Abstract
Chromatin-modifying factors have essential roles in DNA processing pathways that dictate cellular functions. The ability of chromatin modifiers, including the INO80 and SWR1 chromatin-remodelling complexes, to regulate transcriptional processes is well established. However, recent studies reveal that the INO80 and SWR1 complexes have crucial functions in many other essential processes, including DNA repair, checkpoint regulation, DNA replication, telomere maintenance and chromosome segregation. During these diverse nuclear processes, the INO80 and SWR1 complexes function cooperatively with their histone substrates, γ-H2AX and H2AZ. This research reveals that INO80 and SWR1 ATP-dependent chromatin remodelling is an integral component of pathways that maintain genomic integrity.
The structure of chromatin is inherently stable. Therefore, to allow DNA processing and metabolism, chromatin-modifying factors are needed to create a dynamic chromatin environment that facilitates these activities. The modification of chromatin can occur through several mechanisms, including histone post-translational modification by histone-modifying enzymes as well as variation of nucleosome composition and positioning by ATP-dependent chromatin-remodelling complexes.
The ATPase subunits of chromatin-remodelling com-plexes belong to the SWI/SNF family1, which is part of a large superfamily of helicases and translocases called superfamily 2 (SF2). The SWI/SNF family is named after the first identified chromatin-remodelling complex2,3. Members of the SWI/SNF chromatin-remodelling family include the SWI/SNF, ISWI, CHD and INO80 subfamilies in Saccharomyces cerevisiae and have similarities in their DEAD/H box-containing ATPase subunits. However, they also have divergent characteristics that further distinguish them into different subfamilies. The INO80 subfamily is the most recent addition to the SWI/SNF family of chromatin remodellers, and ATPase orthologues and homologues of Ino80 have been identified in yeast4–7, flies8,9, plants10,11 and mammals12–14 (TABLE 1). The chromatin-remodelling complexes of the INO80 subfamily are: INO80 and SWR1 in S. cerevisiae; INO80, Snf2-related CBP activator protein (SRCAP) and p400 in mammals; and INO80 and p400 in Drosophila melanogaster. The INO80 subfamily seems to be the most evolutionarily conserved of all of the chromatin -remodelling subfamilies owing to the high degree of homology in the ATPase subunit (both in the ATPase domain and the surrounding sequences), the presence of a large number of orthologous sub-units in many organisms and the relatively conserved composition of individual complexes.
Table 1 |.
Homologous INO80, SWR1 and SRCAP chromatin-remodelling complexes
| Subunit type | INO80 complex | SWR1 and SRCAP complexes | |||
|---|---|---|---|---|---|
| Saccharomyces | Homo sapiens | Drosophila | Saccharomyces | Homo sapiens | |
| cerevisiae | melanogaster | cerevisiae | |||
| ATPase | Ino80 | INO80 | INO80 | Swr1 | SRCAP |
| RuvB-like | Rvb1 and Rvb2 | RUVBL1 and RUVBL2 | Reptin and Pontin | Rvb1 and Rvb2 | RUVBL1 and RUVBL2 |
| Actin | Act1 | β-Actin | Actin | Act1 | β-Actin |
| Actin-related protein | Arp4, Arp5 and Arp8 | BAF53A, ARP5 and | ARP5 and ARP8 | Arp4 and Arp6 | BAF53A and ARP6 |
| ARP8 | |||||
| YEATS protein | Taf14 | Not known | Not known | Yaf9 | GAS41 |
| Non-conserved | Ies1, Ies2, Ies3, Ies4, | Amida, CCDC95, | Pleiohomeotic | Bdf1, Swc2, Swc3, | DMAP1, GAS41, |
| subunits* | Ies5, Ies6 and Nhp10 | FLJ20309, IES2, IES6, | Swc4, Swc5, Swc6 and | tubulin, XPG, YL1 and | |
| MCRS1, NFRKB, | Swc7 | ZnF-HIT1 | |||
| UCH37 and YY1 | |||||
Subunits in all other rows of this table are conserved. Act, actin; Arp, actin-related protein; BAF53A, BRG1-associated factor 53A; Bdf1, bromodomain factor; CCDC95, coiled-coil domain-containing 95; DMAP1, DNA methyltransferase 1-associated protein 1; GAS41, glioma amplified sequence 41; Ies, INO80 subunit; MCRS1, microspherule protein 1; NFRKB, nuclear factor related to κB-binding protein; Nhp10, non-histone protein 10; Rvb, RuvB-like; RUVBL, RuvB-like; SRCAP, SNF2-related CBP activator protein; Swc, SWR1 complex; Taf14, TBP-associated factor 14; UCH37, ubiquitin C-terminal hydrolase 37; XPG, xeroderma pigmentosa group G; Yaf9, yeast AF9; YEATS, Yaf9, ENL, AF9, Taf14, Sas5; YY1, yin yang 1; Znf-HIT1, zinc finger-His triad protein 1.
Like most chromatin-remodelling complexes, INO80 subfamily complexes have been identified as transcriptional regulators. The ability of the S. cerevisiae INO80gene product to regulate inositol-responsive gene expression was the reason for its initial discovery15. Since their characterization, INO80 subfamily complexes have been found to directly regulate the expression of genes in yeast4–7,16, mammals17,18, flies9 and plants10,11 . The effect of the INO80 complex on transcription is undoubtedly linked to the function of the transcription factors yin yang 1 (YY1; in mammals)17 and Pleiohomeotic (in D. melanogaster)9, which are found in INO80 complexes (TABLE 1). These trans-cription factors, which are involved in cell proliferation, differentiation and embryonic development, can serve to specify the genes that are targeted for INO80-mediated chromatin remodelling. Unlike genes that are regulated by the INO80 complex, SWR1-regulated genes in S. cerevisiae are enriched near telo-meres, suggesting that the chromatin-remodelling activity of the SWR1 complex might be needed to regulate transcription in particularly heterochromatic chromosomal regions5,6. In addition, the chromatin- remodelling activity of homologous SWR1 and SRCAP complexes results in increased expression of genes in plants and mammals10,18.
The purified S. cerevisiae INO80 and SWR1 complexes contain 15 and 14 subunits (TABLE 1), respectively, and are approximately 1.2–1.5 MDa each in mass4–6. A distinguishing characteristic of the INO80 subfamily, including the INO80 and SWR1 complexes, is the presence of RuvB-like helicases in the complexes4,5,14. The RuvB helicase is a DNA repair factor in bacteria19,20, and thus the presence of RuvB-like subunits in the complexes suggests a role for the INO80 subfamily in DNA damage responses. Indeed, recent research shows that DNA damage responses are impaired when components of the INO80 subfamily are mutated in yeast21–23, plants11, flies8 and mammals14,24. These findings changed the perception of chromatin-remodelling complexes, which were typically considered to be transcriptional regulators, and initiated pioneering research that has revealed multiple roles for chromatin- remodelling complexes in additional genome stability pathways, including cell cycle checkpoint activation, DNA replication, telomere regulation, centromere stability and chromosome segregation.
The activity of homologous INO80 and SWR1 complexes in these diverse nuclear processes is a composite of the function of several subunits in these complexes. essential subunits that form the catalytic core include the ATPase (INO80, Swr1 and SRCAP), RuvB-like proteins and the actin-related protein (Arp) subunits (TABLE 1). Other subunits have regulatory functions that are specific to particular processes, such as the yeast INO80 complex components non-histone protein 10 (Nhp10; an HMG-like protein) in DNA repair22, INO80 subunit 4 (Ies4) in the DNA damage checkpoint response25 and Ies3 in telomere regulation26 (TABLE 1). One function of the regulatory subunits is to facilitate the association of unique chromatin substrates with the INO80, SWR1 and SRCAP complexes. Interestingly, complexes of the INO80 subfamily have a prominent and distinct affinity for the histone variants γ-H2AX and H2AZ. (Phosphorylated H2AX is often referred to as γ-H2AX and will be used in this article for consistent nomenclature.) γ-H2AX and H2AZ are not found in canonical nucleosomes and function to direct the activities of the INO80 subfamily in specific processes, such as DNA repair and DNA damage checkpoint responses5,8,21–23.
In this Review, we discuss the roles of homologous INO80 and SWR1 chromatin-remodelling complexes beyond transcription, and elaborate on their roles in pathways that maintain genomic stability, such as DNA repair, cell cycle checkpoint regulation, DNA replication, chromosome segregation and telomere maintenance. Moreover, using the INO80 and SWR1 complexes as examples, we aim to show that the major pathways of chromatin modification, such as chromatin remodelling, histone post-translational modification and histone variant deposition are connected. Furthermore, INO80 and SWR1 also exemplify the distinct mechanisms that regulate chromatin-remodelling complexes. Other chromatin-remodelling complexes, including the INO80 subfamily member and SRCAP paralogue p400 (of the TIP60, or p400, complex), are not mentioned. However, we discuss the regulatory mechanisms that direct the activities of these complexes in such varied nuclear processes, such as the functional relationship between histone substrates and the INO80, SWR1 and SRCAP complexes.
Unique components of the INO80 subfamily
The ATPase subunits of the INO80 subfamily are distinguished from other ATPases in the ISWI, SWI/SNF and CHD subfamilies owing to the presence of a spacer region that splits the conserved ATPase domain (FIG. 1a,b). Deletion of this spacer region in S. cerevisiae Swr1 ATPase results in loss of the association of several subunits from the SWR1 complex, including the RuvB-like Rvb1 and Rvb2 subunits27. Rvb1 and Rvb2 are also present in the S. cerevisiae INO80 complex4 and are homologous to human RUvBL1 and RUvBL2 and D. melanogaster Reptin and Pontin, which are also present in other INO80 subfamily complexes (TABLE 1).
Figure 1 |. Chromatin-remodelling mechanisms of the INO80 and SWR1 complexes.
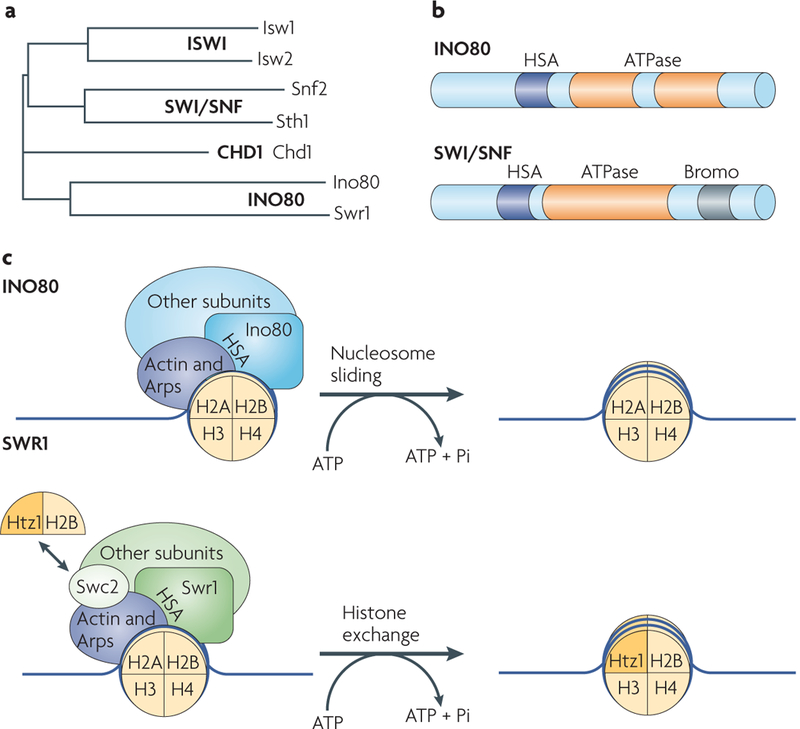
| a | Phylogeny of Saccharomyces cerevisiae chromatin-remodelling ATPase subunits. Members of the ISWI, SWI/SNF, CHD1 and INO80 subfamilies are shown. b | ATPase subunits of the INO80 and SWI/SNF subfamilies are shown. The ATPase domain of the INO80 subfamily is split in the middle by a spacer region. The SWI/SNF ATPase subunit contains a bromodomain (Bromo), which binds to acetylated histones. Both subunits contain a helicase-SANT-associated (HSA) domain that mediates the association of the subunit with actin and actin-related proteins (Arps). c | Distinct ATP-dependent chromatinremodelling mechanisms of INO80 and SWR1 complexes. The INO80 complex repositions nucleosomes to a central position from the end of the DNA. The SWR1 complex incorporates the histone variant Htz1 into the nucleosome. Both mechanisms require ATP hydrolysis and the function of actin and Arp subunits in the complexes.
The RuvB-like proteins found in homologous INO80 and SWR1 complexes are essential for viability in eukaryotes28,29 and are functionally related to the bacte-rial RuvB helicase, which forms a double hexamer and mediates Holliday junction migration during homologous recombination in double-strand break (DSB) repair 30. Interestingly, these RuvB-like proteins are each found at a 1:6 stoichometry in the INO80 subfamily complexes4,5, which is reminiscent of the bacterial RuvB hexameric ring surrounding a Holliday junction, and is suggestive of a role for the INO80 subfamily in DSB repair.
Remodelling mechanisms and substrates
Homologous INO80 and SWR1 complexes have distinc-tive features that are indicative of specialized chromatin-remodelling activities. In this section, we discuss the activities of homologous INO80 and SWR1 complexes, with emphasis on their chromatin substrates. Notably, these complexes associate with histone variants (BOX 1) and this association is evolutionarily conserved and distinct from other chromatin-remodelling subfamilies. These unique chromatin substrates have important roles in specifying the chromatin-remodelling activities of homologous INO80 and SWR1 complexes.
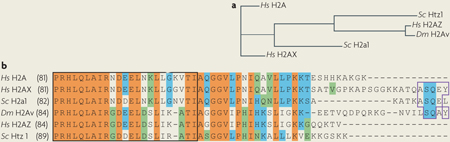
Box 1 |. Histone H2a variants are substrates of the INO80 subfamily.
Of the four core histones, variants of histone H2A are the most common114 and can be incorporated into the nucleosome to signal for specific DNA processing events, such as DNA repair and transcription. Members of the INO80 subfamily bind to H2A variants to facilitate chromatin remodelling. H2AZ is a highly conserved histone variant that has been found in a range of organisms, including protozoans, fungi and metazoans (see the figure, part a, which shows the phylogeny of histone H2A variants in Homo sapiens (Hs), Saccharomyces cerevisiae (Sc) and Drosophila melanogaster (Dm)). These H2AZ-like variants are more homologous to each other across species than to canonical H2A, which suggests that they arose relatively early during evolution and have specialized conserved functions114. Mammalian histone H2AX, as well as the D. melanogaster homologue H2Av, is a variant found in approximately 10% of nucleosomes throughout the genome115,116. H2a1 and H2a2 (only H2a1 is shown), the predominant canonical histones in yeast, are found in most nucleosomes and are orthologous to the mammalian H2AX variant. Unlike H2AZ-like histones, the H2AX-like histone variants have a large degree of homology with canonical human H2A, with the main sequence deviation located in their carboxyl termini. During the activation of DNA damage responses, ataxia telangiectasia (A-T) mutated (ATM) and A-T and RAD3-related (ATR) kinases (Tel1 and Mec1 in yeast) phosphorylate the H2AX-like histones on the Ser of the C-terminal (S/T)Q consensus motif55,56 (see the figure, part b). In addition to being homologous to human H2AZ, D. melanogaster H2Av is also phosphorylated by ATM and ATR on its C terminus consensus sequences. Thus, H2Av is a unique hybrid that contains similarities to both H2AX and H2AZ. Identical amino acids are shown in orange, conserved amino acids in blue, similar amino acids in green and unique amino acids are not shaded. Amino acids in the core histone domain are boxed with a black line and C terminus residues that contain phosphorylation sites are highlighted in purple.The numbers in parentheses indicate the position of amino acids in the sequence.
Chromatin-remodelling mechanisms of the INO80 complex.
Most of the chromatin-remodelling complexes in the SWI/SNF family have helicase domains in their respec-tive ATPase subunits but do not exhibit helicase activity. However, the S. cerevisiae INO80 complex is unique because it does exhibit in vitro ATP-dependent DNA helicase activity4. The mammalian INO80 complex preferentially binds two specialized DNA structures in vitro — a four-way junction that mimics a Holliday junction and a three-way junction that resembles a replication fork24.
The in vitro ATPase activity of IN080 is stimulated not only by DNA but also by nucleosomes4,31. Furthermore, chromatin-remodelling assays show that the INO80 complex repositions (or slides) nucleosomes in vitro on a reconstituted chromatin template4,31,32 (FIG. 1c). In vivo , INO80 influences nucleosome evic-tion33,34, a rather dramatic chromatin-remodelling event in which the nucleosomes are completely removed from the DNA, thereby leaving the DNA accessible to participate in nuclear functions without the pres-ence of histones. INO80-mediated nucleosome sliding might precede the resulting nucleosome eviction that is observed in vivo, thereby linking these two mechanisms in the same process. However, if distinct, these activities might functionally facilitate different DNA processing events. For instance, nucleosome sliding can reposition the nucleosome to expose or mask a specific DNA sequence to regulate its recognition to a DNA-binding factor, such as a transcription factor. Alternatively, his-tone eviction might be necessary when the presence of nucleosomes severely impedes the DNA processing event, such as DNA repair and replication.
The chromatin-remodelling activity of the S. cer-evisiae INO80 and SWR1 complexes is dependent on the function of actin and Arps27,31,35,36 (TABLE 1). Studies in S. cerevisiae show that the evolutionarily conserved helicase-SANT-associated domain (HSA domain) in the ATPase subunits of the INO80 subfamily complexes are required for the binding of Arps and actin components to the complex36. Mutants of Arps in S. cerevisiae diminish the chromatin-remodelling activity of the INO80 com-plex, as these factors mediate interactions between the nucleosome and chromatin-remodelling complexes21,31,37. Therefore, Arps and actin provide a conserved func-tional module that links the chromatin-remodelling enzyme and its substrate (FIG. 1b,c).
Chromatin-remodelling mechanisms of the SWR1 complex.
The S. cerevisiae SWR1 complex co-purifies with the histone variant Htz1 (H2AZ in mammals)5 (BOX 1). Histone variants are often incorporated into nucleo-somes at site-specific locations to accommodate cell-ular functions, such as transcription and replication, that require particular chromatin templates. The SWR1 and SRCAP complexes facilitate ATP-dependent histone replacement of canonical H2A with the H2AZ variant in the nucleosome5,6,10,12. H2AZ has essential functions in a range of cellular processes, such as transcriptional regulation5, heterochromatin establishment38,39, chromo- some segregation40,41, cell cycle progression42 and DNA damage responses34,43. A clue to the function of H2AZ in these diverse processes might be found in the structural analyses of this histone variant. Analyses of chromatin fibres in vitro suggest that H2AZ promotes different conformational states of the nucleosome that might facilitate varied chromatin functions44. Specifically, the crystal structure of H2AZ-containing nucleosomes suggests that the nucleosome is destabilized by a region that includes the carboxyl-terminal α-helix of H2AZ, which contains a high degree of sequence divergence from H2A45 (BOX 1). Loss of this H2AZ C-terminal region results in poor survival in yeast27 and impaired development in D. melanogaster46. Importantly, this region of H2AZ is also essential for recognition by the SWR1 chromatin-remodelling complex for histone exchange27.
This H2AZ histone exchange mechanism is a multistep process that uses the function of the SWR1 complex 2 (Swc2), Swc6 and Arp6 subunits of the complex. These are responsible for binding to the histone variant, associating with the nucleosome and facilitating histone exchange27. In particular, the conserved Swc2 subunit that binds the histone variant contains clusters of acidic residues, a feature that is reminiscent of other histone chaperones27 (FIG. 1c). This H2AZ histone exchange process is specific to the SWR1 and SRCAP complexes and requires unique subunits that are found only in these homologous complexes. However, there are some similarities between the S. cerevisiae SWR1 and INO80 complexes in that the in vitro remodelling activity of both complexes is dependent on their Arps and actin subunits27,31,36 (FIG. 1b,c). Therefore, both common and distinct mechanisms of chromatin remodelling exist among the different members of the INO80 subfamily.
Regulation of the INO80 complex
The function of the chromatin-remodelling complexes can be modulated by multiple mechanisms in response to cell stimuli and signalling pathways. One mechanism for regulating the function of the INO80 complex is the posttranslational modification of specific subunits of the complex. For instance, in response to activation of DNA damage response pathways, the Ies4 subunit of the S. cerevisiae INO80 complex is phosphorylated by the ataxia-telangiectasia (A-T) mutated (ATM) and A-T and RAD3-related (ATR) kinases in vivo and in vitro25. The ATM and ATR kinases (Tel1 and Mec1 in yeast) are essential regulators of the DNA damage responses47, as mutations in these genes in humans result in disorders that are characterized by DNA damage sensitivity and cancer predisposition48,49. Phosphorylation of Ies4 modulates the cellular response to DNA-damaging agents but does not alter the composition of the complex or its ability to localize to sites of DNA damage. Although not formally proven, the chromatin-remodelling activity of the INO80 complex at sites of DNA damage might be modified by Ies4 phosphorylation. Indeed, a similar scenario has been identified for the chromatin-remodelling activities of SWI/SNF in humans, which are inactivated in a cell cycle-dependent manner following phosphorylation50.
The activities of the INO80 complex are also regu-lated by the inositol signalling pathway. Inositol lipids and inositol polyphosphates modulate the activities of chromatin-remodelling complexes of the INO80, ISWI and SWI/SNF subfamilies51–53. Inositol signalling is initiated by a range of external stimuli, such as growth factors and hormones, and regulates a number of crucial cellular functions, including cell growth, apoptosis and differentiation54. By an unknown mechanism, the inositol polyphosphate kinase Arg82 (also known as Ipk2) promotes the in vivo association of the S. cerevisiae INO80 complex to the promoters of the phosphate responsive genes PHO5 and PHO84 (REF. 52). Decreased binding of the INO80 complex to these promoters in arg82 mutant cells results in reduced chromatin remodelling and reduced expression of these genes. Furthermore, inositol hexakisphosphate (InsP6) inhibits the ATP-dependent nucleosome sliding activity of the S.cerevisiae INO80 complex in vitro51. Thus, small signalling molecules can modulate the activity of the INO80 complex.
Remodelling beyond transcription
Many of the chromatin-remodelling mechanisms that facilitate the roles of the INO80, SWR1 and SRCAP complexes beyond transcription are unknown. However, the functional relationship between the complexes and their chromatin substrates, such as γ-H2AX and H2AZ histone variants, is emerging in some cases. As pre-viously mentioned, the INO80 subfamily prominently and specifically associates with γ-H2AX and H2AZ, which have important roles in specifying the functions of homologous INO80 and SWR1 complexes in processes that maintain genome stability.
γ-H2AX is a substrate of INO80 and SWR1.
The histone variant H2AX is phosphorylated on its C terminus by ATM and ATR (or Tel1 and Mec1) in chromatin regions that surround damaged DNA55,56 (BOX 1). γ-H2AX is also a crucial component of DNA damage responses, as defects in the regulation of H2AX phosphorylation lead to alterations in the DNA damage checkpoint, genomic instability and cancer predisposition in mice57–60. γ-H2AX participates in the maintenance of genome integrity by serving as docking sites for several DNA damage response proteins61–64, such as the INO80 and SWR1 complexes, thereby focusing the activity of these factors to regions that are proximal to the damage site.
Two S. cerevisiae INO80 subfamily components, Nhp10 and Arp4, have been implicated in conferring an association of the complex with γ-H2AX. Deletion of the Nhp10 subunit greatly reduces the association of the INO80 subunit Ies3 and γ-H2AX with the complex, and eliminates recruitment of the S. cerevisiae INO80 complex to DSBs22. Nhp10 and Ies3 are unique to the INO80 complex, demonstrating that this chromatin-remodelling complex has specialized components that facilitate its activity in DNA damage response pathways through association with γ-H2AX. Additionally, Arp4 (BRG1-associated factor 53a (BAF53A) in mammals), a subunit of the INO80, SWR1 and SRCAP complexes, binds to γ-H2AX in vitro and its deletion reduces recruitment of Rvb- containing complexes, such as INO80 and SWR1, to DSBs in yeast21.
INO80 and SWR1 assist DSB repair.
DSBs caused by genotoxic stress are particularly dangerous lesions that can result in mutations owing to error-prone repair or cell death if left unrepaired. The major pathways of DSB repair include homologous recombination (HR) and non-homologous end joining (NHEJ)65. The Mre11–Rad50– Xrs2 (MRX) complex, which contains exonuclease activity, collaborates with other factors to promote the production of single-stranded DNA, a process known as resection66. Repair and checkpoint factors, such as the Mec1 kinase, then localize to break sites. During HR, RAD52 epistasis group proteins (Rad50, Rad51, Rad52, Rad54 and Rad55) promote homology search, strand invasion and synapsis between the invading recipient strand and donor DNA, leading to the formation of Holliday junctions. DNA repair is complete once DNA synthesis has finished and Holliday junctions have been resolved. Alternatively, during NHEJ, the Ku70–Ku80 complex facilitates tethering and ligation of the broken DNA ends.
As previously mentioned, S. cerevisiae INO80 and SWR1 complexes bind directly to sites of DNA DSBs through their association with γ-H2AX22,23,34. In particular, these complexes are required for the proper processing of the DNA ends that are involved in the DSB (FIG. 2). Specifically, the S. cerevisiae INO80 com-plex influences the proximal eviction of nucleosomes surrounding DSBs, including nucleosomes that contain γ-H2AX and H2AZ. Deletion of ARP8, which reduces the in vitro chromatin-remodelling activity of the INO80 complex31, or deletion of NHP10, which decreases the recruitment of the INO80 complex to DSB22, result in defective nucleosome eviction in regions proximal to the DSB33,34,67 and in chromatin of the homologous donor locus67.
Figure 2 |. INO80 and SWR1 complexes regulate double-strand break repair.
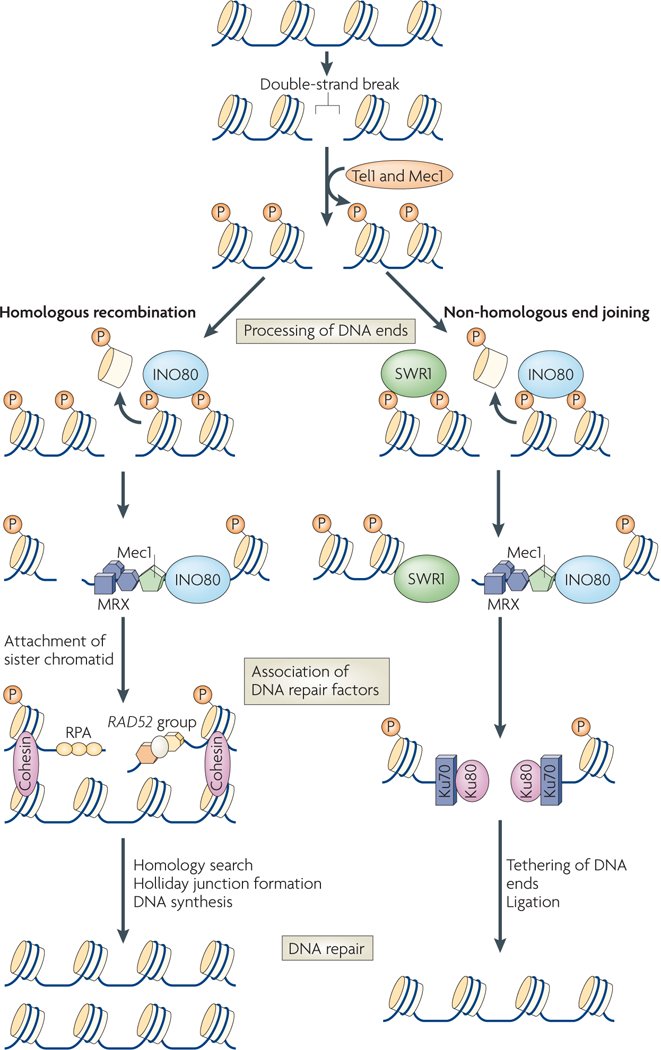
The Saccharomyces cerevisiae kinases Tel1 and Mec1 (ataxia telangiectasia (A-T) mutated (ATM) and A-T and RAD3-related (ATR) in mammals) phosphorylate H2AX after the creation of a double-strand break, which can be repaired by homologous recombination (HR) or non-homologous end joining (NHEJ). During HR and NHEJ, the INO80 and SWR1 complexes bind to phosphorylated H2AX. The INO80 complex is involved in nucleosome eviction proximal to the break site. The DNA ends are then recognized by the Mre11–Rad50–Xrs2 (MRX) complex. The Mre11 nuclease is involvedin the production of single-stranded DNA. During HR, the single-stranded DNA-binding protein replication protein A (RPA) and the Mec1 checkpoint kinase bind to resected DNA. The cohesin complex assists in holding the sister chromatids together64,117. The Rad52 epistasis group, which includes Rad51, Rad52 and Rad54, facilitate the search for and synapsis of homologous DNA sequences. A Holliday junction is formed between the two DNA strands, followed by DNA synthesis and resolution of the junction. During NHEJ, the SWR1 complex promotes the association of Ku80 to the DNA ends, a component of the Ku70–Ku80 complex that is required for NHEJ. Repair is then completed following ligation of the DNA ends. Mutants of the INO80 complex have defects in HR and NHEJ, whereas mutants of the SWR1 complex have defects in error-free NHEJ.
Impaired nucleosome eviction seems to alter the subsequent steps that facilitate the DNA damage response owing to the reduced association of repair and checkpoint factors with the site of the DSB. Thus, this suggests that the presence of nucleosomes at repair sites impedes the association of proteins that facilitate these processes. For example, mutants of the INO80 complex in S. cerevisiae have defects in the association of the Mre11 nuclease with DSBs, and defects in the Mre11 -mediated promotion of single-stranded DNA, which is a prerequisite for repair through HR23,25,34. The finding that the INO80 complex directly influencessingle-stranded DNA resection is controversial — a sep-arate study did not observe this defect in single-stranded DNA production33. Moreover, another study has questioned whether nucleosome eviction is a determinant or a consequence of single-stranded DNA production because these two events are tightly linked and are difficult to separate experimentally68. Nevertheless, a downstream event of DNA resection, namely the invasion of the single-stranded DNA into the homologous donor locus, is impaired in an arp8 mutant67. The association of other DNA damage response factors, such as Mec1 and Rad51, is also decreased in this arp8 mutant33,34,67.
Conversely, the yeast SWR1 complex does not affect nucleosome eviction at DSBs34. However, deletion of its chromatin substrate HTZ1, which is transiently enriched at DSB sites, results in decreased production of single stranded DNA and reduced association of Rad51 to DSB proximal regions69. The SWR1 complex is also needed for efficient recruitment of Mec1 and Ku80 to DSBs, which are required for NHEJ34 (FIG. 2). In addition, deletion of HTZ1 results in the inability of a persistent DSB to local-ize to the nuclear periphery69, a rather enigmatic event that promotes DNA repair70.
Defects in the chromatin-remodelling activity of the INO80 subfamily complexes ultimately result in deficient DNA repair. For instance, mutants of the Arp subunits in the S. cerevisiae INO80 complex have defects in NHEJ23,34 as well as the HR pathway71. In arp8 mutants, when HR repair does occur, gene conversion often consists of large and discontinuous DNA tracts that might result from unstable heteroduplex DNA that forms during strand invasion and branch migration67. Indeed, mutants of the INO80 complex in plants and mammals also display defects in DSB repair11,24, suggesting conserved mechanisms for the INO80 complex in this pathway. By contrast, the S. cerevisiae Swr1 ATPase subunit does not seem to function in HR, but rather participates in the error-free NHEJ pathway34,71. These results show that different complexes in the INO80 sub-family can contribute to distinct repair mechanisms, in part owing to the function of specialized subunits in each complex.
INO80 and SWR1 influence checkpoint pathways.
Checkpoint pathways function cooperatively with DNA repair pathways by altering cell cycle kinetics, which allows for repair of damaged DNA and re-entry into the cell cycle72,73. For example, the production of singlestranded DNA during DNA repair is required for the recruitment and activation of the checkpoint Mec1 kinase74–76. Mec1 then activates downstream effector kinases that target proteins to arrest the cell cycle77. Additional checkpoint proteins, such as S. cerevisiae Rad9 (53BP1 in mammals), bind to dam-age sites in a γ-H2AX-dependent manner and also assist in the activation of downstream checkpoint signalling components56,61,62,78.
The Ies4 subunit of the INO80 complex is phosphorylated by the Tel1 and Mec1 kinases to modulate DNA replication checkpoint responses without altering DSB repair processes25. Activation of the S phasecheckpoint delays replication origin firing, and cells that carry mutations that mimic phosphorylated Ies4 display inappropriately elevated S phase checkpoint activation. This results in decreased viability when exposed to DNA-damaging agents. In cells that have mutations that prevent Ies4 phosphorylation, deletion of the TOF1 checkpoint factor (which mediates the DNA damage checkpoint response during exposure to replication stress79,80) causes dramatic defects in the ability to resume the progression of the cell cycle when the replication stress is removed. These results show redundant or compensating roles between Tof1 and phosphorylated Ies4 as components of the cell cycle checkpoint pathway. Thus, the INO80 complex is capable of many distinct activities in DNA damage response pathways, such as the repair of DSBs and the regulation of the replication checkpoint, in part by using specific subunits, such as Nhp10 and Ies4 (FIG. 3).
Figure 3 |. The INO80 complex is a component of the Tel1 and Mec1 pathway.
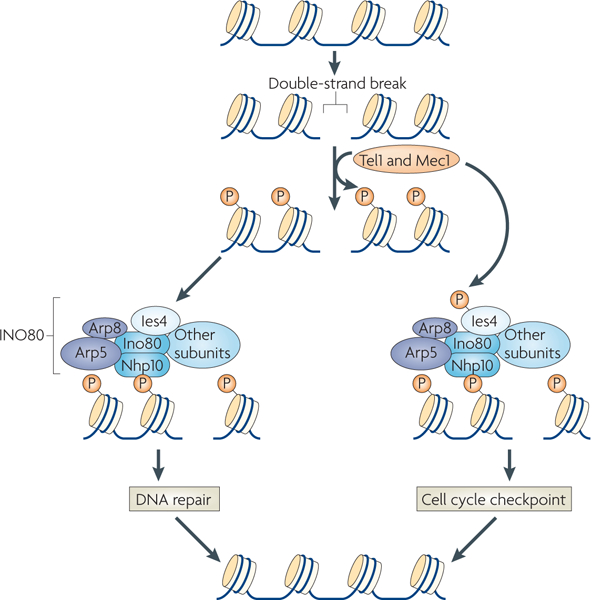
Following the creation of a double-strand break, the Saccharomyces cerevisiae kinases Tel1 and Mec1 (ataxia telangiectasia (A-T) mutated (ATM) and A-T and RAD3-related (ATR) in mammals) phosphorylate H2AX. The non-histone protein 10 (Nhp10) subunit of the INO80 complex is needed for the complex to associate with phosphorylated H2AX (γ-H2AX). Once localized to the DNA break site, core catalytic subunits, such as actin-related protein 5 (Arp5) and Arp8, participate in DNA repair processes. The Tel1 and Mec1 kinases also phosphorylate INO80 subunit 4 (Ies4) to regulate the cell cycle checkpoint response. Therefore, Tel1 and Mec1 regulate two aspects of the INO80 complex when cells are exposed to DNA-damaging agents: the phosphorylation of H2AX, leading to the recruitment of the INO80 complex at DNA damage sites, and phosphorylation of the Ies4 subunit to influence the cell cycle checkpoint response.
Although the precise mechanisms of INO80 subfamily-mediated checkpoint regulation are unknown, evidence suggests that the roles of INO80 and SWR1 complexes in checkpoint pathways are directly linked to the function of the H2AZ and H2AX histone variants.
Both INO80 and SWR1 complexes in S. cerevisiae regulate the abundance of H2A variants in chromatin following exposure to DNA-damaging agents43, and this influences checkpoint adaptation — a particularly rare occurrence in which the cell survives despite the presence of a persistently unrepaired DSB43. Moreover, a recent report shows that deletion of SWR1 or the HTZ1 histone variant in S. cerevisiae results in delayed checkpoint activation in response to a single persistent DSB69.
Chromatin-remodelling activities that alter the levels of γ-H2AX might indirectly regulate the abundance of DNA damage proteins that bind to γ-H2AX and activate cell cycle checkpoints at sites of DNA damage, as well as the subsequent dissociation of these proteins to facilitate checkpoint recovery. Alternatively, the chromatin-remodelling activity of the INO80 subfamily complexes might produce DNA substrates that activate checkpoint factors. Indeed, mutants of the INO80 subfamily in S. cerevisiae that have reduced single-stranded DNA also have decreased recruitment of the checkpoint factor Mec1 to DSBs along with delayed checkpoint activation34.
The INO80 complex participates in DNA replication.
Chromatin modulation is a crucial step in DNA replication processes, particularly when challenged with replicative stress that impedes the progression of the replication fork81. Local chromatin structure influences the function of the replication origin82,83 as well as DNA damage responses during replication stress68,84. Stalled replication forks arise when the replication machinery encounters a DNA lesion or when nucleotide levels are low. If the damage is left unrepaired, or if nucleotide levels are not restored, disassembly of the replication machinery might occur at the same time as replication fork collapse, and this can result in DNA breaks72. The S phase DNA damage response attempts to resolve the DNA lesion by activating cell cycle checkpoint arrest and assembling repair proteins at the DNA lesion.
Checkpoint alterations occur in mutants of the yeast INO80 complex in response to replication stress, such as low dNTP levels and DNA crosslinks caused by genotoxic agents16,25,43. As previously described, phosphorylated Ies4 specifically influences S phase checkpoint responses25. Furthermore, global genetic screens in S. cerevisiae implicate both INO80 and SWR1 complexes in the maintenance of DNA integrity, including replicative damage response pathways85,86.
Recent reports show that the S. cerevisiae INO80 complex is enriched at replication origins16,87–89. This enrichment is further increased at stalled replication forks that are under replicative stress, a pattern that is similar for other S phase DNA damage response factors at these sites16,87,88,90. Specifically, the INO80 complex is needed for sustained replication and survival of yeast cells that are transiently exposed to agents that cause stalling of the replication fork16,87,88. Mutation of the INO80 ATPase results in replisome integrity defects, as components of the replication machinery dissociate from the stalled replication fork87. This defect in recovery of DNA replication after replication stress is also accompanied by anincrease and persistent accumulation of checkpoint and repair protein foci, which is indicative of elevated DNA damage levels16. As a result, mutants of the INO80 complex, such as ies4, arp8, arp5 and ino80 mutants, have defects in checkpoint recovery and decreased viability in the presence of replication stress16,25,87 (FIG. 4). SWR1 is not enriched at replication origins16 and swr1 mutants do not have decreased viability when exposed to replication stress5. Therefore, the role of the SWR1 complex in DNA replication is distinct from that of the S. cerevisiae INO80 complex.
Figure 4 |. The INO80 complex promotes recovery of stalled replication forks.
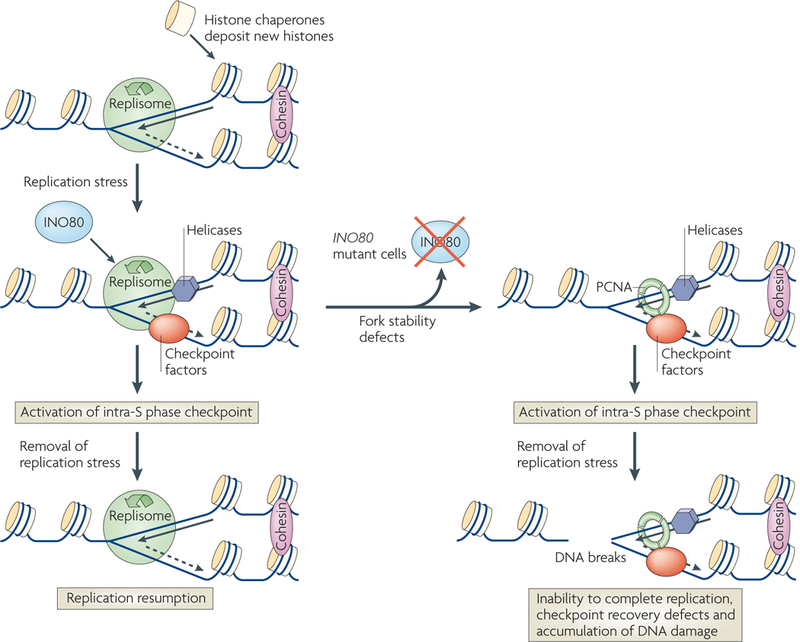
During replication, DNA synthesis is catalysed by the replisome, which contains polymerases, primases and helicases118. Histone chaperones deposit histones on to newly synthesized DNA. Replication forks stall when exposed to replication stress, such as depleted dNTP pools. When this happens, the replisome is stabilized by DNA damage response factors, such as the Saccharomyces cerevisiae INO80 complex and the Tof1 and Mrc1 checkpoint factors, which activate the intra-S phase checkpoint to prevent replication origin firing. On the removal of replication stress, the replication fork recovers and DNA synthesis resumes. In the absence of the INO80 complex, fork stability defects occur as the replisome is destabilized and some of its components dissociate, leaving others, such as proliferating cell nuclear antigen (PCNA), at the replication fork. In this case, replication does not restart following the removal of replication stress. Accordingly, checkpoint recovery is delayed and DNA damage accumulates.
Research on the roles of the INO80 and SWR1 complexes in DNA replication is in its infancy, and recent studies present conflicting information on the role of the INO80 complex during DNA replication16,87. The precise mechanisms and pathways through which these complexes influence DNA replication are unknown. Moreover, additional research is needed to determine if the function of the INO80 subfamily is dependent on an interaction with its H2A histone variant substrates. It should be noted that mutants of γ-H2AX and H2AZ in yeast display decreased viability when exposed to genotoxic agents that impede replication5,21, and deletion of HTZ1 in S. cerevisiae results in slower kinetics of S phase progression compared with wild type42. Furthermore, replication processes are dramatically impaired when the Nhp10 subunit of INO80 complex, which confers an association of the complex with γ-H2AX22, and the ATPase of the ISW2 chromatin-remodelling complex are both mutated in S. cerevisiae88. These results suggest that multiple chromatin-remodelling complexes from different subfamilies, as well as histone variants, cooperatively facilitate chromatin modulation during DNA replication. Alternatively, it might be possible that a particular DNA structure stimulates the activity of the INO80 complex (for example, a three-way junction that mimics a stalled replication fork and has been shown to associate with the INO80 complex24).
The INO80 complex is needed for telomere regulation.
Telomeres are chromosomal loci at the ends of chromosomes that contain repetitive DNA sequences, as well as specialized protein and DNA structures that stabilize the chromosome and avoid recognition by the DNA repair machinery of the chromosome end as a DNA DSB91. The telomerase enzyme adds repetitive DNA sequences on to the ends of chromosomes, and these are progressively lost during each DNA replication cycle as a result of the incomplete replication of linear chromosomes.
Genetic screens implicate both INO80 and SWR1 complexes in the process of telomere length regulation26,86. Subunits of the S. cerevisiae INO80 and SWR1 complexes, including Ino80, Arp4, Nhp10 and Ies3, associate with yeast telomeric regions26,92. Specifically, the Ies3 subunit of the INO80 complex interacts with the telomerase complex26. Subunits of the INO80 complex, including Ies3, Arp8 and Nhp10, regulate telomere length in cells that contain active telomerase. Inactivation of telomerase and deletion of Ies3 results in delayed growth of surviving cells, as well as severely impaired genomic stability in these survivors, such as the fusion of telomere ends between two chromosomes26. These results show that specific subunits of the INO80 subfamily are specialized in regulating telomere structure and function.
The DNA damage response is intricately involved in telomere regulation, as both repair and cell cycle checkpoint factors associate with telomere ends to facilitate DNA end processing as well as telomere protection mechanisms91. For instance, the ATM and ATR kinases (and Tel1 and Mec1) and γ-H2AX are present on telomeres of normal length93–95. The presence of these kinases on telomeres influences the recruitment of the telomerase and presumably regulates telomere length during normal replication94,96. ATM and ATR kinases and γ-H2AX become further enriched at crucially shortening telo meres in an attempt to repair the telomere end that resembles a DSB97. As previously discussed, the subunits of the INO80 subfamily that mediate the interaction between the chromatin-remodelling complexes and γ-H2AX to localize to DSBs in S. cerevisiae — Arp4 of both INO80 and SWR1 complexes and Nhp10 of the INO80 complex — are enriched at telomeres26,92. Furthermore, deletion of S. cerevisiae NHP10 results in altered lengthening of telomeres26. In D. melanogaster, loss of the histone variant H2Av, which is homologous to both H2AX and H2AZ in other organisms (BOX 1), results in telomere fusions and decreased association of telomere capping proteins that prevent the recognition of a telomere end as a DSB98. These observations raise the interesting possibility that the INO80 subfamily might use a common mechanism of associating with modified histone variants, such as γ-H2AX and H2AZ, to accumulate at both DSBs and telomeres. As such, the INO80 subfamily might function at telomeres in a manner similar to that of DSBs, such as the regulation of DNA repair and recombination. Alternatively, specific subunits that have not been found to affect DSB repair, such as Ies3, might facilitate specialized activities of the INO80 complex that are unique to telomere regulation.
Centromere stability and chromosome segregation.
Proper chromosome segregation during mitosis ensures the faithful transmission of genetic information to daughter cells. Chromosome segregation occurs through a rather elaborate process that includes accurate sister chromatid cohesion, attachment of micro tubules to chromosomes through the kinetochore complex, and separation of the replicated chromosomes. Global genetic screens in S. cerevisiae show that the INO80 and SWR1 complexes are involved in chromosome stability mechanisms through a role in chromosome segregation40,99,100. Subunits of the S. cerevisiae INO80 and SWR1 complexes, such as Arp4 and the Ino80 and Swr1 ATPases, bind directly to centromeres92. Mutation of ARP4 in S. cerevisiae causes defects in the assembly of kinetochore components, such as the centromeric histone H3 variant chromosome segregation protein 4 (Cse4), resulting in mitotic cell cycle arrest92. Overexpression of ARP4 partially suppresses the increase in chromosomal ploidy owing to the defects in chromosome segregation that are observed in histone H2A yeast mutant strains101. In addition, ARP8 deletion in the S. cerevisiae INO80 complex results in decreased association of the sister chromatid cohesion component Ctf18 (chromosome transmission fidelity 18) to chromatin and increased rates of sister chromatin separation during mitosis compared with wild-type cells90.
The chromatin-remodelling activities of the INO80 and SWR1 complexes that facilitate centromere stability and chromosome segregation are unknown. However, it might be possible that the function of these complexes in chromosome segregation is directly linked to the role of histone H2A variants, as both yeast and mammalian Htz1 and H2AZ variants participate in establishing the specialized chromatin structures in centromeric chromo somal regions40,41,102.
Conclusions and perspectives
Chromatin-remodelling complexes contribute to many essential cellular processes beyond their roles in transcription. Specifically, the roles of INO80 and SWR1 complexes in nuclear activities are quite diverse and range from DSB repair to chromosome segregation. The involvement of these complexes in such varied processes suggests that they might function in different and specialized chromatin-remodelling activities in each process. If so, sophisticated regulatory mechanisms must exist to manage and facilitate the chromatinremodelling activity of each complex. Potential modes of regulation include the targeting of the complex to specific genes by transcription factors, association with different histone and DNA substrates, the participation of specialized subunits in the diverse processes103 and the regulation of the complex in signalling pathways by post-translational modification. Additional modes of regulation include swapping of functional subunits and modulation by small molecules, such as inositol lipids and inositol polyphosphates (FIG. 5). Taken together, a single chromatin-remodelling complex can be regulated by multiple mechanisms to generate considerable functional diversity.
Figure 5 |. Regulation of aTP-dependent chromatin remodelling.
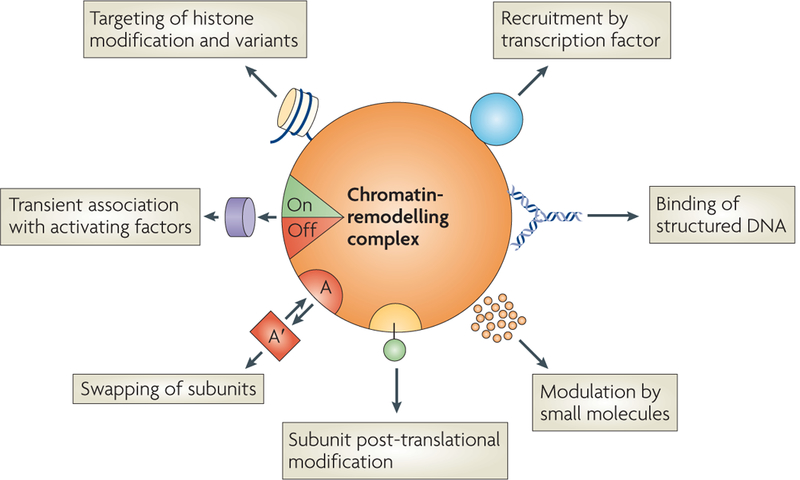
Seven distinct mechanisms in combination can generate numerous structural and functional diversities for a single chromatin-remodelling complex. Transcription factors can target the complex for recruitment to specific target genes. The binding of structured DNA, such as three-way and four-way junctions that represent DNA replication and repair intermediates, can also recruit and potentially alter the activity of the complex. Small molecules, such as inositol polyphosphates and phosphoinositides, can modulate the activity of the complex. Additionally, subunits in the complex can be post-translationally modified or exchanged (swapped)119 to alter the function of the complex. The activity of specific subunits can also be regulated by transient association of the complex with activating factors, such as the activation of the deubiquitylating enzyme ubiquitin carboxy-terminal hydrolase 37 (UCH37) following association of this 19S proteasome component to the INO80 complex. Finally, the association of unique chromatin substrates, such as histone variants and post-translationally modified histones, can further specify the chromatin-remodelling mechanisms of the complex.
The precise chromatin-remodelling mechanisms of the INO80 and SWR1 complexes in diverse nuclear processes are largely unknown. However, both the INO80 and SWR1 complexes associate with histone variants in vivo and specialized DNA structures in vitro21–24. In certain instances, the association of these specific substrates with the INO80 and SWR1 complexes recruits their activity to a specific nuclear pro cess, such as DNA repair22,23. An interesting possibility is that these substrates also stimulate different chromatin-remodelling activities of the complexes, such as nucleosome sliding or helicase activity. Additionally, it is likely that future research will expand the number of known chromatin substrates to which the INO80 subfamily complexes associate. For instance, it might be possible that acetylated and methylated residues on histones also direct the recruitment and/or activity of the complex to facilitate distinct nuclear processes104. As exemplified by the INO80 subfamily of chromatinremodelling complexes, the three major mechanisms of chromatin modification — post-translationally modified histones, ATP-dependent chromatin remodeling and histone variants — are intimately and specifically connected. Such connections are expanding, for instance the WICH chromatin-remodelling complex in the ISWI subfamily has recently been linked to a modified histone variant (H2AX with phosphorylated Tyr142) to regulate DNA damage responses105.
Studies discussed in this Review show that the participation of specific subunits in the function of the complex is dynamic, with different subunits influencing particular processes in which the complex is involved, such as Nhp10 of the S. cerevisiae INO80 complex in mediating an interaction with γ-H2AX22 and Ies4 in influencing checkpoint responses. Recent studies reveal that the deubiquitylating enzyme ubiquitin C-terminal hydrolase 37 (UCH37), a component of the 19S regulatory particle of the proteasome, is also present in the mammalian INO80 complex106,107. The transient association of the INO80 complex with UCH37 regulates the deubiquitylating activity of this subunit. As both the INO80 complex and ubiquitylated histones are involved in transcription and repair processes108, it is possible that chromatin components are the substrates of the deubiquitylation-associated activity of INO80 in these pathways. Thus, for each nuclear process that requires chromatin remodelling, a functional hier archy of subunits, including core subunits and regulatory subunits, might be invoked to regulate the activities of that complex.
Currently, most research investigating the role of the INO80 and SWR1 complexes in DNA repair processes has focused on DSB repair. Recent research also implicates the INO80 subfamily in other repair pathways in both yeast and mammals, such as repair of ultraviolet-induced lesions through nucleotide excision repair4,5,24,109. Additional research is needed to determine the exact chromatin-remodelling mechanisms that are required for these different DNA repair processes. Indeed, it might be possible that each different DNA damage pathway elicits the function of specific chromatin-remodelling subunits, or chromatin substrates, to facilitate certain mechanisms of DNA repair.
The roles of INO80 and SWR1 complexes outside of transcription are largely found in genome stability pathways, such as DNA repair, replication, telomere regulation and centromere stability. The consequences of alterations in these chromatin-remodelling complexes are only beginning to be investigated. Gross chromosomal defects have been found in mutants of the yeast INO80 complex110 and it will be interesting to see if these defects contribute to diseases, such as cancer, in mammals. A precedent for the designation of a chromatin-remodelling complex as a tumour suppressor has been previously demonstrated, as haploin sufficiency of the mammalian TIP60 complex accelerates oncogene-induced tumorigenesis111 and mutation of the SWI/SNF chromatin-remodelling complex results in carcinogenesis112. Interestingly, like the SWI/SNF complex, the mammalian INO80 complex also contributes to chromosomal polyploidy when mutated24,113. Indeed, the role of the INO80 subfamily in health and human disease has been largely un explored. As research investigating the function of the INO80 chromatin-remodelling subfamily emerges, it is becoming clear that mechanisms of DNA integrity maintenance rely on the activity of these complexes to regulate chromatin architecture.
Helicase.
An enzyme that unwinds double-stranded nucleic acids in an energy-dependent manner.
DEAD/H box.
An evolutionarily conserved domain found in proteins that are a part of the super family 2 helicases, which are essential for RNA metabolism and processing.
RuvB.
A bacterial ATP-dependent DNA helicase that forms a ring-shaped double hexamer complex around a Holliday junction and promotes its migration during homologous recombination.
Centromere.
Region of a chromosome that is attached to the spindle during cell division.
HMG.
(High mobility group). A large protein family of small, non-histone components of chromatin that function in chromatin structure
Holliday junction.
A mobile junction that contains four strands of DNA and is generated during the synaptic phase of homologous recombination.
Homologous recombination.
A DNA recombination pathway, which includes the repair of double-strand DNA breaks, that uses a homologous double-stranded DNA as a template for the repair of the broken DNA.
Helicase-SANT-associateddomain.
The region in the ATPase subunit of the INO80 and SWI/SNF subfamilies that serves as the primary binding site for nuclear actin and actin-related proteins within the chromatin-remodelling complex.
Heterochromatin.
Dense chromatin that is less transcriptionally active than euchromatin. Heterochromatin includes structural regions of the chromosome, such as centromeres and telomeres constitutive heterochromatin), as well as genes that are silenced in a given cell type (facultative heterochromatin).
Non-homologous end joining.
(NHEJ). A DNA repair pathway for double-strand breaks that is often error-prone because it leads to the joining of breaks without a template.
Mre11 nuclease.
A subunit of the Mre11–Rad50–Xrs2 (MRX) complex that has 3′ to 5′ exonuclease activity and assists in resecting the DNA ends of a double-strand break.
Replication origin.
A site where replication is initiated during S phase.
Replisome.
A multiprotein complex at the junction of the DNA replication fork that contains all the enzymes that are required for DNA replication.
Kinetochore.
A multiprotein complex that assembles on centromeric DNA and mediates the attachment and movement of chromosomes along the microtubules of the mitotic spindle.
Acknowledgements
The authors apologize for the omission of many interesting research articles that could not be described in this Review owing to the focused scope of the article and space limitations. We thank R. Herrera and J. Holcomb for assistance in developing figures. We also thank C. Wu for helpful discussions regarding the content of this manuscript. Funding for A.J.M. was provided by a National Institute of General Medical Sciences (NIGMS)-sponsored ‘Pathways to Independence’ K99 award.
Footnotes
DATABASES
BioGRID — Repository for Interaction Datasets: www.thebiogrid.org INO80 | SWR1 | SRCAP
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query fcgi?db=genehttp://www.ncbi.nlm.nih.gov/entrez/query%20fcgi?db=gene PHO5 | PHO84
Human Protein Reference Database: www.hprd.org BAF53A | RUVBL1 | RUVBL2 | SRCAP | UCH37 | YY1
Saccharomyces Genome Database: www.yeastgenome.org Arp4 | Arp6 | aRP8 | Ctf18 | Ies3 | Ies4 | INO80 | Ku70 | Mec1 | Nhp10 | Rvb1 | Rvb2 | Swc2 | Swc6 | Swr1 | Tel1 | TOF1
UniProtKB: http://www.uniprot.org ATM | ATR | Htz1 | Ku80 | Pleiohomeotic | Pontin | Reptin | RuvB
FUrthEr inFormation
Xuetong Shen’s homepage: http://gsbs.uth.tmc.edu/tutorial/shen_x.html
Ashby Morrison’s homepage: http://www.stanford.edu/ group/morrison
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Gorbalenya AE & Koonin EV Helicases: amino acid sequence comparisons and structure–function relationships. Curr. Opin. Struct. Biol. 3, 419–429 (1993). [Google Scholar]
- 2.Cairns BR, Kim YJ, Sayre MH, Laurent BC & Kornberg RD A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl Acad. Sci. USA 91, 1950–1954 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peterson CL, Dingwall A & Scott MP Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl Acad. Sci. USA 91, 2905–2908 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen X, Mizuguchi G, Hamiche A & Wu C A chromatin remodelling complex involved in transcription and DNA processing. Nature 406, 541–544 (2000).Identifies and characterizes the yeast INO80 chromatin-remodelling complex, the founding member of the INO80 subfamily.
- 5.Mizuguchi G et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Krogan NJ et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12, 1565–1576 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Kobor MS et al. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2, e131 (2004).References 5–7 show that that the SWR1 complex catalyses the exchange of the H2AZ histone variant into chromatin.
- 8.Kusch T Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306, 2084–2087 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Klymenko T et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 20, 1110–1122 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deal RB, Topp CN, McKinney EC & Meagher RB Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19, 74–83 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fritsch O, Benvenuto G, Bowler C, Molinier J & Hohn B The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol. Cell 16, 479–485 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Ruhl DD et al. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z. into nucleosomes. Biochemistry 45, 5671–5677 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Jin J et al. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J. Biol. Chem. 280, 41207–41212 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Ikura T et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102, 463–473 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Ebbert R, Birkmann A & Schuller HJ The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol. Microbiol. 32, 741–751 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Shimada K et al. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr. Biol. 18, 566–575 (2008).This report, along with references 87 and 88, reveals a role for the yeast INO80 complex in replication processes.
- 17.Cai Y et al. YY1 functions with INO80 to activate transcription. Nature Struct. Mol. Biol. 14, 872–874 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Wong MM, Cox LK & Chrivia JC The chromatin remodeling protein, SRCAP, is critical for deposition of the histone variant H2A.Z at promoters. J. Biol. Chem. 282, 26132–26139 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Kanemaki M et al. Molecular cloning of a rat 49-kDa TBP-interacting protein (TIP49) that is highly homologous to the bacterial RuvB. Biochem. Biophys. Res. Commun. 235, 64–68 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Tsaneva IR, Muller B & West SC ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell 69, 1171–1180 (1992). [DOI] [PubMed] [Google Scholar]
- 21.Downs J et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16, 979–990 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Morrison AJ et al. INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119, 767–775 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Van Attikum H, Fritsch O, Hohn B & Gasser S Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119, 777–788 (2004).References 21–23 reveal that the INO80 complex is involved in DNA damage responses through an association with γ-H2AX.
- 24.Wu S et al. A YY1–INO80 complex regulates genomic stability through homologous recombinationbased repair. Nature Struct. Mol. Biol. 14, 1165–1172 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison AJ et al. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell 130, 499–511 (2007).Shows that the INO80 complex is phosphorylated by the Tel1 and Mec1 kinases as a component of the checkpoint signalling pathway.
- 26.Yu EY et al. Regulation of telomere structure and functions by subunits of the INO80 chromatin remodeling complex. Mol. Cell. Biol. 27, 5639–5649 (2007).Reveals that the INO80 complex associates with telomerase and has a role in regulating telomere length and stability.
- 27.Wu W et al. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nature Struct. Mol. Biol 12, 1064–1071 (2005).Shows the mechanism by which the SWR1 chromatinremodelling complex facilitates histone exchange in the nucleosome.
- 28.Qiu XB et al. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J. Biol. Chem. 273, 27786–27793 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Kanemaki M et al. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J. Biol. Chem. 274, 22437–22444 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Holliday R Molecular aspects of genetic exchange and gene conversion. Genetics 78, 273–287 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen X, Ranallo R, Choi E & Wu C Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell 12, 147–155 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Cai Y et al. Purification and assay of the human INO80 and SRCAP chromatin remodeling complexes. Methods 40, 312–317 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukuda T, Fleming A, Nickoloff J & Osley MA Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438,379–383 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Attikum H, Fritsch O & Gasser S Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 26, 4113–4125 (2007).This study differentiates the roles of the S. cerevisiae INO80 and SWR1 complexes in the homologous recombination and NHEJ repair pathways.
- 35.Boyer LA & Peterson CL Actin-related proteins (Arps): conformational switches for chromatinremodeling machines? Bioessays 22, 666–672 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Szerlong H et al. The HSA domain binds nuclear actin-related proteins to regulate chromatinremodeling ATPases. Nature Struct. Mol. Biol 15, 469–476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harata M et al. The nuclear actin-related protein of Saccharomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol. Biol. Cell 10, 2595–2605 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rangasamy D, Berven L, Ridgway P & Tremethick DJ Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 22, 1599–607 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swaminathan J, Baxter EM & Corces VG The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 19, 65–76 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krogan NJ et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl Acad. Sci. USA 101, 13513–13518 (2004).Shows the functional relationship between the SWR1 complex and modified histones in centromere stability and chromosome segregation.
- 41.Rangasamy D, Greaves I & Tremethick DJ RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nature Struct. Mol. Biol. 11, 650–655 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Dhillon N, Oki M, Szyjka SJ, Aparicio OM & Kamakaka RT H2A.Z functions to regulate progression through the cell cycle. Mol. Cell. Biol. 26, 489–501 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papamichos-Chronakis M, Krebs JE & Peterson C Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 20, 2437–2449 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan JY, Gordon F, Luger K, Hansen JC & Tremethick DJ The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nature Struct. Biol. 9, 172–176 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Suto RK, Clarkson MJ, Tremethick DJ & Luger K Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nature Struct. Biol. 7, 1121–1124 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Clarkson MJ, Wells JR, Gibson F, Saint R & Tremethick DJ Regions of variant histone His2AvD required for Drosophila development. Nature 399, 694–697 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Shiloh Y ATM and related protein kinases: safeguarding genome integrity. Nature Rev. Cancer 3, 155–168 (2003). [DOI] [PubMed] [Google Scholar]
- 48.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA & Goodship JA A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nature Genet. 33, 497–501 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Savitsky K et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268, 1749–1753 (1995). [DOI] [PubMed] [Google Scholar]
- 50.Sif S, Stukenberg PT, Kirschner MW & Kingston RE Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 12, 2842–2851 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen X, Xiao H, Ranallo R, Wu WH & Wu C Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science 299, 112–114 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Steger DJ, Haswell ES, Miller AL, Wente SR & O’Shea EK Regulation of chromatin remodeling by inositol polyphosphates. Science 299, 114–116 (2003).References 51 and 52 show that inositol polyphosphates can modulate the activity of chromatin-remodelling complexes.
- 53.Zhao K et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95, 625–636 (1998). [DOI] [PubMed] [Google Scholar]
- 54.York J Regulation of nuclear processes by inositol polyphosphates. Biochim. Biophys. Acta 1761, 552–559 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Burma S, Chen BP, Murphy M, Kurimasa A & Chen DJ ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276, 42462–42467 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Ward IM & Chen J Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276, 47759–47762 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Bassing CH et al. Histone H2AX: a dosagedependent suppressor of oncogenic translocations and tumors. Cell 114, 359–370 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Celeste A et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114, 371–383 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Downs JA, Lowndes NF & Jackson SP A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408, 1001–1004 (2000).References 57–59 show the crucial role of γ-H2AX in evolutionarily conserved genome stability pathways.
- 60.Keogh MC et al. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature 439, 497–501 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Celeste A et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature Cell Biol. 5, 675–679 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Nakamura TM, Du LL, Redon C & Russell P Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol. Cell. Biol. 24, 6215–6230 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paull TT et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10, 886–895 (2000). [DOI] [PubMed] [Google Scholar]
- 64.Unal E et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16, 991–1002 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Haber JE Partners and pathwaysrepairing a double-strand break. Trends Genet. 16, 259–264 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Mimitou EP & Symington LS Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsukuda T et al. INO80-dependent chromatin remodeling regulates early and late stages of mitotic homologous recombination. DNA Repair (Amst.) 8, 360–369 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Chen CC et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134, 231–243 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kalocsay M, Hiller NJ & Jentsch S Chromosomewide Rad51 spreading and SUMO–H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell 33, 335–343 (2009). [DOI] [PubMed] [Google Scholar]
- 70.Nagai S et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322, 597–602 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kawashima S et al. The INO80 complex is required for damage-induced recombination. Biochem. Biophys. Res. Commun. 355, 835–841 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Branzei D & Foiani M Regulation of DNA repair throughout the cell cycle. Nature Rev. Mol. Cell Biol. 9, 297–308 (2008). [DOI] [PubMed] [Google Scholar]
- 73.Harrison JC & Haber JE Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40, 209–235 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Lisby M, Barlow JH, Burgess RC & Rothstein R Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713 (2004). [DOI] [PubMed] [Google Scholar]
- 75.Nakada D, Hirano Y & Sugimoto K Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol. Cell. Biol. 24, 10016–10025 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zou L & Elledge SJ Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science 300, 1542–1548 (2003). [DOI] [PubMed] [Google Scholar]
- 77.Sweeney FD et al. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr. Biol. 15, 1364–1375 (2005). [DOI] [PubMed] [Google Scholar]
- 78.Gilbert CS, Green CM & Lowndes NF Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8, 129–136 (2001). [DOI] [PubMed] [Google Scholar]
- 79.Katou Y et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424, 1078–1083 (2003). [DOI] [PubMed] [Google Scholar]
- 80.Tourriere H, Versini G, Cordon-Preciado V, Alabert C & Pasero P Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol. Cell 19, 699–706 (2005). [DOI] [PubMed] [Google Scholar]
- 81.Zhou J et al. Cell cycle regulation of chromatin at an origin of DNA replication. EMBO J. 24, 1406–1417 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aggarwal BD & Calvi BR Chromatin regulates origin activity in Drosophila follicle cells. Nature 430, 372–376 (2004). [DOI] [PubMed] [Google Scholar]
- 83.Vogelauer M, Rubbi L, Lucas I, Brewer BJ & Grunstein M Histone acetylation regulates the time of replication origin firing. Mol. Cell 10, 1223–1233 (2002). [DOI] [PubMed] [Google Scholar]
- 84.Masumoto H, Hawke D, Kobayashi R & Verreault A A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436, 294–298 (2005). [DOI] [PubMed] [Google Scholar]
- 85.Pan X et al. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124, 1069–1081 (2006). [DOI] [PubMed] [Google Scholar]
- 86.Collins S et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446, 806–810 (2007). [DOI] [PubMed] [Google Scholar]
- 87.Papamichos-Chronakis M & Peterson C The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nature Struct. Mol. Biol 15,338–345 (2008). [DOI] [PubMed] [Google Scholar]
- 88.Vincent J, Kwong T & Tsukiyama T ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nature Struct. Mol. Biol. 15, 477–484 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rountree MR, Bachman KE & Baylin SB DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nature Genet. 25, 269–277 (2000). [DOI] [PubMed] [Google Scholar]
- 90.Ogiwara H, Enomoto T & Seki M The INO80 chromatin remodeling complex functions in sister chromatid cohesion. Cell Cycle 6, 1090–1095 (2007). [DOI] [PubMed] [Google Scholar]
- 91.d’Adda di Fagagna F, Teo SH & Jackson SP Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 18, 1781–1799 (2004). [DOI] [PubMed] [Google Scholar]
- 92.Ogiwara H et al. Actin-related protein Arp4 functions in kinetochore assembly. Nucleic Acids Res. 35, 3109–3117 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goudsouzian LK, Tuzon CT & Zakian VA S cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol. Cell 24, 603–610 (2006). [DOI] [PubMed] [Google Scholar]
- 94.Takata H, Kanoh Y, Gunge N, Shirahige K & Matsuura A Reciprocal association of the budding yeast ATM-related proteins Tel1 and Mec1 with telomeres in vivo. Mol. Cell 14, 515–522 (2004). [DOI] [PubMed] [Google Scholar]
- 95.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A & Haber JE Heterochromatin is refractory to γ-H2AX modification in yeast and mammals. J. Cell Biol. 178, 209–218 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Takata H, Tanaka Y & Matsuura A Late S phasespecific recruitment of Mre11 complex triggers hierarchical assembly of telomere replication proteins in Saccharomyces cerevisiae. Mol. Cell 17, 573–583 (2005). [DOI] [PubMed] [Google Scholar]
- 97.Takai H, Smogorzewska A & de Lange T DNA damage foci at dysfunctional telomeres. Curr. Biol. 13, 1549–1556 (2003). [DOI] [PubMed] [Google Scholar]
- 98.Rong YS Loss of the histone variant H2A.Z restores capping to checkpoint-defective telomeres in Drosophila. Genetics 180, 1869–1875 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Measday V et al. Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc. Natl Acad. Sci. USA 102, 13956–13961 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ben-Aroya S et al. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol. Cell 30, 248–258 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pinto I & Winston F Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J. 19, 1598–1612 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Greaves I, Rangasamy D, Ridgway P & Tremethick D H2A.Z contributes to the unique 3D structure of the centromere. Proc. Natl Acad. Sci. USA 104, 525–530 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu JI, Lessard J & Crabtree GR Understanding the words of chromatin regulation. Cell 136, 200–206 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morrison AJ & Shen X Chromatin modifications in DNA repair. Results Probl. Cell Differ. 41, 109–125 (2006). [DOI] [PubMed] [Google Scholar]
- 105.Xiao A et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 457, 57–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yao T et al. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol. Cell 31, 909–917 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sardiu ME et al. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc. Natl Acad. Sci. USA 105, 1454–1459 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Weake VM & Workman JL Histone ubiquitination: triggering gene activity. Mol. Cell 29, 653–663 (2008). [DOI] [PubMed] [Google Scholar]
- 109.Jha S, Shibata E & Dutta A Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol. Cell. Biol. 28, 2690–2700 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smith S et al. Mutator genes for suppression of gross chromosomal rearrangements identified by a genomewide screening in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 101, 9039–9044 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gorrini C et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 448, 1063–1067 (2007). [DOI] [PubMed] [Google Scholar]
- 112.Versteege I et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394, 203–206 (1998). [DOI] [PubMed] [Google Scholar]
- 113.Vries RG et al. Cancer-associated mutations in chromatin remodeler hSNF5 promote chromosomal instability by compromising the mitotic checkpoint. Genes Dev. 19, 665–670 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thatcher TH & Gorovsky MA Phylogenetic analysis of the core histones H2A, H2B, H3, and H4. Nucleic Acids Res. 22, 174–179 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rogakou EP, Pilch DR, Orr AH, Ivanova VS & Bonner WM DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 (1998). [DOI] [PubMed] [Google Scholar]
- 116.Palmer D, Snyder LA & Blumenfeld M Drosophila nucleosomes contain an unusual histone-like protein. Proc. Natl Acad. Sci. USA 77, 2671–2675 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Strom L, Lindroos HB, Shirahige K & Sjogren C Postreplicative recruitment of cohesin to doublestrand breaks is required for DNA repair. Mol. Cell 16, 1003–1015 (2004). [DOI] [PubMed] [Google Scholar]
- 118.Aguilera A & Gomez-Gonzalez B Genome instability: a mechanistic view of its causes and consequences. Nature Rev. Genet. 9, 204–217 (2008). [DOI] [PubMed] [Google Scholar]
- 119.Wu JI et al. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56, 94–108 (2007). [DOI] [PubMed] [Google Scholar]


