Abstract
All organisms must regulate the cellular uptake, efflux, and intracellular trafficking of essential elements, including d-block metal ions. In bacteria, such regulation is achieved by the action of metal-responsive transcriptional regulators. Among several families of zinc-responsive transcription factors, the ‘zinc uptake regulator’ Zur is the most widespread. Zur normally represses transcription in its zinc-bound form, in which DNA-binding affinity is enhanced allosterically. Experimental and bioinformatic searches for Zur-regulated genes have revealed that in many cases, Zur proteins govern zinc homeostasis in a much more profound way than merely through the expression of uptake systems. Zur regulons also comprise biosynthetic clusters for metallophore synthesis, ribosomal proteins, enzymes, and virulence factors. In recognition of the importance of zinc homeostasis at the host–pathogen interface, studying Zur regulons of pathogenic bacteria is a particularly active current research area.
Keywords: bacteria, metal ions, zinc-responsive transcription factors, zinc uptake regulator, Zur
Introduction
For a long time, there has been a prevailing assumption that zinc is not a particularly important element for prokaryotes. Indeed, as recently as 2010, the contention that ‘the akaryotic superkingdoms of Archaea and Bacteria eschew Zn’ was highlighted as a finding of a phylogenomic analysis of protein structures [1]. It is certainly true that the proportion of zinc-requiring proteins in prokaryotes (5–6%) is significantly lower than that in eukaryotes (9–10%) [2], but this should not detract us from the fact that zinc is also essential for bacteria, and that bacterial zinc homeostasis plays rather critical roles in a variety of contexts. Much recent attention has focused on pathogenic bacteria [3], where both extremely efficient zinc acquisition and enhanced zinc tolerance contribute to survival in the host, virulence, and overall pathogenicity. In zinc-poor environments, bacterial cells are capable of concentrating zinc by several thousand-fold [4]; such highly efficient zinc acquisition is critical in the face of the host's immune response where the availability of zinc, iron, and manganese is severely restricted [5,6], a phenomenon referred to as ‘nutritional immunity’ [7]. Conversely, high zinc concentrations are part of the toxic cocktail that macrophages use to kill pathogens encapsulated in their phagosomes [8] — but not always successfully. Zinc toxicity is thought to be at least partially due to mis-metallation of other metalloproteins, as Zn2+ forms more stable complexes than most other essential metal ions [9], so intracellular concentrations must be stringently regulated.
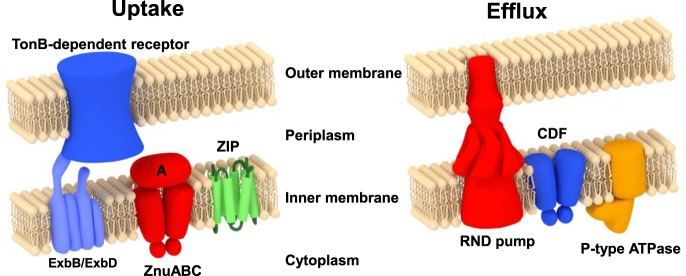
Zinc homeostasis in bacteria is maintained by means of a few critical processes, including zinc import and export (Figure 1), intracellular zinc binding, and zinc-sensing [3,10,11]. The fluxes of Zn2+ in and out of the cell are, in the first instance, controlled by the levels at which importer and exporter proteins are present. In bacteria, these levels are to a large extent controlled at the transcriptional level [12], with available intracellular Zn2+ being sensed by several types of zinc-responsive transcription factors [13]. These include members of the ferric uptake regulator (Fur) family (Zur; [4,14–17]) and the MarR/SlyA family (AdcR) [18–20] to up-regulate zinc import, and members of the MerR (ZntR) and ArsR/SmtB families (SmtB, ZiaR, and CzrA; [21–24]), to increase zinc export and/or intracellular sequestration. Besides Zur, the Fur family (COG0735) [25,26] also includes Fur [27–29], Nur [30,31], and Mur [32,33], which sense other metal cations (Fe2+, Ni2+, and Mn2+, respectively). Further members are PerR [34,35] and Irr [36], which sense cytoplasmic peroxide and haem, respectively.
Figure 1. Overview of the major players in bacterial zinc uptake and efflux, illustrated for a Gram-negative bacterium.
Proteins for import include members of the ZIP (zinc-iron permease) family and members of the ATP-binding cassette (ABC) superfamily. The latter systems consist of a membrane-bound permease, an ATPase, and a protein that is periplasmic in Gram-negative bacteria or on the cell surface in Gram-positive bacteria. These systems are usually named ZnuABC (Gram-negative bacteria) or AdcABC (Gram-positive bacteria), although this distinction is not consistently adhered to. A third label used frequently for such zinc importers is TroABC. Exporters include P-type ATPases, members of the cation–diffusion facilitator (CDF) family, and tripartite RND (root–nodulation–cell division) systems [3,10]. Regulatory proteins and further processes are explained in the main text.
In principle, zinc-sensing transcription factors can work as either repressors or activators. The unifying point is that in each case Zn2+ binding alters the affinity to the cognate DNA. This is achieved by allostery; the bound Zn2+ alters the conformation and/or the structural dynamics of the protein, which can either enhance or decrease the affinity for specific DNA sequences [37]. Like other Fur family proteins, Zur proteins act predominantly as transcriptional repressors when Zn2+ is bound, by blocking the binding site for the RNA polymerase transcription initiation complex (Figure 2). Often, the Zur-specific DNA sequences (Zur boxes) overlap with the −35 position in the promoter regions of regulated genes. This review examines structures and properties of Zur proteins and their typical mode of action, and gives a comprehensive overview of which genes are Zur-regulated. This includes the recent discovery that Zur proteins may also act as transcriptional activators.
Figure 2. Schematic illustration of canonical regulation of transcription by Zur.
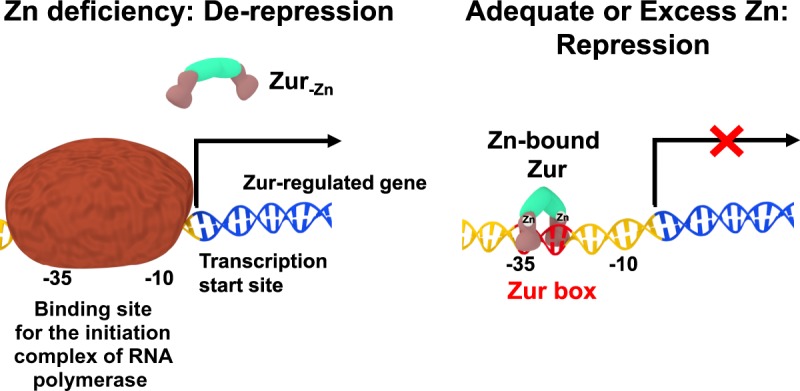
Zinc-bound Zur (right-hand panel; see below for further details) represses transcription by binding to specific DNA sequences (Zur boxes) in the promoter region of Zur-regulated genes and thus inhibits initiation of transcription. When cells are deprived of zinc, demetallated Zur has a dramatically reduced affinity for DNA, allowing transcription to occur.
Structures and mode of action of Zur proteins
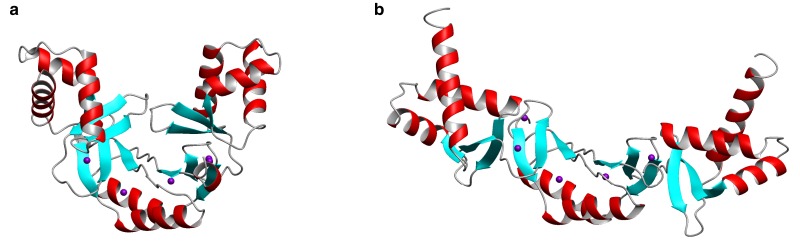
Among many 3D structures for Fur family proteins [26], the structures of three Zur proteins have been determined by X-ray crystallography, namely those of Mycobacterium tuberculosis FurB (MtZur) [15], Streptomyces coelicolor (ScZur; Figure 3) [38], and Escherichia coli (EcZur) [17]. All three proteins are homodimeric in solution (Table 1) and at least dimeric in the crystal. Each monomer has two domains: an N-terminal DNA-binding domain that is predominantly α-helical and a C-terminal dimerisation domain containing a three-stranded β-sheet. Its longest β-strand pairs up with its counterpart in the dimer. The two domains in each monomer are connected by a hinge.
Figure 3. X-ray crystal structure of dimeric S. coelicolor Zur (pdb 3mwm) [38].
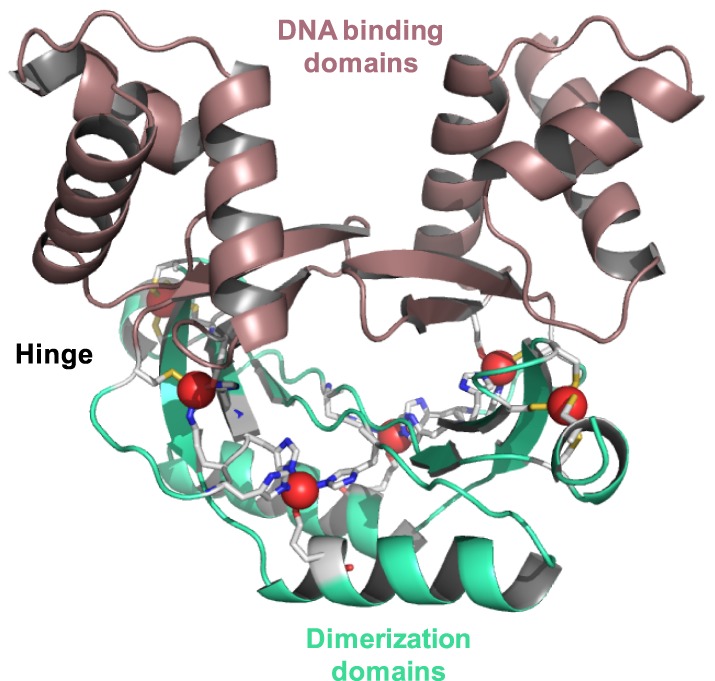
DNA-binding domains are shown in maroon and dimerisation domains in green. Zinc ions are shown as red spheres, nitrogen atoms in blue, sulfur in yellow, and carbon in light grey.
Table 1. Summarised data for expression constructs and purification protocols, oligomerisation states, and Zn stoichiometry of Zur proteins from different organisms.
| Organism | Expression and purification | Oligomerisation | Stoichiometry (Zn per monomer) | Ref. |
|---|---|---|---|---|
| Mycobacterium tuberculosis | GST-fusion; glutathione sepharose 4B resin | n.d.; assumed dimeric | 1.8 ± 0.2 (Bradford assay and FAAS) | [39] |
| Mycobacterium tuberculosis | His-tagged; Ni-IMAC, TEV-cleavage, SEC | Dimer: X-ray crystallography | 3 (X-ray crystallography) 1 (post EDTA treatment; μPIXE) |
[15] |
| Streptomyces coelicolor | His-tagged; Ni-charged Chelex-100, elution with imidazole, dialysis against 5 mM EDTA | Dimer (analytical ultracentrifugation). No indication of self-association. | n.d. | [16] |
| Streptomyces coelicolor | No tag; Ni-charged NTA column; elution with imidazole gradient; dialysis; no EDTA during purification | Dimer (X-ray crystallography and SEC) for WT, D, and M site mutants. C90S mutant monomeric | 3 (X-ray crystallography) 2.4 (WT; ICP-OES) 1.2 (EDTA treated) 1.5 (H84A mutant; D site) 1.0 (C79S mutant; M site) 0.25 (C90S mutant; C site) |
[38] |
| Bacillus subtilis | Cell lysis in presence of 2 mM EDTA and 2 mM DTT; heparin affinity, SEC, AEX, dialysis |
Dimer (SEC) | 0.5–0.8 initially bound; additional 1.4 Zn by titration, giving approximately 2 Zn per monomer ([P] by A277, [M] by PAR assay and titration with FluoZin3) |
[40,41] |
| Streptococcus suis | His- or GST-tag. Ni-IMAC or glutathione Sepharose 4B (PBS). GST-tag removed |
Dimer (chemical cross-linking assays) | n.d. | [42] |
| Escherichia coli | Tag-less expression. Lysis in the presence of 6 M urea, and 100 mM DTT, then refolded in presence of 100 μM Zn2+. AEX, HIC and SEC |
n.d., assumed dimeric | 1.4–1.8 (standardised Bradford assay and ICP-AES) | [43] |
| Escherichia coli | Similar to [43] | Dimer (X-ray crystallography) C103S mutant monomeric |
2 (X-ray crystallography) 2.8 (WT; ICP-MS) 0.7 (EDTA-treated WT) 2.1 (C103S mutant) 0.0 (EDTA-treated C103S mutant) 2.5 (C88S mutant) 0.6 (EDTA-treated C88S mutant) |
[17] |
| Salmonella enterica | His-tagged. Lysis: 1 mM EDTA IMAC, SEC, heparin affinity |
n.d., assumed dimeric | Ca. 1 (ICP-MS) Capacity to bind up to two more per monomer |
[44,45] |
| Paracoccus denitrificans | MBP fusion tag. Lysis: 1 mM EDTA. Amylose resin column. Tag cleaved by Factor Xa protease, AEX. Apo-Zur by dialysis | Dimer (SEC) | Ca. 1 (ICP-OES), capacity to bind one more Zn per monomer | [46] |
| Synechocystis sp. PCC6803 | Tag-less expression. Lysis: 100 mM NaCl, 5 mM DTT, 1 mM EDTA, heparin affinity and SEC |
n.d., assumed dimeric | 1.02 ± 0.15 (ICP-MS) Capacity to bind at least one more Zn per monomer |
[47] |
| Anabaena sp. PCC 7120 | His-tagged. Cell lysis in presence of 2 M GdnHCl (pH 8). Zn-IMAC, dialysis pH 5.5 |
Mostly monomer, some dimer (SDS–PAGE, denaturing conditions) | 1 (ICP-MS) Up to two more binding sites per monomer |
[48] |
Abbreviations: AEX: anion-exchange chromatography; DTT: dithiothreitol; EDTA: ethylenediaminetetraacetic acid; FAAS: flame atomic absorption spectroscopy; GdnHCl: guanidinium hydrochloride; GST: Glutathione S-transferase; HIC: hydrophobic interaction chromatography; His-tag: polyhistidine-tag; ICP-AES/ICP-OES: inductively coupled plasma atomic/optical emission spectroscopy; ICP-MS: inductively coupled plasma mass spectrometry; IMAC: immobilised metal ion affinity chromatography; MBP: maltose-binding protein; n.d: not determined; μPIXE: micro-proton-induced X-ray emission; PAR: pyridyl-azo-resorcinol; SDS–PAGE: sodium dodecyl sulfate–polyacrylamide gel electrophoresis; SEC: size-exclusion chromatography; TEV: tobacco etch virus; WT: wild type.
Like other members of the Fur family, Zur dimers can adopt at least two conformations, a ‘closed’ one with high DNA-binding affinity and an ‘open’ one with a low DNA-binding affinity (Figure 4). It has been suggested that the DNA-binding domains in Fur-family transcription factors are highly mobile in solution [49,50], with the structures found in single crystals representing ‘frozen-out’ states from an entire range of conformations. Metal binding shifts the conformational equilibrium towards the closed, high-affinity conformation. In the case of Zur, this provides the basis for the allosteric sensing of Zn2+.
Figure 4. Conformational flexibility of Zur proteins.
The “closed” vs. “open” conformation of dimeric Zurs is illustrated by the X-ray structures of (A) ScZur (pdb 3mwm) and (B) MtZur (pdb 2o03).
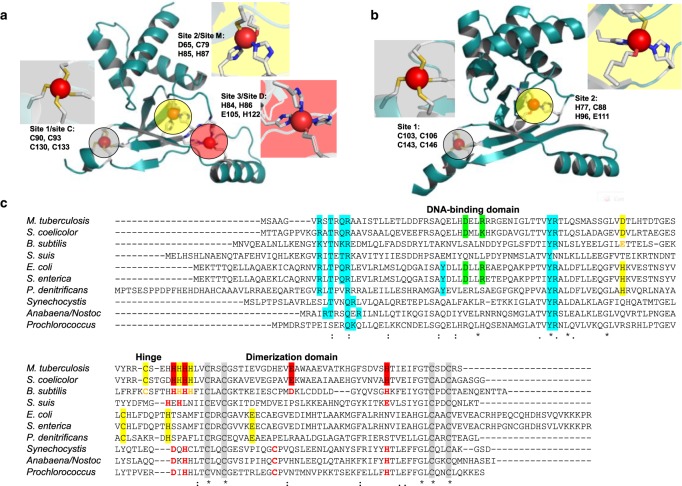
This relatively simple picture is somewhat complicated by the fact that each Zur monomer can bind more than one zinc ion (either two or three). EcZur harbours only two Zn sites per monomer [17], while MtZur [15] and ScZur [38] both contain three sites per monomer (see the next section for stoichiometric data for Zurs from other species). In all cases, the Zn2+ ions are tetrahedrally co-ordinated (Figure 5).
Figure 5. Structural and sensory zinc sites on Zur proteins.
(A) ScZur (pdb 3mwm [38]) and (B) EcZur (pdb 4mtd [17]). The structural sites are highlighted in grey, the single or major sensory site in yellow, and the additional site in ScZur is highlighted in red. (C) Sequence alignment of Zur proteins from a variety of species. Residues confirmed to participate in zinc binding by X-ray crystallography are highlighted by red, yellow, and grey backgrounds. Residues involved in DNA binding are highlighted in cyan. Predicted metal-binding residues or sensory sites in Zur proteins that have not been structurally characterised are printed in red or yellow. The two residues forming a salt bridge in EcZur (see the text) are highlighted in green; they are (semi-)conserved in Zur from Salmonella, M. tuberculosis, and S. coelicolor.
Not all Zn sites are involved in zinc sensing, the Cys4 site 1 (also termed site C; Figure 5A,B) that all three proteins have in common is a non-labile structural site. It is located entirely in the dimerisation domain and stabilises the protein fold by tethering the C-terminus to the β-sheet. Analogous structural Zn sites are also present in a range of other Fur-family proteins that sense other metals [26], and the four Cys residues are also well conserved in Zur proteins that have not yet been structurally characterised (Figure 5C). The single sensory site in EcZur is composed of a mixture of N, S, and O ligands and is derived from residues from both domains and the hinge region, namely a His and a Cys residue from the DNA-binding domain, His77 situated in the hinge region, and a Glu from a short β-strand in the dimerisation domain. Site 2 in MtZur and ScZur sits in an analogous position to this site and also has an N2OS composition, even though none of the zinc-coordinating residues is strictly conserved between EcZur and the actinobacterial Zurs (Figure 5C). Their location between the two domains strongly suggests that Zn binding to these sites affects the mutual orientation of the domains, by essentially providing a relatively rigid cross-link through the four bonds between amino acid side chains and Zn2+. This cross-link thus favours the prevalence of the ‘closed’ conformation of the dimer.
Thus, site 2 is thought to be the major Zn sensory site in MtZur and ScZur, while the significance of the third Zn site, which is entirely located within the dimerisation domain, is less clear. This site is composed of three His and one Glu residues and was only partially occupied in the MtZur structure [15]. For ScZur, a regulatory role of this site has been confirmed [38]; the case of MtZur is unresolved. The residues forming this site are also (semi-)conserved in Bacillus subtilis Zur, but based on metal : protein stoichiometry data, it has been concluded that although the residues are important for dimerisation, they do not bind zinc under physiological conditions [40]. Perplexingly, the position of this site within the protein is very similar to those of the sensory sites in other Fur family proteins. Sequence analysis and structural modelling of cyanobacterial Zur proteins predicted two sites, i.e. the structural site 1, and a sensory site, again with N2OS coordination [51]; the predicted residues are also highlighted in Figure 5C.
EcZur is the only Zur protein which has been crystallised in complex with its cognate DNA (Figure 6); this complex has provided insights about which residues are critical for this interaction.
Figure 6. DNA binding by EcZur.
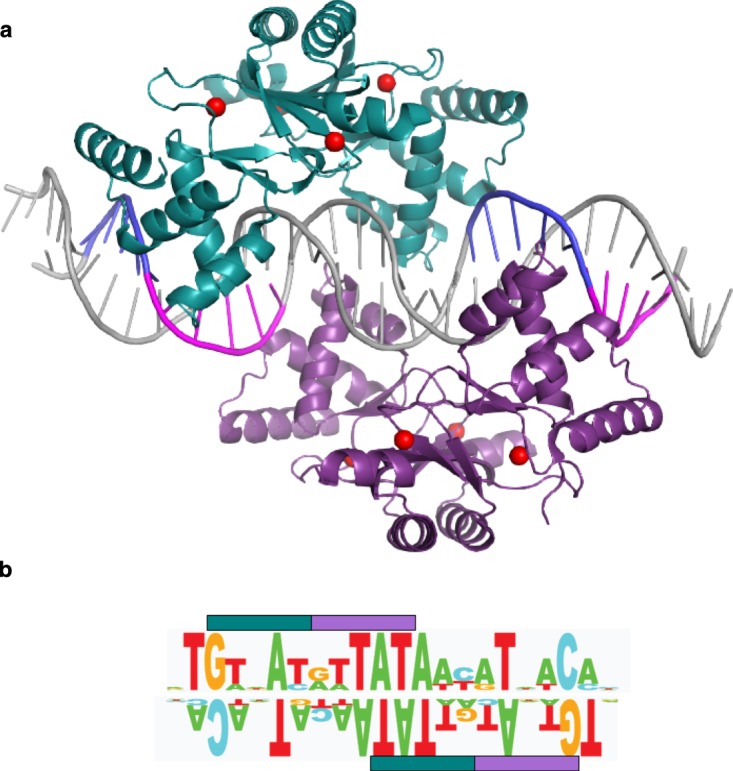
(A) EcZur in complex with DNA (31 base pairs from the znuABC promoter; pdb 4mtd [17]). The two dimers binding to the complete Zur box are shown in green and purple. DNA backbone and bases are shown schematically, with the regions forming interactions with the protein highlighted in blue and magenta. The position of zinc ions is indicated in red. E. coli Zur boxes can bind one or two dimers; for the znuABC promoter, there is a high degree of cooperativity, leading to the overwhelming prevalence of the complex involving two dimers. (B) Illustration of the consensus sequence for the E. coli Zur box, with RNNNY (R = purine; Y = pyrimidine; N = any base) motifs important for Zur–DNA interactions highlighted. Each of the bars corresponds to the interaction motif for one monomer. The sequence logo for the consensus sequence is taken from ref. [17].
Residues involved in DNA binding highlighted in cyan in Figure 5C are well conserved between species, including several residues interacting with the phosphate and sugar backbone, and Arg65 which forms hydrogen bonds to N7 of a guanine or adenine base, and thus provides sequence-specific recognition. Since the overall fold of the DNA-binding domains is predicted to be also well conserved, it is likely that these residues also participate in DNA binding in Zur proteins from other species. However, Tyr45, the second residue that directly interacts with a nucleobase, is only present in enterobacterial Zurs and that of Paracoccus denitrificans. Therefore, it is reasonable to expect that the recognised DNA sequences differ between species, and this is indeed the case (Figure 7).
Figure 7. Examples for computationally assembled Zur boxes.
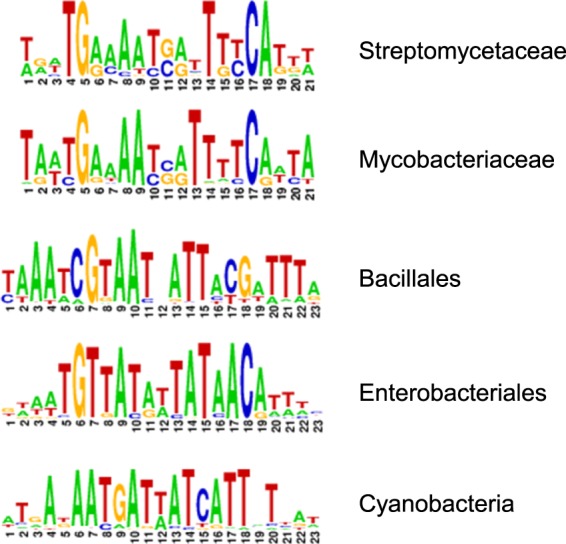
The sequence logos are taken directly from RegPrecise, using manually curated regulons [52]. While certain commonalities are evident for the Zur boxes from actinobacteria (Streptomycetaceae and Mycobacteriaceae) and Bacillales, the Zur boxes for enterobacteria (including E. coli and Salmonella) and cyanobacteria are clearly different.
The Zur box for EcZur is illustrated in Figure 6B. EcZur binds to the znuABC promoter as a dimer of dimers, which is stabilised by a pair of salt bridges between Asp49 and Arg52. These residues are only conserved in enterobacterial and actinobacterial Zurs (Figure 5C). Therefore, the oligomerisation state on DNA is unlikely to be transferrable to other Zurs. Zur boxes for other genomes have been defined from promoter analyses of Zur-regulated genes, and are typically presented as palindromic sequences, for example, 9-1-9 [41], 7-1-7 [53–55], or as 23-bp palindromes [56]. Figure 6B exemplifies that the at least pseudo-palindromic nature of Zur boxes relates to the dimeric nature of the DNA-binding Zur species. It is worth noting that many other metal-sensing transcription factors also bind as dimers, with their cognate DNA sequences also being at least pseudo-palindromes [18–24,57]. Once one or more Zur boxes have been identified for a particular bacterial phylum, genomes can be computationally screened for further putative Zur-binding sites. Definition of a specific motif (TRWNAYRWTATAWYRTNWCA) allowed finding a new Zur-regulated gene in E. coli, pliG [17]. The manually curated database RegPrecise contains a large collection of Zur boxes and Zur-regulated genes [52].
Biophysical and biochemical studies on Zur proteins
A range of Zur proteins from different bacteria have been expressed recombinantly and studied in some detail. Table 1 summarises the two most fundamental parameters, namely oligomerisation state and Zn : protein stoichiometry. Since the methods used for protein production may affect these parameters, these are also given.
In most cases, dimers were isolated, but the production of homogeneously metallated Zur proteins is non-trivial. Considering that the structural work discussed above has demonstrated that different Zur proteins can harbour a different number of zinc-binding sites with different functions, it is clear that the determination of accurate metal : protein stoichiometries is the first crucial step towards the understanding of the mechanism of action of any Zur protein. An important common feature is that even in the many instances where the proteins were treated with the metal chelator EDTA [2,2′,2″,2″′-(ethane-1,2-diyldinitrilo)tetraacetic acid], the purified proteins still contained about one zinc ion per monomer. The EDTA-resistant Zn2+ is bound to the structural Cys4 site. It has remained unclear whether this behaviour is attributable to the site binding zinc with extremely high thermodynamic stability. Given the high affinity (KD (Zn-EDTA) = 2.3×10−14 M at pH 7.4 [58]) and the large excesses in which EDTA is typically administered, the Zn2+ in the structural site would need to be bound with an affinity well below sub-femtomolar, which is unparalleled in any other Cys4 sites in, e.g. zinc fingers or metallothioneins. An alternative explanation may be that the site is kinetically inert. Given that the site is not deeply buried, accessibility is unlikely to be sterically restricted, but it is possible that interaction with EDTA is electrostatically disfavoured.
In those cases where attempts were made to establish the number of total binding sites, stoichiometries of either two or three per monomer were found. In this context, the stoichiometries reported for MtZur and EcZur are particularly instructive. Initial work on MtZur [39] identified only two Zn per monomer, even though three sites were found in the crystal structure. Such discrepancies can have many causes: firstly, metallation during expression may be incomplete or excessive. This can also be a consequence of the chosen tag [59]. Secondly, zinc may be lost during protein purification. Many of the protocols in Table 1 utilise metal-chelating agents including EDTA, DTT, and imidazole. Thirdly, the protein or metal concentrations might not have been determined with sufficient accuracy. In one instance, careful analysis revealed that the conventionally calibrated Bradford assay overestimated EcZur protein concentration by a factor of 2.33 [43]. Finally, it may also be argued that not all metal-binding sites found by X-ray crystallography are necessarily physiologically relevant, but at least in the case of ScZur, the participation of both sites 2 and 3 in zinc sensing has been confirmed (see below) [38]. In contrast, based on stoichiometry data, site 3 in BsZur is believed to be not involved in zinc sensing, even though a site 3 mutant had decreased DNA-binding affinity [12]. The latter was suggested to be due to a decrease in dimerisation tendency of this mutant. Stoichiometric data for enterobacterial Zurs are also not clear-cut: the X-ray structure of E. coli Zur contains two Zn per monomer, even though ICP-MS analysis detected an almost 3 : 1 stoichiometry [17]. Similarly, Salmonella Zur also binds up to three Zn per monomer [45]. The possibility of a third Zn binding to the structurally disordered C-terminus has been raised [17].
To understand the mode of action and operating ranges of metalloregulatory transcription factors, two thermodynamic parameters are of fundamental importance: the affinity to the cognate DNA and the affinity to the cognate metal (Table 2). The two most frequently used approaches to determine protein–DNA affinities are electrophoretic mobility shift assays (EMSAs) [17,53,54,62] and fluorescence anisotropy (FA) [40,45,47,61]. Both methods can be employed in titrations, where typically the DNA concentration is held constant, and the protein concentration is varied. Evidently, the metallation state of the sensor protein in these assays is critical. The perhaps best-controlled approach employs purified protein with clearly defined metal : protein stoichiometry and buffers that avoid loss of metal. A frequently used alternative approach is to work in EDTA-containing buffers, but supplying an excess of Zn2+ [or other metal ion(s)] to ensure saturation of the sensory site(s).
Table 2. Affinity of Zur proteins for DNA and Zn2+.
The DNA-binding affinities refer to fully metallated Zur, except for mutant proteins.
| Organism | KD (DNA) | KD (Zn) |
|---|---|---|
| Bacillus subtilis | 4.3 nM (FA [40]) 6 nM (EMSA [41]) WT: 12.5–25.4 nM, site 2 mutant: 16.5 nM for PrpsNB;* 80.6 nM for PyciC; site 3 mutant: ∼100 nM (EMSA [12]) |
KD1 = 5.5 × 10−14 M (0.9 Zn/monomer) KD2 = 1.2 × 10−12 M (0.5 Zn/monomer) (titration against Quin-2† [40]) KD1 = 4.2 × 10−15 M (0.1 μM BsZur) = 5.9 × 10−16 M (1.0 μM BsZur) (In vitro Zn2+ activation assay [40]) |
| Streptomyces coelicolor | 17.7 nM PrpmG2; 17.6 nM PSCO7682; 74.9 nM PznuA; 68.3 nM PrpmF2 (EMSA [38]) 15 nM PznuA1, 19 nM PznuA2 and PzitB (EMSA [55]) |
7.8–4.5 × 10−16 M (EMSA, Zn titration in the presence of TPEN [38]) |
| E. coli | 8.2 ± 0.7 × 10−18 M2 (PznuC) 0.053 ± 0.01 × 10−18 M2 (PzinT) 0.025 ± 0.01 × 10−18 M2 (Pl31p) 520 ± 90 × 10−18 M2 (PpliG) (Zur2Zn4)2-Pxxxx; EMSA [17] |
9.6 ± 3.0 × 10−17 M (in vitro DNA binding) 2.0 ± 0.1 × 10−16 M (transcription assay PznuC [4]) |
| Salmonella enterica | Zur2Zn6: 54 ± 18 nM Zur2Zn4: 41 ± 10 nM Zur2Zn2: ≥2.7 ± 0.4 × 10−5 M (FA [45]) |
K1–2 6.36 ± 0.41 × 10−13 M (titration against Quin-2) K3 8.04 ± 2.92 × 10−11 M (titration against Quin-2) K4 ≥5 × 10−7 M (titration against MagFura2‡) Indices refer to binding sites on dimer, with structural sites already occupied [44] |
| Anabaena sp. PCC 7120 | 220 ± 10 nM (EMSA [48]) Pall4725: 2.5 nM Pall4723: 7 nM (EMSA [60]) |
Isothermal titration calorimetry (ITC): [48] ZurZn + Zn ↔ ZurZn2, KD ∼3.5 × 10−7 M ITC in the presence of DTT: two sites; KD1 = 6.5 × 10−7 M |
| Synechocystis sp. PCC 6803 | KD ≤ 55 nM (FA [47,61]) | KD1 = 2.3 ± 1.9 × 10−13 M (titration against Quin-2 [47,61]) |
| Paracoccus denitrificans | n.d. |
KD1 = 4.0 ± 0.4 × 10−8 M (titration against MagFura2) KD2 > 1×10−6 M [46] |
Pxxxx: Promoter for gene xxxx. †Quin-2: 2-[(2-amino-5-methylphenoxy)methyl]-6-methoxy-8-aminoquinoline-N,N,N′,N′-tetraacetic acid. ‡MagFura2: 2-[6-[bis(carboxymethyl)amino]-5-(carboxymethoxy)-2-benzofuranyl]-5-oxazolecarboxylic acid.
The detected species in EMSAs is the DNA, in both its free and protein-bound forms. The DNA employed may be of different length (typically 25–450 bp) depending on how well-defined the location and size of the Zur-box is. For EMSAs to yield unambiguous affinity data, the gels should contain exactly two bands, one for the free DNA, and one for the bound DNA, which in the case of dimeric transcription factors should in the most straightforward cases refer to the 1 : 1 Dimer : DNA complex. In practice, the raw data are often more complicated and may include bands with bound monomer as well as bands with more than one dimer bound, and in more unfavourable cases also diffuse intensity not clearly attributable to either form. The latter effect is due to the complex(es) present in the gel wells dissociating while moving through the gel; this is particularly likely when binding is weak and kinetically labile. FA measurements are a true equilibrium method, yielding data that do not show such complexity and problems. FA may require more stringent experimental design: the detection of protein binding is based on the molecular tumbling rate of fluorescently labelled DNA, which is affected by whether or not a protein is bound. To maximise this effect, the oligonucleotides employed in FA are typically quite short (e.g. 24 bp); hence, the Zur-box sequence needs to be well defined. Both EMSA and FA allow the determination of binding stoichiometry. In the case of EMSA, this can be deduced from measuring the mass of the complex(es) from their in-gel mobility (see, e.g. [17]), while FA titrations at high DNA concentration (approximately two orders of magnitude above KD) provide the saturation stoichiometry directly. Both data types require fitting to one or more models. The data given in Table 2 refer in most cases to simple 1 : 1 Dimer : DNA binding models, except for the EcZur data which refer to the binding of two dimers per DNA, i.e. the 2 : 1 complexes. All reported dissociation constants for 1 : 1 complexes are in the low to mid-nanomolar range. Recent work has revealed that different promoters within the same genome can have different affinities (see, e.g. the entries for B. subtilis, S. coelicolor, and E. coli); this allows a graded response to different degrees of zinc limitation [17,38,55]. A direct correlation between in vivo -fold repression and KD was proposed for E. coli [17]. In cases of single-dimer binding, KDs vary over less than two orders of magnitude; in the case of the dimer-of-dimer-binding mode for E. coli Zur, the strongest and weakest sites differ by four orders of magnitude — but it should be noted that the units of these constants differ, and therefore values and factors are not directly comparable.
There are several cases where promoters contain more than one Zur box; for example, the znuA promoter of S. coelicolor [38] and the yciC promoter of B. subtilis [41] contain at least two Zur boxes, one overlapping with the −35 motif and one with the −10 motif. The significance of this has remained unclear; it is most likely that the two binding sites operate independently, and that this allows for more stringent repression, with the downstream box providing backup in the rare case that repression by the upstream box was not effective [41].
Various methods are available to determine metal–protein affinities. The most direct approach involves mixtures of the protein of interest and a metallochromic dye, into which small aliquots of Zn2+ are titrated. The concentration of either free or zinc-bound dye is the measured quantity. Importantly, the dye must have an affinity in a similar order of magnitude as the protein. Otherwise, the two metal-binding molecules cannot compete, and the respective ‘equilibrium constants’ become meaningless.
Activation assays involve studying the DNA-binding ability at different free Zn2+ concentrations; this can be monitored in vitro by EMSA, or by measuring in vivo transcription via a reporter gene under the control of a promoter containing the Zur box. Free [Zn2+] is controlled by the addition of well-established zinc chelators such as TPEN [N,N,N′,N′-Tetrakis(2-pyridinylmethyl)-1,2-ethanediamine] or EDTA.
The data compiled in Table 2 reflect in most cases the very high affinity of zinc sensor proteins for their cognate metal ion, which is necessary, as intracellular free Zn2+ concentrations in bacteria are picomolar or lower [63]. Accordingly, most Zn–Zur dissociation constants are at least in the picomolar range, with two notable exceptions, namely those for P. denitrificans and Anabaena PCC 7120 Zur, which are likely erroneous due to inappropriate experimental design. In the first case, the detected ‘high’ affinity site — for which KD = 40 nM is still at least four orders of magnitude higher than expected — was suggested to refer to binding of Zn to what was thought to be the apo-protein, which had been generated by extensive dialysis [46]. A second site with micromolar KD was also found. Considering that all known zinc-binding residues are conserved between E. coli and P. denitrificans Zur (Figure 5C), the discrepancies between the data for those two proteins are astonishing. It is also surprising that the structural site in P. denitrificans Zur apparently remains intact during initial treatment with EDTA (Table 1), but dissociates during dialysis. We note that this report [46] does not mention whether care was taken to prevent protein oxidation during purification, while most other studies were conducted either in the presence of reducing agents [DTT or TCEP; Tris(2-carboxyethyl)phosphine] or under an inert atmosphere. In the case of Anabaena PCC 7120 Zur, ITC experiments were conducted both in the presence and absence of DTT. While the presence of DTT enhanced affinity by about one order of magnitude, the KDs all lie within the micromolar range. This range is what is achievable with the reported experimental design: the ITC experiments involved 20 μM protein, but employing micromolar concentrations of protein (and metal) without a competing agent only allows the determination of KDs within the same concentration range (± maximally two orders of magnitude), while binding events with higher affinity cannot be quantified.
For the remaining zinc affinity data, there is a clear trend for the data obtained by in vitro DNA binding and in vitro or in vivo transcription assays reporting significantly higher affinities than data obtained by direct titrations of the purified protein in the absence of DNA. Since metal- and DNA-binding equilibria are coupled allosterically, it follows that not only does Zn2+ bound to the sensory site increase the affinity of Zur to DNA, but also that DNA bound to Zur will enhance the affinity of the sensory site to Zn2+. Such allosteric coupling in metalloregulatory proteins has been quantified and discussed comprehensively by Giedroc and co-workers [64].
EMSA experiments, in conjunction with biophysical characterisation of mutant Zur proteins, have also been instrumental in determining the role of the various Zn-binding sites. When one of the cysteine residues of the structural Zn-binding sites (site 1) was mutated to serine, Zur from both S. coelicolor (C90S) [38] and E. coli (C103S) [17] was unable to form dimers and bind DNA. CD spectroscopy showed drastic changes in the secondary structure of the C90S mutant ScZur, while mutations affecting sites 2 or 3 did not affect the secondary structure or oligomerisation state [38]. The essential role of the structural site to maintain both the tertiary and quaternary structure is evident. The mutation C88S affecting the sensory site in EcZur completely abolished Zn-responsive transcriptional regulation [17], and the site 2 C79S mutation in ScZur virtually abolished binding to all promoters [38], testifying to the essentiality of this site for zinc sensing.
EMSA experiments have also been employed to explore the metal selectivity of Zur proteins. Typically, this involves removal of the sensory metal(s) by EDTA and supplementing the reaction mixtures with different metal dications (e.g. Zn2+, Cd2+, Co2+, Cu2+, Fe2+, Mn2+, and Ni2+). Selectivity is established by assessing whether the supplemented metal can trigger the formation of the DNA–Zur complex. The Zur proteins from P. denitrificans [46], Neisseria meningitidis [65], and Brucella abortus [66] only showed significant complex formation in the presence of Zn2+. In contrast, Zur from Corynebacterium glutamicum [67] formed Zur–DNA complexes in the presence of Mn in addition to Zn, while Zur from M. tuberculosis [68] showed Zur–DNA binding in the presence of Zn, Mn, and Cd. It must be appreciated that sensor proteins do not need to be 100% specific, because in vivo not only allostery or stability constants define the sensed metal, but also the cytosolic free metal concentrations [61]. These differ dramatically for different metal ions, and indeed, the Robinson laboratory has found a close correlation between metal sensor KDs and cytosolic-free concentrations [45,61,63]. Thus, since the described in vitro experiments do not emulate the intracellular environment and concentrations of metal and proteins, they may not necessarily reflect likely interactions in vivo. It should be added that at least in S. coelicolor, Zur is, with a cellular concentration of 3.7 μM, a fairly abundant protein [55].
Zur regulons
After the discovery of Zur proteins as sensors that regulate the expression of high-affinity zinc uptake systems, many studies have elucidated the full complement of Zur-regulated genes in a range of bacteria (Table 3). Searches for Zur-regulated genes often, but not always, involve the generation of zur knockout mutants. This also enables the determination of phenotypes. Δzur mutants often have higher zinc contents, and suffer from increased sensitivity towards elevated concentrations of zinc, although there are exceptions to that rule, for example, in Corynebacterium diphtheriae [73]. The viability and/or virulence of Δzur mutants of pathogenic bacteria in the host is often more or less severely compromised [74,85,86,95], but that is not always the case [75]. The knockout phenotypes highlight that Zur provides tolerance towards both high and low zinc: it is required not only to boost zinc uptake under limiting conditions but also to down-regulate zinc uptake in excess conditions.
Table 3. Genes of the Zur regulons from a range of bacteria.
| Species | Experimental approaches | Zn import | COG0523 | Ribosomal proteins | Enzymes | Other | Activation | Refs. | |
|---|---|---|---|---|---|---|---|---|---|
| Actinobacteria | Streptomyces coelicolor* | Δzur mutant; qRT-PCR. Forty-one putative Zur boxes identified by CHiP [55] | znuABC | yciC | rpmE2, rpmF2, rpmB2, rpmG2, rpsN2, rpsR2 | SCO7676 (Putative fer-redoxin), 7681, 7682 (Coelibactin biosynthesis) | SCO0472, 0474, 3431 (various) | zitB (CDF effluxer) | [16,55,69,70] |
| Mycobacterium tuberculosis | Δzur mutant, microarrays, qRT-PCR. Thirty-two genes (probably 16 operons) up-regulated | znuABC | yciC | rpmB1 and B2, rpmG1, rpsN2, rpsR1 | Secretory proteins involved in virulence | [68] | |||
| Mycobacterium avium ssp. paratuberculosis | Exposure to metal starvation, qRT-PCR, RNAseq. Zinc-responsive genomic islands (ZnGI) |
mptABC, mptDEF |
3× cobW |
rpsR2, rpsN2, rpmG2, rpmE2 rpmB1, rpmG1 |
sidAB and G; metallophore bio-synthesis and export | Secretory proteins involved in virulence; lamB | [71] | ||
| Corynebacterium glutamicum | Δzur mutant, microarrays, qRT-PCR; nine genes differentially regulated in mutant | 2× znuABC |
cg0794 (yciC) |
cg0795 (oxidoreductase) cg3107 (adhA) |
cg0040 (secreted protein) |
zrf (CDF effluxer) zra (ATPase) |
[67,72] | ||
| Corynebacterium diphtheriae | ΔZur mutant, qRT-PCR, reporter assays | troABC | zrg (yciC) | adhA | Several surface-anchored proteins | [73] | |||
| Firmicutes | Bacillus subtilis | Δzur mutant, microarrays, quantitative RT-PCR |
znuABC (annotated as adcABC), ZinT |
yciC | rpsN2, rpmE2 |
folE2 (yciA), yciB ytiB (carbonic anhydrase) |
[12,53] | ||
| Listeria monocytogenes* | Δzur mutant, quantitative RT-PCR | znuABC (2×) | rpsN2 | [74] | |||||
| Staphylococcus aureus* | Transcription assays |
adcBC, adcA-II zur |
yciC | rpsN2, rpmG2 | [75] | ||||
| Enterococcus faecalis* | Exposure of WT to high [Zn]; microarrays, qPCR | adcBC, adcA-II, adcA | yciC | rpmF2 (x2), rpsN2 (x2), rpmG2 (x2) | [76] | ||||
| Streptococcus suis | Δzur mutant, microarrays, 121 genes (72 up-, 49 down-regulated in mutant); qRT-PCR | None of the commonly Zur-regulated genes. Several enzymes and membrane proteins up-regulated in mutant, e.g. Zn-dep. NADPH-quinone reductase and 3-phosphatidyltransferase. Unclear whether genes have Zur boxes in the upstream region. Alternative sensor AdcR more common in Streptococci. | [42] | ||||||
| Gamma-proteobacteria | E. coli | Δzur mutant, qRT-PCR, EMSA | znuABC, zinT | rpmE2, rpmJ2 | pliG | [17,77] | |||
| Pseudomonas aeruginosa* | Δzur mutant, qRT-PCR; WT grown in high and low Zn, microarray, qRT-PCR; RNASeq of ΔznuA mutant | znuABC, znuD (TonB-dR), zrmABCD (tonB-dR) | yciC | rpmE2, rpmJ |
folE2, amiA, can, pyrC2, PA5537 (glutamine synthetase) cntOLMI (metallophore synthesis) |
dksA, zbp | [78–81] | ||
| Pseudomonas protegens* | Exposure to Zn limitation; microarrays (73 genes up-, 28 down-regulated); qRT-PCR | znuABC (3×), 3× tonB-dR | yciC, yciC2 | rpmJ, rpmE1 | folE2, amiC, can, pyrC2, hisI2 | dksA | [82] | ||
| Yersinia pestis | Δzur mutant, microarrays; 154 differentially regulated genes in response to high [Zn] | znuABC (2×) |
rpmJ2 rpmE2 |
[83] | |||||
| Vibrio cholerae | Bioinformatics and biochemical promoter analysis | znuABC, zrgABC(DE) | rpmE2, rpmJ2 | ribA | zrgD and E: hypothetical proteins | [84] | |||
| Acinetobacter baumannii* | Δzur mutant, 76 genes up- and 68 genes down-regulated, qRT-PCR | znuABC, 3× tonB-dR, ExbD, ExbB | zigA | rpmE2 | [85] | ||||
| Xanthomonas campestris | Δzur mutant, microarrays, 64 putative Zur-regulated ORFs; in vitro transcription assays |
znuABC 2× tonB-dR |
XC0267 | folE2, amiC | hrpX; involved in pathogenicity | czcD (CDF effluxer) | [86,87] | ||
| Francisella tularensis and F. novicida | Δzur mutant, RNASeq, qRT-PCR | zupT (FN) | FTN_0880 | FTN_0395; ArsR family transcrip-tional regulator | [88] | ||||
| Alpha-proteobacteria | Caulobacter crescentus | Microarrays, transcription assays. Twenty-eight genes (7 up-, 21 down-) regulated in Δzur mutant |
znuGHI 3× tonB-dR (znuK, L and M) |
zrpX | rpUI, rpmA | zrpW (putative transporter) |
RND systems, ATPase, tonB-dRs |
[89] | |
| Agrobacterium tumefaciens* | Δzur mutant, qRT-PCR | znuABC/troABC, zinT | yciC (2×) | [90,91] | |||||
| Paracoccus denitrificans* | RNASeq (Zn-chelated/depleted/replete), 147 genes (133 up-, 14 down-regulated in low [Zn]), qRT-PCR | znuABC, aztABC | yciC | [46] | |||||
| Beta-proteobacteria | Neisseria meningitidis | Δzur mutant, microarrays, qRT-PCR; 17 genes differentially regulated in mutant |
znuABC, 2× TonB-dR |
rpmE2, rpmJ | queC, queF | adhP | [65] | ||
| Cupriavidus metallidurans | Δzur mutant, microarrays, Zur binding to promoter regions was tested |
zupT tonB-dR |
Op0317f cobW1 W2, W3 |
No | dehH2 | dksA1 | σ-factor fliA; cadA but not zntA | [92,93] | |
| Cyanobacteria | Anabaena PCC 7120 | Promoter mapping, screening for putative Zur boxes, qRT-PCR, 23 genes identified [60] Δzur mutant, semiquant. qRT-PCR, EMSA [62] |
znuABC, 3 tonB-dR, (alr3243, alr4031) |
alr1197, all1751, all4722 |
hemB2, thrS2, folE2, glycosyl-transferase [60]; sodA, catalase, peroxiredoxin [62], several more predicted |
aztR all1474, alr3495 |
[60,62] | ||
| Synechococcus PCC 7002* | Δzur mutant, RNASeq | znuABC | hemB2, folE2 (+2 more) | [94] | |||||
In most cases, Zur regulation has been confirmed experimentally. Some entries have been complemented by data extracted from the RegPrecise database [52]; experimentally confirmed Zur-regulated genes are printed in bold. Actinobacteria and Firmicutes have a single membrane; all other bacterial groups are Gram-negative and have an outer and inner membrane and a periplasm. Asterisks (*) indicate species in which zur expression is subject to autoregulation. Also see Figure 1 regarding uptake/efflux proteins. Abbreviations: znuABC, znuGHI, troABC, adcABC, aztABC, zrgABC are all ABC-type zinc uptake systems; zinT and adcA: periplasmic zinc-binding proteins; the latter has both a ZnuA and ZinT-like domain; zupT: zinc importer of the ZIP family; zitB: zinc exporter of the cation diffusion facilitator (CDF) family; tonB-dR: TonB-dependent receptor; exbB/D: parts of energy transduction system for TonB-dependent receptors; oprD: outer-membrane porin; aztR, smtB: zinc excess sensors; cobW/yciC: frequently used labels for COG0523 proteins; ribosomal proteins: rpmE = L31, rpmJ = L36, rpUI = L21, rpmA = L27, rpmB = L28, rpmG = L33, rpsN2 = S14p, rpsR1 = S18, rpmF2 = L32p; zrpW: zinc-regulated protein; zbp: putative zinc-binding protein; pliG: periplasmic lysozyme inhibitor; dksA: zinc-independent transcription factor; can: gamma-carbonic anhydrase; pyrC2: dihydroorotase; amiA/amiC: N-acetyl-muramoyl-l-alanine amidase; adhA: zinc-dependent alcohol dehydrogenase; adhP: alcohol dehydrogenase; folE2 and ribA: GTP cyclohydrolases; (tetrahydrofolate biosynthesis); dehH2: haloacetate dehydrogenase; hemB: delta-aminolevulinic acid dehydratase (tetrapyrrole biosynthesis); thrS2: threonyl-tRNA synthetase; hrpX: hypersensitivity-pathogenicity regulatory gene; queC: 7-cyano-7-deazaguanine synthase; queF: NADPH-dependent 7-cyano-7-deazaguanine reductase; Other abbreviations such as alr1197 and XC0267 are locus tags.
To determine Zur regulons, wild-type and mutant strains are subjected to low and high zinc concentrations, and the expression of either all or a limited number of pre-selected genes is assessed, typically by microarrays or quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR), under these four conditions. A gene that is Zur-regulated is expected to be regulated by zinc concentration in the wild-type, but not in the Δzur strain. This is a necessary but not sufficient criterion, because the consequences of deleting zur may be more complex: (i) Zur may regulate the expression of other transcription factors (including other urs, SmtB-like excess sensors, and sigma factors; see Table 3), or be itself regulated by other transcription factors. (ii) The deletion of zur may lead to general disturbance of metal homeostasis, with potential indirect consequences, including effects on other metal sensors and on the cellular redox balance. Although zinc is not itself redox-active like copper or iron, its levels may affect redox balance through its frequent association with redox-active thiols [96], and therefore, concurrence of oxidative stress protection and zinc homeostasis is expected. Indeed, higher zinc levels protect B. subtilis from increased H2O2 concentrations [53]. Examples of indirect effects on gene transcription in knockout or overexpressing Zur mutants include potential mis-metallation of Fur at high [Zn2+] in a Δzur mutant causing repression of Fur-regulated genes in Caulobacter crescentus [89] and interplay between zinc homeostasis and disulfide stress via Zur and sigma factor R in S. coelicolor [69]. Furthermore, the pleiotropic phenotype of Δzur and zur-overexpressing mutants, extensive cross-talk between Zur, Fur, and PerR, and up-regulation of antioxidant enzymes (superoxide dismutase sodA, catalase alr0998, and peroxiredoxin gct3) in a Δzur mutant of the cyanobacterium Anabaena sp. PCC 7120 probably also include both direct and indirect effects [48,62,97,98]. In turn, the levels of metal ions other than Zn2+ may also affect the expression of members of Zur regulons, as shown for Enterococcus faecalis [76], where exposure to high levels of Cu2+ activates Zur-regulated zinc uptake, presumably to protect from mis-metallation of Zn-requiring proteins and from oxidative stress.
The identification of a true member of a Zur regulon therefore also requires: (i) the presence of Zur boxes, (ii) proof that Zur binds to the promoter region of the selected gene, and/or (iii) in vivo reporter assays. A recent study illustrates these requirements dramatically: Of the hundreds of genes that were differentially expressed in a Δzur mutant of Cupriavidus metallidurans, only 11 had computationally identified Zur boxes, and of those, only four were experimentally verified to be true Zur boxes [92].
In many bacterial species, besides repressing uptake systems, Zur can also repress genes encoding paralogues of ribosomal proteins. These paralogues generally constitute non-Zn-requiring alternatives of ribosomal proteins that require Zn2+ for folding. The expression of zinc-free alternatives under Zn-depletion conditions liberates Zn2+ from ribosomes, which is then available for other cellular processes in which Zn2+ is indispensable. Since ribosomes are highly abundant in rapidly growing cells, the presence of two or three zinc-containing ribosomal proteins represents a large reservoir of zinc, and this may, in fact, account for the majority of intracellular zinc [16,70]. This was first discovered for B. subtilis [99], but it is now evident that many bacteria utilise this strategy (see Table 3).
Zur-regulated expression of enzymes is also widespread. In many cases, these enzymes are zinc-free paralogues of otherwise zinc-requiring enzymes, for example, alcohol dehydrogenase and delta-aminolevulinic acid dehydratase. These alternatives are thought to decrease the cell's requirements for zinc. A further common occurrence are putative small GTPases of the COG0523 family [100]. These are frequently annotated as yciC, the label for this gene in B. subtilis. Other members of this family are involved in the maturation of metal-requiring enzymes (e.g. Fe-requiring nitrile hydratase) and cofactors (e.g CobW for cobalamin biosynthesis; note that mis-annotation of zinc-related COG0523 members as cobW is frequent), which has led to the suggestion that these proteins may function as metallochaperones. Precisely how they might contribute to zinc homeostasis has remained unclear, although their zinc-binding ability and consequences thereof have been demonstrated [101].
In several actinomycetes and some pathogenic bacteria, biosynthetic clusters for the synthesis of metallophores are under the control of Zur, for example, coelibactin in S. coelicolor [70], ethylenediamine-disuccinate in Amycolatopsis japonicum [54], and pseudopaline in Pseudomonas aeruginosa [78,79]. Typically, such metallophores are secreted to capture scarce zinc, and the resulting complexes are then taken up by TonB-dependent receptors which mediate active transport through the outer membrane of Gram-negative bacteria. In P. aeruginosa, both pseudopaline and TonB-dependent receptor production are under the control of Zur, and there are now many more examples of Zur-regulated TonB-dependent receptors (Table 3). These receptors and the metallophores they transport play a crucial role in zinc acquisition by pathogenic bacteria [5].
In several genomes (e.g. S. coelicolor, Pseudomonads, and P. denitrificans), Zur is subject to autoregulation, i.e. represses its own transcription in zinc-replete conditions. This only seems to be the case when the zur gene is part of an operon, e.g. with znuABC [56]. Conversely, the expression of Zur in Mycobacteria [39] and C. diphtheriae [73] is inducible by high [Zn2+]. In these species, zur is co-transcribed with the gene for an SmtB-like zinc excess sensor.
Beyond repression: Zur as a transcriptional activator
Evidence has been emerging that in at least some bacteria, Zur proteins may also activate the transcription of genes. Two of the best-studied cases concern Zur-regulated expression of metal efflux proteins of the CDF family in Xanthomonas campestris [86] and S. coelicolor [55]. Zur binding to their promoter regions was studied in both cases. In X. campestris, a GC-rich 59-bp sequence that shows no significant similarity with Zur boxes in this organism was identified by DNAse footprinting in the upstream region of the gene xc2976, which codes for a CDF effluxer. The size of the protected region suggests that more than one dimer is required for binding and activation. In S. coelicolor, the promoter region of the zitB gene contains a Zur box upstream of the −35 site, with similar affinity to other Zur boxes in this organism (15–20 nM). However, since the zitB Zur box does not overlap with the RNA polymerase-binding site, this interaction is not expected to lead to repression. Activation occurs at higher [Zn2+], apparently through Zur oligomerisation, as judged from the expansion of the DNA footprint. Higher-order oligomers have recently been detected for several Fur-family proteins [29]. Zinc-dependent activation of gene expression under zinc-rich conditions was also observed in N. meningitidis [65] and C. crescentus [89]. The expression of two efflux proteins in C. glutamicum is also Zur-dependent [72]. Here, a new 10-1-10 direct repeat sequence binds Zur in which sensory sites are not populated. This recognition site overlaps with the −35 site, resulting in repression at low [Zn2+]. Thus, it can be anticipated that there is significant variation between species in activation/de-repression mechanisms that deviate from the canonical mode of action.
Conclusions
Zinc sensors of the Zur family are ubiquitous, present in most Gram-negative and many Gram-positive bacteria, with the number of recognised Zur-regulated genes still increasing. By governing the expression of zinc-supplying and zinc-requiring proteins, they provide regulation of intracellular total and free Zn2+ concentrations. This protects bacteria against both high and low external Zn2+ concentrations, which is of particular interest regarding pathogens [7] but also human microbiomes [102]. Although protein folds are well conserved between Zur proteins from different bacterial phyla, there is surprising diversity regarding the position of the sensory Zn-binding residues in the protein sequence. Yet in all cases where this is known, the tetrahedral coordination sphere of the (major) sensory site consists of two nitrogens, one oxygen and one sulfur. This seems to be particularly suited to Zn2+, providing a very high affinity in the atto- to femtomolar range. Some Zur proteins seem to fine-tune DNA affinity by employing an additional zinc site, which extends the range at which Zn2+ can be sensed, and allows for differential expression of several sets of genes. Zur–DNA interactions can also be modulated by employing different degrees of oligomerisation, and the number and type of Zur boxes. Moreover, Zur proteins may also act as activators of transcription, and there even appear to be cases where the allosteric switch has the opposite effect, namely increased DNA affinity in the absence of zinc. In the face of this variety, it is clear that our understanding of structures and dynamics of Zur and its biomolecular complexes is as yet far from comprehensive.
Acknowledgements
The authors thank the University of Warwick for a Chancellor's International Scholarship (to A.M.).
Abbreviations
- EDTA
2,2′,2″,2″′-(ethane-1,2-diyldinitrilo)tetraacetic acid
- EMSAs
electrophoretic mobility shift assays
- FA
fluorescence anisotropy
- Fur
ferric uptake regulator
- qRT-PCR
quantitative reverse-transcriptase polymerase chain reaction
- TPEN
N,N,N′,N′-Tetrakis(2-pyridinylmethyl)-1,2-ethanediamine
- Zur
zinc uptake regulator
Funding
This work was also supported by NERC grant [NE/F004249/1].
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Dupont C.L. and Caetano-Anolles G. (2010) Mulkidjanian and Galperin: Zn may have constrained evolution during the Proterozoic but not the archean reply. Proc. Natl Acad. Sci. U.S.A. 107, E138 10.1073/pnas.1009565107 [DOI] [Google Scholar]
- 2.Andreini C., Banci L., Bertini I. and Rosato A. (2006) Zinc through the three domains of life. J. Proteome Res. 5, 3173–3178 10.1021/pr0603699 [DOI] [PubMed] [Google Scholar]
- 3.Capdevila D.A., Wang J.F. and Giedroc D.P. (2016) Bacterial strategies to maintain zinc metallostasis at the host-pathogen interface. J. Biol. Chem. 291, 20858–20868 10.1074/jbc.R116.742023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Outten C.E. and O'Halloran T.V. (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292, 2488–2492 10.1126/science.1060331 [DOI] [PubMed] [Google Scholar]
- 5.Neumann W., Gulati A. and Nolan E.M. (2017) Metal homeostasis in infectious disease: recent advances in bacterial metallophores and the human metal-withholding response. Curr. Opin. Chem. Biol. 37, 10–18 10.1016/j.cbpa.2016.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J.Z., Jellbauer S., Poe A.J., Ton V., Pesciaroli M., Kehl-Fie T.E. et al. (2012) Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11, 227–239 10.1016/j.chom.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kehl-Fie T.E. and Skaar E.P. (2010) Nutritional immunity beyond iron: a role for manganese and zinc. Curr. Opin. Chem. Biol. 14, 218–224 10.1016/j.cbpa.2009.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ong C.L.Y., Gillen C.M., Barnett T.C., Walker M.J. and McEwan A.G. (2014) An antimicrobial role for zinc in innate immune defense against group A Streptococcus. J. Infect. Dis. 209, 1500–1508 10.1093/infdis/jiu053 [DOI] [PubMed] [Google Scholar]
- 9.Irving H.M.N.H. and Williams R.J.P. (1953) The stability of transition-metal complexes. J. Chem. Soc. 3192–3210 10.1039/JR9530003192 [DOI] [Google Scholar]
- 10.Choi S.Y. and Bird A.J. (2014) Zinc'ing sensibly: controlling zinc homeostasis at the transcriptional level. Metallomics 6, 1198–1215 10.1039/c4mt00064a [DOI] [PubMed] [Google Scholar]
- 11.Blindauer C.A. (2015) Advances in the molecular understanding of biological zinc transport. Chem. Commun. 51, 4544–4563 10.1039/c4cc10174j [DOI] [PubMed] [Google Scholar]
- 12.Shin J.H. and Helmann J.D. (2016) Molecular logic of the Zur-regulated zinc deprivation response in Bacillus subtilis. Nat. Commun. 7, 9 10.1038/ncomms12612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waldron K.J., Rutherford J.C., Ford D. and Robinson N.J. (2009) Metalloproteins and metal sensing. Nature 460, 823–830 10.1038/nature08300 [DOI] [PubMed] [Google Scholar]
- 14.Patzer S.I. and Hantke K. (1998) The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28, 1199–1210 10.1046/j.1365-2958.1998.00883.x [DOI] [PubMed] [Google Scholar]
- 15.Lucarelli D., Russo S., Garman E., Milano A., Meyer-Klaucke W. and Pohl E. (2007) Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J. Biol. Chem. 282, 9914–9922 10.1074/jbc.M609974200 [DOI] [PubMed] [Google Scholar]
- 16.Shin J.H., Oh S.Y., Kim S.J. and Roe J.H. (2007) The zinc-responsive regulator Zur controls a zinc uptake system and some ribosomal proteins in Streptomyces coelicolor A3(2). J. Bacteriol. 189, 4070–4077 10.1128/jb.01851-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilston B.A., Wang S.N., Marcus M.D., Canalizo-Hernandez M.A., Swindell E.P., Xue Y. et al. (2014) Structural and mechanistic basis of zinc regulation across the E. coli Zur regulon. PLoS Biol. 12, 16 10.1371/journal.pbio.1001987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu R.F., Song Y.Q., Liu H.P., Yang Y.F., Wang S.L., Yi C.Q. et al. (2017) Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation. Proc. Natl Acad. Sci. U.S.A. 114, 13661–13666 10.1073/pnas.1708563115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerra A.J., Dann C.E. and Giedroc D.P. (2011) Crystal structure of the zinc-dependent MarR family transcriptional regulator AdcR in the Zn(II)-bound state. J. Am. Chem. Soc. 133, 19614–19617 10.1021/ja2080532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanson M., Makthal N., Flores A.R., Olsen R.J., Musser J.M. and Kumaraswami M. (2015) Adhesin competence repressor (AdcR) from Streptococcus pyogenes controls adaptive responses to zinc limitation and contributes to virulence. Nucleic Acids Res. 43, 418–432 10.1093/nar/gku1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morby A.P., Turner J.S., Huckle J.W. and Robinson N.J. (1993) SmtB is a metal-dependent repressor of the cyanobacterial metallothionein gene smtA—identification of a Zn inhibited DNA-protein complex. Nucleic Acids Res. 21, 921–925 10.1093/nar/21.4.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondrat F.D.L., Kowald G.R., Scarff C.A., Scrivens J.H. and Blindauer C.A. (2013) Resolution of a paradox by native mass spectrometry: facile occupation of all four metal binding sites in the dimeric zinc sensor SmtB. Chem. Commun. 49, 813–815 10.1039/c2cc38387j [DOI] [PubMed] [Google Scholar]
- 23.Thelwell C., Robinson N.J. and Turner-Cavet J.S. (1998) An SmtB-like repressor from Synechocystis PCC 6803 regulates a zinc exporter. Proc. Natl Acad. Sci. U.S.A. 95, 10728–10733 10.1073/pnas.95.18.10728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arunkumar A.I., Campanello G.C. and Giedroc D.P. (2009) Solution structure of a paradigm ArsR family zinc sensor in the DNA-bound state. Proc. Natl Acad. Sci. U.S.A. 106, 18177–18182 10.1073/pnas.0905558106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagg A. and Neilands J.B. (1987) Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26, 5471–5477 10.1021/bi00391a039 [DOI] [PubMed] [Google Scholar]
- 26.Fillat M.F. (2014) The Fur (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 546, 41–52 10.1016/j.abb.2014.01.029 [DOI] [PubMed] [Google Scholar]
- 27.Pohl E., Haller J.C., Mijovilovich A., Meyer-Klaucke W., Garman E. and Vasil M.L. (2003) Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47, 903–915 10.1046/j.1365-2958.2003.03337.x [DOI] [PubMed] [Google Scholar]
- 28.Sheikh M.A. and Taylor G.L. (2009) Crystal structure of the Vibrio cholerae ferric uptake regulator (Fur) reveals insights into metal co-ordination. Mol. Microbiol. 72, 1208–1220 10.1111/j.1365-2958.2009.06718.x [DOI] [PubMed] [Google Scholar]
- 29.Pérard J., Covès J., Castellan M., Solard C., Savard M., Miras R. et al. (2016) Quaternary structure of Fur proteins, a new subfamily of tetrameric proteins. Biochemistry 55, 1503–1515 10.1021/acs.biochem.5b01061 [DOI] [PubMed] [Google Scholar]
- 30.Ahn B.E., Cha J., Lee E.J., Han A.R., Thompson C.J. and Roe J.H. (2006) Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 59, 1848–1858 10.1111/j.1365-2958.2006.05065.x [DOI] [PubMed] [Google Scholar]
- 31.An Y.J., Ahn B.E., Han A.R., Kim H.M., Chung K.M., Shin J.H. et al. (2009) Structural basis for the specialization of Nur, a nickel-specific Fur homolog, in metal sensing and DNA recognition. Nucleic Acids Res. 37, 3442–3451 10.1093/nar/gkp198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diaz-Mireles E., Wexler M., Sawers G., Bellini D., Todd J.D. and Johnston A.W.B. (2004) The Fur-like protein Mur of Rhizobium leguminosarum is a Mn2+-responsive transcriptional regulator. Microbiology 150, 1447–1456 10.1099/mic.0.26961-0 [DOI] [PubMed] [Google Scholar]
- 33.Bellini P. and Hemmings A.M. (2006) In vitro characterization of a bacterial manganese uptake regulator of the Fur superfamily. Biochemistry 45, 2686–2698 10.1021/bi052081n [DOI] [PubMed] [Google Scholar]
- 34.Traoré D.A.K., El Ghazouani A., Ilango S., Dupuy J., Jacquamet L., Ferrer J.L. et al. (2006) Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis. Mol. Microbiol. 61, 1211–1219 10.1111/j.1365-2958.2006.05313.x [DOI] [PubMed] [Google Scholar]
- 35.Jacquamet L., Traore D.A.K., Ferrer J.L., Proux O., Testemale D., Hazemann J.L. et al. (2009) Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol. Microbiol. 73, 20–31 10.1111/j.1365-2958.2009.06753.x [DOI] [PubMed] [Google Scholar]
- 36.Hamza I., Chauhan S., Hassett R. and O'Brian M.R. (1998) The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. J. Biol. Chem. 273, 21669–21674 10.1074/jbc.273.34.21669 [DOI] [PubMed] [Google Scholar]
- 37.Reyes-Caballero H., Campanello G.C. and Giedroc D.P. (2011) Metalloregulatory proteins: metal selectivity and allosteric switching. Biophys. Chem. 156, 103–114 10.1016/j.bpc.2011.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin J.H., Jung H.J., An Y.J., Cho Y.B., Cha S.S. and Roe J.H. (2011) Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc. Natl Acad. Sci. U.S.A. 108, 5045–5050 10.1073/pnas.1017744108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milano A., Branzoni M., Canneva F., Profumo A. and Riccardi G. (2004) The Mycobacterium tuberculosis Rv2358-furB operon is induced by zinc. Res. Microbiol. 155, 192–200 10.1016/j.resmic.2003.11.009 [DOI] [PubMed] [Google Scholar]
- 40.Ma Z., Gabriel S.E. and Helmann J.D. (2011) Sequential binding and sensing of Zn2+ by Bacillus subtilis Zur. Nucleic Acids Res. 39, 9130–9138 10.1093/nar/gkr625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabriel S.E., Miyagi F., Gaballa A. and Helmann J.D. (2008) Regulation of the Bacillus subtilis yciC gene and insights into the DNA-binding specificity of the zinc-sensing metalloregulator Zur. J. Bacteriol. 190, 3482–3488 10.1128/jb.01978-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng Y.J., Li M., Zhang H.M., Zheng B.W., Han H.M., Wang C.J. et al. (2008) Functional definition and global regulation of Zur, a zinc uptake regulator in a Streptococcus suis serotype 2 strain causing streptococcal toxic shock syndrome. J. Bacteriol. 190, 7567–7578 10.1128/jb.01532-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Outten C.E., Tobin D.A., Penner-Hahn J.E. and O'Halloran T.V. (2001) Characterization of the metal receptor sites in Escherichia coli Zur, an ultrasensitive zinc(II) metalloregulatory protein. Biochemistry 40, 10417–10423 10.1021/bi0155448 [DOI] [PubMed] [Google Scholar]
- 44.Osman D., Piergentili C., Chen J.J., Chakrabarti B., Foster A.W., Lurie-Luke E. et al. (2015) Generating a metal-responsive transcriptional regulator to test what confers metal sensing in cells. J. Biol. Chem. 290, 19806–19822 10.1074/jbc.M115.663427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osman D., Foster A.W., Chen J., Svedaite K., Steed J.W., Lurie-Luke E. et al. (2017) Fine control of metal concentrations is necessary for cells to discern zinc from cobalt. Nat. Commun. 8, 1884 10.1038/s41467-017-02085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neupane D.P., Jacquez B., Sundararajan A., Ramaraj T., Schilkey F.D. and Yukl E.T. (2017) Zinc-dependent transcriptional regulation in Paracoccus denitrificans. Front. Microbiol. 8, 15 10.3389/fmicb.2017.00569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patterson, C.J. (2010) In Vitro Properties and In Vivo Responses of CoaR, ZiaR and Zur (Trans-family Metal-Sensing). Ph.D. Thesis, Newcastle University [Google Scholar]
- 48.Sein-Echaluce V.C., Pallares M.C., Lostao A., Yruela I., Velazquez-Campoy A., Peleato M.L. et al. (2018) Molecular basis for the integration of environmental signals by FurB from Anabaena sp. PCC 7120. Biochem. J. 475, 151–168 10.1042/bcj20170692 [DOI] [PubMed] [Google Scholar]
- 49.Lucarelli D., Vasil M.L., Meyer-Klaucke W. and Pohl E. (2008) The metal-dependent regulators FurA and FurB from Mycobacterium tuberculosis. Int. J. Mol. Sci. 9, 1548–1560 10.3390/ijms9081548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giedroc D.P. (2009) Hydrogen peroxide sensing in Bacillus subtilis: it is all about the (metallo)regulator. Mol. Microbiol. 73, 1–4 10.1111/j.1365-2958.2009.06752.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barnett J.P., Millard A., Ksibe A.Z., Scanlan D.J., Schmid R. and Blindauer C.A. (2012) Mining genomes of marine cyanobacteria for elements of zinc homeostasis. Front. Microbiol. 3, 21 10.3389/fmicb.2012.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Novichkov P.S., Brettin T.S., Novichkova E.S., Dehal P.S., Arkin A.P., Dubchak I. et al. (2012) Regprecise web services interface: programmatic access to the transcriptional regulatory interactions in bacteria reconstructed by comparative genomics. Nucleic Acids Res. 40, W604–W608 10.1093/nar/gks562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaballa A., Wang T., Ye R.W. and Helmann J.D. (2002) Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 184, 6508–6514 10.1128/jb.184.23.6508-6514.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spohn M., Wohlleben W. and Stegmann E. (2016) Elucidation of the zinc-dependent regulation in Amycolatopsis japonicum enabled the identification of the ethylenediamine-disuccinate (S,S-EDDS) genes. Environ. Microbiol. 18, 1249–1263 10.1111/1462-2920.13159 [DOI] [PubMed] [Google Scholar]
- 55.Choi S.H., Lee K.L., Shin J.H., Cho Y.B., Cha S.S. and Roe J.H. (2017) Zinc-dependent regulation of zinc import and export genes by Zur. Nat. Commun. 8, 11 10.1038/ncomms15812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Panina E.M., Mironov A.A. and Gelfand M.S. (2003) Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl Acad. Sci. U.S.A. 100, 9912–9917 10.1073/pnas.1733691100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White A., Ding X.C., van der Spek J.C., Murphy J.R. and Ringe D. (1998) Structure of the metal-ion-activated diphtheria toxin repressor tox operator complex. Nature 394, 502–506 10.1038/28893 [DOI] [PubMed] [Google Scholar]
- 58.Krężel A. and Maret W. (2016) The biological inorganic chemistry of zinc ions. Arch. Biochem. Biophys. 611, 3–19 10.1016/j.abb.2016.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blindauer C.A. (2011) Bacterial metallothioneins: past, present, and questions for the future. J. Biol. Inorg. Chem 16, 1011–1024 10.1007/s00775-011-0790-y [DOI] [PubMed] [Google Scholar]
- 60.Napolitano M., Rubio M.A., Santamaria-Gomez J., Olmedo-Verd E., Robinson N.J. and Luque I. (2012) Characterization of the response to zinc deficiency in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 194, 2426–2436 10.1128/jb.00090-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Foster A.W., Pernil R., Patterson C.J. and Robinson N.J. (2014) Metal specificity of cyanobacterial nickel-responsive repressor InrS: cells maintain zinc and copper below the detection threshold for InrS. Mol. Microbiol. 92, 797–812 10.1111/mmi.12594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sein-Echaluce V.C., Gonzalez A., Napolitano M., Luque I., Barja F., Peleato M.L. et al. (2015) Zur (FurB) is a key factor in the control of the oxidative stress response in Anabaena sp. PCC 7120. Environ. Microbiol. 17, 2006–2017 10.1111/1462-2920.12628 [DOI] [PubMed] [Google Scholar]
- 63.Foster A.W., Osman D. and Robinson N.J. (2014) Metal preferences and metallation. J. Biol. Chem. 289, 28095–28103 10.1074/jbc.R114.588145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guerra A.J. and Giedroc D.P. (2012) Metal site occupancy and allosteric switching in bacterial metal sensor proteins. Arch. Biochem. Biophys. 519, 210–222 10.1016/j.abb.2011.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pawlik M.C., Hubert K., Joseph B., Claus H., Schoen C. and Vogel U. (2012) The zinc-responsive regulon of Neisseria meningitidis comprises 17 genes under control of a Zur element. J. Bacteriol. 194, 6594–6603 10.1128/jb.01091-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheehan L.M., Budnick J.A., Roop R.M. and Caswell C.C. (2015) Coordinated zinc homeostasis is essential for the wild-type virulence of Brucella abortus. J. Bacteriol. 197, 1582–1591 10.1128/jb.02543-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schröder J., Jochmann N., Rodionov D.A. and Tauch A. (2010) The Zur regulon of Corynebacterium glutamicum ATCC 13032. BMC Genomics 11, 12 10.1186/1471-2164-11-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Macia¸g A., Dainese E., Rodriguez G.M., Milano A., Provvedi R., Pasca M.R. et al. (2007) Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189, 730–740 10.1128/jb.01190-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Owen G.A., Pascoe B., Kallifidas D. and Paget M.S.B. (2007) Zinc-responsive regulation of alternative ribosomal protein genes in Streptomyces coelicolor involves Zur and sigma(R). J. Bacteriol. 189, 4078–4086 10.1128/jb.01901-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kallifidas D., Pascoe B., Owen G.A., Strain-Damerell C.M., Hong H.J. and Paget M.S.B. (2010) The zinc-responsive regulator Zur controls expression of the coelibactin gene cluster in Streptomyces coelicolor. J. Bacteriol. 192, 608–611 10.1128/jb.01022-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eckelt E., Jarek M., Frömke C., Meens J. and Goethe R. (2014) Identification of a lineage specific zinc responsive genomic island in Mycobacterium avium ssp. paratuberculosis. BMC Genomics 15, 15 10.1186/1471-2164-15-1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teramoto H., Inui M. and Yukawa H. (2012) Corynebacterium glutamicum Zur acts as a zinc-sensing transcriptional repressor of both zinc-inducible and zinc-repressible genes involved in zinc homeostasis. FEBS J. 279, 4385–4397 10.1111/febs.12028 [DOI] [PubMed] [Google Scholar]
- 73.Smith K.F., Bibb L.A., Schmitt M.P. and Oram D.M. (2009) Regulation and activity of a zinc uptake regulator, Zur, in Corynebacterium diphtheriae. J. Bacteriol. 191, 1595–1603 10.1128/jb.01392-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dowd G.C., Casey P.G., Begley M., Hill C. and Gahan C.G.M. (2012) Investigation of the role of ZurR in the physiology and pathogenesis of Listeria monocytogenes. FEMS Microbiol. Lett. 327, 118–125 10.1111/j.1574-6968.2011.02472.x [DOI] [PubMed] [Google Scholar]
- 75.Lindsay J.A. and Foster S.J. (2001) Zur: a Zn2+-responsive regulatory element of Staphylococcus aureus. Microbiology 147, 1259–1266 10.1099/00221287-147-5-1259 [DOI] [PubMed] [Google Scholar]
- 76.Latorre M., Low M., Garate E., Reyes-Jara A., Murray B.E., Cambiazo V. et al. (2015) Interplay between copper and zinc homeostasis through the transcriptional regulator Zur in Enterococcus faecalis. Metallomics 7, 1137–1145 10.1039/c5mt00043b [DOI] [PubMed] [Google Scholar]
- 77.Patzer S.I. and Hantke K. (2000) The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275, 24321–24332 10.1074/jbc.M001775200 [DOI] [PubMed] [Google Scholar]
- 78.Mastropasqua M.C., D'Orazio M., Cerasi M., Pacello F., Gismondi A., Canini A. et al. (2017) Growth of Pseudomonas aeruginosa in zinc poor environments is promoted by a nicotianamine-related metallophore. Mol. Microbiol. 106, 543–561 10.1111/mmi.13834 [DOI] [PubMed] [Google Scholar]
- 79.Lhospice S., Gomez N.O., Ouerdane L., Brutesco C., Ghssein G., Hajjar C. et al. (2017) Pseudomonas aeruginosa zinc uptake in chelating environment is primarily mediated by the metallophore pseudopaline. Sci. Rep. 7, 10 10.1038/s41598-017-16765-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ellison M.L., Farrow J.M., Parrish W., Danell A.S. and Pesci E.C. (2013) The transcriptional regulator Np20 is the zinc uptake regulator in Pseudomonas aeruginosa. PLoS ONE 8, e75389 10.1371/journal.pone.0075389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pederick V.G., Eijkelkamp B.A., Begg S.L., Ween M.P., McAllister L.J., Paton J.C. et al. (2015) ZnuA and zinc homeostasis in Pseudomonas aeruginosa. Sci. Rep. 5 10.1038/srep13139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lim C.K., Hassan K.A., Penesyan A., Loper J.E. and Paulsen I.T. (2013) The effect of zinc limitation on the transcriptome of Pseudomonas protegens Pf-5. Environ. Microbiol. 15, 702–715 10.1111/j.1462-2920.2012.02849.x [DOI] [PubMed] [Google Scholar]
- 83.Li Y.L., Qiu Y.F., Gao H., Guo Z.B., Han Y.P., Song Y.J. et al. (2009) Characterization of Zur-dependent genes and direct Zur targets in Yersinia pestis. BMC Microbiol. 9, 13 10.1186/1471-2180-9-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sheng Y., Fan F.X., Jensen O., Zhong Z.T., Kan B.A., Wang H. et al. (2015) Dual zinc transporter systems in Vibrio cholerae promote competitive advantages over gut microbiome. Infect. Immun. 83, 3902–3908 10.1128/iai.00447-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mortensen B.L., Rathi S., Chazin W.J. and Skaar E.P. (2014) Acinetobacter baumannii response to host-mediated zinc limitation requires the transcriptional regulator Zur. J. Bacteriol. 196, 2616–2626 10.1128/jb.01650-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang D.L., Tang D.J., Liao Q., Li H.C., Chen Q., He Y.Q. et al. (2008) The Zur of Xanthomonas campestris functions as a repressor and an activator of putative zinc homeostasis genes via recognizing two distinct sequences within its target promoters. Nucleic Acids Res. 36, 4295–4309 10.1093/nar/gkn328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang D.L., Tang D.J., Liao Q., Li X.Q., He Y.Q., Feng J.X. et al. (2009) The Zur of Xanthomonas campestris is involved in hypersensitive response and positively regulates the expression of the hrp cluster via hrpX but not hrpG. Mol. Plant-Microbe Interact. 22, 321–329 10.1094/mpmi-22-3-0321 [DOI] [PubMed] [Google Scholar]
- 88.Moreau G.B., Qin A.P. and Mann B.J. (2018) Zinc acquisition mechanisms differ between environmental and virulent Francisella species. J. Bacteriol. 200, 16 10.1128/jb.00587-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mazzon R.R., Braz V.S., Neto J.F.D. and Marques M.D. (2014) Analysis of the Caulobacter crescentus Zur regulon reveals novel insights in zinc acquisition by TonB-dependent outer membrane proteins. BMC Genomics 15, 14 10.1186/1471-2164-15-734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chaoprasid P., Dokpikul T., Johnrod J., Sirirakphaisarn S., Nookabkaew S., Sukchawalit R. et al. (2016) Agrobacterium tumefaciens Zur regulates the high-affinity zinc uptake system TroCBA and the putative metal chaperone YciC, along with ZinT and ZnuABC, for survival under zinc-limiting conditions. Appl. Environ. Microbiol. 82, 3503–3514 10.1128/aem.00299-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bhubhani S., Sittipo P., Chaoprasid P., Nookabkaew S., Sukchawalit R. and Mongkolsuk S. (2014) Control of zinc homeostasis in Agrobacterium tumefaciens via Zur and the zinc uptake genes znuABC and zinT. Microbiology 160, 2452–2463 10.1099/mic.0.082446-0 [DOI] [PubMed] [Google Scholar]
- 92.Bütof L., Schmidt-Vogler C., Herzberg M., Grosse C. and Nies D.H. (2017) The components of the unique Zur regulon of Cupriavidus metallidurans mediate cytoplasmic zinc handling. J. Bacteriol. 199, 20 10.1128/jb.00372-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schmidt C., Schwarzenberger C., Gross C. and Nies D.H. (2014) FurC regulates expression of zupT for the central zinc importer ZupT of Cupriavidus metallidurans. J. Bacteriol. 196, 3461–3471 10.1128/jb.01713-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ludwig M., Chua T.T., Chew C.Y. and Bryant D.A. (2015) Fur-type transcriptional repressors and metal homeostasis in the cyanobacterium Synechococcus sp PCC 7002. Front. Microbiol. 6, 13 10.3389/fmicb.2015.01217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Navarrete F. and De La Fuente L. (2015) Zinc detoxification is required for full virulence and modification of the host leaf ionome by Xylella fastidiosa. Mol. Plant-Microbe Interact. 28, 497–507 10.1094/mpmi-07-14-0221-r [DOI] [PubMed] [Google Scholar]
- 96.Maret W. (2004) Zinc and sulfur: a critical biological partnership. Biochemistry 43, 3301–3309 10.1021/bi036340p [DOI] [PubMed] [Google Scholar]
- 97.Hernandez J.A., Lopez-Gomollon S., Bes M.T., Fillat M.F. and Peleato M.L. (2004) Three Fur homologues from Anabaena sp. PCC7120: exploring reciprocal protein-promoter recognition. FEMS Microbiol. Lett. 236, 275–282 10.1016/j.femsle.2004.05.050 [DOI] [PubMed] [Google Scholar]
- 98.Lopez-Gomollon S., Sevilla E., Bes M.T., Peleato M.L. and Fillat M.F. (2009) New insights into the role of Fur proteins: FurB (all2473) from Anabaena protects DNA and increases cell survival under oxidative stress. Biochem. J. 418, 201–207 10.1042/bj20081066 [DOI] [PubMed] [Google Scholar]
- 99.Nanamiya H., Akanuma G., Natori Y., Murayama R., Kosono S., Kudo T. et al. (2004) Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol. Microbiol. 52, 273–283 10.1111/j.1365-2958.2003.03972.x [DOI] [PubMed] [Google Scholar]
- 100.Haas C.E., Rodionov D.A., Kropat J., Malasarn D., Merchant S.S. and de Crecy-Lagard V. (2009) A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10, 21 10.1186/1471-2164-10-470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blaby-Haas C.E., Flood J.A., de Crecy-Lagard V. and Zamble D.B. (2012) YeiR: a metal-binding GTPase from Escherichia coli involved in metal homeostasis. Metallomics 4, 488–497 10.1039/c2mt20012k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gielda L.M. and DiRita V.J. (2012) Zinc competition among the intestinal microbiota. mBio 3, 7 10.1128/mBio.00171-12 [DOI] [PMC free article] [PubMed] [Google Scholar]