The United Nations Environment Programme (UNEP) convened the Minamata Convention on Mercury in 2013 to ban mercury-containing products in order to ensure human and environmental health. It will be effectuated in 2020 to discontinue use of low-pressure mercury lamps and new UV-emitting sources have to replace this conventional technology. However, the UV germicidal irradiation (UVGI) system still uses conventional UV lamps, and no research has been conducted for air disinfection using UVC LEDs. The research reported here investigated the inactivation effect of aerosolized microorganisms, including viruses, bacteria, and fungi, with an UVC LED module. The results can be utilized as a primary database to replace conventional UV lamps with UVC LEDs, a novel type of UV emitter. Implementation of UVC LED technology is truly expected to significantly reduce the extent of global mercury contamination, and this study provides important baseline data to help ensure a healthier environment and increased health for humanity.
KEYWORDS: UVC LED, air disinfection, aerosolized microorganism, pathogens, viruses, fungi, nebulizing, inactivation rate constant
ABSTRACT
In this study, the possibility of inactivating viral, bacterial, and fungal aerosols in a chamber-type air disinfection system by using a UVC light-emitting-diode (LED) array was investigated and inactivation rate constants of each microorganism were calculated in fitting curves of surviving populations. UVC LED array treatment effectively inactivated viral infectivity, achieving 5-log reductions within 45 mJ/cm2 for MS2, Qβ, and ϕX174 viruses. UVC LED array effectiveness in inactivating Escherichia coli O157:H7, Salmonella enterica serovar Typhimurium, Listeria monocytogenes, and Staphylococcus aureus aerosols achieved 2.5- to 4-log reductions within 1.5 to 4.6 mJ/cm2. Also, 4-log reductions of Aspergillus flavus and Alternaria japonica were achieved at a dosage of 23 mJ/cm2 using UVC LED array irradiation. The highest UV susceptibility, represented by the inactivation rate constant, was calculated for bacteria, followed by fungi and viruses. UVC LED, an innovative technology, can effectively inactivate microorganisms regardless of taxonomic classification and can sufficiently substitute for conventional mercury UV lamps.
IMPORTANCE The United Nations Environment Programme (UNEP) convened the Minamata Convention on Mercury in 2013 to ban mercury-containing products in order to ensure human and environmental health. It will be effectuated in 2020 to discontinue use of low-pressure mercury lamps and new UV-emitting sources have to replace this conventional technology. However, the UV germicidal irradiation (UVGI) system still uses conventional UV lamps, and no research has been conducted for air disinfection using UVC LEDs. The research reported here investigated the inactivation effect of aerosolized microorganisms, including viruses, bacteria, and fungi, with an UVC LED module. The results can be utilized as a primary database to replace conventional UV lamps with UVC LEDs, a novel type of UV emitter. Implementation of UVC LED technology is truly expected to significantly reduce the extent of global mercury contamination, and this study provides important baseline data to help ensure a healthier environment and increased health for humanity.
INTRODUCTION
Microorganism transmission via air holds an important position in nosocomial infections with viruses, bacteria, and fungi (1, 2). Exposure to airborne fungi, such as Aspergillus, Alternaria, and Cladosporium, is related to several respiratory diseases (3–5). Fungal contamination of indoor air is continuous irrespective of seasonal and regional change (6–9). Severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 and swine influenza virus H1N1 in 2009 stimulated numerous research studies of indoor air disinfection and development of air purification systems to control pathogenic microorganisms, including viruses, bacteria, and fungi (10). In order to ensure microbial safety of air, many studies utilizing UV irradiation have been performed (11–14). The Centers for Disease Control and Prevention recommended UV germicidal irradiation (UVGI) as a supplementary process in order to prevent transmission of bacteria causing tuberculosis (15). UVC light is well known to possess a very powerful germicidal effect capable of inactivating a wide spectrum of microorganisms, such as viruses, bacteria, protozoa, fungi, yeasts, and algae, through the formation of pyrimidine dimers, the photoproducts of genetic materials (16, 17). Dimerization of pyrimidine disturbs DNA replication and transcription, which leads to cell death (18, 19). Until now, UV irradiation has mostly been performed with conventional low-pressure mercury UV lamps (LP lamps), which emit a 254-nm peak wavelength.
Approximately 140 representatives from the United Nations Environment Programme (UNEP) in 2013 approved the Minamata Convention on Mercury, an international treaty to maintain public health and protect the environment from mercury pollution. This treaty regulates the manufacture of mercury-containing products and the import/export of mercury, reducing the amount of mercury released into the environment in order to prevent the spread of mercury contamination. When the Minamata Convention becomes implemented in 2020, the use of conventional low-pressure mercury UV lamps will be prohibited and new alternative UV emission technologies must be utilized by industry.
UVC light-emitting diodes (LEDs) are gaining popularity as an alternative technology which can overcome the limitations of conventional mercury-containing UV lamps. For example, their small size makes them easy to incorporate into a sterilization system and, above all, they do not contain mercury, thus alleviating risks of human and environmental toxicity (20). Shin et al. reported that UVC LED intensity was not influenced by temperature change and no warm-up time was required for maximum intensity output, whereas LP lamps had decreased output intensity at low temperature and a warm-up time of about 5 min was required (21). In addition, UVC LEDs showed much higher inactivating efficacy against Escherichia coli O157:H7, Salmonella enterica serovar Typhimurium, and Listeria monocytogenes than LP lamps at the same dosages with intensity adjustment (22). MS2, Qβ, and ϕX174, human enteric virus surrogates, effectively lost their infectivity in batch and continuous-type water disinfection systems incorporating UVC LED arrays (23).
In this study, we investigated the inactivation of nebulized microorganisms, including viruses, bacteria, and fungi, by using an UVC LED array in a chamber-type air disinfection system. Also, inactivation rate constants (k values) were calculated in order to analyze the inactivation efficacy of this novel UVC irradiation system.
RESULTS AND DISCUSSION
Nebulizing efficacy of microorganisms.
Viral populations after nebulization but without UVC treatment (Table 1) were evaluated to help determine the optimum nebulization time for each microbial type, because particle size is an important factor affecting nebulization efficacy. For viruses, including MS2, Qβ, and ϕX174, a gradual increase in population was observed as nebulization time increased up to 5 min, beyond which no statistically significant further increase occurred (P > 0.05). Approximately 8, 7, and 5.5 log PFU/vol (27 liters) were measured after 5 min of nebulization for MS2, Qβ, and ϕX174, respectively. Therefore, 5 min of nebulization was chosen for further research with viruses. Optimum nebulizing times for bacteria and fungi were assessed in the same way and were determined to be 5 min and 15 min, respectively, for each of these groups of microorganisms (data not shown).
TABLE 1.
Aerosolization efficacy associated with nebulization time in viruses
| Virus | Viral population relative to nebulization time (log PFU/vola)b |
||||
|---|---|---|---|---|---|
| 1 min | 3 min | 5 min | 7 min | 10 min | |
| MS2 | 4.88 ± 0.21 A | 7.07 ± 0.34 B | 8.23 ± 0.60 C | 8.15 ± 0.04 C | 8.30 ± 0.17 C |
| Qβ | 3.84 ± 0.23 A | 5.66 ± 0.31 B | 7.22 ± 0.03 C | 7.17 ± 0.05 C | 7.00 ± 0.26 C |
| ϕX174 | 3.95 ± 0.18 A | 4.64 ± 0.12 AB | 5.52 ± 0.38 C | 5.34 ± 0.14 BC | 5.35 ± 0.37 BC |
Log PFU per unit volume of the chamber (27 liters).
Data represent means ± standard deviations from three replications. Values followed by the same uppercase letters within rows are not significantly different (P > 0.05).
Because of differences in particle or cell sizes, nebulizing efficacy differed among microorganism types: viruses > bacteria > fungi. Mass median aerodynamic diameter (MMAD) is the level of aerodynamic diameter in which half of an aerosol is associated with particles smaller than the MMAD and half is associated with particles larger than the MMAD. Virus diameters were approximately 30 nm, and the MMAD of the system was 2.44 μm, so the viruses were easily nebulized by capture with water aerosols almost 100 times larger. Bacterial cells and fungal spores had much larger diameters, about 2 μm and 3 to 5 μm, respectively. Nebulizing efficacy against bacteria and fungi was dramatically decreased, to 0.1 to 0.01% of its efficacy against viruses.
Microbial populations of nontreated controls.
Table 2 shows surviving populations of each microorganism relative to nebulization time without UVC LED irradiation. Levels of nebulized populations of each microorganism in accordance with circulating times remained constant in the air circulating chamber, and there were no significant population differences relative to circulation time (P > 0.05). Populations of MS2 and Qβ viruses ranged from 7 to 8 log PFU/vol, and that of ϕX174 was 5 log PFU/vol with 0- to 10-min circulation times. Populations of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium were 4 log CFU/vol, while those of Listeria monocytogenes and Staphylococcus aureus were 5 to 6 log CFU/vol. Aspergillus flavus and Alternaria japonica maintained populations of 4 to 5 log CFU/vol, which were similar to bacterial population levels.
TABLE 2.
Surviving populations of viruses, bacteria, and fungi subjected to nebulization and circulation without UVC LED irradiation
| Organism | Surviving population of microorganisms relative to nebulization time without UVC LED treatment (log PFU/vola)b | ||||
|---|---|---|---|---|---|
| Viruses | 0 min | 1 min | 3 min | 5 min | 10 min |
| MS2 | 8.23 ± 0.60 A | 8.00 ± 0.46 A | 7.97 ± 0.30 A | 8.06 ± 0.42 A | 7.93 ± 0.34 A |
| Qβ | 7.22 ± 0.03 A | 6.97 ± 0.38 A | 6.90 ± 0.42 A | 6.81 ± 0.57 A | 6.85 ± 0.64 A |
| ϕX174 | 5.22 ± 0.08 A | 5.16 ± 0.08 A | 5.20 ± 0.11 A | 5.19 ± 0.40 A | 5.27 ± 0.47 A |
| Bacteria | 0 s | 15 s | 30 s | 45 s | 60 s |
| E. coli O157:H7 | 4.20 ± 0.17 A | 4.26 ± 0.16 A | 4.25 ± 0.31 A | 4.21 ± 0.10 A | 4.17 ± 0.15 A |
| S. Typhimurium | 4.06 ± 0.10 A | 4.13 ± 0.16 A | 4.12 ± 0.11 A | 4.15 ± 0.09 A | 4.22 ± 0.11 A |
| L. monocytogenes | 5.54 ± 0.15 A | 5.39 ± 0.26 A | 5.44 ± 0.11 A | 5.45 ± 0.21 A | 5.53 ± 0.51 A |
| S. aureus | 6.03 ± 0.25 A | 6.15 ± 0.16 A | 6.02 ± 0.18 A | 6.01 ± 0.22 A | 5.95 ± 0.20 A |
| Fungi | 0 min | 1 min | 2 min | 3 min | 5 min |
| As. flavus | 4.87 ± 0.03 A | 4.78 ± 0.12 A | 4.63 ± 0.24 A | 4.77 ± 0.27 A | 4.51 ± 0.37 A |
| Al. japonica | 5.23 ± 0.24 A | 5.21 ± 0.49 A | 5.32 ± 0.60 A | 5.20 ± 0.45 A | 5.12 ± 0.14 A |
Log PFU per unit volume of the chamber (27 liters).
Data represent means ± standard deviations from three replications. Values followed by the same letters within rows are not significantly different (P > 0.05).
The forced airflow generated by the fan was ca. 96.8 m3/h (26.9 liters/s), in which the whole volume (27 liters) of air in the isolated chamber system was convected and circulated each second. This powerful air circulation prevented nebulized cells from settling onto the chamber floor.
Surviving populations after UVC LED irradiation.
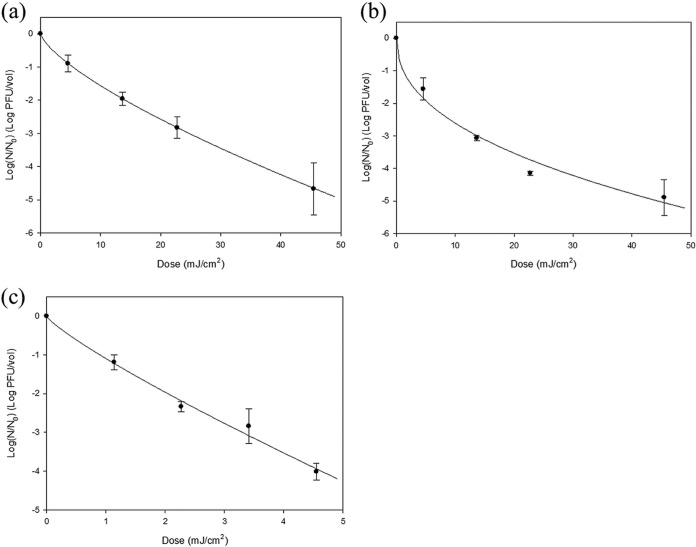
Surviving populations of the three viruses, MS2, Qβ, and ϕX174, after UVC LED array irradiation and their fitting curves are presented in Fig. 1. As UV dosage increased, infectious populations of the three viruses decreased. MS2 lost its infectivity almost linearly with increased dosages, and a 4.7-log reduction was achieved at 46 mJ/cm2 of UVC LED irradiation. A similar result was obtained for Qβ at the same irradiation dosage, achieving a 4.9-log reduction, while much upward concavity developed. ϕX174 lost its infectivity at a much lower dosage of UV irradiation, as a 4-log reduction was achieved with 4.6 mJ/cm2. These viral reductions accomplished by UVC LED array irradiation were much more efficacious than in prior research studies using conventional UV lamps. For MS2, a 0.5-log reduction (70% reduction) was achieved after dosage of 2.6 mJ/cm2 using a low-pressure mercury lamp at 50% relative humidity (RH) (14). Griffiths et al. reported a 1.6-log reduction of MS2 by a heating, ventilation, and air conditioning (HVAC) system incorporating 6 UV lamps (no irradiation dose specified) (24). Also, an approximately 3-log reduction of MS2 dispersed in beef extract medium solution and artificial saliva occurred when treated with 7,200 mJ/cm2 in an air disinfection system (25).
FIG 1.
Plotting and analysis of the surviving infectivity of nebulized MS2 (a), Qβ (b), and ϕX174 (c) after UVC LED array irradiation using the Weibull model equation. The experimental procedure included 5 min of nebulization, UVC treatment, and 5 min of air sampling with an AGI-30 glass impinger. The error bars indicate SDs.
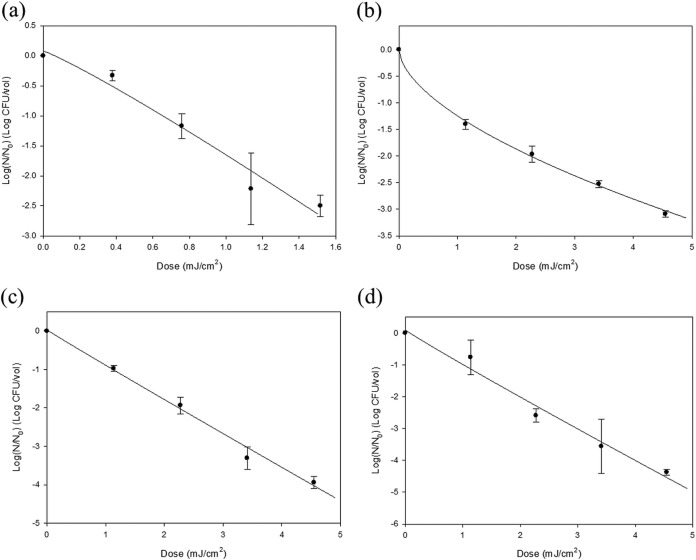
Surviving populations and fitted curves for E. coli O157:H7, S. Typhimurium, L. monocytogenes, and S. aureus are presented in Fig. 2. Like for the viruses, increased UVC irradiation dosages resulted in decreased bacterial populations. An approximately 2.5-log reduction was achieved at a dosage of 1.5 mJ/cm2 for E. coli O157:H7, with 3- to 4.5-log reductions at 4.6 mJ/cm2 for the other bacterial pathogens. Only the survival curve of S. Typhimurium showed upward concavity among the pathogenic bacteria, and an almost linear decrease was observed for the other bacterial pathogens. In the case of bacterial inactivation by UVC LED array irradiation, there was superior efficacy in dose-response value. Lin and Li reported that a 2-log reduction of nebulized E. coli was achieved by conventional UV lamp (UVGI system) irradiation at 2.4 mJ/cm2 at 80% RH (26). Also, aerosols of S. aureus were reduced by 3 logs at a dosage of 2,300 mJ/cm2 with a UVGI system, 30-liter/min airflow, and 87.3 to 90% RH (27). These bacteria are associated with foodborne illness; our results provide a significant baseline of data to help ensure biological food safety in processing.
FIG 2.
Plotting and analysis of the surviving populations of nebulized E. coli O157:H7 (a), S. Typhimurium (b), L. monocytogenes (c), and S. aureus (d) after UVC LED array irradiation using the Weibull model equation. The experimental procedure included 5 min of nebulization, UVC treatment, and 5 min of air sampling with an AGI-30 glass impinger. The error bars indicate SDs.
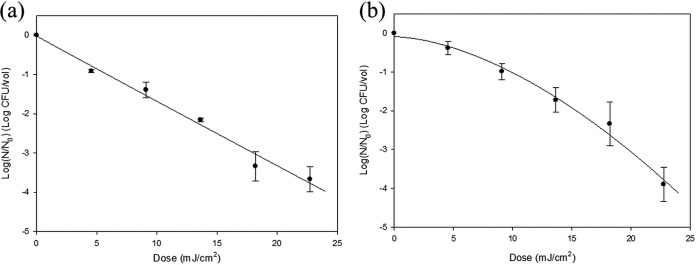
Figure 3 shows surviving populations of As. flavus and Al. japonica relative to UVC LED irradiation dosage. Both fungi needed a dosage of 23 mJ/cm2 for achieving approximately 4-log reductions. A linear fitting line was developed for As. flavus, while a downward concave curve was constructed for Al. japonica. Fungi required irradiation dosages approximately 5 times higher than for bacteria for accomplishing 4-log reductions, because eukaryotic cells show resistance to UVC treatment (28–30). As with other microorganisms, inactivating fungal aerosols by UVC LED irradiation showed higher efficiency than with conventional UV lamps. For 2-log reduction of Penicillium citrinum, low-pressure mercury UV lamp dosages ranging from 12.9 to 17.5 mJ/cm2 were needed (26). Brickner et al. reported that 0.03- to 0.08-log reductions of several types of fungal spores in air were achieved with a LP lamp dosage of 1 mJ/cm2 in their review article (31).
FIG 3.
Plotting and analysis of the surviving populations of nebulized As. flavus (a) and Al. japonica (b) after UVC LED array irradiation using the Weibull model equation. The experimental procedure included 15 min of nebulization, UVC treatment, and 5 min of air sampling with an AGI-30 glass impinger. The error bars indicate SDs.
Inactivation rate constant measurement.
The inactivation rate constant, k, the level of reduction after treatment with unit irradiation dosage (1 mJ/cm2), was calculated from the slopes of the log linear-tail fitting equations and presented in Table 3.
TABLE 3.
Inactivation rate constant and calculated D5d values for microorganisms following UVC LED irradiationa
| Organism | Inactivation rate constant, k (cm2/mJ)b | D5dc |
|---|---|---|
| Viruses | ||
| MS2 | 0.28 ± 0.02 D | 51.55 ± 10.20 A |
| Qβ | 0.44 ± 0.05 D | 42.16 ± 4.95 B |
| ϕX174 | 2.02 ± 0.22 C | 6.07 ± 0.43 DE |
| Bacteria | ||
| E. coli O157:H7 | 4.71 ± 0.90 A | 2.60 ± 0.16 E |
| S. Typhimurium | 1.90 ± 0.17 C | 10.61 ± 1.02 D |
| L. monocytogenes | 2.23 ± 0.19 BC | 5.60 ± 0.13 DE |
| S. aureus | 2.64 ± 0.38 B | 4.99 ± 0.18 DE |
| Fungi | ||
| As. flavus | 0.40 ± 0.03 D | 30.42 ± 3.57 C |
| Al. japonica | 0.38 ± 0.05 D | 26.88 ± 1.37 C |
Data represent means ± standard deviations from three replications. Values followed by the same letters within columns are not significantly different (P > 0.05).
k was determined from the first linear stage of surviving population lines applied to the log linear-tail model equation.
D5d values indicate UV dosages necessary for a 5-log reduction obtained from the Weibull model equation.
For MS2, Qβ, and ϕX174, k values of 0.28, 0.44, and 2.02 cm2/mJ were calculated, respectively, and ϕX174 showed the highest UV sensitivity among the studied viruses. This result is consistent with a study investigating loss of viral infectivity following UVC LED water and air disinfection in which ϕX174 demonstrated higher sensitivity than MS2 (23, 25, 32, 33). Dimerization of thymine in DNA and uracil in RNA by UVC irradiation occurred not to the same degree, because absorption spectra of the nucleotides showed different peak wavelengths. Also, viruses do not contain a DNA repair system. Therefore, DNA viruses are more vulnerable to UVC irradiation than are RNA viruses (34).
Statistically much higher k values were calculated for bacteria than for MS2 and Qβ viruses (P < 0.05). E. coli O157:H7 had the highest k value (4.7 cm2/mJ) and S. Typhimurium had the lowest k value (1.9 cm2/mJ), and significant differences between those values (P < 0.05) were observed. Inactivation rate constants of L. monocytogenes and S. aureus were 2.23 and 2.64 cm2/mJ (P > 0.05), respectively, and these values fell between those of the Gram-negative bacteria in our study. Except for ϕX174, MS2 and Qβ had inactivation rate constants 5 to 10% that of bacteria (P < 0.05), which means that it was 10 to 20 times more difficult to inactivate viruses than bacteria. MS2 and Qβ, which have particle diameters ranging from 20 to 30 nm, could easily evade the 280-nm UVC wavelength, the germicidal wavelength of the UVC LED array, while the bacteria, having sizes ranging up to 2 μm, were fully exposed to UVC irradiation. Therefore, the inactivation rate constants had size-dependent characteristics.
Fungal k values, as well, were 0.38 and 0.40 cm2/mJ, which were 10 to 20% of bacterial k values and similar to MS2 and Qβ k values. As. flavus has conidia measuring 3 to 6 μm and Al. japonica has conidia measuring 18 to 83 by 7 to 18 μm (35, 36), so these fungi have substantially larger cells than the other microorganisms in our study and thus cannot avoid UVC irradiation. However, eukaryotes demonstrated greater UV resistance than prokaryotes (bacteria), and as a consequence, inactivation rate constants had cell classification-dependent characteristics.
In this study, investigating the microbicidal effect of UVC LEDs, a novel inactivation method, was performed to control aerosolized viruses, bacteria, and fungi. UVC LEDs effectively inactivated nebulized microorganisms in the chamber, and different UV susceptibilities were observed relative to taxonomic classification of microorganisms. These results can be utilized as a fundamental database for ensuring microbial air safety.
MATERIALS AND METHODS
UVC LED array.
Figure 4a shows the UVC LED arrays utilized in this study. Sixteen UVC LED package chips (LG Innotek Co., Seoul, Republic of Korea) were connected and arrayed linearly to electronic printed circuit boards (PCB; 250 by 25 mm). The chips were attached at an 11-mm distance from each other, and a 40-mm blank space on both sides was retained for connection to a cooling panel. Approximately 1.6 A was applied so that a voltage of 12 V was obtained at the PCB. Irradiation flow rate was measured with a spectrophotometer (Avaspec-ULS2048-USB2-UA-50; Avantes, Netherlands) which was calibrated for the entire UV spectrum from 200 to 400 nm. For averaging the UV irradiation intensity, distance between the probe and UVC LED array was maintained at 15 cm, which was half the height of the chamber. Even distribution of UV intensity on a hypothetical square area (15 cm away from the UVC LED array) was quantified by petri factor calculation. Intensities of the 16 spots covering the area were scanned by the spectrometer and measured. The value of each point was divided by the maximum fluence rate and averaged to obtain the petri factor. Modified UV intensity was calculated by multiplying the maximum intensity by the petri factor, so that averaged UVC LED fluence rate was represented by the calculated value (21–23).
FIG 4.
Photograph of the UVC LED array (a) and schematic diagram of the chamber-type air disinfection system utilizing an UVC LED array for nebulization (b) and air sampling (c).
Experimental setup.
A schematic diagram of an air disinfection system is presented in Fig. 4b and c. An acrylic chamber (30 by 30 by 30 cm) was constructed to contain the system. The UVC LED array was attached to the center of the lid, and a fan was also attached to one end of the UVC module to generate forced convection current in the isolated system. The nominal airflow of the fan was 96.8 m3/h, which replaced the whole volume of air in the chamber each second. For effective circulation of the microbially contaminated air, the lower section of the chamber had a ball-type valve to insert or extract nebulized microorganisms and water vapor.
Preparation of microorganisms.
Bacteriophage MS2 (ATCC 15597-B1), Qβ (ATCC 23631-B1), and ϕX174 (ATCC 13706-B1) and the host strains, Escherichia coli C3000 (ATCC 15597) and Escherichia coli CN13 (ATCC 700609), were obtained from the Culture Collection at Seoul National University (Seoul, Republic of Korea). The phages were propagated by inoculating 50 ml of tryptic soy broth (TSB) (Difco, Becton Dickinson and Company, Sparks, MD) with 100 μl of each stock coliphage and 500 μl of a late-exponential or early-stationary-phase host strain (E. coli C3000 for MS2 and Qβ and E. coli CN13 for ϕX174). After incubation overnight at 37°C, cultures were centrifuged for 20 min at 4,000 × g, and the supernatant was carefully collected into sterile falcon tubes and stored at −70°C until investigation.
Three strains each of Escherichia coli O157:H7 (ATCC 35150, ATCC 43889, and ATCC 43890), Salmonella enterica serovar Typhimurium (ATCC 19585, ATCC 43971, and DT 104), Listeria monocytogenes (ATCC 19111, ATCC 19115, and ATCC 15313), and Staphylococcus aureus (ATCC 25923, ATCC 27213, and ATCC 29273) were obtained from the Food Science and Human Nutrition culture collection at Seoul National University (Seoul, Republic of Korea). Stock cultures were kept frozen at −80°C in 0.7 ml of TSB (Difco) and 0.3 ml of 50% glycerol. Working cultures were streaked onto tryptic soy agar (TSA; MB Cell, Seoul, Republic of Korea), incubated at 37°C for 24 h, and stored at 4°C.
Aspergillus flavus ATCC 46110 and Alternaria japonica ATCC 44897, which are often detected in indoor environments, were obtained from the Korean Culture Center of Microorganisms (KCCM). Working cultures of fungi were streaked onto yeast mold agar medium (YM agar; Difco) adjusted to pH 3.0 with lactic acid (Daejung, Gyeonggi-do, Republic of Korea), incubated at 25°C for 3 to 7 days, and stored at 4°C.
Preparation of microbial suspensions for nebulization.
Four-milliliter aliquots of bacteriophage suspensions prepared as described in the previous section were dispensed into the nebulizing container (atomizer).
All bacterial strains were cultured in 5 ml of TSB at 37°C for 24 h and harvested by centrifugation at 4,000 × g for 20 min at 4°C. Pelleted cells were washed three times with sterile 0.2% peptone water (PW; Bacto; Becton, Dickinson and Company; Sparks, MD), and the final pellets were resuspended in 9 ml of PW, corresponding to approximately 108 to 109 CFU/ml. The mixed-pathogen culture cocktail was produced by combining the resuspended cell pellets, and 4 ml of the bacterial suspension was delivered to the atomizer.
Conidial (spore) suspensions of each fungal strain were prepared using glass beads (425 to 600 μm; Sigma-Aldrich Corp., St. Louis, MO) and 0.1% Tween 80 (Sigma-Aldrich Corp.) solution. Three grams of glass beads and 20 ml of 0.1% Tween 80 solution were transferred to pH 3.0 YM agar cultures of each fungus, and the agar media were vigorously agitated using the Spindle, an apparatus that detaches microorganisms from surfaces with rotational and vibrational force (37, 38). After 2 min of agitation, the fungal suspension was transferred to a sterile 50-ml centrifuge tube and 4 ml of the solution was delivered to the atomizer for nebulization.
Nebulization and UVC LED treatment.
Microbial nebulization was conducted with an air jet piston compressor nebulizer (BD5002; Bremed, Kowloon, Hong Kong) in which high-velocity-compressed airflow generated aerosols from liquid suspensions. This nebulizer produced aerosolized particles with a mass median aerodynamic diameter (MMAD) of 2.44 μm. Air pressure for the nebulizer was maintained at 150 kPa (21.8-lb/in.2 gauge; approximately 1.5-fold higher than atmospheric pressure). The 4 ml of microbial suspension in the atomizer was nebulized for 5, 10, or 15 min for viruses, bacteria, or fungi, respectively, because nebulizing efficacy differed according to the criterion of microorganism cell size. During nebulization, the fan next to the UVC LED array was operated to generate forced convection to prevent aerosols from settling on the chamber floor. The air recirculated during UVC LED treatment.
After the nebulization step, the nebulizer was stopped and the air containing microbial particles was irradiated with the UVC LED array for up to 10 min for viruses, 1 min for bacteria, and 5 min for fungi with the fan still running.
The treated air-microorganism mixture was drawn from the air disinfection chamber into a sampling unit, which consisted of a 30-ml glass impinger (AGI-30) and mini vacuum pump (AS30; Haosheng Pneumatic, Ningbo Zheniang, China). Vacuum was maintained at 650 mm Hg so that a pumping airflow of 35 to 40 liters/min was generated. The impinger contained 10 ml of sterile PW for sampling the nebulized microorganisms by resuspending them in suspension. After a 5-min sampling time, 10-fold serial dilutions were performed in 9.0 ml of PW.
Between every nebulizing-treatment-sampling operation, decontamination by spraying 70% alcohol onto the inside and outside of the chamber was conducted. The sanitizer was removed with sterile Kimtech wipes (Kimberly-Clark, UK), and the decontaminated chamber was placed in the hood for 10 min to evaporate remaining alcohol.
The viruses were analyzed using the soft agar overlay plaque assay method (23, 39, 40). Quantities of 0.01 ml of host bacterial cultures (E. coli C3000 and E. coli CN13) incubated overnight were transferred to 5 ml of TSB and incubated for 4 h at 37°C. Early-exponential-phase host bacteria were used to prepare a lawn on the bottom agar layer (TSA), and LB broth (Difco) with 1% (wt/vol) agar (Difco) was used for the soft agar overlay. Then selected sample diluents were aliquoted to 5 ml of soft agar tempered to 48°C in which 100 μl of the corresponding log-phase host bacterium was added, and the soft agar was gently vortexed and poured onto the bottom agar layer. After solidification, the plates were incubated at 37°C for 24 h and typical plaques were enumerated.
Selective media were used for analyzing surviving populations of nebulized bacteria: sorbitol MacConkey agar (SMAC; Oxoid) for E. coli O157:H7, xylose lysine desoxycholate agar (XLD; Oxoid) for S. Typhimurium, Oxford agar base with antimicrobial supplement (OAB; MB Cell) for L. monocytogenes, and Baird-Parker agar (BPA; Difco) for S. aureus. After UVC LED treatment and air sampling, 0.1-ml quantities of appropriate diluents were spread plated onto each selective media. The plates were incubated at 37°C for 24 h to 48 h.
The same portions (0.1 ml) of fungal sample diluents were spread onto pH 3.0-adjusted YM agar medium (Difco) and incubated at 25°C for 3 to 5 days, and typical colonies were enumerated following incubation.
Control samples of all nebulized microorganisms were collected after recirculating for selected treatment times but without UVC LED treatment in order to ascertain if microbial inactivation occurred during air circulation.
Fitting to model equations.
All experiments were duplicated and replicated three times, and surviving microorganism populations following air disinfection treatment were fitted with the Weibull and log linear-tail model equations using GInaFiT (23, 41). The Weibull model equation (42) is equation 1, described as
| (1) |
where α represents the treatment dosage to achieve a 1-log reduction of a certain microorganism at the first stage of the inactivation process and β represents the shape of the surviving population line, such as upward concavity of a curve when β < 1, downward concavity when β > 1, and a linear curve when β = 1.
The log linear-tail model equation (43) is equation 2, described as
| (2) |
where kmax is a specific inactivation rate constant in the linear section and Nres refers to the remaining population cell density after treatment.
In order to evaluate wellness of the model, equations fitting root mean square error (RMSE) and R2 were analyzed and presented in Table 4 (44). The UV dosage necessary for achieving a 5-log reduction by UVC LED irradiation from the Weibull equation (D5d) was assessed by Microsoft Excel 2010. Description of the model equation in terms of independent variables followed by calculating dosage for the 5-log reduction by the “Goal seek” function (23) was performed.
TABLE 4.
Goodness of fit of kinetic model equations for surviving populations of viruses, bacteria, and fungi in a chamber-type air disinfection system using UVC LED array irradiation
| Organism | Weibull |
Log linear-tail |
||
|---|---|---|---|---|
| RMSEa | R2 | RMSE | R2 | |
| Viruses | ||||
| MS2 | 0.1354 | 0.9972 | 0.2062 | 0.9935 |
| Qβ | 0.4058 | 0.9710 | 0.4184 | 0.9775 |
| ϕX174 | 0.2166 | 0.9847 | 0.2595 | 0.9858 |
| Bacteria | ||||
| E. coli O157:H7 | 0.3074 | 0.9557 | 0.2768 | 0.9688 |
| S. Typhimurium | 0.0742 | 0.9978 | 0.2854 | 0.9710 |
| L. monocytogenes | 0.2295 | 0.9874 | 0.1695 | 0.9945 |
| S. aureus | 0.5059 | 0.9599 | 0.3116 | 0.9857 |
| Fungi | ||||
| As. flavus | 0.2442 | 0.9823 | 0.2179 | 0.9859 |
| Al. japonica | 0.2188 | 0.9835 | 0.4175 | 0.9489 |
RMSE, root mean square error.
Also, UV sensitivity (inactivation rate constant k), which means the level of log reduction when 1 mJ/cm2 of irradiation is imposed within the linear range of surviving population curves, was calculated. The early stage of the log linear-tail model was used for obtaining the k value.
Statistical analysis.
All experiments were duplicate plated and replicated three times. All data were analyzed with analysis of variance (ANOVA) using the Statistical Analysis System (SAS Institute, Cary, NC) and Duncan's multiple-range test to determine if there were significant differences (P < 0.05) in mean values of surviving populations of microorganisms, inactivation rate constants, or D5d values.
ACKNOWLEDGMENTS
We are grateful for technical support from LG Innotek.
This work was financially supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) through the Agriculture, Food and Rural Affairs Research Center Support Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (710012-03-1-HD220), and also supported by a National Research Foundation of Korea (NRF) grant funded by the Korea government (NRF-2018R1A2B2008825).
REFERENCES
- 1.Ferroni A, Werkhauser-Bertrand A, Le Bourgeois M, Beauvais R, Vrielynck S, Durand C, Lenoir G, Berche P, Sermet-Gaudelus I. 2008. Bacterial contamination in the environment of hospitalised children with cystic fibrosis. J Cyst Fibros 7:477–482. doi: 10.1016/j.jcf.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Spencker F, Haupt S, Claros M, Walter S, Lietz T, Schille R, Rodloff A. 2000. Epidemiologic characterization of Pseudomonas aeruginosa in patients with cystic fibrosis. Clin Microbiol Infect 6:600–607. doi: 10.1046/j.1469-0691.2000.00171.x. [DOI] [PubMed] [Google Scholar]
- 3.Alberti C, Bouakline A, Ribaud P, Lacroix C, Rousselot P, Leblanc T, Derouin F. 2001. Relationship between environmental fungal contamination and the incidence of invasive aspergillosis in haematology patients. J Hosp Infect 48:198–206. doi: 10.1053/jhin.2001.0998. [DOI] [PubMed] [Google Scholar]
- 4.Faure O, Fricker-Hidalgo H, Lebeau B, Mallaret M, Ambroise-Thomas P, Grillot R. 2002. Eight-year surveillance of environmental fungal contamination in hospital operating rooms and haematological units. J Hosp Infect 50:155–160. doi: 10.1053/jhin.2001.1148. [DOI] [PubMed] [Google Scholar]
- 5.Perdelli F, Cristina M, Sartini M, Spagnolo A, Dallera M, Ottria G, Lombardi R, Grimaldi M, Orlando P. 2006. Fungal contamination in hospital environments. Infect Control Hosp Epidemiol 27:44–47. [DOI] [PubMed] [Google Scholar]
- 6.Górny RL, Dutkiewicz J. 2002. Bacterial and fungal aerosols in indoor environment in Central and Eastern European countries. Ann Agric Environ Med 9:17–23. [PubMed] [Google Scholar]
- 7.Sautour M, Sixt N, Dalle F, L'Ollivier C, Fourquenet V, Calinon C, Paul K, Valvin S, Maurel A, Aho S. 2009. Profiles and seasonal distribution of airborne fungi in indoor and outdoor environments at a French hospital. Sci Total Environ 407:3766–3771. doi: 10.1016/j.scitotenv.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 8.Topbaş M, Tosun I, Çan G, Kaklikkaya N, Aydin F. 2006. Identification and seasonal distribution of airborne fungi in urban outdoor air in an eastern Black Sea Turkish town. Turk J Med Sci 36:31–36. [Google Scholar]
- 9.de Ana SG, Torres-Rodríguez J, Ramírez E, García S, Belmonte-Soler J. 2006. Seasonal distribution of Alternaria, Aspergillus, Cladosporium and Penicillium species isolated in homes of fungal allergic patients. J Investig Allergol Clin Immunol 16:357–363. [PubMed] [Google Scholar]
- 10.Xu Z, Wu Y, Shen F, Chen Q, Tan M, Yao M. 2011. Bioaerosol science, technology, and engineering: past, present, and future. Aerosol Sci Technol 45:1337–1349. doi: 10.1080/02786826.2011.593591. [DOI] [Google Scholar]
- 11.Kim J, Jang J. 2018. Inactivation of airborne viruses using vacuum ultraviolet photocatalysis for a flow-through indoor air purifier with short irradiation time. Aerosol Sci Technol 52:557–566. doi: 10.1080/02786826.2018.1431386. [DOI] [Google Scholar]
- 12.Ko G, First MW, Burge HA. 2002. The characterization of upper-room ultraviolet germicidal irradiation in inactivating airborne microorganisms. Environ Health Perspect 110:95. doi: 10.1289/ehp.02110s195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kujundzic E, Hernandez M, Miller SL. 2007. Ultraviolet germicidal irradiation inactivation of airborne fungal spores and bacteria in upper-room air and HVAC in-duct configurations. J Environ Eng Sci 6:1–9. doi: 10.1139/s06-039. [DOI] [Google Scholar]
- 14.Walker CM, Ko G. 2007. Effect of ultraviolet germicidal irradiation on viral aerosols. Environ Sci Technol 41:5460–5465. doi: 10.1021/es070056u. [DOI] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 2005. Guidelines for preventing the transmission of Mycobacterium tuberculosis in health-care settings, 2005. MMWR Recommend Rep 54(RR17):1–141. [PubMed] [Google Scholar]
- 16.Bintsis T, Litopoulou-Tzanetaki E, Robinson RK. 2000. Existing and potential applications of ultraviolet light in the food industry—a critical review. J Sci Food Agric 80:637–645. doi:. [DOI] [PubMed] [Google Scholar]
- 17.Yaun BR, Sumner SS, Eifert JD, Marcy JE. 2004. Inhibition of pathogens on fresh produce by ultraviolet energy. Int J Food Microbiol 90:1–8. doi: 10.1016/S0168-1605(03)00158-2. [DOI] [PubMed] [Google Scholar]
- 18.Franz CM, Specht I, Cho G-S, Graef V, Stahl MR. 2009. UV-C-inactivation of microorganisms in naturally cloudy apple juice using novel inactivation equipment based on Dean vortex technology. Food Control 20:1103–1107. doi: 10.1016/j.foodcont.2009.02.010. [DOI] [Google Scholar]
- 19.Guerrero-Beltrán J, Barbosa-Cánovas G. 2004. Advantages and limitations on processing foods by UV light. Food Sci Technol Int 10:137–147. doi: 10.1177/1082013204044359. [DOI] [Google Scholar]
- 20.Song K, Mohseni M, Taghipour F. 2016. Application of ultraviolet light-emitting diodes (UV-LEDs) for water disinfection: a review. Water Res 94:341–349. doi: 10.1016/j.watres.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Shin J-Y, Kim S-J, Kim D-K, Kang D-H. 2016. Fundamental characteristics of deep-UV light-emitting diodes and their application to control foodborne pathogens. Appl Environ Microbiol 82:2–10. doi: 10.1128/AEM.01186-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S-J, Kim D-K, Kang D-H. 2016. Using UVC light-emitting diodes at wavelengths of 266 to 279 nanometers to inactivate foodborne pathogens and pasteurize sliced cheese. Appl Environ Microbiol 82:11–17. doi: 10.1128/AEM.02092-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D-K, Kim S-J, Kang D-H. 2017. Inactivation modeling of human enteric virus surrogates, MS2, Qβ, and ΦX174, in water using UVC-LEDs, a novel disinfecting system. Food Res Int 91:115–123. doi: 10.1016/j.foodres.2016.11.042. [DOI] [PubMed] [Google Scholar]
- 24.Griffiths W, Bennett A, Speight S, Parks S. 2005. Determining the performance of a commercial air purification system for reducing airborne contamination using model micro-organisms: a new test methodology. J Hosp Infect 61:242–247. doi: 10.1016/j.jhin.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Woo M-H, Grippin A, Anwar D, Smith T, Wu C-Y, Wander JD. 2012. Effects of relative humidity and spraying medium on UV decontamination of filters loaded with viral aerosols. Appl Environ Microbiol 78:5781–5787. doi: 10.1128/AEM.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin C-Y, Li C-S. 2002. Control effectiveness of ultraviolet germicidal irradiation on bioaerosols. Aerosol Sci Technol 36:474–478. doi: 10.1080/027868202753571296. [DOI] [Google Scholar]
- 27.Chang CW, Li SY, Huang SH, Huang CK, Chen YY, Chen CC. 2013. Effects of ultraviolet germicidal irradiation and swirling motion on airborne Staphylococcus aureus, Pseudomonas aeruginosa, and Legionella pneumophila under various relative humidities. Indoor Air 23:74–84. doi: 10.1111/j.1600-0668.2012.00793.x. [DOI] [PubMed] [Google Scholar]
- 28.Anderson JG, Rowan NJ, MacGregor SJ, Fouracre RA, Farish O. 2000. Inactivation of food-borne enteropathogenic bacteria and spoilage fungi using pulsed-light. IEEE Trans Plasma Sci 28:83–88. doi: 10.1109/27.842870. [DOI] [Google Scholar]
- 29.Kim D-K, Kim S-J, Kang D-H. 2017. Bactericidal effect of 266 to 279 nm wavelength UVC-LEDs for inactivation of Gram positive and Gram negative foodborne pathogenic bacteria and yeasts. Food Res Int 97:280–287. doi: 10.1016/j.foodres.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Lonnen J, Kilvington S, Kehoe S, Al-Touati F, McGuigan K. 2005. Solar and photocatalytic disinfection of protozoan, fungal and bacterial microbes in drinking water. Water Res 39:877–883. doi: 10.1016/j.watres.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 31.Brickner PW, Vincent RL, First M, Nardell E, Murray M, Kaufman W. 2003. The application of ultraviolet germicidal irradiation to control transmission of airborne disease: bioterrorism countermeasure. Public Health Rep 118:99. doi: 10.1016/S0033-3549(04)50225-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aoyagi Y, Takeuchi M, Yoshida K, Kurouchi M, Yasui N, Kamiko N, Araki T, Nanishi Y. 2011. Inactivation of bacterial viruses in water using deep ultraviolet semiconductor light-emitting diode. J Environ Eng 137:1215–1218. doi: 10.1061/(ASCE)EE.1943-7870.0000442. [DOI] [Google Scholar]
- 33.Hijnen W, Beerendonk E, Medema GJ. 2006. Inactivation credit of UV radiation for viruses, bacteria and protozoan (oo)cysts in water: a review. Water Res 40:3–22. doi: 10.1016/j.watres.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 34.Kowalski W. 2010. Ultraviolet germicidal irradiation handbook: UVGI for air and surface disinfection. Springer Science & Business Media, Berlin, Germany. [Google Scholar]
- 35.Al-Doory Y, Domson JF. 1984. Mould allergy. Lea & Febiger, Philadelphia, PA. [Google Scholar]
- 36.Gravesen S, Wilken-Jensen K, Foundation for Allergy Research in Europe. 1984. Atlas of moulds in Europe causing respiratory allergy. Ask Publishing, Copenhagen, Denmark. [Google Scholar]
- 37.Kim D-K, Kim S-J, Kang D-H. 2015. Comparison of a four-section spindle and stomacher for efficacy of detaching microorganisms from fresh vegetables. J Food Prot 78:1380–1386. doi: 10.4315/0362-028X.JFP-15-003. [DOI] [PubMed] [Google Scholar]
- 38.Kim SJ, Kim DK, Kang DH. 2016. Evaluation of micro-organism-detaching efficacy from meat samples by spindle or stomacher treatment and quality analysis of suspensions. J Appl Microbiol 120:946–954. doi: 10.1111/jam.13075. [DOI] [PubMed] [Google Scholar]
- 39.Cho M, Gandhi V, Hwang T-M, Lee S, Kim J-H. 2011. Investigating synergism during sequential inactivation of MS-2 phage and Bacillus subtilis spores with UV/H2O2 followed by free chlorine. Water Res 45:1063–1070. doi: 10.1016/j.watres.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 40.Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. 2009. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol Biol 501:69–76. doi: 10.1007/978-1-60327-164-6_7. [DOI] [PubMed] [Google Scholar]
- 41.Geeraerd A, Valdramidis V, Van Impe J. 2005. GInaFiT, a freeware tool to assess non-log-linear microbial survivor curves. Int J Food Microbiol 102:95–105. doi: 10.1016/j.ijfoodmicro.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 42.van Boekel MA. 2002. On the use of the Weibull model to describe thermal inactivation of microbial vegetative cells. Int J Food Microbiol 74:139–159. doi: 10.1016/S0168-1605(01)00742-5. [DOI] [PubMed] [Google Scholar]
- 43.Geeraerd A, Herremans C, Van Impe J. 2000. Structural model requirements to describe microbial inactivation during a mild heat treatment. Int J Food Microbiol 59:185–209. doi: 10.1016/S0168-1605(00)00362-7. [DOI] [PubMed] [Google Scholar]
- 44.Chen Z, Zhu C. 2011. Modelling inactivation by aqueous chlorine dioxide of Dothiorella gregaria Sacc. and Fusarium tricinctum (Corda) Sacc. spores inoculated on fresh chestnut kernel. Lett Appl Microbiol 52:676–684. [DOI] [PubMed] [Google Scholar]