologues have been observed. In Helicobacter pylori, Slt70 homologue knockouts produced significantly less peptidoglycan monomer (40% of wild-type) resulting in reduced induction of IL-8 in human (HEK293) cells (Viala et al., 2004), and likewise in Shigella flexneri an Slt70 homologue is required for full virulence (Bartoleschi et al., 2002).
Family 1B
Membrane-bound lytic transglycosylase C (MltC; ECK2958) of E. coli was first reported by Dijkstra and Keck in 1996 on the basis of sequence alignment with E. coli Slt70 (Dijkstra & Keck, 1996). E. coli MltC gene encodes an N-terminal signal peptide and Cys17 lipoyl anchor (LISCSTT) lipobox sequence to enable transport of the enzyme to the inner leaflet of the outer membrane for subsequent incorporation as an S-lipidated lipoprotein. MltC has the catalytic SLT domain (Pfam: PF01464) as its C-terminus and a DUF3393 (Domain of Unknown Function) domain (Pfam: PF11873) as its N-terminus. The three-dimensional structure of apo MltC (Figure 5) revealed that the DUF3393 domain comprised a five-stranded anti-parallel β-sheet, flanked by a single α-helix on one side and two α-helices on the other (Artola-Recolons et al., 2014). The catalytic acid is E218. Using the sacculus as a substrate, MltC processed both crosslinked and non-crosslinked peptidoglycan. Furthermore, MltC turned over oligomeric sugars (endolytic product) and also formed hydrolytic reaction products (lacking the 1,6-anhydro moiety). Both reactions were minor contributors to the overall reaction products (Lee et al., 2013). N. gonorrhoeae LtgB (NGO1033) is homologous to E. coli MltC (29% sequence identity and 54% query coverage), yet lacks an N-terminal DUF3393 domain (Kohler et al., 2005). The ltgB-knockout N. gonorrhoeae strain was viable and was unaffected with respect to release of peptidoglycan fragments, notwithstanding previous studies that implicated a role in peptidoglycan degradation. LtgB mutant analysis demonstrated that loss of the “catalytic residue” (E117A, inferred from homology) did not reduce activity, while loss of a (seemingly) non-conserved glutamate (E115A) in LtgB homologues was inactive (Kohler et al., 2005). Understanding the apparent deviation from conserved sequence of the protein, and the identification of the specific role of LtgB in N. gonorrhoeae, awaits further study.
Family 1C
The membrane-bound lytic transglycosylase E (MltE; ECK1181) of E. coli was first reported as a 22 kDa single-domain LT, named initially after its gene emtA (Kraft et al., 1998). The MltE gene encodes an N-terminal signal peptide, a lipobox sequence (LAGCSSK), and an SLT transglycosylase domain (Pfam: PF01464). MltE is the primary endolytic LT of E. coli with oligomeric saccharides containing a 1,6-anhydroMurNAc terminus comprising more than 33% of its reaction products (Lee et al., 2013). Its endolytic ability is attributed to a fully accessible peptidoglycan-binding groove that lacks the steric clash provided by a transverse loop at the +2 binding site or provided by a separate domain (van Asselt et al., 1999b; van Asselt et al., 2000; van Straaten et al., 2007; Fibriansah et al., 2012). The small size of MltE likely contributes to its endolytic activity, as the enzyme is small enough to navigate the complex peptidoglycan meshwork. E. coli MltE processes crosslinked, non-crosslinked, and peptide-free peptidoglycan. It appears to favor substrates that lack peptide crosslinking. The cellular role of MltE in a pathogenic E. coli strain has been elucidated. E. coli MltE is recruited by the periplasmic domain of TssM, a structural component of the Type VI secretion system, for excavation of the cell wall at the Type VI secretion system insertion site (Santin & Cascales, 2017). The structure of MltE (Figure 5) was first reported in apo form (Artola-Recolons et al., 2011a; Artola-Recolons et al., 2011b) and subsequently (as the E64Q catalytic mutant) in complex with bulgecin A and a murodipeptide, and in complex with chitopentaose (Fibriansah et al., 2012). These structures reveal the ability of MltE to accommodate long oligomeric saccharides. Its structural simplicity (compared to its Family 1 counterparts) and the availability of these structures distinguish MltE for mechanistic study. The other Gram-negative organisms included within the focus of this review (P. aeruginosa, S. maltophilia, and N. gonorrhoeae) do not encode an MltE homologue.
Family 1D
The structure of a Family 1D LT, exemplified by membrane-bound lytic transglycosylase D (MltD; ECK0211) of E. coli, has yet to be solved. However, there is sequence homology between Slt70 and MltD (gene product of yafG) (Koonin & Rudd, 1994). E. coli MltD is a 49 kDa protein that has an N-terminal SLT domain (Pfam: PF01464) and two C-terminal LysM domains (Pfam: PF01476). Note, however, that some Family 1D LTs encode one or three lysM domains (Figure 3). MltD of E. coli is a lipoprotein (anchor residue Cys16), in contrast to P. aeruginosa MltD (PA1812) that encodes a presumptive Lol avoidance lipobox (LAGCQGSG) sequence that would retain the protein in the outer leaflet of the inner membrane (Lewenza et al., 2008). A structure of the LysM domain of MltD (~40 residues, Figure 5) was reported in 2000 by Bateman and Bycroft as a representative LysM domain structure. LysM domains are peptidoglycan-binding modules that are common in proteins that facilitate bacterial pathogenesis and degrade the murein sacculus (Bateman & Bycroft, 2000; Mulder et al., 2006). Although the cellular role of E. coli MltD is not known, its interaction with peptidoglycan has been investigated. E. coli MltD has the uncommon ability to act on non-crosslinked peptidoglycan preferentially, but not exclusively, by exolytic cleavage. MltD reactions also form a significant proportion of hydrolytic products (Lee et al., 2013). E. coli MltD is homologous to the aforementioned MltD of P.aeruginosa (38% sequence identity, 96% query coverage), two LTs of S. maltophilia, MltD1 (Smlt0994: 29% sequence identity, 72% query coverage) and MltD2 (Smlt3434: 33% sequence identity, 44% query coverage), and N. gonorrhoeae LtgE (NGO0608: 35% sequence identity, 86% query coverage). The LtgE-knockout of N. gonorrhoeae showed no change in peptidoglycan monomer release, similar to the LtgB-knockout (Cloud-Hansen et al., 2008). An MltD-knockout of H. pylori showed reduced autolysis in stationary-phase cells. MltD is believed to act primarily toward rearrangement of the peptidoglycan layer (Chaput et al., 2007). However, the precise cellular function of the Family 1D LTs is unknown.
Family 1E
A distinguishing characteristic of the Family 1E LTs is their regulation by an allosteric mechanism. This property is likely true (but has yet to be demonstrated) for the other LTs that have secondary peptidoglycan-binding domains, such as the SPOR domain of RlpA and the LysM domain of MltD. Family 1E includes the membrane-bound lytic transglycosylase F (MltF; ECK2556) of E. coli. The E. coli MltF protein was first reported as a putative LT under the gene name yfhD (Koonin & Rudd, 1994) and was proven so in 2008 (Scheurwater & Clarke, 2008). E. coli MltF is a 58 kDa lipoprotein of the outer membrane (Scheurwater & Clarke, 2008). While the MltF of P. aeruginosa (38% sequence identity, 86% query coverage) is homologous, it appears to have a Lol avoidance lipobox sequence (Tokuda & Matsuyama, 2004), possibly resulting in its retention in the outer leaflet of the inner membrane. P. aeruginosa has a second Family 1E LT, MltF2 (PA2865), whose sequence closely resembles that of E. coli MltF, and thus is likely a lipoprotein of the inner leaflet of the outer membrane. MltF2 (26%, sequence identity, 72% query coverage compared to E. coli MltF) was studied recently (Lee et al., 2017). The domain architecture of E. coli MltF consists of an N-terminal SBP_bac_3 (Bacterial extracellular solute-binding proteins, Family 3) domain (Pfam: PF00497) and a C-terminal classic Family 1 SLT domain (Pfam: PF01464). MltF acts entirely on non-crosslinked peptidoglycan, both exo- and endolytically, consistent with an assignment as a specialized LT activated through its allosteric mechanism (Lee et al., 2013). MltF lacks function either in side-wall growth or in septum formation in both E. coli and P. aeruginosa (Scheurwater & Clarke, 2008; Lamers et al., 2015). Accordingly, its assignment may be for cell-wall excavation so as to enable the passage of the pilli, fimbriae, and other macromolecular complexes through the peptidoglycan (Dominguez-Gil et al., 2016). Structures of the apo form of P. aeruginosa MltF (PA3764), and its complexes with bulgecin, muropeptide, and various amino acids (cysteine, valine, leucine, and isoleucine) in its ABC-transporter domain were deposited by Thunnissen in 2015, but without accompanying publication (Madoori & Thunnissen, 2010). Complementary structures of active and inactive P. aeruginosa MltF (Figure 5) in complex with NAG-anhNAM-pentapeptide, NAM-pentapeptide, tetrasaccharide and tetrapeptide, and tetrapeptide gave insight into its allosteric regulation (Dominguez-Gil et al., 2016). The structure of the N-terminal SBP_bac_3 regulatory module of MltF, comprising of two subdomains connected by a flexible linker, resembles that of an ABC-transporter domain. Structural studies accompanied by mass-spectrometry analysis demonstrated that binding of a muropeptide (L-Ala-γ-D-Glu-L-Lys-D-Ala) to this regulatory module prompted a dramatic and long-distance (40 Å) conformational change (55 Å movement) across its RM-linker-CM domains, exposing the active site of the Family 1 SLT domain of MltF for catalysis. Analysis of the domain composition of other LTs suggests that this regulation may not be unique to Family 1E.
Family 1F
The one current member of Family 1F is the soluble lytic transglycosylase F (SltF; not a soluble form of MltF) of Rhodobacter sphaeroides (de la Mora et al., 2007). No homologous protein to SltF is found in E. coli, P. aeruginosa, N. gonorrhoeae, or S. maltophilia. The protein was discovered from a phenotypic screen in which an N-terminal deletion mutant of SltF lacked motility. Further studies revealed that SltF interacts with FlgJ to excavate the cell wall for the insertion of the flagellum protein edifice through the sacculus (de la Mora et al., 2007). Herlihey and Clarke assigned SltF to the new subfamily 1F of Family 1 based on sequence analysis (Herlihey & Clarke, 2016). The Pfam database recognizes the catalytic domain of SltF as a Family 1 SLT domain (Pfam: PF01464). However, the catalytic domain of SltF is seemingly larger than that of the other Family 1 LTs, possibly due to additional amino acids within disordered loop regions. As noted earlier, the glutamate in Motif I of R. sphaeroides SltF is flanked by a threonine rather than a serine. Homologous SltF proteins in other Gram-negative organisms retain the more common ES Motif I (Herlihey & Clarke, 2016). The E57A and E83A mutants of SltF lacked activity (de la Mora et al., 2007). Three other consensus motifs are moderately conserved and give further credence to the Family 1F subfamily assignment. Moreover, a homology model of SltF displayed the classic Family 1 LT catalytic fold. More recently, its catalytic activity was elucidated (confirming it as an endolytic LT) through MS analysis of the products released from the sacculus by a C-terminal SLT domain construct (Herlihey et al., 2016). Its LT activity is regulated by two proteins (FlgB and FlgF), which stabilize SltF and increase its catalytic activity.
Family 1G
The 1G subfamily of the LTs exemplifies the smallest (typically a mere 17 kDa) of the LT enzymes. Their single domain is identified as an SLT domain (Pfam: PF01464). Family 1G LTs include the EtgA enzyme of enteropathogenic E. coli (EPEC) and the LtgX enzyme of N. gonorrhoeae. LtgX is one of two LTs (with AtlA in Family 4) that is encoded on the gonococcal genetic island (GGI) acquired by horizontal gene transfer (Kohler et al., 2007). This 57 kb gonococcal chromosome is found in 80% of N. gonorrhoeae strains for the collective purpose of facilitating chromosomal DNA secretion through a type IV secretion system (Chan et al., 2012). Knockout of the LtgX activity severely decreased DNA secretion (Kohler et al., 2007). The locus for enterocyte effacement (LEE) plasmid of EPEC encodes EtgA, a LT that functions in partnership with 20 other proteins encoded by the LEE plasmid to assemble a Type 3 secretion system assembly (Kubori et al., 1998; Blocker et al., 2001; Mueller et al., 2005; Mueller et al., 2008; Garcia-Gomez et al., 2011). EPEC bacteria adhere to the epithelium of the small intestine, with effacement of the microvilli and formation of actin pedestals at the attachment site. Since the E. coli peptidoglycan creates a barrier for proteins exceeding a molecular weight of 50 kDa, EtgA seemingly excavates the cell wall for the insertion of the entire secretion system, to enable delivery of virulence factors from the bacterium to the host. The structure of the EtgA LT domain (D60A mutant, Figure 5) resembles the SLT domain of Family 1 LTs (Burkinshaw et al., 2015). EtgA localizes to the periplasm and forms a 1:1 complex with EscI, a protein involved in the assembly of the inner rod component of the type 3 secretion system (Garcia-Gomez et al., 2011; Burkinshaw et al., 2015). The catalytic activity of EtgA increases significantly upon complexation with EscI. This purpose of EscI may be to recruit EtgA to secretion system assembly, thus protecting the cell from nonspecific peptidoglycan degradation. The reaction products of the Family 1G LTs with respect to the sacculus have not been determined. LtgX and EtgA are homologous to geneX (encoded in the E. coli F-plasmid on orf169) and VirB (also known as VirB1). Both are presumptive Family 1G LTs (Llosa et al., 2000; Ward et al., 2002; Höppner et al., 2005; Zahrl et al., 2005; Zupan et al., 2007; Kohler et al., 2017).
Family 1H
GP144 of the pseudomonal bacteriophage ΦKZ is a Family 1 LT encoding an SLT domain (Pfam: PF01464), as supported by structural comparisons of GP144 and the Family 1 LTs (Fokine et al., 2008). Previously, GP144 was assigned as a Family 4 LT only on the basis of its bacteriophage origin (Domínguez-Gil et al., 2016). The distinction between Family 1 LTs and Family 4 LTs (such as E. coli bacteriophage lambda lysozyme) is evident from structure analysis of the catalytic domains. In addition to its C-terminal SLT domain, GP144 has an N-terminal PG_binding_1 domain (Pfam: PF01471) that is homologous to the peptidoglycan-binding domain of Family 3B LTs, which is discussed later. GP144 was discovered in 2007 and demonstrated as a LT (Paradis-Bleau et al., 2007). Its structure was solved shortly thereafter (Figure 5). The catalytic acid residue is E115 (Fokine et al., 2008). The catalytic groove of GP144 accommodates five saccharides, as reflected by successful complexation of chitotetraose as a substrate mimic. Interestingly, other evidence suggests that GP144 has two active sites and identifies both E115 and E178 as catalytic residues (Chertkov et al., 2017). GP144 interacts with anionic membranes and causes membrane disruption and eventual lysis, effects not seen in a zwitterionic membrane model (Cloutier et al., 2010). Furthermore, in solution the protein exists in a monomer, dimer, and trimer equilibrium (Miroshnikov et al., 2006). Interestingly, bacteriophage lysozymes that attack E. coli, N. gonorrhoeae, and S. maltophilia encode instead a Phage_lysozyme LT domain (Pfam: PF00959). How these catalytic domains function differentially, both mechanistically and physiologically, is unknown. At this time, GP144 is the sole member of Family 1H LTs.
Gram-negative Family 2 Lytic Transglycosylases
Overview
The LT Family 2 is less expansive than the LT Family 1. In the four Gram-negative species that are the focus of this analysis, a single Family 2 LT is present. This LT is named MltA in E. coli, P. aeruginosa, and S. maltophilia; and LtgC in N. gonorrhoeae. The sequence conservation of Family 2 LTs is less evident than that of the Family 1 LTs (Figure 6). Although the specific residues corresponding to the identification to each motif are less clearly defined compared to their Family 1 counterparts, Blackburn and Clarke identified six consensus motifs in the Family 2 LTs (Blackburn & Clarke, 2001). These motifs are Motif I (Residues Q–G–X8–G); Motif II (G); Motif III (no conserved residues); Motif IV (N–X5–F); Motif V (P–X5–A–X1–D); Motif VI (D–X1–G–X1–A–X6–D–X3–D–X3–G–X–G–X3–G–X2–A–G). MltA is a member of the glycoside hydrolase GH102 Family. LtgC of N. gonorrhoeae is differentiated from the other MltA enzymes by an amino-acid insertion (residues 172–205) of unknown function (Figure 7). This insertion may warrant the creation of a new Family 2 subfamily (family 2B). However, as the solitary example of this insertion among the LTs covered (and therefore not identifying with a domain in the Pfam database) and lacking a functional designation for this insertion, LtgC here is kept as a Family 2 LT.
Figure 6.
Multiple-sequence alignment displaying the LT domain of Gram-negative Family 2 LTs. The asterisk denotes the position of the catalytic residue. A color version of this figure is available at www.tandfonline.com/ibmg.
Figure 7.
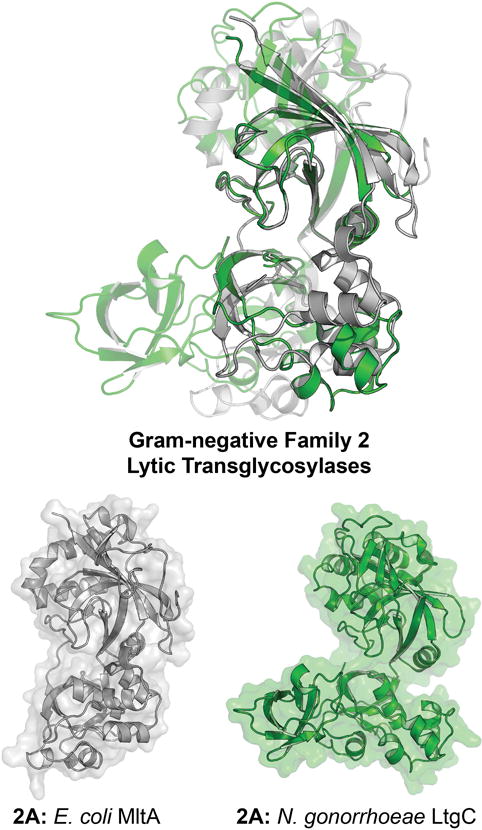
X-ray structure alignment of Gram-negative Family 2 LTs, displaying conservation of the 3D domain (Pfam: PF06725). The 3D domain is comprised of two subdomains linked by a hinge. The two structures show an open (E. coli MltA) and a closed (N. gonorrhoeae LtgC) protein conformation. The structure overlay aligns the top sub-domain. The bottom sub-domains are also conserved, but align differently in the open and closed states. The ribbon representation of each apo LT crystal structure is displayed to the right with a transparent surface representation. A color version of this figure is available at www.tandfonline.com/ibmg.
Family 2A
E. coli MltA (ECK2809) was identified and named Mlt38 (38 kDa in size, after signal peptide removal) in 1994 (Ursinus & Höltje, 1994). It was later renamed as MltA (Ehlert et al., 1995). MltA is a two-domain lipoprotein of the periplasm. MltA shares with the RlpA LT of Family 6 (discussed later) the absence of the quintessential catalytic glutamate (homologous to E64 of E. coli MltE). Its first domain is an N-terminal MltA domain (Pfam: PF03562, and conserved in all four MltAs) that assists in peptidoglycan binding (Figure 3). The catalytic 3D domain (Pfam: PF06725) at the C-terminus has two aspartates (E. coli numbering: D317 and D328) assigned as a catalytic pair (Powell et al., 2006). P. aeruginosa MltA (PA1222), N. gonorrhoeae LtgC (NGO2048, and previously named GNA33 as the genome-derived Neisseria antigen-33), and S.maltophilia MltA (Smlt0155) have respective sequence similarities of 31%, 28%, and 30% (and query coverage of 97%, 97%, and 93%), compared to MltA of E. coli.
The apo-crystal structure of E. coli MltA was solved in 2005 (Van Straaten et al., 2004; van Straaten et al., 2005) (Figure 7), and re-solved subsequently as a catalytically-inactivate mutant (labeled D308A; residue D328 of the full-length protein) in complex with chitohexaose as a substrate mimetic (van Straaten et al., 2007). The catalytic domain has a double-psi β-barrel fold. Unlike the lysozyme-like fold of the Family 1 LTs, the double-psi β-barrel fold of Family 2 LTs is similar to the catalytic domain of the barwin-like endoglucanase superfamily (van Straaten et al., 2005). Solution of the structure of N. gonorrhoeae LtgC allowed comparison to E. coli MltA (Figure 7) (Powell et al., 2006). While both catalytic and substrate-binding residues are conserved in the active site, the morphology of the domains differs. The cleft of the catalytic domain of MltA is much wider than the cleft seen in the N. gonorrhoeae LtgC. This difference defines the breadth of movement available to MltA during substrate binding (Powell et al., 2006). A shift in subdomain 2 relative to subdomain 1 opens the active site to allow the peptidoglycan to bind. However, the shift is so large (37° rotation) that domain closure once the peptidoglycan has bound may occur. This rotation results in a narrowing of the active-site cleft and a synchronous increase in cleft depth (as seen in MltA bound to chitohexaose) (van Straaten et al., 2007). Both E. coli MltA and N. gonorrhoeae LtgC engage in a protein-protein interaction with penicillin-binding proteins. E. coli MltA immobilized on a Sepharose column retained four PBPs: 1b, 1c, 2 and 3 as well as five different non-PBP proteins (von Rechenberg et al., 1996). Surface-plasmon resonance analysis of the MltA interaction with PBP1b and MipA (MltA-interacting protein) suggested a possible 1:2:1 stoichiometry, respectively, for the multiprotein complex (Vollmer et al., 1999). LtgC interacts with PBP2 of N. gonorrhoeae as well (Jennings et al., 2002). However, no interaction of LtgC with a MipA homologue was detected. Powell proposed that the difference in affinity for MipA between the two LTs may be due to an insertion (referred to as Domain 3 by Powell and colleagues) in LtgC. Based on the sequence conservation between PBP2 of N. gonorrhoeae and PBP3 of E. coli, the LtgC-PBP2 complex may localize to the septum of N. gonorrhoeae (the location for E. coli PBP3) (Park & Burman, 1973; Schmidt et al., 1981). This conclusion is supported by the inability of LtgC-knockout strains to complete separation into daughter cells during bacterial division (Adu-Bobie et al., 2004; Cloud & Dillard, 2004). However, this phenotype was not seen for the E. coli MltA-knockout strain (Lommatzsch et al., 1997). The interaction network for the MltA LTs of P. aeruginosa and S. maltophilia has not been studied.
Studies of MltA as a catalyst in E. coli are limited to a single analysis comparing the ability of seven LTs to digest the E. coli sacculus. E. coli MltA catalyzes, with near equal ability, the release of both cross-linked and non-crosslinked muropeptides from the E. coli sacculus as a substrate. This enzyme showed exolytic activity, and (in contrast to the other LTs) there was no evidence for the formation of hydrolytic products (Lee et al., 2013). This observation opens the possibility that the mechanism of MltA is different from the Family 1 LTs.
Gram-negative Family 3 Lytic Transglycosylases
Overview
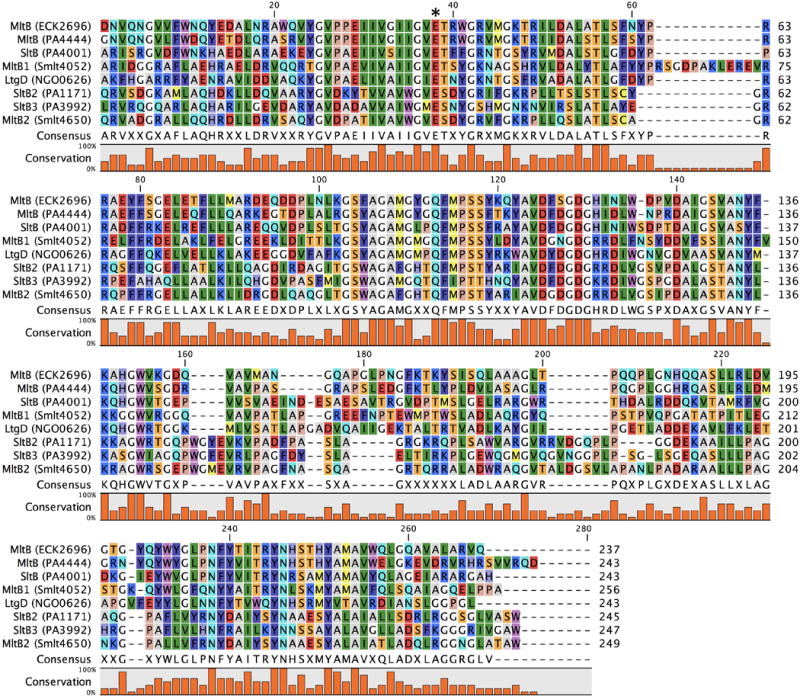
Family 3 LTs encompass both soluble and membrane-bound enzymes possessing a range of cellular functions. Although the Family 3 LTs share the same consensus motifs of the Family 1 LTs, numerous amino-acid insertions within the catalytic module (Figure 8) distinguish Family 3 structurally, and also likely functionally. The representative Family 3 LT is the MltB lipoprotein of E. coli, assigned by the CAZy database as a member of the glycoside hydrolase 103 (GH103) Family. This MltB undergoes proteolytic truncation in the periplasm to yield a soluble enzyme named Slt35. Individual point mutations of either R187 or R188 to an Ala significantly impaired the ability of peptidoglycan to bind to the catalytic site. R188 (10-fold decrease in catalytic activity of R188A) is suggested to hydrogen bond with the peptidoglycan lactyl carbonyl, and so complement the salt-bridge interaction between R187 (100-fold decrease in catalytic activity of R187A) and the substrate at the –1 subsite (Reid et al., 2006). Sequence alignments of MltB homologues in more than 40 organisms revealed that R188 is conserved in all examined enzymes, while R187 is less conserved (~70%). Reid et al. propose that the presence of R187 should distinguish two Family 3 subfamilies. Family 3A has arginine at position 187, whereas Family 3B LTs would not. All MltB homologues with the absent of R187 (~30%) are Gram-negative. It is suggested that these enzymes are predatory enzymes, functioning akin to a bacteriophage endolysin (Kadurugamuwa & Beveridge, 1996; Kadurugamuwa & Beveridge, 1997; Kadurugamuwa et al., 1998; Li et al., 1998; Kadurugamuwa & Beveridge, 1999). However, division of Family 3 LTs into two subfamilies based on a few residues is inconsistent with the nomenclature used to distinguish the subfamilies of the LT Family 1. Family 1 LTs have a conserved core catalytic domain and are divided into subfamilies on the basis of their peripheral domains (Figure 3). Applying this same criterion to Family 3 keeps all of the MltBs (and SltB1 of P. aeruginosa) in one sub-family, and would separate the two P. aeruginosa LTs, SltB2 and SltB3 (and the S. maltophilia MltB2), into the sub-family 3B. These latter three LTs have a C-terminal putative peptidoglycan-binding domain (annotated as PG_binding_1, Pfam: PF01471), whereas E. coli MltB does not (Figure 9). Our preference is this sub-family division. In this resulting sub-family 3B, only P. aeruginosa SltB1 lacks R187 (R187P), therefore the mechanistic implications of this absence will not be representative of its own subfamily. The recent discovery of Family 3 LTs in other bacteria, including plant bacteria, may at a later time allow for expansion of Family 3 (Guglielmetti et al., 2014; Neudorf & Yost, 2017). The consensus motifs of Family 3 LTs were defined by Blackburn and Clarke as Motif I (Residues V–X11–E–S, 3A as defined herein and V–X11–E–T, 3B as defined herein); Motif II (L); Motif III (G–S– X1–A–X1–A–X1–G–X3–F); Motif IV (EF-hand) (D); Motif V (S–X2–N–X5–G–W). Structural differences, resulting from amino-acid insertions in the Family 3 primary structure as compared to Family 1, are believed to have mechanistic consequences (possibly invoking an oxazolinium intermediate; described later), notwithstanding the shared substrate-binding and catalytic residues between the two families.
Figure 8.
Multiple-sequence alignment displaying the LT domain of Gram-negative Family 3 LTs. The asterisk denotes the position of the catalytic residue. A color version of this figure is available at www.tandfonline.com/ibmg.
Figure 9.
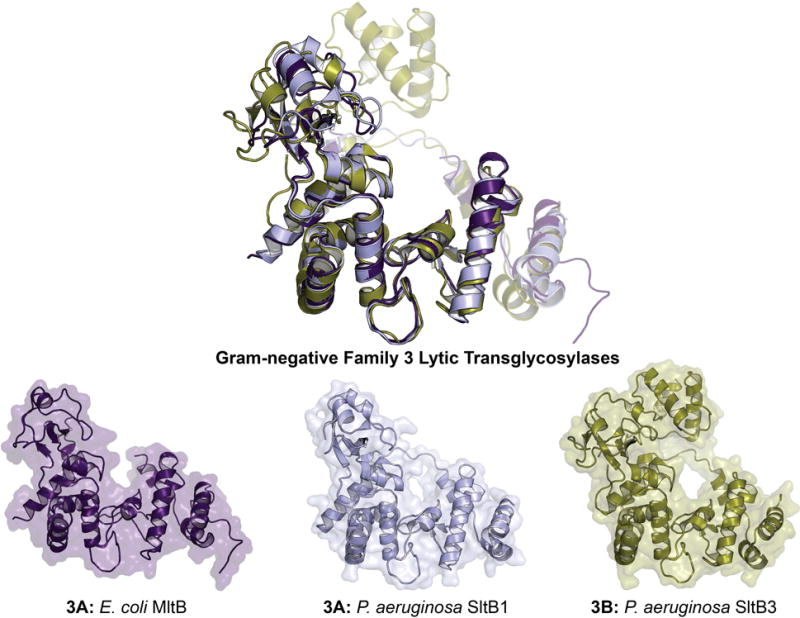
X-ray structure alignment of Gram-negative Family 3 LTs, displaying the conservation of the Slt_2 domain (Pfam: PF13406). The ribbon representation of each apo LT crystal structure is displayed below with a transparent surface representation. A color version of this figure is available at www.tandfonline.com/ibmg.
Family 3A
MltB of E. coli (ECK2696) is an S-lipidated (LAACSS lipobox sequence, anchored at Cys19 to the inner leaflet of the outer membrane) lipoprotein of the periplasm (Ehlert et al., 1995). Its proteolytically-processed form (releasing a single-domain soluble enzyme of 35 kDa, named Slt35) was identified prior to the lipoprotein form (Engel et al., 1992). The site of proteolytic cleavage is believed to be between residues F39 and L40 of MltB (Dijkstra et al., 1995). The single domain of MltB is a SLT_2 (Pfam: PF13406) domain containing a catalytic glutamate at position 162. E162 is homologous to the catalytic glutamate of Family 1 LTs (E64 of MltE) and likewise demarcates the –1 and +1 saccharide-binding subsites (Reid et al., 2006). The structure of Slt35 was first reported in 1998 (shown in Figure 9 as MltB) (van Asselt et al., 1998). Although prediction tools recognize Slt35 as single-domain LT, van Asselt et al. subdivide the structure into three; an α-domain, a core (LT) domain, and a β-domain (van Asselt et al., 1999a). The core domain sits between the α and β domains, includes the substrate-binding site, and resembles the lysozyme-like fold of the goose egg-white lysozyme. At the C-terminal end of the core domain is an EF-hand motif that binds a calcium ion. The calcium ion is presumed to have a structural, rather than regulatory or catalytic role. E. coli MltB has broad specificity with respect to digestion of the E. coli sacculus (Lee et al., 2013), accommodating both crosslinked and non-crosslinked peptidoglycan substrates. Although MltB is primarily exolytic, it processes oligomeric substrates at a similar rate to MltD of E. coli. Catalysis of turnover of peptidoglycan by MltB is disciplined, as the hydrolytic reaction product is only rarely formed. An early study of the binding partners of E. coli MltB revealed protein-protein interactions with PBP 1b, 1c, and 3 (von Rechenberg et al., 1996). The other Family 3A LTs include P. aeruginosa MltB (PA4444) and SltB1 (PA4001), N. gonorrhoeae LtgD (NGO0626), and S. maltophilia MltB1 (Smlt4052). These four have, respectively, 68%, 47%, 35%, and 41% sequence identity (and, respectively, 89%, 77%, 90%, and 74% query coverage) to MltB of E. coli. The structure of SltB1 was solved by Nikolaidis et al. in 2012 and showed high similarity to that of E. coli MltB (Figure 9).
Blackburn and Clarke reported that P. aeruginosa SltB1 is more catalytically efficient than its membrane-anchored partner MltB, notwithstanding the findings that both proteins produce identical reaction products (Blackburn & Clarke, 2002). SPR analysis of P. aeruginosa SltB1 demonstrated that it binds to PBP2 (also of P. aeruginosa) with 1:1 stoichiometry (Nikolaidis et al., 2012). Complex formation required calcium. Cloud-Hansen and colleagues studied the N. gonorrhoeae lipoprotein LtgD, the homologue of MltB. The enzyme functions as an exo-muramidase, similar to MltB (Cloud-Hansen et al., 2008). LtgD produces identical reaction products to N. gonorrhoeae LtgA (the Slt70 homologue, previously described), though it is less active. Interestingly, the products from each of the two LT have distinct fates: the reaction products of LtgA are recycled into the cytoplasm, while the reaction products of LtgD are released from the bacterium for the purpose of virulence via the inflammatory response initiated by their binding to NOD1 (Mavrogiorgos et al., 2014; Schaub et al., 2016). LtgA and LtgD mediate bacterial survival in neutrophils by reinforcing the N. gonorrhoeae cell-wall envelope so as to improve its resistance to lysozyme degradation, independent of their roles in peptidoglycan monomer release (Ragland et al., 2017).
Family 3B
Blackburn and Clarke discovered the enzymes SltB2 (PA1171) and SltB3 (PA3992) in P. aeruginosa in 2002, marking a second subfamily of the Family 3 LTs (Blackburn & Clarke, 2002). Although Family 3B lacks a counterpart protein in either E. coli or N. gonorrhoeae, sequence analysis reveals a Family 3B representative in S. maltophilia (MltB2, Smlt4650) (Wu et al., 2016). Family 3B LTs have the following sequence identity (query coverage) to E. coli MltB: P. aeruginosa SltB2 33% (75%), P. aeruginosa SltB3 34% (58%), and S. maltophilia MltB2 33% (66%). The catalytic glutamate was assigned to residue 172 of SltB3 based on structural alignments. The catalytic activity of P. aeruginosa SltB3 was similar to that of E. coli MltA, showing exolytic digestion of the sacculus without formation of endolytic products (Lee et al., 2016b). The crystal structures of apo SltB3 (Figure 9), and SltB3 in complex with NAG-anhNAM-pentapeptide and NAG-NAM-pentapeptide were reported as well (Lee et al., 2016b). The catalytic domain of SltB3 displays folds that are nearly identical to E. coli MltB. The C-terminal domain of SltB3 is a PG_binding_1 domain (previously described). The peptidoglycan-binding domain of Family 3B LTs are homologous to the peptidoglycan-binding domain of the peptidoglycan amidases, including P. aeruginosa AmpDh2 and AmpDh3, among others (Lee et al., 2013; Martínez-Caballero et al., 2013). However, no functional regulation has been attributed to this domain in Family 3B LTs and the cellular role(s) of these LTs remains unknown.
Gram-negative Family 4 Lytic Transglycosylases
Overview
Family 4 LTs are commonly the LT enzymes of the bacteriophages. There are, as yet, no chromosomally encoded Family 4 LT enzymes. These LTs are presumed to function in the assembly of a secretion system to facilitate host-cell pathogenicity (Walmagh et al., 2013). Bacteriophages encoding LTs with this domain that infect E. coli, S. maltophilia, and N. gonorrhoeae are known. The Family 4 LTs are single-domain enzymes annotated as a Phage_Lysozyme (Pfam: PF00959) catalytic domain. The four consensus motifs of this domain, as identified by Blackburn and Clarke, are Motif I (Residues A–X7–S–E); Motif II (Y–X4–G–X5–D–X–S–X–HP); Motif III (S–T– X4–G–R–Y–Q–X5–W); and Motif IV (W–X–S) (Blackburn & Clarke, 2001). The catalytic glutamate in Motif I has a preceding serine, in contrast to Family 1 LTs, where the serine follows the catalytic glutamate (Figure 10). All known Family 4 LTs are in a single subfamily, termed Family 4A.
Figure 10.
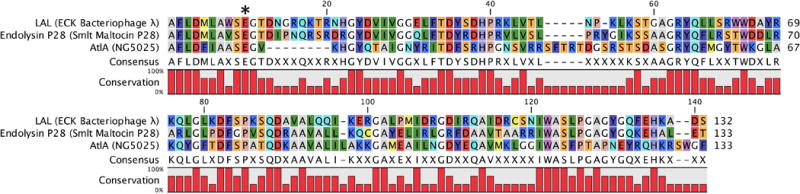
Multiple-sequence alignment displaying the LT domain of Gram-negative Family 4 LTs. The asterisk denotes the position of the catalytic residue. A color version of this figure is available at www.tandfonline.com/ibmg.
Family 4A
The LT of bacteriophage endolysin λ (“λ-lysozyme” or “LAL”) was one of the earliest LTs discovered (Taylor et al., 1975). The structure of apo λ-lysozyme (Evrard et al., 1998) and its complex with chitohexasaccharide as a substrate mimic (Leung et al., 2001) are shown in Figure 11. Family 4 LTs have different structural folds from the other LT families. A search for homologous proteins to the bacteriophage endolysin λ in N. gonorrhoeae revealed a second Family 4 LT (38% sequence identity, 84% query coverage) encoded by the GGI of N. gonorrhoeae. This LT (AtlA, a λ-lysozyme homologue) was first identified as an autolysin and named autolysin A (Dillard & Seifert, 1997). AtlA is involved in type IV secretion system assembly and hence has a similar function to the GGI LT LtgX. Mutation of the catalytic E48 of AtlA significantly reduced type IV secretion system dependent DNA secretion (Kohler et al., 2007). In this respect, it is similar to endolysin λ of E. coli. The gonococci expressing AtlA, however, do not experience cell lysis. A bacteriophage (maltocin P28) isolated from S. maltophilia encodes among its 23 ORFs a Family 4 LT (Endolysin P28) in its ORF8 having 57% sequence identity, 84% query coverage, compared to endolysin λ of E. coli. Endolysin P28 degrades peptidoglycan with sufficient catalytic competency as to show bactericidal activity against many Gram-negative bacteria (Dong et al., 2015).
Figure 11.
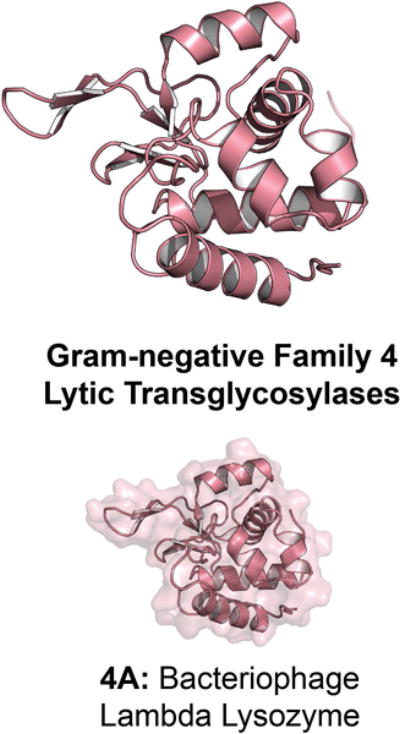
X-ray structure of the Gram-negative Family 4 LT bacteriophage endolysin λ; the Phage_Lysozyme domain (Pfam: PF00959) comprises most of the solved structure. The ribbon representation of the apo LT crystal structure is displayed with a transparent surface representation. A color version of this figure is available at www.tandfonline.com/ibmg.
Gram-negative Family 5 Lytic Transglycosylases
Overview
The recently discovered Family 5 LTs are the first examples of an LT family found in both Gram-negative and Gram-positive bacteria. They are commonly called MltG. The presumptive function of MltG is glycan-chain termination during the biosynthesis of the cell-wall peptidoglycan (Yunck et al., 2016). It has long been believed that LTs function in the chain termination of nascent peptidoglycan, given the relative scarcity of reducing saccharide termini in muropeptides obtained from amidase-catalyzed degradation of sacculi. The seven consensus motifs of the Family 5 LTs are Motif I (residues K–X7–G–T–Y); Motif II (L–X4–G–K–E–X–Q–X6–E–G); Motif III (E–G–X3–P–X–T–X2–Y–X5–D–X3–L–X); Motif IV (A–S–I–X–E–K–E); Motif V (E–R–X2–V–X–S–V–F–X2–N–R–L–X3–M–X–L–Q–T–D–V–I–Y–G–X–g); Motif VI (G–X5–D–L–X5–Y–N–T–Y–X–I–X–G–L–P–P); and Motif VII (L–X–A–X–A–X–P–X2–T–X3–Y–F–V–A–D–G–X3–G–G–H–X–F–X–L–X2–H–N) (Herlihey & Clarke, 2016). The catalytic glutamate in Motif IV is preceded by a conserved EK pair and is followed commonly by a threonine residue (Figure 12), similar to Family 3 LTs. All known Family 5 LTs form the single Family 5A subfamily.
Figure 12.
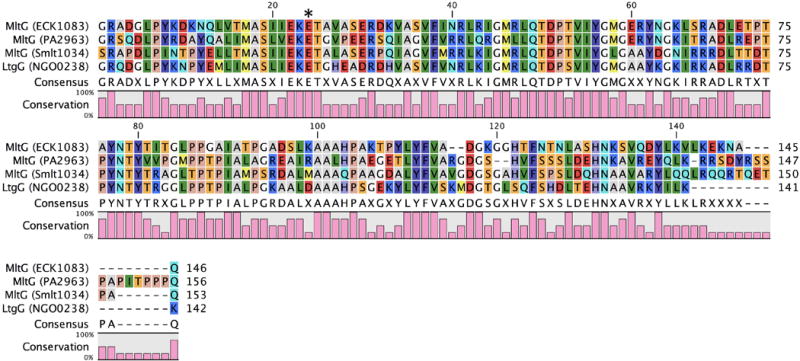
Multiple-sequence alignment displaying the LT domain of Gram-negative Family 5 LTs. The asterisk denotes the position of the catalytic residue. A color version of this figure is available at www.tandfonline.com/ibmg.
Family 5A
The crystal structure of MltG was determined prior to its recognition as a LT (Figure 13). A PDB-deposited structure of MltG was solved in 2007 by Patskovsky et al. and annotated as an aminodeoxychorismate lyase based on the protein sequence. MltG is 38 kDa in size, has E218 as its catalytic acid, and is located in the outer leaflet of the inner membrane (Yunck et al., 2016). The catalytic domain of E. coli MltG is structurally alike to the catalytic domain of the Gram-positive LT SleB (Figure 13). However, Pfam database annotates its catalytic domain as a YceG (Pfam: PF02618) domain, while the domain of SleB is annotated as a PG_Hydrolase_2 (Pfam: PF07486) domain. Further elucidation of the reaction products is required to understand whether or not the two domains are mechanistically identical. Yunck et al. first reported E. coli MltG as an LT when a multi-copy MltG-encoding (gene yceG) plasmid exhibited a lethal phenotype in a PBP1b-knockout strain (Yunck et al., 2016). MltG is the first LT from E. coli localized in the inner membrane. In this location, MltG is suitably located to act in peptidoglycan chain termination. Strong circumstantial evidence that MltG regulates glycan strand length was obtained from an MltG-knockout strain where the average length of the glycan strands (obtained upon sacculus degradation through amidase cleavage of the peptide crosslinks of this polymer) were longer than for the wild-type strain. Although the specific reaction products of E. coli MltG have not been elucidated, the enzyme has been shown to be endolytic. E. coli MltG interacts with PBP1b, lending credence to the initial finding that PBP1b was required for the non-lethal phenotype. MltG is a widely conserved protein. Homologues to E. coli MltG are identified by sequence analysis in P. aeruginosa (38% sequence identity, 97% query coverage), S. maltophilia (46% sequence identity, 77% query coverage), and N. gonorrhoeae (42% sequence identity, 83% query coverage). The N. gonnorhoeae homologue may be named as LtgG (rather than as an MltG) for consistency with the previous LT nomenclature developed for the N. gonorrhoeae LTs by the Dillard group. An MltG homologue was recently identified in the Gram-positive bacterium Streptococcus pneumoniae from sequence analysis (Tsui et al., 2016). The assignment of this protein as an LT awaits experimental verification.
Figure 13.
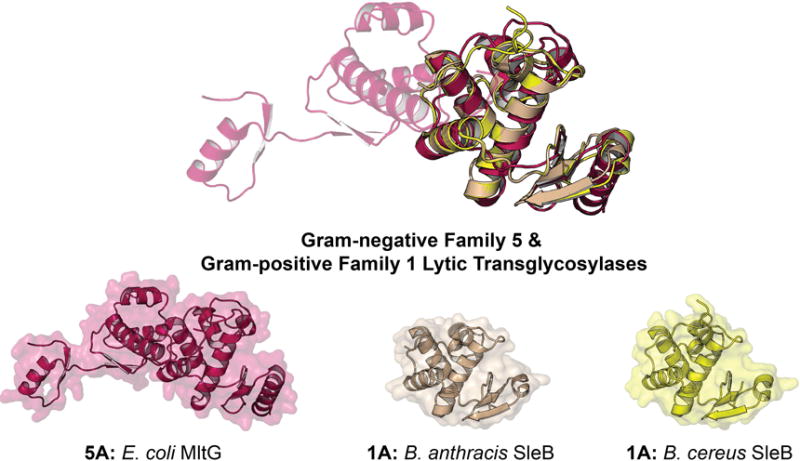
X-ray structure alignment of Gram-negative Family 5 and Gram-positive Family 1 LTs, displaying the similar folds of the YceG domain (Pfam: PF02618) of Gram-negative Family 5 and the PG_Hydrolase_2 domain (Pfam: PF07486) of Gram-positive Family 1. Notably, the amino-acid sequence between the two domains is not similar. The ribbon representation of each apo LT crystal structure is displayed with a transparent surface representation. A color version of this figure is available at www.tandfonline.com/ibmg.
Family 6 Lytic Transglycosylases
Overview
Family 6 LTs have three conserved aspartates, including two believed to be involved in catalysis, akin to MltA of the Family 2 LTs (previously described). The four consensus motifs of the Family 6 LTs are Motif I (residues T–X2–G–E–X2–D); Motif II (A–A–H–X–T–L–P–X–P–S–X4–T–N–X2–N–G); Motif III (R–X–N–D–R–G–P); and Motif IV (R–X–I–X–L–S–X–A–X–A–X2–L) (Herlihey & Clarke, 2016). Although a mutant of the second aspartate (D157A) abolished catalytic activity, sequence alignment to the Family 2 LTs suggests that the third aspartate (D168) may be the primary catalytic residue in P. aeruginosa (Jorgenson et al., 2014). Notably, in E. coli the third aspartate is a serine (S147) (Figure 14). Family 6 LTs have a catalytic DPBB_1 domain (Pfam: PF03330) and a structure of a domain with this designation is solved (PDB code: 4AVR) for a P. aeruginosa protein (locus tag PA4485 in strain PAO1) (Moynie et al., 2013). However, a Family 6 LT structure has not yet been solved, and the catalytic activity of PA4485 is yet unproven. The DPBB_1 domain of PA4485 shows distant similarity to cellulose-binding domains, and to the 3D domain of MltA described earlier. The PA4485 protein is only 13 kDa in size. Although it lacks the SPOR domain of Family 6A proteins, should future studies confirm it as a Family 6 LT, it would be assigned to a new subfamily (Family 6B).
Figure 14.
Multiple-sequence alignment displaying the LT domain of Gram-negative Family 6 LTs. The asterisk denotes the position of the catalytic residue. A color version of this figure is available at www.tandfonline.com/ibmg.
Family 6A
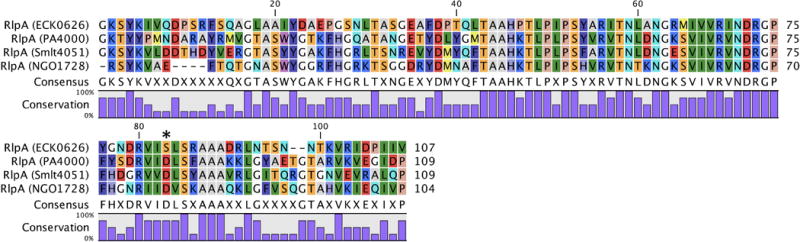
A highly-specialized LT, the so-called rare-lipoprotein A (RlpA; PA4000) from P. aeruginosa, is a 37 kDa protein involved in cell division. P. aeruginosa RlpA is an outer-membrane lipoprotein (lipobox motif: LSSCSS) comprised of two domains, a catalytic N-terminal DPBB_1 (Pfam: PF03330) domain and a C-terminal Sporulation-related (SPOR) domain (Pfam: PF05036). RlpA localization experiments showed that the protein localizes to the septal ring during cell division (Gerding et al., 2009; Arends et al., 2010). This finding is expected as all known proteins possessing SPOR domains are either involved in cell division or morphogenesis. SPOR domains are widely recognized as peptidoglycan-binding domains (Mishima et al., 2005; Gode-Potratz et al., 2011; Yahashiri et al., 2015, Yahashiri et al., 2017). In contrast to the three other cell-wall proteins encoding a SPOR domain in E. coli, RlpA is the only one that is non-critical (Gerding et al., 2009; Arends et al., 2010). Interestingly, E. coli rlpA shares an operon with the peptidoglycan-synthesizing proteins PBPa and RodA (Matsuzawa et al., 1989; Mohammadi et al., 2011; Banzhaf et al., 2012). Neighboring this operon is the gene dacA, which encodes a peptidoglycan hydrolase (PBP5) also implicated in cell division (Potluri et al., 2012). P. aeruginosa RlpA is a bona fide LT with preference for glycan strands lacking peptide stems, sometimes referred to as “naked glycan” (Jorgenson et al., 2014). Comparison of E. coli RlpA to P. aeruginosa RlpA presents a dilemma. Although the two proteins have sequence similarity (45% sequence identity, 36% query coverage), LT activity has not been shown for the E. coli RlpA (Jorgenson et al., 2014). A possible explanation is the presence of a serine (S147) in E. coli RlpA in contrast to an aspartate (D168)—the presumptive catalytic aspartate—at the equivalent position in P. aeruginosa RlpA. Careful assay will be required to show whether this difference abolishes catalytic activity.
P. aeruginosa RlpA-knockout strains contain longer naked glycan strands than the wild-type strain (Jorgenson et al., 2014). This finding suggests that RlpA activity is linked to periplasmic activity of the P. aeruginosa AmpDh2 and AmpDh3 amidases. RlpA-knockout strains displayed no phenotype when grown in lysogeny broth (LB). However, in LB without NaCl the knockout failed to produce single colonies when plated. The knockout grew slowly and formed long chains of cells, each with a visible septum, but with incomplete cell division. In addition, these cells were 50% shorter in length and were 20% wider. Replacing NaCl with proline (or sucrose) in the media recovered the wild-type phenotype, suggesting that the long-chain phenotype of the knockout was osmotic-stress dependent. However, induction of RlpA is not linked to osmotic-stress response. Sequence analysis reveals RlpA homologues in both N. gonorrhoeae (39% sequence identity, 46% query coverage) and S. maltophilia (45% sequence identity, 50% query coverage to E. coli RlpA). The low query coverage may be the result of comparably low sequence identity of the SPOR domains among the species. More experiments will be required to understand the role of Family 6A LTs in their respective organisms.
The Gram-positive LTs
The cell wall of Gram-positive bacteria is very different than its Gram-negative counterpart. Although the Gram-positive and Gram-negative peptidoglycan have very similar gross structural chemistries, the Gram-positive cell wall is a thicker, multi-layered exoskeleton. In addition, other polymeric structures (notably those of the lipoteichoic acids, the wall teichoic acids, and the capsular polysaccharide) are covalently linked to the Gram-positive cell wall. Although remodeling of the cell wall of Gram-positive organisms is documented, only recently has the contribution of muropeptide recycling been assessed (Reith & Mayer, 2011). The Gram-positive B. subtilis has a chromosomally encoded six-gene cluster implicated in cell-wall recycling (Litzinger et al., 2010). However, none of the proteins encoded by these genes is an LT. The LT activities of the Gram-positive bacteria appear to be limited to glycan-strand sizing during peptidoglycan biosynthesis (MltG activity) (Tsui et al., 2016) and for the Gram-positive spore-forming bacteria, the separate processes of spore formation and of germination. Spore-forming bacteria (including the Bacillus and Clostridium species) undergo sporulation in the face of nutrient challenge (Moir & Cooper, 2015). The bacterial spore preserves the genetic identity of the bacterium in the face of extreme temperatures, dehydration, environmental chemicals, and radiation (Setlow, 2006). Bacterial spores are the ultimate survival mechanism. They retain a highly transmissible capacity even after a dormancy period of years. When a favorable environment for growth is encountered, the spores transform by germination (or desporulation) for the restoration of their bacterial state (Setlow, 2014). Spores are a challenge in the food industry as they are primary agents for spoilage and the spread of disease. Equally important is the threat of bioterrorism through release of the spores of a pathogenic Gram-positive bacterium, such as Bacillus anthracis. The transition from the spore state of dormancy to the bacterium (and the reverse process) involves extensive remodeling of the cell-wall peptidoglycan. The unique structure of the peptidoglycan in the spore is named the peptidoglycan cortex, wherein a portion of the NAM saccharides of the (NAG-NAM)n glycan chains are converted to the muramic-δ-lactam structure and are primarily linked by L-Ala (in contrast to the oligopeptide crosslinking of the Gram-positive bacterium) (Atrih & Foster, 1999; Setlow, 2003; Bernhards et al., 2015). As such, the peptidoglycan cortex contains significantly fewer cross-linked peptides relative to the bacterial cell-wall peptidoglycan (Popham, 2002). During germination the peptidoglycan cortex of the spore degrades (Moir, 2006).
Gram-positive Family 1 Lytic Transglycosylases
Overview
The initiation of cell-wall degradation during peptidoglycan outgrowth of Bacillus species requires the two enzymes, SleB and CwlJ (Paidhungat et al., 2001; Chirakkal et al., 2002; Hu et al., 2007; Giebel et al., 2009; Heffron et al., 2009; Setlow et al., 2009; Heffron et al., 2010). SleB is a LT. The specific activity of CwlJ is unknown. Both enzymes cleave muramic-δ-lactam-containing peptidoglycan, and are therefore peptidoglycan cortex-specific. A spore containing a single knockout of either protein will germinate, although at a much slower rate (Ishikawa et al., 1998; Moriyama et al., 1999; Boland et al., 2000; Giebel et al., 2009; Heffron et al., 2009). Spores with knockouts of both SleB and CwlJ exhibit poor germination and were less viable. The SleB enzyme was the first LT identified in Gram-positive bacteria, and its structure defines a new Family of bacterial LTs (Figure 15). We assign SleB as a Gram-positive Family 1 LT. The LT domains of Gram-positive Family 1 LTs are structurally similar to the LT domain of Gram-negative Family 5 LTs (as previously described; Figure 13). Sequence analysis identifies a homologue to the Bacilli SleB in C. difficile (Sebaihia et al., 2006; Burns et al., 2010). This homologue shares a C-terminal Hydrolase_2 domain, but lacks the N-terminal PG_binding_1 domain of SleB. Importantly, recent studies suggest that the molecular events of Bacillus germination may be very different from those of C. difficile, and therefore these homologous proteins may not share identical functions (Paredes-Sabja et al., 2008; Paredes-Sabja et al., 2009). This conclusion is consistent with the discovery that C. difficile sleB gene knockout mutants exhibited no ill effects, where instead SleC (discussed below) has assumed the functional attribution of both SleB and CwlJ (Kumazawa et al., 2007). Regardless, if the sleB homologue in C. difficile is shown to have LT activity, it should be assigned to the Gram-positive Family 1 LTs. The sequence of the Hydrolase_2 domain from this protein is included in Figure 16 as SleB (C. difficile) for comparative purpose. Catalysis by Gram-positive Family 1 LTs uses a conserved catalytic Glu, flanked by a Ser in the Bacillus species and by an Ala in C. difficile.
Figure 15.
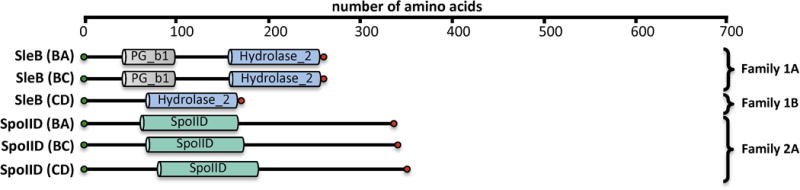
Domain architecture of Gram-positive LTs. Domains were assigned based on consensus analysis of Pfam Database and NCBI conserved-domain database. Protein names and those of the organisms are given for B. anthracis (BA), B. cereus (BC) and C. difficile (CD). Locus tags are not given. Notably, it is not known if the SleB homologue of C. difficile, which lacks the N-terminal PG_binding_1 (PG_b1) domain, is an LT, therefore it is not included. A color version of this figure is available at www.tandfonline.com/ibmg.
Figure 16.
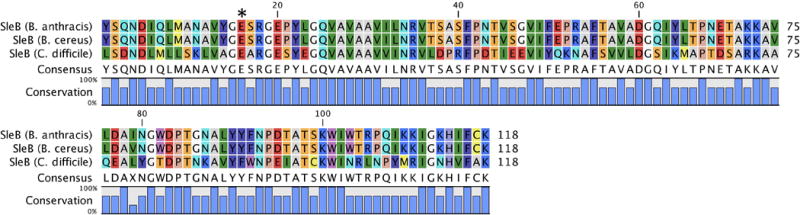
Multiple-sequence alignment displaying the LT domain of Gram-positive Family 1 LTs. The sequence of a C. difficile SleB homologue is included for comparison, although it is not known whether this protein is an LT. The asterisk denotes the position of the catalytic residue. A color version of this figure is available at www.tandfonline.com/ibmg.
Family 1A
The founding member of the Gram-positive LTs and herein assigned to Family 1A is SleB of B. cereus. The gene for this enzyme encodes a 28 kDa protein having an N-terminal signal peptide for initial delivery to the peptidoglycan-membrane interstitial space (Moriyama et al., 1996). SleB remains inactive until the spore begins germination (Boland et al., 2000; Heffron et al., 2009). The specific localization of SleB within the spore has been debated (Moriyama et al., 1999; Atrih & Foster, 2001; Chirakkal et al., 2002). As previously described, SleB has an N-terminal PG_binding_1 domain (Pfam: PF01471) and a C-terminal Hydrolase_2 domain (Pfam: PF07486). The peptidoglycan-binding domain increases the protein’s affinity for peptidoglycan and the rate of peptidoglycan cleavage. However, it is not necessary for catalysis (Heffron et al., 2009). A mutation study performed with the Bacillus megaterium SleB demonstrated that germination and outgrowth proceeded correctly in the absence of the C-terminal hydrolase domain. Interestingly, mutants encoding only the N-terminal peptidoglycan-binding domain were more efficient at facilitating outgrowth and germination than mutants encoding only the C-terminal hydrolase domain. The authors infer that both domains have LT activity, an intriguing observation considering the prevalence of the PG_binding_1 domain in the LT superfamily (Christie et al., 2010). This phenomenon has also been reported in the protein SleC from Clostridium perfringens. SleC only encodes a single C-terminal PG_binding_1 domain, yet recent reports demonstrate by MALDI mass spectrometry analysis of sacculus digestion products that SleC functions as an exolytic LT (Gutelius et al., 2014). Notably, this activity has not been assigned to this same PG_binding_1 domain of the Gram-negative Family 1H or 3B LTs. The SleB gene-knockout strain failed to produce in the germination exudate the anhydromuropeptide products that are characteristic of the LT reaction (Boland et al., 2000; Atrih & Foster, 2001; Heffron et al., 2009; Christie et al., 2010). Furthermore, LT activity of purified recombinant SleB was demonstrated in vitro (Heffron et al., 2011; Li et al., 2013). The structure of the C-terminal domain of SleB reported in 2012, revealed the position of the catalytic glutamate (E157 of B. cereus) in the active site (Li et al., 2012). The topological arrangement of this LT is different from that of any previous LT discovered at that time (Li et al., 2013). However, recent identification of the Gram-negative Family 5 LTs reveals a near identical catalytic domain to that of the MltG protein from E. coli (Figure 13). Conservation of this secondary structure is also seen in the structure of the B. anthracis SleB (Jing et al., 2012). In this structure, a metal-binding site at the entrance of the substrate-binding pocket was discerned. The identity of the metal was not identified, but is believed to be either a Ca2+ or Na+ ion. SleB from B. anthracis and B. cereus are nearly identical (97% sequence identity, 100% query coverage). SleB is believed to localize with the protein YbeP. Although protein-protein interactions were not seen in vitro, YbeP-mediated inhibition of SleB has been shown (Li et al., 2013). The intricacies of YbeP regulation of SleB are not well understood.
Gram-positive Family 2 Lytic Transglycosylases
Overview
Gram-positive Family 2 LTs have also found a niche among the spore-forming Gram-positive organisms. Particularly, Gram-positive Family 2 LTs are involved in sporulation (spore formation), in contrast to the Gram-positive Family 1 LTs, which are involved in germination. Sporulation occurs following the formation of a septum near a pole of the cell, dividing the cell into one large and one small cell structures. The smaller structure—the forespore—ultimately becomes the spore. The larger compartment (named the mother cell) is doomed to lysis. The forespore engulfs the desired components of the cell and pinches closed, thereby releasing itself from the mother cell. Processing by the mother cell coats the forespore with protective proteins, whereby it can be exported as a mature spore. Formation of the spore requires large-scale remodeling of the cellular membrane and sacculus. Family 2 LTs (in collaboration with other cell-wall remodeling enzymes) participate in this process (Gutierrez et al., 2010). Gram-positive Family 2 LTs each encode a single SpoIID catalytic domain (Figure 15) that has a unique structural topography (Figure 18) in comparison to the previous LT structures that we have discussed (Nocadello et al., 2016). They possess a catalytic glutamate (E87 of B. anthracis) flanked by a conserved methionine (Figure 17).
Figure 18.
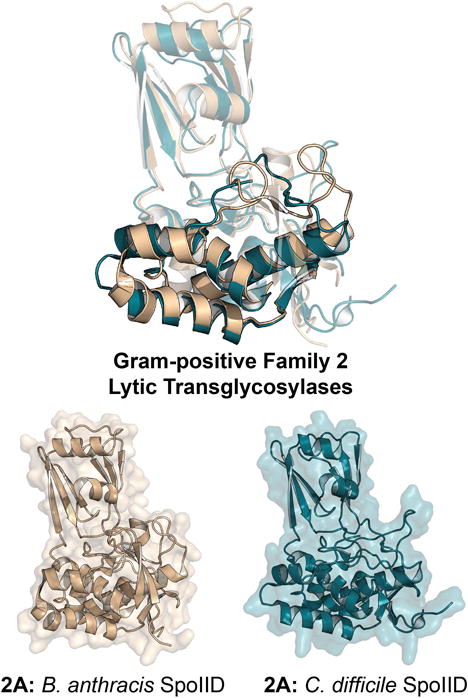
X-ray structure alignment of Gram-positive Family 2 LTs, displaying the conservation of the SpoIID domain (Pfam: PF08486). The ribbon representation of each apo LT crystal structure is displayed below with a transparent surface representation. A color version of this figure is available at www.tandfonline.com/ibmg.
Figure 17.
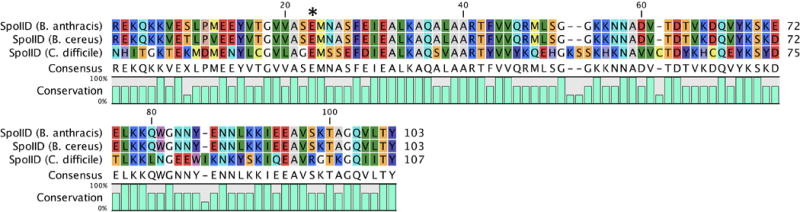
Multiple-sequence alignment displaying the LT domain of Gram-positive Family 2 LTs. The asterisk denotes the position of the catalytic residue. A color version of this figure is available at www.tandfonline.com/ibmg.
Family 2A
It was widely assumed that Gram-positive bacteria did not encode a LT until the discovery of SleB (see Gram-positive Family 1 LTs) in 2012. A second LT, Stage II sporulation protein D (SpoIID), was discovered subsequently in Gram-positive spore-forming bacteria. SpoIID has a single catalytic SpoIID domain (Pfam: PF08486). SpoIID forms macromolecular complexes with a series of related proteins, similar to Gram-negative Family 1C and 1G, among others. SpoIID interacts with both SpoIIM and SpoIIP to breakdown peptidoglycan and to allow for spore engulfment (Abanes-De Mello et al., 2002; Gutierrez et al., 2010). In this process, SpoIIM serves as a scaffold protein that congregates both SpoIID and SpoIIP (Chastanet & Losick, 2007; Gutierrez et al., 2010; Morlot et al., 2010). SpoIIP is a bifunctional enzyme that performs both amidase and endopeptidase activities. Subsequently, SpoIID cleaves the glycosidic bond of the resulting peptide-free glycan strands, forming the 1,6-anhydroMurNAc LT reaction products (Gutierrez et al., 2010; Morlot et al., 2010; Nocadello et al., 2016). SpoIIP and SpoIID are single-pass transmembrane proteins. In C. difficile, the catalytic residue E101 has been verified in a single-mutant study resulting in a complete loss of enzyme activity. This residue is located in the signature position for the catalytic glutamate of Gram-negative Family 1 LTs between the –1 and +1 subsites. The apo structure of C. difficile and B. anthracis SpoIID were reported in 2016 by Nocadello et al. Crystals of the C. difficile SpoIID only formed in complex with triacetylchitotriose (Nocadello et al., 2016). The two homologous proteins (43% sequence identity, 79% query coverage between the B. anthracis and C. difficile enzymes) share features in their catalytic domain, but also differences. Notably, (and similar to Family 3 LTs) C. difficile SpoIID coordinates with zinc, while a zinc coordination site is not seen in B. subtilis SpoIID. Sequence analysis found that the four amino acids (H134, C140, H145, and C146) involved in zinc coordination by C. difficile SpoIID are not conserved either in the B. anthrancis or B. subtilis SpoIID LTs. Interestingly, single mutations of C140, H145, and C146 debilitated the C. difficile SpoIID. Why C. difficile SpoIID requires metal coordination but the Bacillus SpoIID does not remains unclear (Nocadello et al., 2016). Nonetheless, we can infer from the Gram-negative Family 3 LTs that the zinc-ion coordination by the protein in this case is likely involved in stabilization of the folded enzyme.
LTs Catalyze a Unique Transacetalization Reaction
Families 1, 3, and 4 of the Gram-negative LTs have been classified by the CAZy database as belonging to the enigmatic goose-type (G-type) lysozyme sub-families. Family 1 and 4 belong to the GH23 sub-family, and Family 3 belongs to the GH103 subfamily. Family 2 belongs to the non-G-type GH102 sub-family. The well-studied chicken-type (C-type) lysozyme belongs to the GH22 sub-family (Gloster & Davies, 2010; Wohlkonig et al., 2010; Domínguez-Gil et al., 2016). The C-type lysozymes use a dual catalytic acid-catalytic base mechanism. The acid facilitates departure of the GlcNAc leaving group, resulting in an oxocarbenium transition state, that is trapped by the base (Asp) as a nucleophile forming a covalent acyl-enzyme intermediate. Catalysis is completed by water hydrolysis, in a net stereochemistry-retaining mechanism (Vocadlo et al., 2001). In contrast, the mechanism of the LTs is uncertain (Gloster & Davies, 2010). The Gram-negative GH23/GH103 LTs have a general acid catalytic residue (Glu in Gram-negative families 1, 3, 4, 5 and Asp in families 2, 6), and also show retention of stereochemistry. As such their mechanism likely uses a similar activation for breaking of the glycosidic bond and GlcNAc departure, with formation of an oxocarbenium species. However, in the GH23 and GH103 Families, no residue equivalent to the GH22 carboxylate nucleophile appears (Weaver et al., 1995). Possible LT mechanisms have been proposed without covalent intermediate formation, invoking substrate-assisted catalysis from the MurNAc N-acetyl group whereby the oxygen of the N-acetyl group on C2 stabilizes the oxocarbenium species through bonding to C1 with formation of an oxazolinium intermediate (Figure 19) (Reid et al., 2007; Fibriansah et al., 2012). Efforts to mimic this oxazolinium intermediate using a NAG-thiazoline analog are discussed in the following section (Reid et al., 2004a; Reid et al., 2004b). Whether this oxazolinium intermediate is formed, or if stabilization of the oxcarbenium occurs following near-synchronous formation of the 1,6-anhydro species and forgoing the oxazolinium intermediate, awaits further study. Regardless, the reaction concludes with deprotonation of the C6-hydroxyl, assisted by the catalytic glutamate now serving as a general base, enabling bond formation between the C6 oxygen and C1 to form the 1,6-anhydroMurNAc product. It is by no means clear whether a single universal mechanism pertains to all LTs. The reaction mechanism of the Gram-positive LTs on the muramic-δ-lactam of the peptidoglycan cortex has not been studied.
Figure 19.
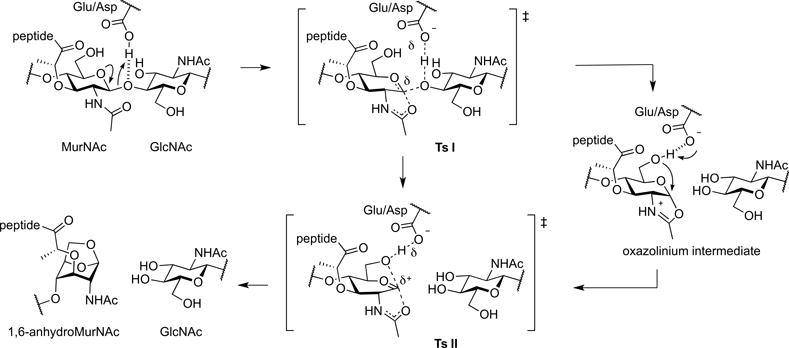
Proposed LT reaction mechanisms.
Methods for LT analysis
The idealized sine qua non for the study of an enzyme is an assay method that is reliable, quantifiable, sensitive, and continuous. Although several LT assays exist, none encompasses all of these criteria. Two reasons explain this limitation. The first reason is the substantial synthetic effort required to prepare the (NAG-NAM)n oligosaccharide as a discrete substrate. This difficulty is further compounded by the certainty (as established by the existing assay methods) that the different LTs respond to changes to this structure differently (for example, the value for n in terms of preferential endolytic or exolytic cleavage; the presence or absence of the peptide stem; whether the stem is or is not crosslinked). Moreover, the simple (NAG-NAM)n oligosaccharide lacks a reporting chromophore. While such a chromophore could surely be introduced (for example, in the departing NAG saccharide, or alternatively as a pair of chormophores for FRET assay), these additional synthetic objectives would add to an already herculean synthetic commitment. Given the incomplete understanding with respect to substrate recognition by these enzymes, multiple chromophore-labeled substrates might well be required to unmask the particular (NAG-NAM)n structure matching to recognition by the particular LT under study. The second reason is the proven multi-protein regulation of the membrane-bound LTs. It is highly probable that all membrane-bound LTs use protein-protein interaction both for localization and catalytic activation. We are only just now beginning to grasp (as opposed to understand) the role of LT interaction networks in the control of their function (Garcia-Gomez et al., 2011; Santin & Cascales, 2017). Although the soluble LTs may be exceptional (compared to the membrane-bound LTs) in having a simpler regulation by protein-protein interaction, their ability to complex with PBPs (Romeis & Höltje, 1994; Höltje, 1996; von Rechenberg et al., 1996; Legaree et al., 2007; Legaree & Clarke, 2008; Gutierrez et al., 2010; Nikolaidis et al., 2012) strongly suggests that their activity too is regulated by protein-protein interaction. The reliable (but again, exceedingly laborious) synthesis of (NAG-NAM)n structures is now possible (Wang et al., 2017). In this concise section, we summarize the strengths and limitations of the five LT assay methods in current practice. These five assays are the bacterial phenotypic assay, the qualitative gel-based assay, the semi-qualitative continuous sacculus-based turbidity assay, the sacculus-based fluorescence-perturbation continuous (or release, discontinuous) assay, and the synthetic substrate (and sacculus) quantitative and discontinuous chromatographic-mass spectrometry (LC-MS) assay.
The use of the bacterial sacculus, either as isolated from bacteria or reporter group-labeled, as the substrate is common to many of these assays. The procedure for the isolation of sacculi following bacterial culture is straightforward (but painstaking). Sacculus preparation involves dissolution of the non-polymeric compounds of the bacterium using hot aqueous SDS, repetitive wash and high-speed centrifugal sedimentations to remove the SDS (essential!), and possibly with intervening HF or protease digestion to remove the proteins and lipids covalently-attached to the peptidoglycan. Once these steps are practiced to perfection (for example, the thinner and therefore lighter P. aeruginosa-derived sacculus requires an entirely different centrifugal wash procedure than is required for the E. coli sacculus), isolation of an SDS-free sacculus is a reliable preparation. An alternative SDS-free procedure for the preparation of Gram-negative sacculi is described (Kühner et al., 2014), but comparative data on these sacculi for LT analysis is not available. Three caveats require emphasis. For E. coli the peptidoglycan structure is extensively remodeled in the transition from log growth to stationary phase. Accordingly, sacculi isolated from early log-phase, late-log phase, and stationary phase must be regarded as chemically distinct entities. Second, even the well-prepared sacculus is chemically heterogeneous. Muropeptide release from sacculi that have been radiolabeled (typically by glucosmine or diaminopimelate incorporation) was a historical method of LC assay. This assay method is now largely supplanted by the simpler fluorescent assays, or the much more informative LC-MS assay of released muropeptides, as discussed below. While the value of all of these sacculus-based assays must not be underestimated, the use of “bulk” sacculus as the substrate may obscure mechanistically important distinction with respect to muropeptide recognition by the membrane-bound LTs. The presentation of the entire (that is, intact single-molecule) sacculus to a solubilized LT that in the bacterium localizes to the septum, likely will competitively release simultaneously septal-, cap-, and sidewall-derived muropeptides. Moreover, the assay leaves invisible possibly significant glycan cleavage events that do not release muropeptides. With respect to the MltG (Gram-negative Family 5) LTs and the soluble LTs in particular, the sacculus polymer may be a misleading substrate. Without further acknowledging the potential limitations of the sacculus as the assay substrate, the principle LT assay methods are summarized.
Phenotypic Evaluation
The application of microscopy for the phenotypic characterization of LT function, typically as a consequence of gene knockout, is a powerful means of analyzing LT function. Numerous studies affirm the interpretive value of microscopic analysis, as exemplified by a series of reports (Heidrich et al., 2002; Meisel et al., 2003; Viollier & Shapiro, 2003; Reid et al., 2004b; Hett et al., 2010; Roure et al., 2012; Jorgenson et al., 2014; Yunck et al., 2016). Important complementary information is also provided from the measurement of the MIC values for β-lactam antibiotics, as exemplified by the additional publications (Templin et al., 1999; Costa & Anton, 2006; Cavallari et al., 2013; Lamers et al., 2015; Zeng et al., 2015). Fluorescent microscopy of LT fusion proteins can provide information (in rod-shaped bacteria) as to septal or sidewall localization (Schaub et al., 2016; Yunck et al., 2016). While these studies have powerful interpretive value as to LT function, they are not LT assay methods.
Zymographic Assays
Impregnation of a gel with sacculi enables the zymographic detection of LT activity as measured by the clearing that follows digestion of the polymer (Strating & Clarke, 2001). Although the generally very slow turnover of LTs (as compared to muraminidases) demands typically long incubation times (and thus careful control assays), the assay is reliable (Watt & Clarke, 1994; Blackburn & Clarke, 2002; de la Mora et al., 2012). For example, this assay differentiates easily between catalytically active and inactive mutant LTs (Weadge & Clarke, 2006). The limitations of this assay are its extensive time requirement and its essentially binary (active/inactive) output.
Suspended-Sacculus Turbidity Assay
This assay is a widely used spectrophotometric variation of the zymographic assay. Sacculi are suspended in solution and the progressive change in light scattering upon addition of the LT (generally seen as a clearance of turbidity) is measured over time (≤ 15 min, as the effect of the settling of the sacculi on the light scattering contributes increasingly at longer reaction times) (Hash, 1967; Scheurwater & Clarke, 2008). This assay validated successfully the Ivy protein as an LT inhibitor (Clarke et al., 2010), gave confirmation that O-acetylation of the peptidoglycan was protective against LT degradation (Weadge & Clarke, 2006; Clarke et al., 2010; Moynihan & Clarke, 2011), and enabled generation of a pH-rate profile for LT catalysis (Herlihey et al., 2014). The strength of the assay is its relative ease of performance and its rapidity. Nonetheless, in our hands this assay has significant shortcomings with respect to reproducibility (reflecting perhaps a high degree of sensitivity to the method used for sacculus preparation) and interpretation. Importantly, the assay is often performed with commercially available Gram-positive Micrococcus luteus sacculus. Although the use of this commercial sacculus improves experimental reproducibility, it otherwise does not address the challenge of interpreting the activity of a Gram-negative enzyme on a Gram-positive substrate (Scheurwater & Clarke, 2008; Fibriansah et al., 2012). The transmittance/absorbance change in the spectrophotometric signal does not translate as a quantitative measure of LT catalytic activity. Like the zymographic assay, the interpretation of the spectrophotometric turbidity assay does not extend beyond interpretation against a carefully matched control.
Suspended Sacculus Fluorescent Response Assay
This further variation of the turbidity assay, now available as a commercial assay kit, is the most reliable turbidity assay. In the commercial kit sacculi obtained from the Gram-positive bacterium Micrococcus luteus are purified and impregnated with a fluorescent reporter. The catalytic action of an LT on the suspended sacculi gives a time-dependent perturbation of the emissive intensity of the fluorophore by continuous assay. Effects of sacculus sedimentation are reduced compared to the non-fluorescent methodology due to its use of a lower concentration of suspended sacculus. An alternative assay uses discontinuous measurement of fluorophore release (Zhou et al., 1988; Nocadello et al., 2016; Yunck et al., 2016). This latter assay is generally applicable to all sacculi (from different bacteria, or from the same bacterium as obtained under different growth stages or from different enzyme expression levels). As is the case with the other turbidity assays, these fluorescence assays are proportional rather than quantitative. For example, in the fluorescence perturbation assay the slope is not proportional to amount of enzyme added; the slope is not quantifiable with respect to the substrate, and neither v nor KM values may be extracted from the line shape. A further limitation of this assay is its poor translation from fluorescence measurement within a cuvette (good reproducibility) to fluorescent measurement using a microplate (poor reproducibility).
LC-MS Evaluation of Sacculus and Synthetic Peptidoglycan Substrate Degradation
The analysis of the muropeptide composition of the sacculus by LC-MS analysis is unquestionably the dominant assay method for the quantitative evaluation of LT activity. Although the sacculus lacks an intrinsic chromophore, the sensitivities and resolving powers of both the LC and MS methods give great value to this assay. The assay is appropriate to the use of both the sacculus and synthetic muropeptides as substrates. The assay is reliable, robust, quantifiable, sensitive, and interpretable. Its limitations are its discontinuity, cost, and low volume. Nonetheless, this assay method has evolved from its origins (Glauner et al., 1988; Glauner, 1988; Blackburn & Clarke, 2000) to high levels of sophistication and automation (Kühner et al., 2014; Desmarais et al., 2015; Alvarez et al., 2016; Espaillat et al., 2016; Bern et al., 2017). Its value to LT analysis is showcased by its application to the comparative study of the LTs of E. coli (Lee et al., 2013), N. gonorrhoeae (Schaub et al., 2016) and P. aeruginosa (Lee et al., 2017), wherein the assay distinguishes between exolytic and endolytic preference, stem peptide or stem-peptide-free preference, and tolerance for the presence of cross-link. The assay can be adopted to provide v and KM values.
Opportunities for LT Assay Development
The absent assay method is the one that encompasses the strengths of the LC-MS assay, but is rapid, sensitive, and continuous. The equivalent assay for the muraminidases has a visible light or fluorescent reporter as the leaving group. Although the LTs very likely share with the muraminidases a requirement for oligosaccharide presentation for optimal catalytic efficiency, in principle the presentation on a MurNAc monosaccharide of a reactive leaving group (as is customary to many reporter groups in such assays) that exploits the sensitivity of fluorescence detection might address this need. A possible limitation (and key difference between the muraminidases and the LTs) is the likelihood that the membrane-bound LTs are not, as the soluble enzyme and in the absence of their regulatory protein partner, robust catalysts. Accordingly, even the provision of a good leaving group for sensitive detection may prove inadequate. As attention turns to assessing the value of LT inhibitors as adjuvants of the β-lactam antibiotics, efforts toward the development of new LT assays will continue.
LTs as Targets for Antibiotic Development
The combination of β-lactam antibiotics and β-lactamase inhibitors has extended by decades the clinical efficacy of the β-lactams. In the current time of rampant bacterial resistance, when the efficacy of this combination is threatened by the exchange of increasingly powerful β-lactamases between and among species, the identification of new adjuvants for the β-lactams is highly desirable. One very attractive possibility is a combination of an LT inhibitor with the β-lactam. The value of this strategy for further extending the clinical life of β-lactams arises from the appreciation that LTs and PBPs cooperate during cell-wall biosynthesis, in a homeostatic balancing act between construction and demolition of the cell wall (Johnson et al., 2013). The inhibition of PBPs by β-lactams disturbs this equilibrium, and in many pathogenic Gram-negative bacteria the resulting change in the pool of the recycling cell-wall fragments triggers β-lactamase expression (Jacobs et al., 1994; Fisher & Mobashery, 2014; Vadlamani et al., 2015; Lee et al., 2016a; Dik et al., 2017) These observations raise two central questions with respect to LT inhibition. The first is whether an LT inhibitor might suppress the ability of these Gram-negative bacteria to induce β-lactamases. The second is whether LT inhibition might further synergize with PBP inhibition to secure the ultimately bactericidal compromise of peptidoglycan integrity. Compelling evidence already in the literature points to a positive answer to both questions.
LT-knockout studies yield insight into the enzymatic roles of the LTs and suggest the possible clinical value of LT inhibition. With respect to the answer for the first question, the triple LT (MltA, MltB and Slt70) knockout in an ampR-reconstituted E. coli strain showed unimpaired in vitro growth, but substantially reduced cell-wall turnover (by approximately 70%) and removing these proteins reduced the bacterium’s ability to induce β-lactamase expression (Kraft et al., 1998; Korsak et al., 2005). The Slt70-deletion strain of E. coli likewise lacked a growth defect, yet was more sensitive to the β-lactam mecillinam (Templin et al., 1992). An explanation for this result was offered by Cho et al. (Cho et al., 2014) in their assignment of a “quality control” role to Slt70. Slt70 is suggested to remove the non-crosslinked peptidoglycan that is the result of β-lactam inhibition of PBP transpeptidation, thus preventing misincorporation of peptidoglycan into the cell wall. Therefore, Slt70 may be responsible in part for repairing cell-wall damage caused by β-lactam antibiotics. In such a role, this LT could be an advantageous drug target. Simultaneous genetic deletion of six LT genes (slt70, mltA-E) still did not inhibit cell growth (Heidrich et al., 2002), yet a viable strain could not be isolated with a seventh (mltF) LT-knockout (Scheurwater & Clarke, 2008). As a clear functional redundancy exists among the LTs (Lee et al., 2013), a broad-spectrum LT inhibitor might be necessary.
Additional studies in other bacteria, however, suggest that even when such an inhibitor is in hand, the value of its activity may not be general. Both insertional and deletion knockouts of the LTs of Pseudomonas aeruginosa identified MltB and SltB1 (both family 3) knockout strains as showing increased resistance to β-lactam drugs and further suggested that SltB3 (SltH), MltD, or MltF2 knockout strains displayed a similar effect (Cavallari et al., 2013; Lamers et al., 2015). The loss of both MltB and SltB1 together resulted in an even higher MIC of β-lactams (although this effect was unrelated to β-lactamase induction). These studies suggest that impairing the activity of (at least certain) LTs could be “protective” by reducing the ability of the bacterium to undergo lysis following β-lactam exposure. In contrast, both analyses showed that Slt-knockout strains had increased susceptibility to β-lactams (identical to the earlier studies with Slt70 in E. coli). The strain with insertional inactivation of MltF also displayed lower MICs for β-lactams. Moreover, knocking out the Family 1 LTs (MltD, MltF, MltF2, Slt) or several Mlts (MltA, MltB, MltD, MltF, MltF2) increased susceptibility to β-lactams. Finally, two single LT-knockout strains, (separately MltA and SltB2 (SltG)) did not affect the MICs of β-lactam antibiotics. While the deletion of specific LTs in E. coli reduced β-lactamase expression, single-knockout strains of Pseudomonas gave a different conclusion. Individual deletion of MltB, MltF, Slt, and SltB1 mutant strains had no significant effect on either basal or cefoxitin-induced AmpC expression compared to the wild-type strain (Cavallari et al., 2013). Similarly, LT-gene knockouts in Stenotrophomonas maltophilia (including Slt, MltA, MltB1, MltB2, MltD1, and MltD2) explored the effects of loss of LT function in this species (Huang et al., 2015; Wu et al., 2016). Removal of one LT (MltD1) yielded high levels of uninduced β-lactamase activity, an undesirable phenotype. Yet, inactivation of this single LT as well as MltB1, MltD2, or Slt resulted in strains that were more susceptible to macrolide antibiotics compared to the wild-type strain. Deletions of slt or mltB1 had a similar effect with aminoglycoside antibiotics, suggesting an increase in outer-membrane permeability (and thus antibiotic susceptibility) as a result of loss of LT function (Wu et al., 2016; Wu et al., 2016). An “appropriate” LT inhibitor could thus be beneficial for combination with antimicrobials outside of the β-lactam family. LT mutational analysis has also studied LT function in N. gonorrhoeae, where extracellular anhydroMurNac muropeptides directly contribute to pathogenicity (Cloud & Dillard, 2002; Chan et al., 2012). Two LTs (LtgA, a homolog of E. coli Slt70; and LtgD, a homolog of E. coli MltB) are involved in the release of the toxic muropeptides (Kohler et al., 2005; Cloud-Hansen et al., 2008). The remaining (known) LTs [LtgC (MltA), LtgB (MltC), LtgE (MltD), LtgX, and AltA] do not contribute. Although LtgA and LtgD are not essential for in vitro planktonic growth (Cloud-Hansen et al., 2008), their inhibition could reduce the virulence of N. gonorrhoeae infections.
Despite their potential value, the development of inhibitors targeting the LTs has been slow, reflecting in part the perception of extensive LT functional redundancy and the lack of a simple assay for LT activity. However, one natural product is a distinguished LT inhibitor (Williams et al., 2017). In the early 1980s, the exploration of the monobactam antibiotics produced by soil-dwelling Pseudomonas strains at Takeda Chemical Industries coincided with the observation that these bacteria also produced compounds that gave a “bulge” phenotype in E. coli cells when co-administered with a β-lactam (Imada et al., 1982). These aptly named compounds, the “bulgecins” (Figure 20A) (Shinagawa et al., 1985), were strongly synergistic with β-lactam antibiotics (Nakao et al., 1986). The nearly identical observation—bulge formation—was made subsequently with an E. coli Slt70-deletion strain, which also showed increased sensitivity to β-lactams (Templin et al., 1992). The bulgecins were postulated (and then confirmed) as Slt70 inhibitors (IC50 of 0.4 μM). The additional observation that “a crude preparation of solubilized membrane-bound LT was not inhibited by bulgecin,” suggested specificity of the bulgecins for Slt70 (Templin et al., 1992). The particular targeting of Slt70 by the bulgecins is further supported by the observation that the bulgecins no longer displayed synergy with β-lactams in the Slt70 deletion strain (Kraft et al., 1999). However, bulgecin-A-complexed crystal structures were solved subsequently for several LTs including Slt70 (Thunnissen et al., 1995b), Slt35 (van Asselt et al., 2000), and MltE (Fibriansah et al., 2012). The seeming discrepancy between the in vivo (apparent) selectivity and the in vitro ability to bind to multiple LTs may reflect both different individual affinities and how the LT activity is regulated in vivo. In these LT structures, bulgecin A acts as an apparent standard iminosaccharide GH inhibitor, wherein both the hydroxymethyl moiety and the nitrogen of the pyrrolidine are in near proximity to the catalytic glutamic acid, substantiating a competitive mode of inhibition. The mode of inhibition might be characterized as mimicry of the transient oxocarbenium species in the mechanism of substrate turnover. In addition to the previously cited studies in E. coli, crude bulgecin extracts have demonstrated potential when combined with β-lactams in vitro against clinical strains of both Pseudomonas aeruginosa and Acinetobacter baumannii (Skalweit & Li, 2016). Bulgecin A likewise synergizes with amoxicillin against Helicobacter pylori (Bonis et al., 2012) and is effective at reestablishing β-lactam vulnerability in resistant Neisseria species (Williams et al., 2017).
Figure 20.
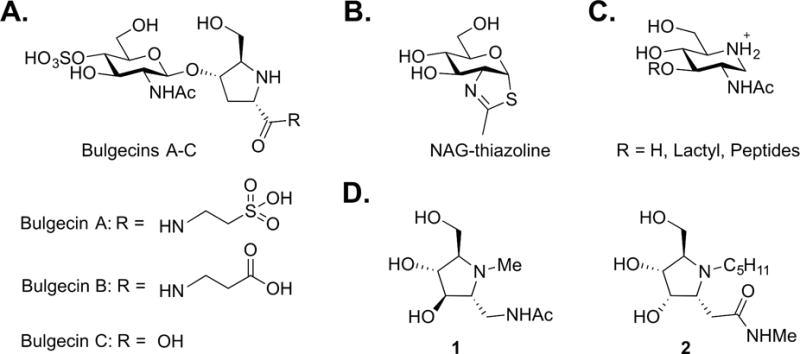
LT inhibitors: (A) the natural products bulgecin A-C, (B) NAG-thiazoline, (C) iminosaccharides, and (D) iminocyclitols.
The Family 3 LTs are proposed to use a substrate-assisted mechanism involving an oxazolinium intermediate (Reid et al., 2004a; Reid et al., 2004b). A GlcNAc-based thiazoline inhibitor, commonly understood to mimic the oxazolinium intermediate (Knapp et al., 2009) (Figure 20B), inhibited MltB from P. aeruginosa with a millimolar IC50 (Reid et al., 2004b). At a similar concentration (1 mg/mL, equal to 4.6 mM), this same NAG-thiazoline induced morphological changes in E. coli, but did not prevent growth (Reid et al., 2004a). In fact, NAG-thiazoline prevented β-lactam-induced cell lysis, consistent with the LT-housekeeping hypothesis (Cho et al., 2014). A similar phenotype was noted also in certain Pseudomonas LT knockout strains, including the MltB and SltB1 LT deletions mentioned above (Lamers et al., 2015). Regardless, NAG-thiazoline appears insufficiently potent for consideration as a β-lactam adjuvant. Several piperidine-based iminosaccharides based on MurNAc (Figure 20C) were evaluated as inhibitors of E. coli MltB (Yamaguchi et al., 2012). The nitrogen of the piperidine in these inhibitors serves the same function as the nitrogen of the pyrrolidine of bulgecin: as a (possibly) positively charged mimic of an oxocarbenium transition-state entity. The best compound had saturable binding with a 0.2 mM Kd value. As successful iminosaccharide inhibition of GH enzymes coincides with precise complementation of the inhibitor (ring size, stereochemistry, and substitution) and enzyme target (Stütz & Wrodnigg, 2011), the possibility of further exploration of the iminosaccharide class for LT inhibition remains open. Iminocyclitols 1 and 2 (Figure 20D) have nanomolar Ki values against hexosaminidases (Liu et al., 2001) and O-GlcNAcase (Bergeron-Brlek et al., 2015), offering exemplification of the level of potency that can be achieved upon optimization of the iminosaccharide structure. As the idealized LT inhibitor might interfere with the induction of the AmpC β-lactamases; synergize both with β-lactams and antimicrobials outside of the β-lactam family; and suppress the virulence of muropeptide-producing bacteria, future chemistry efforts toward the identification of LT inhibitors offers considerable opportunity.
Conclusion
The LTs defy generalization. The eleven LT enzyme ensemble encoded by P. aeruginosa (as an exemplary bacterium) includes enzymes that are small and single domain, and large and multidomain; lipoproteins and soluble enzymes; enzymes of peptidoglycan synthesis and enzymes of peptidoglycan processing; enzymes presumptively of the sidewall and of the septum; enzymes without direct evidence of allosteric regulation and enzymes with direct evidence of allosteric regulation; and enzymes with (apparently) either one or two critical catalytic residues. The LTs accomplish a diversity of cellular functions that credibly may be surmised to correspond to selective assignment within the ensemble, yet have proven functional redundancy. Strong circumstantial evidence argues that the inability to accomplish at least some of these functions is lethal. We have a credible perspective on the breadth of the LT landscape.
Nonetheless, within this landscape key answers are missing with respect to their structure, mechanism, and function. A partial enumeration of unanswered questions follows. Which LTs match to which function? Which function(s) are essential LT function(s)? Which LT functions synergize with β-lactam inactivation of the PBPs, and which antagonize? Here, of course, the diversity of β-lactam structure within a breath of PBP functions may exclude a simple answer. What is the true substrate(s) recognized by each LT? Here, there is a structural chasm among the peptidoglycan structures embedded in a polymeric bacterial exoskeleton (the sacculus) and the peptidoglycan structures of bacterial catabolism, and the uncertainty whether the select structures that may be crafted by de novo synthesis, align with either. If MltG is an enzyme of peptidoglycan synthesis, how do we reconcile the presumptive neighboring presence of other LTs, as suggested by the ability of these LTs to form protein-protein complexes with PBPs? Which LTs have regulatory partners, and what is the identity of these partners? For which LTs is their catalysis regulated by simultaneous contact with a regulatory partner and allosteric occupancy of a peripheral domain? Are the extents of LT structures (whether with or without ligand) only approximations of the true catalytic conformations, given the ability of their partners to stimulate their turnover chemistry? What value might be offered by an LT inhibitor, and does this value rest with an inhibitor that targets preferentially LT participation in peptidoglycan synthesis, peptidoglycan recycling, or peptidoglycan-dependent virulence?
Recent clever interdisciplinary study has clarified the identities of the proteins and enzymes that assemble to enable cytoskeleton-driven peptidoglycan synthesis (Bisson-Filho et al., 2017; Egan et al., 2017; Rued et al., 2017; Yang et al., 2017). Their identities are captured in imaginative cartoons suggesting the complexity of the protein-protein interaction network (Egan & Vollmer, 2013; Massidda et al., 2013; Szwedziak & Lowe, 2013; Fleurie et al., 2014; Egan & Vollmer, 2015; Leclercq et al., 2017; Lewis, 2017). A credible measure of the inchoate understanding of LT function is the absence of an LT in all of these cartoons. Our observation of this absence is not criticism. We simply do not know where, in this specific function, the LTs properly fit. Efforts toward finding this fit (while yet to supply the answer) have been no less clever, and the foundational efforts of Bernhardt, Boneca, Clarke, Dillard, Dijkstra, Hermoso, Höltje, Keck, Thunnissen, Vollmer and others have magnified into an explosion of recent studies on LT identity, structure, and function. This review is, therefore, a timely assimilation of these studies into a coherent framework, to support future efforts to place properly each member into the intriguing LT enzyme superfamily landscape.
Table 2.
Summary of the Gram-negative Family 2 LTs.
| Length (AA) | MW (Da) | Apo Structure | Complex Structure | |
|---|---|---|---|---|
| Family 2A | ||||
| MltA (ECK2809) | 365 | 40410.6 | 2AE0 (Apo) (van Straaten et al., 2005) 2GAE (Apo) (Powell et al., 2006) 2PIC (Catalytic Mutant) (van Straaten et al., 2007) 2PJJ (Catalytic Mutant) (van Straaten et al., 2007) |
2PI8 (Chitohexaose) (van Straaten et al., 2007) |
| MltA (PA1222) | 385 | 41896.6 | ||
| MltA (Smlt0155) | 388 | 41492.3 | ||
| LtgC (NGO2048) | 441 | 47975.6 | 2G6G (Apo) (Powell et al., 2006) 2G5D (Apo) (Powell et al., 2006) |
Table 3.
Summary of the Gram-negative Family 3 LTs.
| Length (AA) | MW (Da) | Apo Structure | Complex Structure | |
|---|---|---|---|---|
| Family 3A | ||||
| MltB (ECK2696) | 361 | 40255.7 | 1LTM (Slt35, Apo) (van Asselt et al., 1998) 1QDR (Slt35, Apo) (van Asselt & Dijkstra, 1999) 1QUS (Slt35, Apo) (van Asselt et al., 1999a) |
1QDT (Slt35, Calcium) (van Asselt & Dijkstra, 1999) 1QUT (Slt35, NAG) (van Asselt et al., 1999a) 1D0K (Slt35, GLCNAC-MURNAC-L-ALA-D-GLU) (van Asselt et al., 2000) 1D0L (Slt35, Bulgecin A) (van Asselt et al., 2000) 1D0M (Slt35, Bulgecin A & (GLCNAC)2) (van Asselt et al., 2000) |
| MltB (PA4444) | 367 | 41404.1 | ||
| SltB1 (PA4001) | 340 | 37835.6 | 4ANR (Apo) (Nikolaidis et al., 2012) | |
| MltB1 (Smlt4052) | 381 | 41821.6 | ||
| LtgD (NGO0626) | 363 | 39821.2 | ||
| Family 3B | ||||
| SltB2 (PA1171) | 398 | 42682.5 | ||
| SltB3 (PA3992) | 448 | 47760.9 | 5ANZ (Apo) (Lee et al., 2016b) | 5AO7 (NAG-anhNAM-pentapeptide) (Lee et al., 2016b) 5AO8 (NAG-NAM-pentapeptide) (Lee et al., 2016b) |
| MltB2 (Smlt4650) | 422 | 44588.4 |
Table 4.
Summary of the Gram-negative Family 4 LTs.
| Length (AA) | MW (Da) | Apo Structure | Complex Structure | |
|---|---|---|---|---|
| Family 4A | ||||
| LAL (Bacteriophage λ) | 154 | 17368.7 | 1AM7 (Evrard et al., 1998) | 3D3D (Chitohexasaccharide) (Leung et al., 2001) 1D9U (Chitohexasaccharide) (Leung et al., 2001) |
| Endolysin P28 (Maltocin P28) | 161 | 17283.6 | ||
| AtlA (GGI) | 181 | 20464.0 |
Table 5.
Summary of the Gram-negative Family 5 LTs.
| Length (AA) | MW (Da) | Apo Structure | Complex Structure | |
|---|---|---|---|---|
| Family 5A | ||||
| MltG (ECK1083) | 340 | 38247.4 | 2R1F (unpublished) | |
| MltG (PA2963) | 349 | 39562.3 | ||
| MltG (Smlt1034) | 353 | 38644.1 | ||
| LtgG (NGO2038) | 331 | 36805.2 |
Table 6.
Summary of the Gram-negative Family 6 LTs.
| Length (AA) | MW (Da) | Apo Structure | Complex Structure | |
|---|---|---|---|---|
| Family 6A | ||||
| RlpA (ECK0626) | 362 | 37527.9 | ||
| RlpA (PA4000) | 342 | 36482.6 | ||
| RlpA (Smlt4051) | 422 | 43784.4 | ||
| RlpA (NGO1728) | 228 | 24667.8 |
Table 7.
Summary of the Gram-positive Family 1 LTs.
| Length (AA) | MW (Da) | Apo Structure | Complex Structure | |
|---|---|---|---|---|
| Family 1A | ||||
| SleB (B. anthracis) | 253 | 27644.56 | 4FET(Jing et al., 2012) | |
| SleB (B. cereus) | 259 | 28247.20 | 4F55(Li et al., 2012) |
Table 8.
Summary of the Gram-positive Family 2 LTs.
| Length (AA) | MW (Da) | Apo Structure | Complex Structure | |
|---|---|---|---|---|
| Family 2A | ||||
| SpoIID (B. anthracis) | 339 | 37338.83 | 4RWR(Nocadello et al., 2016) | |
| SpoIID (B. cereus) | 344 | 37864.34 | ||
| SpoIID (C. difficile) | 354 | 40005.60 | 5I1T (Triacetylchitotriose) (Nocadello et al., 2016) |
Acknowledgments
We thank Dr. Byungjin Byun for his assistance in the creation of the X-ray structure alignments in the manuscript.
Biographies
David Dik is an alumnus of Hillsdale College (B.Sc., 2013). He is now working toward his doctoral degree in Chemical Biology at the University of Notre Dame under the supervision of Shahriar Mobashery with particular interest in the biochemical signaling pathways that manifest bacterial resistance to β-lactam antibiotics.
Daniel Marous received his undergraduate degree from Wittenberg University before completing his doctoral studies with Craig Townsend at the Johns Hopkins University. During his postdoctoral fellowship with Shahriar Mobashery at the University of Notre Dame, he seeks to develop innovative treatments of Gram-negative pathogens.
Four decades after defining (with Jeremy Knowles) the acyl-enzyme mechanism for catalysis by, and inactivation of, the Class A β-lactamases, Fisher has returned to the compelling enzymology of the micrometer-sized carnivores.
Shahriar Mobashery did his undergraduate and doctoral studies, followed by postdoctoral work, at the University of Southern California, the University of Chicago, and the Rockefeller University, respectively. He holds the Navari Chair in the Department of Chemistry and Biochemistry at the University of Notre Dame.
Footnotes
Declaration of Interest:
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article. This work was supported by a grant to S.M. from the National Institute of Health (NIH Grant GM61629). D.A.D. is a Fellow of the Chemistry-Biochemistry-Biology Interface (CBBI) Program (NIH Training Grant T32GM075762) and a Fellow of the ECK Institute of Global Health at the University of Notre Dame.
Query Coverage: The percent of the query sequence that is aligned based on an E Value optimization.
References
- Abanes-De Mello A, Sun YL, Aung S, et al. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 2002;16:3253–3264. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adin DM, Engle JT, Goldman WE, et al. Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri. J Bacteriol. 2009;191:2012–2022. doi: 10.1128/JB.01547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adu-Bobie J, Lupetti P, Brunelli B, et al. GNA33 of Neisseria meningitidis is a lipoprotein required for cell separation, membrane architecture, and virulence. Infect Immun. 2004;72:1914–1919. doi: 10.1128/IAI.72.4.1914-1919.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcorlo M, Martínez-Caballero S, Molina R, et al. Carbohydrate recognition and lysis by bacterial peptidoglycan hydrolases. Curr Opin Struct Biol. 2017;44:87–100. doi: 10.1016/j.sbi.2017.01.001. [DOI] [PubMed] [Google Scholar]
- Alvarez L, Hernandez SB, de Pedro MA, et al. Ultra-sensitive, high-resolution liquid chromatography methods for the high-throughput quantitative analysis of bacterial cell wall chemistry and structure. Methods Mol Biol. 2016;1440:11–27. doi: 10.1007/978-1-4939-3676-2_2. [DOI] [PubMed] [Google Scholar]
- Arends SJR, Williams K, Scott RJ, et al. Discovery and characterization of three new Escherichia coli septal ring proteins that contain a SPOR domain: DamX, DedD, and RlpA. J Bacteriol. 2010;192:242–255. doi: 10.1128/JB.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artola-Recolons C, Carrasco-López C, Llarrull LI, et al. High-resolution crystal structure of an outer membrane-anchored endolytic peptidoglycan lytic transglycosylase (MltE) from Escherichia coli. Biochemistry. 2011a;50:2384–2386. doi: 10.1021/bi200085y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artola-Recolons C, Lee M, Bernardo-Garcia N, et al. Structure and cell wall cleavage by modular lytic transglycosylase MltC of Escherichia coli. ACS Chem Biol. 2014;9:2058–2066. doi: 10.1021/cb500439c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artola-Recolons C, Llarrull LI, Lastochkin E, et al. Crystallization and preliminary X-ray diffraction analysis of the lytic transglycosylase MltE from Escherichia coli. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011b;67:161–163. doi: 10.1107/S1744309110049171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atrih A, Foster SJ. The role of peptidoglycan structure and structural dynamics during endospore dormancy and germination. Antonie Van Leeuwenhoek. 1999;75:299–307. doi: 10.1023/a:1001800507443. [DOI] [PubMed] [Google Scholar]
- Atrih A, Foster SJ. In vivo roles of the germination-specific lytic enzymes of Bacillus subtilis 168. Microbiology. 2001;147:2925–2932. doi: 10.1099/00221287-147-11-2925. [DOI] [PubMed] [Google Scholar]
- Balasubramanian D, Kumari H, Mathee K. Pseudomonas aeruginosa AmpR: An acute-chronic switch regulator. FEMS Pathog Dis. 2015;73:12208. doi: 10.1111/2049-632X.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcewich MD, Reeve TM, Orlikow EA, et al. Crystal structure of the AmpR effector binding domain provides insight into the molecular regulation of inducible ampC β-lactamase. J Mol Biol. 2010;400:998–1010. doi: 10.1016/j.jmb.2010.05.040. [DOI] [PubMed] [Google Scholar]
- Banzhaf M, van den Berg van Saparoea B, Terrak M, et al. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol Microbiol. 2012;85:179–194. doi: 10.1111/j.1365-2958.2012.08103.x. [DOI] [PubMed] [Google Scholar]
- Baron C, Coombes B. Targeting bacterial secretion systems: benefits of disarmament in the microcosm. Infect Disord Drug Targets. 2007;7:19–27. doi: 10.2174/187152607780090685. [DOI] [PubMed] [Google Scholar]
- Barreteau H, Kovac A, Boniface A, et al. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:168–172. doi: 10.1111/j.1574-6976.2008.00104.x. [DOI] [PubMed] [Google Scholar]
- Bartoleschi C, Pardini MC, Scaringi C, et al. Selection of Shigella flexneri candidate virulence genes specifically induced in bacteria resident in host cell cytoplasm. Cell Microbiol. 2002;4:613–626. doi: 10.1046/j.1462-5822.2002.00216.x. [DOI] [PubMed] [Google Scholar]
- Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD) J Mol Biol. 2000;299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- Beachey EH, Keck W, de Pedro MA, et al. Exoenzymatic activity of transglycosylase isolated from Escherichia coli. Eur J Biochem. 1981;116:355–358. doi: 10.1111/j.1432-1033.1981.tb05342.x. [DOI] [PubMed] [Google Scholar]
- Bergeron-Brlek M, Goodwin-Tindall J, Cekic N, et al. A convenient approach to stereoisomeric iminocyclitols: Generation of potent brain-permeable OGA Inhibitors. Angew Chem Int Ed. 2015;54:15429–15433. doi: 10.1002/anie.201507985. [DOI] [PubMed] [Google Scholar]
- Bern M, Beniston R, Mesnage S. Towards an automated analysis of bacterial peptidoglycan structure. Anal Bioanal Chem. 2017;409:551–560. doi: 10.1007/s00216-016-9857-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhards CB, Chen Y, Toutkoushian H, et al. HtrC is involved in proteolysis of YpeB during germination of Bacillus anthracis and Bacillus subtilis spores. J Bacteriol. 2015;197:326–336. doi: 10.1128/JB.02344-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsche U, Mayer C, Götz F, et al. Peptidoglycan perception—Sensing bacteria by their common envelope structure. Int J Med Microbiol. 2015;305:217–223. doi: 10.1016/j.ijmm.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Betzner AS, Ferreira LC, Höltje JV, et al. Control of the activity of the soluble lytic transglycosylase by the stringent response in Escherichia coli. FEMS Microbiol Lett. 1990;55:161–164. doi: 10.1016/0378-1097(90)90187-u. [DOI] [PubMed] [Google Scholar]
- Betzner AS, Keck W. Molecular cloning, overexpression and mapping of the slt gene encoding the soluble lytic transglycosylase of Escherichia coli. Mol Gen Genet. 1989;219:489–491. doi: 10.1007/BF00259625. [DOI] [PubMed] [Google Scholar]
- Biberstine KJ, Darr DS, Rosenthal RS. Tolerance to appetite suppression induced by peptidoglycan. Infect Immun. 1996;64:3641–3645. doi: 10.1128/iai.64.9.3641-3645.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson-Filho AW, Hsu YP, Squyres GR, et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science. 2017;355:739–743. doi: 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn NT, Clarke AJ. Assay for lytic transglycosylases: a family of peptidoglycan lyases. Anal Biochem. 2000;284:388–393. doi: 10.1006/abio.2000.4707. [DOI] [PubMed] [Google Scholar]
- Blackburn NT, Clarke AJ. Identification of four families of peptidoglycan lytic transglycosylases. J Mol Evol. 2001;52:78–84. doi: 10.1007/s002390010136. [DOI] [PubMed] [Google Scholar]
- Blackburn NT, Clarke AJ. Characterization of soluble and membrane-bound family 3 lytic transglycosylases from Pseudomonas aeruginosa. Biochemistry. 2002;41:1001–1013. doi: 10.1021/bi011833k. [DOI] [PubMed] [Google Scholar]
- Blocker A, Jouihri N, Larquet E, et al. Structure and composition of the Shigella flexneri “needle complex”, a part of its type III secreton. Mol Microbiol. 2001;39:652–663. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- Boland FM, Atrih A, Chirakkal H, et al. Complete spore-cortex hydrolysis during germination of Bacillus subtilis 168 requires SleB and YpeB. Microbiology. 2000;146:57–64. doi: 10.1099/00221287-146-1-57. [DOI] [PubMed] [Google Scholar]
- Bonis M, Williams A, Guadagnini S, et al. The effect of bulgecin A on peptidoglycan metabolism and physiology of Helicobacter pylori. Microb Drug Resist. 2012;18:230–239. doi: 10.1089/mdr.2011.0231. [DOI] [PubMed] [Google Scholar]
- Boudreau MA, Fisher JF, Mobashery S. Messenger functions of the bacterial cell wall-derived muropeptides. Biochemistry. 2012;51:2974–2990. doi: 10.1021/bi300174x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhss A, Trunkfield AE, Bugg TD, et al. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev. 2008;32:208–233. doi: 10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- Burkinshaw BJ, Deng W, Lameignère E, et al. Structural analysis of a specialized type III secretion system peptidoglycan-cleaving enzyme. J Biol Chem. 2015;290:10406–10417. doi: 10.1074/jbc.M115.639013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns DA, Heap JT, Minton NP. SleC is essential for germination of Clostridium difficile spores in nutrient-rich medium supplemented with the bile salt taurocholate. J Bacteriol. 2010;192:657–664. doi: 10.1128/JB.01209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler EK, Davis RM, Bari V, et al. Structure-function analysis of MurJ reveals a solvent-exposed cavity containing residues essential for peptidoglycan biogenesis in Escherichia coli. J Bacteriol. 2013;195:4639–4649. doi: 10.1128/JB.00731-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso R, Núñez G. Innate immunity: ER stress recruits NOD1 and NOD2 for delivery of inflammation. Curr Biol. 2016;26:R508–11. doi: 10.1016/j.cub.2016.05.021. [DOI] [PubMed] [Google Scholar]
- Cavallari JF, Lamers RP, Scheurwater EM, et al. Changes to its peptidoglycan-remodeling enzyme repertoire modulate β-lactam resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:3078–3084. doi: 10.1128/AAC.00268-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JM, Dillard JP. Neisseria gonorrhoeae crippled its peptidoglycan fragment permease to facilitate toxic peptidoglycan monomer release. J Bacteriol. 2016;198:3029–3040. doi: 10.1128/JB.00437-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YA, Hackett KT, Dillard JP. The lytic transglycosylases of Neisseria gonorrhoeae. Microb Drug Resist. 2012;18:271–279. doi: 10.1089/mdr.2012.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CI, Chelliah Y, Borek D, et al. Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science. 2006;311:1761–1764. doi: 10.1126/science.1123056. [DOI] [PubMed] [Google Scholar]
- Chaput C, Labigne A, Boneca IG. Characterization of Helicobacter pylori lytic transglycosylases Slt and MltD. J Bacteriol. 2007;189:422–429. doi: 10.1128/JB.01270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastanet A, Losick R. Engulfment during sporulation in Bacillus subtilis is governed by a multi-protein complex containing tandemly acting autolysins. Mol Microbiol. 2007;64:139–152. doi: 10.1111/j.1365-2958.2007.05652.x. [DOI] [PubMed] [Google Scholar]
- Chertkov OV, Armeev GA, Uporov IV, et al. Dual Active Site in the Endolytic Transglycosylase gp144 of Bacteriophage phiKZ. Acta Naturae. 2017;9:81–87. [PMC free article] [PubMed] [Google Scholar]
- Chirakkal H, O’Rourke M, Atrih A, et al. Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology. 2002;148:2383–2392. doi: 10.1099/00221287-148-8-2383. [DOI] [PubMed] [Google Scholar]
- Cho H, Uehara T, Bernhardt TG. β-Lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell. 2014;159:1300–1311. doi: 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie G, Ustok FI, Lu Q, et al. Mutational analysis of Bacillus megaterium QM B1551 cortex-lytic enzymes. J Bacteriol. 2010;192:5378–5389. doi: 10.1128/JB.00830-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke CA, Scheurwater EM, Clarke AJ. The vertebrate lysozyme inhibitor Ivy functions to inhibit the activity of lytic transglycosylase. J Biol Chem. 2010;285:14843–14847. doi: 10.1074/jbc.C110.120931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud-Hansen KA, Hackett KT, Garcia DL, et al. Neisseria gonorrhoeae uses two lytic transglycosylases to produce cytotoxic peptidoglycan monomers. J Bacteriol. 2008;190:5989–5994. doi: 10.1128/JB.00506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud KA, Dillard JP. A lytic transglycosylase of Neisseria gonorrhoeae is involved in peptidoglycan-derived cytotoxin production. Infect Immun. 2002;70:2752–2757. doi: 10.1128/IAI.70.6.2752-2757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud KA, Dillard JP. Mutation of a single lytic transglycosylase causes aberrant septation and inhibits cell separation of Neisseria gonorrhoeae. J Bacteriol. 2004;186:7811–7814. doi: 10.1128/JB.186.22.7811-7814.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier I, Paradis-Bleau C, Giroux AM, et al. Biophysical studies of the interactions between the phage varphiKZ gp144 lytic transglycosylase and model membranes. Eur Biophys J. 2010;39:263–276. doi: 10.1007/s00249-009-0530-1. [DOI] [PubMed] [Google Scholar]
- Cookson BT, Tyler AN, Goldman WE. Primary structure of the peptidoglycan-derived tracheal cytotoxin of Bordetella pertussis. Biochemistry. 1989;28:1744–1749. doi: 10.1021/bi00430a048. [DOI] [PubMed] [Google Scholar]
- Costa CS, Anton DN. High-level resistance to mecillinam produced by inactivation of soluble lytic transglycosylase in Salmonella enterica serovar Typhimurium. FEMS Microbiol Lett. 2006;256:311–317. doi: 10.1111/j.1574-6968.2006.00133.x. [DOI] [PubMed] [Google Scholar]
- Davies GJ, Wilson KS, Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem J. 1997;321:557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mora J, Ballado T, González-Pedrajo B, et al. The flagellar muramidase from the photosynthetic bacterium Rhodobacter sphaeroides. J Bacteriol. 2007;189:7998–8004. doi: 10.1128/JB.01073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mora J, Osorio-Valeriano M, Gonzalez-Pedrajo B, et al. The C-terminus of the flagellar muramidase SltF modulates the interaction with FlgJ in Rhodobacter sphaeroides. J Bacteriol. 2012;194:4513–4520. doi: 10.1128/JB.00460-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmarais SM, Tropini C, Miguel A, et al. High-throughput, highly sensitive analyses of bacterial morphogenesis using ultra-performance liquid chromatography. J Biol Chem. 2015;290:31090–31100. doi: 10.1074/jbc.M115.661660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra AJ, Hermann F, Keck W. Cloning and controlled overexpression of the gene encoding the 35 kDa soluble lytic transglycosylase from Escherichia coli. FEBS Lett. 1995;366:115–118. doi: 10.1016/0014-5793(95)00505-4. [DOI] [PubMed] [Google Scholar]
- Dijkstra AJ, Keck W. Identification of new members of the lytic transglycosylase family in Haemophilus influenzae and Escherichia coli. Microb Drug Resist. 1996;2:141–145. doi: 10.1089/mdr.1996.2.141. [DOI] [PubMed] [Google Scholar]
- Dijkstra BW, Thunnissen AM. ‘Holy’ proteins. II: The soluble lytic transglycosylase. Curr Opin Struct Biol. 1994;4:810–813. doi: 10.1016/0959-440x(94)90261-5. [DOI] [PubMed] [Google Scholar]
- Dik DA, Domínguez-Gil T, Lee M, et al. Muropeptide binding and the x-ray structure of the effector domain of the transcriptional regulator AmpR of Pseudomonas aeruginosa. J Am Chem Soc. 2017;139:1448–1451. doi: 10.1021/jacs.6b12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillard JP, Seifert HS. A peptidoglycan hydrolase similar to bacteriophage endolysins acts as an autolysin in Neisseria gonorrhoeae. Mol Microbiol. 1997;25:893–901. doi: 10.1111/j.1365-2958.1997.mmi522.x. [DOI] [PubMed] [Google Scholar]
- Dominguez-Gil T, Lee M, Acebrón-Avalos I, et al. Activation by allostery in cell-wall remodeling by a modular membrane-bound lytic transglycosylase from Pseudomonas aeruginosa. Structure. 2016;24:1729–1741. doi: 10.1016/j.str.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Gil T, Molina R, Alcorlo M, et al. Renew or die: the molecular mechanisms of peptidoglycan recycling and antibiotic resistance in Gram-negative pathogens. Drug Resist Updates. 2016;28:91–104. doi: 10.1016/j.drup.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Dong H, Zhu C, Chen J, et al. Antibacterial activity of Stenotrophomonas maltophilia endolysin P28 against both Gram-positive and Gram-negative bacteria. Front Microbiol. 2015;6:1299. doi: 10.3389/fmicb.2015.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan AJF, Cleverley RM, Peters K, et al. Regulation of bacterial cell wall growth. FEBS J. 2017 doi: 10.1111/febs.13959. [DOI] [PubMed] [Google Scholar]
- Egan AJF, Vollmer W. The physiology of bacterial cell division. Ann N Y Acad Sci. 2013;1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- Egan AJF, Vollmer W. The stoichiometric divisome: a hypothesis. Front Microbiol. 2015;6:455. doi: 10.3389/fmicb.2015.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert K, Höltje JV, Templin MF. Cloning and expression of a murein hydrolase lipoprotein from Escherichia coli. Mol Microbiol. 1995;16:761–768. doi: 10.1111/j.1365-2958.1995.tb02437.x. [DOI] [PubMed] [Google Scholar]
- Engel H, Kazemier B, Keck W. Murein-metabolizing enzymes from Escherichia coli: sequence analysis and controlled overexpression of the slt gene, which encodes the soluble lytic transglycosylase. J Bacteriol. 1991;173:6773–6782. doi: 10.1128/jb.173.21.6773-6782.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel H, Smink AJ, van Wijngaarden L, et al. Murein-metabolizing enzymes from Escherichia coli: existence of a second lytic transglycosylase. J Bacteriol. 1992;174:6394–6403. doi: 10.1128/jb.174.20.6394-6403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espaillat A, Forsmo O, El Biari K, et al. Chemometric analysis of bacterial peptidoglycan reveals atypical modifications that empower the cell wall against predatory enzymes and fly innate immunity. J Am Chem Soc. 2016;138:9193–9204. doi: 10.1021/jacs.6b04430. [DOI] [PubMed] [Google Scholar]
- Evrard C, Fastrez J, Declercq JP. Crystal structure of the lysozyme from bacteriophage λ and its relationship with V and C-type lysozymes. J Mol Biol. 1998;276:151–164. doi: 10.1006/jmbi.1997.1499. [DOI] [PubMed] [Google Scholar]
- Fibriansah G, Gliubich FI, Thunnissen AM. On the mechanism of peptidoglycan binding and cleavage by the endo-specific lytic transglycosylase MltE from Escherichia coli. Biochemistry. 2012;51:9164–9177. doi: 10.1021/bi300900t. [DOI] [PubMed] [Google Scholar]
- Fisher JF, Mobashery S. The sentinel role of peptidoglycan recycling in the β-lactam resistance of the Gram-negative Enterobacteriaceae and Pseudomonas aeruginosa. Bioorg Chem. 2014;56:41–48. doi: 10.1016/j.bioorg.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming TJ, Wallsmith DE, Rosenthal RS. Arthropathic properties of gonococcal peptidoglycan fragments: implications for the pathogenesis of disseminated gonococcal disease. Infect Immun. 1986;52:600–608. doi: 10.1128/iai.52.2.600-608.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurie A, Manuse S, Zhao C, et al. Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet. 2014;10:e1004275. doi: 10.1371/journal.pgen.1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Miroshnikov KA, Shneider MM, et al. Structure of the bacteriophage phiKZ lytic transglycosylase, gp144. J Biol Chem. 2008;283:7242–7250. doi: 10.1074/jbc.M709398200. [DOI] [PubMed] [Google Scholar]
- Garcia-Gomez E, Espinosa N, de la Mora J, et al. The muramidase EtgA from enteropathogenic Escherichia coli is required for efficient type III secretion. Microbiology. 2011;157:1145–1160. doi: 10.1099/mic.0.045617-0. [DOI] [PubMed] [Google Scholar]
- Gerding MA, Liu B, Bendezu FO, et al. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol. 2009;191:7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebel JD, Carr KA, Anderson EC, et al. The germination-specific lytic enzymes SleB, CwlJ1, and CwlJ2 each contribute to Bacillus anthracis spore germination and virulence. J Bacteriol. 2009;191:5569–5576. doi: 10.1128/JB.00408-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- Glauner B, Höltje JV, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- Gloster TM, Davies GJ. Glycosidase inhibition: assessing mimicry of the transition state. Org Biomol Chem. 2010;8:305–320. doi: 10.1039/b915870g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gode-Potratz CJ, Kustusch RJ, Breheny PJ, et al. Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol Microbiol. 2011;79:240–263. doi: 10.1111/j.1365-2958.2010.07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell EW. Recycling of murein by Escherichia coli. J Bacteriol. 1985;163:305–310. doi: 10.1128/jb.163.1.305-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell EW, Schwarz U. Release of cell wall peptides into culture medium by exponentially growing Escherichia coli. J Bacteriol. 1985;162:391–397. doi: 10.1128/jb.162.1.391-397.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway DL, Perkins HR. Turnover of the cell wall peptidoglycan during growth of Neisseria gonorrhoeae and Escherichia coli. Relative stability of newly synthesized material. J Gen Microbiol. 1985;131:253–263. doi: 10.1099/00221287-131-2-253. [DOI] [PubMed] [Google Scholar]
- Guglielmetti S, Balzaretti S, Taverniti VA, et al. TgaA, a VirB1-like component belonging to a putative Type IV secretion system of Bifidobacterium bifidum MIMBb75. Appl Environ Microbiol. 2014;80:5161–5169. doi: 10.1128/AEM.01413-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutelius D, Hokeness K, Logan SM, et al. Functional analysis of SleC from Clostridium difficile: an essential lytic transglycosylase involved in spore germination. Microbiology. 2014;160:209–216. doi: 10.1099/mic.0.072454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez J, Smith R, Pogliano K. SpoIID-mediated peptidoglycan degradation is required throughout engulfment during Bacillus subtilis sporulation. J Bacteriol. 2010;192:3174–3186. doi: 10.1128/JB.00127-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hash JH. Measurement of bacteriolytic enzymes. J Bacteriol. 1967;93:1201–1202. doi: 10.1128/jb.93.3.1201-1202.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron JD, Lambert EA, Sherry N, et al. Contributions of four cortex lytic enzymes to germination of Bacillus anthracis spores. J Bacteriol. 2010;192:763–770. doi: 10.1128/JB.01380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron JD, Orsburn B, Popham DL. Roles of germination-specific lytic enzymes CwlJ and SleB in Bacillus anthracis. J Bacteriol. 2009;191:2237–2247. doi: 10.1128/JB.01598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron JD, Sherry N, Popham DL. In vitro studies of peptidoglycan binding and hydrolysis by the Bacillus anthracis germination-specific lytic enzyme SleB. J Bacteriol. 2011;193:125–131. doi: 10.1128/JB.00869-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich C, Ursinus A, Berger J, et al. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J Bacteriol. 2002;184:6093–6099. doi: 10.1128/JB.184.22.6093-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss LN, Lancaster JR, Corbett JA, et al. Epithelial autotoxicity of nitric oxide: role in the respiratory cytopathology of pertussis. Proc Natl Acad Sci USA. 1994;91:267–270. doi: 10.1073/pnas.91.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlihey FA, Clarke AJ. Controlling autolysis during flagella insertion in Gram-negative bacteria. Adv Exp Med Biol. 2016 doi: 10.1007/5584_2016_52. [DOI] [PubMed] [Google Scholar]
- Herlihey FA, Moynihan PJ, Clarke AJ. The essential protein for bacterial flagella formation FlgJ functions as a β-N-acetylglucosaminidase. J Biol Chem. 2014;289:31029–31042. doi: 10.1074/jbc.M114.603944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlihey FA, Osorio-Valeriano M, Dreyfus G, et al. Modulation of the lytic activity of the dedicated autolysin for flagellum formation SltF by flagellar rod proteins FlgB and FlgF. J Bacteriol. 2016;198:1847–1856. doi: 10.1128/JB.00203-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hett EC, Chao MC, Rubin EJ. Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria. PLoS Pathog. 2010;6:e1001020. doi: 10.1371/journal.ppat.1001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höltje JV. Molecular interplay of murein synthases and murein hydrolases in Escherichia coli. Microb Drug Resist. 1996;2:99–103. doi: 10.1089/mdr.1996.2.99. [DOI] [PubMed] [Google Scholar]
- Höltje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höltje JV, Heidrich C. Enzymology of elongation and constriction of the murein sacculus of Escherichia coli. Biochimie. 2001;83:103–108. doi: 10.1016/s0300-9084(00)01226-8. [DOI] [PubMed] [Google Scholar]
- Höltje JV, Mirelman D, Sharon N, et al. Novel type of murein transglycosylase in Escherichia coli. J Bacteriol. 1975;124:1067–1076. doi: 10.1128/jb.124.3.1067-1076.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höppner C, Carle A, Sivanesan D, et al. The putative lytic transglycosylase VirB1 from Brucella suis interacts with the type IV secretion system core components VirB8, VirB9 and VirB11. Microbiology. 2005;151:3469–3482. doi: 10.1099/mic.0.28326-0. [DOI] [PubMed] [Google Scholar]
- Hu K, Yang H, Liu G, et al. Cloning and identification of a gene encoding spore cortex-lytic enzyme in Bacillus thuringiensis. Curr Microbiol. 2007;54:292–295. doi: 10.1007/s00284-006-0430-x. [DOI] [PubMed] [Google Scholar]
- Huang Y-W, Wu C-J, Hu R-M, et al. Interplay among membrane-bound lytic transglycosylase D1, the CreBC two-component regulatory system, the AmpNG-AmpDI-NagZ-AmpR regulatory circuit, and L1/L2 β-lactamase expression in Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2015;59:6866–6872. doi: 10.1128/AAC.05179-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imada A, Kintaka K, Nakao M, et al. Bulgecin, a bacterial metabolite which in concert with β-lactam antibiotics causes bulge formation. J Antibiot. 1982;35:1400–1403. doi: 10.7164/antibiotics.35.1400. [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Yamane K, Sekiguchi J. Regulation and characterization of a newly deduced cell wall hydrolase gene (cwlJ) which affects germination of Bacillus subtilis spores. J Bacteriol. 1998;180:1375–1380. doi: 10.1128/jb.180.6.1375-1380.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C, Huang LJ, Bartowsky E, et al. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 1994;13:4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings GT, Savino S, Marchetti E, et al. GNA33 from Neisseria meningitidis serogroup B encodes a membrane-bound lytic transglycosylase (MltA) Eur J Biochem. 2002;269:3722–3731. doi: 10.1046/j.1432-1033.2002.03064.x. [DOI] [PubMed] [Google Scholar]
- Jing X, Robinson HR, Heffron J, et al. The catalytic domain of the germination-specific lytic transglycosylase SleB from Bacillus anthracis displays a unique active site topology. Proteins. 2012;80:2469–2475. doi: 10.1002/prot.24140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsen L. Biological properties of bacterial peptidoglycan. APMIS. 1993;101:337–344. doi: 10.1111/j.1699-0463.1993.tb00119.x. [DOI] [PubMed] [Google Scholar]
- Johnson JW, Fisher JF, Mobashery S. Bacterial cell-wall recycling. Ann N Y Acad Sci. 2013;1277:54–75. doi: 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson MA, Chen Y, Yahashiri A, et al. The bacterial septal ring protein RlpA is a lytic transglycosylase that contributes to rod shape and daughter cell separation in Pseudomonas aeruginosa. Mol Microbiol. 2014;93:113–128. doi: 10.1111/mmi.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J Bacteriol. 1996;178:2767–2774. doi: 10.1128/jb.178.10.2767-2774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J Antimicrob Chemother. 1997;40:615–621. doi: 10.1093/jac/40.5.615. [DOI] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Membrane vesicles derived from Pseudomonas aeruginosa and Shigella flexneri can be integrated into the surfaces of other Gram-negative bacteria. Microbiology. 1999;145:2051–2060. doi: 10.1099/13500872-145-8-2051. [DOI] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Mayer A, Messner P, et al. S-layered Aneurinibacillus and Bacillus spp. are susceptible to the lytic action of Pseudomonas aeruginosa membrane vesicles. J Bacteriol. 1998;180:2306–2311. doi: 10.1128/jb.180.9.2306-2311.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki A, Karasudani Y, Otsuka Y, et al. Synthesis of diaminopimelic acid containing peptidoglycan fragments and tracheal cytotoxin (TCT) and investigation of their biological functions. Chem Eur J. 2008;14:10318–10330. doi: 10.1002/chem.200801121. [DOI] [PubMed] [Google Scholar]
- Keck W, Wientjes FB, Schwarz U. Comparison of two hydrolytic murein transglycosylases of Escherichia coli. Eur J Biochem. 1985;148:493–497. doi: 10.1111/j.1432-1033.1985.tb08866.x. [DOI] [PubMed] [Google Scholar]
- Keep NH, Ward JM, Cohen-Gonsaud M, et al. Wake up! Peptidoglycan lysis and bacterial non-growth states. Trends Microbiol. 2006;14:271–276. doi: 10.1016/j.tim.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Keestra-Gounder AM, Byndloss MX, Seyffert N, et al. NOD1 and NOD2 signalling links ER stress with inflammation. Nature. 2016;532:394–397. doi: 10.1038/nature17631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S, Fash D, Abdo M, et al. GlcNAc-thiazoline conformations. Bioorg Med Chem. 2009;17:1831–1836. doi: 10.1016/j.bmc.2009.01.066. [DOI] [PubMed] [Google Scholar]
- Kohler PL, Cloud KA, Hackett KT, et al. Characterization of the role of LtgB, a putative lytic transglycosylase in Neisseria gonorrhoeae. Microbiology. 2005;151:3081–3088. doi: 10.1099/mic.0.28125-0. [DOI] [PubMed] [Google Scholar]
- Kohler PL, Hamilton HL, Cloud-Hansen K, et al. AtlA functions as a peptidoglycan lytic transglycosylase in the Neisseria gonorrhoeae type IV secretion system. J Bacteriol. 2007;189:5421–5428. doi: 10.1128/JB.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler V, Probst I, Aufschnaiter A, et al. Conjugative type IV secretion in Gram-positive pathogens: TraG, a lytic transglycosylase and endopeptidase, interacts with translocation channel protein TraM. Plasmid. 2017 doi: 10.1016/j.plasmid.2017.02.002. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Rudd KE. A conserved domain in putative bacterial and bacteriophage transglycosylases. Trends Biochem Sci. 1994;19:106–107. doi: 10.1016/0968-0004(94)90201-1. [DOI] [PubMed] [Google Scholar]
- Koraimann G. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell Mol Life Sci. 2003;60:2371–2388. doi: 10.1007/s00018-003-3056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsak D, Liebscher S, Vollmer W. Susceptibility to antibiotics and β-lactamase induction in murein hydrolase mutants of Escherichia coli. Antimicrob Agents Chemother. 2005;49:1404–1409. doi: 10.1128/AAC.49.4.1404-1409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft AR, Prabhu J, Ursinus A, et al. Interference with murein turnover has no effect on growth but reduces β-lactamase induction in Escherichia coli. J Bacteriol. 1999;181:7192–7198. doi: 10.1128/jb.181.23.7192-7198.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft AR, Templin MF, Höltje JV. Membrane-bound lytic endotransglycosylase in Escherichia coli. J Bacteriol. 1998;180:3441–3447. doi: 10.1128/jb.180.13.3441-3447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger JM, Karnovsky ML, Martin SA, et al. Peptidoglycans as promoters of slow-wave sleep. II. Somnogenic and pyrogenic activities of some naturally occurring muramyl peptides; correlations with mass spectrometric structure determination. J Biol Chem. 1984;259:12659–12662. [PubMed] [Google Scholar]
- Kubori T, Matsushima Y, Nakamura D, et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- Kühner D, Stahl M, Demircioglu DD, et al. From cells to muropeptide structures in 24 h: peptidoglycan mapping by UPLC-MS. Sci Rep. 2014;4:7494. doi: 10.1038/srep07494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuk AC, Mashalidis EH, Lee SY. Crystal structure of the MOP flippase MurJ in an inward-facing conformation. Nat Struct Mol Biol. 2017;24:171–176. doi: 10.1038/nsmb.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazawa T, Masayama A, Fukuoka S, et al. Mode of action of a germination-specific cortex-lytic enzyme, SleC, of Clostridium perfringens S40. Biosci Biotechnol Biochem. 2007;71:884–892. doi: 10.1271/bbb.60511. [DOI] [PubMed] [Google Scholar]
- Kusser W, Schwarz U. Escherichia coli murein transglycosylase. Purification by affinity chromatography and interaction with polynucleotides. Eur J Biochem. 1980;103:277–281. doi: 10.1111/j.1432-1033.1980.tb04312.x. [DOI] [PubMed] [Google Scholar]
- Lamers RP, Nguyen UT, Nguyen Y, et al. Loss of membrane-bound lytic transglycosylases increases outer membrane permeability and β-lactam sensitivity in Pseudomonas aeruginosa. MicrobiologyOpen. 2015;4 doi: 10.1002/mbo3.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Derouaux A, Olatunji S, et al. Interplay between penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci Rep. 2017;7:43306. doi: 10.1038/srep43306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Artola-Recolons C, Carrasco-López C, et al. Cell-wall remodeling by the zinc-protease AmpDh3 from Pseudomonas aeruginosa. J Am Chem Soc. 2013;135:12604–12607. doi: 10.1021/ja407445x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Dhar S, De Benedetti S, et al. Muropeptides in Pseudomonas aeruginosa and their role as elicitors of β-lactam-antibiotic resistance. Angew Chem Int Ed. 2016a;55:6882–6886. doi: 10.1002/anie.201601693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Domínguez-Gil T, Hesek D, et al. Turnover of bacterial cell wall by SltB3, a multidomain lytic transglycosylase of Pseudomonas aeruginosa. ACS Chem Biol. 2016b;11:1525–1531. doi: 10.1021/acschembio.6b00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Hesek D, Dik DA, et al. From genome to proteome to elucidation of reactions for all eleven known lytic transglycosylases from Pseudomonas aeruginosa. Angew Chem Int Ed. 2017;56:2735–2739. doi: 10.1002/anie.201611279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Hesek D, Llarrull LI, et al. Reactions of all Escherichia coli lytic transglycosylases with bacterial cell wall. J Am Chem Soc. 2013;135:3311–3314. doi: 10.1021/ja309036q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Hesek D, Suvorov M, et al. A mechanism-based inhibitor targeting the DD-transpeptidase activity of bacterial penicillin-binding proteins. J Am Chem Soc. 2003;125:16322–16326. doi: 10.1021/ja038445l. [DOI] [PubMed] [Google Scholar]
- Lee W, McDonough MA, Kotra L, et al. A 1.2 Å snapshot of the final step of bacterial cell wall biosynthesis. Proc Natl Acad Sci USA. 2001;98:1427–1431. doi: 10.1073/pnas.98.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legaree BA, Adams CB, Clarke AJ. Overproduction of penicillin-binding protein 2 and its inactive variants causes morphological changes and lysis in Escherichia coli. J Bacteriol. 2007;189:4975–4983. doi: 10.1128/JB.00207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legaree BA, Clarke AJ. Interaction of penicillin-binding protein 2 with soluble lytic transglycosylase B1 in Pseudomonas aeruginosa. J Bacteriol. 2008;190:6922–6926. doi: 10.1128/JB.00934-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Duewel HS, Honek JF, et al. Crystal structure of the lytic transglycosylase from bacteriophage λ in complex with hexa-N-acetylchitohexaose. Biochemistry. 2001;40:5665–5673. doi: 10.1021/bi0028035. [DOI] [PubMed] [Google Scholar]
- Lewenza S, Mhlanga MM, Pugsley AP. Novel inner membrane retention signals in Pseudomonas aeruginosa lipoproteins. J Bacteriol. 2008;190:6119–6125. doi: 10.1128/JB.00603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RJ. The GpsB files: the truth is out there. Mol Microbiol. 2017;103:913–918. doi: 10.1111/mmi.13612. [DOI] [PubMed] [Google Scholar]
- Li Y, Butzin XY, Davis A, et al. Activity and regulation of various forms of CwlJ, SleB, and YpeB proteins in degrading cortex peptidoglycan of spores of Bacillus species in vitro and during spore germination. J Bacteriol. 2013;195:2530–2540. doi: 10.1128/JB.00259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jin K, Setlow B, et al. Crystal structure of the catalytic domain of the Bacillus cereus SleB Protein Important in cortex peptidoglycan degradation during spore germination. J Bacteriol. 2012;194:4537–4545. doi: 10.1128/JB.00877-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Clarke AJ, Beveridge TJ. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J Bacteriol. 1998;180:5478–5483. doi: 10.1128/jb.180.20.5478-5483.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litzinger S, Duckworth A, Nitzsche K, et al. Muropeptide rescue in Bacillus subtilis involves sequential hydrolysis by β-N-acetylglucosaminidase and N-acetylmuramyl-L-alanine amidase. J Bacteriol. 2010;192:3132–3143. doi: 10.1128/JB.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Shikhman AR, Lotz MK, et al. Hexosaminidase inhibitors as new drug candidates for the therapy of osteoarthritis. Chem Biol. 2001;8:701–711. doi: 10.1016/s1074-5521(01)00045-x. [DOI] [PubMed] [Google Scholar]
- Llosa M, Zupan J, Baron C, et al. The N- and C-terminal portions of the Agrobacterium VirB1 protein independently enhance tumorigenesis. J Bacteriol. 2000;182:3437–3445. doi: 10.1128/jb.182.12.3437-3445.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommatzsch J, Templin MF, Kraft AR, et al. Outer membrane localization of murein hydrolases: MltA, a third lipoprotein lytic transglycosylase in Escherichia coli. J Bacteriol. 1997;179:5465–5470. doi: 10.1128/jb.179.17.5465-5470.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker KE, Collier JL, Kolodziej EW, et al. Bordetella pertussis tracheal cytotoxin and other muramyl peptides: distinct SAR for respiratory epithelial cytopathology. Proc Natl Acad Sci USA. 1993;90:2365–2369. doi: 10.1073/pnas.90.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madoori PK, Thunnissen AM. Purification, crystallization and preliminary x-ray diffraction analysis of the lytic transglycosylase MltF from Escherichia coli. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:534–538. doi: 10.1107/S1744309110010596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Caballero S, Lee M, Artola-Recolons C, et al. Reaction products and the x-ray structure of AmpDh2, a virulence determinant of Pseudomonas aeruginosa. J Am Chem Soc. 2013;135:10318–10321. doi: 10.1021/ja405464b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massidda O, Novakova L, Vollmer W. From models to pathogens: how much have we learned about Streptococcus pneumoniae cell division? Environ Microbiol. 2013;15:3133–3157. doi: 10.1111/1462-2920.12189. [DOI] [PubMed] [Google Scholar]
- Matsuzawa H, Asoh S, Kunai K, et al. Nucleotide sequence of the rodA gene, responsible for the rod shape of Escherichia coli: rodA and the pbpA gene, encoding penicillin-binding protein 2, constitute the rodA operon. J Bacteriol. 1989;171:558–560. doi: 10.1128/jb.171.1.558-560.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrici D, Prigozhin DM, Alber T. Mycobacterium tuberculosis RpfE crystal structure reveals a positively charged catalytic cleft. Protein Sci. 2014;23:481–487. doi: 10.1002/pro.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrogiorgos N, Mekasha S, Yang Y, et al. Activation of NOD receptors by Neisseria gonorrhoeae modulates the innate immune response. Innate Immun. 2014;20:377–389. doi: 10.1177/1753425913493453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Sham LT, Kimsey H, et al. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci USA. 2015;112:6437–6442. doi: 10.1073/pnas.1504967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel U, Höltje JV, Vollmer W. Overproduction of inactive variants of the murein synthase PBP1B causes lysis in Escherichia coli. J Bacteriol. 2003;185:5342–5348. doi: 10.1128/JB.185.18.5342-5348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mett H, Keck W, Funk A, et al. Two different species of murein transglycosylase in Escherichia coli. J Bacteriol. 1980;144:45–52. doi: 10.1128/jb.144.1.45-52.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroshnikov KA, Faizullina NM, Sykilinda NN, et al. Properties of the endolytic transglycosylase encoded by gene 144 of Pseudomonas aeruginosa bacteriophage phiKZ. Biochemistry (Mosc) 2006;71:300–305. doi: 10.1134/s0006297906030102. [DOI] [PubMed] [Google Scholar]
- Mishima M, Shida T, Yabuki K, et al. Solution structure of the peptidoglycan binding domain of Bacillus subtilis cell Wall lytic enzyme CwlC: characterization of the sporulation-related repeats by NMR. Biochemistry. 2005;44:10153–10163. doi: 10.1021/bi050624n. [DOI] [PubMed] [Google Scholar]
- Mohammadi T, Sijbrandi R, Lutters M, et al. Specificity of the transport of lipid II by FtsW in Escherichia coli. J Biol Chem. 2014;289:14707–14718. doi: 10.1074/jbc.M114.557371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, van Dam V, Sijbrandi R, et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011;30:1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir A. How do spores germinate? J Appl Microbiol. 2006;101:526–530. doi: 10.1111/j.1365-2672.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- Moir A, Cooper G. Spore germination. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.TBS-0014-2012. TBS0014-2012. [DOI] [PubMed] [Google Scholar]
- Moriyama R, Fukuoka H, Miyata S, et al. Expression of a germination-specific amidase, SleB, of Bacilli in the forespore compartment of sporulating cells and its localization on the exterior side of the cortex in dormant spores. J Bacteriol. 1999;181:2373–2378. doi: 10.1128/jb.181.8.2373-2378.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama R, Hattori A, Miyata S, et al. A gene (sleB) encoding a spore cortex-lytic enzyme from Bacillus subtilis and response of the enzyme to L-alanine-mediated germination. J Bacteriol. 1996;178:6059–6063. doi: 10.1128/jb.178.20.6059-6063.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot C, Uehara T, Marquis KA, et al. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Genes Dev. 2010;24:411–422. doi: 10.1101/gad.1878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynie L, Schnell R, McMahon SA, et al. The AEROPATH project targeting Pseudomonas aeruginosa: crystallographic studies for assessment of potential targets in early-stage drug discovery. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69:25–34. doi: 10.1107/S1744309112044739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan PJ, Clarke AJ. O-Acetylated peptidoglycan: controlling the activity of bacterial autolysins and lytic enzymes of innate immune systems. Int J Biochem Cell Biol. 2011;43:1655–1659. doi: 10.1016/j.biocel.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Mueller CA, Broz P, Cornelis GR. The type III secretion system tip complex and translocon. Mol Microbiol. 2008;68:1085–1095. doi: 10.1111/j.1365-2958.2008.06237.x. [DOI] [PubMed] [Google Scholar]
- Mueller CA, Broz P, Müller SA, et al. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science. 2005;310:674–676. doi: 10.1126/science.1118476. [DOI] [PubMed] [Google Scholar]
- Mulder L, Lefebvre B, Cullimore J, et al. LysM domains of Medicago truncatula NFP protein involved in Nod factor perception. Glycosylation state, molecular modeling and docking of chitooligosaccharides and Nod factors. Glycobiology. 2006;16:801–809. doi: 10.1093/glycob/cwl006. [DOI] [PubMed] [Google Scholar]
- Müller S, Wolf AJ, Iliev ID, et al. Poorly cross-linked peptidoglycan in MRSA due to mecA induction activates the inflammasome and exacerbates immunopathology. Cell Host Microbe. 2015;18:604–612. doi: 10.1016/j.chom.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao M, Yukishige K, Kondo M, et al. Novel morphological changes in Gram-negative bacteria caused by combination of bulgecin and cefmenoxime. Antimicrob Agents Chemother. 1986;30:414–417. doi: 10.1128/aac.30.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudorf KD, Yost CK. An uncharacterized gene coding a conserved lytic transglycosylase domain (RL4716) is required for proper cell envelope function in Rhizobium leguminosarum. FEMS Microbiol Lett. 2017;364 doi: 10.1093/femsle/fnx035. fnx035. [DOI] [PubMed] [Google Scholar]
- Nikolaidis I, Izore T, Job V, et al. Calcium-dependent complex formation between PBP2 and lytic transglycosylase SltB1 of Pseudomonas aeruginosa. Microb Drug Resist. 2012;18:298–305. doi: 10.1089/mdr.2012.0006. [DOI] [PubMed] [Google Scholar]
- Nocadello S, Minasov G, Shuvalova LS, et al. Crystal structures of the SpoIID lytic transglycosylases essential for bacterial sporulation. J Biol Chem. 2016;291:14915–14926. doi: 10.1074/jbc.M116.729749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J. Review on antimicrobial resistance 2016 [Google Scholar]
- Paidhungat M, Ragkousi K, Setlow P. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca(2+)-dipicolinate. J Bacteriol. 2001;183:4886–4893. doi: 10.1128/JB.183.16.4886-4893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis-Bleau C, Cloutier I, Lemieux L, et al. Peptidoglycan lytic activity of the Pseudomonas aeruginosa phage phiKZ gp144 lytic transglycosylase. FEMS Microbiol Lett. 2007;266:201–209. doi: 10.1111/j.1574-6968.2006.00523.x. [DOI] [PubMed] [Google Scholar]
- Paredes-Sabja D, Setlow P, Sarker MR. SleC is essential for cortex peptidoglycan hydrolysis during germination of spores of the pathogenic bacterium Clostridium perfringens. J Bacteriol. 2009;191:2711–2720. doi: 10.1128/JB.01832-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes-Sabja D, Torres JA, Setlow P, et al. Clostridium perfringens spore germination: characterization of germinants and their receptors. J Bacteriol. 2008;190:1190–1201. doi: 10.1128/JB.01748-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JT. Turnover and recycling of the murein sacculus in oligopeptide permease-negative strains of Escherichia coli: indirect evidence for an alternative permease system and for a monolayered sacculus. J Bacteriol. 1993;175:7–11. doi: 10.1128/jb.175.1.7-11.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JT. Why does Escherichia coli recycle its cell wall peptides? Mol Microbiol. 1995;17:421–426. doi: 10.1111/j.1365-2958.1995.mmi_17030421.x. [DOI] [PubMed] [Google Scholar]
- Park JT, Burman L. FL-1060: a new penicillin with a unique mode of action. Biochem Biophys Res Commun. 1973;51:863–868. doi: 10.1016/0006-291x(73)90006-5. [DOI] [PubMed] [Google Scholar]
- Pazos M, Peters K, Vollmer W. Robust peptidoglycan growth by dynamic and variable multi-protein complexes. Curr Opin Microbiol. 2017;36:55–61. doi: 10.1016/j.mib.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Philpott DJ, Sorbara MT, Robertson SJ, et al. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol. 2014;14:9–23. doi: 10.1038/nri3565. [DOI] [PubMed] [Google Scholar]
- Pisabarro AG, de Pedro MA, Vazquez D. Structural modifications in the peptidoglycan of Escherichia coli associated with changes in the state of growth of the culture. J Bacteriol. 1985;161:238–242. doi: 10.1128/jb.161.1.238-242.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham DL. Specialized peptidoglycan of the bacterial endospore: the inner wall of the lockbox. Cell Mol Life Sci. 2002;59:426–433. doi: 10.1007/s00018-002-8435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potluri LP, de Pedro MA, Young KD. Escherichia coli low-molecular-weight penicillin-binding proteins help orient septal FtsZ, and their absence leads to asymmetric cell division and branching. Mol Microbiol. 2012;84:203–224. doi: 10.1111/j.1365-2958.2012.08023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AJ, Liu ZJ, Nicholas RA, et al. Crystal structures of the lytic transglycosylase MltA from N. gonorrhoeae and E. coli: Insights into interdomain movements and substrate binding. J Mol Biol. 2006;359:122–136. doi: 10.1016/j.jmb.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Pratt RF. β-Lactamases: why and how. J Med Chem. 2016;59:8207–8220. doi: 10.1021/acs.jmedchem.6b00448. [DOI] [PubMed] [Google Scholar]
- Ragland SA, Schaub RE, Hackett KT, et al. Two lytic transglycosylases in Neisseria gonorrhoeae impart resistance to killing by lysozyme and human neutrophils. Cell Microbiol. 2017;19:e12662. doi: 10.1111/cmi.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid CW, Blackburn NT, Clarke AJ. The effect of NAG-thiazoline on morphology and surface hydrophobicity of Escherichia coli. FEMS Microbiol Lett. 2004a;234:343–348. doi: 10.1016/j.femsle.2004.03.047. [DOI] [PubMed] [Google Scholar]
- Reid CW, Blackburn NT, Clarke AJ. Role of arginine residues in the active site of the membrane-bound lytic transglycosylase B from Pseudomonas aeruginosa. Biochemistry. 2006;45:2129–2138. doi: 10.1021/bi052342t. [DOI] [PubMed] [Google Scholar]
- Reid CW, Blackburn NT, Legaree BA, et al. Inhibition of membrane-bound lytic transglycosylase B by NAG-thiazoline. FEBS Lett. 2004b;574:73–79. doi: 10.1016/j.febslet.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Reid CW, Legaree BA, Clarke AJ. Role of Ser216 in the mechanism of action of membrane-bound lytic transglycosylase B: further evidence for substrate-assisted catalysis. FEBS Lett. 2007;581:4988–4992. doi: 10.1016/j.febslet.2007.09.037. [DOI] [PubMed] [Google Scholar]
- Reith J, Mayer C. Peptidoglycan turnover and recycling in Gram-positive bacteria. Appl Microbiol Biotechnol. 2011;92:1–11. doi: 10.1007/s00253-011-3486-x. [DOI] [PubMed] [Google Scholar]
- Roeder W, Somerville RL. Cloning the trpR gene. Mol Gen Genet. 1979;176:361–368. doi: 10.1007/BF00333098. [DOI] [PubMed] [Google Scholar]
- Romeis T, Höltje JV. Specific interaction of penicillin-binding proteins 3 and 7/8 with soluble lytic transglycosylase in Escherichia coli. J Biol Chem. 1994;269:21603–21607. [PubMed] [Google Scholar]
- Rosenthal RS, Nogami W, Cookson BT, et al. Major fragment of soluble peptidoglycan released from growing Bordetella pertussis is tracheal cytotoxin. Infect Immun. 1987;55:2117–2120. doi: 10.1128/iai.55.9.2117-2120.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roure S, Bonis M, Chaput C, et al. Peptidoglycan maturation enzymes affect flagellar functionality in bacteria. Mol Microbiol. 2012;86:845–856. doi: 10.1111/mmi.12019. [DOI] [PubMed] [Google Scholar]
- Rozeboom HJ, Dijkstra BW, Engel H, et al. Crystallization of the soluble lytic transglycosylase from Escherichia coli K12. J Mol Biol. 1990;212:557–559. doi: 10.1016/0022-2836(90)90221-7. [DOI] [PubMed] [Google Scholar]
- Rued BE, Zheng JJ, Mura A, et al. Suppression and synthetic-lethal genetic relationships of ΔgpsB mutations indicate that GpsB mediates protein phosphorylation and penicillin-binding protein interactions in Streptococcus pneumoniae D39. Mol Microbiol. 2017;103:931–957. doi: 10.1111/mmi.13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero A, Squeglia F, Romano M, et al. The structure of resuscitation promoting factor B from M. tuberculosis reveals unexpected ubiquitin-like domains. Biochim Biophys Acta. 2016;1860:445–451. doi: 10.1016/j.bbagen.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Ruiz N. Filling holes in peptidoglycan biogenesis of Escherichia coli. Curr Opin Microbiol. 2016;34:1–6. doi: 10.1016/j.mib.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santin YG, Cascales E. Domestication of a housekeeping transglycosylase for assembly of a type VI secretion system. EMBO Rep. 2017;18:138–149. doi: 10.15252/embr.201643206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub RE, Chan YA, Lee M, et al. Lytic transglycosylases LtgA and LtgD perform distinct roles in remodeling, recycling and releasing peptidoglycan in Neisseria gonorrhoeae. Mol Microbiol. 2016;102:865–881. doi: 10.1111/mmi.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers DJ, Tol MB. Lipid II: just another brick in the wall. PLoS Pathog. 2015;11:e1005213. doi: 10.1371/journal.ppat.1005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheurwater E, Reid CW, Clarke AJ. Lytic transglycosylases: bacterial space-making autolysins. Int J Biochem Cell Biol. 2008;40:586–591. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Scheurwater EM, Burrows LL. Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiol Lett. 2011;318:1–9. doi: 10.1111/j.1574-6968.2011.02228.x. [DOI] [PubMed] [Google Scholar]
- Scheurwater EM, Clarke AJ. The C-terminal domain of Escherichia coli YfhD functions as a lytic transglycosylase. J Biol Chem. 2008;283:8363–8373. doi: 10.1074/jbc.M710135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt LS, Botta G, Park JT. Effects of furazlocillin, a β-lactam antibiotic which binds selectively to penicillin-binding protein 3, on Escherichia coli mutants deficient in other penicillin-binding proteins. J Bacteriol. 1981;145:632–637. doi: 10.1128/jb.145.1.632-637.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebaihia M, Wren BW, Mullany P, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- Setlow B, Peng L, Loshon CA, et al. Characterization of the germination of Bacillus megaterium spores lacking enzymes that degrade the spore cortex. J Appl Microbiol. 2009;107:318–328. doi: 10.1111/j.1365-2672.2009.04210.x. [DOI] [PubMed] [Google Scholar]
- Setlow P. Spore germination. Curr Opin Microbiol. 2003;6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- Setlow P. Germination of spores of Bacillus species: what we know and do not know. J Bacteriol. 2014;196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton DL, St-Onge RJ, Haiser HJ, et al. Resuscitation-promoting factors are cell wall-lytic enzymes with important roles in the germination and growth of Streptomyces coelicolor. J Bacteriol. 2015;197:848–860. doi: 10.1128/JB.02464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham LT, Butler EK, Lebar MD, et al. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345:220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa S, Maki M, Kintaka K, et al. Isolation and characterization of bulgecins, new bacterial metabolites with bulge-inducing activity. J Antibiot. 1985;38:17–23. doi: 10.7164/antibiotics.38.17. [DOI] [PubMed] [Google Scholar]
- Sieger B, Schubert K, Donovan C, et al. The lipid II flippase RodA determines morphology and growth in Corynebacterium glutamicum. Mol Microbiol. 2013;90:966–982. doi: 10.1111/mmi.12411. [DOI] [PubMed] [Google Scholar]
- Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalweit MJ, Li M. Bulgecin A as a β-lactam enhancer for carbapenem-resistant Pseudomonas aeruginosa and carbapenem-resistant Acinetobacter baumannii clinical isolates containing various resistance mechanisms. Drug Des Devel Ther. 2016;10:3013–3020. doi: 10.2147/DDDT.S110193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorbara MT, Philpott DJ. Peptidoglycan: a critical activator of the mammalian immune system during infection and homeostasis. Immunol Rev. 2011;243:40–60. doi: 10.1111/j.1600-065X.2011.01047.x. [DOI] [PubMed] [Google Scholar]
- Squeglia F, Romano M, Ruggiero A, et al. Carbohydrate recognition by RpfB from Mycobacterium tuberculosis unveiled by crystallographic and MD analyses. Biophys J. 2013;104:2530–2539. doi: 10.1016/j.bpj.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strating H, Clarke AJ. Differentiation of bacterial autolysins by zymogram analysis. Anal Biochem. 2001;291:149–154. doi: 10.1006/abio.2001.5007. [DOI] [PubMed] [Google Scholar]
- Stütz AE, Wrodnigg TM. Imino sugars and glycosyl hydrolases: historical context, current aspects, emerging trends. Adv Carbohydr Chem Biochem. 2011;66:187–298. doi: 10.1016/B978-0-12-385518-3.00004-3. [DOI] [PubMed] [Google Scholar]
- Suarez G, Romero-Gallo J, Piazuelo MB, et al. Modification of Helicobacter pylori peptidoglycan enhances NOD1 activation and promotes cancer of the stomach. Cancer Res. 2015;75:1749–1759. doi: 10.1158/0008-5472.CAN-14-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Lowe J. Do the divisome and elongasome share a common evolutionary past? Curr Opin Microbiol. 2013;16:745–751. doi: 10.1016/j.mib.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Taylor A, Das BC, Van Heijenoort J. Bacterial cell wall peptidoglycan fragments produced by phage λ or Vi II endolysin and containing 1,6-anhydro-N-acetylmuramic acid. Eur J Biochem. 1975;53:47–54. [Google Scholar]
- Templin MF, Edwards DH, Höltje JV. A murein hydrolase is the specific target of bulgecin in Escherichia coli. J Biol Chem. 1992;267:20039–20043. [PubMed] [Google Scholar]
- Templin MF, Ursinus A, Höltje JV. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 1999;18:4108–4117. doi: 10.1093/emboj/18.15.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thunnissen AM, Dijkstra AJ, Kalk KH, et al. Doughnut-shaped structure of a bacterial muramidase revealed by x-ray crystallography. Nature. 1994;367:750–753. doi: 10.1038/367750a0. [DOI] [PubMed] [Google Scholar]
- Thunnissen AM, Isaacs NW, Dijkstra BW. The catalytic domain of a bacterial lytic transglycosylase defines a novel class of lysozymes. Proteins. 1995a;22:245–258. doi: 10.1002/prot.340220305. [DOI] [PubMed] [Google Scholar]
- Thunnissen AM, Rozeboom HJ, Kalk KH, et al. Structure of the 70-kDa soluble lytic transglycosylase complexed with bulgecin A. Implications for the enzymatic mechanism. Biochemistry. 1995b;34:12729–12737. doi: 10.1021/bi00039a032. [DOI] [PubMed] [Google Scholar]
- Tokuda H, Matsuyama S. Sorting of lipoproteins to the outer membrane in E. coli. Biochim Biophys Acta. 2004;1693:5–13. doi: 10.1016/j.bbamcr.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Tsui HC, Zheng JJ, Magallon AN, et al. Suppression of a deletion mutation in the gene encoding essential PBP2b reveals a new lytic transglycosylase involved in peripheral peptidoglycan synthesis in Streptococcus pneumoniae D39. Mol Microbiol. 2016;100:1039–1065. doi: 10.1111/mmi.13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, et al. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Park JT. Growth of Escherichia coli: significance of peptidoglycan degradation during elongation and septation. J Bacteriol. 2008;190:3914–3922. doi: 10.1128/JB.00207-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Suefuji K, Valbuena N, et al. Recycling of the anhydro-N-acetylmuramic acid derived from cell wall murein involves a two-step conversion to N-acetylglucosamine-phosphate. J Bacteriol. 2005;187:3643–3649. doi: 10.1128/JB.187.11.3643-3649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursinus A, Höltje JV. Purification and properties of a membrane-bound lytic transglycosylase from Escherichia coli. J Bacteriol. 1994;176:338–343. doi: 10.1128/jb.176.2.338-343.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamani G, Thomas MD, Patel TR, et al. The β-lactamase gene regulator AmpR Is a tetramer that recognizes and binds the D-Ala-D-Ala motif of its repressor UDP-N-acetylmuramic Acid (MurNAc)-pentapeptide. J Biol Chem. 2015;290:2630–2643. doi: 10.1074/jbc.M114.618199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Asselt EJ, Dijkstra AJ, Kalk KH, et al. Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand. Structure. 1999a;7:1167–1180. doi: 10.1016/s0969-2126(00)80051-9. [DOI] [PubMed] [Google Scholar]
- van Asselt EJ, Dijkstra BW. Binding of calcium in the EF-hand of Escherichia coli lytic transglycosylase Slt35 is important for stability. FEBS Lett. 1999;458:429–435. doi: 10.1016/s0014-5793(99)01198-9. [DOI] [PubMed] [Google Scholar]
- van Asselt EJ, Kalk KH, Dijkstra BW. Crystallographic studies of the interactions of Escherichia coli lytic transglycosylase Slt35 with peptidoglycan. Biochemistry. 2000;39:1924–1934. doi: 10.1021/bi992161p. [DOI] [PubMed] [Google Scholar]
- van Asselt EJ, Perrakis A, Kalk KH, et al. Accelerated x-ray structure elucidation of a 36 kDa muramidase/transglycosylase using wARP. Acta Crystallogr D Biol Crystallogr. 1998;54:58–73. doi: 10.1107/s0907444997010330. [DOI] [PubMed] [Google Scholar]
- van Asselt EJ, Thunnissen AM, Dijkstra BW. High resolution crystal structures of the Escherichia coli lytic transglycosylase Slt70 and its complex with a peptidoglycan fragment. J Mol Biol. 1999b;291:877–898. doi: 10.1006/jmbi.1999.3013. [DOI] [PubMed] [Google Scholar]
- van Heijenoort J. Peptidoglycan hydrolases of Escherichia coli. Microbiol Mol Biol Rev. 2011;75:636–663. doi: 10.1128/MMBR.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Straaten KE, Barends TR, Dijkstra BW, et al. Structure of Escherichia coli lytic transglycosylase MltA with bound chitohexaose: Implications for peptidoglycan binding and cleavage. J Biol Chem. 2007;282:21197–21205. doi: 10.1074/jbc.M701818200. [DOI] [PubMed] [Google Scholar]
- Van Straaten KE, Dijkstra BW, Thunnissen AM. Purification, crystallization and preliminary x-ray analysis of the lytic transglycosylase MltA from Escherichia coli. Acta Crystallogr D Biol Crystallogr. 2004;60:758–760. doi: 10.1107/S0907444904002574. [DOI] [PubMed] [Google Scholar]
- van Straaten KE, Dijkstra BW, Vollmer W, et al. Crystal structure of MltA from Escherichia coli reveals a unique lytic transglycosylase fold. J Mol Biol. 2005;352:1068–1080. doi: 10.1016/j.jmb.2005.07.067. [DOI] [PubMed] [Google Scholar]
- Viala J, Chaput C, Boneca IG, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- Viollier PH, Shapiro L. A lytic transglycosylase homologue, PleA, is required for the assembly of pili and the flagellum at the Caulobacter crescentus cell pole. Mol Microbiol. 2003;49:331–345. doi: 10.1046/j.1365-2958.2003.03576.x. [DOI] [PubMed] [Google Scholar]
- Vocadlo DJ, Davies GJ, Laine R, et al. Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature. 2001;412:835–838. doi: 10.1038/35090602. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta. 2008;1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Seligman SJ. Architecture of peptidoglycan: more data and more models. Trends Microbiol. 2010;18:59–66. doi: 10.1016/j.tim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Vollmer W, von Rechenberg M, Höltje JV. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J Biol Chem. 1999;274:6726–6734. doi: 10.1074/jbc.274.10.6726. [DOI] [PubMed] [Google Scholar]
- von Rechenberg M, Ursinus A, Höltje JV. Affinity chromatography as a means to study multienzyme complexes involved in murein synthesis. Microb Drug Resist. 1996;2:155–157. doi: 10.1089/mdr.1996.2.155. [DOI] [PubMed] [Google Scholar]
- Walmagh M, Boczkowska B, Grymonprez B, et al. Characterization of five novel endolysins from Gram-negative infecting bacteriophages. Appl Microbiol Biotechnol. 2013;97:4369–4375. doi: 10.1007/s00253-012-4294-7. [DOI] [PubMed] [Google Scholar]
- Wang N, Hasegawa H, Huang C-y, et al. Synthesis of peptidoglycan fragments from Enterococcus faecalis with Fmoc-strategy for glycan elongation. Chem–Asian J. 2017;12:27–30. doi: 10.1002/asia.201601357. [DOI] [PubMed] [Google Scholar]
- Ward DV, Draper O, Zupan JR, et al. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc Natl Acad Sci USA. 2002;99:11493–11500. doi: 10.1073/pnas.172390299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt SR, Clarke AJ. Initial characterization of two extracellular autolysins from Pseudomonas aeruginosa PAO1. J Bacteriol. 1994;176:4784–4789. doi: 10.1128/jb.176.15.4784-4789.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weadge JT, Clarke AJ. Identification and characterization of O-acetylpeptidoglycan esterase: a novel enzyme discovered in Neisseria gonorrhoeae. Biochemistry. 2006;45:839–851. doi: 10.1021/bi051679s. [DOI] [PubMed] [Google Scholar]
- Weaver LH, Grütter MG, Matthews BW. The refined structures of goose lysozyme and its complex with a bound trisaccharide show that the “goose-type” lysozymes lack a catalytic aspartate residue. J Mol Biol. 1995;245:54–68. doi: 10.1016/s0022-2836(95)80038-7. [DOI] [PubMed] [Google Scholar]
- Wheeler R, Chevalier G, Eberl G, et al. The biology of bacterial peptidoglycans and their impact on host immunity and physiology. Cell Microbiol. 2014;16:1014–1023. doi: 10.1111/cmi.12304. [DOI] [PubMed] [Google Scholar]
- Wiedemann B, Pfeifle D, Wiegand I, et al. β-Lactamase induction and cell wall recycling in Gram-negative bacteria. Drug Resist Updat. 1998;1:223–226. doi: 10.1016/s1368-7646(98)80002-2. [DOI] [PubMed] [Google Scholar]
- Wientjes FB, Woldringh CL, Nanninga N. Amount of peptidoglycan in cell walls of gram-negative bacteria. J Bacteriol. 1991;173:7684–7691. doi: 10.1128/jb.173.23.7684-7691.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AH, Wheeler R, Thiriau C, et al. Bulgecin A: the key to a broad spectrum inhibitor that targts lytic translgycosylases. Antibiotics. 2017;6:8. [Google Scholar]
- Wohlkonig A, Huet J, Looze Y, et al. Structural relationships in the lysozyme superfamily: significant evidence for glycoside hydrolase signature motifs. PLoS One. 2010;5:e15388. doi: 10.1371/journal.pone.0015388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-J, Huang Y-W, Lin Y-T, et al. Inactivation of lytic transglycosylases increases susceptibility to aminoglycosides and macrolides by altering the outer membrane permeability of Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2016;60:3236–3239. doi: 10.1128/AAC.03026-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahashiri A, Jorgenson MA, Weiss DS. Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proc Natl Acad Sci USA. 2015;112:11347–11352. doi: 10.1073/pnas.1508536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahashiri A, Jorgenson MA, Weiss DS. The SPOR domain: A widely-conserved peptidoglycan binding domain that targets proteins to the site of cell division. J Bacteriol. 2017;199:e00118–17. doi: 10.1128/JB.00118-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Blázquez B, Hesek D, et al. Inhibitors for bacterial cell-wall recycling. ACS Med Chem Lett. 2012;3:238–242. doi: 10.1021/ml2002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lyu Z, Miguel A, et al. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science. 2017;355:744–747. doi: 10.1126/science.aak9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD. Bacterial shape: two-dimensional questions and possibilities. Annu Rev Microbiol. 2010;64:223–240. doi: 10.1146/annurev.micro.112408.134102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunck R, Cho H, Bernhardt TG. Identification of MltG as a potential terminase for peptidoglycan polymerization in bacteria. Mol Microbiol. 2016;99:700–718. doi: 10.1111/mmi.13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahrl D, Wagner M, Bischof K, et al. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology. 2005;151:3455–3467. doi: 10.1099/mic.0.28141-0. [DOI] [PubMed] [Google Scholar]
- Zeng X, Gillespie B, Lin J. Important role of a putative lytic transglycosylase Cj0843c in β-lactam resistance in Campylobacter jejuni. Front Microbiol. 2015;6:1292. doi: 10.3389/fmicb.2015.01292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Chen S, Recsei P. A dye release assay for determination of lysostaphin activity. Anal Biochem. 1988;171:141–144. doi: 10.1016/0003-2697(88)90134-0. [DOI] [PubMed] [Google Scholar]
- Zou Y, Lei W, He Z, et al. The role of NOD1 and NOD2 in host defense against chlamydial infection. FEMS Microbiol Lett. 2016;363 doi: 10.1093/femsle/fnw170. fnw170. [DOI] [PubMed] [Google Scholar]
- Zupan J, Hackworth CA, Aguilar J, et al. VirB1* promotes T-pilus formation in the vir-Type IV secretion system of Agrobacterium tumefaciens. J Bacteriol. 2007;189:6551–6563. doi: 10.1128/JB.00480-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


