Summary
Rtt109 is a unique histone acetyltransferase acetylating histone H3 lysine 56 (H3K56), a modification critical for DNA replication-coupled nucleosome assembly and genome stability. In cells, histone chaperone Asf1 is essential for H3K56 acetylation, yet the mechanisms for H3K56 specificity and Asf1 requirement remain unknown. We have determined the crystal structure of the Rtt109-Asf1-H3-H4 complex, and found that unwinding of histone H3 αN, where K56 is normally located, and stabilization of the very C-terminal β-strand of histone H4 by Asf1 are prerequisites for H3K56 acetylation. Unexpectedly, an interaction between Rtt109 and the central helix of histone H3 is also required. The observed multiprotein, multisite substrate recognition mechanism among histone modification enzymes provides mechanistic understandings of Rtt109 and Asf1 in H3K56 acetylation, as well as valuable insights into substrate recognition by histone modification enzymes in general.
Keywords: Histone, acetylation, chaperone, crystal structure, DNA replication, nucleosome assembly, DNA damage
Graphical Abstract
A multiprotein complex of histone chaperone Asf1 and H3 and H4 as an intact substrate for a histone-modifying enzyme
Introduction
Genomic DNA in eukaryotic cells is packaged into a highly ordered chromatin structure. The basic repeating unit of chromatin is the nucleosome, which consists of ~146bp DNA wrapped around a histone octamer assembled from two copies of histones H2A, H2B, H3 and H4 each (Luger et al., 1997). As nucleosomes pose as barriers for cellular machineries carrying out gene transcription and DNA replication, they need to be first disassembled and then reassembled during these processes. This dynamic nucleosome disassembly/assembly is important for genome integrity, epigenetic inheritance and maintenance of cell fate commitment (Bellush and Whitehouse, 2017; MacAlpine and Almouzni, 2013; Serra-Cardona and Zhang, 2017; Teves et al., 2014).
Histone chaperones play important roles in nucleosome assembly during DNA replication and gene transcription (Gurard-Levin et al., 2014; Smith and Stillman, 1989; Verreault et al., 1996). For instance, Asf1, a histone H3-H4 chaperone conserved from yeast to humans, facilitates CAF-1-mediated DNA replication-coupled nucleosome assembly (Le et al., 1997; Mello et al., 2002; Tyler et al., 1999; Tyler et al., 2001). Structural studies revealed that Asf1 binds to the H3 surface at the interface of the H3-H4 heterotetramer and disrupts the formation of the H3-H4 tetramer (English et al., 2006; Natsume et al., 2007). During DNA replication-coupled nucleosome assembly, Asf1 functions by transferring H3-H4 heterodimers to the CAF-1 complex (Liu et al., 2012b; Serra-Cardona and Zhang, 2017). Asf1 is also involved in DNA replication-independent nucleosome assembly in both yeast and human cells. Yeast cells lacking Asf1 are highly sensitive to DNA damaging agents and exhibit increased levels of cryptic transcription (Le et al., 1997; Schwabish and Struhl, 2006; Tyler et al., 1999). Asf1a, one of the two human Asf1 isoforms, interacts with histone H3.3 chaperone HIRA and hands off the H3.3-H4 heterodimer to HIRA during replication-independent nucleosome assembly (Tagami et al., 2004). Asf1a is also required for maintenance of pluripotency of human embryonic stem (ES) cells (Gonzalez-Munoz et al., 2014). Moreover, Asf1b, another Asf1 isoform in human cells, is involved in proliferation and is overexpressed in breast cancer cells (Corpet et al., 2011). Thus, Asf1 functions in both DNA replication dependent and independent nucleosome assembly, and plays an important role in maintaining genome integrity and cell fate determination.
Post-translational modifications of histones, including acetylation and ubiquitination, play important roles in regulating nucleosome assembly. For instance, acetylation of lysine 56 of histone H3 (H3K56ac), a mark on newly synthesized H3 (Das et al., 2009; Masumoto et al., 2005; Xu et al., 2005), is crucial for nucleosome assembly following DNA replication and DNA repair (Chen et al., 2008; Li et al., 2008). In budding yeast, H3K56ac promotes H3 ubiquitination catalyzed by Rtt101Mms1, an ortholog of mammalian Cul4/DDB1 E3 ubiquitin ligase (Han et al., 2013). The ubiquitination of H3 weakens its interaction with Asf1 and enables efficient handoff of H3-H4 to CAF-1. Furthermore, H3K56ac increases the binding affinity of histone chaperones CAF-1 and Rtt106 with H3-H4 (Li et al., 2008; Su et al., 2012). Yeast cells lacking H3K56ac are highly sensitive to DNA damaging agents and exhibit a significant increase in spontaneous chromosome breaks (Han et al., 2007a; Masumoto et al., 2005; Ozdemir et al., 2005). H3K56ac also regulates histone exchange outside of S phase in budding yeast cells (Rufiange et al., 2007), and regulates expression homeostasis during S phase (Voichek et al., 2016). In human cells, while H3K56ac levels are considerably lower than in yeast cells, it is crucial for DNA damage response (Das et al., 2009), and is linked to the core transcription network involving key regulators of pluripotency in ES cells (Tan et al., 2013; Xie et al., 2009). Therefore, H3K56ac, a rare modification at the histone globular domain, extends its functions from chromatin integrity to regulation of gene expression.
In budding yeast, H3K56ac is catalyzed by a unique histone acetyltransferase (HAT), Rtt109 (Driscoll et al., 2007; Han et al., 2007a; Tsubota et al., 2007). In human cells, H3K56ac is catalyzed by p300/CBP (Das et al., 2009), which bears no sequence similarity with fungal Rtt109 but their catalytic domains share a similar overall three-dimensional fold (Lin and Yuan, 2008; Park et al., 2008; Stavropoulos et al., 2008; Tang et al., 2008). Besides H3K56, Rtt109 also acetylates H3K9 and H3K27 at the N-terminal tail of H3 (Burgess et al., 2010; Fillingham et al., 2008). A marked difference between these acetylation sites is that H3K9 and H3K27 are located in the unstructured N-terminal tail, whereas H3K56 is located in the N-terminal helix (αN) of H3. H3K56 interacts with DNA at the entry and exit points of the nucleosome, and also with histone chaperone DAXX (Elsasser et al., 2012; Liu et al., 2012a; Luger et al., 1997). Although numerous previous studies have revealed how HATs bind and acetylate lysine residues located at unstructured N-terminal tails of histones (Marmorstein and Zhou, 2014), it remains not understood how H3K56 located in a helical context may fit into the catalytic site of Rtt109.
S. cerevisiae Rtt109 forms a stable complex with Vps75, a Nap1-like chaperone, in the absence of bound histones. In yeast cells, Vps75 is important for acetylation of H3K9 and H3K27 along with Gcn5 but not required for H3K56ac (Burgess et al., 2010; Fillingham et al., 2008). In contrast, Asf1 is essential for H3K56ac (Driscoll et al., 2007; Han et al., 2007a; Han et al., 2007b; Tsubota et al., 2007). A prevailing view is that Rtt109 forms two distinct complexes, an Rtt109-Asf1 complex for H3K56ac, and an Rtt109-Vps75 complex acetylates H3K9 and H3K27 (D’Arcy and Luger, 2011; Tsubota et al., 2007). Finally, since Rtt109 is unique to fungal species, it may serve as an effective target for antifungal infection treatments including those from Aspergillus, which causes infection, morbidity and mortality of immune-compromised patients like organ transplant recipients and AIDS and cancer patients (Marr et al., 2002). Indeed, genetic deletion of Rtt109 in C. albicans results in increased sensitivity towards antifungal agents and reduced virulence in a mouse model (Lopes da Rosa et al., 2010; Wurtele et al., 2010).
To understand the molecular mechanism of Asf1 in H3K56ac, we have determined the structures of A. fumigatus Rtt109 alone and in complex with Asf1-H3-H4 and Coenzyme A (CoA). Our analysis reveals that αN of H3 is unwound during H3K56 acetylation. Remarkably, Asf1 does not contact Rtt109 directly. Instead, it holds the C-terminal tail of H4 in a rigid conformation to facilitate binding to Rtt109, establishing that the Asf1-H3-H4 complex functions as an intact substrate. This rare view of a histone-modifying enzyme in complex with a multiprotein substrate reveals intricate contacts between the enzyme and the substrate to achieve specificity. Since many histone-modifying enzymes use macromolecular complexes as substrates, such as the nucleosome, the multivalent enzyme-substrate interaction features learned here should shed light on mechanisms of substrate recognition under these settings.
Results
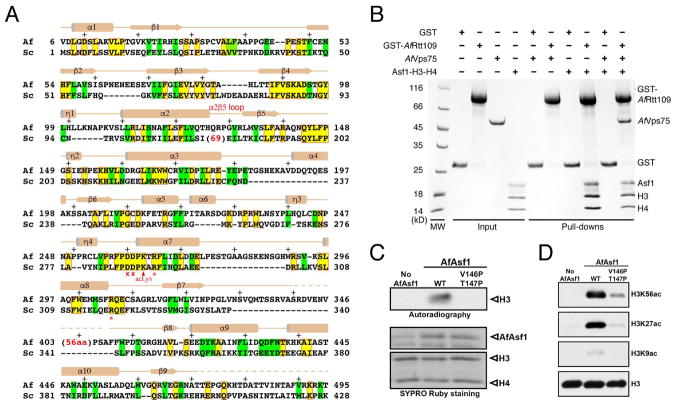
Structural and biochemical properties of A. fumigatus Rtt109
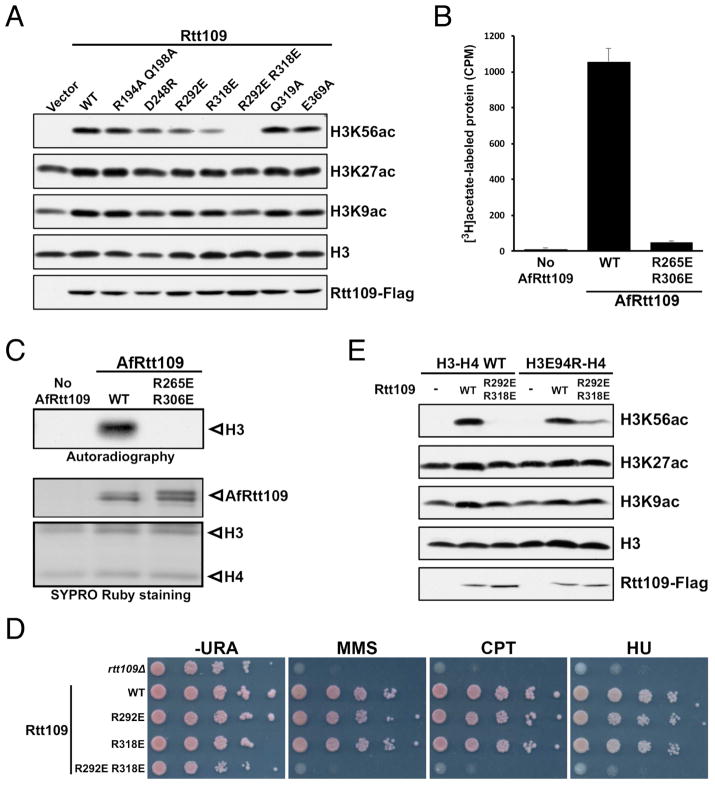
Rtt109 from the pathogenic fungus A. fumigatus (AfRtt109) shares approximately 20% sequence identity and 35% similarity with S. cerevisiae Rtt109 (ScRtt109) in the catalytic core (Figure 1A). Noticeably missing in AfRtt109 is a ~60–70-residue insertion between α2 and β5 that is important for Vps75 binding in ScRtt109 (Kolonko et al., 2010; Su et al., 2011; Tang et al., 2011). Indeed, while AfRtt109 interacts with the AfAsf1-H3-H4 complex avidly, the uncharacterized A. fumigatus Vps75 homolog (GenBank: EDP51141.1) shows no interaction with AfRtt109 (Figure 1B). However, the Vps75 homolog is capable of joining the AfRtt109-AfAsf1-H3-H4 complex, presumably through interaction with histones. Importantly, as with ScRtt109, AfRtt109 can acetylate H3 at high concentrations (data not shown), and its HAT activity toward H3K56 is greatly stimulated by Asf1, which, interestingly, also stimulates AfRtt109 to acetylate H3K27 and H3K9 (Figures 1C, 1D and S6). The latter observation is consistent with a published report that Asf1 is required for H3K9 acetylation in budding yeast (Adkins et al., 2007). Therefore, AfRtt109, in spite of lacking physical interaction with an ScVps75 homolog, fully recapitulates the H3K56 acetylation property of ScRtt109 in vitro, and provides a simplified system for investigation of the conserved mechanism of H3K56ac regulation.
Figure 1. Sequence and functional conservation of Rtt109.
(A) Structure-based alignment of A. fumigatus and S. cerevisiae Rtt109 catalytic core sequences. Identical and similar residues are highlighted yellow (boxed in red) and green, respectively. The asterisks and crosses indicate residues interacting with H3E94 and those lining the H3K56 entry channel, respectively. Auto-acetylated Lys263 is also labeled. Above the sequences, secondary structural elements of AfRtt109 are superimposed; “+” signs mark every ten residues. (B) GST-pulldown of AfRtt109 in the presence of the AfAsf1-H3-H4 complex or a putative AfVps75 homolog. (C) Autoradiograph of the HAT activities of AfRtt109 using WT or mutant AfAsf1 in complex with histones H3-H4 as the substrate. (D) Detection of H3K56ac, H3K27ac and H3K9ac in the in vitro HAT assay by Western blot analysis using AfRtt109 as the enzyme and WT or mutant AfAsf1 in complex with histones H3-H4 as the substrate. See also Figure S6; Table S3.
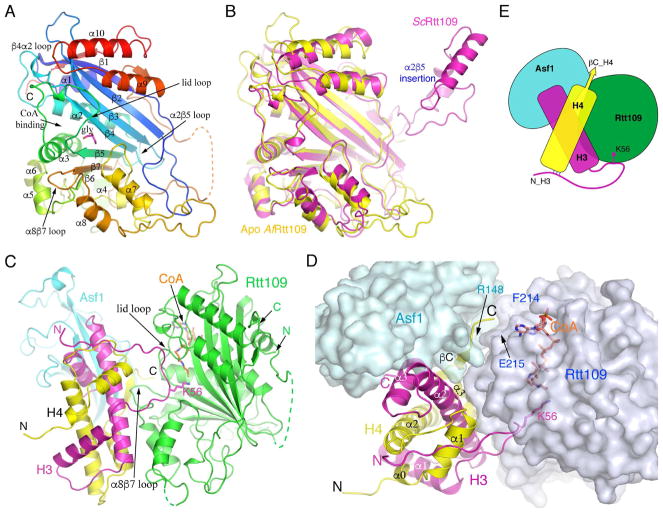
We first solved a 2.5 Å structure of AfRtt109 alone. The structure shows a globular catalytic core principally composed of a seven-stranded β-sheet, which is packed by ten well-defined helices, several short helical turns and a number of loops are located on both sides of the β-sheet (Figure 2A; Table S1). As expected, the structure of AfRtt109 closely resembles that of ScRtt109 (PDB id: 3QM0), as evidenced by a 1.94-Å root-mean-squared deviation (rmsd) between the two. Notable differences include the connection between α2 and β5, which in ScRtt109 has a ~60–70 residue insertion important for interaction with Vps75 (Figures 1A and 2B). A ~65 residue segment between β7 and the penultimate helix in AfRtt109 is disordered, while the corresponding region in ScRtt109 is significantly shorter and ordered (Figures 1A and 2A). Other differences include a loop segment (~a.a. 142–149) immediately following β5 that serves as a “lid” covering the acetyl-CoA (AcCoA) binding pocket at the end of the acetyl group. The “lid” loop exhibits an “open” conformation in the apo AfRtt109 structure. A glycerol molecule is found to stabilize the loop conformation by making multiple hydrogen bonds (Figure 2A). All in all, AfRtt109 biochemically behaves like ScRtt109, and despite limited sequence identity, it shares a conserved catalytic core with the yeast counterpart, with structural divergence occurring at none-conserved and surface-located regions where regulatory proteins may bind.
Figure 2. Structures of AfRtt109 and its complex with AfAsf1-H3-H4.
(A) Apo structure of AfRtt109 colored in rainbow spectrum. A glycerol molecule (magenta) sits near the active site and is shown in a stick model. (B) Comparison of the apo AfRtt109 structure (yellow) with the structure of ScRtt109 (magenta) from its complex with Vps75 (PDB id: 3Q66). The nonconserved α2β5 insertion in ScRtt109 (see Figure 1A) important for Vps75 binding is indicated. (C) Front view of the structure of AfRtt109 (green) in complex with AfAsf1 (cyan), histones H3 (magenta), H4 (yellow) and CoA. (D) A top view of the structure with AfRtt109 and AfAsf1 shown in a semi-transparent surface representation colored in pale blue and light green, respectively. The sidechains of H3K56, Glu215 of AfRtt109 and Arg148 of AfAsf1 are superimposed and labeled. (E) A cartoon drawing schematizing protein-protein interactions of the AfRtt109 complex. See also Figures S1–S3; Table S1.
Overall structure of the Rtt109-Asf1-H3-H4 complex
We have determined the crystal structures of AfRtt109 in complex with the conserved globular domain of AfAsf1 (a.a. 1–154) and full-length histones H3 and H4 from the budding yeast, in the absence and presence of CoA at 3.6 Å and 3.5 Å resolutions, respectively (Table S1). There is one protein complex per asymmetric unit in both cases, and, despite the medium resolutions, high quality electron density maps allowed unambiguous building of the structure model (Figures S1 and S2A). Despite the unequivocal electron density, however, it should be cautioned that, at this resolution range, precise interpretation of the structural feature would benefit from additional verifications, such as a combination of structural and functional approaches. The yeast histones used here have the same lengths and share a high degree of sequence conservation with that of corresponding A. fumigatus histones (Figure S3A). Residues 42–134 of histone H3 and 21–101 of histone H4 are ordered in the structure. The unobserved portions of H3 and H4 are present but disordered, as detected by MALDI-TOF analysis of washed crystals (Figure S3B). For convenience of description, hereafter we shall omit specification of origins of proteins wherever no confusions are expected.
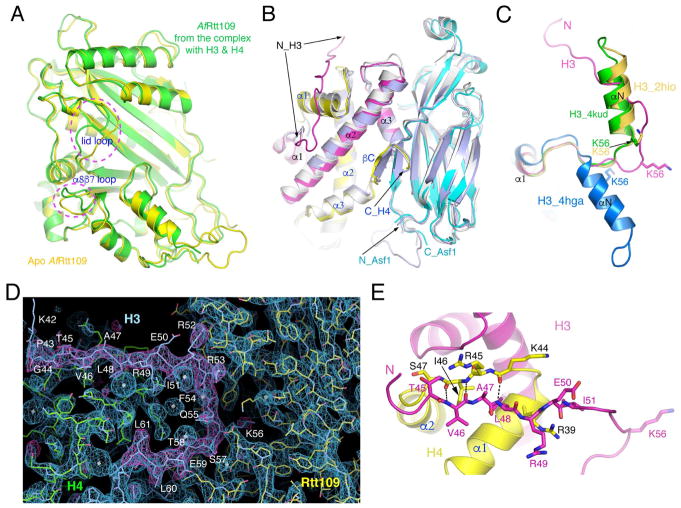
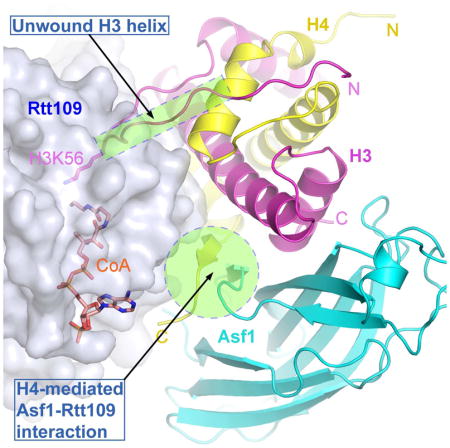
In the complex, Rtt109 makes major contacts with both histones H3 and H4, and inconsequential direct contact with Asf1 (Figures 2C and 2D). A schematic drawing illustrating essential features of the protein-protein interaction is depicted in Figure 2E. Since the CoA-bound and unbound structures are virtually identical (rmsd 0.38 Å), we shall only use the CoA-bound structure for analysis hereafter. The structure of the protein substrate-bound Rtt109 remains essentially the same as that of the apo form, with notable exceptions at two places. First, the tip of the α8β7 loop, which directly interacts with histone H4, tilted toward the “lid” loop by as much as 4 Å. Second, the “lid” loop adopts a different conformation (Figure 3A). The Asf1-H3-H4 module is bound to Rtt109 as an integral unit, in which Asf1 binds histone-fold domain (HFD) helices α2 and α3 of H3 and the very C-terminal short strand of H4 in the same manner as in stand-alone Asf1-H3-H4 complexes (Figure 3B) (English et al., 2006; Natsume et al., 2007). However, in previously determined Asf1-H3-H4 structures, the H3 region encompassing K56 was either not included or disordered. H3K56 is normally located at the C-terminal end of an α-helix (a.a. 45–56) N-terminal to the HFD, termed αN, in all occasions H3K56 has been visualized thus far (Figure 3C) (Arents et al., 1991; Elsasser et al., 2012; Liu et al., 2012a; Luger et al., 1997). In the present structure, surprisingly, αN is unwound and H3K56 is found in an extended region (Figures 2C, 2D and 3C), as evidenced by clear, continuous electron density maps (Figure 3D). The unwinding of αN is necessary for H3K56 acetylation, as placing a helix-located H3K56 into the substrate binding channel of Rtt109 would introduce severe steric conflict. Stretched αN residues 45 to 51 are stabilized by interactions with histone H4 (Figure 3E), and residues Arg52 to Thr58 interact directly with Rtt109, details of which will be described later.
Figure 3. Structural changes of Rtt109 and histones.
(A) Comparison of AfRtt019 structures between the apo and substrate-bound forms. Two regions of conspicuous differences, the “lid” and the α8β7 loops are indicated. (B) Superposition of Asf1-H3-H4 structures. The AfAsf1 complex is colored the same as before, and the yeast (PDB id: 2HUE) and human (PDB id: 2IO5) complexes are colored light blue and white, respectively. (C) Conformational differences of the N-terminal region of H3 in the structures of the AfRtt109 complex (magenta), NCP (PDB id: 4KUD, green), histone octamer (PDB id: 2HIO, goldenrod) and the DAXX complex (PDB id: 4HGA, blue). (D) Superposition of 2Fo-Fc (blue, 1.5σ) and Fo-Fc (magenta, 3σ) omit electron density maps calculated from leaving out residues 43–61 of histone H3. (E) Interactions between residues 45–51 of histone H3 (magenta) and the α1-α2 region of histone H4 (yellow) in the Rtt109-Asf1-H3-H4 complex. The involved residues are shown in a stick representation. The dashed lines indicate hydrogen bonds.
Equally surprising is the revelation that, despite Asf1’s essential role in H3K56ac, there is little contact between Asf1 and Rtt109 (Figures S1, S2B, S2C). The only notable direct interaction is a salt bridge between Arg148 of Asf1 and Glu215 of Rtt109 (Figure 2D). In contrast, the C-terminal tail of H4 makes significant interaction with Rtt109. This region of histone H4 is stabilized by antiparallel β-pairing with the C-terminal strand of Asf1. The C-terminal tail of H4 is normally disordered except in nucleosome/histone octamer structures where it adopts a distinct conformation to interact with a histone H2A-H2B heterodimer (Arents et al., 1991; Luger et al., 1997). It is also ordered in the ternary complex of H3.3-H4-DAXX (Elsasser et al., 2012; Liu et al., 2012a), but again the conformation is different from that in the Asf1 bound structures (Figure S4). The adjacent helix α3 of histone H4 is engaged in extensive interaction with Rtt109, but the conformation of α3 is little affected by Asf1 binding (Figures 4A and S4).
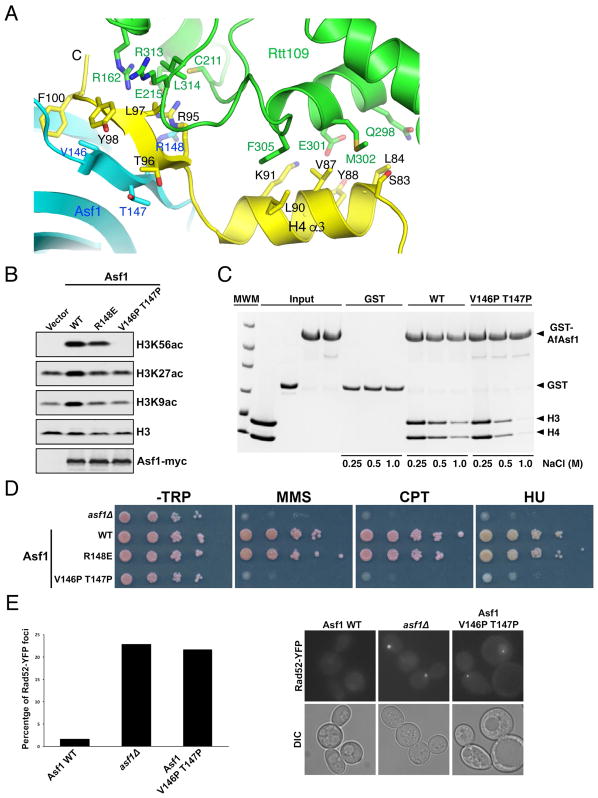
Figure 4. Asf1 stimulates H3K56 acetylation through stabilization of the C-terminal end of H4.
(A) The interface between AfAsf1 (cyan), H4 (yellow) and AfRtt109 (green). Involved residues are labeled. (B) Western blot detection of H3K56, H3K27 and H3K9 acetylation in budding yeast cells expressing indicated Asf1 mutants. (C) GST-pulldown of WT and the V146P T147P mutant of AfAsf1 in the presence of H3-H4 with at the indicated salt concentrations detected by coomassie blue staining. (D) Cell growth analysis with ten-fold serial dilutions of budding yeast cells expressing the indicated Asf1 proteins in the absence and presence of indicated DNA-damaging agents. (E) Live cell fluorescence images showing that the Asf1 V146P T147P mutant budding yeast cells exhibit a higher percentage (left panel, quantification from two independent experiments) of cells containing spontaneous Rad52-YFP foci (right panel, representative images) than WT cells. See also Figures S4, S5; Tables S2, S3.
Asf1 shapes histone H4 for optimal interaction with Rtt109
The observed interaction between Rtt109 and Asf1 appears to be not important for H3K56ac, as a R148E substitution in AfAsf1 lowered the binding affinity by less than 2 fold compared to the wild-type (WT) complex (Figure S5). Importantly, the corresponding R148E mutation of ScAsf1 has little effect on H3K56ac in yeast cells (Figure 4B). The structure suggests that Asf1 functions in H3K56ac by holding the C-terminal tail of histone H4 in a rigid conformation for optimal interaction with Rtt109 (Figure 4A). The interactions appear to be mostly through surface complementation, van der Waals and hydrophobic interactions, all of which are less sequence-stringent than polar interactions as those between histone H4 and Asf1. This feature accounts for the involvement of non-strictly-conserved residues of AfRtt109. A notable exception is the charge interaction between H4R95 and Glu215 of AfRtt109, although the latter is not strictly conserved either (Figures 1A and 4A).
To test our structure-based hypothesis, we substituted Val146 and Thr147, two Asf1 residues residing in the C-terminal β-strand and involved in antiparallel β-pairing with the C-terminal strand of H4, with a pair of prolines. The intention was to disrupt the β-pairing between Asf1 and histone H4 without changing H4 residues, thus not affecting H4-Rtt109 interaction due to amino acid differences (Figure 4A). Significantly, the V146P T147P double mutant of ScAsf1 completely abolished H3K56ac in vivo (Figure 4B). Furthermore, in vitro HAT activity assays show that the V146P T147P mutant AfAsf1-H3-H4 complex is a poor substrate for AfRtt109, as evidenced by a dramatic reduction of acetylation of histone H3 and H3K56 levels in the in vitro assays (Figures 1C, 1D and S6A). This defect in H3K56ac is not because of the AfAsf1 mutant’s inability to form a complex with histones, although the mutant does bind H3 and H4 with a reduced affinity (Figure 4C). Budding yeast cells lacking Asf1 are sensitive to DNA-damaging agents and display an elevated level of spontaneous chromosome breaks, and these defects are correlated with loss of H3K56ac (Han et al., 2007a; Le et al., 1997; Tyler and Kadonaga, 1999). A cell growth assay in the presence of DNA-damaging agents shows that yeast cells expressing the R148E mutant of Asf1 have no obvious growth defect under these conditions, whereas the ones carrying the Asf1 V146P T147P double mutation behave almost like Asf1-null cells (Figure 4D). Furthermore, the double mutant cells have a higher rate of Rad52-YFP foci, an indication of spontaneous chromosome breaks (Lisby et al., 2001), again at a level similar to that of asf1Δ cells (Figure 4E). The above in vitro and in vivo results firmly support the notion that Asf1 contributes to Rtt109-dependent acetylation of H3K56 through formation of a suitable substrate complex by stabilizing the C-terminus of H4, not by functioning as a productive component of an enzyme complex.
Histone H3 recognition and architecture of the active site
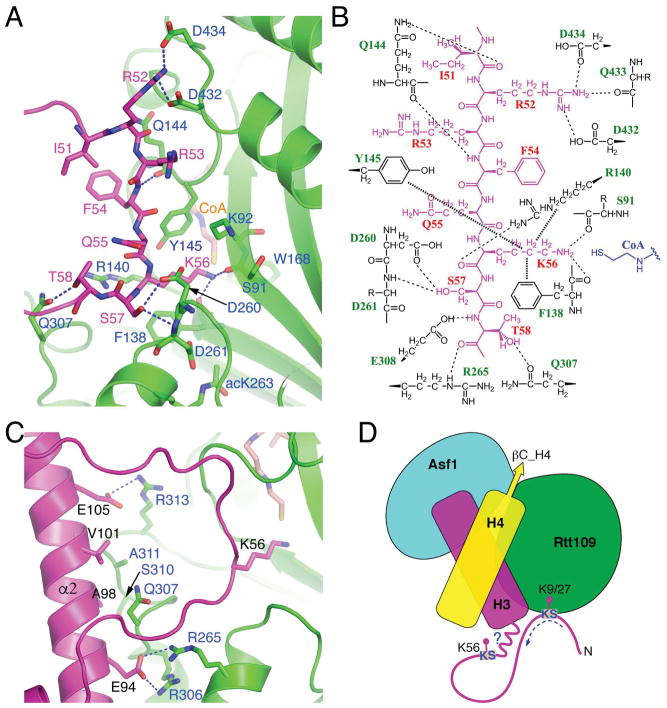
Besides interactions involving histone H4, interactions between the Asf1-H3-H4 complex and Rtt109 mainly occur at two distinct regions of histone H3: an unwound αN segment encompassing Lys56, and surprisingly also the central helix (α2) of H3 HFD (Figures 5A–5C). In the first region, the sidechain of H3K56 is positioned in the lysine-binding channel formed mainly by Tyr145, Arg140, Phe138 and Asp260 of AfRtt109 (Figures 5A and 5B). Connected to the lysine-binding channel is the tunnel where the pantetheine arm of CoA binds. The Rtt109 residues involved in AcCoA binding are highly conserved, and the structural basis for their interaction has been described in detail previously (Lin and Yuan, 2008; Stavropoulos et al., 2008; Tang et al., 2011; Tang et al., 2008). The junction of the lysine-binding channel and the AcCoA binding tunnel where AcCoA comes in close contact with the lysine ε-amino group forms the active site of the acetyltransfer reaction. Within ~4.5Å radius of the ε-amino group of H3K56 and the sulfhydryl group of CoA lies Trp168, Tyr145, and the mainchain carbonyl groups of Ser91 and Phe138 (Figure 5A). The ε-amino group of H3K56 forms hydrogen bonds with the carbonyl groups of Ser91 and Phe138 of AfRtt109, and is approximately 4 Å away from the sulfhydryl group of CoA (Figures 5A and 5B).
Figure 5. H3-Rtt109 interaction.
(A) Interactions with Rtt109 (green) involving H3 (magenta) residues surrounding Lys56. Involved residues are shown in a stick representation, and blue dashed lines indicate hydrogen bonds. (B) Schematic drawing of H3-Rtt109 interactions. Thin dashed lines represent hydrogen bonds, while thick dashed lines indicate van der Waals contacts. Rtt109 residues interacting with H3 via mainchain groups only are shown with their sidechains labeled with “R”. (C) Interactions between H3 residues located on HFD helix α2 and AfRtt109. (D) A model of how Rtt109 may accommodate the acetylation of H3K56, H3K27 and H3K9, all of which belong to a consensus KS motif as indicated. Please note that Vps75 is also needed for H3K27 and H3K9 acetylation.
The active site of AfRtt109, as that of ScRtt109, lacks a negatively charged residue as the general base as in well-characterized HATs. While it is clear that Trp168 and Tyr145 are critical for catalysis by coordinating the binding of AcCoA and substrate lysine, their positions and conformation preclude their direct participation in the transfer of acetyl group. Our earlier study identified that Asp287 and Asp288 in ScRtt109, which correspond to Asp260 and Asp261 in AfRtt109, together are important for H3K56 acetylation (Han et al., 2007a). Structural and enzymatic studies of ScRtt109 have argued against the catalytic role of Asp287 and Asp288 (Albaugh et al., 2010; Tang et al., 2008). In our substrate bound structure, Asp260 is located at the entry of the lysine-binding channel, making one hydrogen bond with the hydroxyl group of Ser57 of H3, a reported phosphorylatable residue (Aslam and Logie, 2010), and one hydrogen bond each to the amide groups of Lys56 and Ser57 of H3. Asp261 makes one hydrogen bond with the autoacetylated Lys263, and exposes one side of the carboxylate group to the inner surface of the lysine-binding channel (Figure 5A). An enzymatic study with pH-rate analysis reveals that the ε-amino group of H3K56 is in a neutral form upon entering the active site (Albaugh et al., 2010). Thus, the presence of Asp260 and Asp261 inside the lysine-binding channel may provide a negatively charged environment energetically favorable for the dissociation of non-covalently associated proton from the ε-amino group of H3K56. Our structure supports the proposal of a direct acetyltransfer mechanism by Rtt109 (Albaugh et al., 2010). In this scenario, AcCoA and the substrate bind Rtt109 independently. Once the substrate lysine is in place, the ε-amino group directly attacks the scissile bond of AcCoA. Upon breakage of the scissile bond, the close proximity between the sulfur ion and the ε-amino group may allow the former to directly “hijack” a hydrogen atom from the amino group, concomitant with the transfer of the acetyl group to H3K56.
Determinants of H3K56 specificity
In addition to the binding of H3K56 and the hydrogen bond between Ser57 of H3 and Asp260 of Rtt109 described above, additional interactions between the αN region of histone H3 and Rtt109 involve a pair of hydrogen bonds between H3R52 and Asp432 and Asp434 of AfRtt109, and a hydrogen bond between H3T58 and Gln307 of AfRtt109, as well as various hydrogen bonds involving mainchain groups (Figures 5A and 5B). However, mutational studies of Rtt109 to be described later show that none of the above interactions are essential for H3K56ac in vivo. This observation is consistent with Rtt109 being able of acetylating H3K9 and H3K27, as the sequences preceding K56, K27 and K9 of histone H3 are not conserved. The only common feature for these three acetylation sites is the presence of a serine following the acetylable lysine, namely, a consensus KS motif. A conceivable model accounting for the above observation is shown in Figure 5D. Interestingly, a histone H3 region far away from H3K56, namely, residues located on one side of α2, ranging from Glu94 to Glu105, directly interact with AfRtt109 (Figure 5C). In particular, H3E105 makes a hydrogen bond with Arg313, and H3E94 forms hydrogen bonds with the conserved Arg265 and Arg306 of AfRtt109. Among the three arginines, Arg265 and Arg306 are conserved across species but Arg313 is not. Thus, altogether three histone regions from the Asf1-H3-H4 substrate complex are important for interaction with Rtt109: 1) C-terminal half of αN of H3, including H3K56; 2) α2 of histone H3; and 3) C-terminal tail and α3 of histone H4.
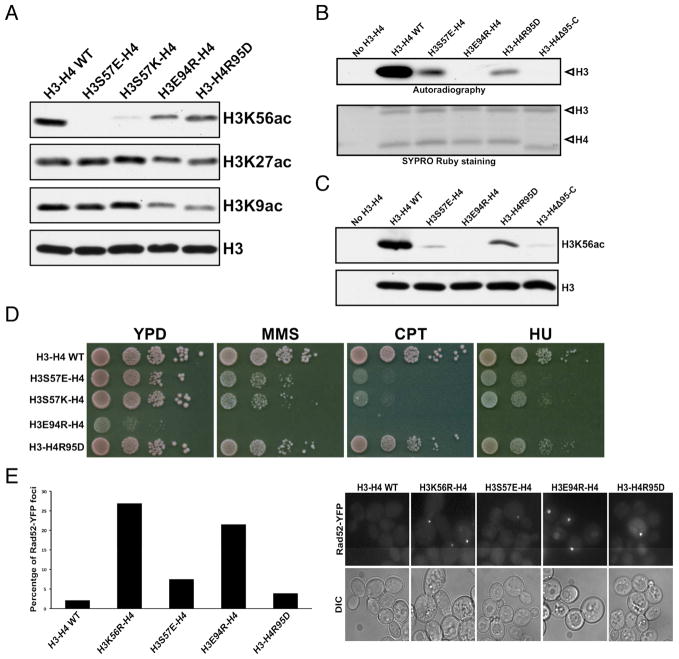
To assess the importance of the involved histone regions for H3K56ac, we chose representative residues from these regions for mutagenesis and functional analyses. Yeast cells expressing H3S57 mutants showed a marked reduction in H3K56ac based on Western blot analysis of H3K56ac levels in cell extracts (Figure 6A). Western blot analysis of the in vitro HAT assay products also detected a pronounced loss of H3K56ac (Figure 6C). However, in the in vitro HAT assay using [3H]acetyl-CoA the H3S57E mutant displayed a significantly reduced but noticeable level of H3 acetylation (Figures 6B and S6B). The discrepancy between the H3K56ac results detected by Western blot and autoradiography suggests that substitution of Ser57, which is immediately next to the acetylated-Lys56 antigenic determinant, compromises immunodetection of H3K56ac. A E94R substitution of histone H3 resulted in moderate decreases of H3K56ac in yeast cells (Figure 6A). Curiously, the H3E94R mutant showed no detectable level of H3 acetylation in the in vitro HAT assays (Figures 6B and 6C). The H3E94R mutation appears to affect H3K56ac more severely in vitro than in cells, but a caveat is that cells with H3E94R grow very poorly even in the absence of any stress (Figure 6D), signaling potentially compounded effects of this mutation besides on H3K56ac. We also substituted Arg95 of histone H4 with an aspartate or deleted the entire segment of histone H4 from Arg95 to the C-terminus (H4Δ95-C). The H4R95D mutation has only minor effect on H3K56ac in cells and in in vitro HAT assays (Figures 6A–C). This is perhaps because, as pointed out earlier, the Rtt109 residues involved in interaction with the H4 region are not strictly conserved and are more tolerant to variations in cognate histone residues. In contrast, yeast cells expressing the truncation variant of H4 were unable to grow (data not shown). The in vitro assay also revealed that the H4Δ95-C deletion jeopardized H3K56ac (Figures 6B and 6C), an observation that fully supports our conclusion that Asf1 functions to stabilize the C-terminal tail of H4 in Rtt109-catalyzed H3K56 acetylation. Finally, we assessed the impact of these mutants on DNA damage sensitivity and spontaneous chromosome break using Rad52-YFP. Both H3S57 mutants are markedly more sensitive to DNA-damaging agents than WT H3 (Figure 6D). Moreover, cells with the H3E94R mutation show an extremely high degree of spontaneous chromosome break, at a level similar to the unacetylable H3K56R cells (Figure 6E). By comparison, percentages of cells with Rad52-YFP foci in the H3S57E and H4R95D strains are approximately 3 and 2 fold higher than that of WT cells, respectively. The above observations highlight the critical importance of the C-terminal region of histone H4, which conveys the Asf1-dependence of H3K56 acetylation, and α2 of the HFD of H3 in the determination of Rttt109’s specificity toward H3K56, in addition to the H3K56-containing KS motif.
Figure 6. Histone residues important for H3K56 acetylation.
(A) Western blot analyses of impacts on H3K56, H3K27 and H3K9 acetylation by indicated histone mutations in budding yeast cells. (B) Autoradiography detection of H3 acetylation in an in vitro HAT assay using purified wild-type AfRtt109, and the indicated mutant histone-AfAsf1 complexes. The bottom panel: SYPRO Ruby staining of the histones used. (C) H3K56 acetylation of wild-type or histone mutants by AfRtt109 in vitro in the presence of AfAsf1 detected by Western blot. (D) Cell growth analysis of budding yeast cells expressing the indicated histone mutants in the absence and presence of the denoted DNA-damaging agents. (E) Spontaneous chromosome breaks in budding yeast cells expressing indicated histone mutants determined by the Rad52-YFP foci. Left panel: percentage of cells with Rad52-YFP foci from two independent experiments. Right panel: representative fields of live cell images. See also Figure S6; Tables S2, S3.
Rtt109 residues important for interaction with histones and H3K56ac
Next, we analyzed the role of Rtt109 residues in H3K56 acetylation. Near the H3K56 binding site, Arg140 and Gln144 of AfRtt109 each makes one hydrogen bond to mainchain carbonyls of H3, but mutating the corresponding ScRtt109 residues, Arg194 and Gln198, to a pair of alanines has little effect on H3K56ac (Figure 7A). The same is true for alanine substitution of ScRtt109 Gln319 and Glu369, which, respectively, correspond to Gln307 and Asp434 of AfRtt109 (Figure 1A). In the structure, Gln307 makes a hydrogen bond with H3T58 and engages α2 of H3 via van der Waals interaction, while Asp434 interacts with H3R52 (Figures 5A–C). Proximal to the C-terminus of H4, Rtt109 residues located in this region are sparsely conserved. Asp212 of AfRtt109 is one of the few residues in this region that can be aligned with Asp248 of ScRtt109 (Figure 1A). However, a D248R substitution has little effect on H3K56ac (Figure 7A). The most dramatic reduction in H3K56ac occurs with the R292E R318E double mutant of ScRtt109, while individual single mutants only have minuscule effects (Figure 7A). Arg292 and Arg318 of ScRtt109 correspond to Arg265 and Arg306 of AfRtt109, respectively (Figure 1A), and the two AfRtt109 arginines are involved in interaction with H3E94 located in H3 HFD (Figure 5C). In vitro HAT assays reveal that recombinant R265E R306E mutant of AfRtt109 has no detectable activity toward H3 nor H3K56ac (Figures 7B, 7C and S7A). In yeast cells, while R292E or R318E single mutation causes no evident sensitivity to DNA-damaging agents, the double mutant results in a severe growth defect (Figure 7D). Consistently, yeast cells expressing the double mutant display a significantly increased number of Rad52-YFP foci, at a level comparable to rtt109Δ cells (Figure S7B). The above data reveal that the interaction between Arg292, Arg318 and H3E94 is essential for H3K56 acetylation. To further validate the importance of this H3-Rtt109 interaction for H3K56ac, we attempted to repair the interaction disrupted by the histone or Rtt109 mutations through swapping the charged residues. Indeed, yeast cells expressing both H3E94R and the Rtt109 R292E R318E double mutant partially restore H3K56ac in budding yeast (Figure 7E).
Figure 7. Rtt109 residues important for H3K56 acetylation.
(A) Western blot analysis of H3K56, H3K27 and H3K9 acetylation in budding yeast cells expressing the indicated Rtt109 mutants. (B) In vitro HAT activity assay of the corresponding AfRtt109 mutants detected by autoradiography. (C) Quantitation of the in vitro HAT activities of the WT and mutant AfRtt109 proteins using the AfAsf1-H3-H4 complex as the substrate. (D) Budding yeast cells expressing the R292E R318E mutant of ScRtt109 are sensitive to DNA-damaging agents like rtt1093Δ cells. (E) Partial rescue of H3K56ac in yeast cells by co-expressing indicated charge-swap mutants of ScRtt109 and H3. See also Figure S7; Tables S2, S3.
Discussion
Our study addressed a long-standing puzzle in the field, namely, how histone chaperone Asf1 controls the H3K56 acetylase activity of Rtt109. In S. cerevisiae, Rtt109 forms an obligatory complex with another histone chaperone, Vps75. A school of thought proposes that Asf1 functions in H3K56ac through direct binding to Rtt109, perhaps via an allosteric mechanism much like that of Vps75 (Berndsen and Denu, 2008; D’Arcy and Luger, 2011; Kolonko et al., 2010; Tsubota et al., 2007). An alternative proposal is that Asf1 functions to present the H3-H4 complex for acetylation by Rtt109, but a precise mechanism has been elusive (Han et al., 2007b). Our Rtt109-Asf1-H3-H4 structure clearly shows that Asf1 has inconsequential direct contacts with Rtt109. Nevertheless, the very C-terminal 12-residue segment of ScRtt109 was reported to interact with Asf1 (Lercher et al., 2017; Radovani et al., 2013). The reported interaction affects H3K56ac by ScRtt109 in vitro and in yeast cells only in the absence of Vps75, suggesting a specialized role of this interaction in the budding yeast, as Vps75 is not a conserved partner of Rtt109 in fungi. The physiological significance of this interaction in S. cerevisiae is also not clear, as Vps75 is present together with Rtt109. On the other hand, our in-depth biochemical and genetic analyses corroborated the structural finding that Asf1 functions in H3K56ac through stabilization of the C-terminal region of histone H4, which is oriented differently in the nucleosome structure. This region of H4 is disordered in most structures of H3 and H4 bound to histone chaperones, except in the complexes with Asf1 and DAXX, but its conformation bears no similarity in the two structures (Figure S4). In summary, the critical role of Asf1 in H3K56 acetylation is attributed to its function in specifying the unique conformation of the C-terminal tail of H4. Thus, the discovery of Asf1’s role in reshaping the H3-H4 complex for H3K56 acetylation unearths a dynamic function of the versatile histone chaperone in addition to its traditional capacity as a histone escort.
A surprising finding of this work is that αN of H3 is unwound and H3K56 is located in a loop region. αN is found in the structures of histone octamer and nucleosomes, where it occupies the space between two gyres of DNA near the entry/exit points and is important for nucleosome stability (Arents et al., 1991; Luger et al., 1997). However, the αN region is disordered in most chaperone-H3-H4 complexes including those with Asf1, Spt2 and Mcm2 (Chen et al., 2015; English et al., 2006; Huang et al., 2015; Natsume et al., 2007; Richet et al., 2015). The only other occasion this region of H3 was visualized was in the complex of H3.3 and H4 with DAXX (Elsasser et al., 2012; Liu et al., 2012a). There it also adopts a helical conformation but with an entirely different orientation (Figure 3C). In all cases, αN is unstructured in the absence of interacting partners. The sole appearance of the αN region in a loop conformation thus far suggests that the binding of Rtt109 induces the unwinding of αN, although we cannot rule out the possibility of a preexisting random coil conformation, possibly in an equilibrial mixture with helices. In any event, the observed loop conformation of a presumed α-helical region resolves the long-standing puzzle of how H3K56 may be easily accessed for acetylation by Rtt109.
Once it is understood that H3K56ac occurs when it resides in a loop, it is appealing to conjecture that a similar substrate-binding mode governs H3K27 and H3K9 acetylation. This notion is consistent with our observation that the sequence-dependent interactions with Rtt109 involving local H3 residues preceding K56 are not crucial for H3K56ac. Because a serine is found next to H3K56, H3K27 and H3K9, it is possible that a common KS local motif is important for Rtt109-mediated acetylation. If this is the case, it is interesting to wonder whether Asf1 is important for H3K27 and H3K9 acetylation. Our in vitro HAT assay indicates that Asf1 is also important for H3K27 and H3K9 acetylation (Figure 1D), a result consistent with the observation of H3K9 acetylation in the budding yeast (Adkins et al., 2007). However, more studies are needed due to the involvement of Gcn5, in addition to Rtt109, in the acetylation of H3K27 and H3K9, and the participation of histone chaperone Vps75. Thus, a careful analysis is needed to tease apart the contribution and roles of the involved enzymes and chaperones. A possible model of Rtt109-catalyzed H3 acetylation depicted in Figure 5D may guide further in-depth investigations.
It is also interesting to wonder whether the H3K56 region forms a helix following acetylation, as this would have important implications. For example, it was shown that histone H3-H4 with an acetylated H3K56 binds tighter to histone chaperones CAF-1 and Rtt106, two factors important for replication-coupled nucleosome assembly (Li et al., 2008; Su et al., 2012; Verreault et al., 1996). It will be interesting to know if an extended conformation of the H3K56 region is favorable for such binding. Furthermore, H3K56 acetylation facilitates nucleosome “breathing”, i.e., transient opening of the nucleosomal DNA ends, and partial unwrapping of the nucleosome (Neumann et al., 2009). The dynamic feature of the acetylated H3K56 nucleosome has been attributed to the possibility that an acetylated H3K56 weakens the interaction with DNA. However, nucleosome structures with H3 K56Q, which mimics an acetylated H3K56, show that DNA is fully wrapped (Watanabe et al., 2010). In view of our observation that αN can be unwound, the stability of αN upon H3K56 acetylation should be investigated.
To date, most, if not all, structural studies of substrate recognition by histone modification enzymes were done with histone peptides. A lesson learned from this study is that when it involves a complex substrate, usage of the whole substrate complex is necessary for understanding the complete picture of substrate recognition. The involvement of histone H3 residues proximal to H3K56, H3E94 in the central histone fold domain distal to the acetylation site and the Asf1-stabilized C-terminal region of H4 in substrate binding by Rtt109 indicates that components of the Asf1-H3-H4 complex function as an integral substrate unit. This fascinating multisite, multiprotein substrate recognition mode governing H3K56 acetylation revealed here is likely to be prevalent in various types of post-translational modifications of histones involving multiprotein substrate and/or enzyme complexes. Therefore, care must be taken in uncovering the complete picture in future mechanistic studies. Finally, the structural information of the H3K56 acetyltransferase from the pathogenic fungus A. fumigatus should also facilitate the discovery of therapeutic agents against fungal infections.
STAR METHODS
Contact for reagent and resource sharing
Additional information and requests for resources and reagents should be directed to the Lead Contact, Rui-Ming Xu (rmxu@ibp.ac.cn).
Experimental model and subject details
E. coli strains DH5α, BL21(DE3) and BL21(DE3) CodonPlus RIL were used for molecular cloning, expression of AfRtt109 and the AfAsf1-H3-H4 complex, respectively. All yeast strains used were isogenic to W303 (leu2-3, 112 ura3-1 his3-11, trp1-1, ade2-1 can1-100), and are listed in Table S2.
Method details
Protein expression, purification and crystallization
Full-length A. fumigatus Rtt109 was produced as a polyhistidine and SUMO-tagged fusion protein in the BL21(DE3) strain of E. coli using the pET28a-Smt3 vector, with the cDNA inserted between the BamHI and NotI sites. E. coli culture was first grown at 37°C until cell density reached OD600 ~0.6–0.8, and then protein expression was induced with the addition of 0.25 mM IPTG at 16°C for 16–20 h. Harvested cells were ruptured by sonication and/or pressure cell in a buffer containing 20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 5% glycerol and 20 mM imidazole. The cell lysate was cleared by centrifugation, and the supernatant was loaded onto a Ni-NTA column (Novagen). The loaded column was washed with 10 column-volume of lysis buffer, followed by elution of the bound proteins with 250 mM imidazole added to the lysis buffer. Then, cleavage of the polyhistidine and SUMO tag was carried out by the addition of SUMO protease at 4°C for 30 minutes. The cleavage products were separated using a HiTrap Heparin column (GE Healthcare) using a 0.1–1.0 M NaCl concentration gradient in 20 mM Tris-HCl, pH 8.0, and 5% glycerol. The eluted fractions were analyzed by coomassie blue-stained SDS-PAGE, and the first of the two peaks enriched with AfRtt109 was chosen for further purification through a HiLoad Superdex 200 16/60 gel-filtration column (GE Healthcare) in 20 mM Tris-HCl, pH 8.0, 100 mM NaCl and 5% glycerol. High purity fractions of AfRtt109 were pooled and concentrated to ~20 mg/ml using a 50 kD-cutoff Amicon concentrator. The concentrated sample was flash frozen in liquid nitrogen and stored at −80°C for later use.
The complex of A. fumigatus Asf1 fragment (a.a. 1–154) and histones H3 and H4 of S. cerevisiae origin were produced by coexpression in the BL21(DE3) CodonPlus RIL strain of E. coli. Because all in vitro studies here use the 1–154 fragment of AfAsf1 except in GST-pull down experiments described later, we shall refer to this fragment as AfAsf1 unless otherwise specified, for convenience of description. The AfAsf1-expressing plasmid was constructed by cloning the cDNA fragment between the BamHI and SalI sites of the pET28a vector, and histones H3 and H4 were expressed using a bicistronic pETDuet vector (Novagen). 35 μM of kanamycin, 100 μM of ampicillin and 35 μM of chloramphenicol were applied to prevent plasmid loss despite utilization of the same incompatibility group of replicons by the two pET vectors. Purification of the complex of polyhistidine-tagged AfAsf1 and full-length, authentic histones H3 and H4 was carried out following the same procedure as that for AfRtt109, except that the protease cleavage and gel-filtration column chromatography steps were omitted, and that a 0.5–2.0 M NaCl gradient was used for column chromatography with heparin resins. Purified AfAsf1-H3-H4 complex was concentrated to ~10 mg/ml and stored at −80°C. Protein mutants were generated by PCR using primers listed in Table S3, and purified following the same procedures as outlined above.
AfRtt109 at a concentration of 10 mg/ml was crystallized in 100 mM sodium cacodylate, pH 6.4, and 16% PEG-1500 at 16°C by hanging drop vapor diffusion. The AfRtt109-AfAsf1-H3-H4 complex was assembled by mixing purified AfRtt109 and the AfAsf1-H3-H4 complex at an approximately 1:1 molar ratio first in a buffer containing 20 mM Tris-HCl, pH 8.0, 1 M sodium chloride, 5% glycerol, followed by lowering the salt concentration to 150 mM through dialysis at 4°C. Then, the sample was loaded onto a HiLoad Superdex 200 16/60 column (GE Healthcare) pre-equilibrated with the same buffer. Eluted fractions enriched with stoichiometric components were pooled and concentrated to ~4 mg/ml. The best diffracting crystals were grown at 4°C in a condition with 100 mM sodium citrate, pH 5.0, 22% PEG-1500, 400 mM sodium iodide. The same crystallization condition gives rise to cocrystals with CoA, which is added at 1 mM concentration.
Crystallographic data collection and structure determination
X-ray diffraction data for the AfRtt109 crystal were collected at beamline BL18U1 of Shanghai Synchrotron Radiation Facility (SSRF) at a wavelength of 0.9786 Å using a Pilatus 6M detector, and processed with HKL3000 (Otwinowski and Minor, 1997). The structure was solved by molecular replacement with PHASER (McCoy et al., 2007), using the structure of S. cerevisiae Rtt109 (PDB code: 2ZFN) as the search model. The final model was generated by iterative cycles of refinement and rebuilt using PHENIX (Adams et al., 2010) and COOT (Emsley and Cowtan, 2004). The AfRtt109-Asf1-H3-H4 complex data were collected at beamline BL17U of SSRF at a wavelength of 0.9788 Å using an ADSC Quantum 315r detector, and processed with HKL2000. The AfRtt109-Asf1-H3-H4-CoA complex data were collected at beamline BL19U1 of SSRF at a wavelength of 0.9785 Å using a Pilatus 6M detector, and processed with HKL3000. The complex structures were solved by molecular replacement with PHASER, using the AfRtt109 structure and the structure of yeast Asf1-H3-H4 complex (PDB code: 2HUE) as the search models. The final models were obtained through refinement and rebuilt with PHENIX and COOT. Detailed statistics for crystallographic analyses are shown in Table S1.
GST-pull downs
To test the interaction between AfRtt109 and an Aspergillus Nap1-like putative Vps75 homolog (AfVps75, GenBank id: EDP51141.1), GST-tagged AfRtt109 was expressed using a pGEX-6P-1 vector in the BL21(DE3) strain of E. coli, and purified by successive glutathione-affinity, anion-exchange and gel-filtration column chromatography steps. N-terminal his-tagged AfVps75 was expressed in E. coli using a pCDFDuet vector (Novagen), and purified by successive Ni-NTA column (Novagen), anion-exchange and gel-filtration column chromatography steps. GST-pull down experiments were carried out by mixing 50 μg of GST-AfRtt109 with 50 μg of AfVps75 or AfAsf1-H3-H4 complex or both in a buffer containing 20 mM Tris-HCl at pH 8.0, 150 mM sodium chloride and 5% glycerol. The mixture was incubated on ice for ~12 h, followed by the addition of 20 μl glutathione-agarose resin and incubation for 3 h at 4ºC with shaking. The glutathione resin was then recovered by gentle centrifugation for 30 s, washed three times with the binding buffer, and the bound proteins were analyzed by SDS-PAGE.
To assess the effect of Asf1 mutation on the binding between Rtt109 and the Asf1-H3-H4 complex, GST-fused WT and the V146P T147P mutant of full-length AfAsf1 were expressed in E. coli using the pGEX-6P-1 vector. The fusion proteins were purified first by glutathione-agarose resins, followed by successive chromatography steps with Q and HiLoad Superdex 200 16/60 columns (GE Healthcare). Approximately 50 μg of purified GST-fused WT or mutant AfAsf1 was mixed with ~50 μg of histone H3-H4 in buffers containing 20 mM Tris-HCl, pH 8.0, 50 mM potassium chloride, 5% glycerol, and 0.25, 0.5 or 1.0 M NaCl and incubated on ice for ~5 h. Then, 20 μl of glutathione-agarose resins were added and incubated for 30 minutes at 4 °C with shaking. Afterwards, the supernatant was removed by centrifugation, and the resins were washed with the binding buffer three times before analysis by SDS-PAGE. As a control, GST bound beads were subjected to the same procedure.
Yeast strains and plasmids
All yeast strains used are listed in Table S2, and standard yeast media and manipulations were used. Mutations in RTT109, ASF1, and HHT2 and HHF2 genes were generated by site-directed mutagenesis using plasmids pZG301 (pRS416-RTT109), pZG557 (pRS313-ASF1), and pZG724 (pRS314-HHT2-HHF2). Sequences of DNA primers used for mutagenesis are listed in Table S3.
Preparation of yeast protein extracts and immunoblotting
To prepare whole cell protein extracts, 5 ml of exponentially growing yeast culture were harvested by centrifugation, washed with cold water, and resuspended in 50 μl of cold TBS buffer containing 1 mM DTT and 1 mM PMSF. An equal amount of glass beads was added to each tube and cells were shaken in a bead beater five times for 1 minute. Lysates were transferred to new tubes, mixed with 50 μl of 2xSDS-PAGE loading buffer, and boiled for 3 minutes. For immunodetection of proteins, 5 μl of protein extract were resolved with 15% SDS-PAGE and transferred onto a nitrocellulose membrane. Histone H3 and acetylation of K56, K27 and K9 of H3 were detected using specific antibodies against these modifications. Antibodies against Flag (F1804, Sigma-Aldrich) and Myc were used to detect Flag-tagged Rtt109 and Myc-tagged Asf1, respectively.
In vitro HAT assays
HAT activity was determined using a filter-binding assay as previously described, with some modifications (Han et al., 2007a; Tanner et al., 1999). Samples were incubated at 30°C for 30 minutes in a 30 μl reaction mixture containing 50 mM Tris-HCl, pH 8.0, 5% glycerol, 0.1 mM EDTA, 50 mM KCl, 1 mM DTT, 10 mM sodium butyrate, 250 mM NaCl, 1 mM PMSF, 1.6 μM [3H]acetyl-CoA (8.6 Ci/mmol, PerkinElmer), and recombinant WT or mutant forms of AfRtt109, AfAsf1 and histones H3 and H4, as specified. 7.5 μl of each reaction were spotted onto P-81 phosphocellulose paper (Whatman), air dried, and washed five times with 50 ml of buffer containing 50 mM NaHCO3 at pH 9.0, and once with 50 ml of acetone. The amount of radioactivity of each air-dried filter paper was measured using a liquid scintillation counter. To visualize the amount of each protein and detect acetylated ones, each reaction mixture was resolved by 15% SDS-PAGE and the gels were either stained with SYPRO Ruby (BioRad) or dried and exposed to photo films after incubation with Amplify (Amersham) for 30 minutes. To detect H3K56 acetylation, HAT assays were performed as before but using unlabeled acetyl-CoA and analyzed by Western blot using antibodies recognizing acetylated H3K56.
DNA-damaging agent sensitivity assay
Yeast strains were grown overnight at 30°C in YPD (H3 and H4 mutants), SCM-URA (Rtt109 mutants), or SCM-TRP (Asf1 mutants). Cultures were diluted to 6×106 cells/ml followed by four ten-fold serial dilutions and spotted onto media containing the indicated DNA-damaging agents: MMS (methylmethane sulfate, 0.005% for SCM or 0.01% for YPD), CPT (camptothecin, 1 mg/ml for SCM or 5 mg/ml for YPD), or HU (hydroxyurea, 50 mM for SCM or 100 mM for YPD).
Determination of Rad52-YFP foci
Counting of Rad52-YFP foci was performed as previously described (Han et al., 2007a). Briefly, indicated yeast strains expressing Rad52-YFP were grown at 26°C, and cells in exponential phase of growth were harvested, washed twice with SCM-TRP, and re-suspended in SCM-TRP. Live cells were deposited on glass slides and images were acquired using a Zeiss Axio Observer Z1 inverted microscope equipped with a Plan-Apochromat 100X/1.4 Oil lens. For each field, ten images were obtained along the z-axis at 0.49 μm intervals. All focal planes were analyzed and cells with Rad52-YFP foci from one imaging field were counted as positive. A total of 450 cells from two independent experiments were counted for each strain and the percentage of cells with Rad52-YFP foci was calculated for each strain.
Quantification and statistical analysis
Quantitation of in vitro HAT activity is represented as the mean of 3 independent experiments (Figures S6A and 7C) or 4 independent experiments (Figure S6B). The error bars indicate the standard error of the mean (SEM).
Data and software availability
The Protein Data Bank (PDB) accession codes for the coordinates and structure factors of apo AfRtt109 and the AfRtt109-Asf1-H3-H4 complex in the presence and absence of CoA are 5ZB9, 5ZBA and 5ZBB, respectively.
Supplementary Material
(A) A front view showing the interface between Rt109 and H3-H4. The superimposed yellow, blue and green Cα chains are that of Rtt109, H3 and H4, respectively. The map was contoured at 1.5σ. (B) A back view showing the region surrounding the C-terminal β-strand of H4. The Cα chain of Asf1 is shown in magenta, and the CoA molecule is shown in a bond model.
(A) A section of the 2Fo-Fc electron density map surrounding the active site of Rtt109 contoured at 1.3σ allows unambiguous placement of protein sidechains. Rtt109 residues were shown with the carbon bonds colored yellow, while the pale blue and green carbon bonds indicate that of hitone H3 and H4, respectively. Red and blue bonds represent that of oxygen and nitrogen, and the white and pale green lines in CoA indicate bonds from phosphorus and sulfur atoms. (B) An Fo-Fc map calculated from omitting the residues 91–101 of histone H4 shows strong mainchain density and most of the sidechains of the C-terminal segment of histone H4. The map is displayed at 3σ level, and the refined structure is superimposed as a bond model, with the carbon bond of histone H4 colored in green. (C) Superposition of the 2Fo-Fc (blue) and Fo-Fc (magenta) omit maps surrounding the C-terminal β-strand of histone H4. The same region of H4 as in (B) was omitted in the map calculation, and the Fo-Fc and 2Fo-Fc maps were contoured at 3σ and 1.3σ, respectively. The refined structure is superimposed.
(A) Alignment of A. fumigatus (Af) and S. cerevsiae (Sc) histone H3 and H4 sequences. Vertical lines, colons and commas indicate identical, highly similar and similar residues, respectively. Residues highlighted in yellow and enclosed in solid-line boxes belong to conserved α-helices; and the H3 residues enclosed in the dashed-line box are part of the unwound αN. The H4 residues highlighted in green belong to the C-terminal β-strand formed when bound to Asf1. Terminal residues colored grey are those disordered in the Rtt109-Asf1-H3-H4 complex structure. (B) MALDI-TOF/TOF results of the protein sample prepared from washed crystals indicate the presence of singly ionized species of 15218.57 and 11233.6 Da, which denote intact yeast histones H3 and H4, respectively.
Structures of histone H4 from the complexes with Asf1 in the presence and absence of Rtt109 are shown in yellow and beige, respectively. Nucleosomal H4 (H4_NCP) and H4 from the complex with histone chaperone DAXX (H4_DAXX; PDB id: 4HGA) are shown in green and blue, respectively.
Thermal titration and fitted data from experiments between AfRtt109 and indicated AfAsf1-H3-H4 complexes. Experimentrs were performed at 16°C in a buffer containing 20 mM Tris-HCl, pH 8.0, and 100 mM NaCl. WT AfRtt109 at a concentration of 30 μM was placed in the sample cell, and 20 injections of 2 μl of WT or mutanat AfAsf1 complexes with H3-H4 in the same buffer were carried out with an injection time of 4 s and a 2 min interval between successive injections. AfAsf1-H3-H4 and AfAsf1(R148E)-H3-H4 complexes at concentrations of 0.26 mM and 0.39 mM, respectively, were used.
(A) Quantitation of in vitro H3 acetylation by measuring incorporation of [3H]acetate through scintillation counting. The in vitro HAT assay was carried out using reconstituted complexes of WT and mutant AfAsf1 with WT histones H3 and H4, in the presence of recombinant, purified WT AfRtt109 and [3H]acetyl-CoA. (B) Quantitation of in vitro H3 acetylation with H3 and H4 or their mutants, together with WT AfRtt109 and AfAsf1.
(A) Detection of H3K56 acetylation by Western blot using antibodies against H3K56ac. The HAT assay was performed using AfAsf1-H3-H4 as the substrate in a non-radioactive reaction. (B) The R292E R318E mutant of ScRtt109 displays a high percentage of cells containing spontaneous chromosome breaks (top panel) as indicated by the presence of Rad52-YFP foci (bottom panel).
Highlights.
We solved the crystal structure of Rtt109 bound to Asf1-H3-H4
The N-terminal helix of histone H3 unwinds for K56 acetylation
Asf1 controls H3K56 acetylation by stabilizing the C-terminal tail of H4
Distant H3 residues are crucial for K56 acetylation and chromosomal integrity
Acknowledgments
We thank Drs. Limper and Dahlin for providing AfRtt109 and AfAsf1 plasmids, staff scientists at SSRF beamlines BL17U, BL18U1 and BL19U1 for assistance with data collection. This study is supported by grants from the Natural Science Foundation of China (NSFC) (31430018 to RMX), the Ministry of Science and Technology (MOST) of China (2015CB856200 to NY and 2017YFA0103304 to RMX) and the NIH (GM118015 to ZZ). The study is further augmented by team grants from NSFC (31521002), the Strategic Priority Research Program of Chinese Academy of Sciences (XDB08010100), and a Beijing Municipal Science and Technology Project (Z171100000417001).
Footnotes
Declaration of interests
The authors declare no competing interests.
Author contributions
RMX and ZZ conceived and supervised the project. LZ performed protein purification, crystallization, in vitro binding and ITC experiments. AS performed all histone acetyltransferase assays in vitro and in vivo, and cell growth and Rad52 foci assays. MW solved the structures. HZ purified proteins used for enzymatic activity assays. NY supervised protein preparation, mutagenesis and ITC experiments. LZ, AS, ZZ and RMX wrote the paper. All authors read and edited the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins MW, Carson JJ, English CM, Ramey CJ, Tyler JK. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J Biol Chem. 2007;282:1334–1340. doi: 10.1074/jbc.M608025200. [DOI] [PubMed] [Google Scholar]
- Albaugh BN, Kolonko EM, Denu JM. Kinetic mechanism of the Rtt109-Vps75 histone acetyltransferase-chaperone complex. Biochemistry. 2010;49:6375–6385. doi: 10.1021/bi100381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci U S A. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam A, Logie C. Histone H3 serine 57 and lysine 56 interplay in transcription elongation and recovery from S-phase stress. PLoS One. 2010;5:e10851. doi: 10.1371/journal.pone.0010851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellush JM, Whitehouse I. DNA replication through a chromatin environment. Philos Trans R Soc Lond B Biol Sci. 2017;372 doi: 10.1098/rstb.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen CE, Denu JM. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr Opin Struct Biol. 2008;18:682–689. doi: 10.1016/j.sbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RJ, Zhou H, Han J, Zhang Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell. 2010;37:469–480. doi: 10.1016/j.molcel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Rufiange A, Huang H, Rajashankar KR, Nourani A, Patel DJ. Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2. Genes Dev. 2015;29:1326–1340. doi: 10.1101/gad.261115.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet A, De Koning L, Toedling J, Savignoni A, Berger F, Lemaitre C, O’Sullivan RJ, Karlseder J, Barillot E, Asselain B, et al. Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 2011;30:480–493. doi: 10.1038/emboj.2010.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arcy S, Luger K. Understanding histone acetyltransferase Rtt109 structure and function: how many chaperones does it take? Curr Opin Struct Biol. 2011;21:728–734. doi: 10.1016/j.sbi.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser SJ, Huang H, Lewis PW, Chin JW, Allis CD, Patel DJ. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature. 2012;491:560–565. doi: 10.1038/nature11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol. 2008;28:4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Munoz E, Arboleda-Estudillo Y, Otu HH, Cibelli JB. Cell reprogramming. Histone chaperone ASF1A is required for maintenance of pluripotency and cellular reprogramming. Science. 2014;345:822–825. doi: 10.1126/science.1254745. [DOI] [PubMed] [Google Scholar]
- Gurard-Levin ZA, Quivy JP, Almouzni G. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu Rev Biochem. 2014;83:487–517. doi: 10.1146/annurev-biochem-060713-035536. [DOI] [PubMed] [Google Scholar]
- Han J, Zhang H, Wang Z, Zhou H, Zhang Z. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell. 2013;155:817–829. doi: 10.1016/j.cell.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007a;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J Biol Chem. 2007b;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- Huang H, Stromme CB, Saredi G, Hodl M, Strandsby A, Gonzalez-Aguilera C, Chen S, Groth A, Patel DJ. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat Struct Mol Biol. 2015;22:618–626. doi: 10.1038/nsmb.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolonko EM, Albaugh BN, Lindner SE, Chen Y, Satyshur KA, Arnold KM, Kaufman PD, Keck JL, Denu JM. Catalytic activation of histone acetyltransferase Rtt109 by a histone chaperone. Proc Natl Acad Sci U S A. 2010;107:20275–20280. doi: 10.1073/pnas.1009860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Lercher L, Danilenko N, Kirkpatrick J, Carlomagno T. Structural characterization of the Asf1-Rtt109 interaction and its role in histone acetylation. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yuan YA. Structural insights into histone H3 lysine 56 acetylation by Rtt109. Structure. 2008;16:1503–1510. doi: 10.1016/j.str.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci U S A. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CP, Xiong C, Wang M, Yu Z, Yang N, Chen P, Zhang Z, Li G, Xu RM. Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX. Nat Struct Mol Biol. 2012a;19:1287–1292. doi: 10.1038/nsmb.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WH, Roemer SC, Port AM, Churchill ME. CAF-1-induced oligomerization of histones H3/H4 and mutually exclusive interactions with Asf1 guide H3/H4 transitions among histone chaperones and DNA. Nucleic Acids Res. 2012b;40:11229–11239. doi: 10.1093/nar/gks906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes da Rosa J, Boyartchuk VL, Zhu LJ, Kaufman PD. Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proc Natl Acad Sci U S A. 2010;107:1594–1599. doi: 10.1073/pnas.0912427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Almouzni G. Chromatin and DNA replication. Cold Spring Harb Perspect Biol. 2013;5:a010207. doi: 10.1101/cshperspect.a010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R, Zhou MM. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol. 2014;6:a018762. doi: 10.1101/cshperspect.a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr KA, Carter RA, Boeckh M, Martin P, Corey L. Invasive aspergillosis in allogeneic stem cell transplant recipients: changes in epidemiology and risk factors. Blood. 2002;100:4358–4366. doi: 10.1182/blood-2002-05-1496. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello JA, Sillje HH, Roche DM, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Ozdemir A, Spicuglia S, Lasonder E, Vermeulen M, Campsteijn C, Stunnenberg HG, Logie C. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J Biol Chem. 2005;280:25949–25952. doi: 10.1074/jbc.C500181200. [DOI] [PubMed] [Google Scholar]
- Park YJ, Sudhoff KB, Andrews AJ, Stargell LA, Luger K. Histone chaperone specificity in Rtt109 activation. Nat Struct Mol Biol. 2008;15:957–964. doi: 10.1038/nsmb.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radovani E, Cadorin M, Shams T, El-Rass S, Karsou AR, Kim HS, Kurat CF, Keogh MC, Greenblatt JF, Fillingham JS. The carboxyl terminus of Rtt109 functions in chaperone control of histone acetylation. Eukaryot Cell. 2013;12:654–664. doi: 10.1128/EC.00291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richet N, Liu D, Legrand P, Velours C, Corpet A, Gaubert A, Bakail M, Moal-Raisin G, Guerois R, Compper C, et al. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res. 2015;43:1905–1917. doi: 10.1093/nar/gkv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Serra-Cardona A, Zhang Z. Replication-Coupled Nucleosome Assembly in the Passage of Epigenetic Information and Cell Identity. Trends Biochem Sci. 2017 doi: 10.1016/j.tibs.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- Stavropoulos P, Nagy V, Blobel G, Hoelz A. Molecular basis for the autoregulation of the protein acetyl transferase Rtt109. Proc Natl Acad Sci U S A. 2008;105:12236–12241. doi: 10.1073/pnas.0805813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, Hu Q, Li Q, Thompson JR, Cui G, Fazly A, Davies BA, Botuyan MV, Zhang Z, Mer G. Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106. Nature. 2012;483:104–107. doi: 10.1038/nature10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, Hu Q, Zhou H, Thompson JR, Xu RM, Zhang Z, Mer G. Structure and histone binding properties of the Vps75-Rtt109 chaperone-lysine acetyltransferase complex. J Biol Chem. 2011;286:15625–15629. doi: 10.1074/jbc.C111.220715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Tan Y, Xue Y, Song C, Grunstein M. Acetylated histone H3K56 interacts with Oct4 to promote mouse embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 2013;110:11493–11498. doi: 10.1073/pnas.1309914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Holbert MA, Delgoshaie N, Wurtele H, Guillemette B, Meeth K, Yuan H, Drogaris P, Lee EH, Durette C, et al. Structure of the Rtt109-AcCoA/Vps75 complex and implications for chaperone-mediated histone acetylation. Structure. 2011;19:221–231. doi: 10.1016/j.str.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Holbert MA, Wurtele H, Meeth K, Rocha W, Gharib M, Jiang E, Thibault P, Verreault A, Cole PA, et al. Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP. Nat Struct Mol Biol. 2008;15:738–745. doi: 10.1038/nsmb.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner KG, Trievel RC, Kuo MH, Howard RM, Berger SL, Allis CD, Marmorstein R, Denu JM. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J Biol Chem. 1999;274:18157–18160. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- Teves SS, Weber CM, Henikoff S. Transcribing through the nucleosome. Trends Biochem Sci. 2014;39:577–586. doi: 10.1016/j.tibs.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- Tyler JK, Collins KA, Prasad-Sinha J, Amiott E, Bulger M, Harte PJ, Kobayashi R, Kadonaga JT. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol Cell Biol. 2001;21:6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Kadonaga JT. The “dark side” of chromatin remodeling: repressive effects on transcription. Cell. 1999;99:443–446. doi: 10.1016/s0092-8674(00)81530-5. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Voichek Y, Bar-Ziv R, Barkai N. Expression homeostasis during DNA replication. Science. 2016;351:1087–1090. doi: 10.1126/science.aad1162. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Resch M, Lilyestrom W, Clark N, Hansen JC, Peterson C, Luger K. Structural characterization of H3K56Q nucleosomes and nucleosomal arrays. Biochim Biophys Acta. 2010;1799:480–486. doi: 10.1016/j.bbagrm.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtele H, Tsao S, Lepine G, Mullick A, Tremblay J, Drogaris P, Lee EH, Thibault P, Verreault A, Raymond M. Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy. Nat Med. 2010;16:774–780. doi: 10.1038/nm.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT, et al. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell. 2009;33:417–427. doi: 10.1016/j.molcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) A front view showing the interface between Rt109 and H3-H4. The superimposed yellow, blue and green Cα chains are that of Rtt109, H3 and H4, respectively. The map was contoured at 1.5σ. (B) A back view showing the region surrounding the C-terminal β-strand of H4. The Cα chain of Asf1 is shown in magenta, and the CoA molecule is shown in a bond model.
(A) A section of the 2Fo-Fc electron density map surrounding the active site of Rtt109 contoured at 1.3σ allows unambiguous placement of protein sidechains. Rtt109 residues were shown with the carbon bonds colored yellow, while the pale blue and green carbon bonds indicate that of hitone H3 and H4, respectively. Red and blue bonds represent that of oxygen and nitrogen, and the white and pale green lines in CoA indicate bonds from phosphorus and sulfur atoms. (B) An Fo-Fc map calculated from omitting the residues 91–101 of histone H4 shows strong mainchain density and most of the sidechains of the C-terminal segment of histone H4. The map is displayed at 3σ level, and the refined structure is superimposed as a bond model, with the carbon bond of histone H4 colored in green. (C) Superposition of the 2Fo-Fc (blue) and Fo-Fc (magenta) omit maps surrounding the C-terminal β-strand of histone H4. The same region of H4 as in (B) was omitted in the map calculation, and the Fo-Fc and 2Fo-Fc maps were contoured at 3σ and 1.3σ, respectively. The refined structure is superimposed.
(A) Alignment of A. fumigatus (Af) and S. cerevsiae (Sc) histone H3 and H4 sequences. Vertical lines, colons and commas indicate identical, highly similar and similar residues, respectively. Residues highlighted in yellow and enclosed in solid-line boxes belong to conserved α-helices; and the H3 residues enclosed in the dashed-line box are part of the unwound αN. The H4 residues highlighted in green belong to the C-terminal β-strand formed when bound to Asf1. Terminal residues colored grey are those disordered in the Rtt109-Asf1-H3-H4 complex structure. (B) MALDI-TOF/TOF results of the protein sample prepared from washed crystals indicate the presence of singly ionized species of 15218.57 and 11233.6 Da, which denote intact yeast histones H3 and H4, respectively.
Structures of histone H4 from the complexes with Asf1 in the presence and absence of Rtt109 are shown in yellow and beige, respectively. Nucleosomal H4 (H4_NCP) and H4 from the complex with histone chaperone DAXX (H4_DAXX; PDB id: 4HGA) are shown in green and blue, respectively.
Thermal titration and fitted data from experiments between AfRtt109 and indicated AfAsf1-H3-H4 complexes. Experimentrs were performed at 16°C in a buffer containing 20 mM Tris-HCl, pH 8.0, and 100 mM NaCl. WT AfRtt109 at a concentration of 30 μM was placed in the sample cell, and 20 injections of 2 μl of WT or mutanat AfAsf1 complexes with H3-H4 in the same buffer were carried out with an injection time of 4 s and a 2 min interval between successive injections. AfAsf1-H3-H4 and AfAsf1(R148E)-H3-H4 complexes at concentrations of 0.26 mM and 0.39 mM, respectively, were used.
(A) Quantitation of in vitro H3 acetylation by measuring incorporation of [3H]acetate through scintillation counting. The in vitro HAT assay was carried out using reconstituted complexes of WT and mutant AfAsf1 with WT histones H3 and H4, in the presence of recombinant, purified WT AfRtt109 and [3H]acetyl-CoA. (B) Quantitation of in vitro H3 acetylation with H3 and H4 or their mutants, together with WT AfRtt109 and AfAsf1.
(A) Detection of H3K56 acetylation by Western blot using antibodies against H3K56ac. The HAT assay was performed using AfAsf1-H3-H4 as the substrate in a non-radioactive reaction. (B) The R292E R318E mutant of ScRtt109 displays a high percentage of cells containing spontaneous chromosome breaks (top panel) as indicated by the presence of Rad52-YFP foci (bottom panel).