Abstract
The cellular processes underpinning life are orchestrated by proteins and their interactions. Investigating the structural and dynamic heterogeneity of proteins, despite being key to their function, and their interaction with drugs, poses a fundamental challenge to existing analytical and structural methodologies. We use interferometric scattering microscopy to mass-image single biomolecules in solution with less than 2% mass error, up to 19-kDa resolution, and 1-kDa precision. We resolve oligomeric distributions at high dynamic range, detect small-molecule binding, and mass-image proteins with associated lipids and sugars. These capabilities enable us to characterize the molecular dynamics of processes as diverse as glycoprotein cross-linking, amyloidogenic protein aggregation, and actin polymerization. Interferometric scattering mass spectrometry provides spatially-resolved access to a broad range of biomolecular interactions, one molecule at a time.
Biomolecular interactions and assembly are central to a wide range of physiological and pathological processes spanning length scales from small complexes (1) to the mesoscale (2, 3). Despite considerable developments in techniques capable of providing high-resolution structural information (4), they are typically static and involve averaging over many molecules in the sample, and therefore often do not fully capture the diversity of structures and interactions made. Solution-based ensemble methods enable dynamic studies but lack the resolution of separation required to distinguish different species (5–7). Single molecule methods offer a means to circumvent heterogeneity in both structure and dynamics, and significant progress has been made in terms of characterizing interactions (8) and mechanisms (9, 10). There exists no single-molecule approach, however, capable of quantifying and following the diversity of interactions made by biomolecules with sufficient spatiotemporal accuracy and resolution.
Given sufficient sensitivity, light scattering is an ideal means for detecting and characterizing molecules in low-scattering in vitro conditions because of its universal applicability. In an interferometric detection scheme (Fig. 1A), the scattering signal scales with the polarizability, which is a function of the refractive index and proportional to the particle volume (11). Combining the approximation that single amino acids effectively behave like individual nano-objects with the observation that the specific volumes of amino acids and refractive indices of proteins vary by only ~1% (Fig. S1; Table S1) suggests that the number of amino acids in a polypeptide, and thus its mass, is proportional to its scattering signal. This close relationship between mass and interferometric contrast, which has been predicted (12, 13) and observed (14, 15) to hold coarsely even at the single molecule level, could thus in principle be used to achieve high mass accuracy.
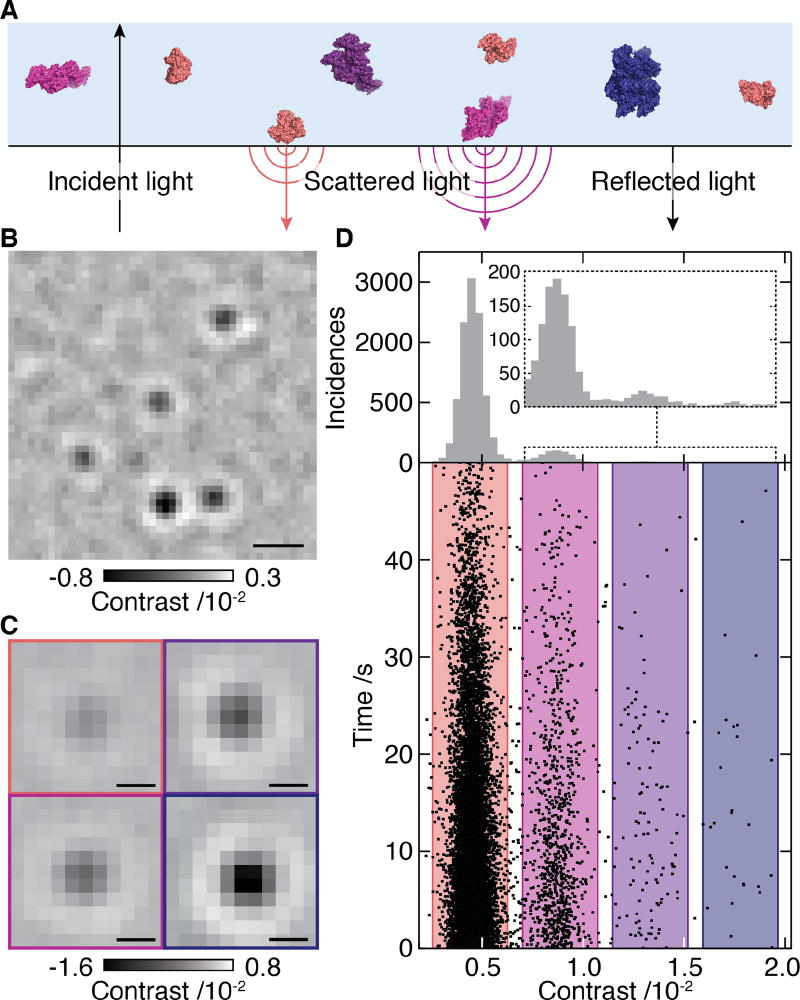
Fig. 1. Concept of interferometric scattering mass spectrometry (iSCAMS).
(A) Schematic of the experimental approach relying on immobilization of individual molecules near a refractive index interface. Oligomeric states are coloured differently for clarity (B) Differential interferometric scattering image of BSA. Scale bar: 0.5 µm. (C) Representative images of monomers, dimers, trimers and tetramers of BSA. Scale bar: 200 nm. (D) Scatter plot of single molecule binding events and their scattering contrasts for 12 nM BSA from 14 movies (lower). Corresponding histogram (N=12209) and zoom of the region for larger species (upper). The reduction in landing rate results from a drop in BSA concentration with time due to the large surface-to-volume ratio of our sample cell (see Supplementary Information).
Building on recent advances in the experimental approach (Fig. S2) that improve imaging contrasts for interferometric scattering microscopy (15, 16), we could obtain high quality images of single proteins as they diffuse from solution to bind non-specifically near the interface consisting of a microscope coverslip and the solution (Fig. 1B, Movie S1). Reaching signal-to-noise ratios >10, even for small proteins such as bovine serum albumin (BSA), combined with an optimized data analysis approach (16), allowed us to extract the scattering contrast for each molecular binding event with high precision (Fig. 1C, Fig. S3). These led to clear signatures of different oligomeric states, shown here for BSA with relative abundances of 88.63%, 9.94%, 1.18% and 0.25% of the detected particles (Fig. 1D). For non-specific binding to an unfunctionalized microscope cover glass as used here, surface attachment was effectively irreversible (12209 binding vs 372 unbinding events). As a result, we could determine (bulk) binding rate constants, which generally exhibited only small variations with oligomeric state and could be accounted for to obtain minor corrections to the recorded mass spectra that yield the solution distribution (Fig. S4). These results, including the detection and quantification of rare complexes such as BSA tetramers, demonstrate the ability of interferometric scattering mass spectrometry (iSCAMS) to characterize solution distributions of oligomeric species and molecular complexes at high dynamic range.
The regular spacing in the contrast histogram of BSA tentatively confirms the expected linear scaling between mass and interferometric contrast. Repeating these measurements for eight different proteins, spanning 53 – 803 kDa, revealed a linear relationship (Fig. 2A, Fig. S5A). The deviation between measured and sequence mass was <5 kDa, resulting in an average error of 1.9%, and no detectable correlation with refractivity in relation to the overall shape of the molecule (Fig. S6A). Even for large structural differences, such as those between the extended and folded conformation of smooth-muscle myosin (530.6 kDa, Fig. 2A and Figs. S5B and S7), we did not find measurable differences in the molecular mass beyond the mass increase expected for addition of glutaraldehyde molecules (Extended conformation: 528.4 ± 16.2 kDa, folded conformation: 579.4 ± 14.8 kDa, Fig. S5B) used to crosslink myosin into the folded conformation. The resolution, as defined by the full-width at half-maximum (FWHM) of the measured contrast reached 19 kDa for streptavidin. In all cases, the resolution was limited by photon shot noise and influenced by molecular mass, increasing from 19 kDa for streptavidin to 102 kDa for thyroglobulin (Fig. S6B,C). The sub-0.5% deviation from sequence mass for species of >100 kDa compares well to native mass spectrometry (17), and demonstrates the intrinsic utility of iSCAMS for the accurate mass measurement of biomolecules with oligomeric resolution.
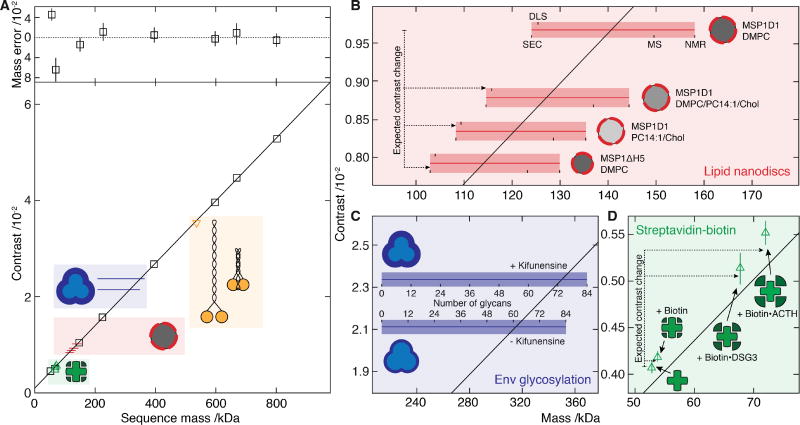
Fig. 2. Characterization of iSCAMS accuracy, precision, and dependence on molecular shape and identity.
(A) Contrast vs molecular mass including proteins used for mass calibration (black), and characterization of shape dependence (yellow), protein-ligand binding (green), lipid nanodisc composition (red) and glycosylation (blue). Mass error (upper panel) is given as a percentage of the sequence mass relative to the given linear fit. (B) Nanodisc mass-measurement for different lipid compositions and protein belts. Masses obtained by alternative methodologies for MSP1D1/DMPC are marked and extrapolated to the other compositions. The horizontal bars indicate the expected mass range as a function of characterization technique, with the thin bar indicating the contrast measured, and the thick bar representative of the measurement uncertainty in terms of the standard error of the mean for repeated experiments. For each sample, the upper text denotes the membrane scaffold protein (MSP) used, and the lower the lipids in the nanodisc. (C) Recorded differential contrast for Env expressed in the presence or absence of kifunensine, and associated mass ranges expected for different glycosylation levels as defined for C. (D) Mass-sensitive detection of ligand binding using the biotin-streptavidin system according to the sequence mass of streptavidin and the masses of biotin and two biotinylated peptides relative to the calibration obtained from A. Abbreviations used are summarized in Supplementary Table S8.
Moving beyond species composed solely of amino acids, lipid nanodiscs represent an ideal system for testing the broad applicability of iSCAMS due to their flexibility in terms of polypeptide and lipid content (18). For nanodiscs composed of the MSP1D1 belt protein and DMPC lipids, we obtained a mass of 141.0 ± 1.6 kDa, in good agreement with the range of masses reported by other methods, spanning 124 – 158 kDa (Fig. 2B and Fig. S5D). Replacing MSP1D1 with the smaller MSP1ΔH5 reduces the nanodisc diameter and the lipid content by ~20%, after accounting for the thickness of the protein belt (19). Given the masses of MSP1D1 and MSP1ΔH5 (47 and 42 kDa, respectively), we predicted a mass for the MSP1ΔH5 nanodisc of 113.6 kDa, in excellent agreement with our measurement (114.1 ± 1.9 kDa). Notably, mass shifts associated with changes in lipid composition, such as those introduced by partially unsaturated lipids and cholesterol, matched those predicted from the assembly ratios (Fig. 2B, Tables S2–S6).
To see whether our approach also applies to solvent-exposed moieties that experience a different dielectric environment to those buried within a protein, we selected the HIV envelope glycoprotein complex (Env), which is a trimer of gp41–gp120 heterodimers. Env is extensively N-glycosylated, with the carbohydrates contributing to almost half of its mass (20). For an Env trimer mimic expressed in the presence of kifunensine, a mannosidase inhibitor that leads predominantly to unprocessed Man9GlcNAc2 glycans (Fig. S8), we recorded a mass of 350.0 ± 5.7 kDa. Making the crude approximation that glycans and amino-acids have similar polarizabilities, this corresponds to a glycan occupancy of 74 ± 3 out of 84 possible sites (Fig. 2C and Fig. S5E), consistent with recent observations of high occupancy for gp120 expressed with kifunensine (21). For Env expressed without kifunensine we recorded a lower mass of 315.3 ± 10.5 kDa. The mass difference can only in part be attributed to the lower average mass of the processed glycans (Fig. S8) and yields a total N-glycan occupancy of 61 ± 6. While the exact values for occupancy are beholden to our calibration (Fig. 2A), the presence of unoccupied sites is consistent with their observation in proteomics data (22).
The high precision of 1.8 ± 0.5% with respect to the protein mass (Fig. 2A), indicates the potential for direct detection of small-molecule binding. To probe the current limits of iSCAMS in terms of precision, we therefore examined the biotin-streptavidin system (Fig. 2D, Fig. S5C), and measured masses for streptavidin in the absence (55.7 ± 1.1 kDa) and presence (57.4 ± 0.9 kDa) of biotin. This corresponds to a difference of 1.7 ± 1.4 kDa, in good agreement with the expected 0.98 kDa for complete occupancy of the four binding sites. Upon addition of two different biotinylated peptides (3705.9 Da and 4767.4 Da), we obtained increases of 16.1 ± 2.8 kDa and 22.0 ± 2.2 kDa (compared to 14.8 kDa and 19.1 kDa expected) (Fig. 2D, Fig. S5C). These data show that iSCAMS can detect the association of kDa-sized ligands, demonstrating its suitability for highly sensitive ligand-binding studies in solution.
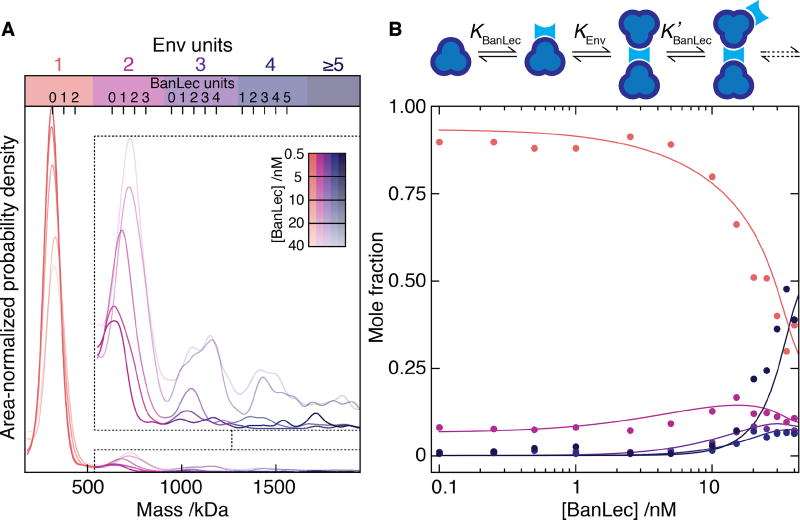
After having established the capabilities of iSCMAS, we sought to test it on more complex systems that are difficult to quantitatively assess with existing techniques as a consequence of heterogeneity and multi-step assembly mechanisms (Fig. 3). In addition, we aimed to monitor nucleation and polymerization dynamics of mesoscopic structures down to the single molecule level, which are challenging because of the simultaneous requirement for high dynamic range, imaging speed and direct correlation between the observed signals and the associated molecular events. The biotin-streptavidin system exhibits nearly covalent binding, raising the question whether iSCAMS is capable of not only determining mass distributions but also of quantifying weaker equilibria, as often encountered for protein-protein interactions. We therefore investigated the interaction of Env with the anti-viral lectin BanLec, which neutralises HIV by binding to surface N-glycans(23, 24) via an unknown mechanism. We could monitor the interactions and short-lived complexes prior to aggregation, with the addition of BanLec to Env resulting in a reduction of single Env units coupled to the appearance of dimers and higher-order assemblies (Fig. 3A). The experimental oligomeric evolution coupled with a simple model (Fig. 3B) enabled us to extract the underlying association constants (KBanLec = 0.12 nM−1, KEnv = 8 nM−1, K'BanLec = 0.4 nM−1), in good agreement with recent bulk studies (KBanLec = 0.19 nM−1), which also observed signatures of and estimated the energetics of a secondary binding event (K2 = 2.85 nM−1) (25). Our ability to follow and model the evolution of different oligomeric species allowed us to directly extract the interaction mechanism and the energetics underlying the lectin-glycoprotein interaction, despite the heterogeneity of this multi-component system. As a result, we can show that binding of Env to BanLec that is already bound to Env is much stronger than to free BanLec, a key characteristic of cooperative behavior. Moreover, the mass resolution of our approach enabled us to quantify the number of BanLecs bound per dimer (1–2), trimer (2–3) and tetramer (3–4) of Env, demonstrating bivalent activity. These results are directly relevant to the characterization and optimization of anti-retrovirals, given that multivalency and aggregation have been proposed to be directly linked to neutralization potency(25). We anticipate similar quantitative insights to be achievable for other therapeutic target proteins and protein-protein interactions in general.
Fig. 3. Single molecule mass analysis of heterogeneous protein assembly.
(A) Mass distributions for Env in the presence of 0.5 – 40 nM BanLec monomer. Inset: zoom alongside expected positions for multiples of bound BanLec tetramers. (B) Oligomeric fractions colored according to A vs BanLec concentration including predictions (solid) using the given cooperative model.
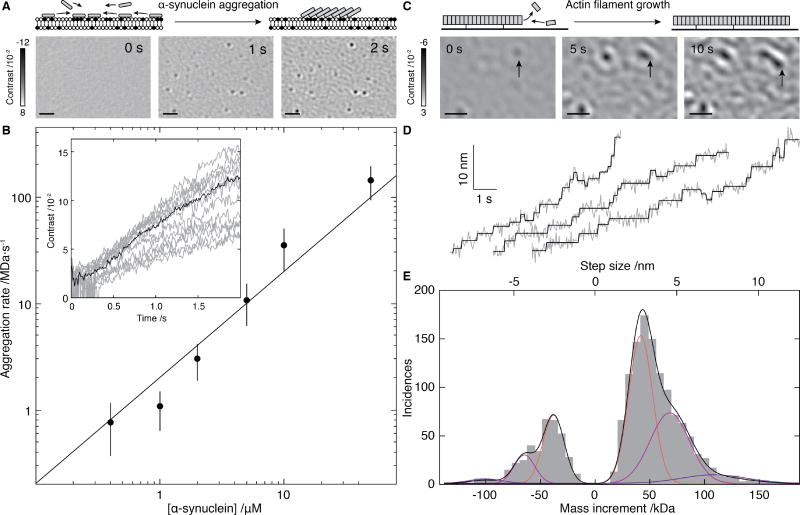
An advantage of our imaging-based approach stems from its ability to time-resolve mass changes in a position- and local concentration-sensitive manner. This enables us to examine surface-catalyzed nucleation events that may eventually lead to amyloid formation (26). Previous studies using fluorescence labeling, found aggregates of ~0.6 µm diameter within a minute of addition of the amyloidogenic protein α-synuclein at 10 µM to an appropriately charged bilayer (27). Upon adding α-synuclein to a planar, negatively charged DOPC/DOPS (3:1) membranes at physiological pH, we observed the appearance and growth of nanoscopic objects within seconds, even at low µM concentrations (Fig. 4A, Movie S2). While we were unable to determine the sizes of initial nucleating species or individual assembly steps, given the low molecular mass of α-synuclein (14 kDa), we could nevertheless monitor the nanoscale formation of associated structures in the range of the hundreds of kDa in time and determine the kinetics (Fig. 4B). Growth of these clusters was uniform across the field of view, with the initial rates following expectations for a first-order process (Fig. 4B and Fig. S9A), pointing towards a simple growth mechanism. We did not detect such structures on neutral, DOPC-only bilayers, and found evidence for thioflavin-T positive aggregates after overnight incubation (Fig. S9B), suggesting that our assay probes early stages of amyloid assembly.
Fig. 4. Mass-imaging of mesoscopic dynamics.
(A) Schematic of and iSCAMS images for α-synuclein (1 µM) aggregation on a negatively charged bilayer membrane. (B) Initial growth rate vs. α-synuclein concentration alongside the best fit assuming first order kinetics (solid). Inset: Individual growth trajectories (grey) and average (black) for 21 particles from A. (C) Schematic and iSCAMS images of actin polymerization. The arrow highlights a growing filament. (D) Representative traces of actin filament tip position (grey) and corresponding detected steps (black). (E) Step and mass histogram from 1523 steps and 33 filaments including a fit to a Gaussian mixture model (black) and individual contributions (colored). Scale bars: 1 µm. In these experiments, background correction involved removal of the static background prior to acquisition, rather than continuous differential imaging as in Figs. 2 and 3 (see Supplementary Information).
At the extremes of our current sensitivity, iSCAMS enables mass-imaging of mesoscopic self-assembly, molecule-by-molecule. In an actin polymerization assay, subtraction of the constant background revealed growth of surface-immobilized filaments. In contrast to α-synuclein, where the growth of interest took place within a diffraction-limited spot, here we could quantify length changes of filaments larger than the diffraction limit upon the attachment and detachment of actin subunits (Fig. 4C, Fig. S10C, Movie S3). We observed distinct, step-wise changes in the filament length (Fig. 4D, Supplementary Fig. S10D–F and Movie S4), the most frequent forward and backward step sizes in the traces being 3.0 ± 0.8 nm and 2.7 ± 0.7 nm, respectively, remarkably close to the expected length increase of 2.7 nm upon binding of a single actin subunit to a filament (Fig. 4E). Detection of larger step sizes represents the addition of multiple actin subunits within our detection time window. The contrast changes associated with the different step sizes corresponded to mass changes of one, two, or three actin monomers binding to and unbinding from the tip of the growing filaments during acquisition (Fig. S10G,H). Even though we cannot yet distinguish between models invoking monomer (28) or oligomer (29) addition to a growing filament at our current level of spatio-temporal resolution, these results demonstrate the capability of iSCAMS for quantitatively imaging mesoscopic dynamics and how they are influenced by associated proteins at the single molecule level.
We anticipate that combining iSCAMS with established surface modifications (30) will dramatically expand its capabilities. Passivation decreases surface binding probabilities and thereby should provide access to much higher analyte concentrations (>µM), while surface activation will reduce measurement times at low concentrations (<nM). Specific functionalization and immobilization of individual subunits or binding partners could also allow for the determination of on and off rates in addition to equilibrium constants, and enable targeted detection in the presence of other analytes. Although studies within complex three-dimensional environments like the cell may prove to be beyond reach, these advances will make iSCAMS a powerful approach for dynamic in vitro studies of biomolecular interactions, assembly and structure at the single molecule level.
Supplementary Material
Movie S1 First 30 s (real time) of a representative ratiometric movie of BSA molecules binding non-specifically to a glass coverslip. The field of view is 3.2 × 9.7 µm, and the contrast scale is from −0.0075 to +0.003. The raw frames were saved at 100 Hz, and the sliding ratiometric processing was applied with a frame averaging of 5 frames. The movie is played back at 30 Hz.
Movie S2 Representative background-divided movie of addition of 1 µM α-synuclein to a supported lipid bilayer, showing the initial perturbation of the sample due to manual addition of the α-synuclein followed by growth of aggregates on the bilayer. The field of view is 11.9 × 11.9 µm, and the contrast scale is from −0.12 to +0.08. The frames were recorded at 100 Hz, and are played back at 33 Hz.
Movie S3 500 nM actin (+1% biotin-actin) polymerizing on avidin-biotin-PEG-coated glass. The size is 3.1 × 4.4 µm (scale bar = 1 µm), and the contrast scale is from −0.05 to +0.04. Images were recorded at 468 Hz then averaged to 3 Hz, and are played back at 25 Hz.
Movie S4 Growing actin filament tips corresponding to the traces shown in Fig 4D. The actin concentration was 300 nM, the scale bar is 250 nm, and the contrast scale is from −0.05 to +0.04. Images were recorded at 468 Hz then averaged to 58.5 Hz (as for the analysis performed for Fig 4D), and are played in real time.
Acknowledgments
JRS thanks Fang Zhang for technical assistance and the NHLBI electron microscopy core facility. GY was supported by a Zvi and Ofra Meitar Magdalen Graduate Scholarship; NH by a DFG research fellowship (HU 2462/1-1). EGM thanks the Swedish Research Council and the European Commission for a Marie Skłodowska Curie International Career Grant (2015-00559). MPC is a Clarendon Scholar supported by the Oxford University Press. SAC is supported by the Biotechnology and Biological Sciences Research Council and Waters Corp by the iCASE studentship BB/L017067/1 to JLPB. JA and MC were supported by the National Institute of Allergy and Infectious Diseases (Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery grant UM1AI100663). JRS was supported by NHLBI Intramural program HL0001786. CE is supported by a SNSF advanced postdoctoral mobility fellowship (P300PA160979). PS is funded by an ERC Consolidator Grant (NeuroInCellNMR, 647474). JLPB thanks the Engineering and Physical Sciences Research Council for EP/J01835X/1. PK was supported by an ERC Starting Investigator Grant (Nanoscope, 337577).
Conceptualization, WBS, JLPB and PK; Methodology GY, NH, DC, JA, EGM, CE, PS, MRG, WBS, JLPB, PK; Software GY, NH; Validation GY, NH, JLPB, PK; Formal analysis, GY, NH, AT, AA, AO, JA, EGM, MRG; Investigation GY, NH, DC, AF, JA, AT, AA, NB, YT, CE; Resources MPC, SAC, OT, JA, MC, NB, YT, JRS, CE, PS, LF, RR, WS; Writing - original draft GY, JLPB, PK; Writing - review and editing GY, NH, AF, AO, EGM, MPC, OT, MC, JRS, CE, PS, RR, MRG, WBS, JLPB, PK; Visualization GY, NH, JLPB, PK; Supervision PK.
DC, JLPB and PK have filed a patent on the contrast enhancement methodology and its application to mass measurement of single biomolecules.
Footnotes
All other authors declare no competing interests.
All data is available in the main text or the supplementary information with raw data provided at https://doi.org/10.5287/bodleian:PmA5Va0a2
References
- 1.Ahnert SE, Marsh JA, Hernandez H, Robinson CV, Teichmann SA. Principles of assembly reveal a periodic table of protein complexes. Science. 2015;350:aaa2245. doi: 10.1126/science.aaa2245. [DOI] [PubMed] [Google Scholar]
- 2.Rottner K, Faix J, Bogdan S, Linder S, Kerkhoff E. Actin assembly mechanisms at a glance. J. Cell Sci. 2017;130:3427–3435. doi: 10.1242/jcs.206433. [DOI] [PubMed] [Google Scholar]
- 3.Hemmat M, Castle BT, Odde DJ. Microtubule dynamics: moving toward a multi-scale approach. Curr. Opin. Cell Biol. 2018;50:8–13. doi: 10.1016/j.ceb.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H-W, Wang J-W. How cryo-electron microscopy and X-ray crystallography complement each other. Protein Sci. 2017;26:32–39. doi: 10.1002/pro.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanno DM, Levitus M. Protein Oligomerization Equilibria and Kinetics Investigated by Fluorescence Correlation Spectroscopy: A Mathematical Treatment. J. Phys. Chem. B. 2014;118:12404–12415. doi: 10.1021/jp507741r. [DOI] [PubMed] [Google Scholar]
- 6.Chakraborty M, et al. Protein Oligomerization Monitored by Fluorescence Fluctuation Spectroscopy: Self-Assembly of Rubisco Activase. Biophys. J. 2012;103:949–958. doi: 10.1016/j.bpj.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen SIA, Vendruscolo M, Dobson CM, Knowles TPJ. From Macroscopic Measurements to Microscopic Mechanisms of Protein Aggregation. J. Mol. Biol. 2012;421:160–171. doi: 10.1016/j.jmb.2012.02.031. [DOI] [PubMed] [Google Scholar]
- 8.Jain A, et al. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuler B, Eaton WA. Protein folding studied by single-molecule FRET. Curr. Opin. Struct. Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenleaf WJ, Woodside MT, Block SM. High-resolution, single-molecule measurements of biomolecular motion. Annu. Rev. Biophys. Biochem. Struct. 2007;36:171–190. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohren CF, Huffman DR. Absorption and scattering of light by small particles. Wiley Interscience; New York: 1983. [Google Scholar]
- 12.Kukura P, et al. High-speed nanoscopic tracking of the position and orientation of a single virus. Nat. Meth. 2009;6:923–927. doi: 10.1038/nmeth.1395. [DOI] [PubMed] [Google Scholar]
- 13.Ortega Arroyo J, et al. Label-free, all-optical detection, imaging, and tracking of a single protein. Nano Lett. 2014;14:2065–2070. doi: 10.1021/nl500234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piliarik M, Sandoghdar V. Direct optical sensing of single unlabelled proteins and super-resolution imaging of their binding sites. Nat. Commun. 2014;5:4495. doi: 10.1038/ncomms5495. [DOI] [PubMed] [Google Scholar]
- 15.Liebel M, Hugall JT, van Hulst NF. Ultrasensitive Label-Free Nanosensing and High-Speed Tracking of Single Proteins. Nano Lett. 2017;17:1277–1281. doi: 10.1021/acs.nanolett.6b05040. [DOI] [PubMed] [Google Scholar]
- 16.Cole D, Young G, Weigel A, Sebesta A, Kukura P. Label-Free Single-Molecule Imaging with Numerical-Aperture Shaped Interferometric Scattering Microscopy. ACS Photonics. 2017;4:211–216. doi: 10.1021/acsphotonics.6b00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van de Waterbeemd M, et al. High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat. Meth. 2017;14:283–286. doi: 10.1038/nmeth.4147. [DOI] [PubMed] [Google Scholar]
- 18.Denisov IG, Sligar SG. Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev. 2017;117:4669–4713. doi: 10.1021/acs.chemrev.6b00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bibow S, et al. Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I. Nat. Struct. Mol. Biol. 2017;24:187–193. doi: 10.1038/nsmb.3345. [DOI] [PubMed] [Google Scholar]
- 20.Lasky LA, et al. Neutralization of the Aids Retrovirus by Antibodies to a Recombinant Envelope Glycoprotein. Science. 1986;233:209–212. doi: 10.1126/science.3014647. [DOI] [PubMed] [Google Scholar]
- 21.Struwe WB, Stuckmann A, Behrens A-J, Pagel K, Crispin M. Global N-Glycan Site Occupancy of HIV-1 gp120 by Metabolic Engineering and High-Resolution Intact Mass Spectrometry. ACS Chem. Biol. 2017;12:357–361. doi: 10.1021/acschembio.6b00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao L, et al. Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat. Commun. 2017;8:14954. doi: 10.1038/ncomms14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanson MD, Winter HC, Goldstein IJ, Markovitz DM. A lectin isolated from bananas is a potent inhibitor of HIV replication. J. Biol. Chem. 2010;285:8646–8655. doi: 10.1074/jbc.M109.034926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopper JTS, et al. The Tetrameric Plant Lectin BanLec Neutralizes HIV through Bidentate Binding to Specific Viral Glycans. Structure. 2017;25:773–782. doi: 10.1016/j.str.2017.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lusvarghi S, et al. Binding Site Geometry and Subdomain Valency Control Effects of Neutralizing Lectins on HIV-1 Viral Particles. ACS Infect. Dis. 2016;2:882–891. doi: 10.1021/acsinfecdis.6b00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galvagnion C, et al. Lipid vesicles trigger alpha-synuclein aggregation by stimulating primary nucleation. Nat. Chem. Biol. 2015;11:229–234. doi: 10.1038/nchembio.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer A, Schilderink N, Claessens MMAE, Subramaniam V. Membrane-Bound Alpha Synuclein Clusters Induce Impaired Lipid Diffusion and Increased Lipid Packing. Biophys. J. 2016;111:2440–2449. doi: 10.1016/j.bpj.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasai M, Oosawa F, Asakura S. Cooperative Nature of G-F Transformation of Actin. Biochim. Biophys. Acta. 1962;57:22–31. doi: 10.1016/0006-3002(62)91073-9. [DOI] [PubMed] [Google Scholar]
- 29.Erickson HP. Co-Operativity in Protein-Protein Association - the Structure and Stability of the Actin Filament. J. Mol. Biol. 1989;206:465–474. doi: 10.1016/0022-2836(89)90494-4. [DOI] [PubMed] [Google Scholar]
- 30.Hua B, et al. An improved surface passivation method for single-molecule studies. Nat. Meth. 2014;11:1233–1236. doi: 10.1038/nmeth.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1 First 30 s (real time) of a representative ratiometric movie of BSA molecules binding non-specifically to a glass coverslip. The field of view is 3.2 × 9.7 µm, and the contrast scale is from −0.0075 to +0.003. The raw frames were saved at 100 Hz, and the sliding ratiometric processing was applied with a frame averaging of 5 frames. The movie is played back at 30 Hz.
Movie S2 Representative background-divided movie of addition of 1 µM α-synuclein to a supported lipid bilayer, showing the initial perturbation of the sample due to manual addition of the α-synuclein followed by growth of aggregates on the bilayer. The field of view is 11.9 × 11.9 µm, and the contrast scale is from −0.12 to +0.08. The frames were recorded at 100 Hz, and are played back at 33 Hz.
Movie S3 500 nM actin (+1% biotin-actin) polymerizing on avidin-biotin-PEG-coated glass. The size is 3.1 × 4.4 µm (scale bar = 1 µm), and the contrast scale is from −0.05 to +0.04. Images were recorded at 468 Hz then averaged to 3 Hz, and are played back at 25 Hz.
Movie S4 Growing actin filament tips corresponding to the traces shown in Fig 4D. The actin concentration was 300 nM, the scale bar is 250 nm, and the contrast scale is from −0.05 to +0.04. Images were recorded at 468 Hz then averaged to 58.5 Hz (as for the analysis performed for Fig 4D), and are played in real time.