Bacillus thuringiensis Cry toxins rely on receptor binding to exert toxicity. Cry1Ca is toxic to different populations of S. frugiperda, a major corn pest in America. Nevertheless, the S. frugiperda midgut proteins that are involved in Cry1Ca toxicity have not been identified. Here we identified aminopeptidase N1 (APN1) as a functional receptor of Cry1Ca. Moreover, we showed that Cry1Ca domain III β16 is involved in APN1 binding. These results give insights on potential target sites for improving Cry1Ca toxicity to S. frugiperda.
KEYWORDS: Bacillus thuringiensis, Cry1Ca, Spodoptera frugiperda, aminopeptidase, mode of action, receptor
ABSTRACT
Bacillus thuringiensis Cry1Ca is toxic to different Spodoptera species. The aims of this work were to identify the Cry1Ca-binding proteins in S. frugiperda, to provide evidence on their participation in toxicity, and to identify the Cry1Ca amino acid residues involved in receptor binding. Pulldown assays using Spodoptera frugiperda brush border membrane vesicles (BBMV) identified aminopeptidase N (APN), APN1, and APN2 isoforms as Cry1Ca-binding proteins. Cry1Ca alanine substitutions in all residues of domain III β16 were characterized. Two β16 nontoxic mutants (V505A and S506A) showed a correlative defect on binding to the recombinant S. frugiperda APN1 (SfAPN1). Finally, silencing the expression of APN1 transcript, by double-stranded RNA (dsRNA) feeding, showed that silenced larvae are more tolerant of the Cry1Ca toxin, which induced less than 40% mortality in silenced larvae whereas nonsilenced larvae had 100% mortality. Overall, our results show that Cry1Ca relies on APN1 binding through domain III β16 to impart toxicity to S. frugiperda.
IMPORTANCE Bacillus thuringiensis Cry toxins rely on receptor binding to exert toxicity. Cry1Ca is toxic to different populations of S. frugiperda, a major corn pest in America. Nevertheless, the S. frugiperda midgut proteins that are involved in Cry1Ca toxicity have not been identified. Here we identified aminopeptidase N1 (APN1) as a functional receptor of Cry1Ca. Moreover, we showed that Cry1Ca domain III β16 is involved in APN1 binding. These results give insights on potential target sites for improving Cry1Ca toxicity to S. frugiperda.
INTRODUCTION
Spodoptera frugiperda, the fall armyworm, is a polyphagous pest that is the major corn pest in North and South America (1). In addition, S. frugiperda is an invasive pest that in recent years invaded cornfields in Africa, jeopardizing corn production in this continent (2). Bacillus thuringiensis (Bt) produces Cry toxins that are specifically active against larval stages of different insect pests. Among the Cry toxins, Cry1Ab and Cry1Ac have been introduced into several plant genomes, creating transgenic plants that are specifically active against different lepidopteran species (3). However, S. frugiperda is not effectively controlled by Cry1Ab or Cry1Ac toxins. To control S. frugiperda, corn plants expressing Cry1Fa have been commercialized (3). Nevertheless, different S. frugiperda populations have evolved resistance to Cry1Fa corn plants in Brazil, Argentina, Puerto Rico, and the United States (4–7). Thus, the use of alternative insecticidal proteins for S. frugiperda control that have a mode of action distinct from that of Cry1Fa is necessary.
Different Bt strains produce different Cry toxins that form crystal inclusions during the sporulation phase of growth. Among them, the three-domain Cry (3d-Cry) toxins, such as Cry1Ab, Cry1Ac, and Cry1Fa, are the best studied and best characterized (8). To kill the insect larvae, Bt crystals are solubilized in the midgut lumen of the larvae and activated by midgut proteases, yielding 60-to-65-kDa proteins with a three-domain structure. Domain I, a seven-α-helix bundle, has been shown to be involved in toxin oligomerization, membrane insertion, and pore formation, while domains II and III, mainly composed of β-sheets, are involved in binding to different insect midgut proteins that are important for triggering toxin oligomerization and pore formation (8). It was proposed that Cry toxins bind first to glycosylphosphatidylinositol (GPI)-anchored proteins such as aminopeptidase N (APN) and alkaline phosphatase (ALP) through domain III β16 attaching the activated toxin to the midgut microvilli (9, 10). After this interaction, Cry toxins bind to cadherin (CAD) protein through the exposed loop regions of domain II, which induces oligomerization of the toxin (11–13). Toxin oligomers bind to APN or ALP through domain II loop 2 to facilitate the insertion of the oligomer into the membrane to finally form pores that kill the cells by osmotic shock (13). In addition to these insect proteins, the ABCC2 transporters have also been shown to play important roles as functional receptors for different Cry toxins (14). It was shown that in Plutella xylostella larvae, the ABCC2 facilitates Cry1Ac oligomerization and also insertion of the oligomer into the membrane (15).
In addition to the Cry1Fa toxin, the Cry1Ca toxin has been shown to be effective for the control of S. frugiperda populations (16), since Cry1Ca toxins do not share binding sites with Cry1Fa toxins or Cry1A toxins (17). However, the mode of action of Cry1Ca, specifically, its interaction with midgut proteins, is unknown. In the case of Manduca sexta, an APN of 106 kDa, different from 115-kDa APN1, was shown to bind Cry1Ca (18, 19). In other Spodoptera species such as S. litura, APN1 has been shown to function as a receptor since a Cry1Ca-resistant population showed reduced levels of the APN1 transcript (20). In addition, Cry1Ca is known to interact with APN4 from S. litura expressed in Sf21 cells (21). Finally, CAD was shown to be involved in the toxicity of Cry1Ca to S. exigua by gene silencing using RNA interference (RNAi) (22).
Cry1Ca domain II loop 2 and 3 and domain III have been shown to be involved in receptor binding since different mutations diminish binding to S. exigua brush border membrane vesicles (BBMV) and showed reduced toxicity to S. exigua (23). Here we identified two Cry1Ca mutations that abolished toxicity and showed reduced binding to recombinant S. frugiperda APN1. In addition, pulldown experiments identified S. frugiperda APN1 and APN2 as Cry1Ca-binding proteins. Silencing the expression in S. frugiperda of APN1 by double-stranded RNA (dsRNA) showed that APN1 is involved in conferring Cry1Ca susceptibility in S. frugiperda. These data indicate that APN1 is a functional receptor of Cry1Ca in S. frugiperda.
RESULTS
Detection of Cry1Ca-binding proteins in S. frugiperda BBMV.
Qualitative binding of biotinylated Cry1Ca to S. frugiperda BBMV showed that binding was specific since unlabeled toxin competed with the binding of biotin-labeled Cry1Ca (data not shown). To identify the S. frugiperda Cry1Ca-binding proteins, the activated Cry1Ca was purified by ion-exchange chromatography after protoxin solubilization and digestion with trypsin. The anti-Cry1Ca antibody was coupled with protein A Sepharose beads and used to perform the pulldown assay. BBMV from third instar larvae of S. frugiperda were solubilized and incubated with Cry1Ca. This sample was incubated afterward with anti-Cry1Ca antibody coupled to protein A Sepharose beads. The protein A Sepharose beads were separated by centrifugation, and the unbound proteins were removed by extensive washing. The BBMV proteins that bind to Cry1Ca toxin were resolved in SDS-PAGE (Fig. 1). The whole-protein extract obtained by the pulldown procedure was concentrated by precipitation, treated with trypsin, and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Fig. 1). Table 1 shows the proteins from M. sexta, Bombyx mori, and S. frugiperda that were identified against the database. APN1, APN2, and trypsin were identified as Cry1Ca-binding proteins where APN1 showed 44% coverage with APN1 from M. sexta, while APN2 showed only 2.5% coverage with APN2 from B. mori (Table 1). For this reason, we decided to analyze whether APN1 has a function as receptor for Cry1Ca.
FIG 1.
SDS-PAGE analysis of S. frugiperda proteins after pulldown performed with Cry1Ca toxin. Cry1Ca was incubated with or without soluble BBMV proteins and then incubated with protein A Sepharose beads that were loaded with anti-Cry1Ca antibody. The beads were separated by centrifugation and washed. Cry1Ca-binding proteins were dissociated with acidic pH as described in Materials and Methods, This figure shows a sample of the eluate that was subjected to SDS-PAGE and the polyacrylamide gel that was subjected to silver staining. The numbers on the left refer to molecular weight standards (in kilodaltons). All of the proteins in the rest of the eluate were precipitated, trypsin digested, and sent for identification by LC-MS/MS as indicated in Materials and Methods. The Cry1Ca-binding proteins that were identified are listed in Table 1.
TABLE 1.
Identification of Cry1Ca-binding proteins in S. frugiperda by mass spectrometrya
| Sample | % query coverage | % identity | Accession no. | Peptide identified | Description |
|---|---|---|---|---|---|
| Cry1Ca-Sf | 44 | 100 | Q11001 | (1) TSTPLVMNQEYIIR | Aminopeptidase N1 (Manduca sexta) |
| (2) SWYVDR | |||||
| (3) QAFPCYDEPGFK | |||||
| (4) ATFDITMNR | |||||
| (5) EADFSPTISNMPIR | |||||
| (6) ATTTLTNGR | |||||
| (7) NNVGSQGDWSLEMGEK | |||||
| (8) QAAIPDFSAGAMENWGLLTYR | |||||
| (9) EALILYDPLNSNHHYRQ | |||||
| (10) MTQHLLSYDTFVK | |||||
| (11) GITIDAYFR | |||||
| (12) AGHPLLSVTVDHESGR | |||||
| (13) MTLVQAR | |||||
| (14) FPGLWHIPITWTR | |||||
| (15) AGAPDFENLKPSQVMTGQSLVIDR | |||||
| (16) QVSGFYR | |||||
| (17) SQIVDDVFQLAR | |||||
| (18) SGVMSYQR | |||||
| (19) ALNILSYLR | |||||
| (20) RFAHDAANLQTLQNQIIGLSEAVVARL | |||||
| (21) GFTEVSGGTYMTDLQR | |||||
| (22) LHVMQFLCNVGHQQCIDAGRQ | |||||
| (23) NFLNWRN | |||||
| (24) YGSAEDFNYFWNR | |||||
| (25) YIVEDLSNEK | |||||
| (26) VVMLEAAGCTR | |||||
| (27) FLNAIVSGNDDVRPQDHSSALSSAITSNDVNTMR | |||||
| (28) AFDWLTK | |||||
| (29) TLGSITSPLNTITSR | |||||
| (30) LLTEAQMTQVQTWLDANR | |||||
| (31) NTIGAAYNTGVNGIATSR | |||||
| (32) MSEFLR | |||||
| Cry1Ca-Sf | 4.3 | 100 | NP_001037013 | (1) QAFPCYDEPGFKATFDIMNR | Aminopeptidase N1 (Bombyx mori) |
| (2) QAAIPDFSAGAMENWGLLTYR | |||||
| Cry1Ca-Sf | 2.5 | 100 | AFK85018 | (1) QAFPSFDEPGFK | Aminopeptidase N2 (Bombyx mori) |
| (2) AQIVDDVFALMR | |||||
| Cry1Ca-Sf | 24 | 100 | ACR25157 | (1) IVGGSVTTIDR | Trypsin (Spodoptera frugiperda) |
| (2) AILTAAHCTVGDAANR | |||||
| (3) IINQATCR | |||||
| (4) GITITDNMLCSGWPTGGR | |||||
| (5) YTAWISSNA |
The proteins from the S. frugiperda BBMV that were pulled down with Cry1Ca (Cry1Ca-Sf) were precipitated, trypsin digested, and sent for identification by LC-MS/MS as indicated in Materials and Methods.
Cry1Ca domain III mutants affected in APN1 binding and in toxicity to S. frugiperda.
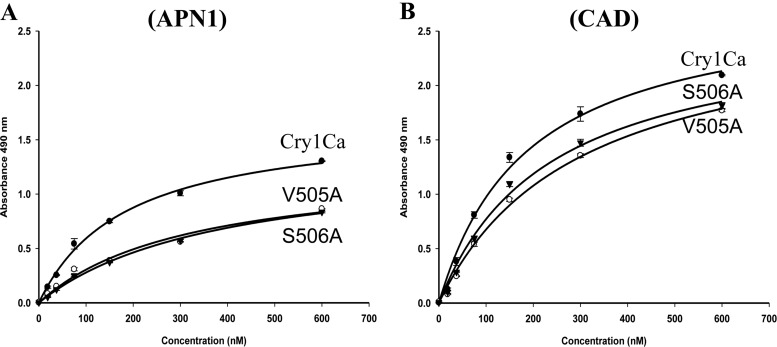
Previous work showed that a Cry1Ab mutant with a mutation in strand β16 of domain III was affected in binding to M. sexta ALP and APN and in toxicity to M. sexta (10). To gain more evidence on the role of APN1 in Cry1Ca toxicity to S. frugiperda, all β16 505VSLQVN510 residues of Cry1Ca were replaced by alanine. All mutants were produced as 130-kDa proteins that yielded a 60-kDa toxin band upon trypsin treatment, suggesting that the mutations did not affect the structure of the toxin (see Fig. S1 in the supplemental material). Table 2 shows that Cry1Ca V505A and S506A mutants were not toxic to S. frugiperda but were still effective against M. sexta, in contrast to all the other Cry1Ca mutants, which were not affected in toxicity. To determine if the loss of toxicity of the Cry1Ca V505A and S506A mutants was related to defects in binding to SfAPN1, the interactions of binding of Cry1Ca and these two mutants to the recombinant SfAPN1 and to a S. frugiperda CAD repeat fragment (SfCR7 to SfCR12) was analyzed by enzyme-linked immunosorbent assay (ELISA). The SfCR7–12 fragment contains the regions of CAD protein involved in Cry1Ab binding. Figure 2A shows that Cry1Ca bound SfAPN1 in a saturable manner with an apparent dissociation constant (Kd) of 173 ± 14.5 nM, confirming the pulldown assays that identified APN1 as a Cry1Ca-binding protein. In contrast, both the V505A and S506A mutants showed reduced binding to SfAPN1, with a 2-fold-higher apparent Kd value (Kd for V505A = 318 ± 22.5 nM; Kd for S506A = 388 ± 26.2 nM) showing extremely statistically significant differences from the Kd value determined for Cry1Ca (P values < 0.0007). Regarding binding to the SfCR7–12 fragment, Fig. 2B shows that Cry1Ca and the two Cry1Ca mutants also bound the CAD fragment in a saturable binding interaction (Kd for Cry1Ca = 188 ± 18.8 nM; Kd for V505A = 293 ± 61.2 nM; Kd for S506A = 233 ± 41.8 nM). The apparent Kd values with respect to CAD of the Cry1Ca mutant toxins (V505A and S506A) showed minimal or not statistically significant differences from the Kd value determined for Cry1Ca (P values of 0.05 and 0.16, respectively). These results show that Cry1Ca domain III mutants affected SfAPN1 binding.
TABLE 2.
Toxicity of Cry1Ca and Cry1Ca mutants against M. sexta and S. frugiperda larvae
| Strain/mutation | LC50 in ng/cm2 (95% confidence interval) |
|
|---|---|---|
| M. sexta | S. frugiperda | |
| Cry1Ca | 25 (18–35) | 122 (81–174) |
| V505A | 44 (27–100) | >2,500 |
| S506A | 90 (56–222) | >2,500 |
| L507A | 42 (27–83) | 335 (250–444) |
| Q508A | 43 (30–70) | 241 (141–416) |
| V509A | 16 (11–22) | 118 (36–218) |
| N510A | 33 (25–45) | 79 (54–109) |
FIG 2.
ELISAs of binding of Cry1Ca toxins to recombinant APN1 and CAD receptors. Totals of 2.5 μg each of APN1 (A) or CAD (B) proteins were fixed to ELISA plates independently and incubated with different concentrations of Cry1Ca or Cry1Ca mutants (V505A and S506A), and bound toxins were revealed with anti-Cry1Ca antibody. Nonspecific binding controls were run in parallel where the APN1 or CAD ligands were not included in the assays. The data shown here were obtained after subtracting the nonspecific binding controls. Results represent means of three repetitions. Binding to APN1 of Cry1Ca mutants showed statistically significant (P < 0.0007) differences from Cry1Ca as analyzed by t test.
Silencing S. frugiperda APN1.
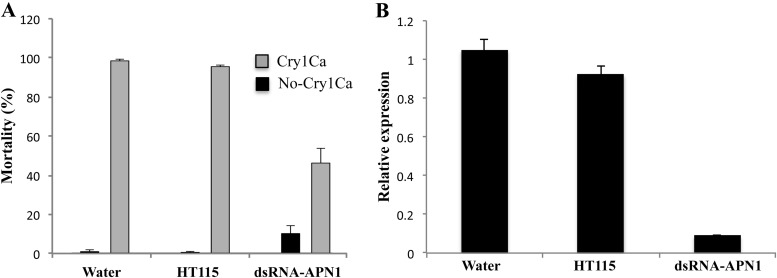
To analyze the potential role of APN1 as receptor of Cry1Ca in S. frugiperda, neonate larvae were fed with lysed HT115 Escherichia coli cells producing a dsRNA specific to the S. frugiperda APN1 gene sequence. After 12 h of APN1-dsRNA feeding, the larvae were exposed to 25 μg/cm2 of Cry1Ca. As a negative control, larvae were exposed to water or to lysed HT115 E. coli cells that did not express the APN1-dsRNA. Figure 3A shows that the larvae exposed to water or lysed HT115 E. coli cells showed more than 95% mortality when exposed to Cry1Ca toxin. In contrast, larvae fed with lysed HT115 E. coli cells producing the APN1-dsRNA showed a mortality rate that was reduced by 60% in the presence of Cry1Ca. To analyze the efficiency of APN1 silencing, the expression of APN1 protein was analyzed in individual larvae by real-time qPCR (Fig. 3B) or by Western blotting using an anti-APN1 antibody (Fig. 4). Larvae treated with APN1 dsRNA showed a reduction in the level of APN1 transcript of 80% in comparison to control larvae (Fig. 3B). Figure 4 shows that all larvae treated with water (Fig. 4A) or with HT115 E. coli cells (Fig. 4B) expressed a 170-kDa protein that cross-reacted with the anti-APN1 antibody. In the case of larvae fed with APN1-dsRNA and treated with water, three of seven larvae analyzed showed the 170-kDa APN1 band (Fig. 4C). In contrast, among the larvae that had been fed with APN1-dsRNA and that survived after Cry1Ca exposure, only one larva of seven analyzed showed the 170-kDa APN1 band (Fig. 4D). These results show that most larvae that survived the treatment with Cry1Ca had reduced expression of APN1.
FIG 3.
Silencing APN1 expression results in tolerance of S. frugiperda to Cry1Ca. (A) Mortality of S. frugiperda larvae after feeding with APN1 dsRNA. A total of 48 larvae were treated with APN1 dsRNA as described in Materials and Methods. After feeding with lysed E. coli HT115 cells producing dsRNA-APN1 as indicated in Materials and Methods, the larvae were divided in two groups, one treated with a diet without Cry1Ca toxin (No-Cry1Ca) and the other with a diet with 25 μg/cm2 of Cry1Ca toxin (Cry1Ca). Mortality was recorded after 7 days. As controls, larvae were treated with water or with lysed E. coli HT115 cells. Data show results of three replicates with significant differences between treatments. (B) APN1 gene transcript abundance was determined using quantitative reverse transcription-PCR (qRT-PCR) and Sybr green as described in Materials and Methods. The data were normalized by analyzing the ribosomal protein S3 (rps3) gene. In these experiments, the larvae were also fed with lysed E. coli HT115 producing dsRNA-APN1 as indicated in Materials and Methods. As controls, larvae were treated with water or with lysed E. coli HT115 cells. Bars represent the means and standard errors of results from the analysis of 8 individual midguts for each group.
FIG 4.
Detection of APN1 expression in individual larvae after dsRNA feeding. (A) Control larvae fed with water. (B) Larvae fed with lysed HT115 bacteria. (C) Larvae fed with lysed HT115 bacteria producing dsRNA-APN1. (D) Larvae fed with lysed HT115 bacteria producing dsRNA-APN1 and treated with Cry1Ca. The midgut tissue was dissected for individual larvae. Seven larvae were dissected per treatment. Lanes 1 to 7 correspond to different individual larvae. Individual midgut tissues were homogenized in PBS, and the supernatant samples recovered after centrifugation were loaded on 10% SDS-PAGE gels. Proteins were electrotransferred to PVDF, and the 170-kDa APN1 protein was detected by the use of anti-APN1 antibody and Western blot assays. The numbers on the left refer to molecular weight standards (in kilodaltons).
DISCUSSION
One of the limiting steps in the toxicity of Bt toxins is binding to larval midgut proteins. The aim of this work was to identify the Cry1Ca-binding proteins in S. frugiperda to provide evidence on their participation in toxicity and to identify the Cry1Ca amino acid residues involved in receptor binding.
The S. frugiperda APN1 was identified as a functional receptor of Cry1Ca based on the following data. (i) APN1 was identified as Cry1Ca-binding protein in pulldown assays of Cry1Ca with S. frugiperda BBMV proteins. (ii) Cry1Ca domain III mutants affected in toxicity showed a correlative defect on binding to the recombinant SfAPN1 but not to CAD SfCR7–12. (iii) Moreover, silencing the expression of APN1 transcript, by APN1-dsRNA feeding, showed that silenced larvae were at least two times more tolerant of Cry1Ca intoxication than nonsilenced larvae.
Role of APN in sequential binding model of Cry toxicity.
Different APN isoforms have been shown to act as receptors for different Cry toxins (24). It has been proposed that APN fulfils two roles in the toxicity of Cry1A toxins against lepidopteran larvae such as M. sexta: first, as an initial binding step to attach the activated Cry1A toxin to midgut apical membranes; second, to facilitate the insertion of toxin oligomers into the membrane (9, 10, 13). The experimental setup for the pulldown assays could facilitate the identification of gut proteins that are involved in the initial steps of Cry1Ca since activated monomeric toxin was used for finding Cry1Ca-binding proteins. It is probable that pulldown assays using oligomeric forms could reveal other proteins involved in later stages. We cannot exclude the possibility that other proteins involved in the later stages, such as ABCC2 (15), which is important for oligomer membrane insertion, were not identified because of the experimental setup. Pulldown assays identified APN2 in addition to APN1 as a Cry1Ca binding molecule, although the sequence coverage of APN2 was low. We still need to analyze if APN2 is a functional receptor for Cry1Ca.
Cry1Ca domain III mutants affected in toxicity show a defect in APN1 binding.
Site-directed mutagenesis of Cry1Ca domain III β16 showed that Cry1Ca V505A and S506A mutants were not toxic to S. frugiperda. Our data indicated that mutants showed two-times-lower binding affinity to APN1 than Cry1Ca, supporting the conclusion that APN1 is a Cry1Ca-binding protein as suggested by the pulldown assay results. However, the reduced binding of the V505A and S506A mutants to APN1 could not totally explain their loss of toxicity, suggesting that domain III β16 could also be involved in binding to other Cry1Ca receptors such as ALP or APN2. This remains to be analyzed. Nevertheless, these data indicate that domain III β16 of Cry1Ca is involved in APN1 binding and that this interaction is important for toxicity. In the case of Cry1Ab and Cry1Fa toxins, it was shown that certain β16 amino acid substitutions, such as Cry1AbN514A or Cry1FaN507A, showed 4-fold-to-10-fold-higher toxicity to different S. frugiperda populations (I. Gómez, J. Ocelotl, J. Sanchez, C. Lima, E. Martins, A. Rosales-Juarez, S. Aguilar-Medel, A. Abad, H. Dong, R. Monnerat, G. Peña, J. Zhang, M. Nelson, G. Wu, A. Bravo, M. Soberón, submitted for publication). The conserved Cry1Ca N510A mutation did not show an effect on Cry1Ca toxicity against the S. frugiperda population used in this study but showed 4-fold-higher toxicity than Cry1Ca in another S. frugiperda population from Mexico City (Gómez et al., submitted). It remains to be determined if this mutation increases toxicity of Cry1Ca in other S. frugiperda populations. Also, we analyzed the effect of Cry1Ca mutations reported here on CAD binding (Fig. 2). Binding to CAD SfCR7–12 was not substantially affected by these Cry1Ca mutations since the Kd values showed minimal or not statistically significant differences from the Kd value of Cry1Ca (P = 0.05 and P = 0.16). In S. exigua, CAD has been shown to be a functional receptor of Cry1Ca (21). It remains to be analyzed if CAD is a functional receptor of Cry1Ca in S. frugiperda by dsRNA feeding.
Silencing of APN1 shows its role in Cry1Ca toxicity against S. frugiperda.
Finally, we determined if APN1 was involved in Cry1Ca toxicity against S. frugiperda by silencing the expression of APN1 in S. frugiperda larvae and analyzed its effect in larval toxicity. The data showed that larvae with low APN1 expression were at least two times more tolerant of Cry1Ca than nonsilenced larvae. There have been few reports of successful gene silencing in lepidopteran insects by feeding dsRNA, especially in Spodoptera species (25). It was recently reported that efficient gene silencing was achieved in S. litura by feeding lysed E. coli cells that produce the specific dsRNA to the insects after sonication of the bacterial culture before feeding the insects (26). We showed here that delivering APN1-dsRNA by feeding, using E. coli cells that produce the dsRNA, could be used for gene silencing in S. frugiperda. Quantitative PCR analysis of the APN1 transcript levels (Fig. 3B) showed a reduction of more than 80% in the expression of APN1, indicating the efficacy of this method in silencing the expression of the targeted gene. However, although unlikely, another explanation is that exposure of the larvae to the toxin affects APN1 expression (Fig. 4D). This hypothesis remains to be evaluated.
The method presented here could be useful for analyzing the role of different insect proteins in the mode of action of other Cry toxins and for analyzing the role of specific insect genes in the physiology and development of S. frugiperda.
Overall, our results show that Cry1Ca relies on APN1 binding to impart toxicity to S. frugiperda.
MATERIALS AND METHODS
Spodoptera frugiperda and Manduca sexta populations.
The S. frugiperda and M. sexta populations were maintained in the Instituto de Biotecnología (IBT), Universidad Nacional Autónoma de Mexico (UNAM), without exposure to Cry toxins (16).
Site-directed mutagenesis.
A QuikChange multisite-directed mutagenesis system was used for mutagenesis using plasmid pHT315-Cry1Ca as the template following the instructions of the manufacturer (Stratagene). Mutagenic oligonucleotides are shown in Table 3. Plasmids were purified from E. coli cells and verified by DNA sequencing at the Instituto de Biotecnología, UNAM. Plasmids were propagated in E. coli dam dcm mutant strains and used to transform Bt strain 407cry− by electroporation as previously described (27, 28).
TABLE 3.
Oligonucleotide sequences
| Primer description | Primer sequence (5′ to 3′)a |
|---|---|
| APN1 S. frugiperda | |
| Forward | 5′-ATG ACC ATG GCG AAT CGC TGG TTT AGC CTC-3′ |
| Reverse | 5′-ATT AGG ATC CTT AAC CAT GTT GAT GAT AAC TGT GAT-3′ |
| APN1 dsRNA | |
| Forward | 5′-TCA GGA ATT CAG TGA GAA TCA ACC AAT CAA CGA A-3′ |
| Reverse | 5′-AAC GAA GCT TGG CGT AGT GTA GTA TAT GTC CTC T-3′ |
| qPCR APN1 | |
| Forward | 5′-TAT GCT CGA GGT AAC GTT GG-3′ |
| Reverse | 5′-TGT GGG AAG CCA TGT GTA GT-3′ |
| qPCR RPS3 | |
| Forward | 5′-ACA GAG TGT GCT CGG AGA GA-3′ |
| Reverse | 5′-GGC AAG ACC TCC AAT GAG TT-3′ |
| Mutagenic primers for mutation: | |
| V505A | 5′-ACC TTT GGT GAT TTT GCA TCT CTA CAA GTC AAT-3′ |
| S506A | 5′-TTT GGT GAT TTT GTA GCT CTA CAA GTC AAT ATT-3′ |
| L507A | 5′-GGT GAT TTT GTA TCT GCA CAA GTC AAT ATT AAT-3′ |
| Q508A | 5′-GAT TTT GTA TCT CTA GCA GTC AAT ATT AAT TCA-3′ |
| V509A | 5′-TTT GTA TCT CTA CAA GCC AAT ATT AAT TCA CCA-3′ |
| N510A | 5′-GTA TCT CTA CAA GTC GCT ATT AAT TCA CCA ATT-3′ |
For, forward; Rev, reverse. Bold letters indicate the codon that is changed during mutagenesis.
Expression, purification, and proteolytic activation of Cry1Ca, and β16 mutants.
Bt strains containing Cry1Ca or Cry1Ca mutants were grown in nutrient sporulation medium with erythromycin (10 μg/ml) for 72 h at 30°C with shaking at 250 rpm until sporulation and autolysis were completed (29). Solubilization of crystals was performed in 50 mM Na2CO3–NaHCO3 buffer containing 0.02% mercaptoethanol (pH 10.5) at 37°C for 1 h. The protein concentration of protoxins and toxins was determined by the Bradford method using bovine serum albumin (BSA) as a standard. Soluble protoxins were subjected to proteolytic activation with trypsin (Sigma) (1:50 [trypsin/protoxin]) at 37°C for 1 h. Trypsin was inactivated with 1 mM phenylmethylsulfonyl fluoride (PMSF), and samples were analyzed by SDS-PAGE using a 10% acrylamide gel.
Preparation of brush border membrane vesicles (BBMV).
Midgut tissue was dissected from third instar S. frugiperda larvae (from IBT) and used to prepare BBMV using a differential precipitation method with MgCl2 as previously reported (30). BBMV were stored at −70°C until use. For pulldown assays, BBMV were solubilized at 4°C for 2 h in 20 mM Tris-HCl (pH 7.4)–150 mM NaCl–5 mM EDTA–1 mM PMSF containing 1% (vol/vol) CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}. Undissolved material was removed by centrifugation at 100,000 × g for 1 h at 4°C. The protein concentration in the supernatant was determined by the procedure of Lowry using the DC protein assay dye method (Bio-Rad) and BSA as a protein standard.
Immunoprecipitation of Cry1Ca-binding proteins.
A 2-μl volume of serum containing the anti-Cry1Ca antibody was incubated with 100 μl of protein A Sepharose CL-4B beads (Pharmacia) for 1 h at 4°C. For washing, loaded beads were centrifuged at 5,000 rpm at 4°C and suspended in phosphate-buffered saline (PBS) buffer, and the washing steps were done five times. Soluble BBMV protein (100 μg) was incubated with 10 μg soluble Cry1Ca in a total volume of 250 μl for 1 h; after the incubation, the sample was incubated with 100 μl anti-Cry1Ca antibody-loaded protein A Sepharose beads for 12 h at 4°C. Samples were centrifuged for 5 min at 5,000 rpm, and pellets containing the beads were washed six times with PBS buffer. Cry1Ca-binding proteins were dissociated with 50 μl 0.2 M glycine (pH 2.6) by incubating 10 min with frequent agitation before gentle centrifugation was performed. A sample of the eluate was loaded in SDS-PAGE gel and silver stained using a Pierce silver stain kit (Thermo Scientific, Rockford, IL). The eluate was neutralized by adding an equal volume of Tris-Cl (pH 8). Trichloroacetic acid (TCA) was added to the extract to reach a final concentration of 13% to allow proteins to precipitate at −80°C for 24 h. Samples were centrifuged at 4°C for 30 min, after which the supernatant was discarded. Pellets were washed with precooled acetone, incubated 10 min on ice, and centrifuged at 14,000 rpm for 10 min. The final pellets were subjected to trypsin digestion in solution and sent for identification by LC-MS/MS to the Proteomics Discovery Research Platform of the Institut de Recherches Cliniques de Montréal (IRCM) at the University of Montreal. The results were searched against B. mori, M. sexta, and S. frugiperda full-proteome databases.
Heterologous expression of APN1 and CAD from S. frugiperda in E. coli cells.
We have analyzed the cloning of APN1 and a CAD fragment containing CAD repeats 7 to 12 (CR7 to -12) from S. frugiperda in plasmid pET22b, which was used to transform E. coli strain BL21(DE3) (Gómez et al., submitted). For protein production and purification, transformed E. coli cells were grown overnight with agitation at 37°C in 5 ml of LB broth containing 50 μg/ml ampicillin. A 100-μl volume of culture was used to inoculate 250-ml flasks with 100 ml of 2× tryptone-yeast extract (TY) broth with 100 μg/ml ampicillin. The 100-ml culture was incubated at 37°C with agitation until an OD600 of 0.6 was reached, expression of the APN1 or CAD fragment was induced by adding 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Sigma), and further incubation was performed for 5 h at 30°C. Cells were collected by centrifugation at 5,000 rpm for 15 min at 4°C, and the pellet was suspended in STE buffer (10 mM Tris-HCl, 1 mM EDTA, 8 M urea, pH 8). After sonication for 5 min on ice, cell debris was eliminated by centrifugation at 70,000 rpm for 30 min at 15°C, and the supernatant was subjected to purification with nickel-agarose beads (Qiagen). After washing was performed with 35 mM imidazole–PBS buffer (pH 7.5), the recombinant proteins were eluted with 250 mM imidazole and dialyzed against PBS buffer.
Cry1Ca or domain III mutants binding to APN1 or CAD analyzed by ELISA binding assays.
Recombinant APN1 or CAD (0.5 μg/well) was used to coat ELISA plates (Nunc). Activated Cry1Ca or Cry1Ca mutants at different concentrations were incubated with the receptor-coated ELISA plates. Unbound toxin was washed out with three washes with phosphate-buffered saline (PBS) and three washes with PBS supplemented with 0.1% Tween 20, and toxins bound to the ELISA plates were detected using anti-Cry1Ca antibody (1:20,000 dilution) and then with a secondary anti-rabbit antibody conjugated with horseradish peroxidase (HRP) enzyme. Finally, o-phenylenediamine (Sigma) and H2O2 were used as substrates for peroxidase activity detection. The reaction was stopped by adding 50 μl of 5 M HCl, and the OD was measured at 490 nm using an ELISA microplate reader. Nonspecific binding controls were run in parallel where the APN1 or CAD ligands were not included in the assays and wells were blocked and treated with Cry1Ca proteins as stated above. The data shown here were obtained after subtracting the data from the nonspecific binding controls from the data from the samples containing the receptors. All experiments were done in triplicate. Comparisons of binding data were analyzed by t test using GraphPad Prism 7 (version 5.0b), and data from Scatchard plots were used for obtaining relative binding affinity (Kd) values by using Scatchard analysis with SigmaPlot (Systat Software).
Silencing S. frugiperda APN1.
Primers were designed to clone a 378-bp fragment of the S. frugiperda APN1 gene containing restriction sites for enzymes EcoRI-HindIII by PCR (Table 3). The apn1 fragment was subcloned into pLITMUS28i vector (New England BioLabs), which contains two T7 promoters in an inverted orientation. Recombinant pLITMUS28i:apn1 plasmid was transformed into E. coli HT115 cells. To prepare samples of APN1 dsRNA, HT115/pLITMUS28i:apn1 was inoculated into 5 ml of LB with 100 μg/ml of ampicillin and incubated at 37°C overnight. A 50-ml volume of the same medium was inoculated with 1 ml of the overnight culture and grown until an OD600 of 0.6 was reached. Synthesis of dsRNA was induced by adding 0.1 mM IPTG followed by an additional incubation for 4 h at 30°C. A 1-ml volume was used for dsRNA extraction by Quick RNA MiniPrep (Qiagen), samples were run on agarose gels, and the dsRNA concentration was estimated by comparison with the RNA molecular weight marker. From the rest of the cell culture, cells were harvested by centrifugation at 5,000 rpm for 15 min at 4°C and suspended in 5 ml of PBS buffer (pH 7.4). Cell samples were sonicated at 95% intensity with 10 cycles of 1-min bursts separated by 2 min in ice as previously reported (25). A 35-μl volume of the sonicated sample was applied to one well containing an artificial diet, and one neonate larvae was placed in one well. Control larvae were fed with water or with strain HT115 cultured as described for dsRNA induction. After 12 h, the larvae subjected to dsRNA feed treatments were randomly divided into two groups. One group was placed on an untreated control diet, and the other group was placed on a diet containing 25 μg/cm2 of purified Cry1Ca crystals. Individual larvae were placed into separate wells of a 24-well plate with at least three replicates, and larval mortality was evaluated after 7 days.
Western blotting.
Midgut tissue was isolated from individual larvae from the groups consisting of control and dsRNA-treated larvae. Midgut tissue was homogenized in PBS buffer, and samples were centrifuged 2 min at 14,000 rpm. The supernatant was recovered, the samples were loaded on 10% SDS-PAGE gels, and proteins were electrotransferred to a polyvinylidene difluoride (PVDF) membrane (Millipore). Blots were blocked with 5% nonfat dry milk in PBS–0.1% Tween 20 (PBST) and incubated with 1:25,000 anti-APN antibody/PBST. Subsequently, blots were incubated with HRP-conjugated goat anti-rabbit IgG (Santa Cruz) diluted 1:30,000 in PBST and developed with luminol (Pierce) as indicated by the manufacturer.
Quantitative real-time PCR.
At 7 days postfeeding with the dsRNA, larvae that were not exposed to toxin were evaluated for expression of APN1. Eight larval guts from each of the control or dsRNA-fed groups were dissected and placed in RNAlater, and total RNA was isolated (RNeasy kit; Qiagen). Quantitative real-time PCR was performed on each template using primers listed in Table 3, a LightCycler 480 instrument (Roche), and a Sybr green I detection system (Fermentas, Life Sciences). Relative fold calculations were made with duplicates for each treatment group, and the expression data were normalized by analyzing the ribosomal protein S3 (rps3) gene using the primers described in Table 3.
Insect bioassay.
Different doses (0.1 to 2,500 ng/cm2) of crystal proteins were applied onto the diet surface contained in 24-well polystyrene plates (Cell Wells; Corning Glass Works, Corning, New York). A total of 24 larvae per plate (one larva was used per well to avoid cannibalism) were fed with same dose of Cry1Ca proteins, and six different doses were used for the bioassay of each Cry1Ca protein in three repetitions. The plates were incubated at 28°C with 65% ± 5% relative humidity and light and dark photoperiods of 16 h and 8 h, and mortality was recorded after 7 days. The 50% lethal concentration (LC50) was determined using Probit software.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Blanca-Ines Garcia-Gomez and Lizbeth Cabrera for technical assistance.
This research was supported in part by DGAPA/UNAM IN209011 and IN201016, CONACyT 83135 and CONACyT Fronteras de la Ciencia 008, and Pioneer Hi Bred.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01089-18.
REFERENCES
- 1.Blanco CA, Portilla M, Jurat-Fuentes JL, Sanchez JF, Viteri D, Vega-Aquino P, Teran-Vargas AP, Azuara-Dominguez A, Lopez JD Jr, Arias RS, Zhu Y-C, Lugo-Barreras D, Jackson R. 2010. Susceptibility of isofamilies of Spodoptera frugiperda (Lepidoptera: Noctuidae) to Cry1Ac and Cry1Fa proteins of Bacillus thuringiensis. Southwest Entomol 35:409–415. doi: 10.3958/059.035.0325. [DOI] [Google Scholar]
- 2.Goergen G, Kumar PL, Sankung SB, Togola A, Tamo M. 2016. First report of outbreaks of the fall armyworm Spodoptera frugiperda (JE Smith) (Lepidoptera Noctuidae), a new alien invasive pest in west and central Africa. PLoS One 11:e0165632. doi: 10.1371/journal.pone.0165632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanahuja G, Banakar R, Twyman RM, Capell T, Christou P. 2011. Bacillus thuringiensis: a century of research development and commercial applications. Plant Biotechnol J 9:283–300. doi: 10.1111/j.1467-7652.2011.00595.x. [DOI] [PubMed] [Google Scholar]
- 4.Storer NP, Kubiszak ME, King JE, Thompson GD, Santos AC. 2012. Status of resistance to Bt maize in Spodoptera frugiperda: lessons from Puerto Rico. J Invertebr Pathol 110:294–300. doi: 10.1016/j.jip.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Huang F, Qureshi JA, Meagher RL Jr, Reisig DD, Head GP, Andow DA, Ni X, Kerns D, Buntin GD, Niu Y, Yang F, Dangal V. 2014. Cry1F resistance in fall armyworm Spodoptera frugiperda: single gene versus pyramided Bt maize. PLoS One 9:e112958. doi: 10.1371/journal.pone.0112958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farias JR, Andow DA, Horikoshi RJ, Sorgatto RJ, Fresia P, dos Santos AC, Omoto C. 2014. Field-evolved resistance to Cry1F maize by Spodoptera frugiperda (Lepidoptera: Noctuidae) in Brazil. Crop Prot 64:150–158. doi: 10.1016/j.cropro.2014.06.019. [DOI] [Google Scholar]
- 7.Monnerat R, Martins E, Macedo C, Queiroz P, Praça L, Soares CM, Moreira H, Grisi I, Silva J, Soberón M, Bravo A. 2015. Evidence of field-evolved resistance of Spodoptera frugiperda to Bt corn expressing Cry1F in Brazil that is still sensitive to modified Bt toxins. PLoS One 10:e0119544. doi: 10.1371/journal.pone.0119544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bravo A, Likitvivatanavong S, Gill SS, Soberón M. 2011. Bacillus thuringiensis: a story of a successful bioinsecticide. Insect Biochem Mol Biol 41:423–431. doi: 10.1016/j.ibmb.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gómez I, Arenas I, Benitez I, Miranda-Ríos J, Becerril B, Grande G, Almagro JC, Bravo A, Soberón M. 2006. Specific epitopes of domains II and III of Bacillus thuringiensis Cry1Ab toxin involved in the sequential interaction with cadherin and aminopeptidase-N receptors in Manduca sexta. J Biol Chem 281:34032–34039. doi: 10.1074/jbc.M604721200. [DOI] [PubMed] [Google Scholar]
- 10.Arenas I, Bravo A, Soberón M, Gomez I. 2010. Role of alkaline phosphatase from Manduca sexta in the mechanism of action of Bacillus thuringiensis Cry1Ab toxin. J Biol Chem 285:12497–12503. doi: 10.1074/jbc.M109.085266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gómez I, Sánchez J, Miranda R, Bravo A, Soberón M. 2002. Cadherin-like receptor binding facilitates proteolytic cleavage of helix α-1 in domain I and oligomer pre-pore formation of Bacillus thuringiensis Cry1Ab toxin. FEBS Lett 513:242–246. doi: 10.1016/S0014-5793(02)02321-9. [DOI] [PubMed] [Google Scholar]
- 12.Gómez I, Dean DH, Bravo A, Soberón M. 2003. Molecular basis for Bacillus thuringiensis Cry1Ab toxin specificity: two structural determinants in the Manduca sexta Bt-R1 receptor interact with loops a-8 and 2 in domain II of Cy1Ab toxin. Biochemistry 42:10482–10489. doi: 10.1021/bi034440p. [DOI] [PubMed] [Google Scholar]
- 13.Pacheco S, Gomez I, Arenas I, Saab-Rincon G, Rodriguez-Almazan C, Gill SS, Bravo A, Soberón M. 2009. Domain II loop 3 of Bacillus thuringiensis Cry1Ab toxin is involved in a “ping-pong” binding mechanism with Manduca sexta aminopeptidase-N and cadherin receptors. J Biol Chem 284:32750–32757. doi: 10.1074/jbc.M109.024968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heckel DG. 2012. Learning the ABCs of Bt: ABC transporters and insect resistance to Bacillus thuringiensis provide clues to a crucial step in toxin mode of action. Pest Biochem Physiol 104:103–110. doi: 10.1016/j.pestbp.2012.05.007. [DOI] [Google Scholar]
- 15.Ocelotl J, Sanchez J, Gómez I, Tabashnik BE, Bravo A, Soberón M. 2017. ABCC2 is associated with Bacillus thuringiensis Cry1Ac toxin oligomerization and membrane insertion in diamondback moth. Sci Rep 7:2386. doi: 10.1038/s41598-017-02545-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aranda E, Sanchez J, Perferoen M, Güereca L, Bravo A. 1996. Interactions of Bacillus thuringiensis crystal proteins with the mid-gut epithelial cells of Spodoptera frugiperda (Lepidoptera: Noctuidae). J Invertbr Pathol 68:203–212. doi: 10.1006/jipa.1996.0087. [DOI] [PubMed] [Google Scholar]
- 17.Banerjee R, Hasler J, Meagher R, Nagoshi R, Hietala L, Huang F, Narva K, Jurat-Fuentes JL. 2017. Mechanism and DNA-based detection of field-evolved resistance to transgenic Bt corn in fall armyworm (Spodoptera frugiperda). Sci Rep 7:10877. doi: 10.1038/s41598-017-09866-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo K, Lu Y-J, Adang MJ. 1996. A 106 kDa form of aminopeptidase is a receptor for Bacillus thuringiensis Cry1C δ-endotoxin in the brush border membrane of Manduca sexta. Insect Biochem Mol Biol 26:783–791. doi: 10.1016/S0965-1748(96)00027-6. [DOI] [Google Scholar]
- 19.Masson L, Lu Y-J, Mazza A, Brousseau R, Adang MJ. 1995. The CryIA(c) receptor purified from Manduca sexta displays multiple specificities. J Biol Chem 270:20309–20315. doi: 10.1074/jbc.270.35.20309. [DOI] [PubMed] [Google Scholar]
- 20.Herrero S, Gechev T, Bakker PL, Moar WJ, de Maagd RA. 2005. Bacillus thuringiensis Cry1Ca-resistant Spodoptera exigua lacks expression of one of four aminopeptidase N genes. BMC Genomics 6:96. doi: 10.1186/1471-2164-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajagopal R, Sivakumar S, Agrawal N, Malhorta P, Bhatnagar RK. 2002. Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J Biol Chem 277:46849–46851. doi: 10.1074/jbc.C200523200. [DOI] [PubMed] [Google Scholar]
- 22.Ren X-L, Chen R-R, Zhang Y, Ma Y, Cui J-J, Han Z-J, Mu L-L, Li G-K. 2013. A Spodoptera exigua cadherin serves as a putative receptor for Bacillus thuringiensis Cry1Ca toxin and shows differential enhancement of Cry1Ca and Cry1Ac toxicity. Appl Environ Microbiol 79:5576–5583. doi: 10.1128/AEM.01519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrero H, Gonzalez-Cabrera J, Ferré J, Bakker PL, de Maagd R. 2004. Mutations in the Bacillus thuringiensis Cry1Ca toxin demonstrate the role of domains II and III in specificity towards Spodoptera exigua larvae. Biochem J 384:507–513. doi: 10.1042/BJ20041094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pigott CR, Ellar DJ. 2007. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol Mol Biol Rev 71:255–281. doi: 10.1128/MMBR.00034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, Kanginakudru S, Albrechtsen M, An C, Aymeric JL, Barthel A, Bebas P, Bitra K, Bravo A, Chevalier F, Collinge DP, Crava CM, de Maagd RA, Duvic B, Erlandson M, Faye I, Felföldi G, Fujiwara H, Futahashi R, Gandhe AS, Gatehouse HS, Gatehouse LN, Giebultowicz JM, Gómez I, Grimmelikhuijzen CJ, Groot AT, Hauser F, Heckel DG, Hegedus DD, Hrycaj S, Huang L, Hull JJ, Iatrou K, Iga M, Kanost MR, Kotwica J, Li C, Li J, Liu J, Lundmark M, Matsumoto S, Meyering-Vos M, Millichap PJ, Monteiro A, Mrinal N, Niimi T, et al. . 2011. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol 57:231–245. doi: 10.1016/j.jinsphys.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 26.Kim E, Park Y, Kim Y. 2015. A transformed bacterium expressing double-stranded RNA specific to integrin b1 enhances Bt toxin efficacy against a polyphagous insect pest, Spodoptera exigua. PLoS One 10:e0132631. doi: 10.1371/journal.pone.0132631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lereclus D, Arantès O, Chaufaux J, Lecadet M. 1989. Transformation and expression of a cloned delta-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol Lett 51:211–217. doi: 10.1111/j.1574-6968.1989.tb03448.x. [DOI] [PubMed] [Google Scholar]
- 28.Arantes O, Lereclus D. 1991. Construction of cloning vectors from Bacillus thuringiensis. Gene 108:115–119. doi: 10.1016/0378-1119(91)90495-W. [DOI] [PubMed] [Google Scholar]
- 29.Schaeffer P, Millet J, Aubert J-P. 1965. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A 54:704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfersberger MG. 1993. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval mid-gut of the gypsy moth (Lymantria dispar). Arch Insect Biochem Physiol 24:139–147. doi: 10.1002/arch.940240304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.