Prochlorococcus, the most abundant and smallest known free-living photosynthetic microorganism, plays a key role in marine ecosystems and biogeochemical cycles. Prochlorococcus genome evolution is a fundamental issue related to how Prochlorococcus clades adapted to different ecological niches. Recent studies revealed that the gene gain and loss is crucial to the clade differentiation. The significance of our research is that we interpreted the Prochlorococcus genome evolution from the perspective of genome structure and associated the genome rearrangement with the Prochlorococcus clade differentiation and subsequent ecological adaptation.
KEYWORDS: Prochlorococcus, genome evolution, genome rearrangement, genomic backbone
ABSTRACT
Prochlorococcus is the most abundant and smallest known free-living photosynthetic microorganism and is a key player in marine ecosystems and biogeochemical cycles. Prochlorococcus can be broadly divided into high-light-adapted (HL) and low-light-adapted (LL) clades. In this study, we isolated two low-light-adapted clade I (LLI) strains from the western Pacific Ocean and obtained their genomic data. We reconstructed Prochlorococcus evolution based on genome rearrangement. Our results showed that genome rearrangement might have played an important role in Prochlorococcus evolution. We also found that the Prochlorococcus clades with streamlined genomes maintained relatively high synteny throughout most of their genomes, and several regions served as rearrangement hotspots. Backbone analysis showed that different clades shared a conserved backbone but also had clade-specific regions, and the genes in these regions were associated with ecological adaptations.
IMPORTANCE Prochlorococcus, the most abundant and smallest known free-living photosynthetic microorganism, plays a key role in marine ecosystems and biogeochemical cycles. Prochlorococcus genome evolution is a fundamental issue related to how Prochlorococcus clades adapted to different ecological niches. Recent studies revealed that the gene gain and loss is crucial to the clade differentiation. The significance of our research is that we interpreted the Prochlorococcus genome evolution from the perspective of genome structure and associated the genome rearrangement with the Prochlorococcus clade differentiation and subsequent ecological adaptation.
INTRODUCTION
Prochlorococcus, a marine cyanobacterium, is the most abundant and smallest known free-living photosynthetic microorganism (1–3). Prochlorococcus is mainly distributed in the euphotic zone of nutrient-poor tropical and subtropical waters and is a key player in marine ecosystems and biogeochemical cycles (2, 4, 5). With a cell concentration of up to 105 cells/ml in the oligotrophic surface ocean, Prochlorococcus accounts for 40% to 60% of photosynthetic biomass and produces approximately 4 gigatons of fixed carbon each year (2, 6–10).
In recent decades, at least 12 different Prochlorococcus clades were discovered in the world's oceans, which can be broadly divided into two major groups, high-light-adapted (HL) and low-light-adapted (LL) groups (11, 12), but only five clades have been cultivated: HLI, HLII, LLI, LLII/III, and LLIV (13–19). With the rapid development of sequencing technologies, dozens of Prochlorococcus genomes were published (20–25). The HL and LLII/III clades have genomes as small as 1.6 to 1.7 Mbp that encode 1,800 to 1,900 genes, which makes them the smallest known genomes among nonsymbiotic photosynthetic autotrophs (26). The LLI clade genomes are slightly larger (1.8 to 1.9 Mbp) and encode 2,100 to 2,200 genes. In contrast, the LLIV genome is approximately 2.4 to 2.6 Mbp, which is similar to that of the marine Synechococcus (24, 25).
Comparative genomics reveal that genome streamlining is a striking feature of Prochlorococcus genomes (22, 24, 27, 28). Highly compressed genomes allow Prochlorococcus to thrive in oligotrophic oceans by requiring less energy and fewer nutrients (3, 19, 24). The HL, LLI, and LLII/III clades, which have undergone substantial genome reduction compared with the LLIV clade and Synechococcus, have significantly different physiological characteristics, ecological distributions, and genomic content (3, 24). The HLI and HLII clades are distinguished by their temperature optima, account for most of the known Prochlorococcus, and are distributed in the upper euphotic zone (5). The LLI clade, which is usually found in the middle euphotic zone, has characteristics that are intermediate between HL and other LL clades (4, 5, 29). It is well known that the LLI clade contains more HL-inducible genes (hli), which encode proteins that protect cells from light shock as well as other stresses, and is the only LL clade that encodes photolyase, which is a photoprotective enzyme (19, 24). Currently, there are only four published genomes of this clade (24, 25). In contrast, the LLII/III clade, which was originally defined as two separate clades (LLII and LLIII) (13), is more restricted to the deep euphotic zone (4). In terms of genome reduction, one hypothesis is that early genome streamlining resulted in cell size reduction and facilitated light absorption of cells in low-light environments (3), and the LLII/III clade might diverge from this event. Previous studies also found that genome reduction was initiated when the most recent common ancestor (MRCA) of the HL, LLI, and LLII/III clades diverged from the nonstreamlined LLIV clade (24, 27). Thus, comparative genomic analysis of these streamlined but substantially different Prochlorococcus clades may provide insight into the key evolutionary processes associated with genome reduction, clade differentiation, and ecological adaptation.
Previous studies on Prochlorococcus genome evolution mainly focused on local mutations that only affected individual genes; for example, genomic content was compared between LL and HL genomes (22), hypervariable genomic islands (23), and gene gain/loss among different clades (24, 27). Those investigations reconstructed the history of vertical inheritance and horizontal gene transfer. In contrast to local mutations, genomes can also undergo large-scale mutation events, such as genome rearrangement, which can result in substantial changes in gene order and genomic content (30). Genome rearrangements are rare events and include inversions, deletions, duplications, and translocations (30, 31), and they have the potential to elucidate genome evolution (32, 33). In the case of Prochlorococcus, only a couple of studies were related to genome rearrangement (33, 34).
Here, we compared Prochlorococcus genomes, including two newly isolated LLI strains from the western Pacific Ocean. We reconstructed Prochlorococcus phylogeny based on genome rearrangement and showed that genome rearrangement might have played an important role in Prochlorococcus evolution. We also performed backbone analysis and showed that different clades shared a conserved backbone but also had clade-specific regions, and we revealed that the genes in these regions were associated with ecological adaptation.
RESULTS AND DISCUSSION
Genomes of the two newly isolated LLI strains.
In this study, two Prochlorococcus strains were obtained from the western Pacific Ocean, which is an undersampled region for Prochlorococcus. These two strains were both isolated at 150-m depths but from two different stations. XMU1408 was isolated from the Luzon strait, which is the main channel between the South China Sea and the western Pacific Ocean, whereas XMU1403 was isolated from open water of the western Pacific Ocean (detailed locations are described in Table 1). Preliminary internal transcribed spacer (ITS) analysis showed that these two strains belonged to the LLI clade, which was further confirmed by the phylogenomic analysis based on 31 core genes (see below). The completeness and contamination of the recovered genomes were estimated by CheckM (35). The unialgal status was confirmed by obtaining a single Prochlorococcus ITS sequence from the assembled contigs.
TABLE 1.
Prochlorococcus strain isolation location, genome characteristics, and assembly statistics
| Genome feature | Value for strain: |
|
|---|---|---|
| XMU1403 | XMU1408 | |
| Ecotype/clade | LLI | LLI |
| Isolation location | ||
| Longitude (E) | 124 | 121 |
| Latitude (N) | 20 | 20 |
| Isolation depth (m) | 150 | 150 |
| Sequencing method | HiSeq | HiSeq |
| Read length (bp) | 150 × 2 | 150 × 2 |
| Assembly size (bp) | 1,746,033 | 1,795,147 |
| G+C content (%) | 35.2 | 34.1 |
| No. of contigs | 7 | 6 |
| N50 (bp) | 335,809 | 389,747 |
| No. of protein-coding sequences | 2,042 | 2,059 |
| Completenessa (%) | >98 | >98 |
| NCBI accession no. | QJUF00000000 | QJUE00000000 |
Completeness is estimated by CheckM (35).
Detailed genome analysis showed that XMU1403 and XMU1408 had genome sizes of 1.746 Mb (GC content, 35.2%) and 1.795 Mb (GC content, 34.1%), respectively, which contained 2,042 and 2,059 protein-coding sequences, respectively (Table 1). These genomic features were similar to those of the LLI strains isolated from other regions (see Table 3). Because there are only four LLI genomes published at present (24, 25) and a previous study found that LL strains contain a high number of novel genes (19), the genomic data of these first two LLI strains isolated from the western Pacific Ocean may enhance the understanding of the genetic diversity of this clade. Based on the six known LLI genomes, we calculated pan-, core, and accessory genome sizes of the LLI clade (2.74 Mb, 1.35 Mb, and 1.39 Mb, respectively). Clustering analysis of core and accessory genome fragments showed that each strain has its own unique accessory genome regions, and XMU1408 had greater differences than other strains (see Fig. S1 in the supplemental material), which is consistent with the results of the phylogenomic analysis (see below).
TABLE 3.
Reference Prochlorococcus genomes used in this study
| Strain | Ecotype | Assembly size (bp) | %GC | No. of coding sequences | NCBI accession no. |
|---|---|---|---|---|---|
| MIT9515 | HLI | 1,704,176 | 30.8 | 1,964 | CP000552 |
| EQPAC1 | HLI | 1,654,739 | 30.8 | 1,958 | JNAG00000000 |
| MED4 | HLI | 1,657,990 | 30.8 | 1,962 | BX548174 |
| MIT0604 | HLII | 1,780,061 | 31.2 | 2,092 | CP007753 |
| AS9601 | HLII | 1,669,886 | 31.3 | 1,938 | CP000551 |
| GP2 | HLII | 1,624,310 | 31.2 | 1,878 | JNAH00000000 |
| MIT9107 | HLII | 1,699,937 | 31 | 1,994 | JNAI00000000 |
| MIT9116 | HLII | 1,685,398 | 31 | 1,984 | JNAJ00000000 |
| MIT9123 | HLII | 1,697,748 | 31 | 1,999 | JNAK00000000 |
| MIT9201 | HLII | 1,672,416 | 31.3 | 1,987 | JNAL00000000 |
| MIT9202 | HLII | 1,691,453 | 31.1 | 2,019 | DS999537 |
| MIT9215 | HLII | 1,738,790 | 31.1 | 2,043 | CP000825 |
| MIT9301 | HLII | 1,641,879 | 31.3 | 1,927 | CP000576 |
| MIT9302 | HLII | 1,745,343 | 31.1 | 2,016 | JNAM00000000 |
| MIT9311 | HLII | 1,711,064 | 31.2 | 1,978 | JNAN00000000 |
| MIT9312 | HLII | 1,709,204 | 31.2 | 1,979 | CP000111 |
| MIT9314 | HLII | 1,690,556 | 31.2 | 1,979 | JNAO00000000 |
| MIT9321 | HLII | 1,658,664 | 31.2 | 1,962 | JNAP00000000 |
| MIT9322 | HLII | 1,657,550 | 31.2 | 1,961 | JNAQ00000000 |
| MIT9401 | HLII | 1,666,808 | 31.2 | 1,969 | JNAR00000000 |
| SB | HLII | 1,669,823 | 31.5 | 1,932 | JNAS00000000 |
| MIT0801 | LLI | 1,929,203 | 34.9 | 2,278 | CP007754 |
| NATL1A | LLI | 1,864,731 | 35 | 2,248 | CP000553 |
| NATL2A | LLI | 1,842,899 | 35.1 | 2,209 | CP000095 |
| PAC1 | LLI | 1,841,163 | 35.1 | 2,254 | JNAX00000000 |
| LG | LLII/III | 1,754,063 | 36.4 | 1,989 | JNAT00000000 |
| MIT0601 | LLII/III | 1,707,342 | 37 | 1,936 | JNAU00000000 |
| MIT0602 | LLII/III | 1,750,918 | 36.3 | 2,002 | JNAV00000000 |
| MIT0603 | LLII/III | 1,752,482 | 36.3 | 2,012 | JNAW00000000 |
| MIT9211 | LLII/III | 1,688,963 | 38 | 1,952 | CP000878 |
| SS35 | LLII/III | 1,751,015 | 36.4 | 1,988 | JNAZ00000000 |
| SS52 | LLII/III | 1,754,053 | 36.4 | 1,985 | JNBE00000000 |
| SS120 | LLII/III | 1,751,080 | 36.4 | 1,982 | AE017126 |
| SS2 | LLII/III | 1,752,772 | 36.4 | 1,986 | JNAY00000000 |
| SS51 | LLII/III | 1,746,977 | 36.4 | 1,977 | JNBD00000000 |
| MIT0701 | LLIV | 2,592,571 | 50.6 | 3,082 | JNBA00000000 |
| MIT0702 | LLIV | 2,583,057 | 50.6 | 3,084 | JNBB00000000 |
| MIT0703 | LLIV | 2,575,057 | 50.6 | 3,078 | JNBC00000000 |
| MIT9303 | LLIV | 2,682,675 | 50 | 3,243 | CP000554 |
| MIT9313 | LLIV | 2,410,873 | 50.7 | 3,002 | BX548175 |
Prochlorococcus phylogenomics.
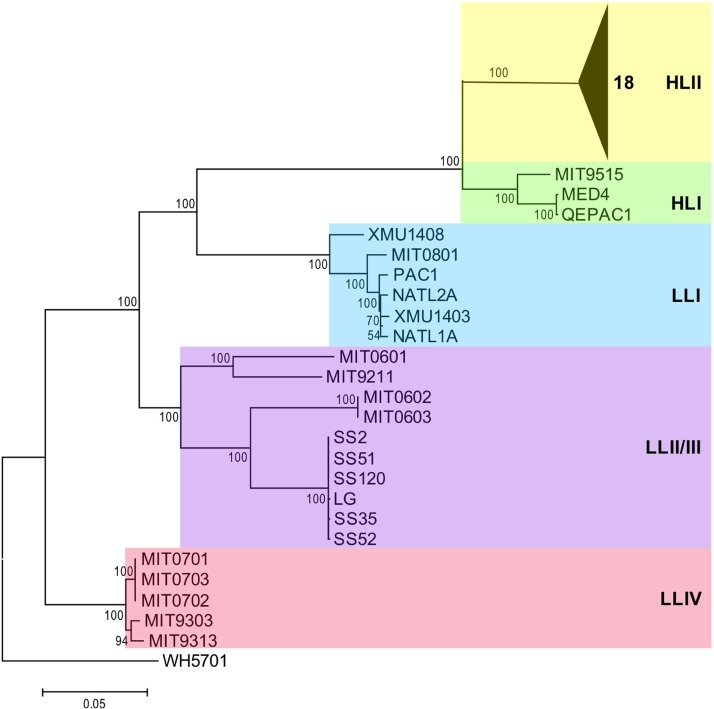
To reconstruct a robust phylogeny of Prochlorococcus for genomic study, we analyzed concatenated protein sequences of 31 core genes. The resulting phylogenetic tree, which is consistent with previous phylogenomic studies that used other methods (24, 28), showed that Prochlorococcus consists of at least five clades with relatively large phylogenetic distances. The HLI and HLII clades formed a monophyletic HL clade; in contrast, LL clades were more divergent (Fig. 1). It is also worth noting that the streamlined HL, LLI, and LLII/III clades shared a most recent common ancestor (MRCA), which had a large phylogenetic distance from the nonstreamlined LLIV clade. The LLI clade clustered with the HL clade, which is consistent with its intermediate-light adaptation. At the subclade level, XMU1408 was separated from other strains in the LLI clade, whereas the 10 LLII/III strains could also be divided into at least two subclades, which indicates that the LLI and LLII/III clades have complex evolutionary histories and different subclades.
FIG 1.
Phylogenomic relationships of Prochlorococcus. Shown is a phylogenetic tree reconstructed by concatenated protein sequences of 31 core genes with the maximum likelihood method, using Synechococcus sp. strain WH5701 as an outgroup. Numbers at the nodes represent bootstrap values (1,000 resamplings).
Genome rearrangement of streamlined clades.
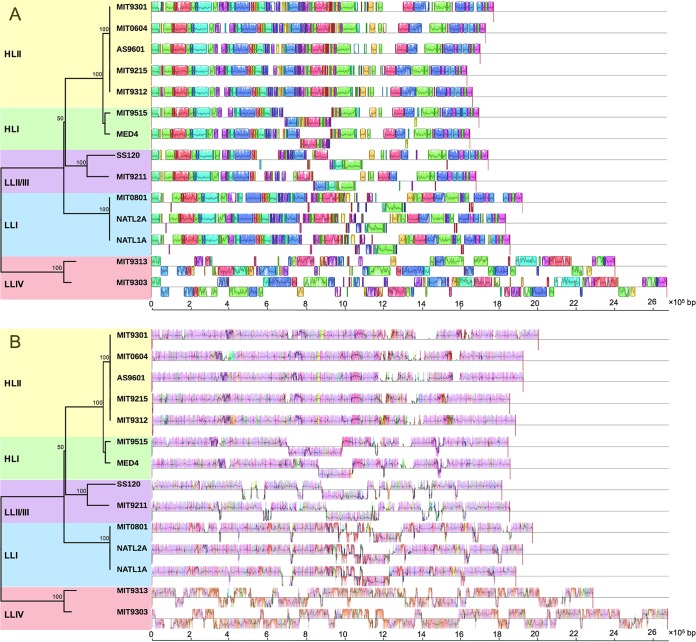
It is believed that genome streamlining is the result of Prochlorococcus adaptation to a relatively stable ocean surface environment (3, 19, 24). Comparative genomics of these streamlined but substantially different genomes may provide insight into the key evolutionary processes that shaped ecological adaptations of Prochlorococcus. In this study, we attempted to compare the Prochlorococcus genomes based on genome rearrangement. Sixty-nine LCBs were detected across 14 complete genomes of Prochlorococcus (Fig. 2A), which indicates a long and complex evolutionary history of Prochlorococcus. After excluding the LLIV clade, the number of LCBs of the three streamlined clades (HL, LLI, and LLII/III) was substantially reduced to 39, which signifies that the LLIV clade had a large phylogenetic distance from other clades. It is worth noting that most parts of the streamlined LLI, LLII/III, and HL genomes maintained relatively high synteny but substantially differed from the LLIV genomes (Fig. 2A and Fig. S2). Interestingly, the average LCB lengths of these streamlined clades were similar but significantly shorter than that of the LLIV clade (P < 0.001 by one-sample t test, n = 34; null hypothesis was set as 55,187 bp [the average LCB length of the LLIV clade]) (Fig. S3 to S7 and Table S1). These results indicate that the major genome rearrangement events coincided with genome reduction and occurred before divergence of the LLI, LLII/III, and HL clades. Previous studies on gene gain and loss also revealed that genome reduction occurred just after divergence of the LLIV clade and the MRCA of other clades (24, 27).
FIG 2.
Genome comparison among 14 Prochlorococcus complete genomes. The genome comparison was generated by Mauve (53). (A) Rearrangements of locally colinear blocks (LCBs) among Prochlorococcus genomes. LCBs are color coded. LCBs below the black line have an inverse orientation relative to that of MIT9301. (B) Visualization of the backbone and clade-specific regions among Prochlorococcus genomes. Pink-colored segments are conserved among all strains (backbone), whereas other colored segments are conserved in a subset of strains. Segments below the black line have an inverse orientation relative to that of MIT9301. For both panels A and B, the large white regions indicate strain-specific content. The phylogenetic tree was reconstructed based on permutations of 69 LCBs with the maximum likelihood method. Numbers at the nodes represent bootstrap values (1,000 resamplings).
Phylogenomic analysis based on genome rearrangement reconstructs important evolutionary events of genome structure. In this study, the genome rearrangement-based phylogenomic tree showed a topology similar to that of the tree based on concatenated protein sequences of 31 core genes but with one discrepancy: the HL and LLII/III clades clustered into one group in the genome rearrangement-based tree (Fig. 2). This result is consistent with a previous study that reconstructed Prochlorococcus phylogeny based on gene order (33). It is possible that this discrepancy reflects a higher rate of genome rearrangement in the LLI clade. In detail, the LLII/III clade, which is usually found in deep waters and possesses more DNA repair genes (24), resulting in a low mutation rate, may have retained more of the genome structure of the MRCA of these streamlined clades; in contrast, the HL and LLI clades, which possess fewer DNA repair genes (24), could have less genome structure similarity because of a higher mutation rate of genome rearrangement in the middle to upper euphotic zone, although the latter two may share an MRCA and more similar genomic content. Our explanation does not contradict another hypothesis, which states that increase in mutation rate is the primary cause of genome reduction in Prochlorococcus (36), because the loss of many DNA repair genes in the streamlined HL and LLI genomes (24) could increase the local mutation rate and also the probability of genome rearrangement and thereby enhance Prochlorococcus clade differentiation.
Genome rearrangement is a major genomic mutation that can result from inversions, deletions, duplications, and translocations. Generally, these events are triggered by DNA double-helix breaks at two different sites, followed by rejoining of the broken ends that produce large changes in gene order and genomic content among closely related genomes. Genome rearrangement can result in destruction of gene structure when a breakpoint occurs inside a gene and can also have substantial influence on gene expression (30, 37, 38). Changes in genome structure are almost always destructive; however, in very rare cases, they provide significant benefits. In the case of Prochlorococcus evolution, the right genome rearrangement may provide significant advantages for occupying a new ecological niche (e.g., different light intensity and temperature), promoting clade differentiation and destroying dispensable genes, which results in genome reduction.
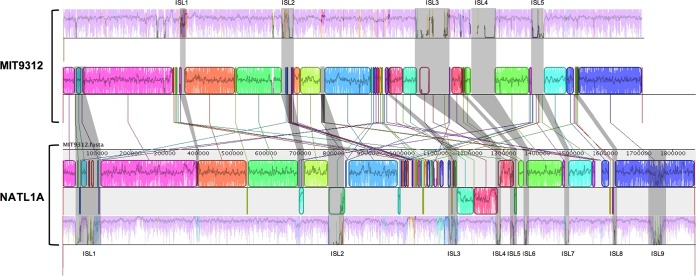
To better elucidate genome rearrangement, we compared two complete genomes from the HLII clade (MIT9312) and the LLI clade (NATL1A) in detail. Based on genome alignment, nine major genomic islands were found in the LLI genomes (we define major genomic islands as highly nonconserved regions that appeared in at least half of the genomes within a clade) (Fig. 3). By comparing the HLII and LLI genomes, we found that most LCB rearrangements and clade-specific segments (see below) were located in the vicinity of the genomic islands (Fig. 3). For example, in the NATL1A genome, most LCBs that have an inverse orientation relative to MIT9312 (LCB inversions) locate at or near the genome islands (Fig. 3). Genomic islands 1 and 2 (ISL 1 and 2) in the MIT9312 genome have multiple rearrangements (LCB translocations, shown as LCB linking to different locations in NATL1A in Fig. 3). These results indicate that the genomic islands serve as hotspots that induce genome rearrangements.
FIG 3.
Comparison of the backbone, genomic islands, and locally colinear blocks (LCBs) between the HLII (MIT9312) and LLI (NATL1A) clades. The genome comparison was generated by Mauve (53). Pink-colored segments are conserved among all strains within a clade (backbone), and other colored segments are conserved in a subset of strains. Shaded regions indicate genomic islands (ISL). LCBs are color coded. LCBs below the black line have an inverse orientation relative to that of MIT9312. Connecting lines between the two genomes indicate corresponding LCBs.
Genomic backbone and clade-specific segments.
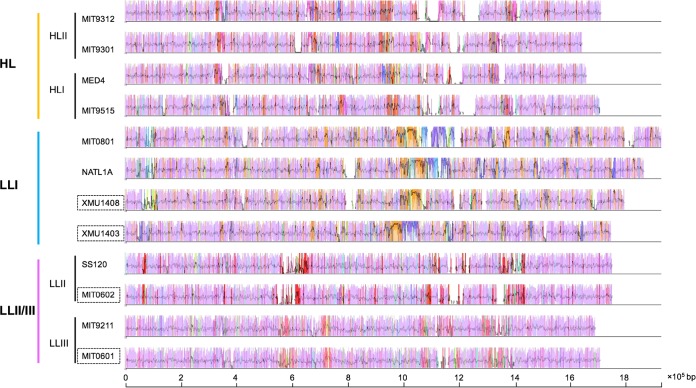
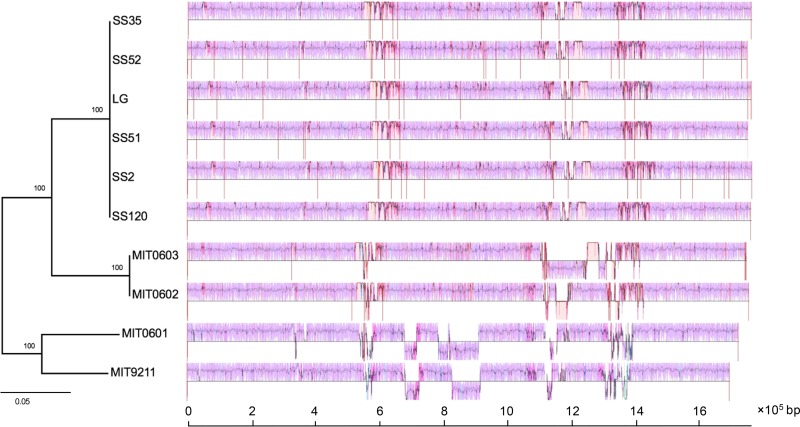
Comparison of closely related genomes has revealed the existence of highly conserved regions that form a genomic backbone, which is interrupted by many, less conserved genome regions (39). To investigate genome evolution, genome segmentation into backbone and variable regions is very useful. Thus, we performed genomic backbone analysis of streamlined clades and attempted to find the regions that may have served as hotspots of Prochlorococcus clade differentiation and ecological adaptation. The genome regions shared between all clades (backbones) and the regions shared between a subset of clades (clade-specific segments) were detected using genome alignment. Our results showed that the HL, LLI, and LLII/III genomes shared a conserved backbone, which was substantially different from that of LLIV clade (Fig. 2B), and each clade contained several clade-specific segments (Fig. 2B and 4). Compared to genome rearrangement, we found that these regions seemed to occur predominantly near regions that underwent genome rearrangement (Fig. 2B). In addition, we extended backbone analysis to the subclade level and found that there are substantial subclade-specific segments within clades. For example, XMU1408 has a different backbone pattern from other LLI strains (Fig. 5). In the LLII/III clade, the subclades also have different subclade-specific regions and genome rearrangement patterns (Fig. 6), which indicates that there are complex subclades within the LLII/III clade. A recent study grouped LLII and LLIII into a single clade (LLII/III) because they do not have a resolved phylogenetic relationship (19); however, our results indicate that LLII and LLIII belong to at least two separate clades based on genome structure differences (although, in this study, we still treated them as two subclades in the LLII/III clade for coherence).
FIG 4.
Visualization of the backbone and clade-specific regions among streamlined Prochlorococcus genomes. The genome comparison was generated by Mauve (53). Pink-colored segments are conserved among all strains (backbone), whereas segments in other colors are conserved in a subset of strains. All segments are above the black line because orientation was set relative to each genome for easier visualization. The large white regions indicate strain-specific content. Genome names in dashed boxes indicate draft genomes.
FIG 5.
Visualization of the backbone and subclade-specific regions among six Prochlorococcus LLI clade genomes. The genome comparison was generated by Mauve (53). Pink-colored segments are conserved among all strains (backbone), whereas segments in other colors are conserved in a subset of strains. Segments below the black line have an inverse orientation relative to PAC1. The large white regions indicate strain-specific content. The phylogenetic tree was reconstructed using concatenated protein sequences of 31 core genes with the maximum likelihood method using MIT9313 as an outgroup (not shown). Numbers at the nodes represent bootstrap values (1,000 resamplings). Vertical red lines indicate contig breaks.
FIG 6.
Visualization of the backbone and subclade-specific regions among 10 Prochlorococcus LLII/III clade genomes. The genome comparison was generated by Mauve (53). Pink-colored segments are conserved among all strains (backbone), whereas other segments are conserved in a subset of strains. Segments below the black line have an inverse orientation relative to that of SS120. The large white regions indicate strain-specific content. The phylogenetic tree was reconstructed using concatenated protein sequences of 31 core genes with the maximum likelihood method using MIT9313 as an outgroup (not shown). Numbers at the nodes represent bootstrap values (1,000 resamplings). Vertical red lines indicate contig breaks.
Genes in clade/subclade-specific segments.
Clade-specific segments are conserved regions shared within a clade but not with other clades; in contrast, genomic islands are hypervariable regions between different strains within a clade. Thus, the genes in clade-specific segments reflect clade differentiation and ecological adaptation, whereas genes in genomic islands reflect adaptations of individual strains to local niches. In this study, we attempted to find genes in these clade-specific regions and interpret the Prochlorococcus clade differentiation. Generally, most of the genes with known functions in clade-specific segments are related to light shock protection, DNA repair, and transporters, which correspond to the most dominant factors associated with Prochlorococcus evolution: light, DNA damage, and nutrient limitations (Table 2). At the clade level, the clade-specific segments of the LLI clade contain several genes that encode DNA ligase, high-light-induced proteins (HLIPs), and transporters. The unique regions of the HL clade have several genes for phosphorus transporters. It is worth noting that the HL and LLI clades share a segment that contains a urea assimilation gene cluster (although individual HL strains, such as MIT9515, lost this segment) (Table 2). The specific regions of the LLII/III clade contain genes for HLIP and DNA repair. We also extended this analysis to the subclade level. For example, compared to other LLI genomes, XMU1408 lacks a region that contains HLIP and DNA repair genes, while the LLII subclade has a region that contains DNA repair genes compared to the LLIII subclade (Table 2).
TABLE 2.
Examples of genes found in clade-specific regions
| Section | Gene name | Product | Locus |
|---|---|---|---|
| HL clade-specific region | Possible phosphate-binding protein | MED4_665181_665417 | |
| pstC | Phosphate transport system permease protein PstC | MED4_685879_686826 | |
| pstA | Phosphate transport system permease protein PstA | MED4_686833_687726 | |
| pstB | Phosphate transport ATP-binding protein PstB | MED4_687728_688537 | |
| LLI clade-specific region | pstB | Phosphate transport ATP-binding protein PstB | NATL2A_847867_847064 |
| pstA | Phosphate transport system permease protein PstA | NATL2A_848813_847899 | |
| pstC | Phosphate transport system permease protein PstC | NATL2A_849674_848814 | |
| DNA ligase | NATL2A_955315_954713 | ||
| DNA ligase | NATL2A_959226_959912 | ||
| hli11 | HLI protein hli11 | NATL2A_967126_966956 | |
| Ferric siderophore transport system, periplasmic binding protein TonB | NATL2A_1209792_1210433 | ||
| LLI subclade-specific regiona | hli11 | HLI protein hli11 | NATL2A_1052907_1052761 |
| Endonuclease VIII | NATL2A_1060998_1061843 | ||
| LLII/III clade-specific region | Helicase, SNF2/RAD54 family | SS120_49788_46600 | |
| hli | Phage-encoded HLI protein, HliP | SS120_1097431_1097565 | |
| hli | Possible HLI protein | SS120_1097580_1097789 | |
| LLII subclade-specific region | DNA double-strand break repair Rad50 ATPase | SS120_645533_642819 | |
| DNA double-strand break repair protein Mre11 | SS120_646724_645540 | ||
| Possible DNA gyrase/topoisomerase IV, subunit | SS120_647907_647629 | ||
| HL and LLI shared regionb | urtE | Urea ABC transporter, ATPase protein UrtE | NATL2A_1539531_1538824 |
| urtD | Urea ABC transporter, ATPase protein UrtD | NATL2A_1540283_1539531 | |
| urtC | Urea ABC transporter, permease protein UrtC | NATL2A_1541395_1540280 | |
| urtB | Urea ABC transporter, permease protein UrtB | NATL2A_1542556_1541399 | |
| urtB | Urea ABC transporter, substrate binding protein UrtA | NATL2A_1543910_1542633 | |
| ureG | Urease accessory protein UreG | NATL2A_1544660_1544043 | |
| ureF | Urease accessory protein UreF | NATL2A_1545331_1544660 | |
| ureE | Urease accessory protein UreE | NATL2A_1545747_1545352 | |
| ureD | Urease accessory protein UreD | NATL2A_1545895_1546818 |
XMU1408 does not contain this region.
MIT9515 does not contain this region.
Our results indicate that the streamlined Prochlorococcus clades share a conserved genomic backbone and have flexible regions that facilitate clade-specific ecological adaptations. In detail, after divergence from the MRCA, the LLII/III clade, which thrives in the deep euphotic zone, might face temporal light shock, whereas the LLI clade might face light shock, nutrient limitations, and DNA damage from UV in the middle to upper euphotic zone. The HL clade, which is well adapted to high-light conditions, might mainly face the problem of nutrient limitations in the oligotrophic surface oceans. These results of clade-specific segment analysis are consistent with a previous finding that associated clade differentiation with gene gain and loss at the whole-genome scale (24). However, our results indicate that genome evolution or clade differentiation is not evenly distributed in the Prochlorococcus genome: most parts of the genome were relatively conserved, but several regions served as evolutionary hotspots, which indicates that genome structure played an important role in Prochlorococcus evolution. Our results also indicate that the ability of the HL and LLI clades to adapt to intermediate- and high-light environments might have been, at least partially, acquired along with genome structure changes after they diverged from the MRCA of all streamlined clades. Thus, our results support the idea that genome streamlining, which resulted in reduced cell volume and increased ability to capture limited available light, might occur in low-light deep water rather than high-light surface water (3). A recent study also found that different environmental conditions (e.g., variation in nutrient chemistry) may influence transcription of transposases and may impact the genomic structure of the cyanobacterium Microcystis aeruginosa (40). Although the exact relations for the various shifts in ecological niches and subsequent adjustments of genome structure are still being determined, our results indicate a mechanism that can drive Prochlorococcus genome evolution.
Conclusions.
Prochlorococcus has rapidly become the focus of marine microbiology research, since it was discovered 30 years ago, because of its enormous biomass and important role in marine biogeochemical cycles. In this study, we reported genomes of two LLI strains isolated from the western Pacific Ocean and attempted to expand our understanding of Prochlorococcus genome evolution based on genome structure. Using genome rearrangement and backbone analysis, we reconstructed Prochlorococcus phylogeny and found that genome rearrangement coincided with genome reduction and might have played an important role in Prochlorococcus clade differentiation. We also found that the streamlined Prochlorococcus clades shared a conserved genomic backbone, but clade-specific regions facilitated ecological adaptations. Because our study only compared genomes from five cultivated clades that have whole-genome data, there is a large knowledge gap regarding the differentiation of the nonstreamlined LLIV genomes and the MRCA of the highly streamlined clades. In the future, a clearer picture of Prochlorococcus genome evolution will be produced as an increasing number of Prochlorococcus strains are isolated, especially from other LL clades that may have partially streamlined genomes and different genome structures.
MATERIALS AND METHODS
Isolation and culture conditions.
Two LLI strains were isolated in the western Pacific Ocean in 2014 (details regarding the isolation locations are shown in Table 1). Briefly, seawater collected with a Niskin bottle was filtered by gravity through two polycarbonate filters with 0.6-μm pore sizes (1), and Pro2 medium stock solution was added to the filtrates (41). These filtrates then were placed in an incubator on board for initial enrichment under constant light flux of 5 to 10 μmol Q m−2 s−1 at 22°C. After 4 to 8 weeks, the successful initial Prochlorococcus cultures were enriched again by the serial dilution method (42). Briefly, Prochlorococcus cells in the cultures were counted by flow cytometry and then serially diluted into 96-well plates at final concentrations of 1 to 10 cells per well, and a modified ProMM medium (an f/2 vitamin mix instead of the 1× Va vitamin mix) was used. The wells that appeared green were immediately transferred to Pro99 medium and were further confirmed by flow cytometry and 16S-23S rRNA internal transcribed spacer (ITS) sequence analysis. The Prochlorococcus strains were maintained under constant light flux of 10 to 20 μmol Q m−2 s−1 at 22°C. The presence of heterotrophic bacteria was routinely tested with ProMM (42) and ProAC (43) medium.
DNA sequencing, assembly, and annotation.
Genomic DNA was collected from 25 ml of the laboratory cultures by centrifugation (10,000 × g, 15 min). The QIAamp DNA minikit (Qiagen, Germany) was used to extract genomic DNA. One microgram of genomic DNA was used to construct libraries for Illumina sequencing. Ten nanograms of library DNA was bidirectionally sequenced using a HiSeq 2500 sequencer (Illumina, USA) with a read length of 150 bp. Library construction and sequencing were performed at Shanghai Hanyu Biotechnology Co. (Shanghai, China). Sequencing data of each sample were first trimmed by Trimmomatic v0.32 (44). The clean data with high quality then were assembled using IDBA-UD (45) with a customized kmer (from 21 to 121 with a 10-mer step size). Because the cultures sequenced are not axenic, genome binning was performed using a modified workflow from a well-known method (46) to recover cyanobacterial genomes. Briefly, the binning was based on GC content, sequencing depth, and tetranucleotide frequency to separate each contig by species. To improve the assembly, reads associated with the putative genome bins were extracted for reassembly using SPAdes v3.33.1 (kmer from 21 to 121 with a 10-mer step size) (47) after decreasing the coverage of reads to 100× by BBTools (https://jgi.doe.gov/data-and-tools/bbtools/). Finally, the reassembled bins were estimated by CheckM (35) to assess the completeness and contamination of the recovered genomes. Assembled genomic sequences were annotated using the RAST server against FIGfam, release 70 (48). For comparison, we also reannotated the previously published Prochlorococcus genomes (Table 3) using the same method.
Phylogenomic analysis.
To infer robust phylogenetic relationships between distinct clades, a phylogenetic tree was reconstructed using concatenated protein sequences of 31 core genes (49). Briefly, protein sequences of 31 core genes were concatenated and then aligned with MFFAT v7 (50). Phylogenetic trees were reconstructed using MEGA v7.0 with the maximum likelihood method and 1,000 bootstrap replicates (51). The pan-, core, and accessory genomes of six LLI genomes were calculated using Panseq (52), and the following values for the parameters were set: fragmentation size, 500 bp; homology fragment similarity percent identity cutoff, >70%; blast size, 20.
Genome rearrangement and backbone analysis.
Genome rearrangement analyses were performed on 14 complete genomes with or without draft genomes. For complete genomes, genome alignment was conducted using the progressiveMauve algorithm in Mauve v2.4.0 (53, 54). Locally colinear block (LCB) detection and backbone analysis were performed using the default parameters in the progressiveMauve algorithm, and a permutation matrix of LCBs was exported for phylogeny reconstruction. MLGO (55, 56) was used to reconstruct the phylogenetic tree based on the permutation matrix of LCBs using maximum likelihood with 1,000 bootstrap replicates. For draft genomes, contigs were first reordered using the Move Contigs tool (57), which reorders all contigs based on whole-genome comparison to the closest complete genome defined by the phylogenomic tree. All contig positions were also manually checked. Accession numbers of all reference genomes used in this study are shown in Table 3.
Accession number(s).
The genomic sequences of the two newly isolated LLI strains were deposited in GenBank under BioProject number PRJNA474570. The genome accession number of each strain is shown in Table 1.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Public Science and Technology Research Fund Projects for Ocean Research (201505003-3), National Natural Science Foundation of China (41522603, 31570172). R.Z. was partially supported by Qingdao National Laboratory for Marine Science and Technology (QNLM2016ORP0303).
We thank the captains and crews of the RV Kexue for their excellent support during the cruise (KEXUE2018G07).
We have no competing interests to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01178-18.
REFERENCES
- 1.Chisholm SW, Frankel SL, Goericke R, Olson RJ, Palenik B, Waterbury JB, West-Johnsrud L, Zettler ER. 1992. Prochlorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b. Arch Microbiol 157:297–300. doi: 10.1007/BF00245165. [DOI] [Google Scholar]
- 2.Partensky F, Hess WR, Vaulot D. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev 63:106–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Partensky F, Garczarek L. 2010. Prochlorococcus: advantages and limits of minimalism. Annu Rev Mar Sci 2:305–331. doi: 10.1146/annurev-marine-120308-081034. [DOI] [PubMed] [Google Scholar]
- 4.Zinser ER, Coe A, Johnson ZI, Martiny AC, Fuller NJ, Scanlan DJ, Chisholm SW. 2006. Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl Environ Microbiol 72:723–732. doi: 10.1128/AEM.72.1.723-732.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. 2006. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 6.Campbell L, Liu H, Nolla HA, Vaulot D. 1997. Annual variability of phytoplankton and bacteria in the subtropical North Pacific Ocean at Station ALOHA during the 1991-1994 ENSO event. Deep Sea Res Part I Oceanogr Res Pap 44:167–192. doi: 10.1016/S0967-0637(96)00102-1. [DOI] [Google Scholar]
- 7.Vaulot D, Marie D, Olson RJ, Chisholm SW. 1995. Growth of Prochlorococcus, a photosynthetic prokaryote, in the Equatorial Pacific Ocean. Science 268:1480–1482. doi: 10.1126/science.268.5216.1480. [DOI] [PubMed] [Google Scholar]
- 8.DuRand MD, Olson RJ, Chisholm SW. 2001. Phytoplankton population dynamics at the Bermuda Atlantic time-series station in the Sargasso Sea. Deep-Sea Res Part II 48:1983–2003. doi: 10.1016/S0967-0645(00)00166-1. [DOI] [Google Scholar]
- 9.Partensky F, Blanchot J, Vaulot D. 1999. Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. Bull Inst Océanogr (Monaco) 19:457–476. [Google Scholar]
- 10.Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N, Karl DM, Li WKW, Lomas MW, Veneziano D, Vera CS, Vrugt JA, Martiny AC. 2013. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci U S A 110:9824–9829. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore LR, Goericke R, Chisholm SW. 1995. Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar Ecol Prog Ser 116:259–275. doi: 10.3354/meps116259. [DOI] [Google Scholar]
- 12.Moore LR, Rocap G, Chisholm SW. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 13.Rocap G, Distel DL, Waterbury JB, Chisholm SW. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol 68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rusch DB, Martiny AC, Dupont CL, Halpern AL, Venter JC. 2010. Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proc Natl Acad Sci U S A 107:16184–16189. doi: 10.1073/pnas.1009513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.West NJ, Lebaron P, Strutton PG, Suzuki MT. 2011. A novel clade of Prochlorococcus found in high nutrient low chlorophyll waters in the South and Equatorial Pacific Ocean. ISME J 5:933–944. doi: 10.1038/ismej.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang S, Wilhelm SW, Harvey HR, Taylor K, Jiao N, Chen F. 2012. Novel lineages of Prochlorococcus and Synechococcus in the global oceans. ISME J 6:285–297. doi: 10.1038/ismej.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martiny AC, Tai APK, Veneziano D, Primeau F, Chisholm SW. 2009. Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Environ Microbiol 11:823–832. doi: 10.1111/j.1462-2920.2008.01803.x. [DOI] [PubMed] [Google Scholar]
- 18.Lavin P, González B, Santibáñez JF, Scanlan DJ, Ulloa O. 2010. Novel lineages of Prochlorococcus thrive within the oxygen minimum zone of the eastern tropical South Pacific. Environ Microbiol Rep 2:728–738. doi: 10.1111/j.1758-2229.2010.00167.x. [DOI] [PubMed] [Google Scholar]
- 19.Biller SJ, Berube PM, Lindell D, Chisholm SW. 2015. Prochlorococcus: the structure and function of collective diversity. Nat Rev Microbiol 13:13–27. doi: 10.1038/nrmicro3378. [DOI] [PubMed] [Google Scholar]
- 20.Kelly L, Huang KH, Ding H, Chisholm SW. 2012. ProPortal: a resource for integrated systems biology of Prochlorococcus and its phage. Nucleic Acids Res 40:D632–D640. doi: 10.1093/nar/gkr1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dufresne A, Salanoubat M, Partensky F, Artiguenave F, Axmann IM, Barbe V, Duprat S, Galperin MY, Koonin EV, Le Gall F, Makarova KS, Ostrowski M, Oztas S, Robert C, Rogozin IB, Scanlan DJ, Tandeau de Marsac N, Weissenbach J, Wincker P, Wolf YI, Hess WR. 2003. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc Natl Acad Sci U S A 100:10020–10025. doi: 10.1073/pnas.1733211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, Johnson ZI, Land M, Lindell D, Post AF, Regala W, Shah M, Shaw SL, Steglich C, Sullivan MB, Ting CS, Tolonen A, Webb EA, Zinser ER, Chisholm SW. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 23.Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, DeLong EF, Chisholm SW. 2006. Genomic islands and the ecology and evolution of Prochlorococcus. Science 311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- 24.Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S, Chen F, Lapidus A, Ferriera S, Johnson J, Steglich C, Church GM, Richardson P, Chisholm SW. 2007. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet 3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biller SJ, Berube PM, Berta-Thompson JW, Kelly L, Roggensack SE, Awad L, Roache-Johnson KH, Ding H, Giovannoni SJ, Rocap G, Moore LR, Chisholm SW. 2014. Genomes of diverse isolates of the marine cyanobacterium Prochlorococcus. Sci Data 1:140034. doi: 10.1038/sdata.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hess WR. 2011. Cyanobacterial genomics for ecology and biotechnology. Curr Opin Microbiol 14:608–614. doi: 10.1016/j.mib.2011.07.024. [DOI] [PubMed] [Google Scholar]
- 27.Luo H, Friedman R, Tang J, Hughes AL. 2011. Genome reduction by deletion of paralogs in the marine cyanobacterium Prochlorococcus. Mol Biol Evol 28:2751–2760. doi: 10.1093/molbev/msr081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo H, Huang Y, Stepanauskas R, Tang J. 2017. Excess of non-conservative amino acid changes in marine bacterioplankton lineages with reduced genomes. Nat Microbiol 2:17091. doi: 10.1038/nmicrobiol.2017.91. [DOI] [PubMed] [Google Scholar]
- 29.Ahlgren NA, Rocap G, Chisholm SW. 2006. Measurement of Prochlorococcus ecotypes using real-time polymerase chain reaction reveals different abundances of genotypes with similar light physiologies. Environ Microbiol 8:441–454. doi: 10.1111/j.1462-2920.2005.00910.x. [DOI] [PubMed] [Google Scholar]
- 30.Darling AE, Miklós I, Ragan MA. 2008. Dynamics of genome rearrangement in bacterial populations. PLoS Genet 4:e1000128. doi: 10.1371/journal.pgen.1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moret BM, Wang LS, Warnow T, Wyman SK. 2001. New approaches for reconstructing phylogenies from gene order data. Bioinformatics 17(Suppl 1):S165–S173. doi: 10.1093/bioinformatics/17.suppl_1.S165. [DOI] [PubMed] [Google Scholar]
- 32.Duchemin W, Daubin V, Tannier E. 2015. Reconstruction of an ancestral Yersinia pestis genome and comparison with an ancient sequence. BMC Genomics 16:S9. doi: 10.1186/1471-2164-16-S10-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo H, Shi J, Arndt W, Tang J, Friedman R. 2008. Gene order phylogeny of the genus Prochlorococcus. PLoS One 3:e3837. doi: 10.1371/journal.pone.0003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prabha R, Singh DP, Gupta SK, Rai A. 2014. Whole genome phylogeny of Prochlorococcus marinus group of cyanobacteria: genome alignment and overlapping gene approach. Interdiscip Sci Comput Life Sci 6:149–157. doi: 10.1007/s12539-013-0024-9. [DOI] [PubMed] [Google Scholar]
- 35.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marais GAB, Calteau A, Tenaillon O. 2008. Mutation rate and genome reduction in endosymbiotic and free-living bacteria. Genetica 134:205–210. doi: 10.1007/s10709-007-9226-6. [DOI] [PubMed] [Google Scholar]
- 37.Couturier E, Rocha EPC. 2006. Replication-associated gene dosage effects shape the genomes of fast-growing bacteria but only for transcription and translation genes. Mol Microbiol 59:1506–1518. doi: 10.1111/j.1365-2958.2006.05046.x. [DOI] [PubMed] [Google Scholar]
- 38.Sousa C, de Lorenzo V, Cebolla A. 1997. Modulation of gene expression through chromosomal positioning in Escherichia coli. Microbiology (Reading, Engl) 143(Part 6):2071–2078. doi: 10.1099/00221287-143-6-2071. [DOI] [PubMed] [Google Scholar]
- 39.Devillers H, Chiapello H, Schbath S, Karoui ME. 2011. Robustness assessment of whole bacterial genome segmentations. J Comput Biol 18:1155–1165. doi: 10.1089/cmb.2011.0115. [DOI] [PubMed] [Google Scholar]
- 40.Steffen MM, Dearth SP, Dill BD, Li Z, Larsen KM, Campagna SR, Wilhelm SW. 2014. Nutrients drive transcriptional changes that maintain metabolic homeostasis but alter genome architecture in Microcystis. ISME J 8:2080–2092. doi: 10.1038/ismej.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore LR, Coe A, Zinser ER, Saito MA, Sullivan MB, Lindell D, Frois-Moniz K, Waterbury J, Chisholm SW. 2007. Culturing the marine cyanobacterium Prochlorococcus. Limnol Oceanogr Methods 5:353–362. doi: 10.4319/lom.2007.5.353. [DOI] [Google Scholar]
- 42.Berube PM, Biller SJ, Kent AG, Berta-Thompson JW, Roggensack SE, Roache-Johnson KH, Ackerman M, Moore LR, Meisel JD, Sher D, Thompson LR, Campbell L, Martiny AC, Chisholm SW. 2015. Physiology and evolution of nitrate acquisition in Prochlorococcus. ISME J 9:1195–1207. doi: 10.1038/ismej.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. 2008. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl Environ Microbiol 74:4530–4534. doi: 10.1128/AEM.02479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 46.Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KARL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 47.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75–15. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu M, Eisen JA. 2008. A simple, fast, and accurate method of phylogenomic inference. Genome Biol 9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laing C, Buchanan C, Taboada EN, Zhang Y, Kropinski A, Villegas A, Thomas JE, Gannon VP. 2010. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics 11:461–414. doi: 10.1186/1471-2105-11-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin Y, Hu F, Tang J, Moret BME. 2013. Maximum likelihood phylogenetic reconstruction from high-resolution whole-genome data and a tree of 68 eukaryotes. Pac Symp Biocomput 2013:285–296. [PMC free article] [PubMed] [Google Scholar]
- 56.Hu F, Lin Y, Tang J. 2014. MLGO: phylogeny reconstruction and ancestral inference from gene-order data. BMC Bioinformatics 15:3340–3347. doi: 10.1186/s12859-014-0354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. 2009. Reordering contigs of draft genomes using the Mauve Aligner. Bioinformatics 25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.