Abstract
Background:
Oral cancer is a frequently encountered neoplasm of the head and neck region, being the eight most common type of human malignancy worldwide. Despite improvement in its control, morbidity and mortality rates have improved little in the past decades. Therefore, prevention and/or early detection are a high priority. Proteomics with network analysis have emerged as a powerful tool to identify important proteins associated with cancer development and progression that can be potential targets for early diagnosis. In the present study, network- based protein- protein interactions (PPI) for oral cancer were identified and then analyzed for use as key proteins/potential biomarkers.
Material and Methods:
Gene expression data in articles which focused on saliva proteomics of oral cancer were collected and 74 candidate genes or proteins were extracted. Related protein networks of differentially expressed proteins were explored and visualized using cytoscape software. Further PPI analysis was performed by Molecular Complex Detection (MCODE) and BiNGO methods.
Results:
Network analysis of genes/proteins related to oral cancer identified kininogen-1, angiotensinogen, annexin A1, IL-8, IgG heavy variable and constant chains, CRP, collagen alpha-1 and fibronectin as 9 hub-bottleneck proteins. In addition, based on clustering with the MCODE tool, vitronectin, collagen alpha-2, IL-8 and integrin alpha-v were established as 5 distinct seed proteins.
Conclusion:
A hub-bottleneck protein panel may offer a potential /candidate biomarker pattern for diagnosis and treatment of oral cancer disease. Further investigation and validation of these proteins are warranted.
Keywords: Protein-protein interaction, oral cancer, hub-bottleneck, Biomarker, Cluster
Introduction
Oral cancer is a global public health problem, accounting for 2%-3% of all types of malignancies. Annually, 300,000 new cases of oral cancer are reported worldwide (Parkin et al., 2005). Oral squamous cell carcinoma(OSCC) represents the most prevalent malignant neoplasm in the oral cavity (Epstein et al., 2002). Five-year survival rate of patients with oral cancer is as low as 50%. Despite advances in treatment methods, the survival rates for patient with oral cancer did not show improvement (Canto and Devesa, 2002). Therefore, early detection of oral malignancy lesions can be promising in reducing mortality and morbidity-associated with this disease. Oral cancer affects about 360,000 individuals’ worldwide and smoking, age, alcohol consumption, lack of oral health, and human papilloma virus are among oral cancer risk factors (Hu et al., 2007). Squamous cells carcinoma is the most common type of oral cancer with 40,000 new cases and 10,000 deaths every year in the USA (Jemal et al., 2010). About 15% of patients with oral cancer have also other types of cancers such as larynx and lung cancers (Gonsalves et al., 2007). The oral cancer symptoms especially OSCC are often presented at the late stage of the disease and the rate of recurrence after treatment is high. Delayed diagnosis of the disease is one of the major reason leading to high percent of morbidity in this population. Since oral cancer develops in the oral cavity and there is a direct contact between saliva and oral cavity, the use of saliva fluid for identification of oral cancer-related biomarkers has attracted a lot of attention among researchers (Shah et al., 2011). Saliva with several advantages rather than blood and tissues can be a promising and effective source for early diagnosis, prognosis, and treatment of oral cancers. Oral cancer proceed from multi-stages from hyperplasia to an increasing degree of dysplasia to carcinoma in situ, and finally followed by invasive squamous cell carcinoma. Despite advances in oral cancer diagnostic tools, the disease continues to face the challenge of early detection; therefore, identifying tumor markers in saliva as an emerging methodology can play a critical role in identifying disease-related diagnostic and therapeutic biomarkers. Most of biomarkers have been identified from different biological fluids in various human disorders such as cancers (Streckfus and Dubinsky, 2007; Wu et al., 2009). Meanwhile, the advantages of saliva over serum and tissue include the following: noninvasive collection, simplicity of shipping and storing, and facility of handling. Hence, the development of disease biomarker identification based on saliva is a necessity, especially in high risk group and individuals with a history of cancer. The proteomics-based investigations have been used to identify proteins with differential expressions and protein biomarkers for different diseases such as oral cancer. Proteins do not function in isolation and there are interactions between proteins which mediate all biological processes such as metabolic and signaling transduction. In addition, their interactions can control the mechanisms leading to healthy and diseased conditions in organisms due to central role of proteins biological functions (Gonzalez and Kann, 2012). However, the analysis of protein-protein interactions serves as an essential tool for identifying molecular basis of a disease. Consequently, knowledge on identification of potential drug targets and hubs (proteins with larger connectivity in network) (Singh et al., 2007) can be translated and applied into effective diagnostic, therapeutic, and immunotherapeutic strategies (Atan et al., 2014; Safari-Alighiarloo et al., 2014; Keyvani et al., 2016) . Meanwhile, proteomics along with PPI networks analysis can be a powerful approach to identify disease-associated diagnostic biomarkers and physiopathology basis of the disease (Rakshit et al., 2014; Ghamari et al., 2015; Rezaei-Tavirani et al., 2016). Recently, khayer et al., reported oral Squamous Cell Cancer protein-protein interaction network interpretation in comparison to esophageal adenocarcinoma and concluded that biomarker set of OSCC is similar to biomarker panel of esophageal adenocarcinoma. They stated that P53 played an important role in OSCC-related network (Khayer et al., 2017). This study aimed to assess the PPI network and enrichment analysis on extracted proteins from salivary proteomics to introduce some related molecular biomarkers/ key proteins for oral cancer.
Materials and Methods
Saliva-based proteomics associated with oral cancer especially oral squamous cell carcinoma (OSCC) were searched in PubMed, Google scholar, and Science Direct. Then differentially expressed proteins were collected in oral cancer patients and matched with healthy participants. According to the conducted search, totally 74 candidate proteins were selected. UniProt accession numbers (http://www.uniprot.org) of these proteins were extracted using Cytoscape 3.0.2 to analyze protein- protein interaction of oral cancer. “Cytoscape is an open source software project for integrating bimolecular interaction networks with high-throughput expression data”(Shannon et al., 2003). In this study, we integrated the databases and networks and used Molecular Complex Detection (MCODE) to analyze the characteristics of the networks. The MCODE is a cytoscape plugin that is able to determine densely connected regions. Interactomes with a score greater than 2.0 and at least two nodes were selected as significant predictions by MCODE. The second stage of MCODE analysis was to recognize seed nodes as a complex with the highest weighted vertex (forward and outward)(Bader and Hogue, 2003). Gene ontology categories were analyzed to identify the function of each cluster that was generated by the MCODE tool. In fact, functional GO enrichment analysis of each highly connected region (clusters) was performed using BiNGO and Cytoscape plug-in. BiNGO maps the predominant functional themes of a given gene set on the Go hierarchy, and outputs this mapping as a Cytoscape graph (Maere et al., 2005)
Results
The PPI networks of the significantly expressed proteins between oral cancer patients and matched healthy participants contained 1,144 nodes and 2,867 edges (Figure 1). In each network, nodes represent the proteins and edges represent physical or functional interaction between two proteins. The topological analysis of the network was performed using Network Analyzer in Cytoscape. The degree of the node in the network is defined as the number of connections associated with that node. Nodes with high degree are known as hub proteins which are essential in the network structure and also functionally point. The power law of node degree distribution and betweenness centrality distribution were also performed that the distribution implies on the presents of proteins with high centrality values computed by Network Analyzer (Figure 2).
Figure 1.

Whole Connected Component of the PPI Network of Oral Cancer Obtained from Reactome Database by Cytoscape 3.0.2 for 74 Selected Proteins. The network consists of 1144 nodes and 2867 edges. The nodes are lay out by degree and betweenness centrality values (bigger and darker circles corresponded to higher degree and betweenness centrality). The hub-bottleneck proteins in our analysis are shown in numbers 1 to 9, which include Kininogen-1(P01042), Angiotensinogen (P01019), Annexin A1(P04083), Interleukin-8 (P10145), Immunoglobulin heavy variable 3-23(P01764), C-reactive protein (P02741), Immunoglobulin heavy constant alpha 2 (P01877), Collagen alpha-1(I) chain (P02452), and Fibronectin (P02751); respectively.
Figure 2.
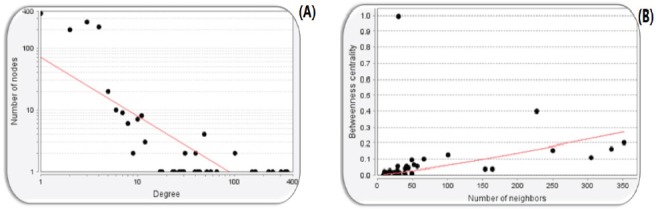
Degree and betweenness Centrality Distribution Curve of the PPI Network of Oral Cancer is Illustrated. The red line indicates the power law. (A) Degree distribution: The degree distribution in the scale-free network in logarithmic scale represents the existence of a small number of nodes with high degree (hubs) and a large of nodes with a low degree. The statistical parameters are determined as R-square on logharithmized values equal to 0.617 and the correlation equals to 0.905. (B) Betweenness centrality distribution: The R-square on logharithmized value is equal to 0.565 and the correlation equals to 0.302.
A scale-free network is a network which has a power-law degree distribution. Protein-protein interaction networks are scale-free networks. The majority of nodes (proteins) in scale-free networks have only a few connections to other nodes, whereas some nodes (hubs) are connected to many other nodes in the network.
The high degree nodes were presented as hub proteins and high betweenness centrality nodes were presented as bottleneck proteins. According to Figure 3, common proteins (9 proteins) were selected in two states (hub and bottleneck states) and represent hub-bottleneck proteins. These proteins included Kininogen-1, angiotensinogen, Annexin A1, Interleukin-8, Immunoglobulin heavy variable 3-23, C-reactive protein, Immunoglobulin heavy constant alpha 2, Collagen alpha-1(I) chain, and Fibronectin.
Figure 3.
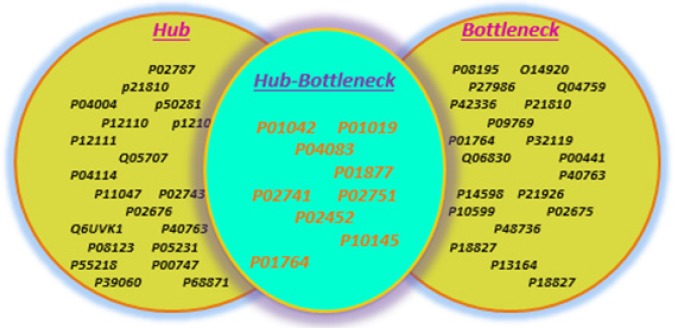
The List of Proteins with High Centrality Measurement, Including Nodes with High Degree Score (as Hub Proteins in Left of Figure) and High betweenness Centrality Score (as Bottleneck Proteins in Right of Figure). The common proteins in two lists are presented as hub-bottleneck proteins (crucial nodes) which are important proteins in pathogenesis and disease therapy.
Protein complexes (clusters) of our biological data were identified by MCODE algorithm. In this method, all nodes in whole weights by local neighborhood density and determines highly connected regions. MCODE is available as a default plugin for Cytoscape network visualization and analytical tool (Killcoyne et al., 2009). MCODE revealed 5 sub-networks that are described in Table 2.
Table 1.
Nine Crucial Nodes Related to the PPI Network of Oral Cancer
| ID | Gene name | Protein name | D | BC |
|---|---|---|---|---|
| P01042 | KNG1 | Kininogen-1 | 334 | 0.16 |
| P01019 | AGT | Angiotensinogen | 325 | 0.2 |
| P04083 | ANXA1 | Annexin A1 | 305 | 0.1 |
| P10145 | CXCL8 | Interleukin-8 | 250 | 0.15 |
| P01764 | IGHV3-23 | Immunoglobulin heavy variable 3-23 | 227 | 0.39 |
| P02741 | CRP | C-reactive protein | 164 | 0.03 |
| P01877 | IGHA2 | Immunoglobulin heavy constant alpha 2 | 154 | 0.03 |
| P02452 | COL1A1 | Collagen alpha-1(I) chain | 101 | 0.12 |
| P02751 | FN1 | Fibronectin | 57 | 0.05 |
ID, D, and BC are abbreviations of Uniprot number, degree, and betweenness centrality, respectively.
Table 2.
Five Significant Clusters Related to the PPI Network of Oral Cancer and Their Properties
| clusters | details |
|---|---|
 |
Nodes: 5 (P12110, P04004, P02461, P12111, P12109) Edges: 13 Cluster Score: 5 Seed node: p04004 Seed Protein: Vitronectin Seed MCODE score: 4.15 |
 |
Nodes: 11 (P29400, P02458, P08572, P05556, P02452, P08123, P02462, P53420, P21810, Q01955, Q14031) Edges: 23 Cluster Score: 4 Seed node: P08123 Seed Protein: Collagen alpha-2(I) chain Seed MCODE score: 6.37 |
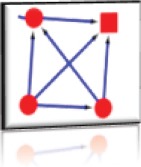 |
Nodes: 4 (P01042, P01019, P10145, P04083) Edges: 7 Cluster Score: 4 Seed node: P10145 Seed Protein: Interleukin-8 Seed MCODE score: 0.15 |
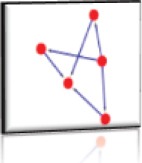 |
Nodes: 5 (P61812, P10600, P01137, Q14766, Q9NS15) Edges: 6 Cluster Score: 3 Seed node: - Seed Protein: - Seed MCODE score: 2 |
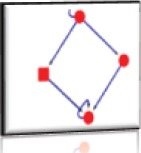 |
Nodes: 4 (P39060, P06756, P24821, P05106) Edges: 6 Cluster Score: 2.6 Seed node: P06756 Seed Protein: Integrin alpha-V Seed MCODE score: 1.2 |
Functional distribution of biological process of oral cancer clusters was shown in Figure 4. According to this Figure, the majority of presented hub-bottleneck proteins in Table 1 were involved in extracellular matrix and structure organization, organ development, cell adhesion, and collagen fibril organization.
Figure 4.
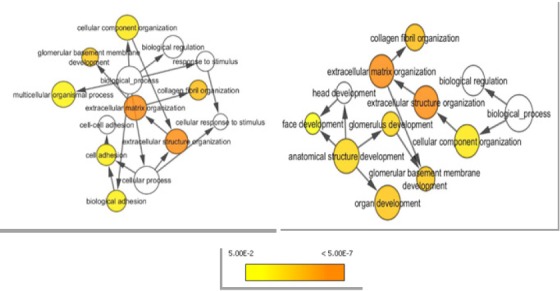
The Biological Process Enrichment Analysis Related to Oral Cancer PPI Network Clusters which are Extracted from BiNGO Plugin in Cytoscape. The colors are corresponded on the biological pathways and the dark circles arerelated to main biological pathways.
Discussion
To form complex assemblies, proteins interact with each other in cellular pathways. Disorders such as cancers are resulted from deregulations of proteins. Molecular mapping is one of methods to evaluate cellular and molecular pathways (Ficenec et al., 2003). Protein- protein interaction network analysis is a useful tool in the field of biomarker discovery in many diseases including cancer (Gulmann et al., 2006; Nguyen and Ho, 2012). In recent years, various cancers are targeted by the researchers in bioinformatics and network analysis. There are several reports in saliva-based proteomics in oral cancer, but there is no investigations analyze the network of this cancer. Therefore, PPI network of 74 proteins related to oral cancer were constructed and assessed in the current study to predict key proteins that are potential drug targets in this disease. Here, the proteins were evaluated based on the importance of their roles in the network. One of the advantages of network analysis is the discrimination of a few nodes among too many nodes in a PPI network (Taylor IW et al., 2009). As represented in Table 1; 9 crucial nodes (bottlenecks) which mostly interacted with the other nodes of the network and controlled them were introduced. These proteins were introduced as the essential elements in PPI network of oral cancer which also play important roles in pathophysiology of oral cancer. For better interpretation, 9 key nodes were classified in two groups based on the presence or absence of any of these hubs in network clusters. The first group included KNG1, AGT, ANXA1, CXCL8, and COLA1 which all of them were presented in clusters number 3 except COLA1 which was located in cluster 2. Therefore, it seems that cluster 3 is tightly related to oral cancer. The proteins in this group are known as potent crucial nodes. In addition, further analysis revealed CXCL8 as seed node in cluster 3, indicating its high importance in this disease. Each of the proteins in the first group had correlation with oral cancer separately or in combination with others. The second category involved IGHV3-23, CRP, IGHA2, and FN1 which are referred to as weak crucial nodes. Due to their absence in clusters, it seems that these proteins are less important than those in the first group. The first potent crucial node with highest score of degree and centrality belonged to Kininogen-1 (KNG1). Kininogen-1 is a multifunctional protein that is expressed in almost all tumors and plays an important role in several pathophysiologic processes, including fibrinolysis, thrombosis, and inflammation, as well as in oncogenesis (Yousef and Diamandis, 2001; Wang et al., 2013). In addition, kininogen-1 is identified as serum biomarker for the early detection of colorectal cancer (Wang et al., 2013). Since the sensitivity and specificity of a suggested biomarker are based on two important indexes (Nickolas et al., 2008), it seems that using this marker alone as diagnostic biomarker in the field of oral cancer cannot be beneficial. Concerning oral cancer, the microarray analysis data indicated that mRNA expression of kininogen-1 gene had correlation with survival rate of patients with tumors. In other words, patients with high level expression of kininogen-1 tend to have a shorter overall survival and recurrence-free survival. Consequently, Kininogen-1 can be a prognostic biological marker for patients with oral cancer (Rastogi et al., 2016).
Angiotensinogen (AGT) is another crucial hub-bottleneck protein. Various factors such as genetic mutations and polymorphisms are associated with oral malignancies. According to literature, angiotensinogen is related to inflammation and also inhibition of human endothelial cell proliferation, cell migration, and angiogenesis in vitro. However, angiotensinogen gene polymorphism is associated with increased risk of oral cancer (Vairaktaris et al., 2008). The correlation between angiotensinogen and oral cancer indicates that the method chosen this crucial node was appropriate in the present study. Annexin A1 was the next crucial hub in oral cancer-related protein network. The important of this protein is reported in several process, including the inflammation response, cell proliferation, cell signaling, and specially in carcinogenesis (Lin et al., 2008). The significant correlation between Annexin A1 expression and pathologic differentiation grade in oral cancer individuals is established ; therefore, Annexin A1 can be a potential biomarker for pathologic differentiation grade of oral cancer (Zhang et al., 2009). Recent studies have reported dysregulation (Overexpression and reduced levels) of ANXA1 expression in several types of cancers (Bai et al., 2004; Wang et al., 2006) such as gastric cancer (Silistino-Souza et al., 2007). Therefore, Annexin A1 cannot be applied alone as a specific biomarker for oral cancer but it can be a useful biomarker panel in oral cancer when combined with other crucial proteins. In most cells, ANXA1 is localized in cytoplasm, but it may translocate to nuclei upon stimulation. Furthermore, the nuclear localization of this crucial node can be used as a promising prognostic factor in oral cancer (Lin et al., 2008).
Another hub-bottleneck and also seed protein (in cluster3) in the first group was IL-8. IL-8 is a pro-inflammatory cytokine that can be detected in tissues and body fluids (blood and saliva) with angiogenic activity. The high expression of IL-8 in cancer cells play an important role in development of various malignancies such as oral cancer (Lisa Cheng et al., 2014; Sahibzada et al., 2017). Due to the high morbidity and mortality rates associated with oral cancer especially OSCC, early detection, prevention, and prediction of this disease is likely to be vital important. The association between expressions of IL-6 and IL-8 and oral cancer can contribute to the pathogenesis of this disease. This association was also observed in this study and interleukin-8 was detected at higher concentrations in saliva that has been linked with increased tumor growth and metastasis. Our findings indicated that salivary IL-8 level can be as a promising biomarker for detecting OSCC. The latest crucial node in the first group was Collagenalpha-1 (I) chain in oral cancer-related network. It is widely acknowledged that the Extracellular Matrix (ECM) of connective tissues plays active roles in numerous biological processes such as cell differentiation, life/death promotion, and carcinogenesis. The relationship between collagen and cancers has been investigated in several studies (Luparello, 2013). For example, in a study, tumor progression reflected a change in collagen present, from type I to type III (Kumari et al., 2017). Furthermore, collagen was introduced as crucial hub node according to our results; therefore, determining the type of collagen in different grade of oral cancer can facilitate therapeutic targeting of molecules responsible for invasion and progression of this cancer.
The second category comprised IGHV3-23, CRP, IGHA2, and FN1 as weak crucial nodes. Khanna et al., studied serum immunoglobulins (IgG, IgA, and IgM) levels in 70 patients with oral cancer and in 40 age-matched normal controls and reported the correlation between clinical stage, tumor size, lymph node status, tumor differentiation, and treatment modalities and Ig serum levels. Moreover, a significant elevation was reported in serum IgM and IgA levels in oral cancer patients as compared to healthy participants (Khanna et al., 1982). C-reactive protein (CRP) was another weak crucial node which is an inflammation marker. In many types of human cancers, CRP plays the role of a prognostic predicting factor. The association between serum CRP level and cancer survival rate has been established in different types of cancers. The presence of an elevated serum CRP level preoperatively is an independent prognostic indicator in oral cancer. Furthermore, determining CRP levels before surgery is useful for better management of cancers treatment such as oral cancer (Kruse et al., 2010; Chen et al., 2011). According to our results, CRP is a hub and can be used in combination with other hubs as a biomarker for oral cancer due to the lack of specificity of this protein for oral cancer. The latest weak crucial hub in this study was fibronection (FN). Fibronectin is a glycoprotein which correlates with cancer (Nagai, 1990). Some investigations have shown that inhibition of fibronectin cleavage could be of significant benefit for the management of oral diseases, including periodontal disease and oral cancer (Stanley et al., 2008). With respect to oral cancer, oral squamous cell carcinoma biopsies exhibit decreased laminin content and increased fibronectin, depending on the aggressiveness and the location of the tumor (de Oliveira Ramos et al., 2016). Hence, the invasive behavior of OSCC not only relies on intrinsic factors (i.e. mutations and abnormal expression of proteins) but also on extrinsic factors (such as the ECM composition), which can help understand the failure of some tumor therapies and development of new anti-tumorigenic approaches (de Oliveira Ramos et al., 2016). We also identified the seed nodes in each cluster. Vitronectin, Collagen alpha-2(I) chain, and integrin alpha-V were seed proteins in number 1, 2, and 5 clusters. Seed nodes are the highest weighted nodes in a graph. In the human protein-protein network, each node (protein) with corresponding gene expression value is regarded as a “seed node”. For a seed node i, this node and its neighbors j within the shortest distance k, form a connected subnetwork with n nodes (Zhuang et al., 2014). Vitronectin (VTN or VN) is a glycoprotein of the hemopexin family which is abundantly found in serum, the extracellular matrix, and bone(Boron and Boulpaep, 2012). Vitronectin has been speculated to be involved in hemostasis and tumor malignancy(Hurt et al., 2010). Therefore, manipulation of this protein may be helpful in predicting the treatment of oral cancer. Seed node in cluster 5 is integrin alpha-V. Integrins are heterodimeric transmembrane proteins formed by non-covalent association of alpha and beta subunits. Both subunits are type 1 membrane proteins with large extracellular ectodomains and short cytoplamic tails (Hynes, 1992). The integrin alpha V (CD51) subunit can form heterodimers with at least five distinct beta subunits. The integrin alpha V beta 3, also known as the vitronectin receptor, has been the focus of intensive research because of its major role in several distinct processes, particularly osteoclast mediated bone resorption, angiogenesis and pathological neovascularization, and tumor metastasis (Teitelbaum, 2000; Felding-Habermann et al., 2001). Expression of the alpha(v)beta6 integrin is strikingly upregulated in several types of carcinoma, including human oral squamous cell carcinoma.
Networks provide a systems-level understanding of the mechanisms underlying diseases by serving as a model for data integration and analysis and they play an important role in system biomedicine (Sevimoglu et al., 2014). They have been used to gain insight into disease mechanisms, analyze therapeutic drugs and their targets, and discover novel network-based biomarkers (Goh et al., 2007). In this study, the interactions between proteins in oral cancer were identified using network and GO biological process that can be useful in identifying biomarker candidates as well as drug targets and investigating possible disease mechanisms that can be helpful in clinic. Network analysis of oral cancer proteins can also help diagnose and predict treatments for oral cancer disease. According to obtained results concerning network analysis of oral cancer, hub-bottleneck proteins (particularly those proteins that are also referred to as hub-bottleneck and also as seed nodes in clusters) had critical role and high importance in pathophysiology and progression of oral cancer. These proteins can be used as potential biomarker for early diagnosis of the disease and determination of appropriate treatment response. On the other hand, drugs designed against these proteins can be important in controlling and managing of oral malignancies. Through network analysis, 5 clusters and 3 seed nodes were also determined, including VTN, COL1A2, and CXCL8 which can serve as candidate biomarkers for oral cancer disease. However, further investigations are needed to evaluate these proteins in detail.
Acknowledgements
This study was financially supported by grant of the Vice Chancellor for Research in Shahid Beheshti University of Medical Sciences, Iran, hereby is highly appreciated.
References
- 1.Atan NAD, Yekta RF, Nejad MR, et al. Pathway and network analysis in primary open angle glaucoma. J Paramed Sci. 2014;5:92–101. [Google Scholar]
- 2.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Biochem. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai X-F, Ni X-G, Zhao P, et al. Overexpression of annexin 1 in pancreatic cancer and its clinical significance. World J Gastroenterol. 2004;10:1466. doi: 10.3748/wjg.v10.i10.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boron WF, Boulpaep EL. Medical physiology, 2e updated edition e-book:with student consult online access, Elsevier health sciences. 2012 [Google Scholar]
- 5.Canto MaT, Devesa SS. Oral cavity and pharynx cancer incidence rates in the United States, 1975–1998. Oral Oncol. 2002;38:610–7. doi: 10.1016/s1368-8375(01)00109-9. [DOI] [PubMed] [Google Scholar]
- 6.Chen HH, Chen IH, Liao CT, et al. Preoperative circulating C-reactive protein levels predict pathological aggressiveness in oral squamous cell carcinoma:A retrospective clinical study. Clin Otolaryngol. 2011;36:147–53. doi: 10.1111/j.1749-4486.2011.02274.x. [DOI] [PubMed] [Google Scholar]
- 7.de Oliveira Ramos G, Bernardi L, Lauxen I, et al. Fibronectin modulates cell adhesion and signaling to promote single cell migration of highly invasive oral squamous cell carcinoma. PLoS One. 2016;11:e0151338. doi: 10.1371/journal.pone.0151338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epstein JB, Zhang L, Rosin M. Advances in the diagnosis of oral premalignant and malignant lesions. J Can Dent Assoc. 2002;68:617–21. [PubMed] [Google Scholar]
- 9.Felding-Habermann B, O'Toole TE, Smith JW, et al. Integrin activation controls metastasis in human breast cancer. Proc Natl Acad Sci U S A. 2001;98:1853–8. doi: 10.1073/pnas.98.4.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ficenec D, Osborne M, Pradines J, et al. Computational knowledge integration in biopharmaceutical research. Brief Bioinform. 2003;4:260–78. doi: 10.1093/bib/4.3.260. [DOI] [PubMed] [Google Scholar]
- 11.Ghamari E, Zali H, Rezaie Tavirani M, et al. Proteomic study in the rat hippocampus as a measure of human Alzheimer's disease. Koomesh. 2015;16:611–20. [Google Scholar]
- 12.Goh K, Cusick ME, Valle D, et al. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonsalves WC, Chi AC, Neville BW. Common oral lesions:Part I. Superficial mucosal lesions. Am Fam Physician. 2007;75:501–6. [PubMed] [Google Scholar]
- 14.Gonzalez MW, Kann MG. Protein interactions and disease. PLoS Comput Biol. 2012;8:e1002819. doi: 10.1371/journal.pcbi.1002819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gulmann C, Sheehan K, Kay E, et al. Array-based proteomics:mapping of protein circuitries for diagnostics, prognostics, and therapy guidance in cancer. J Pathol. 2006;208:595–606. doi: 10.1002/path.1958. [DOI] [PubMed] [Google Scholar]
- 16.Hu S, Yu T, Xie Y, et al. Discovery of oral fluid biomarkers for human oral cancer by mass spectrometry. Cancer Genomics Proteomics. 2007;4:55–64. [PubMed] [Google Scholar]
- 17.Hurt EM, Chan K, Duhagon Serrat MA, et al. Identification of vitronectin as an extrinsic inducer of cancer stem cell differentiation and tumor formation. Stem Cells. 2010;28:390–8. doi: 10.1002/stem.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hynes RO. Integrins:versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 19.Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 20.Keyvani H, Ahmadi NA, Ranjbar MM, et al. Immunoinformatics Study of gp120 of Human Immunodeficiency Virus Type 1 Subtype CRF35_AD Isolated from Iranian Patients. Arch Clin Infect Dis. 2016;11:e36270. [Google Scholar]
- 21.Khayer N, Azodi MZ, Mansouri V, et al. Oralsquamous cell cancer protein-protein interaction network interpretation in comparison to esophageal adenocarcinoma. Gastroenterol Hepatol Bed Bench. 2017;10:118–24. [PMC free article] [PubMed] [Google Scholar]
- 22.Khanna N, Das S, Khanna S. Serum immunoglobulins in squamous cell carcinoma of the oral cavity. J Surg Oncol. 1982;20:46–8. doi: 10.1002/jso.2930200111. [DOI] [PubMed] [Google Scholar]
- 23.Killcoyne S, Carter GW, Smith J, et al. Cytoscape:a community-based framework for network modeling. J Theor Biol. 2009;563:219–39. doi: 10.1007/978-1-60761-175-2_12. [DOI] [PubMed] [Google Scholar]
- 24.Kruse AL, Luebbers HT, Grätz KW. C-reactive protein levels:a prognostic marker for patients with head and neck cancer? Head Neck Oncol. 2010;2:21. doi: 10.1186/1758-3284-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumari K, Ghosh S, Patil S, et al. Expression of type III collagen correlates with poor prognosis in oral squamous cell carcinoma. J Investig Clin Dent. 2017;8:e12253. doi: 10.1111/jicd.12253. [DOI] [PubMed] [Google Scholar]
- 26.Lin CY, Jeng YM, Chou HY, et al. Nuclear localization of annexin A1 is a prognostic factor in oral squamous cell carcinoma. J Surg Oncol. 2008;97:544–50. doi: 10.1002/jso.20992. [DOI] [PubMed] [Google Scholar]
- 27.Lisa Cheng Y-S, Jordan L, Gorugantula LM, et al. Salivary interleukin-6 and-8 in patients with oral cancer and patients with chronic oral inflammatory diseases. J Periodontol. 2014;85:956–65. doi: 10.1902/jop.2013.130320. [DOI] [PubMed] [Google Scholar]
- 28.Luparello C. Aspects of collagen changes in breast cancer. J Carcinog Mutagen. 2013;13:007. [Google Scholar]
- 29.Maere S, Heymans K, Kuiper M. BiNGO:a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–9. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 30.Nagai S. Experimental studies on dynamics of fibronectin and fibrinogen in oral cancer. J Oral Maxillofac Surg Med Pathol. 1990;36:465–83. [Google Scholar]
- 31.Nguyen TP, Ho TB. Detecting disease genes based on semi-supervised learning and protein- protein interaction networks. Artif Intell Med. 2012;54:63–71. doi: 10.1016/j.artmed.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Nickolas TLO, Rourke MJ, Yang J, et al. Sensitivity and specificity of a single emergecy department measurment of urinary neutrophil geatinase-associated lipocalin for diagnosing acute kidney injury. Ann Int Med. 2008;148:810–9. doi: 10.7326/0003-4819-148-11-200806030-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 34.Rakshit H, Rathi N, Roy D. Construction and analysis of the protein-protein interaction networks based on gene expression profiles of Parkinson's disease. PLoS One. 2014;9:e103047. doi: 10.1371/journal.pone.0103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rastogi N, Kumar S, Kapooor V, et al. Is there a need for adjuvant chemoradiation in periampullary carcinoma after pancreaticoduodenectomy? Eur J Cancer. 2016;60:e10. [Google Scholar]
- 36.Rezaei-Tavirani M, Zamanian-Azodi M, Rajabi S, et al. Protein clustering and interactome analysis in Parkinson and Alzheimer's diseases. Arch Iran Med. 2016;19:101–9. [PubMed] [Google Scholar]
- 37.Safari-Alighiarloo N, Taghizadeh M, Rezaei-Tavirani M, et al. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol Hepatol Bed Bench. 2014;7:17–31. [PMC free article] [PubMed] [Google Scholar]
- 38.Sahibzada HA, Khurshid Z, Khan RS, et al. Salivary IL-8, IL-6 and TNF-αas potential diagnostic biomarkers for oral cancer. Diagnostics. 2017;7:21. doi: 10.3390/diagnostics7020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sevimoglu T, Arga KY. The role of protein interaction networks in systems biomedicine. Comput Struct Biotechnol J. 2014;11:22–7. doi: 10.1016/j.csbj.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah FD, Begum R, Vajaria BN, et al. A review on salivary genomics and proteomics biomarkers in oral cancer. Indian J Clin Biochem. 2011;26:326–34. doi: 10.1007/s12291-011-0149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shannon P, Markiel A, Ozier O, et al. Cytoscape:a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silistino-Souza R, Rodrigues-Lisoni FC, Cury PM, et al. Annexin 1:differential expression in tumor and mast cells in human larynx cancer. Int J Cancer. 2007;120:2582–9. doi: 10.1002/ijc.22639. [DOI] [PubMed] [Google Scholar]
- 43.Singh GP, Ganapathi M, Dash D. Role of intrinsic disorder in transient interactions of hub proteins. Proteins. 2007;66:761–5. doi: 10.1002/prot.21281. [DOI] [PubMed] [Google Scholar]
- 44.Stanley CM, Wang Y, Pal S, et al. Fibronectin fragmentation is a feature of periodontal disease sites and diabetic foot and leg wounds and modifies cell behavior. J Periodontol. 2008;79:861–75. doi: 10.1902/jop.2008.070492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Streckfus CF, Dubinsky WP. Proteomic analysis of saliva for cancer diagnosis. Expert Rev Proteomics. 2007;4:329–32. doi: 10.1586/14789450.4.3.329. [DOI] [PubMed] [Google Scholar]
- 46.Taylor IW, Linding R, Warde-Farley D, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 47.Teitelbaum SL. Osteoclasts, integrins, and osteoporosis. J Bone Miner Res. 2000;18:344–9. doi: 10.1007/s007740070007. [DOI] [PubMed] [Google Scholar]
- 48.Vairaktaris E, Yapijakis C, Vylliotis A, et al. Angiotensinogen polymorphism is associated with risk for malignancy but not for oral cancer. Anticancer Res. 2008;28:1675–9. [PubMed] [Google Scholar]
- 49.Wang J, Wang X, Lin S, et al. Identification of kininogen-1 as a serum biomarker for the early detection of advanced colorectal adenoma and colorectal cancer. PLoS One. 2013;8:e70519. doi: 10.1371/journal.pone.0070519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang KL, Wu T-T, Resetkova E, et al. Expression of annexin A1 in esophageal and esophagogastric junction adenocarcinomas:association with poor outcome. Clin Cncer Res. 2006;12:4598–604. doi: 10.1158/1078-0432.CCR-06-0483. [DOI] [PubMed] [Google Scholar]
- 51.Wong DT. Salivary diagnostics powered by nanotechnologies, proteomics and genomics. J Am Dent Assoc. 2006;137:313–21. doi: 10.14219/jada.archive.2006.0180. [DOI] [PubMed] [Google Scholar]
- 52.Wu Z-Z, Wang J-G, Zhang X-L. Diagnostic model of saliva protein finger print analysis of patients with gastric cancer. World J Gastroenterol. 2009;15:865–70. doi: 10.3748/wjg.15.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yousef GM, Diamandis EP. The new human tissue kallikrein gene family:structure, function, and association to disease. Endocr Rev. 2001;22:184–204. doi: 10.1210/edrv.22.2.0424. [DOI] [PubMed] [Google Scholar]
- 54.Zhang L, Yang X, Zhong Lp, et al. Decreased expression of Annexin A1 correlates with pathologic differentiation grade in oral squamous cell carcinoma. J Oral Pathol Med. 2009;38:362–70. doi: 10.1111/j.1600-0714.2008.00678.x. [DOI] [PubMed] [Google Scholar]
- 55.Zhuang L, Wu Y, Han J, et al. A network biology approach to discover the molecular biomarker associated with hepatocellular carcinoma. Biomed Res Int. 2014;2014:6. doi: 10.1155/2014/278956. [DOI] [PMC free article] [PubMed] [Google Scholar]


