Biorenewable energy sources are of growing interest to mitigate climate change, but like other commodities with nominal value, require innovation to maximize yields. Energetic considerations constrain production of many biofuels, such as molecular hydrogen (H2) because of the competing needs for cell mass synthesis and metabolite formation. Here we describe cell lines of the extremophile Thermotoga maritima that exceed the physiologic limits for H2 formation arising from genetic changes in fermentative metabolism. These cell lines were produced using a novel method called transient gene inactivation combined with adaptive laboratory evolution. Genome resequencing revealed unexpected changes in a maltose transport protein. Reduced rates of sugar uptake were accompanied by lower rates of growth and enhanced productivity of H2.
KEYWORDS: Hyperthermophile, MalK, hydrogen, transient gene inactivation, adaptive laboratory evolution, bacterial genetics, fermentation, hydrogen, Thermotoga maritima
ABSTRACT
When carbohydrates are fermented by the hyperthermophilic anaerobe Thermotoga maritima, molecular hydrogen (H2) is formed in strict proportion to substrate availability. Excretion of the organic acids acetate and lactate provide an additional sink for removal of excess reductant. However, mechanisms controlling energy management of these metabolic pathways are largely unexplored. To investigate this topic, transient gene inactivation was used to block lactate production as a strategy to produce spontaneous mutant cell lines that overproduced H2 through mutation of unpredicted genetic targets. Single-crossover homologous chromosomal recombination was used to disrupt lactate dehydrogenase (encoded by ldh) with a truncated ldh fused to a kanamycin resistance cassette expressed from a native PgroESL promoter. Passage of the unstable recombinant resulted in loss of the genetic marker and recovery of evolved cell lines, including strain Tma200. Relative to the wild type, and considering the mass balance of fermentation substrate and products, Tma200 grew more slowly, produced H2 at levels above the physiologic limit, and simultaneously consumed less maltose while oxidizing it more efficiently. Whole-genome resequencing indicated that the ABC maltose transporter subunit, encoded by malK3, had undergone repeated mutation, and high-temperature anaerobic [14C]maltose transport assays demonstrated that the rate of maltose transport was reduced. Transfer of the malK3 mutation into a clean genetic background also conferred increased H2 production, confirming that the mutant allele was sufficient for increased H2 synthesis. These data indicate that a reduced rate of maltose uptake was accompanied by an increase in H2 production, changing fermentation efficiency and shifting energy management.
IMPORTANCE Biorenewable energy sources are of growing interest to mitigate climate change, but like other commodities with nominal value, require innovation to maximize yields. Energetic considerations constrain production of many biofuels, such as molecular hydrogen (H2) because of the competing needs for cell mass synthesis and metabolite formation. Here we describe cell lines of the extremophile Thermotoga maritima that exceed the physiologic limits for H2 formation arising from genetic changes in fermentative metabolism. These cell lines were produced using a novel method called transient gene inactivation combined with adaptive laboratory evolution. Genome resequencing revealed unexpected changes in a maltose transport protein. Reduced rates of sugar uptake were accompanied by lower rates of growth and enhanced productivity of H2.
INTRODUCTION
Among the various methods for commercial H2 production, biological synthesis offers a carbon-neutral, economic and sustainable approach (1, 2). Fermentative H2 production, also called “dark fermentation,” is an attractive biological route because of higher H2 production yield, the availability of significant sources of saccharified biomass feedstocks, and a lower requirement for energy input (3, 4). Among various mesophilic H2 producers, hyperthermophiles offer significant benefits because they have superior productivity and yields (5, 6). Certain thermodynamic constraints on H2 productivity per glucose molecule are relieved in hyperthermophiles. Under these conditions, the H2 synthesis reaction becomes more favorable due to entropic effects not found under mesophilic conditions (7). In addition to favoring the energetics of H2 production, growth at high temperature promotes solubilization and hydrolysis of complex carbohydrates to fermentable sugars while inhibiting growth of H2 consumers present in raw feedstocks (8, 9). However, the physiologic yield is limited to a maximum of 4 mol of H2 per mol of glucose and accumulation of H2 acts as a metabolic inhibitor, representing major obstacles for commercialization of biological H2 production (6, 10, 11). In vitro, 12 mol of H2 can be formed per mole of glucose, and this has been verified in vitro when recombinant enzymes, ATP and NADH, were evaluated (12, 13). However, in a biological system this reaction does not provide enough Gibbs free energy (G) to sustain growth. Consequently, for whole cells, Thauer et al. proposed a physiologic limit of 33%, or 4 mol of H2 per mol of glucose, to enable sufficient energy formation to support cell growth (14). In growing cells the energy cost of metabolite formation competes with the cost of forming new cell mass. For this reason, uncoupling microbial growth from product formation seeks to maximize the amount of raw material used for end product synthesis while minimizing by-product formation consisting primarily of cell mass (15, 16). Slow growth rather than no growth may be required to support the needs of energy-coupled reactions and to maintain oxidation-reduction balance (16). Nutrient starvation has been explored as a growth uncoupler for production of methane by methanogens, protein by Escherichia coli, and H2 by E. coli (17–20). Studies of bacterial growth under conditions of limited ATP production suggest that continued cell division can be accomplished with limited substrate uptake, in effect achieving an uncoupling of growth from metabolite formation (20, 21). Accomplishing this end in a reproducible manner would relieve a glycolytic bottleneck and support commercial biological H2 production from a biological system.
Thermotoga maritima is a hyperthermophilic anaerobic bacterium that grows optimally at 80°C and produces H2 to levels that approach the thermodynamic limit using simple and complex carbohydrates (6, 10, 22, 23). Seven percent of its genome encodes carbohydrate-utilizing genes, along with abundant transporters that are mostly ATP-binding cassettes, placing it high among sequenced bacterial and archaeal genomes for transporter gene content (24, 25; see also http://www.membranetransport.org). While the majority of the T. maritima transporters are of bacterial origin, 37% are thought to be of archaeal origin (24). These frequently were annotated as oligopeptide transporters but were subsequently found to bind carbohydrates (26). T. maritima employs the Embden-Meyerhof-Parnas (EMP) pathway (85% relative contribution) and the Entner-Doudoroff (ED) pathway (15% relative contribution) to convert glucose into H2 and organic acids (lactate and acetate) (5) (see Fig. S1 in the supplemental material). High levels of H2 synthesis arise from the action of a bifurcating hydrogenase that accepts electrons from both NADH and reduced ferrodoxin (27). Accumulation of H2 in batch culture without headspace exchange results in loss of redox homeostasis that shifts metabolism toward the production of organic acids, presumably through hydrogenase inhibition (28, 29). Availability of a predicted central metabolic network, in silico metabolic knockouts, fermentation studies, and structural genomics and proteomics provide the tools to intentionally uncouple growth from metabolite formation (6, 30–34). Here we present the use of a genetic approach for the creation of new cell lines that overproduce H2 through changes in sugar transport.
RESULTS
Isolation of H2-resistant and H2-overproducing cell lines by transient gene inactivation.
The growth-inhibitory effect of H2 has been reported for T. maritima as well as for other H2-producing organisms (6, 10, 29, 35). A linear relationship between growth and H2 could be established when T. maritima was cultivated in the presence of various amounts of added H2 (Fig. 1A). To assess the magnitude of this effect and its dependence on maltose concentration, T. maritima was cultivated with various amounts of added maltose and the relationship between cell and H2 yields was determined. Cell yields saturated rapidly with increasing maltose concentration (Fig. 1B) due to the growth inhibition caused by increased number of H2-producing cells of T. maritima. H2 production was normalized to cell mass to compare levels of H2 production at different concentrations of maltose. The apparent inverse relationship between H2 accumulation and maltose concentration suggested that H2 became toxic under these conditions. If this toxicity was sufficient to inhibit growth, it could be used to select for mutants that overcame this effect. Since an increase in H2 partial pressure shifts the metabolism of T. maritima toward lactate synthesis (6, 36, 37) and not ethanol, transient inactivation of lactate dehydrogenase (encoded by ldh) could exacerbate H2 toxicity by blocking lactate formation. This would remove one pathway for excretion of excess reductant and thereby create selective pressure to recover such mutants without having to add exogenous H2.
FIG 1.
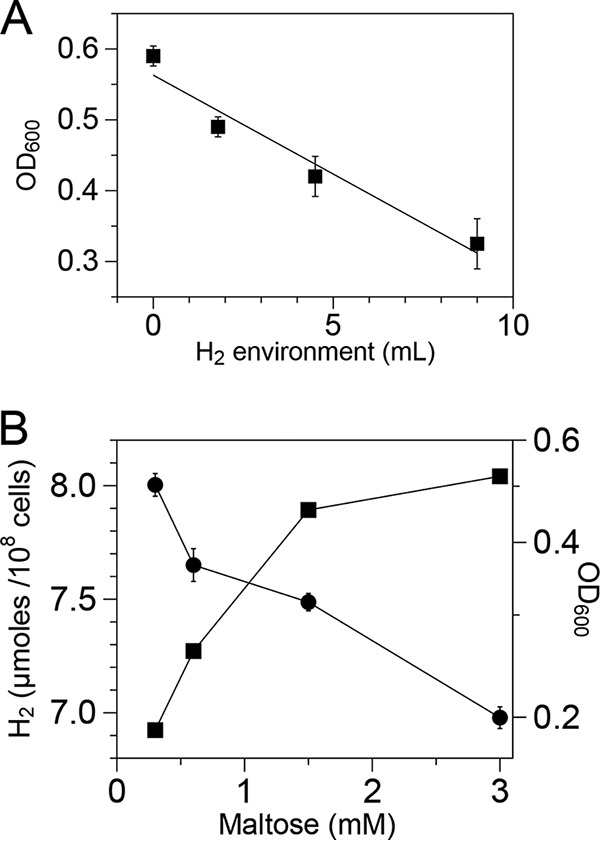
Growth inhibition by hydrogen and relationship between maltose concentration and H2 production. (A) Tubes inoculated at an initial OD600 of 0.02 were incubated at 80°C in the presence of various amounts of H2 and OD changes (growth) were observed after 24 h of incubation. (B) H2 production (●) and growth (■) of the wild type (T. maritima) grown in various amounts of maltose in batch culture in 20 h. H2 that accumulated at each sugar concentration was normalized to 108 cells/ml. The error bars represent the SDs from two biological replicates.
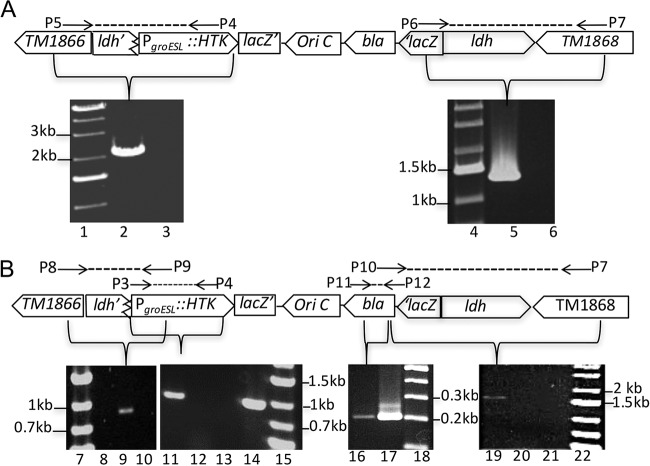
T. maritima is transformable using replicating plasmids (38). Therefore, chromosomal recombination was pursued as a means to generate useful mutations as demonstrated previously (39, 40). As efforts to disrupt ldh by double crossover were not successful, indicating that the gene was essential and therefore unlike its homologs in other H2-producing microbes (41), transient gene disruption was used instead. Cells were transformed with a 3′ terminally truncated copy of ldh lacking a promoter that was fused upstream of a thermostable kanamycin resistance gene (htk) expressed from the T. maritima PgroESL promoter (42). Electroporated cells were enriched for antibiotic-resistant recombinants in liquid culture using various concentrations of added drug depending on the selection process. Total genomic DNA was then screened for the presence of novel chromosomal fusions arising from targeted recombination at ldh. The predicted unique 5′ amplicon (∼2.0 kb) was evident following PCR amplification using a primer complementary to sequences upstream of ldh (P5) and the 3′ end of htk (P4) (Fig. 2A). Similarly, the predicted unique 3′ PCR amplicon (∼1.3 kb) was also detected using lacZ and primers complementary to sequences downstream of ldh (P6 and P7). Both amplicons were detected using genomic DNA from enrichments using 375 μg/ml and 500 μg/ml of kanamycin, and their composition was verified by DNA sequencing. These results confirmed that directed chromosome recombination at the ldh locus was successful and prompted efforts to recover recombinant clonal cell lines.
FIG 2.
Targeted disruption of lactate dehydrogenase gene (ldh). Schematic of ldh disruption by single crossover and PCR amplification of predicted amplicons (liquid enrichment [A] and clonal population of Tma100 [B]). Lanes 1, 4, 7, 15, 18, and 22, molecular markers; lanes 2 and 9, unique 5′ fusion joint at ldh locus in liquid enrichment and for Tma100, respectively; lanes 3, 8, and 10, unique 5′ fusion joint at the ldh locus in the wild type, Kanr mutant, and wild type, respectively; lanes 5 and 19, 3′ unique fusion joint at the ldh locus in liquid enrichment and Tma100, respectively; lanes 6, 20, and 21, unique 3′ fusion joint at the ldh locus for the wild type, Kanr mutant, and wild type, respectively; lanes 11 and 14 and lanes 16 and 17, selectable marker (PgroESL::htk) and bla gene in Tma100 and positive control (pBL1292), respectively; lanes 12 and 13, PgroESL::htk in the Kanr mutant and wild type, respectively.
Direct plating of cells transformed and enriched in an identical manner yielded a 34-fold-higher transformation efficiency than that of untransformed cells. Following clonal purification with selection, PCR screening indicated the presence of the PgroESL::htk transgene in 60% of isolates. One transgene-containing isolate, named Tma100, was pursued for additional analysis (Fig. 2B). In Tma100, all predicted amplicons were evident using the indicated primers, including the genetic marker (PgroESL::htk; 946 bp) (P3 and P4), a 5′ unique fusion joint between the upstream gene and within the genetic marker (TM1866::ldh::PgroESL::htk; 966 bp) (P8 and P9), a unique fusion joint between the 3′ end of ldh and the nonreplicating plasmid vector (bla::ldh::TM1868; 1,730 bp) (P10 and P7), and the vector-borne genetic marker (bla; 200 bp) (P11 and P12). In addition, recombination at the PgroESL locus was excluded by the apparent absence of a PCR amplicon encoding the ldh disruption construct fused to this region. The identity of the genetic marker (PgroESL::htk) and 5′ and 3′ unique fusion joints in Tma100 mutant was confirmed by sequencing. Together these data confirmed targeted integration at the chromosomal ldh locus.
Tma100 exhibited poor growth on maltose plates, resulting in small colonies. As this may have resulted from an effect from the ldh disruption, selection for improved growth was performed in the absence of antibiotic selection. Fifty clonal isolates derived from Tma100 were passaged in maltose medium lacking added kanamycin. PCR analysis of the selected 10 isolates (Tma200 to Tma209) showed segregation that restored the native ldh gene. However, all 10 were found to be resistant to kanamycin. A mutation at the 3′ end (A to G at nucleotide [nt] 1420) in 16S rRNA was evident in strains Tma200 through Tma209. These mutations may have occurred in Tma100 during its cultivation in the presence of a larger amount of kanamycin (500 μg/ml). Such mutations likely improved survival under selection despite the transient presence of the introduced genetic marker. These 10 isolates were then tested for H2 production in liquid culture and one, named Tma200, exhibited levels that exceeded those of the wild type. H2 resistance was then tested in the evolved strains in the presence of H2 supplementation. An increased growth rate in the H2 supplemented environment relative to the wild type indicated that both Tma100 and Tma200 exhibited enhanced H2 resistance (Fig. S2).
Metabolic analysis.
Analysis of organic acid and hydrogen metabolites was conducted first using small batch cultures after a single growth cycle at 80°C for 20 h. Since the wild type (T. maritima) grew to a higher cell density than Tma100 and Tma200 during this period, comparison of H2 production among these stains was established on a per-cell basis by normalizing the concentration of H2 to cell number (Fig. S3). On this basis, levels of H2 were higher for Tma100 and Tma200 than the wild type, by 18% and 46%, respectively, while the other Tma100 derivatives (Tma201 to Tma209) were not significantly different from Tma100.
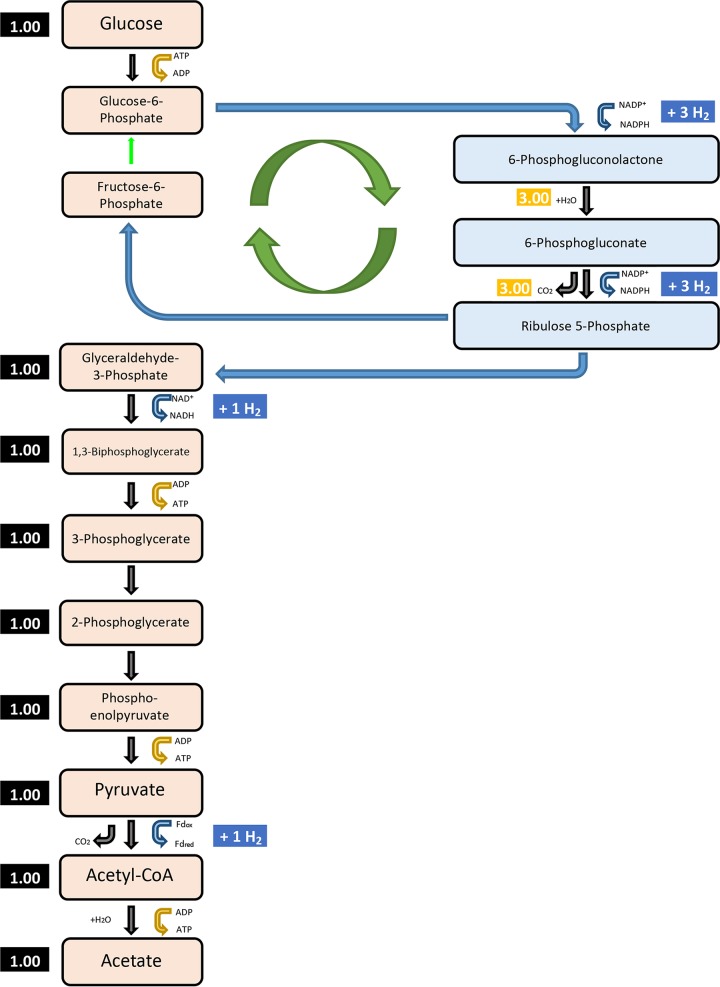
In small volume batch cultures, Tma100 and Tma200 overproduced H2 relative to the wild type while growing more slowly (Fig. S3). To determine if H2 accumulation played a role in this process, additional studies were conducted using bioreactors (3 liters) in which the headspace of the bioreactors was replaced with N2 to avoid H2-associated growth inhibition. Under these conditions, the wild type still grew the fastest and had the highest cell yield, followed by Tma200 and then Tma100 (Fig. 3A; see also Table S1). In contrast, maltose consumption was greatly reduced in Tma100 and Tma200, revealing a defect in catabolism of this sugar. H2 levels produced by Tma100 and Tma200 during exponential growth remained higher than with the wild-type strain, by 18% and 48%, respectively (Fig. 3B). CO2 production by Tma100 and Tma200 was 20% and 40% higher than that of the wild-type strain, respectively. Acetate levels produced during the exponential growth phase also were higher for both Tma100 (39%) and Tma200 (44%) than for the wild type. During exponential phase, traces of lactate (0.17 ± 0.05 mM) were detected in the wild type. After 30 h, levels of lactate in Tma100 were not detectable and in Tma200 were reduced by 71% relative to those of the wild type (12.32 ± 0.05 mM) (Table S2). These data indicated that in the passaged strains, the molar yield of H2 relative to maltose consumed had increased on a per-cell basis (Fig. 3C; Table 1). The stoichiometry of maltose catabolism was calculated for each strain during cultivation in bioreactors. As described in Materials and Methods, the measured metabolites included maltose, H2, CO2, lactate, and acetate. Based on these values, stoichiometric equations for maltose oxidation were established (chemical equations 1 and 2) that distinguished the unique metabolism of Tma100 and Tma200. Production of H2 at levels greater than 4 mol/mol of glucose (Glc) is possible if carbon is redirected through the pentose phosphate pathway (PPP). In this case the H2 yield could be up to 8 mol of H2/mol of Glc (31, 43). The involvement of the PPP along with the glycolytic pathway offer an explanation for the apparent stoichiometry of metabolites formed from Tma100 and Tma200 (Fig. 4). Additionally, the overall carbon recovery was over 90% throughout the fermentation, confirming that consumed carbon primarily formed acetate, lactate, CO2, and biomass. Furthermore, the oxidation-reduction balance of oxidized and reduced products close to the theoretical value of 1.0 shows that the products were in balance and were accurately determined without any indication of new products present in significant quantities (Fig. S4). To determine the H2 produced from the unidentified sugars in complex medium, fermentation studies were carried out without maltose and H2 analysis was performed. All strains grew poorly (optical density [OD] of 0.08/ml) and produced H2 in the micromolar (400-fold lower H2 than H2 with maltose) range, which did not result in change of H2 yield in all strains. This demonstrated that the majority of the carbon derived from maltose contributed to the observed metabolites and cellular biomass. The overall reaction of fermentation can be written as shown below (equations 1 and 2).
| 1 |
| 2 |
FIG 3.
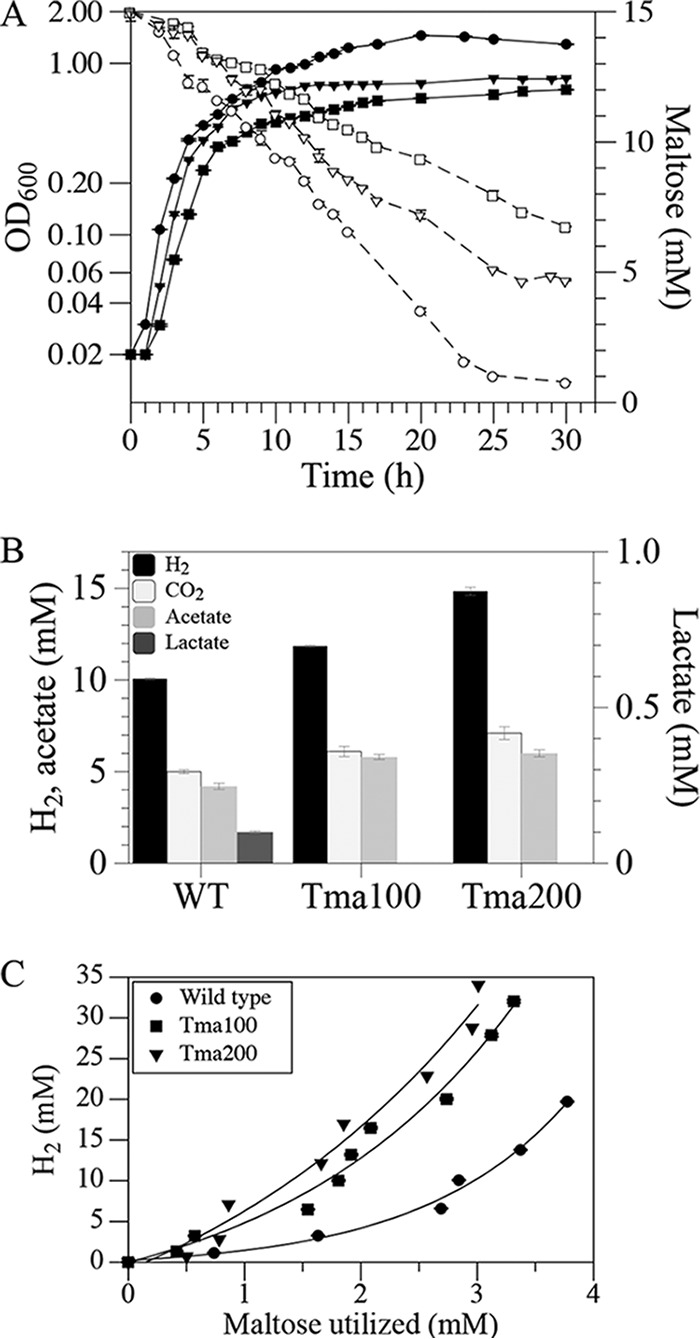
Fermentation profiles of T. maritima strains. Strains were cultivated in 3-liter fermentors at 80°C under anaerobic conditions. (A) The relationship between growth (filled symbols; ●, wild type; ■, Tma100; ▼, Tma200) and maltose utilization (open symbols; ○, wild type; □, Tma100; ▽, Tma200). (B) Comparison of H2, CO2 (cumulative), and organic acids (lactate and acetate) produced in the growth phase normalized to wild-type biomass produced in 5 h. (C) Relationship between H2 production and maltose utilization in the wild-type, Tma100, and Tma200 strains. The error bar represents the SD from triplicate analyses.
TABLE 1.
Yield coefficientsa
| Coefficient | Value (mol mol−1) for: |
||
|---|---|---|---|
| Wild type | Tma100 | Tma200 | |
| YH2/maltose | 6.22 ± 0.13 | 9.69 ± 0.20 | 11.54 ± 0.22 |
| Yacetate/maltose | 1.82 ± 0.02 | 2.66 ± 0.02 | 2.87 ± 0.02 |
| Ylactate/maltose | 0.87 ± 0.001 | NDb | 0.28 ± 0.008 |
| YCO2/maltose | 3.28 ± 0.03 | 4.60 ± 0.05 | 5.44 ± 0.03 |
Yield coefficients were derived from studies using 3-liter bioreactors.
ND, none detected.
FIG 4.
A proposed model showing the redirected carbon flux through the pentose phosphate pathway (PPP) in evolved strains of T. maritima for additional H2 production. The PPP is responsible for generation of glycolytic intermediates and reductants. A combination of the PPP and glycolytic pathway can lead to production of up to 8 mol of H2/mol of Glc.
Genetic basis for increased H2 yield.
Genome resequencing was conducted to identify the genetic basis for H2 overproduction in the passaged strains. A tabulated summary of the confirmed mutations is presented (Table S3). Based on genome alignments of the wild type and derived strains, only malK3 (THMA_1301), a functional malK gene identified previously (39), was identified as a genetic hot spot for mutation formation. Two additional malK genes (THMA_0427 and THMA_1258) have been annotated in T. maritima, and these genes exhibited no mutations relative to the parental strain. In light of the defect in maltose catabolism evident in the passaged strains, mutations in malK3 could play a critical role. Tma100 had a missense mutation (G148D) in malK3 located in close proximity to the signature motif of the ATP binding domain (Fig. S5). A second mutation (D345L) was located near the C terminus. In contrast, the malK3 allele present in Tma200, lacked both G148D and D345L and instead had a mutation located outside the predicted domains (V233S). This position was also altered in the other Tma100-derived isolates (Tma201 to Tma209), but with a different mutation (V233F), and these strains lacked the mutations identified in Tma100. The unexpectedly high mutation frequency in malK3 may have resulted from the use of a strong purifying selection based on maltose catabolism. Since strains Tma201 to Tma209 all had an identical malK3 mutation (V233F), strain Tma201 was included in experiments to establish the rates of maltose uptake.
To test whether maltose is transported only by a maltose transporter, a nonfunctional maltose transporter was created by disrupting malK3, as reported previously (39). A complete loss of maltose uptake in the malK3 mutant and the absence of detectable H2 synthesis established a link between malK3 and H2 synthesis (Fig. S6) and excluded any role played by other putative malK genes of T. maritima in maltose transport. While the malK3 mutant exhibited a complete loss of maltose uptake, the alleles encoded in strains Tma100 and Tma200 resulted in only partial loss of maltose transport function. To verify that the mutation in malK3 was the primary genetic reason for excess H2 production, the mutated allele from Tma200 was used to replace the disrupted allele in the otherwise wild-type background. The resulting strain, called Tma300, exhibited a growth pattern and H2 productivity identical to those of Tma200 (Fig. 5). This demonstrated that the mutant malK3 allele in Tma200 was necessary and sufficient for H2 overproduction.
FIG 5.
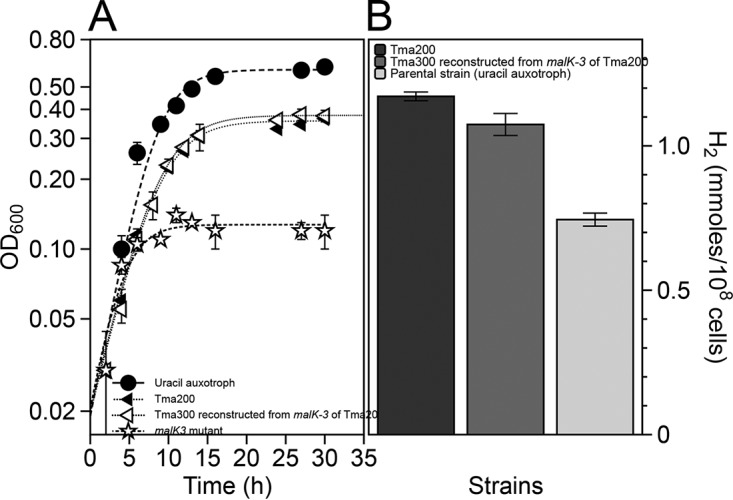
Reconstruction of the excess H2-producing strain. (A) Growth physiology of Tma200, the reconstructed strain (Tma300) containing malK3 of Tma200, parental strain (malK3 mutant), and a uracil auxotrophic strain. (B) H2 production from Tma200, Tma300, and a uracil auxotrophic strain.
Analysis of maltose metabolism.
Maltose consumption was reduced in the passaged strains in liquid culture; therefore, colony sizes were examined during growth on plates containing either maltose or cellobiose. While the wild-type strain formed large colonies regardless of the added sugar, Tma100 preferentially formed small colonies using maltose compared to cellobiose (Fig. S7). In contrast, Tma200 formed intermediate-size colonies regardless of the type of added sugar. Together with high residual levels of maltose in the bioreactor studies (Fig. 3A), a defect in maltose catabolism was evident. Since malK3 encodes the ATP-hydrolyzing subunit of a maltose ABC transporter, the malK3 mutations could affect maltose uptake. Maltose uptake was, therefore, measured at 75°C under anaerobic conditions using [14C]maltose. [14C]maltose uptake was rapid, with linear rates of accumulation between 15 and 30 s after addition, except in Tma100, in which maltose uptake was low (44). Rates of uptake for each strain, in nanomoles per minute per milligram (cell dry weight [cdw]), were 38.53 ± 1.80 for the wild type, 18.68 ± 1.20 for Tma200, 3.13 ± 0.60 for Tma100, and 6.81 ± 0.60 for Tma201 at 169 nM [14C]maltose (Fig. 6A). [14C]maltose uptake was significantly reduced by addition of a 100-fold excess of unlabeled maltose (Fig. 6A). The rate of uptake was saturable when wild-type cells were incubated with maltose at concentrations ranging from 100 nM to 1,000 nM [14C]maltose. The apparent Km for the wild type and Tma200 were 680 nM and 649 nM, with Vmaxs of 182 and 84.8 nmol/min/mg(cdw), respectively (Fig. 6B), while kinetic constants for Tma100 could not be determined due to its low rate of uptake. These data indicate that a slower uptake of maltose in Tma100 and Tma200 altered its metabolism and led to excess H2 production.
FIG 6.
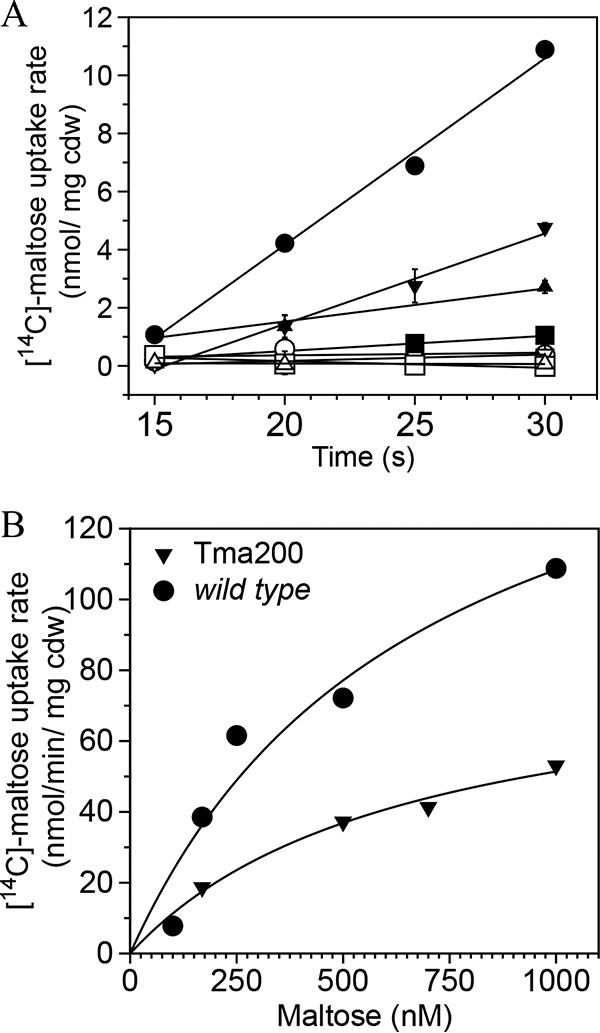
Kinetics of [14C]maltose uptake. (A) [14C]maltose uptake rate in the wild type (●), Tma100 (■), Tma200 (▼), and Tma201 (▲). Uptake was also estimated in the presence of a 100-fold excess unlabeled maltose in the wild type (○), Tma100 (□), Tma200 (▽), and Tma201 (△). (B) Kinetics of [14C]maltose uptake in the wild type and Tma200 with various concentrations (169 nM to 1,000 nM) of maltose. The data in panel B represent the averages from two independent trials and were fitted to the Michaelis-Menten equation. The error bar represents the SD from two biological replicates.
DISCUSSION
Transient inactivation of lactate dehydrogenase (encoded by ldh) and the resulting metabolic stress associated with a buildup of excess reductant enabled the isolation of new evolved T. maritima cell lines that exhibited increased H2 resistance and excess H2 production on a per-cell basis. This increase exceeded the physiologic limit for H2 production (Thauer limit) based on thermodynamic considerations (14). Since the Gibbs free energy of formation of maltose limits the amount of H2 that can be produced, exceeding this level necessitated changes in energy management. Reduced biomass formation and decreased growth rate both offer this option (17, 18, 20). Typically, glucose oxidation leading to the production of 4 mol of H2/mol of glucose represents 33% of the stoichiometric maximum. The amount of H2 that can be produced from a fermentative reaction is determined by the feasibility of the reaction that is predicted by the overall standard Gibbs free energy change of the reaction. The thermodynamic feasibility of this reaction dictates the amount of ATP that can be formed to sustain growth. Further, the thermodynamics of the process is affected by the temperature at which this reaction takes place as the Gibbs free energy change for the overall reaction from glucose to acetate at higher temperature becomes more negative, favoring the reaction. The 80°C growth temperature for Thermotoga maritima changes the ΔG°′ to −274 kJ/mol (45), which is more negative than the ΔG°′ (−216 kJ/mol) under standard conditions (25°C). Fermentative production of more than 4 mol of H2/mol of glucose would result in a decrease of the ΔG°′ because the reaction yielding 12 mol of H2/mol of glucose has a ΔG°′ of 3.2kJ/mol. The calculated ΔG°′ for Tma100 and Tma200 employing the proposed fermentative equations are −258.92 kJ/mol and −220.97 kJ/mol, respectively. Though both strains make more than 4 mol of H2/mol of glucose, the ΔG°′ remains within a range to favor the formation of 4 molecules of ATP (ΔG°′, 175.6 kJ) because −41.8 to −50.2 kJ is required for the synthesis of 1 mol of ATP from ADP and Pi in anaerobic bacteria (14). Therefore, for a hyperthermophile such as T. maritima, the formation of more than 4 mol of H2/mol of Glc is thermodynamically favorable under normal cultivation conditions under low partial pressure. However, fermentative organisms such as wild-type T. maritima preferentially synthesize biomass rather than H2 (7). Metabolic alterations resulting from genetic selection could produce strains that yield more H2 than the normal amount. In the evolved strains described here, a slower uptake of maltose allowed the organism to regenerate additional metabolic intermediates in the PPP that increased the pool of reducing equivalents under conditions where the overall metabolic pathways remained thermodynamically feasible. Normally, in T. maritima the EMP and ED pathways are used to oxidize glucose (5). We propose that in the evolved strains, the PPP is used to generate additional reductant leading to greater levels of H2. This is supported by studies performed by Latif et al. (46) on T. maritima in which in silico metabolic analysis of the wild type and evolved strains was performed. The wild type did not utilize the oxidative branch of the PPP, whereas in both evolved strains the carbon flux was directed to the PPP but not through the EMP pathway (Fig. 4). As per the published literature, glucose is metabolized to glyceraldehyde 3-phosphate (G3P) and fructose-6-phosphate (F6P). G3P is subsequently converted to pyruvate and acetate, and F6P is used for further processing of new glucose molecule. In theory, 8 mol of H2 per glucose will be produced when the PPP is involved (31, 43). Since the PPP generates NAD(P)H, a relevant hydrogenase such as Nfn (47, 48) could yield additional reductants for H2 synthesis and exceed the 4 mol of H2/mol of Glc. Nfn has been proposed to catalyze this reaction (49):
The NfnAB genes are present in T. maritima and have been experimentally validated (H. Huang and R. K. Thauer, unpublished data; 49). This means that PPP intermediates combined with NfnAB could increase the amount of reducing equivalents available for H2 synthesis. More than a 33% stoichiometric conversion of glucose to H2 is therefore likely to increase the amount of CO2 produced, as was observed in these experiments. Increased CO2 production in Tma100 and Tma200 reflected more substrate oxidation that shifted energy management and contributed to increased H2 production. The reduced rate of maltose transport could be sufficient to constrain biomass formation and growth rate. In the current study, wild-type cells transported maltose so rapidly that the maximum levels of H2 synthesis were not achieved, consistent with prior studies in which biological H2 production decreased with excess carbon loading (10, 50). In this study, a direct effect of reduced maltose uptake on lactic acid synthesis was also observed. In Tma100 no lactate was synthesized, and in Tma200 lactic acid synthesis was decreased to 71% of that observed in the wild type. This was also consistent with relative transcript abundance of ldh (Fig. S8). The absence of alternative routes for the recycling of NADH could raise the level of NADH, thereby leading to increased H2 production such as has been reported for an ldh knockout mutant of another lineage (51). Thus, reduced maltose uptake apparently suppressed lactic acid synthesis, while supporting increased H2 formation perhaps shifted away from its use for lactate formation. In both evolved strains, the transcriptional level of ldh was significantly lower than that in the wild type, and perhaps because of the reduced rate of maltose uptake. In the wild type, the maltose uptake rate is rapid and a metabolic response triggers upregulation of the ldh transcript. In the evolved strains, the rate of maltose uptake is lower and this leads to downregulation of ldh transcript abundance.
T. maritima possesses three malK paralogs, all annotated as ABC transporter subunits that catalyze ATP hydrolysis to provide energy necessary to drive maltose import (23, 26). In Tma100 and Tma200, only one of these paralogs, malK3, carried by THMA_1301, underwent mutation, but repeatedly, indicating that it plays a key role in maltose transport. A complete loss of maltose uptake function in the malK3 mutant of T. maritima also corroborates the idea that malK3 is the only functional malK gene of T. maritima (39). Mutation of the E. coli MalK ATP binding domain has been shown to reduce maltose uptake due to reduced ATP hydrolysis (52). A similar observation was made in this study: malK3 suffered mutations in both an ATP-binding domain (G148D) and at its 3′ end (D345K) in Tma100. These mutations were accompanied by a maltose-specific small-colony phenotype of Tma100 (Fig. S7) and a high level of residual maltose during its cultivation in bioreactors (Fig. 3A). Secondary mutations evident in Tma200 also occurred in malK3, including V233S (or V233F, in isolates Tma201 to Tma209). All these secondary mutations were accompanied by reversion of primary mutations G148D and D345K and a gain-of-function phenotype for maltose utilization and colony size, likely in response to the strong purifying selection for maltose utilization. This interpretation was supported by maltose uptake assays indicating that rates of maltose uptake were greatly reduced in the presence of G148D and D345K and rates improved upon their reversion. While V233S correlated with increased H2 production by Tma200, the alternate mutation, V233F, evident in Tma201 to Tma209, did not correspond with H2 overproduction as well as in Tma200. In the current study, a higher Vmax for maltose uptake, 182 nmol/min/mg(cdw), was observed, which is 10- to 20-fold greater than reported for other hyperthermophilic organisms (53, 54). This is consistent with reports that H2 accumulates to unusually high levels in batch culture of T. maritima (10, 29). In contrast, the Km for maltose uptake of 680 nM was significantly less than the KD (equilibrium dissociation constant) (in vitro) values for MalE1 and MalE2 (24 and 8 μM, respectively), perhaps indicating a role for subunit interactions within the entire ABC transporter complex (26). The lack of growth and H2 production in the malK3 mutant established the link between H2 production and the malK3 gene (Fig. S6). Association of the increased H2 production trait with the mutated malK3 allele was confirmed when the mutated malK3 allele of Tma200 increased H2 production in the strain possessing malK3 of Tma200 (Fig. 5). These data confirm that restricted entry of maltose through the defective maltose transporter of T. maritima alters the energy metabolism, leading to excess H2 synthesis rather than biomass formation and thus uncouples biomass formation from H2 formation.
Transient inactivation of ldh enhanced the toxicity of H2 formation arising from maltose fermentation and allowed the recovery of new cell lines that overproduced H2 and acetate on a per-cell basis. Since ldh was reconstituted in the derivative strains, the mechanism causing continued reduction in lactate production remains to be determined. It could be related to a change in the maintenance energy coefficient and or the redirection of reducing equivalents due a change in the rate of maltose catabolism (55, 56). Taken together, the data presented here indicate that predicted limitations governing metabolic processes can be overcome. Strategies such as transient gene inactivation may facilitate efforts to establish biorenewable sources of chemicals using microbial systems.
MATERIALS AND METHODS
Bacterial strains and cultivation.
Thermotoga maritima MSB8 was purchased from the American Type Culture Collection and cultured routinely in a complex medium (CM) (57). CM contained 0.26 M NaCl, 0.05% (wt/vol) tryptone, 0.01% (wt/vol) yeast extract, 14 mM Na2SO4, 9.8 mM MgCl2·6H2O, 3.0 mM NaHCO3, 0.17 mM KBr, 0.12 mM KI, 0.32 mM H3BO3, 9 μM Na2WO4, and 8.4 μM NiCl2 and was adjusted to pH 7.0 before autoclaving using KH2PO4. Autoclaved liquid CM was aliquoted into sterile Hungate tubes or serum bottles and amended by addition of sterile Na2S (42 μM), KH2PO4 (3.4 μM), and sugar (15 mM). T. maritima strains (Table 2) were cultivated in batch culture in biological replicates using Hungate tubes or serum bottles containing 10 ml or 50 ml of CM, respectively, and supplemented with 0.5% (15 mM) maltose unless otherwise indicated. Tubes were sealed with butyl rubber stoppers (Bellco Biotechnology) and crimped with metal collars, and the headspace was exchanged with N2. For direct growth inhibition and testing of H2 resistance by pure H2, tubes were sealed and injected with various volumes of H2 after removing the corresponding volumes (headspace N2) via a syringe to accommodate H2. Growth was monitored spectrophotometrically by the culture absorbance at a wavelength of 600 nm. Sterile 1-ml syringes attached to 20 1/2-gauge needles were used for inoculation at an initial cell density (OD600) of 0.03. All tubes were incubated anaerobically at 80°C overnight unless otherwise specified before chemical analysis.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description or genotype | Source |
|---|---|---|
| T. maritima | Thermotoga maritima MSB8 (wild type) | ATCCa |
| Tma100 | T. maritima; TM0460 (corresponds to THMB_0470) (W229UGA), TM1276 (corresponds to THMB_1301 of Tma100) (G148D and D345L), 16S rRNA (nucleotide A to G at position 1420) | This work |
| Tma200 | T. maritima; TM0459 (corresponds to THMC_0469 of Tma200) (A1045F), TM0460 (corresponds to THMC_0470 of Tma200) (W229UGA), TM1276 (corresponds to THMC_1301 of Tma200) (V233S) and Δ3′TM1322 (corresponds to THMC_1346 of Tma200) to Δ5′TM1332 (corresponds to THMC_1356 of Tma200), 16S rRNA (nucleotide A to G at position 1420) | This work |
| Tma201–Tma209 | T. maritima; TM0459 (A1045F), TM0460 (W229UGA), TM1276 (corresponds to THMA_1301 of wild type) (V233F) and Δ3′TM1322 (corresponds to THMA_1346 of wild type) to Δ5′TM1332 (corresponds to THMA_1356 of wild type), 16S rRNA (nucleotide A to G at position 1420) | This work |
| pBL1292 | pUC57; T. maritima ldhΔ3′::PgroESLp::htk | This work |
ATCC number 43589.
For volumetric H2 productivity measurements, strains were cultivated in 3-liter double-jacketed glass bioreactors (Applicon, MA) with a 1.5-liter working volume with continuous stirring at 200 rpm using dual axial impellers. Bioreactors were equipped with sensors monitoring temperature, pH, and dissolved oxygen while an anaerobic environment was maintained by continuous supply of N2 at 15 ml/min. The pH was maintained at pH 7 by addition of acid (1 M H2SO4)/base (1 M NaOH) via a peristaltic pump. To minimize water loss, water vapor in the outlet headspace gas was condensed using a chilled water supply and returned to the vessel. A solid medium was prepared by combining 0.6% (wt/vol) Gelrite (Research Product Corporation, IL) solubilized by boiling with previously sterilized complex medium components followed by addition of reductant (Na2S), base (KH2PO4), and carbon sources. For preparation of drug plates, kanamycin was added to the medium prior to pouring. Inoculated plates were incubated at 80°C for 2 to 3 days in jars (Almore) under anaerobic conditions using GasPaks (EZ BD). Long-term preservation of cultures was as described previously for other hyperthermophiles (58).
Strain construction.
The chromosomal ldh gene was inactivated by targeted recombination via a single-crossover event using a 3′ terminally truncated promoterless segment of ldh. This segment was cloned into pUC57 (using primers P1 and P2 [Table 3]) along with the kanamycin nucleotidyltransferase gene (htk) under the control of the T. maritima PgroESL heat shock promoter (using primers P3 and P4) (24, 42). The first codon of htk was fused to the 3′ end of PgroESL at nt 532232 (24). To replace the disrupted malK3 gene of the malK3 mutant with the malK3 gene of Tma200, a suicide vector (pUC19) was developed by cloning the malK3 gene of Tma200 at the KpnI/SphI site.
TABLE 3.
Primers used in this study
| Primer | Sequence | Restriction site |
|---|---|---|
| P1 | 5′ ATGAAAATAGGTATCGTAGGACTCG 3′ | EcoRI |
| P2 | 5′ CTTGGAGAAAAGCCGCAGT 3′ | EcoRV |
| P3 | 5′ GCTTCAAGCGCCTTTTTATTT 3′ | BamHI |
| P4 | 5′ TCAAAATGGTATTCTCTTGCTAACG 3′ | BamHI |
| P5 | 5′ TCGGGCAAGATCCCCCATGGA 3′ | NAa |
| P6 | 5′ ATATGCGGTGTGAAATACCGCA 3′ | NA |
| P7 | 5′ ATAGTGCCCCTTCTCATATC 3′ | NA |
| P8 | 5′ GGCTAAACTAATTGAAAGTGACAGA 3′ | NA |
| P9 | 5′ TCGTATGAGAACTCAACACCTTCAGT 3′ | NA |
| P10 | 5′ GGGCGACACGGAAATGTT 3′ | NA |
| P11 | 5′ ATAATACCGCGCCACATAGC 3′ | NA |
| P12 | 5′ CCCTTTTTTGCGGCATTT 3′ | NA |
NA, not applicable.
Ligation, transformation, and selection of E. coli DH5α transformants were performed as described previously (59). The resulting ldh gene disruption construct (pBL1292 [Table 2]) was transformed into wild-type T. maritima spheroplasts prepared as described previously (38), with the following modifications. Proteinase K (10 mg/ml) was used in combination with lysozyme (300 μg/ml) and the efficiency of spheroplast formation was monitored by light microscopy. Spheroplasts (7.5 × 107) were electroporated (1.8 kV, 200 Ω, and 25 μF) with a maximum of 1 μg of plasmid DNA using a Genpulser (Bio-Rad) in chilled 1-mm cuvettes. Electroporated spheroplasts were inoculated into complex liquid medium and incubated anaerobically at 80°C for 18 h. For initial screening, drug selection was imposed on resuspended cells (initial OD600 = 0.02) using 375 μg/ml of kanamycin and incubated until turbidity was observed. Genomic DNA was recovered from the enriched culture and genotyped to verify recombination at ldh. A transformation efficiency of 44 recombinants/μg of DNA was observed on plates containing 500 μg/ml of kanamycin. Transformation efficiency was normalized to plating efficiency without added drug. Colonies were purified on 0.5% (wt/vol) maltose plates containing 500 μg/ml of kanamycin by spot dilution. PCR screening of 5 purified colonies for PgroESL::htk showed the presence of PgroESL::htk in 3 of the purified colonies. One of the purified colonies (Tma100) was selected for further studies.
Genomic DNA from strain Tma100 was prepared as described for other hyperthermophiles and genotyped by PCR and DNA sequencing to confirm the presence of PgroESL::htk, vector sequences, and 5′ and 3′ flanking ldh sequences (58).
For the development of a strain possessing the malK3 gene of Tma200, the malK3 mutant was used as a recipient strain because a colony phenotype can be seen after the repair of the disrupted malK3 (39). Selection of this strain was based on its ability to form bigger colonies than those of the malK3-possessing strain on complex medium plates upon the repair of the disrupted malK3 allele. As shown in Fig. S9, the site of homologous recombination is crucial and can produce either the wild-type malK3 allele or malK3 of Tma200 in the resulting strain. In order to distinguish these strains, the PCR products amplified by outside primers were digested with AciI. The malK3 gene of the Tma200 allele would produce four segments (9 bp, 21 bp, 614 bp, and 836 bp), whereas the wild-type allele would produce three segments (9 bp, 21 bp, and 1,450 bp) on a gel. Based on the restriction digestion pattern, all five isolates were found to possess the malK3 allele of Tma200 (Fig. S9). This was further verified by sequencing the entire malK3 gene of the resultant strain.
Cell line passage and phenotypic analysis.
Passage of Tma100 on CM maltose plates was used to isolate more robust derivatives without concurrent selection for kanamycin resistance. Fifty colonies of Tma100 (from plates with maltose plus kanamycin) were patched on CM maltose (0.1%, wt/vol) plates lacking kanamycin and incubated at 80°C anaerobically. While all 50 isolates grew, 10 were then grown in CM tubes supplemented with maltose (0.5%, wt/vol) without kanamycin and then screened by PCR for the PgroESL::htk transgene and were named Tma200 to Tma209. Colony phenotypes of selected strains (Tma100 and Tma200) were examined on plates with either maltose or cellobiose both at 0.1% (wt/vol) after anaerobic incubation at 80°C for 48 h.
Analytical methods.
Analysis of headspace gas composition was performed by withdrawing 500-μl volumes using a gastight syringe (Hamilton) and injected into a gas chromatograph (GC) (GC 400 series; GOWMAC, PA) fitted with a thermal conductivity detector. A molecular sieve column (GOWMAC), operated at 70°C with a continuous flow of N2 carrier gas, was used to separate H2. For headspace CO2 analysis, a Varian (430) GC equipped with an Alltech Porapak C-5000 column was used. Helium was used as the carrier gas, and column temperature was maintained at 65°C to separate CO2. Calibration curves were obtained by injecting various volumes of pure H2 and CO2, and the amount of H2 and CO2 in the headspace was estimated by comparison to these values. The molar yield of H2 was calculated using the ideal gas law equation at standard temperature and pressure. Since growth varied among different cell lines in small batch cultures, H2 values were normalized to 108 cells/ml. Similarly, in bioreactor studies, the quantity of H2 produced during exponential growth (5-h duration) by Tma100 and Tma200 was normalized to the biomass (milligrams, cdw) produced by the wild-type strain under analogous growth conditions. Rates of H2 production/h in bioreactor studies was calculated for each growth phase and was normalized to 1 g (cdw) for each strain. For biomass measurements, different amounts of washed cell pellets were dried and used to derive the conversion factor between optical density (OD600 of 1.0) and dry weight of 0.2 mg. Potential variation in cell size between respective strains was excluded after examination by light and electron microscopy. Organic acid and maltose concentrations were determined in culture supernatants by high-performance liquid chromatography (HPLC) with comparison to standards. Prior to injection, samples were clarified at 10,000 × g for 10 min and then filtered (AcroDisc, 0.45 μm). Samples (1 μl) were analyzed using an Agilent 1200 HPLC system and an automated sampler equipped with a refractive index detector and a Hi-Plex H column (ChromTech) operated at 65°C. Isocratic separations used 4 mM sulfuric acid at a rate of 0.4 ml per minute per the manufacturer's recommendation. The regression equation was used to calculate aqueous metabolite concentrations. Yield coefficients were expressed as the molar ratio of metabolites produced/maltose consumed. Carbon recovery was calculated as the ratio of maltose consumed to products (acetate, lactate, biomass, and CO2) formed. The micromoles of biomass were calculated based on the empirical formula C5H8O3NS0.05 for T. maritima (60). The proposed equations for Tma100 (equation 1) and Tma200 (equation 2) were used in the Aspen Plus package (v. 10.1) to estimate the Gibbs free energy from ΔG = ΔH − TΔS (G is Gibbs free energy, H is enthalpy, T is temperature, and S is entropy) at 80°C and 1 atm. Nonideality was accounted and temperature-dependent interaction parameters were used by the hybrid thermodynamic model of NRTL-Redlich-Kwong (NRTL-RK).
Uptake assays.
Maltose uptake assays were carried out as described for other anaerobes, with the following modifications (54, 61). Cells were cultivated in 50 ml of CM supplemented with 0.5% (wt/vol) maltose and harvested at early mid-log phase (2.5 × 107 cells/ml), followed by washing using CM twice. Washed cells were maintained under anaerobic conditions prior to uptake assays. Cell suspensions (1.12 × 109 cells/ml) were placed in anaerobic tubes at 75°C and allowed to equilibrate for 35 min. Assays were initiated by addition of 0.169 μM [14C]maltose (American Radiolabeled Chemicals, St. Louis, MO) with a specific activity of 600 mCi/mmol. To determine uptake rates at higher substrate concentrations, unlabeled maltose ranging from 0.25 μM to 1.0 μM was mixed with [14C]maltose (keeping the initial concentration of [14C]maltose constant). [14C]maltose uptake reactions were terminated by filtering 200-μl volumes of cells through a 0.45-μm polycarbonate membrane (Whatman Nuclepore track-etch membrane) with a fabricated and disposable polypropylene filtration apparatus. Cells retained on the filters were washed with 3 ml of CM. Backgrounds were determined using a no-cell sample processed in an otherwise identical fashion. Dried filters were placed in vials prefilled with 5 ml of scintillant (EcoLite) and radioactivity was determined using a scintillation spectrometer (Beckman LSC 6500). For the [14C]maltose uptake calculations, all uptake values were subtracted from the control reaction that was terminated immediately after the addition of [14C]maltose. [14C]maltose uptake rates in all isolates were determined from the slope of the linear regression of total [14C]maltose taken up as a function of time. No sampling was done between 0 and 15 s, as maltose uptake was faster immediately after this time. Km and Vmax values were determined by nonlinear regression fitting to the Michaelis-Menten equation. All rates were verified using biological replicates. The concentration of [14C]maltose stocks was measured experimentally rather than using a theoretical/nominal concentration.
Genome resequencing.
Genome resequencing was performed by DOE-JGI under a Community Sequencing Program (CSP) JGI project ID1011924. DNA was sequenced using a HiSeq Illumina instrument, and the guided sequence assembly was performed to close the genome. IGV (v. 2.3) was used to detect indels and single nucleotide polymorphisms (SNPs) by comparing the genome of laboratory stock of the wild type, Tma100, and Tma200 to the NCBI database wild type (T. maritima MSB8), (accession no. NC_021214.1) (62). Additionally, a laboratory stock of the wild type, Tma100, and Tma200 were compared to three available genomes (accession no. NC_000853.1, NC_023151.1, and NC_021214.1) of T. maritima (Table S4). A laboratory stock of the wild-type strain was used to filter out the common mutations in Tma100 and Tma200. Unique mutations inherited only by Tma100 and Tma200 are shown in Table S3 and Fig. S3. Annotation of the laboratory stock of the wild type, Tma100, and Tma200 was carried out using the NCBI pipeline. Annotated genomes of the wild type, Tma100, and Tma200 are available in GenBank under accession no. CP011107, CP011108, and CP010967, respectively (63). All mutations in Tma100 and Tma200 were verified by PCR and DNA resequencing.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by funds from the U.S. Department of Energy (DE-PS02-08ER08-12) to R.K., K.N., and P.B. and additional funds from DOE-JGI (CSP 1011924) and the UNL Cell Development Facility to P.B.
We thank Karrie Weber and Olivia Healy for providing us the access to instruments for analyzing CO2. We thank Nicole Buan for comments on the manuscript. We thank Christopher Straub, North Carolina University, for providing us with detailed information on the carbon flux redirected through the pentose phosphate pathway and resultant H2 yield.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00998-18.
REFERENCES
- 1.Lee H-S, Vermaas WFJ, Rittmann BE. 2010. Biological hydrogen production: prospects and challenges. Trends Biotechnol 28:262–271. doi: 10.1016/j.tibtech.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Balat H, Kırtay E. 2010. Hydrogen from biomass—present scenario and future prospects. Int J Hydrogen Energy 35:7416–7426. doi: 10.1016/j.ijhydene.2010.04.137. [DOI] [Google Scholar]
- 3.Chen X, Sun Y, Xiu Z, Li X, Zhang D. 2006. Stoichiometric analysis of biological hydrogen production by fermentative bacteria. Int J Hydrogen Energy 31:539–549. doi: 10.1016/j.ijhydene.2005.03.013. [DOI] [Google Scholar]
- 4.Nandi R, Sengupta S. 1998. Microbial production of hydrogen: an overview. Crit Rev Microbiol 24:61–84. doi: 10.1080/10408419891294181. [DOI] [PubMed] [Google Scholar]
- 5.Selig M, Xavier KB, Santos H, Schonheit P. 1997. Comparative analysis of Embden-Meyerhof and Entner-Doudoroff glycolytic pathways in hyperthermophilic archaea and the bacterium Thermotoga. Arch Microbiol 167:217–232. doi: 10.1007/BF03356097. [DOI] [PubMed] [Google Scholar]
- 6.Schröder C, Selig M, Schönheit P. 1994. Glucose fermentation to acetate, CO2 and H2 in the anaerobic hyperthermophilic eubacterium Thermotoga maritima: involvement of the Embden-Meyerhof pathway. Arch Microbiol 161:460–470. [Google Scholar]
- 7.Hallenbeck PC. 2005. Fundamentals of the fermentative production of hydrogen. Water Sci Technol 52:21–29. [PubMed] [Google Scholar]
- 8.Stams AJ. 1994. Metabolic interactions between anaerobic bacteria in methanogenic environments. Antonie Van Leeuwenhoek 66:271–294. doi: 10.1007/BF00871644. [DOI] [PubMed] [Google Scholar]
- 9.Yokoyama H, Waki M, Moriya N, Yasuda T, Tanaka Y, Haga K. 2007. Effect of fermentation temperature on hydrogen production from cow waste slurry by using anaerobic microflora within the slurry. Appl Microbiol Biotechnol 74:474–483. doi: 10.1007/s00253-006-0647-4. [DOI] [PubMed] [Google Scholar]
- 10.Huber R, Langworthy T, König H, Thomm M, Woese C, Sleytr U, Stetter K. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch Microbiol 144:324–333. [Google Scholar]
- 11.Nath K, Das D. 2004. Improvement of fermentative hydrogen production: various approaches. Appl Microbiol Biotechnol 65:520–529. doi: 10.1007/s00253-004-1644-0. [DOI] [PubMed] [Google Scholar]
- 12.Woodward J, Orr M, Cordray K, Greenbaum E. 2000. Enzymatic production of biohydrogen. Nature 405:1014–1015. doi: 10.1038/35016633. [DOI] [PubMed] [Google Scholar]
- 13.Zhang YH, Evans BR, Mielenz JR, Hopkins RC, Adams MW. 2007. High-yield hydrogen production from starch and water by a synthetic enzymatic pathway. PLoS One 2:e456. doi: 10.1371/journal.pone.0000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thauer RK, Jungermann K, Decker K. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41:100–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karel SF, Libicki SB, Robertson CR. 1985. The immobilization of whole cells: engineering principles. Chem Eng Sci 40:1321–1354. doi: 10.1016/0009-2509(85)80074-9. [DOI] [Google Scholar]
- 16.Hatch RT, Hardy R. 1989. Microorganisms as producers of feedstock chemicals, p 28–41. In Marx JL. (ed), A revolution in biotechnology. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 17.Archer DB. 1985. Uncoupling of methanogenesis from growth of Methanosarcina barkeri by phosphate limitation. Appl Environ Microbiol 50:1233–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blum PH, Jovanovich SB, McCann MP, Schultz JE, Lesley SA, Burgess RR, Matin A. 1990. Cloning and in vivo and in vitro regulation of cyclic AMP-dependent carbon starvation genes from Escherichia coli. J Bacteriol 172:3813–3820. doi: 10.1128/jb.172.7.3813-3820.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tunner JR, Robertson CR, Schippa S, Matin A. 1992. Use of glucose starvation to limit growth and induce protein production in Escherichia coli. Biotechnol Bioeng 40:271–279. [DOI] [PubMed] [Google Scholar]
- 20.Keasling JD, Benemann J, Pramanik J, Carrier TA, Jones KL, Van Dien SJ. 1998. A toolkit for metabolic engineering of bacteria, p 87–97. In Zaborsky OR. (ed), BioHydrogen. Springer, New York, NY. [Google Scholar]
- 21.Schink B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61:262–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chhabra SR, Shockley KR, Conners SB, Scott KL, Wolfinger RD, Kelly RM. 2003. Carbohydrate-induced differential gene expression patterns in the hyperthermophilic bacterium Thermotoga maritima. J Biol Chem 278:7540–7552. doi: 10.1074/jbc.M211748200. [DOI] [PubMed] [Google Scholar]
- 23.Conners SB, Montero CI, Comfort DA, Shockley KR, Johnson MR, Chhabra SR, Kelly RM. 2005. An expression-driven approach to the prediction of carbohydrate transport and utilization regulons in the hyperthermophilic bacterium Thermotoga maritima. J Bacteriol 187:7267–7282. doi: 10.1128/JB.187.21.7267-7282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, McDonald L, Utterback TR, Malek JA, Linher KD, Garrett MM, Stewart AM, Cotton MD, Pratt MS, Phillips CA, Richardson D, Heidelberg J, Sutton GG, Fleischmann RD, Eisen JA, White O, Salzberg SL, Smith HO, Venter JC, Fraser CM. 1999. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 25.Vanfossen AL, Verhaart MR, Kengen SM, Kelly RM. 2009. Carbohydrate utilization patterns for the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus reveal broad growth substrate preferences. Appl Environ Microbiol 75:7718–7724. doi: 10.1128/AEM.01959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nanavati DM, Thirangoon K, Noll KM. 2006. Several archaeal homologs of putative oligopeptide-binding proteins encoded by Thermotoga maritima bind sugars. Appl Environ Microbiol 72:1336–1345. doi: 10.1128/AEM.72.2.1336-1345.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schut GJ, Adams MW. 2009. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J Bacteriol 191:4451–4457. doi: 10.1128/JB.01582-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willquist K, Pawar SS, Van Niel EW. 2011. Reassessment of hydrogen tolerance in Caldicellulosiruptor saccharolyticus. Microb Cell Fact 10:111. doi: 10.1186/1475-2859-10-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Niel EW, Claassen PA, Stams AJ. 2003. Substrate and product inhibition of hydrogen production by the extreme thermophile, Caldicellulosiruptor saccharolyticus. Biotechnol Bioeng 81:255–262. doi: 10.1002/bit.10463. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Thiele I, Weekes D, Li Z, Jaroszewski L, Ginalski K, Deacon AM, Wooley J, Lesley SA, Wilson IA, Palsson B, Osterman A, Godzik A. 2009. Three-dimensional structural view of the central metabolic network of Thermotoga maritima. Science 325:1544–1549. doi: 10.1126/science.1174671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nogales J, Gudmundsson S, Thiele I. 2012. An in silico re-design of the metabolism in Thermotoga maritima for increased biohydrogen production. Int J Hydrogen Energy 37:12205–12218. doi: 10.1016/j.ijhydene.2012.06.032. [DOI] [Google Scholar]
- 32.Eriksen N, Riis M, Holm N, Iversen N. 2011. H2 synthesis from pentoses and biomass in Thermotoga spp. Biotechnol Lett 33:293–300. doi: 10.1007/s10529-010-0439-x. [DOI] [PubMed] [Google Scholar]
- 33.Lakhal R, Auria R, Davidson S, Ollivier B, Dolla A, Hamdi M, Combet-Blanc Y. 2010. Effect of oxygen and redox potential on glucose fermentation in Thermotoga maritima under controlled physicochemical conditions. Int J Microbiol 2010:896510. doi: 10.1155/2010/896510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiDonato M, Deacon AM, Klock HE, McMullan D, Lesley SA. 2004. A scaleable and integrated crystallization pipeline applied to mining the Thermotoga maritima proteome. J Struct Funct Genomics 5:133–146. doi: 10.1023/B:JSFG.0000029194.04443.50. [DOI] [PubMed] [Google Scholar]
- 35.Malik B, Su WW, Wald HL, Blumentals II, Kelly RM. 1989. Growth and gas production for hyperthermophilic archaebacterium, Pyrococcus furiosus. Biotechnol Bioeng 34:1050–1057. doi: 10.1002/bit.260340805. [DOI] [PubMed] [Google Scholar]
- 36.Boileau C, Auria R, Davidson S, Casalot L, Christen P, Liebgott P-P, Combet-Blanc Y. 2016. Hydrogen production by the hyperthermophilic bacterium Thermotoga maritima part I: effects of sulfured nutriments, with thiosulfate as model, on hydrogen production and growth. Biotechnol Biofuels 9:269. doi: 10.1186/s13068-016-0678-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janssen PH, Morgan HW. 1992. Heterotrophic sulfur reduction by Thermotoga sp. strain FjSS3.B1. FEMS Microbiol Lett 96:213–217. doi: 10.1111/j.1574-6968.1992.tb05419.x. [DOI] [PubMed] [Google Scholar]
- 38.Yu JS, Vargas M, Mityas C, Noll KM. 2001. Liposome-mediated DNA uptake and transient expression in Thermotoga. Extremophiles 5:53–60. doi: 10.1007/s007920000173. [DOI] [PubMed] [Google Scholar]
- 39.Singh R, White D, Blum P. 2017. Identification of the ATPase subunit of the primary maltose transporter in the hyperthermophilic anaerobe Thermotoga maritima. Appl Environ Microbiol 83:e00930-17. doi: 10.1128/AEM.00930-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White D, Singh R, Rudrappa D, Mateo J, Kramer L, Freese L, Blum P. 2017. Contribution of pentose catabolism to molecular hydrogen formation by targeted disruption of arabinose isomerase (araA) in the hyperthermophilic bacterium Thermotoga maritima. Appl Environ Microbiol 83:e02631-16. doi: 10.1128/AEM.02631-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen A, Hillman JD, Duncan M. 1994. L-(+)-lactate dehydrogenase deficiency is lethal in Streptococcus mutans. J Bacteriol 176:1542–1545. doi: 10.1128/jb.176.5.1542-1545.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoseki J, Yano T, Koyama Y, Kuramitsu S, Kagamiyama H. 1999. Directed evolution of thermostable kanamycin-resistance gene: a convenient selection marker for Thermus thermophilus. J Biochem 126:951–956. doi: 10.1093/oxfordjournals.jbchem.a022539. [DOI] [PubMed] [Google Scholar]
- 43.de Vrije T, Mars AE, Budde MA, Lai MH, Dijkema C, de Waard P, Claassen PA. 2007. Glycolytic pathway and hydrogen yield studies of the extreme thermophile Caldicellulosiruptor saccharolyticus. Appl Microbiol Biotechnol 74:1358–1367. doi: 10.1007/s00253-006-0783-x. [DOI] [PubMed] [Google Scholar]
- 44.Sahm K, Matuschek M, Muller H, Mitchell WJ, Bahl H. 1996. Molecular analysis of the amy gene locus of Thermoanaerobacterium thermosulfurigenes EM1 encoding starch-degrading enzymes and a binding protein-dependent maltose transport system. J Bacteriol 178:1039–1046. doi: 10.1128/jb.178.4.1039-1046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buckel W, Thauer RK. 2018. Flavin-based electron bifurcation, a new mechanism of biological energy coupling. Chem Rev doi: 10.1021/acs.chemrev.7b00707. [DOI] [PubMed] [Google Scholar]
- 46.Latif H, Sahin M, Tarasova J, Tarasova Y, Portnoy VA, Nogales J, Zengler K. 2015. Adaptive evolution of Thermotoga maritima reveals plasticity of the ABC transporter network. Appl Environ Microbiol 81:5477–5485. doi: 10.1128/AEM.01365-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith ET, Tomich JM, Iwamoto T, Richards JH, Mao Y, Feinberg BA. 1991. A totally synthetic histidine-2 ferredoxin: thermal stability and redox properties. Biochemistry 30:11669–11676. doi: 10.1021/bi00114a009. [DOI] [PubMed] [Google Scholar]
- 48.Brandt U. 1996. Bifurcated ubihydroquinone oxidation in the cytochrome bc1 complex by proton-gated charge transfer. FEBS Lett 387:1–6. doi: 10.1016/0014-5793(96)00436-X. [DOI] [PubMed] [Google Scholar]
- 49.Buckel W, Thauer RK. 2013. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na(+) translocating ferredoxin oxidation. Biochim Biophys Acta 1827:94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Van Ginkel SW, Logan B. 2005. Increased biological hydrogen production with reduced organic loading. Water Res 39:3819–3826. doi: 10.1016/j.watres.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 51.Liu S, Nichols N, Dien B, Cotta M. 2006. Metabolic engineering of a Lactobacillus plantarum double ldh knockout strain for enhanced ethanol production. J Ind Microbiol Biotechnol 33:1–7. doi: 10.1007/s10295-005-0001-3. [DOI] [PubMed] [Google Scholar]
- 52.Davidson AL, Sharma S. 1997. Mutation of a single MalK subunit severely impairs maltose transport activity in Escherichia coli. J Bacteriol 179:5458–5464. doi: 10.1128/jb.179.17.5458-5464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silva Z, Sampaio MM, Henne A, Bohm A, Gutzat R, Boos W, da Costa MS, Santos H. 2005. The high-affinity maltose/trehalose ABC transporter in the extremely thermophilic bacterium Thermus thermophilus HB27 also recognizes sucrose and palatinose. J Bacteriol 187:1210–1218. doi: 10.1128/JB.187.4.1210-1218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xavier KB, Martins LO, Peist R, Kossmann M, Boos W, Santos H. 1996. High-affinity maltose/trehalose transport system in the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol 178:4773–4777. doi: 10.1128/jb.178.16.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teixeira de Mattos MJ, Streekstra H, Tempest DW. 1984. Metabolic uncoupling of substrate level phosphorylation in anaerobic glucose-limited chemostat cultures of Klebsiella aerogenes NCTC418. Arch Microbiol 139:260–264. doi: 10.1007/BF00402011. [DOI] [Google Scholar]
- 56.Teixeira de Mattos MJ, Tempest DW. 1983. Metabolic and energetic aspects of the growth of Klebsiella aerogenes NCTC 418 on glucose in anaerobic chemostat culture. Arch Microbiol 134:80–85. doi: 10.1007/BF00429412. [DOI] [PubMed] [Google Scholar]
- 57.Chhabra SR, Shockley KR, Ward DE, Kelly RM. 2002. Regulation of endo-acting glycosyl hydrolases in the hyperthermophilic bacterium Thermotoga maritima grown on glucan- and mannan-based polysaccharides. Appl Environ Microbiol 68:545–554. doi: 10.1128/AEM.68.2.545-554.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maezato Y, Dana K, Blum P. 2011. Engineering thermoacidophilic archaea using linear DNA recombination. Methods Mol Biol 765:435–445. doi: 10.1007/978-1-61779-197-0_26. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 60.Rinker KD, Kelly RM. 2000. Effect of carbon and nitrogen sources on growth dynamics and exopolysaccharide production for the hyperthermophilic archaeon Thermococcus litoralis and bacterium Thermotoga maritima. Biotechnol Bioeng 69:537–547. doi:. [DOI] [PubMed] [Google Scholar]
- 61.Koning SM, Konings WN, Driessen AJ. 2002. Biochemical evidence for the presence of two alpha-glucoside ABC-transport systems in the hyperthermophilic archaeon Pyrococcus furiosus. Archaea 1:19–25. doi: 10.1155/2002/529610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Latif H, Lerman JA, Portnoy VA, Tarasova Y, Nagarajan H, Schrimpe-Rutledge AC, Smith RD, Adkins JN, Lee DH, Qiu Y, Zengler K. 2013. The genome organization of Thermotoga maritima reflects its lifestyle. PLoS Genet 9:e1003485. doi: 10.1371/journal.pgen.1003485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh R, Gradnigo J, White D, Lipzen A, Martin J, Schackwitz W, Moriyama E, Blum P. 2015. Complete genome sequence of an evolved Thermotoga maritima isolate. Genome Announc 3:e00557-15. doi: 10.1128/genomeA.00557-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.