Benzalkonium chlorides (BAC) are biocides broadly used in disinfectant solutions. Disinfectants are widely used in food processing lines, domestic households, and pharmaceutical products and are typically designed to have a different mode of action than antibiotics to avoid interfering with the use of the latter. Whether exposure to BAC makes bacteria more resistant to antibiotics remains an unresolved issue of obvious practical consequences for public health. Using an integrated approach that combines metagenomics of natural microbial communities with gene cloning experiments with isolates and experimental evolution assays, we show that the widely used benzalkonium chloride disinfectants promote clinically relevant antibiotic resistance. Therefore, more attention should be given to the usage of these disinfectants, and their fate in nontarget environments should be monitored more tightly.
KEYWORDS: disinfectants, antibiotics, cross-resistance, metagenomics, Pseudomonas aeruginosa
ABSTRACT
While the misuse of antibiotics has clearly contributed to the emergence and proliferation of resistant bacterial pathogens, with major health consequences, it remains less clear if the widespread use of disinfectants, such as benzalkonium chlorides (BAC), a different class of biocides than antibiotics, has contributed to this problem. Here, we provide evidence that exposure to BAC coselects for antibiotic-resistant bacteria and describe the underlying genetic mechanisms. After inoculation with river sediment, BAC-fed bioreactors selected for several bacterial taxa, including the opportunistic pathogen Pseudomonas aeruginosa, that were more resistant to several antibiotics than their counterparts in a control (no BAC) bioreactor. A metagenomic analysis of the bioreactor microbial communities, confirmed by gene cloning experiments with the derived isolates, suggested that integrative and conjugative elements encoding a BAC efflux pump together with antibiotic resistance genes were responsible for these results. Furthermore, the exposure of the P. aeruginosa isolates to increasing concentrations of BAC selected for mutations in pmrB (polymyxin resistance) and physiological adaptations that contributed to a higher tolerance to polymyxin B and other antibiotics. The physiological adaptations included the overexpression of mexCD-oprJ multidrug efflux pump genes when BAC was added in the growth medium at subinhibitory concentrations. Collectively, our results demonstrated that disinfectants promote antibiotic resistance via several mechanisms and highlight the need to remediate (degrade) disinfectants in nontarget environments to further restrain the spread of antibiotic-resistant bacteria.
IMPORTANCE Benzalkonium chlorides (BAC) are biocides broadly used in disinfectant solutions. Disinfectants are widely used in food processing lines, domestic households, and pharmaceutical products and are typically designed to have a different mode of action than antibiotics to avoid interfering with the use of the latter. Whether exposure to BAC makes bacteria more resistant to antibiotics remains an unresolved issue of obvious practical consequences for public health. Using an integrated approach that combines metagenomics of natural microbial communities with gene cloning experiments with isolates and experimental evolution assays, we show that the widely used benzalkonium chloride disinfectants promote clinically relevant antibiotic resistance. Therefore, more attention should be given to the usage of these disinfectants, and their fate in nontarget environments should be monitored more tightly.
INTRODUCTION
While the inappropriate prescription of antibiotics to humans and the misuse of antibiotics in animal feed are thought to be the leading causes of the increased frequency of antibiotic resistance observed in recent years, there is an increasing concern that widely used disinfectants such as quaternary ammonium compounds (QAC) have contributed to the antibiotic resistance problem (1, 2). However, this issue remains highly debatable because disinfectants typically have a different mode of action than antibiotics to avoid overlap between these two distinct classes of biocides that are used for different purposes. Furthermore, previous studies have yet to elucidate the underlying mechanisms for the reported linkage between disinfectant exposure and increased antibiotic resistance (3–8). For instance, Pseudomonas aeruginosa, an important opportunistic human pathogen, acquired a 12-fold increased resistance to disinfectants upon disinfectant exposure, which was accompanied by a >200-fold increase in ciprofloxacin resistance (4). The increased ciprofloxacin resistance was attributed to specific mutations in gyrA, but it remained unclear if the mutations were indeed induced by the disinfectant, occurred spontaneously, or were a result of the growth conditions. The different modes of action and target sites between QAC and ciprofloxacin and the fact that QAC do not target the product of the gyrA gene argue in support of the latter interpretation.
Furthermore, other studies reported that antibiotic and disinfectant cross-resistance is absent in environmental, clinical, and industrial isolates, and disinfectant-resistant pathogens often show increased antibiotic susceptibility (9–13). Even the recent restriction in the use of triclosan and triclocarban, another family of disinfectants widely used in antimicrobial soaps, is not accompanied by consistent evidence on the linkage between triclosan exposure and antibiotic resistance (14). For instance, several studies reported decreased susceptibility of Escherichia coli to chloramphenicol, erythromycin, imipenem, tetracycline, trimethoprim, and other biocides as an effect of exposure to sublethal concentrations of triclosan (15). However, other studies suggested that there is no clear link between increased triclosan tolerance and increased antibiotic resistance and actually found that E. coli with increased triclosan tolerance is more sensitive to aminoglycoside antibiotics (16).
Benzalkonium chlorides (BAC) are the most commonly used members of the QAC family of disinfectants, have broad-spectrum (i.e., bacterial, algal, fungal, and viral) biocidal activity, and remain stable for both short- and long-term usage (17). Consequently, BAC are widely used as surface-disinfecting agents in food processing lines (e.g., poultry facilities), dairy/agricultural settings, health care facilities, and domestic households and are popular ingredients in over-the-counter cosmetics, hand sanitizers, and pharmaceutical products (18). Therefore, BAC represent ideal molecules to study the effect of disinfectant exposure on microbial antibiotic resistance. Understanding whether the exposure of bacteria to BAC leads to increased antibiotic resistance and, if so, what the underlying molecular mechanisms might be is important for better regulating the usage of BAC and minimizing public risk.
To obtain insights into this issue, we exposed a microbial community originating from a river sediment inoculum (Calcasieu River, USA) to BAC for 3 years in aerobic fed-batch bioreactors with either dextrin-peptone plus BAC (DPB bioreactor), dextrin-peptone (DP bioreactor), or BAC only (B bioreactor) as the main carbon and energy sources, as described previously (6, 19). The Calcasieu River is heavily contaminated with metals, polycyclic aromatic hydrocarbons, and chlorinated/halogenated organic compounds. Therefore, its sediment represented an ideal inoculum for our purposes to identify organisms with resistance to BAC and other organics, since BAC typically accumulate in nontarget sediments and anoxic habitats (though the exact inoculum used in the bioreactors did not contain detectable concentrations of BAC at the time of sampling) (19). Three years of BAC exposure led to significant changes in the composition of microbial communities compared to that of a control community (DP) under the same laboratory conditions (see Fig. S1A in the supplemental material). The BAC exposure also selected for efficient degraders of BAC, which we reported previously together with the identification of a novel BAC-degrading gene cassette (20). Here, we focused on the effects of BAC exposure on antibiotic resistance. We also assessed the effects of BAC exposure on shorter time scales by exposing isolates from the control (DP) and DPB bioreactors to BAC for approximately 200 generations or less (1 to 2 months).
RESULTS AND DISCUSSION
Microbial community exposure to BAC selected for members with increased resistance to antibiotics.
To examine whether BAC exposure promoted (i.e., increased) antibiotic resistance, pairs of isolates, one originating from the DPB or B bioreactor (i.e., BAC fed) and the other representing its counterpart (same species) from the DP bioreactor (i.e., not BAC fed), were characterized for their resistance to representatives of seven classes of antibiotics. The isolates represented four distinct species that were relatively abundant in the bioreactors and highly represented among the total isolates obtained (see Fig. S1A in the supplemental material). P. aeruginosa strain DPB showed higher MIC values for BAC and several, but not all, antibiotics than P. aeruginosa strain DP (Table 1). The genomes of these two P. aeruginosa isolates differed in their shared genes by five single nucleotide polymorphisms (SNPs), one small (9 bp) insertion in their shared genes and two genomic islands (GIs) (Fig. 1A). This number of mutations was comparable to the number of mutations predicted (n = ∼10) to be fixed under neutral evolution on the basis of the spontaneous mutation rate for a bacterial genome (5.4 × 10−10 per base per generation) (21), the genome size of the P. aeruginosa isolates (∼6.4 Mb), and the estimated number of generations since the establishment of the DPB and DP communities after 3 years (∼3,000 generations). Hence, it is highly likely that these isolates represented descendants of the same ancestor in the original river sediment inoculum. P. aeruginosa can grow in soil, marshes, coastal marine habitats, plants, and animal tissues (22), and it is commonly isolated from sediments (23). Therefore, it was not surprising to recover P. aeruginosa from our sediment inoculum.
TABLE 1.
MIC of BAC and antibiotics for isolates of the same species
| Species | 16S rRNA gene identity (%) | ANIa (%) | MIC (mg/liter)b |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BAC | TET | CIP | CHL | POL | KAN | RIF | AMP | |||
| Achromobacter denitrificans DP | —c | — | 15.6 | <3.1 | <0.8 | 3.1 | <0.4 | 100 | 12.5 | 200 |
| Achromobacter sp. DPB | 97.8 | 87.5 | 31.3 | 25 | 1.6 | 25 | <0.4 | 25 | 3.1 | <6.3 |
| Achromobacter sp. B | 97.7 | 87.5 | 125 | 200 | 12.5 | 100 | 3.1 | 100 | 12.5 | <6.3 |
| Citrobacter freundii (human)d | — | — | <7.8 | 1.6 | 0.4 | 12.5 | 0.2 | 6.3 | 25 | 800 |
| Citrobacter freundii DP | 98.5 | N/A | 62.5 | 1.6 | 1.6 | 12.5 | 0.8 | 6.3 | 25 | 12.5 |
| Citrobacter freundii DPB | 99.5 | 94.2 | 62.5 | 1.6 | 0.8 | 12.5 | 0.4 | 6.3 | 25 | 25 |
| Klebsiella michiganensis (type strain)e | — | — | 15.6 | 1.6 | <0.1 | 6.3 | 1.6 | 200 | 12.5 | 200 |
| Klebsiella michiganensis DP | 99.1 | N/A | 62.5 | 1.6 | 1.6 | 12.5 | 0.8 | 25 | 12.5 | 100 |
| Klebsiella michiganensis DPB | 99.9 | 99.4 | 62.5 | 1.6 | 1.6 | 12.5 | 1.6 | 25 | 25 | 100 |
| Pseudomonas aeruginosa DP | — | — | 50 | 12.5 | 0.1 | 50 | 0.2 | 100 | 12.5 | 3,200 |
| Pseudomonas aeruginosa DPB | 100 | 100 | 200 | 6.25 | 0.2 | 100 | 0.2 | 100 | 25 | 3,200 |
ANI, average nucleotide identity.
A range of antibiotic and BAC concentrations (0.4 to 3,200 mg/liter) was tested. TET, tetracycline; CIP, ciprofloxacin; CHL, chloramphenicol; POL, polymyxin B; KAN, kanamycin; RIF, rifampin; AMP, ampicillin.
—, control isolate of the same or closely related species for comparisons.
C. freundii human strain was 4_7_47_CFAA, accession no. ADLG00000000.1, of the Human Microbiome Project (http://www.hmpdacc.org/).
K. michiganensis type strain was KCTC 1686, accession no. CP003218.1, of the Korean Collection for Type Cultures.
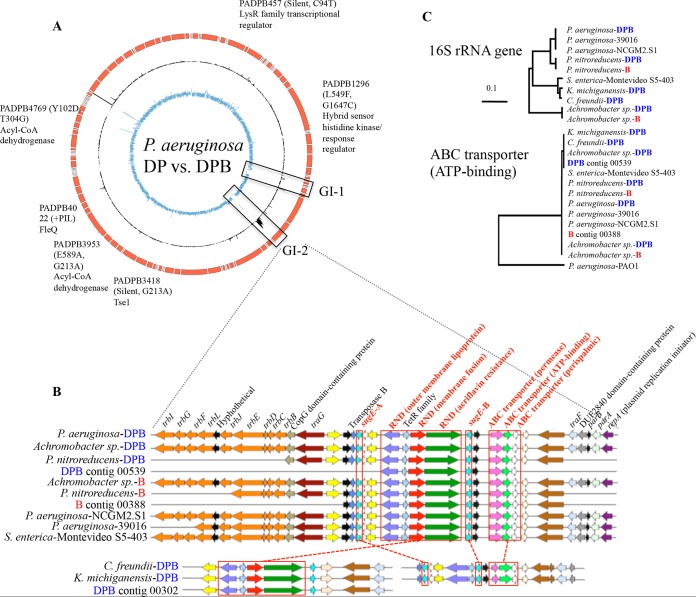
FIG 1.
Horizontal transfer of antibiotic efflux pump genes. (A) Comparison of the draft genomes of P. aeruginosa DPB versus P. aeruginosa DP. Circles represent contigs of P. aeruginosa DPB (red), coverage of contigs by metagenomic reads of DPB reactor (black), and the P. aeruginosa-DP strain (blue). Genes with mutations found between the two genomes are denoted on the graph (ticks on the P. aeruginosa DPB draft genome). (B) Graphic representation of the organization of genes in the ICE recovered from isolate genomes and metagenomes. (C) Phylogenetic tree of 16S rRNA gene sequences and ATP binding gene (of the ABC transporter) sequences based on the maximum likelihood composite model and the neighbor-joining method. Note that phylogenetically diverse isolates shared almost identical (>99% identity) ATP binding gene sequences, suggesting horizontal transfer of the ATP binding gene. Similar results were also observed for sugE-A and sugE-B genes (data not shown). The reference sequences used were P. aeruginosa 39016 (accession no. NZ_CM001020.1), P. aeruginosa NCGM2.S1 (NC_017549.1), P. aeruginosa PAO1 (NC_002516.2), and Salmonella enterica Montevideo S5-403 (AFCS01000001.1).
Similar antibiotic resistance results to those of the P. aeruginosa isolates were observed for Achromobacter sp. strain DPB/B and Achromobacter denitrificans strain DP, which represented more divergent genotypes than the P. aeruginosa isolates (genome aggregate average nucleotide identity [ANI] between the Achromobacter sp. DPB/B and A. denitrificans DP isolates was 87.5%) (Table 1). In contrast, Citrobacter freundii strain DPB and Klebsiella michiganensis strain DPB did not exhibit significant changes in MIC values for antibiotics compared to their unexposed counterparts (Table 1). However, the latter two isolates showed higher MIC values for BAC and several antibiotics than C. freundii and K. michiganensis isolates from culture collections (type strains) or the human gastrointestinal tract, indicating that C. freundii DP and K. michiganensis DP might have been intrinsically resistant to several of the antibiotics tested here before BAC exposure. Overall, although BAC exposure did not result in higher antibiotic resistance in all isolates tested, at least P. aeruginosa showed reduced susceptibilities to BAC by 4-fold and to three antibiotics from the seven tested, including ciprofloxacin, chloramphenicol, and rifampin, by 2-fold, suggesting a resistance link between BAC and antibiotics.
Genetic elements for resistance to both BAC and antibiotics were responsible for the link.
To investigate the molecular mechanisms responsible for the resistance link, we conducted whole-isolate and bioreactor metagenome DNA sequencing. An alignment of the P. aeruginosa DP genomic reads against the P. aeruginosa DPB assembled contigs identified, in addition to the five SNPs and the small insertion mentioned above, two GIs that encoded several integrases and transposases (GI-1) and several (predicted) resistance genes (GI-2) (Fig. 1A). GI-2 harbored four predicted efflux pump systems: two small multidrug-resistant (SMR) family systems (sugE-A and sugE-B), an ATP-binding cassette (ABC) family, and a resistance nodulation division (RND) family member (Fig. 1B). Furthermore, GI-2 harbored all necessary genes for conjugation, such as those for conjugative transfer (trbBCDEJLFGI), plasmid partitioning genes (parAB), and a plasmid replication initiator (repA), in a single assembled Illumina contig. The recruitment of Illumina reads from the DPB metagenome revealed that the coverage of GI-2 (average, 17.7 reads/base pair [17.7×]) was significantly higher (P < 0.001, Student's t test) than for the rest of the P. aeruginosa DPB genome (average, 2.0×), indicating that GI-2 was present in additional members of the DPB community or in multicopy (Fig. 1A). Consistent with this interpretation, a genome sequence analysis of Achromobacter sp., Pseudomonas nitroreducens, C. freundii, K. michiganensis, and P. aeruginosa isolates from DPB or B communities revealed almost identical (>99% nucleotide identity) genetic elements despite the substantial evolutionary divergence of the organisms (Fig. 1B and C). Since GI-2 was the only genomic difference between P. aeruginosa DP and DPB strains that was bioinformatically predicted to encode enzymes potentially relevant for BAC and antibiotic resistance, we hypothesized that GI-2 might have been responsible for the higher antibiotic resistance of strain P. aeruginosa DPB than of strain DP; thus, we studied this genomic island in more detail.
Even though GI-2 was predicted to harbor several of the genes commonly found in conjugative plasmids, long-read Oxford Nanopore sequencing showed that the conjugative element was integrated in the genome of P. aeruginosa DPB. The hybrid assembly of the MinION long reads and Illumina contigs produced a closed genome (single contig), with even coverage across the genome, including the junctions/ends of the Illumina contig harboring the conjugative transfer genes. Further bioinformatics analysis and gene annotation, e.g., the presence of recombinase genes (often referred to as integrases) and conjugation system genes (see Table S1) (24, 25), suggested that GI-2 is most likely an integrative and conjugative element (ICE). ICEs share gene content with conjugative plasmids (e.g., genes for conjugation) and are self-transmissible genetic elements that can be integrated in the chromosome or propagate as independently replicating molecules (see supporting results and discussion in the supplemental material for further details) (24, 25). An alignment of genomic reads of other isolates, i.e., Achromobacter sp. DPB/B, C. freundii DPB, and K. michiganensis DPB, against the P. aeruginosa DPB GI-2 sequence suggested that not all of the isolates shared the whole GI-2 or all integrases, except for Achromobacter sp. DPB (Fig. S2). These results revealed that GI-2 had likely been mobilized and/or integrated into diverse organisms during the bioreactor incubation time (3 years), and various versions of ICEs harboring the same resistance genes were present within the community and were apparently strongly selected for by the BAC exposure conditions.
To test the hypothesis that genes within GI-2 helped organisms to cope with BAC toxicity and were responsible for the high antibiotic resistance of P. aeruginosa strain DPB relative to that of strain DP, the sugE-A, sugE-B, and ABC transporter system genes were cloned into a broad-host range vector (pBBRMCS-4) and transformed into P. aeruginosa strain PAO509. PAO509 lacks several efflux pump systems present in the wild-type P. aeruginosa and our isolates from the bioreactors and hence is a more appropriate strain background to test for antibiotic resistance phenotypes. Plasmid pBBRsugE-A conferred increased resistance to BAC, with an ∼2-fold higher MIC, but not to antibiotics, while plasmid pBBRABC conferred resistance to rifampin (∼2-fold higher MIC) but BAC tolerance was similar to that of the control vector alone (pBBRMCS-4) (see Table S2). It should be noted that even a 2-fold higher MIC can be clinically significant, and efflux pumps genes such as sugE-A and ABC do not often provide much greater changes in MIC values (26–29). The BAC resistance phenotype of the transformant carrying sugE-A (PAO509/pBBRsugE-A) was also in agreement with a previous report that sugE confers resistance to BAC (30). Rifampin is known to act on the RNA polymerase β subunit (RpoB) to interfere with transcription, and rifampin-resistant bacteria frequently emerge due to single point mutations in rpoB (31). To exclude the possibility that the rifampin-resistant phenotype was due to spontaneous mutation (as opposed to the ABC transporter system cloned), rpoB gene sequences of rifampin resistance clusters I, II, and III were PCR amplified from four independent colonies of PAO509 and PAO509/pBBRABC using previously determined primers PAO1rpoB1 (32) and PAOrpoB3 (33) and subsequently sequenced using Sanger chemistry. No mutation was identified between PAO509 and PAO509/pBBRABC, suggesting that the rifampin resistance phenotype was likely not due to spontaneous mutation but to the ABC transporter system. No difference in tetracycline susceptibility was observed in the three recombinants compared to that of the control. These results suggested that two distinct gene cassettes harbored by GI-2 conferred resistance to antibiotics and BAC, but no one gene conferred resistance to both BAC and antibiotics. Therefore, the resistance linkages appeared to be due to the cooccurrence of these two gene cassettes on the same piece of mobile DNA. It still remains unclear at this point if the colocalization of the genes occurred before or after exposure to BAC.
BAC adaptation selected for P. aeruginosa with increased resistance to polymyxin B.
To identify alternative genetic mechanisms that may be responsible for the resistance linkage observed but not elucidated by our genetic manipulations and assess the effects of shorter BAC exposure than that experienced by the microbial communities in the DPB and B bioreactors (∼3 years), the two P. aeruginosa isolates DP and DPB were subjected to increasingly higher BAC concentrations every 24 h, in triplicate batch cultures, until no growth was observed. The last (highest) concentration for which growth was observed was recorded for each replicate population [BAC(+)]. Subsequently, the populations were transferred and maintained in BAC-free medium [BAC(−)] (Fig. 2A and B; Fig. S1B). During this evolution experiment, the two genotypes existed on different adaptive landscapes, because strain DPB possessed the ICE harboring resistance genes and was therefore inherently more resistant to BAC (Table 1). The other pairs of isolates recovered that showed cross-resistance, such as Achromobacter, were not used in the adaptive evolution experiment because these experiments were laborious and the pair of genomes recovered were divergent from each other (i.e., having different genomic backgrounds), which could complicate the results and interpretations.
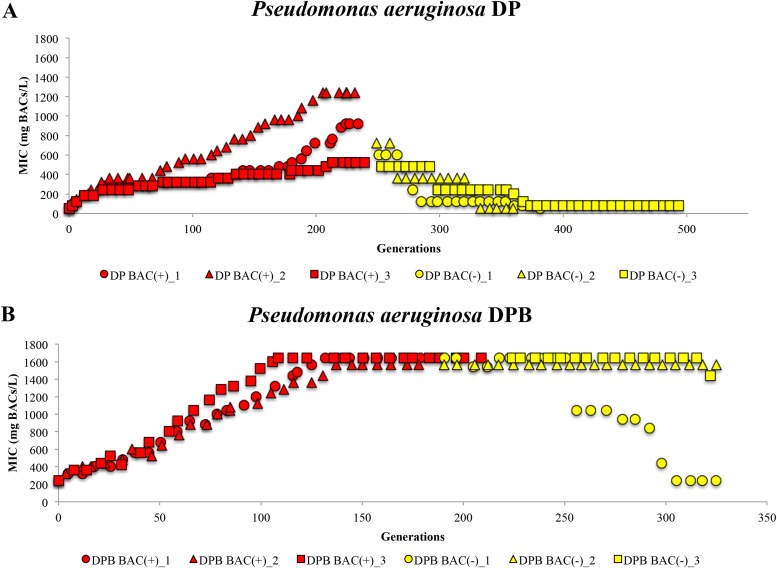
FIG 2.
Changes in MICs of BAC during an adaptive evolution experiment with P. aeruginosa DP (A) and P. aeruginosa DPB (B). The adaptive evolution experiment consisted of two phases: BAC exposure, where BAC concentration was increased after each round until reaching the maximum concentration in which growth was observed (red dots; each dot represents a passage), and BAC free (yellow dots). The MIC for BAC (y axis) was measured during each round of growth, for approximately 320 to 500 generations. The x axis shows the number of generations since the start of the experiment.
All DP and DPB populations showed increased resistance to BAC after growth with BAC for approximately 240 and 180 generations, respectively, compared to that of their ancestor (7-to-25-fold-higher MIC) or the controls (P. aeruginosa DP or DPB population evolving in parallel, under the same laboratory conditions but with no BAC in the growth medium). No BAC biodegradation by any of the populations was observed, while the highest BAC tolerance observed (by the DPB population) was 1,600 mg/liter BAC. After transferring to BAC-free medium, the three DP BAC(−) populations lost their BAC tolerance within 150 generations, and their BAC MIC was similar to that of the ancestor (50 mg/liter BAC), indicating that adaptation to BAC might have been transient, at the level of cell physiology and gene regulation, or due to the rise of compensatory mutations that restored the sensitivity to BAC under conditions of growth in BAC-free medium. In contrast, two of the DPB BAC(−) populations maintained a high BAC tolerance, with MICs for BAC similar to those of the BAC(+) populations and approximately 7-fold higher than that of their ancestor, while the third DPB BAC(−) population lost the acquired BAC tolerance. These results indicated that the two DPB BAC(+) populations might have acquired genetic modifications upon BAC exposure that were maintained in their corresponding DPB BAC(−) populations. Since all DP and DPB BAC(+) populations and two of the DPB BAC(−) populations acquired increased tolerance to BAC, these populations were examined further to assess if the increased BAC tolerance was accompanied by an increased resistance to antibiotics and to determine the underlying mechanism(s).
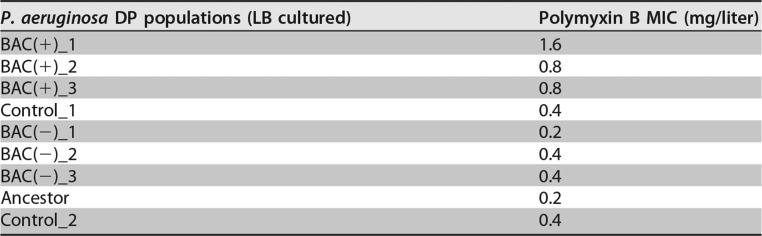
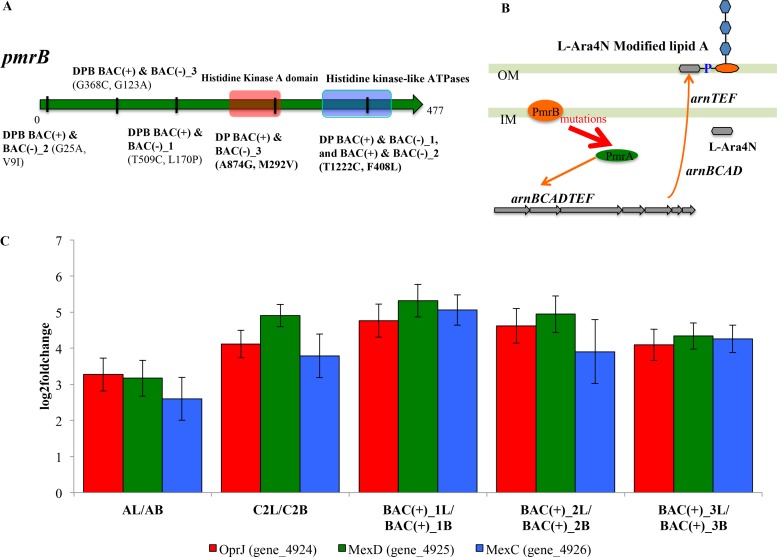
Interestingly, we observed that all DP BAC(+) populations showed higher MICs (2- to 8-fold) to the membrane-active antibiotic polymyxin B than the ancestor or the control (Table 2). Of the remaining antibiotics tested, DP BAC(+) populations did not show higher MICs than their ancestor (Table S3A). To identify the molecular mechanisms for cross-resistance between BAC and polymyxin B, whole-genome sequencing was conducted and identified 29 SNPs and 17 deletions, insertions and other polymorphisms (DIPs) in P. aeruginosa DP BAC(+) and BAC(−) populations relative to the control population and 22 SNPs and 16 DIPs in P. aeruginosa DPB BAC(+) and BAC(−) populations, respectively (see Table S4A to D). Remarkably, all DP BAC(+) and DPB BAC(+) populations had fixed mutations compared to their ancestral populations in only a single gene found in common, pmrB, albeit at different positions of the gene. pmrAB encodes a two-component regulatory system, and mutations in pmrB are known to confer polymyxin resistance in both clinical isolates and a laboratory strain of P. aeruginosa via constitutive activation of the pmrA regulon (34). Expression of pmrA leads to the expression of arnBCADTEF, which is responsible for the addition of 4-amino-l-arabinose (L-Ara4N) to the phosphate groups of lipid A, resulting in the reduction of the net negative charge on the outer membrane (Fig. 3B) (35, 36). Because both polymyxin B and BAC are cationic membrane-disrupting agents (37, 38), the reduction in the net negative charge is expected to increase BAC and polymyxin B tolerance by reducing their adsorption to the outer membrane. Therefore, our results revealed that mutations to the pmrB gene in the DP BAC(+) populations were selected by BAC exposure and led to increased resistance to at least one antibiotic, polymyxin B.
TABLE 2.
MIC values for polymyxin B of all populations of P. aeruginosa DP from the adaptive evolution experiment
FIG 3.
Mutational and transcriptional evidence for the resistance links between BAC and antibiotics. (A) Mutations in pmrB genes at various positions were noted for each population (denoted on the graph). (B) Schematic of the known polymyxin B resistance mechanisms caused by mutations in pmrB (34, 36, 68). (C) Overexpression of mexCD-oprJ operon when BAC is supplemented in the growth medium. AL/AB, P. aeruginosa DPB ancestor in LB growth medium versus LB plus BAC medium; C2L/C2B, P. aeruginosa DPB_Control_2 in LB versus LB plus BAC; BAC(+)_1L/BAC(+)_1B, P. aeruginosa DPB BAC(+)_1 in LB versus LB plus BAC; BAC(+)_2L/BAC(+)_2B, P. aeruginosa DPB BAC(+)_2 in LB versus LB plus BAC; BAC(+)_3L/BAC(+)_3B, P. aeruginosa DPB BAC(+)_3 in LB versus LB plus BAC.
It should be noted that although all the P. aeruginosa populations had fixed mutations in pmrB, only the DP BAC(+) populations showed increased resistance to polymyxin B compared to those of the DP BAC(−), DPB BAC(+) and DPB BAC(−) populations under our experimental conditions (Fig. 3A, Table 2, and Table S3B). A read alignment of the data sets of DP BAC(−) populations against assembled contigs of the ancestors revealed that the populations had different mutations in pmrB and pmrA than DP BAC(+) populations (see Table S5). Transcriptome sequencing (RNA-seq) results suggested that the mutations to pmrB in DPB BAC(+) and DPB BAC(−) populations did not lead to the overexpression of arnBCADTEF relative to that of the ancestor and control (see supporting results and discussion in the supplemental material for further details). These findings presumably explained the lack of increased polymyxin B resistance in the DP BAC(−), DPB BAC(+), and DPB BAC(−) populations, because the location and combination of mutations in pmrB are known to affect the level of expression of arnB (35), as confirmed by our RNA-seq data, and thus polymyxin B resistance. Furthermore, repeated passages without polymyxin in the growth medium have been shown to result in a loss of resistance in some cases, suggesting that the pmrAB locus is not the only determinant of the resistance phenotype (35) (see supporting results and discussion for further details). The lack of increased polymyxin resistance and the different mutations acquired in the pmrB gene in DPB populations are also not surprising given that these populations existed on different adaptive landscapes than DP populations at the beginning of the experiment. Altogether, BAC exposure selected for the mutations in pmrB that conferred increased BAC tolerance in all P. aeruginosa populations, but only specific mutations conferred increased polymyxin B resistance in the DP (but not the DPB) genetic background.
Physiological adaptation to BAC exposure also led to increased antibiotic resistance.
During this evolution experiment, another important resistance link was revealed. All P. aeruginosa DPB ancestor, BAC(+), and control populations showed increased resistances to tetracycline and ciprofloxacin (2- to 16-fold) when BAC was in the growth media at subinhibitory concentrations during the MIC test (Table 3 and Table S3C). All populations tested here reached at least ∼107 cells/ml after 24 h of growth at 35°C in the presence of subinhibitory concentrations of BAC, and these populations were used as the controls for determining MIC values for antibiotics. Therefore, the subinhibitory concentration of BAC used did not inhibit growth, and the MIC values obtained were presumably not due to experimental artifacts, such as absorbance contributed by dead cell debris. Thus, it appeared that copresence of (subinhibitory) BAC in the growth medium of P. aeruginosa DPB induced physiological changes to the exposed cells that also conferred antibiotic resistance regardless of the level of BAC tolerance of the population. RNA-seq analysis revealed, for instance, an overexpression (6- to 40-fold) of the mexCD-oprJ multidrug efflux pump (Fig. 3C) under BAC-supplemented conditions compared to that under BAC-free growth conditions. These findings were also consistent with previous results that mexCD-oprJ contributes to the resistance to several classes of antibiotics, such as fluoroquinolones (e.g., ciprofloxacin and norfloxacin) and tetracycline (39, 40). Furthermore, previous studies have shown that clinically relevant disinfectants such as BAC but not antibiotics (e.g., norfloxacin, tetracycline, or chloramphenicol) can induce the expression of this family of efflux pumps, even though antibiotics such as tetracycline and norfloxacin (in the same class as ciprofloxacin) are substrates for the pump (41, 42), in agreement with the results reported here.
TABLE 3.
MIC values for tetracycline and ciprofloxacin tested with no-BAC and added-BAC growth conditions for P. aeruginosa DPB evolved populations
| P. aeruginosa DPB populations | MIC (mg/liter)a |
|||
|---|---|---|---|---|
| TET | CIP | TET+BAC (100 mg/liter) | CIP+BAC (100 mg/liter) | |
| BAC(+)_1 | 12.5 | 0.2 | 25 | 3.2 |
| BAC(+)_2 | 3.125 | 0.2 | 12.5 | 1.6 |
| BAC(+)_3 | 12.5 | 0.2 | 25 | 0.8 |
| Ancestor | 6.25 | 0.2 | 25 | 1.6 |
| Control_1 | 12.5 | 0.4 | 25 | 1.6 |
TET, tetracycline; CIP, ciprofloxacin.
Conclusions and outlook.
Collectively, our results revealed that BAC exposure can induce antibiotic resistance via multiple genetic mechanisms, including the cooccurrence of BAC tolerance and antibiotic resistance genes on the same mobile DNA molecule, mutations in the pmrB gene, and the induction of efflux pump expression. These results are important because the cross-link between BAC exposure and antibiotic resistance was observed in at least three antibiotics of the seven tested (Table 1), the underlying mechanisms were elucidated (Fig. 1 and 3), and so the cross-link cannot be attributed to spurious findings, and even a 2-fold higher MIC as revealed for polymyxin B and rifampin can be clinically significant on the basis of the pharmacokinetics and pharmacodynamics of these two antibiotics (27, 28). Furthermore, the concentrations of BAC that the adapted P. aeruginosa DP BAC(+) and DPB BAC(+) populations were able to withstand during the adaptive evolution experiments (up to 1,600 mg/liter) were comparable to, or even higher than, those used in practice as a disinfectant (typically between 400 to 500 mg/liter, and almost always below 1,000 mg/liter) (43). Therefore, our results suggested that the accumulation of BAC in any nontarget environment (e.g., freshwater or sediment habitats or the waste stream of hospitals or food processing facilities) should be prevented to limit the spreading of antibiotic resistance determinants. New biotechnologies that employ the recently reported BAC-degrading organisms or their enzymes could be used for the latter purpose (20, 44). Our results also helped to explain why there is a debate in the literature on whether linkage exists between the exposure to disinfectants and increased antibiotic resistance. In particular, multiple adaptive possibilities (e.g., mutations in pmrB) in P. aeruginosa isolates were observed in response to BAC exposure, which is perhaps not unexpected given the nonspecific nature of the disinfectant, and some—but not all—of these mutations also conferred benefits under other conditions, specifically, polymyxin exposure. Similarly, not all bacterial species exposed to BAC acquired increased resistance to antibiotics, but at least several did (e.g., P. aeruginosa). It is also important to note that the list of organisms or growth conditions studied here were by no means exhaustive, and additional mechanisms, which may cause even higher antibiotic resistance levels than noted here, likely exist in natural or clinical/engineered environments. Therefore, more attention should be given to the usage of disinfectants and the better understanding of their fate, especially in nontarget environments. Finally, our experimental design and approach used in the adaptive evolution of P. aeruginosa can be employed to assess whether other important pathogens also show cross-resistance for BAC and antibiotics and to identify the underlying molecular mechanisms.
MATERIALS AND METHODS
Bioreactor development and isolate characterization.
All microbial communities (DP, DPB, and B) analyzed in this study originated from the same inoculum, from a sediment sample collected at the Bayou d'Inde, a tributary of the Calcasieu River, near Lake Charles, LA. The residual concentration of BAC in the sediment was less than the detection limit of the colorimetric method, i.e., 1 μg/g sediment (45). The community development and bioreactor operation (e.g., substrates, feeding cycle, and temperature) were described previously in detail (6). Briefly, the DP microbial community was developed from the sediment inoculum with a dextrin-peptone (50:50 [wt/wt]) mixture as the carbon and energy sources and maintained for 1 year. The DP community was subsequently used as an inoculum for DPB, which in turn was used as an inoculum for the B microbial community. DPB and B communities were fed a dextrin-peptone plus BAC mixture and BAC mixture, respectively, as carbon and energy sources. The BAC mixture consisted of a 60:40 mixture of benzyldimethyldodecylammonium chloride and benzyldimethyltetradecylammonium chloride (C12BDMA-Cl and C14BDMA-Cl, respectively; Sigma-Aldrich). The three communities were maintained for 4 years at room temperature in an aerobic fed-batch 2-liter Pyrex reactor with a total liquid volume of 1.6 liters, a residence time of 14 days, and the previously described mineral medium plus carbon and energy sources (i.e., 2,200 mg/liter of dextrin-peptone and 50 mg/liter of BAC mixture). To obtain isolates, a mixed community suspension from each bioreactor (DP, DPB, and B) was first diluted and plated on agar medium containing dextrin-peptone, salt medium, and 1.5% agar as described previously (no neutralizer for BAC was used during the isolation or transferring of isolates) (46). The taxonomic identification of the isolates was determined by sequencing their 16S rRNA genes.
Construction of plasmids carrying efflux pump genes.
Efflux pump genes found on the integrative and conjugative elements (ICEs) were cloned from the Achromobacter B isolate, and these genes were identical, at the nucleotide level, to those in P. aeruginosa DPB. The genes, including their native promoters, were individually amplified by PCR using primers containing enzyme restriction sites (EcoRI-BamHI for sugE-A and sugE-B: sugE-A-EcoRI, 5′-ATGCGAATTCCAGATAAAGCCAACCTTCC-3′; sugE-A-BamHI, 5′-ATGCGGATCCGACTACGCTACCAATGGAG-3′; sugE-B-EcoRI, 5′-ATGCGAATTCCATTATGGAAAGGGATGGCG-3′; sugE-B-BamHI, 5′-ATGCGGATCCTTGCTCTTCATAATGGGTCTC-3′; and KpnI-EcoRI for the ABC transporter operon: ABC-KpnI, 5′-ATGCGGTACCGTACTAGCGTCATAGTCACGG-3′; ABC-EcoRI, 5′-ATGCGAATTCCAACGTCATTAAAGAGTTCGC-3′). Digested amplicons were ligated into the multiple cloning site of pBBRMCS-4 (47). The resulting constructs were introduced into P. aeruginosa PAO509, which was deficient for several RND-family efflux pump genes (mexAB-oprM, mexCD-oprJ, mexEF-oprN, mexJK, and mexXY) (48), by electroporation as described previously (49). The transformants were selected on LB agar medium supplemented with 100 μg/ml of ampicillin. Recombinant plasmids extracted from the transformants were confirmed by PCR amplification of inserts and enzyme restriction digestion.
Adaptive evolution experiments.
Single colonies of the P. aeruginosa DP and DPB isolates were used as the original inocula for the adaptive evolution experiments. Six test tubes with a range of BAC concentrations were initially inoculated with 1% aliquots of P. aeruginosa DP and DPB cultures grown in 10 ml Luria broth medium (LB) for 24 h in triplicate at 35°C, using an orbital shaker at 225 rpm (see Fig. S1B in the supplemental material). Subsequently, 1% of the adapted population from the tube that showed growth (i.e., ≤80% growth reduction) at the highest concentration of BAC was transferred to new medium daily, followed by similar rounds of increasing BAC concentrations until no growth was observed in any tube [BAC(+) populations]. The MIC was determined as the concentration in which ≥80% inhibition in cell growth was observed by optical density at 600 nm (OD600) values compared to that of the control (no BAC in the medium). A 1% aliquot of inoculum culture was used for each MIC measurement. The generation numbers of the cultures were measured every 24 h by counting CFU. The inoculum from the tube that showed growth in the highest BAC concentration was subsequently transferred to LB [BAC(−) populations], followed by rounds of growth in decreasing BAC concentrations. The BAC used for the experiment consisted of a 60:40 mixture of benzyldimethyldodecylammonium chloride and benzyldimethyltetradecylammonium chloride (C12BDMA-Cl and C14BDMA-Cl, respectively; Sigma-Aldrich).
Antimicrobial susceptibility test.
The antimicrobial susceptibility test was performed with the microdilution procedure (50) in LB medium. All tested inocula were sampled at the end of the exponential phase and diluted in the medium to a final concentration of 5 × 105 CFU/ml before the test. Specifically, we characterized the growth curves for all the tested isolates, populations, and transformants, with optical density values at 600 nm, and sampled at the end of exponential phase to measure CFU and optical density values at 625 nm for 10-fold serially diluted aliquots (i.e., 10−1, 10−2, and 10−3). Based on these data, the relationship between the expected CFU and optical density values at 600 nm was determined, and this relationship was used to obtain aliquots of 5 × 105 CFU/ml on the basis of the optical density values. Therefore, comparable inocula in terms of starting cell numbers were used in all cases. All tests were performed in triplicate on 48-well plates after the growth of the inoculum (5 × 105 CFU/ml) for 24 h at 35°C with mixing on an orbital shaker at 225 rpm. After the incubation, the optical density values at 600 nm were measured and the MIC was determined on the basis of the control cultures with LB only medium for LB plus antibiotics tests and LB plus BAC medium for LB plus BAC plus antibiotics tests.
DNA extraction, RNA extraction, and sequencing.
Mixed community suspensions from bioreactors and isolates were taken for DNA extraction and were processed as previously described (51). These DNA samples were sequenced using the Illumina HiSeq 2000 sequencer at the Los Alamos National Laboratory Genomics Facility and reported previously (51); the ancestor P. aeruginosa DPB genome was closed using data from an in-house MinION instrument and the Illumina assembly. DNA of all P. aeruginosa populations from adaptive evolution experiments was extracted using the QIAamp DNA Blood minikit (Qiagen, Germany). DNA sequencing libraries were prepared using the Nextera XT DNA library preparation kit and sequenced on an in-house Illumina MiSeq instrument (School of Biological Sciences, Georgia Institute of Technology), as described previously (44).
RNA of P. aeruginosa DPB populations from the adaptive evolution experiment was extracted as previously described (52) with slight modifications. In brief, culture suspensions (25 ml of LB culture and 50 ml of LB plus BAC culture) were collected during mid-log-phase growth and centrifuged at 5,000 × g for 10 min. BAC (1,000 mg/liter) was used in the growth medium for all DPB BAC(+) and BAC(−)_2 populations, and 100 mg of BAC/liter was used for the ancestor, control, and BAC(−)_1 populations, which represented subinhibitory concentrations for the corresponding populations. The collected cell pellets were subsequently washed with 1× phosphate-buffered saline (PBS), resuspended with 5 ml of lysis buffer (50 mM Tris-HCl, 40 mM EDTA [pH 8], and 0.75 M sucrose) containing 15 mg/ml of lysozyme, and incubated at 37°C for 3 min. The lysates were then incubated with 0.4 ml of the lysis buffer, containing 1% SDS and 20 mg/ml of proteinase K, for 2 h at 55°C in a rotating hybridization oven. RNA was extracted with acid phenol and chloroform and isolated with the mirVANA RNA isolation kit (Ambion). RNA samples were treated with DNase by using the TURBO DNA-free kit (Ambion, Austin, TX). The elimination of contaminating DNA was confirmed by PCR amplification of the sugE-A gene. rRNA was depleted from total RNA using the Ribo-Zero rRNA removal kit (Illumina, San Diego, CA), and cDNA libraries were constructed using the ScriptSeq v2 RNA-Seq library preparation kit (Illumina, San Diego, CA) according to the manufacturer's instructions. cDNA libraries were quantified and sequenced as described above for P. aeruginosa population DNA samples.
Community metagenome and isolate genome sequence analysis.
Raw Illumina reads were trimmed using a Q value of 15 Phred quality score cutoff using SolexaQA (53) for further analysis. Community and isolate genome reads were assembled using the hybrid protocol previously described (54). Protein-coding genes on assembled metagenomic or isolate genome contigs were annotated with the MetaGene pipeline (55). The phylogenetic affiliation of assembled contigs or unassembled metagenomic reads was determined on the basis of best match searches against all bacterial and archaeal genome sequences available in the GenBank database (as of June 2015; ftp://ftp.ncbi.nih.gov/), using BLASTN (X = 150, q = −1, F = F, and remaining parameters at default settings) with a cutoff of a match with >95% identity and 50% query length coverage. The functional annotation of protein-coding genes was performed using a BLASTP search against the SEED protein database (56), with a cutoff for a match of >40% amino acid sequence identity and >50% query length coverage.
We used Bowtie 2 (57) to map metagenomic reads from the DPB bioreactor against P. aeruginosa DPB assembled contigs to estimate coverage, which was also taken as a proxy for relative abundance. To identify SNPs and genomic modifications, P. aeruginosa DP genomic reads were matched against P. aeruginosa DPB assembled contigs, and vice versa, using breseq (consensus mode and Q = 15 Phred quality score cutoff) (58, 59). The same approach was also applied in comparisons of P. aeruginosa DP and DPB control, BAC(+), and BAC(−) populations against P. aeruginosa DP and DPB assembled contigs (ancestors) in the adaptive evolution experiments. Identified mutations in the evolved control population against its ancestor were considered to be the result of stochastic processes or selection by the growth conditions (e.g., “bottle effect”) and were removed from further analysis. Read recruitment plots were obtained as described previously (60) on the basis of the results of a BLASTN search of reads against the GI-2 sequence of P. aeruginosa DPB and a minimum cutoff for a match of 70% of identity and a 60-bp alignment length.
MinION sequencing and read analysis.
Sequencing libraries for the MinION sequencer were prepared using the SQK-RAD003 rapid sequencing kit, R9 version, lot SR03.10.002 (Oxford Nanopore Technologies, Oxford, UK). Approximately 421 ng of P. aeruginosa strain DPB DNA was tagmented by incubation with 2.5 μl fragmentation mix (FRM) for 1 min at 30°C followed by 1 min at 80°C. Tagmented DNA was then adapter-ligated by incubating with 1.0 μl rapid adapter mix (RAD) for 5 min at room temperature and was stored on ice. A FLO-MIN 107 (R9.5) flow cell was primed as directed by the manufacturer and then loaded with a mixture of 11 μl tagmented library, 30.5 μl running buffer FM (RBF), 26.5 μl library loading beads (LLB), and 7 μl nuclease-free water. P. aeruginosa DPB was sequenced using the SQK-RAD003 MinKNOW protocol script with local basecalling, and the sequencing was stopped after 12 h.
After basecalling of the one-dimensional (1D) MinION reads, the reads were extracted in fastq format using Poretools version 0.6.0 (61). All resulting reads were trimmed using NanoFilt (https://github.com/wdecoster/nanofilt) with a Phred score cutoff of 10 and a minimum fragment length of 1 kbp, and the adapter sequences were removed using Porechop (https://github.com/rrwick/Porechop). Then, the hybrid read set, i.e., Illumina and MinION trimmed reads, was assembled using Unicycler v0.4.2 with the conservative mode (62). Briefly, Unicycler uses a SPAdes assembly of Illumina reads and then scaffolds the assembly graph using MinION reads. Finally, Unicycler uses Illumina reads to polish the final assembly using Pilon (63). To identify genomic islands specific to the P. aeruginosa DPB closed genome from the hybrid Illumina plus MinION assembly, P. aeruginosa DP genomic reads (Illumina) were searched against the P. aeruginosa DPB genome using breseq (consensus mode and Q = 15 Phred quality score cutoff) (58, 59), and the genomic regions not covered by reads were considered strain DPB specific.
Transcriptome sequence analysis.
All transcriptomic reads of the P. aeruginosa DPB ancestor, control, BAC(+), BAC(−)_1, and BAC(−)_2 populations were trimmed using SolexaQA (53) with a Phred score cutoff of 20 and a minimum fragment length of 50 bp, and 3′-end adapter contaminants were removed using Scythe v0.993 (https://github.com/vsbuffalo/scythe). These trimmed reads were filtered using SortMeRNA v2.0 (64) to remove remaining rRNA sequences within all databases in the program. Most of the resulting data sets had >95% of their total reads as non-rRNA except three libraries (see Table S7). One library in particular [i.e., P. aeruginosa DPB BAC(+)_2L_3] had 9.42% of reads shown as non-rRNA, which resulted in too few non-rRNA sequences relative to the remaining libraries (e.g., >2-fold difference in number of non-rRNA reads), and thus was removed from further analysis to avoid biasing the comparisons and results (65). Non-rRNA reads were mapped to P. aeruginosa DPB assembled contigs using Bowtie 2 (57), and read count tables against predicted genes were generated by featureCounts v1.4.6-p3 (66). The output read count tables were used as the input for DESeq2 (67) to obtain the lists of differentially expressed genes in pairwise comparisons. The pairwise comparisons included both the type of population [e.g., ancestor versus BAC(+) populations] and culture condition (i.e., BAC free versus BAC supplemented). All genes that were differentially expressed from all pairwise comparisons with a cutoff adjusted P value of <0.01 and number of read counts >1 were included for further analysis. Differentially expressed genes from the comparison between the control population and the ancestor that were also observed in the comparison of the BAC(+) or BAC(−) population against the ancestor were discarded from further analysis, similar to that for the mutations described above.
Data availability.
The bioreactor metagenome sequences used in this study were deposited in GenBank under the accession numbers SRR643889 (DP), SRR643891 (DPB), and SRR643892 (B). The genome sequences of isolates, all genomic and transcriptomic sequences of P. aeruginosa DP/DPB BAC(+) and BAC(−) populations, and MinION sequencing data can be found under the BioProject PRJNA184698.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by U.S. National Science Foundation award CBET 0967130 to S.G.P. and K.T.K. and award 1241046 to K.T.K.
The authors declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01201-18.
REFERENCES
- 1.Tezel U, Pavlostathis SG. 2015. Quaternary ammonium disinfectants: microbial adaptation, degradation and ecology. Curr Opin Biotechnol 33:296–304. doi: 10.1016/j.copbio.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 2.Gerba CP. 2015. Quaternary ammonium biocides: efficacy in application. Appl Environ Microbiol 81:464–469. doi: 10.1128/AEM.02633-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karatzas KA, Randall LP, Webber M, Piddock LJ, Humphrey TJ, Woodward MJ, Coldham NG. 2008. Phenotypic and proteomic characterization of multiply antibiotic-resistant variants of Salmonella enterica serovar Typhimurium selected following exposure to disinfectants. Appl Environ Microbiol 74:1508–1516. doi: 10.1128/AEM.01931-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mc Cay PH, Ocampo-Sosa AA, Fleming GT. 2010. Effect of subinhibitory concentrations of benzalkonium chloride on the competitiveness of Pseudomonas aeruginosa grown in continuous culture. Microbiology 156:30–38. doi: 10.1099/mic.0.029751-0. [DOI] [PubMed] [Google Scholar]
- 5.Rakic-Martinez M, Drevets DA, Dutta V, Katic V, Kathariou S. 2011. Listeria monocytogenes strains selected on ciprofloxacin or the disinfectant benzalkonium chloride exhibit reduced susceptibility to ciprofloxacin, gentamicin, benzalkonium chloride, and other toxic compounds. Appl Environ Microbiol 77:8714–8721. doi: 10.1128/AEM.05941-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tandukar M, Oh S, Tezel U, Konstantinidis KT, Pavlostathis SG. 2013. Long-term exposure to benzalkonium chloride disinfectants results in change of microbial community structure and increased antimicrobial resistance. Environ Sci Technol 47:9730–9738. doi: 10.1021/es401507k. [DOI] [PubMed] [Google Scholar]
- 7.Romanova N, Wolffs P, Brovko L, Griffiths M. 2006. Role of efflux pumps in adaptation and resistance of Listeria monocytogenes to benzalkonium chloride. Appl Environ Microbiol 72:3498–3503. doi: 10.1128/AEM.72.5.3498-3503.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Condell O, Iversen C, Cooney S, Power KA, Walsh C, Burgess C, Fanning S. 2012. Efficacy of biocides used in the modern food industry to control Salmonella enterica, and links between biocide tolerance and resistance to clinically relevant antimicrobial compounds. Appl Environ Microbiol 78:3087–3097. doi: 10.1128/AEM.07534-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole E, Addison R, Rubino J, Leese K, Dulaney P, Newell M, Wilkins J, Gaber D, Wineinger T, Criger D. 2003. Investigation of antibiotic and antibacterial agent cross-resistance in target bacteria from homes of antibacterial product users and nonusers. J Appl Microbiol 95:664–676. doi: 10.1046/j.1365-2672.2003.02022.x. [DOI] [PubMed] [Google Scholar]
- 10.Kücken D, Feucht H-H, Kaulfers P-M. 2000. Association of qacE and qacE Δ1 with multiple resistance to antibiotics and antiseptics in clinical isolates of Gram-negative bacteria. FEMS Microbiol Lett 183:95–98. doi: 10.1111/j.1574-6968.2000.tb08939.x. [DOI] [PubMed] [Google Scholar]
- 11.Lear J, Maillard J-Y, Dettmar P, Goddard P, Russell A. 2006. Chloroxylenol- and triclosan-tolerant bacteria from industrial sources—susceptibility to antibiotics and other biocides. Int Biodeterior Biodegradation 57:51–56. doi: 10.1016/j.ibiod.2005.11.002. [DOI] [Google Scholar]
- 12.Ortiz S, López P, López V, Martínez-Suárez JV. 2014. Antibiotic susceptibility in benzalkonium chloride-resistant and-susceptible Listeria monocytogenes strains. Foodborne Pathog Dis 11:517–519. doi: 10.1089/fpd.2013.1724. [DOI] [PubMed] [Google Scholar]
- 13.Anderson RL, Carr JH, Bond WW, Favero MS. 1997. Susceptibility of vancomycin-resistant enterococci to environmental disinfectants. Infect Control Hosp Epidemiol 18:195–199. [DOI] [PubMed] [Google Scholar]
- 14.Giuliano CA, Rybak MJ. 2015. Efficacy of triclosan as an antimicrobial hand soap and its potential impact on antimicrobial resistance: a focused review. Pharmacotherapy 35:328–336. doi: 10.1002/phar.1553. [DOI] [PubMed] [Google Scholar]
- 15.Braoudaki M, Hilton A. 2004. Adaptive resistance to biocides in Salmonella enterica and Escherichia coli O157 and cross-resistance to antimicrobial agents. J Clin Microbiol 42:73–78. doi: 10.1128/JCM.42.1.73-78.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cottell A, Denyer SP, Hanlon GW, Ochs D, Maillard JY. 2009. Triclosan-tolerant bacteria: changes in susceptibility to antibiotics. J Hosp Infect 72:71–76. doi: 10.1016/j.jhin.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Marple B, Roland P, Benninger M. 2004. Safety review of benzalkonium chloride used as a preservative in intranasal solutions: an overview of conflicting data and opinions. Otolaryngol Head Neck Surg 130:131–141. doi: 10.1016/j.otohns.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 18.De Saint Jean M, Brignole F, Bringuier A-F, Bauchet A, Feldmann G, Baudouin C. 1999. Effects of benzalkonium chloride on growth and survival of Chang conjunctival cells. Invest Ophthalmol Vis Sci 40:619–630. [PubMed] [Google Scholar]
- 19.Tezel U, Tandukar M, Martinez RJ, Sobecky PA, Pavlostathis SG. 2012. Aerobic biotransformation of n-tetradecylbenzyldimethylammonium chloride by an enriched Pseudomonas spp. community. Environ Sci Technol 46:8714–8722. doi: 10.1021/es300518c. [DOI] [PubMed] [Google Scholar]
- 20.Oh S, Kurt Z, Tsementzi D, Weigand MR, Kim M, Hatt JK, Tandukar M, Pavlostathis SG, Spain JC, Konstantinidis KT. 2014. Microbial community degradation of widely used quaternary ammonium disinfectants. Appl Environ Microbiol 80:5892–5900. doi: 10.1128/AEM.01255-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drake JW, Charlesworth B, Charlesworth D, Crow JF. 1998. Rates of spontaneous mutation. Genetics 148:1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardalo C, Edberg SC. 1997. Pseudomonas aeruginosa: assessment of risk from drinking water. Crit Rev Microbiol 23:47–75. doi: 10.3109/10408419709115130. [DOI] [PubMed] [Google Scholar]
- 23.Pellett S, Bigley D, Grimes D. 1983. Distribution of Pseudomonas aeruginosa in a riverine ecosystem. Appl Environ Microbiol 45:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kung VL, Ozer EA, Hauser AR. 2010. The accessory genome of Pseudomonas aeruginosa. Microbiol Mol Biol Rev 74:621–641. doi: 10.1128/MMBR.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson CM, Grossman AD. 2015. Integrative and conjugative elements (ICEs): what they do and how they work. Annu Rev Genet 49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKinnon P, Davis S. 2004. Pharmacokinetic and pharmacodynamic issues in the treatment of bacterial infectious diseases. Eur J Clin Microbiol Infect Dis 23:271–288. doi: 10.1007/s10096-004-1107-7. [DOI] [PubMed] [Google Scholar]
- 27.Tam VH, Schilling AN, Vo G, Kabbara S, Kwa AL, Wiederhold NP, Lewis RE. 2005. Pharmacodynamics of polymyxin B against Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:3624–3630. doi: 10.1128/AAC.49.9.3624-3630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burman WJ, Gallicano K, Peloquin C. 2001. Comparative pharmacokinetics and pharmacodynamics of the rifamycin antibacterials. Clin Pharmacokinet 40:327–341. doi: 10.2165/00003088-200140050-00002. [DOI] [PubMed] [Google Scholar]
- 29.Chung YJ, Saier MH. 2002. Overexpression of the Escherichia coli sugE gene confers resistance to a narrow range of quaternary ammonium compounds. J Bacteriol 184:2543–2545. doi: 10.1128/JB.184.9.2543-2545.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He G-X, Zhang C, Crow RR, Thorpe C, Chen H, Kumar S, Tsuchiya T, Varela MF. 2011. SugE, a new member of the SMR family of transporters, contributes to antimicrobial resistance in Enterobacter cloacae. Antimicrob Agents Chemother 55:3954–3957. doi: 10.1128/AAC.00094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, Galagan J, Niemann S, Gagneux S. 2011. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet 44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jatsenko T, Tover A, Tegova R, Kivisaar M. 2010. Molecular characterization of Rif(r) mutations in Pseudomonas aeruginosa and Pseudomonas putida. Mutat Res 683:106–114. doi: 10.1016/j.mrfmmm.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 33.Weigand MR, Sundin GW. 2012. General and inducible hypermutation facilitate parallel adaptation in Pseudomonas aeruginosa despite divergent mutation spectra. Proc Natl Acad Sci U S A 109:13680–13685. doi: 10.1073/pnas.1205357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moskowitz SM, Ernst RK, Miller SI. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol 186:575–579. doi: 10.1128/JB.186.2.575-579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moskowitz SM, Brannon MK, Dasgupta N, Pier M, Sgambati N, Miller AK, Selgrade SE, Miller SI, Denton M, Conway SP. 2012. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 56:1019–1030. doi: 10.1128/AAC.05829-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan A, Guan Z, Raetz CR. 2007. An undecaprenyl phosphate-aminoarabinose flippase required for polymyxin resistance in Escherichia coli. J Biol Chem 282:36077–36089. doi: 10.1074/jbc.M706172200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikaido H, Vaara M. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol Rev 49:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaara M. 1992. Agents that increase the permeability of the outer membrane. Microbiol Rev 56:395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srikumar R, Kon T, Gotoh N, Poole K. 1998. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob Agents Chemother 42:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gotoh N, Tsujimoto H, Tsuda M, Okamoto K, Nomura A, Wada T, Nakahashi M, Nishino T. 1998. Characterization of the MexC-MexD-OprJ multidrug efflux system in ΔmexA-mexB-oprM mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother 42:1938–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morita Y, Murata T, Mima T, Shiota S, Kuroda T, Mizushima T, Gotoh N, Nishino T, Tsuchiya T. 2003. Induction of mexCD-oprJ operon for a multidrug efflux pump by disinfectants in wild-type Pseudomonas aeruginosa PAO1. J Antimicrob Chemother 51:991–994. doi: 10.1093/jac/dkg173. [DOI] [PubMed] [Google Scholar]
- 42.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi JI, Li XZ, Nishino T. 1996. Overexpression of the mexC–mexD–oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol 21:713–725. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 43.Tiedink J. 2001. Cationic surfactants: cationic surfactants in biocides, p 318–321. In Holmberg K. (ed), Handbook of applied surface and colloid chemistry. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 44.Ertekin E, Hatt JK, Konstantinidis KT, Tezel U. 2016. Similar microbial consortia and genes are involved in the biodegradation of benzalkonium chlorides in different environments. Environ Sci Technol 50:4304–4313. doi: 10.1021/acs.est.5b05959. [DOI] [PubMed] [Google Scholar]
- 45.Çeçen F, Tezel U (ed). 2017. Hazardous pollutants in biological treatment systems: fundamentals and a guide to experimental research. IWA Publishing, London, United Kingdom. [Google Scholar]
- 46.Oh S, Tandukar M, Pavlostathis SG, Chain PS, Konstantinidis KT. 2013. Microbial community adaptation to quaternary ammonium biocides as revealed by metagenomics. Environ Microbiol 15:2850–2864. doi: 10.1111/1462-2920.12154. [DOI] [PubMed] [Google Scholar]
- 47.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 48.Mima T, Joshi S, Gomez-Escalada M, Schweizer HP. 2007. Identification and characterization of TriABC-OpmH, a triclosan efflux pump of Pseudomonas aeruginosa requiring two membrane fusion proteins. J Bacteriol 189:7600–7609. doi: 10.1128/JB.00850-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi K-H, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7–A7. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 51.Oh S, Caro-Quintero A, Tsementzi D, DeLeon-Rodriguez N, Luo C, Poretsky R, Konstantinidis KT. 2011. Metagenomic insights into the evolution, function, and complexity of the planktonic microbial community of Lake Lanier, a temperate freshwater ecosystem. Appl Environ Microbiol 77:6000–6011. doi: 10.1128/AEM.00107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsementzi D, Poretsky R, Rodriguez-R LM, Luo C, Konstantinidis KT. 2014. Evaluation of metatranscriptomic protocols and application to the study of freshwater microbial communities. Environ Microbiol Rep 6:640–655. doi: 10.1111/1758-2229.12180. [DOI] [PubMed] [Google Scholar]
- 53.Cox MP, Peterson DA, Biggs PJ. 2010. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT. 2011. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci U S A 108:7200–7205. doi: 10.1073/pnas.1015622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noguchi H, Park J, Takagi T. 2006. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res 34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang H-Y, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1,000 genomes. Nucleic Acids Res 33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 59.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez-R LM, Konstantinidis KT. 2016. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints 4:e1900v1. doi: 10.7287/peerj.preprints.1900v1. [DOI] [Google Scholar]
- 61.Loman NJ, Quinlan AR. 2014. Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics 30:3399–3401. doi: 10.1093/bioinformatics/btu555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kopylova E, Noé L, Touzet H. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 65.Rodriguez-R LM, Konstantinidis KT. 2014. Estimating coverage in metagenomic data sets and why it matters. ISME J 8:2349. doi: 10.1038/ismej.2014.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 67.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abraham N, Kwon DH. 2009. A single amino acid substitution in PmrB is associated with polymyxin B resistance in clinical isolate of Pseudomonas aeruginosa. FEMS Microbiol Lett 298:249–254. doi: 10.1111/j.1574-6968.2009.01720.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bioreactor metagenome sequences used in this study were deposited in GenBank under the accession numbers SRR643889 (DP), SRR643891 (DPB), and SRR643892 (B). The genome sequences of isolates, all genomic and transcriptomic sequences of P. aeruginosa DP/DPB BAC(+) and BAC(−) populations, and MinION sequencing data can be found under the BioProject PRJNA184698.