The biology of phages as members of the human oral microbiome is understudied. Here, we report the characterization of xhp1, a novel linear plasmid-like prophage identified from a human oral isolate, Actinomyces odontolyticus subsp. actinosynbacter strain XH001. xhp1 can be induced and reinfect xhp1-cured XH001. The spontaneous induction of xhp1 leads to the lysis of a subpopulation of bacterial hosts and the release of eDNA that promotes biofilm assembly, thus potentially contributing to the persistence of A. odontolyticus within the oral cavity.
KEYWORDS: linear plasmid-like prophage, oral phage, Actinomyces odontolyticus, biofilm assembly, bacteriophages, oral phage biology
ABSTRACT
The human oral cavity is home to a large number of bacteria and bacteriophages (phages). However, the biology of oral phages as members of the human microbiome is not well understood. Recently, we isolated Actinomyces odontolyticus subsp. actinosynbacter strain XH001 from the human oral cavity, and genomic analysis revealed the presence of an intact prophage named xhp1. Here, we demonstrated that xhp1 is a linear plasmid-like prophage, which is a newly identified phage of A. odontolyticus. The prophage xhp1 genome is a 35-kb linear double-stranded DNA with 10-bp single-stranded, 3’ cohesive ends. xhp1 exists extrachromosomally, with an estimated copy number of 5. Annotation of xhp1 revealed 54 open reading frames, while phylogenetic analysis suggests that it has limited similarity with other phages. xhp1 phage particles can be induced by mitomycin C and belong to the Siphoviridae family, according to transmission electron microscopic examination. The released xhp1 particles can reinfect the xhp1-cured XH001 strain and result in tiny blurry plaques. Moreover, xhp1 promotes XH001 biofilm formation through spontaneous induction and the release of host extracellular DNA (eDNA). In conclusion, we identified a linear plasmid-like prophage of A. odontolyticus, which enhances bacterial host biofilm assembly and could be beneficial to the host for its persistence in the oral cavity.
IMPORTANCE The biology of phages as members of the human oral microbiome is understudied. Here, we report the characterization of xhp1, a novel linear plasmid-like prophage identified from a human oral isolate, Actinomyces odontolyticus subsp. actinosynbacter strain XH001. xhp1 can be induced and reinfect xhp1-cured XH001. The spontaneous induction of xhp1 leads to the lysis of a subpopulation of bacterial hosts and the release of eDNA that promotes biofilm assembly, thus potentially contributing to the persistence of A. odontolyticus within the oral cavity.
INTRODUCTION
The human oral microbiome is composed of diverse microbial residents, including bacteria, archaea, eukaryotic organisms, and viruses, that interact with each other as well as with the host and that have been implicated in human health and diseases (1). Compared to studies on other resident members, the study of oral viral biology and ecology is still in its infancy (2–4).
Recent studies have shown that the oral cavity is populated by communities of virus, with approximately 108 virus-like particles (VLPs) per milliliter of saliva (5) and 107 VLPs per milligram of dental plaque (6). The next-generation sequencing technology has significantly increased our knowledge of viromes in the oral cavity by uncovering the huge diversity of oral virus communities (5–10). Most of the virome sequences are identified as bacteriophages (phages) (10). Intriguingly, within the oral cavity, different biogeographic niches have unique phage ecology which could contribute to the shaping of bacterial biota within the same niches (2). Meanwhile, the oral viromes are highly personalized, and the shifted virome may be associated with dysbiosis and potentially promoting oral diseases (7, 9). However, data on the ecological and physiological role of phages in the oral microbiome are still lacking due to the limited number of phages that have been successfully isolated and characterized (2).
One reason for the limited number of isolated oral phages is likely the narrow host range of phages. For example, Streptococcus mutans phage M102AD only infects serotype c strains (11). Meanwhile, next-generation sequencing data revealed that most oral phages are lysogenic (5, 10). These phages may assume an inactive lysogenic form or are prevented from infecting bacterial hosts due to prophage-mediated defense (12). Thus, the traditional plaque assay may not be the ideal approach for isolating these oral phages.
With the increasing number of oral bacterial genomes that have been sequenced, identifying prophages through genomic analysis has allowed for isolation and characterization of new phages (13, 14). Recently, we isolated (15) and performed whole-genome sequencing (16) of an Actinomyces sp. named A. odontolyticus subsp. actinosynbacter strain XH001 from the human oral cavity. Genomic analysis revealed only one intact prophage, xhp1. Here, we characterize A. odontolyticus prophage xhp1 through genomic and physiological approaches.
RESULTS
Genomic analysis of xhp1.
The intact prophage xhp1 was predicted from the XH001 genome (GenBank accession no. LLVT00000000) using the prophage prediction program PHASTER (17). The xhp1 genome is 35 kb, with a GC content of 65.29%, which is similar to that of its host, XH001, which has an overall G+C content of 65.9%. xhp1 encodes a total of 54 putative open reading frames (ORFs), 17 of which are functionally annotated by protein BLAST (Fig. 1; see also Table S1 in the supplemental material). The remaining ORFs share little homology to the database at either the nucleotide or amino acid level.
FIG 1.
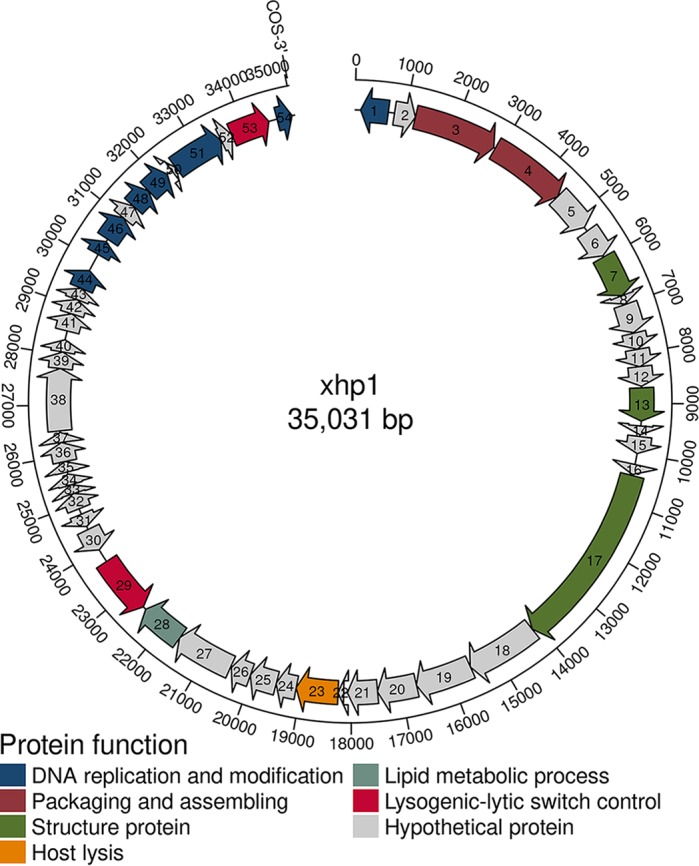
Schematic representation of the organization of xhp1. ORFs that are classified in the same functional categories are in the same color. ORFs in gray are hypothetical proteins.
The annotated proteins can be classified into the following groups: structural proteins (capsid and tail proteins), transcriptional regulators of lysogenic-lytic switch (repressor and integrase), DNA replication and modification (DNA primase and methyltransferase), packaging (terminase), and host cell lysis (phage lysin).
xhp1 is a linear plasmid-like prophage.
Based on the sequencing data, xhp1 was assembled into an independent plasmid element by SPAdes (18), indicating that xhp1 might not be integrated into the host genome but rather exists as a plasmid-like prophage.
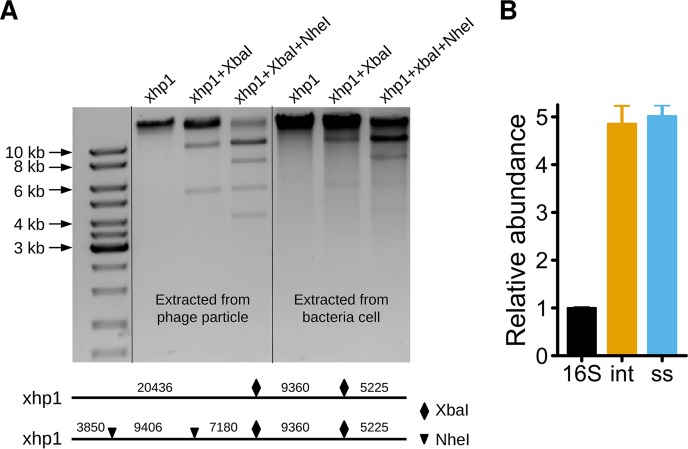
To confirm the bioinformatics finding, phage genomic DNA was extracted using a plasmid extraction kit from log-phase XH001 cells without mitomycin C induction. We showed that the prophage genome can be extracted with a high yield (Fig. 2A). To further determine if xhp1 is circular or linear, we performed diagnostic digestion. According to the xhp1 genome sequence, there are two recognition sites for each of the two restriction enzymes, NheI and XbaI (Fig. 2A). Thus, XbaI alone and NheI/XbaI double digestion will result in three and five bands, respectively, if xhp1 is linear. As shown in Fig. 2A, three bands were observed for the XbaI-digested xhp1 genome, which included two bands with predicted sizes of ∼5.2 kb and 9.5 kb, while the third band of predicted size of 20 kb stayed on the top of the lane in gel electrophoresis. In the NheI/XbaI doubly digested xhp1 genome, in addition to the three predicted fragments of 3.8 kb, 5.2 kb, and 7.2 kb, there is another more intense band close to 10 kb, which is likely due to the fact that the other two predicted fragments, at 9,360 bp and 9,406 bp, are too close to be separated on the gel. The band above the dark 10-kb band in the doubly digested lane is likely the undigested bacterial genomic DNA contamination occurring during the extraction process, which involved mechanical cell breakage using bead-beating.
FIG 2.
xhp1 is a linear plasmid-like prophage with an estimated copy number of 5. (A) The genome of prophage xhp1 was extracted from phage particles or bacterial cells and digested with XbaI or NheI/XbaI. The digestion pattern is consistent with a linear genome inside both the phage particle and the bacterial cell. (B) Estimation of the copy number of xhp1 by qPCR. 16S, bacterial 16S rRNA gene; int, integrase; ss, single-stranded DNA-binding protein.
To further investigate if the xhp1 genome exists as circular or linear DNA within phage particles, log-phase XH001 culture was treated with mitomycin C for 3 h, and the released xhp1 particles in the supernatant were collected and enriched by polyethylene glycol 8000 (PEG-8000) precipitation. Genomic DNA was isolated from phage particles and subjected to the same restriction enzyme digestion analysis. The resultant banding patterns are consistent with that of digested phage genome extracted from host cells. Thus, the xhp1 genome is linear both inside the host and when packaged into phage particles.
The termini of xhp1 were predicted as a cohesive-end site, with 10-bp 3′ single-stranded cohesive ends (CGTCAAGAGC), using PhageTerm (version 1.0.11) (19), which suggests that the prophage genome might become circularized via its cohesive termini during the lytic cycle (20).
Next, the xhp1 copy number in XH001 was estimated through quantitative PCR (qPCR) using total DNA isolated from XH001 culture. Specific primers for genes encoding xhp1 integrase and single-stranded DNA-binding protein were used for qPCR analysis (Table 1). The result showed that the copy numbers of genes encoding integrase (int) and single-stranded DNA-binding protein (ss) were 4.85 ± 0.67-fold and 5.01 ± 0.41-fold that of the XH001 16S rRNA gene. The primer sequences for the XH001 16S rRNA gene (XH-F5 and XH-R3) have only one copy in the sequenced XH001 genome. Thus, the copy number of xhp1 was estimated to be 5 copies per XH001 chromosome, confirming that xhp1 exists as an extrachromosomal element.
TABLE 1.
Primers used in this study
| Gene/protein | Primer | Sequence |
|---|---|---|
| Bacterial 16S rRNA gene | XH-F5 | GCGGAGCATGCGGATTA |
| XH-R3 | AACGTGCTGGCAACATAGGG | |
| Integrase | xhp1-intF | TGCCTCATCCGATAGGTATGCGTCT |
| xhp1-intR | TAGCGAGGGCACCTGGGTCAGT | |
| Single-stranded DNA-binding protein | xhp1-ssF | GTGAGGCGACCCTGAACGATG |
| xhp1-ssR | GACCCCGAGAACCTACGACCG |
Observation of xhp1 particles.
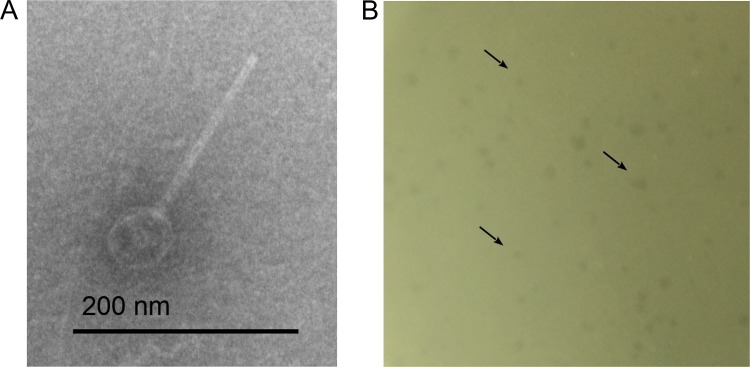
Phage particles were induced from XH001 by mitomycin C and enriched by PEG-8000 precipitation. Then, phage particles were subjected to electron microscope photography. An icosahedral capsid with a long noncontractile tail was observed, suggesting that xhp1 belongs to the Siphoviridae family (Fig. 3A). The sizes for the capsid and the tail were estimated to be 30 ± 1 nm in diameter and 220 ± 3 nm in length, respectively.
FIG 3.
(A) Transmission electron microscopy (TEM) images of phage xhp1. (B) xhp1 forms tiny and blurry plaques on XH001 Δxhp1 in the plaque assay, which are indicated by black arrows.
Prophage xhp1 forms tiny plaques on XH001 Δxhp1.
Prophage usually is very stable, and the rate of spontaneous loss is very low, due to prophage-encoded special stabilization machinery, such as toxin-antitoxin module (20). To facilitate the loss of linear prophage, a heat technique was performed, as described in Materials and Methods. The deletion frequency of xhp1 was quite low. Only one out of 1,000 colonies tested was cured of xhp1, and this was named XH001 Δxhp1. 16S rRNA gene sequencing showed that XH001 Δxhp1 shared 100% identity with XH001, indicating it was derived from XH001. Furthermore, no xhp1 genomic DNA can be isolated from XH001 Δxhp1 using the same isolation method.
We then tested whether xhp1 can reinfect XH001 Δxhp1. The result showed that while no plaque formation was observed when xhp1 was added to XH001 cells, xhp1 resulted in the formation of tiny blurry plaques on XH001 Δxhp1 in the plaque assay (Fig. 3B).
Prophage xhp1 increases biofilm formation.
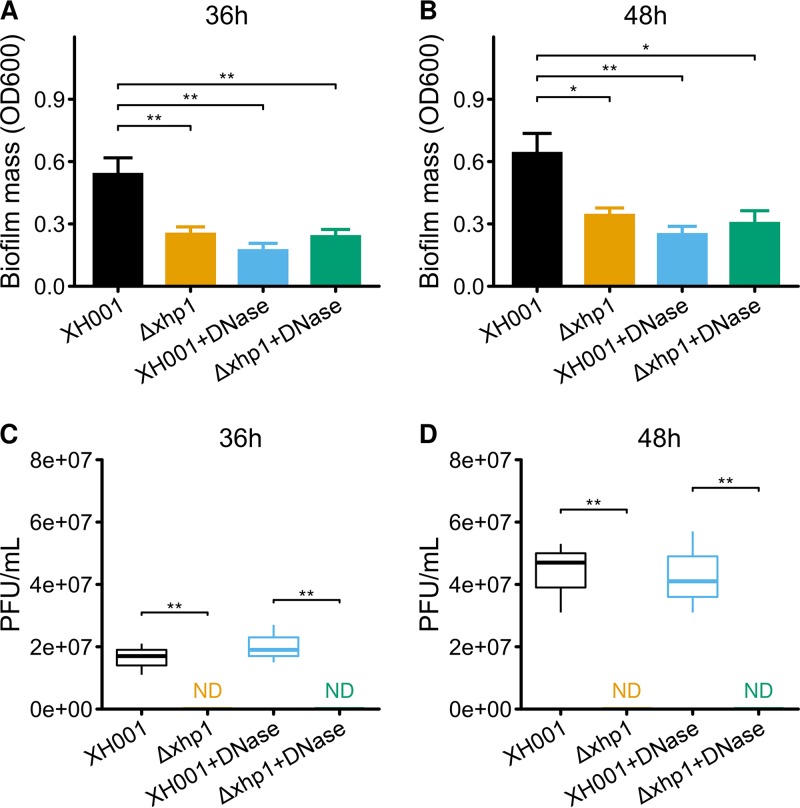
Biofilm is the essential lifestyle of oral bacteria. Increasing lines of evidence suggest that prophages can affect the progression of host cell biofilm formation through all stages (21, 22). Thus, we further tested the impact of xhp1 on its host cell's biofilm formation capability. Data showed that the most drastic difference in biofilm formation was observed at 36 h after incubation. XH001 carrying xhp1 formed better biofilm than XH001 cured of prophage, as reflected by the significantly increased biomass measured by a crystal violet assay (Fig. 4A and B).
FIG 4.
Prophage xhp1 increases biofilm formation. (A and B) Biofilm formation of XH001 and XH001 Δxhp1 in the absence or presence of DNase for 36 h (A) and 48 h (B). Data are the averages of three replicate wells in 96-well plates from three independent cultures. The titer of prophage xhp1 in the supernatant was calculated after 36 h (C) and 48 h (D) of culturing. *, P < 0.05 as calculated by Student's t test; **, P < 0.01. ND, not determined.
The spontaneous induction of prophage often results in the lysis of a subpopulation of bacterial host and the subsequent release of extracellular DNA (eDNA), which can promote biofilm formation (23). To test if a similar mechanism could be involved in the observed xhp1-dependent enhanced biofilm formation, we first measured the titer of the released prophages when host cells were cultured in biofilm versus planktonic form. The result showed that a phage titer of ∼107 PFU/ml was detected in the supernatant of 36-h- and 48-h-old statically grown XH001 biofilms (Fig. 4C and D), while no phage release can be detected in the supernatant of 36-h- or 48-h-old XH001 Δxhp1 biofilm. Our data suggested that xhp1 can be induced in mature biofilm.
Furthermore, while the addition of DNase (final concentration, 100 μg/ml) did not affect the release of prophage by the biofilm cells, it significantly reduced the biomass of XH001 biofilm to a level close to that of XH001 Δxhp1, whose biofilm formation was not significantly affected by the addition of DNase (Fig. 4A and B).
Our data strongly indicate that the biofilm formation process was promoted by the spontaneous activation of xhp1, which could lead to the lysis of a subpopulation of host cells. The released eDNA from lysed cells would then contribute to the observed enhanced biofilm formation.
DISCUSSION
In this study, we reported the characterization of a new A. odontolyticus prophage, xhp1, identified from a human oral isolate, XH001. Our data revealed the uncommon extrachromosomal existence of xhp1 as a plasmid-like prophage based on following evidence: first, XH001 whole-genome sequencing data suggested that xhp1 does not integrate into the host genome; furthermore, the xhp1 genome can be extracted from the bacterial cells without mitomycin C induction (Fig. 2A); last, the estimated copy number of xhp1 is 5 (Fig. 2B), while most integrated prophage should have only one copy per cell. We then demonstrated that xhp1 is a linear plasmid with 10-bp single-stranded cohesive ends (CGTCAAGAGC) at the 3′ of both ends, based on sequencing data analysis (19). The endonuclease digestion analysis further confirmed that xhp1 possesses a linear genome both in phage particles and inside the host cell.
The linear plasmid-like prophage is quite unique, and currently, only a few examples of such phages from bacteria are known. The best-studied linear plasmid-like prophage is N15, the genome of which has 12-bp single-stranded cohesive ends. Mechanisms to ensure the complete replication of linear DNA include protein priming, recombination, covalently closed terminal hairpins, and special telomerase enzymes, which have been reviewed previously (24). Thus, N15 employs telomerase to replicate its genome (20).
Intriguingly, compared to other known linear plasmid-like prophages, such as N15, φKO2, pY54, ΦHAP-1, VHML, VP882, Vp58.5, and vB_VpaM_MAR (20), xhp1 did not contain a protelomerase gene but rather carries an integrase-encoding gene. Furthermore, phylogenetic analysis (data not shown) revealed limited similarity between xhp1 and the other eight linear plasmid-like phages, suggesting that xhp1 might belong to a unique group of phages.
Protein BLAST revealed that the predicted xhp1 integrase contains a C-terminal catalytic domain from bacterial phages and conjugate transposons and has more than 90% similarity at the amino acid level to the integrase from Actinomyces sp. strain ICM54, Xylanimonas cellulosilytica, Bifidobacterium sp. strain MSTE12, and Kytococcus sedentarius. However, the integrase of xhp1 has no similarity to the telomerase of N15, as was revealed by protein BLAST. Thus, we infer that xhp1 might use a different approach to replicate its linear genome through terminal-protein-primed DNA synthesis and subsequent endonucleolytic processing, which leads to a free terminal 3′-overhang in the xhp1 linear genome (25). Bioinformatics also predicted a DNA primase (XHP1_00001) in xhp1, suggesting that primase might synthesize an RNA primer during replication, which leads to a 10-bp 3′-single-strand end in the xhp1 genome after the degradation of leading RNA.
Moreover, the presence of integrase in the xhp1 genome implies that xhp1 might possess two lysogenic lifestyles, where it exists in XH001 as a linear plasmid-like element or integrates into the genomes of different A. odontolyticus strains or even other Actinomyces species as an integrated prophage. For example, circular plasmid-like lysogenic Staphylococcus aureus phages Φs80b and Φs84b can integrate into the chromosome of S. aureus strain s64c but remain extrachromosomal in S. aureus strain 8325-4 (26, 27). Thus, further studies on the host range of xhp1 and its life cycles as well as its lifestyle in other strains or species would shed more light on this intriguing new phage.
For the lysogenic cycle control, we identified a repressor (xhp1_00053) in the xhp1 genome. It encodes a helix-turn-helix motif, which is conserved in all the phage-encoded repressors. The repressor is routinely expressed during bacterial growth phase to repress the expression of other phage genes (20, 28). This might be the reason that xhp1 cannot form any plaque in XH001. Other prophages usually encode antirepressor proteins for better regulation of their lysogenic cycle (29). However, no obvious antirepressor protein can be predicted from the xhp1 genome, which warrants further investigation.
The frequency of spontaneous deletion of plasmid-like prophage is low, mainly due to the stabilization machinery. For example, a toxin-antitoxin module (30) and the partitioning system (31) in linear plasmid-like prophage N15 both contribute to the prophage stabilization. In this study, the deletion rate of xhp1 is likely to be quite low. Nevertheless, we managed to isolate a single xhp1-cured strain, XH001 Δxhp1, through extensive screening of colonies recovered from heat-treated XH001 cells. Thus, XH001 Δxhp1 was used as a host strain to determine the titer of the prophage xhp1 (Fig. 3B). Moreover, XH001 Δxhp1 proved to be an ideal control to study the impact of prophage on host physiology (Fig. 4A and B).
In most habitats, including the human oral cavity, microbes reside within biofilms, which comprise microbes embedded within an extracellular matrix that consists of polysaccharides, proteins, DNA, and lipid (32). Biofilm is an essential lifestyle of oral microbes, and it protects oral bacteria from saliva flow, daily oral hygiene, and the host immune system, allowing them to persist within the oral cavity (32, 33). Our data clearly showed that prophage xhp1 promotes XH001 biofilm assembly by spontaneous induction and release of eDNA, which could serve as a building material for better biofilm formation (34). This is in agreement with previous reports showing that prophage spontaneous activation promotes DNA release that enhances biofilm formation in a variety of bacterial species, including Streptococcus pneumoniae (23), Pseudomonas aeruginosa (35), and Shewanella oneidensis (36). However, the factors and mechanisms involved in xhp1 induction under laboratory and natural conditions require further investigation (37).
The oral cavity is one of the most densely microbe-populated habitats of the human body, with 6 billion bacteria and potentially 30 times more phages (2). Due to their intimate interaction with bacterial hosts, phages have been implicated in shaping the ecology of oral bacterial communities. However, until recently, the study of oral microbiota has been mainly bacterium oriented, while the role of phages in modulating bacterial physiology, impacting bacterial host interaction with other oral residents, and their ecological contribution are understudied (7, 10, 13). Our study provides additional evidence demonstrating the role of prophage in contributing to the persistence of the bacterial host in the oral cavity by enhancing biofilm formation (Fig. 4A and B).
Interestingly, XH001 also serves as a host for TM7x, a recently isolated ultrasmall oral bacterium that belongs to the Saccharibacteria phylum, whose relative abundance has been positively correlated with oral mucosal infectious diseases, such as periodontitis. Intriguingly, TM7x forms a unique epibiotic parasitic relationship with XH001 (15). Thus, the interactions between XH001 and its two “parasites,” the epibiotic parasitic bacterium TM7x and intracellular prophage xhp1, potentially have a significant impact on the physiology and life cycle of all three partners, as well as on local ecology. A model system containing host XH001, prophage xhp1, and the epibiotic parasitic bacterium TM7x is currently being developed in our laboratory for studying the impact of prophage on bacterial interspecies interactions and bacterial ecology.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and prophage used in this work are listed in Table 2. A. odontolyticus subsp. actinosynbacter strain XH001 was isolated from the oral cavity (15) and cultured in brain heart infusion (BHI) medium at 37°C under microaerophilic conditions (5% O2, 5% CO2, balanced with N2).
TABLE 2.
Bacterial strains and phages used in this study
| Strain or phage | Description | Source |
|---|---|---|
| XH001 | Wild-type Actinomyces odontolyticus strain isolated from human oral cavity | 16 |
| XH001 Δxhp1 | Strain cured of prophage xhp1 | This study |
| xhp1 | Prophage induced from XH001 | This study |
Induction of phage particles.
An exponential-phase culture of XH001 (optical density at 600 nm [OD600], 0.5) in BHI medium was treated for 3 h with mitomycin C (Sigma) at a final concentration of 1 μg/ml. Then, the culture was centrifuged at 9,000 rpm for 2 min, and the supernatant was collected to pass through a 0.45-μm-pore-size membrane filter.
Phage particles in the supernatant were concentrated and purified by PEG-8000 precipitation, as previously described (38).
Transmission electron microscopy images.
Phage particles were deposited on carbon-coated copper grids for 10 min, stained with phosphotungstic acid (PTA [pH 7.0]) for 5 s, and examined under a Philips EM 300 electron microscope (39). The sizes for the icosahedral capsid and the tail were estimated using AxioVision LE, based on five randomly selected images.
Bacteriophage genome sequence analysis.
XH001 was sequenced (16), and the sequencing data were assembled by SPAdes (18). The assembly result was manually checked and investigated using Bandage (40). PHASTER was used to predict prophage from the XH001 genome (17). Only one intact prophage, named xhp1, was predicted. After retrieving the phage genome, the phage termini were identified by PhageTerm (19). The xhp1 genome was further annotated by Prokka (41). DNA and protein sequences were scanned for homologs by BLAST.
Isolation of phage DNA from phage particles and bacterial cells.
First, phage particles were enriched by PEG-8000 precipitation after 3 h of mitomycin C induction. Phage DNA was isolated from the phage particles, as described previously (42).
Extraction of prophage DNA from bacterial cells was performed using a plasmid extraction kit (D6943; Omega Bio-Tek), as per the manufacturer's instruction. Since XH001 is Gram positive, mechanical cell breakage using bead-beating for 10 s was used to increase the cell lysis efficiency and DNA yield. The DNA isolated from 1 ml of bacterial culture (OD600, 1.0) was about 36 ng/μl, with a final volume of 40 μl. However, the bead-beating tends to break bacterial genomic DNA, which likely increased the host DNA contamination when extracting prophage genome using a plasmid extraction kit (Fig. 2A).
Digestion of the xhp1 genome.
To determine if the xhp1 genome is linear or circular, phage DNA extracted from phage particles and bacterial cells was digested by XbaI or NheI/XbaI. The mixture was incubated at 37°C for 60 min and visualized by 1% agarose gel electrophoresis, followed by ethidium bromide staining.
Determination of the copy number of xhp1.
The copy number of xhp1 in the host XH001 strain was determined by qPCR, using total genomic DNA isolated from XH001 culture as the template. The reactions were performed using iQ SYBR green supermix (Bio-Rad) and the CFX96 real-time PCR system (Bio-Rad). Primers specific for xhp1 integrase, single-stranded DNA-binding protein, and the XH001 16S rRNA gene were designed and used for qPCR (Table 1). The copy numbers were calculated as the mean quantification cycle (Cq) values of the 16S rRNA gene, compared to the phage-specific genes, using the formula 2ΔCq.
Isolation of XH001 Δxhp1.
To facilitate the loss of linear plasmid prophage, a heat technique was performed. Specifically, a single colony of XH001 was picked and inoculated in 5 ml BHI broth. The BHI tube was incubated at 42°C for 24 h to allow bacterial growth. Then, culture was diluted and spread on BHI agar plates. Plates were incubated at 37°C until individual colonies appeared. One thousand colonies were picked and cultured in BHI medium in 96-well plates. The plates were incubated at 37°C until the cultures become turbid. One microliter of the bacterial culture was used as the template and screened for the loss of xhp1, as indicated by a lack of the predicted PCR product using xhp1-specific primers (xhp1-intF and xhp1-intR; Table 1). The frozen stock of XH001 Δxhp1, which was cured of xhp1, was made and stored at −80°C.
Biofilm assay.
Biofilms were examined by crystal violet staining, as previously described (21). Log-phase bacterial culture was diluted 1:100 in BHI medium. Next, 0.2-ml aliquots were added to 96-well polystyrene microplates and incubated for 36 h or 48 h at 37°C under microaerophilic conditions. Plates were washed with phosphate-buffered saline (PBS), and the biofilm biomass was stained with crystal violet for 15 min. For quantitation, crystal violet was solubilized in 0.2 ml of 95% ethanol, and the optical density at 600 nm (OD600) was determined using a SpectraMax M3 multimode microplate reader. Samples were examined in triplicate, and BHI medium alone was used as the negative control. The OD600 values of the BHI control were subtracted from the average OD600 of each strain.
In the DNase control group, DNase I (Roche) was used at a final concentration of 100 μg/ml in BHI medium when inoculating the cells.
Measuring prophage titers.
XH001 Δxhp1 was used as the host strain to measure the titer of prophage xhp1. The supernatant containing xhp1 was diluted in 10-fold increments to 10−6. Then, 0.1-ml aliquots of each diluted xhp1 supernatant were mixed with 0.1 ml XH001 Δxhp1. Each resultant mixture was added to 3 ml of 0.7% BHI soft agar and plated on 1.5% BHI agar plates. The PFU were calculated after 24 h of incubation at 37°C under microaerophilic conditions. Note that the measurement of prophage xhp1 titers might not be accurate, because the efficiency of plating (EOP) for xhp1 on XH001 Δxhp1 is unknown.
Statistical analyses.
Student's t test was used to compare two-group data. A P value of <0.05 was considered statistically significant.
Accession number(s).
The GenBank accession numbers of Actinomyces odontolyticus subsp. actinosynbacter strain XH001 and prophage xhp1 are LLVT00000000 and MG941013, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NIH (grants 1R01DE023810, 1R01DE026186, and 1R01DE020102 to W. Shi, X. He, and J. S. McLean) and the National Natural Science Foundation of China (NSFC; grant 31501004 to S. Le).
W. Shi is an employee of C3 Jian, Inc., which has licensed technologies from UC Regents that could be indirectly related to this research project.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01263-18.
REFERENCES
- 1.Baker JL, Bor B, Agnello M, Shi WY, He XS. 2017. Ecology of the oral microbiome: beyond bacteria. Trends Microbiol 25:362–374. doi: 10.1016/j.tim.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edlund A, Santiago-Rodriguez TM, Boehm TK, Pride DT. 2015. Bacteriophage and their potential roles in the human oral cavity. J Oral Microbiol 7:12. doi: 10.3402/jom.v7.27423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr JJ. 2017. A bacteriophages journey through the human body. Immunol Rev 279:106–122. doi: 10.1111/imr.12565. [DOI] [PubMed] [Google Scholar]
- 4.Szafranski SP, Winkel A, Stiesch M. 2017. The use of bacteriophages to biocontrol oral biofilms. J Biotechnol 250:29–44. doi: 10.1016/j.jbiotec.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Pride DT, Salzman J, Haynes M, Rohwer F, Davis-Long C, White RA, Loomer P, Armitage GC, Relman DA. 2012. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J 6:915–926. doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naidu M, Robles-Sikisaka R, Abeles SR, Boehm TK, Pride DT. 2014. Characterization of bacteriophage communities and CRISPR profiles from dental plaque. BMC Microbiol 14:13. doi: 10.1186/1471-2180-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abeles SR, Robles-Sikisaka R, Ly M, Lum AG, Salzman J, Boehm TK, Pride DT. 2014. Human oral viruses are personal, persistent and gender-consistent. ISME J 8:1753–1767. doi: 10.1038/ismej.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robles-Sikisaka R, Ly M, Boehm T, Naidu M, Salzman J, Pride DT. 2013. Association between living environment and human oral viral ecology. ISME J 7:1710–1724. doi: 10.1038/ismej.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ly M, Abeles SR, Boehm TK, Robles-Sikisaka R, Naidu M, Santiago-Rodriguez T, Pride DT. 2014. Altered oral viral ecology in association with periodontal disease. mBio 5:e01133-14. doi: 10.1128/mBio.01133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willner D, Furlan M, Schmieder R, Grasis JA, Pride DT, Relman DA, Angly FE, McDole T, Mariella RP, Rohwer F, Haynes M. 2011. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci U S A 108:4547–4553. doi: 10.1073/pnas.1000089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delisle AL, Guo M, Chalmers NI, Barcak GJ, Rousseau GM, Moineau S. 2012. Biology and genome sequence of Streptococcus mutans phage M102AD. Appl Environ Microbiol 78:2264–2271. doi: 10.1128/AEM.07726-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, Maxwell KL. 2016. Prophages mediate defense against phage infection through diverse mechanisms. ISME J 10:2854–2866. doi: 10.1038/ismej.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou W-C, Huang S-C, Chiu C-H, Chen Y-YM. 2017. YMC-2011, a temperate phage of Streptococcus salivarius 57.I. Appl Environ Microbiol 83:e03186-16. doi: 10.1128/AEM.03186-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornuault JK, Petit MA, Mariadassou M, Benevides L, Moncaut E, Langella P, Sokol H, De Paepe M. 2018. Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes. Microbiome 6:65. doi: 10.1186/s40168-018-0452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He XS, McLean JS, Edlund A, Yooseph S, Hall AP, Liu SY, Dorrestein PC, Esquenazi E, Hunter RC, Cheng GH, Nelson KE, Lux R, Shi WY. 2015. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci U S A 112:244–249. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLean JS, Liu Q, Bor B, Bedree JK, Cen L, Watling M, To TT, Bumgarner RE, He X, Shi W. 2016. Draft genome sequence of Actinomyces odontolyticus subsp. actinosynbacter strain XH001, the basibiont of an oral TM7 epibiont. Genome Announc 4:e01685-15. doi: 10.1128/genomeA.01685-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang YJ, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garneau JR, Depardieu F, Fortier LC, Bikard D, Monot M. 2017. PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci Rep 7:8292. doi: 10.1038/s41598-017-07910-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravin NV. 2015. Replication and maintenance of linear phage-plasmid N15. Microbiol Spectr 3:PLAS-0032-2014. doi: 10.1128/microbiolspec.PLAS-0032-2014. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Kim Y, Wood TK. 2009. Control and benefits of CP4-57 prophage excision in Escherichia coli biofilms. ISME J 3:1164–1179. doi: 10.1038/ismej.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Secor PR, Sweere JM, Michaels LA, Malkovskiy AV, Lazzareschi D, Katznelson E, Rajadas J, Birnbaum ME, Arrigoni A, Braun KR, Evanko SP, Stevens DA, Kaminsky W, Singh PK, Parks WC, Bollyky PL. 2015. Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe 18:549–559. doi: 10.1016/j.chom.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrolo M, Frias MJ, Pinto FR, Melo-Cristino J, Ramirez M. 2010. Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS One 5:e15678. doi: 10.1371/journal.pone.0015678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casjens S. 1999. Evolution of the linear DNA replicons of the Borrelia spirochetes. Curr Opin Microbiol 2:529–534. doi: 10.1016/S1369-5274(99)00012-0. [DOI] [PubMed] [Google Scholar]
- 25.Qin Z, Cohen SN. 1998. Replication at the telomeres of the Streptomyces linear plasmid pSLA2. Mol Microbiol 28:893–903. doi: 10.1046/j.1365-2958.1998.00838.x. [DOI] [PubMed] [Google Scholar]
- 26.Utter B, Deutsch DR, Schuch R, Winer BY, Verratti K, Bishop-Lilly K, Sozhamannan S, Fischetti VA. 2014. Beyond the chromosome: the prevalence of unique extra-chromosomal bacteriophages with integrated virulence genes in pathogenic Staphylococcus aureus. PLoS One 9:e100502. doi: 10.1371/journal.pone.0100502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao L, Wu X, Sun B. 2010. Alternative sigma factor sigmaH modulates prophage integration and excision in Staphylococcus aureus. PLoS Pathog 6:e1000888. doi: 10.1371/journal.ppat.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobocka MB, Svarchevsky AN, Rybchin VN, Yarmolinsky MB. 1996. Characterization of the primary immunity region of the Escherichia coli linear plasmid prophage N15. J Bacteriol 178:2902–2910. doi: 10.1128/jb.178.10.2902-2910.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravin NV, Svarchevsky AN, Deho G. 1999. The anti-immunity system of phage-plasmid N15: identification of the antirepressor gene and its control by a small processed RNA. Mol Microbiol 34:980–994. doi: 10.1046/j.1365-2958.1999.01658.x. [DOI] [PubMed] [Google Scholar]
- 30.Dziewit L, Jazurek M, Drewniak L, Baj J, Bartosik D. 2007. The SXT conjugative element and linear prophage N15 encode toxin-antitoxin-stabilizing systems homologous to the tad-ata module of the Paracoccus aminophilus plasmid pAMI2. J Bacteriol 189:1983–1997. doi: 10.1128/JB.01610-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grigoriev PS, Lobocka MB. 2001. Determinants of segregational stability of the linear plasmid-prophage N15 of Escherichia coli. Mol Microbiol 42:355–368. doi: 10.1046/j.1365-2958.2001.02632.x. [DOI] [PubMed] [Google Scholar]
- 32.Bowen WH, Burne RA, Wu H, Koo H. 2018. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol 26:229–242. doi: 10.1016/j.tim.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanz M, Beighton D, Curtis MA, Cury JA, Dige I, Dommisch H, Ellwood R, Giacaman RA, Herrera D, Herzberg MC, Kononen E, Marsh PD, Meyle J, Mira A, Molina A, Mombelli A, Quirynen M, Reynolds EC, Shapira L, Zaura E. 2017. Role of microbial biofilms in the maintenance of oral health and in the development of dental caries and periodontal diseases. Consensus report of group 1 of the Joint EFP/ORCA workshop on the boundaries between caries and periodontal disease. J Clin Periodontol 44(Suppl 18):S5–S11. doi: 10.1111/jcpe.12682. [DOI] [PubMed] [Google Scholar]
- 34.Das T, Sehar S, Manefield M. 2013. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ Microbiol Rep 5:778–786. doi: 10.1111/1758-2229.12085. [DOI] [PubMed] [Google Scholar]
- 35.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol 59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 36.Godeke J, Paul K, Lassak J, Thormann KM. 2011. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J 5:613–626. doi: 10.1038/ismej.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nanda AM, Thormann K, Frunzke J. 2015. Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J Bacteriol 197:410–419. doi: 10.1128/JB.02230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu SG, Le S, Tan YL, Zhu JM, Li M, Rao XC, Zou LY, Li S, Wang J, Jin XL, Huang GT, Zhang L, Zhao X, Hu FQ. 2013. Genomic and proteomic analyses of the terminally redundant genome of the Pseudomonas aeruginosa phage PaP1: establishment of genus PaP1-like phages. PLoS One 8:e62933. doi: 10.1371/journal.pone.0062933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang YH, Lu SG, Shen W, Zhao X, Shen MY, Tan YL, Li G, Li M, Wang J, Hu FQ, Le S. 2016. Characterization of the first double-stranded RNA bacteriophage infecting Pseudomonas aeruginosa. Sci Rep 6:38795. doi: 10.1038/srep38795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 42.Shen MY, Le S, Jin XL, Li G, Tan YL, Li M, Zhao X, Shen W, Yang YH, Wang J, Zhu HB, Li S, Rao XC, Hu FQ, Lu SG. 2016. Characterization and comparative genomic analyses of Pseudomonas aeruginosa phage PaoP5: new members assigned to PAK_P1-like viruses. Sci Rep 6:34067. doi: 10.1038/srep34067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.