ABSTRACT
Long intergenic non-coding RNA 00152 (LINC00152) is aberrantly expressed in various human malignancies and plays an important role in the pathogenesis. Here, we found that LINC00152 is upregulated in hepatocellular carcinoma (HCC) tissues as compared to adjacent non-neoplastic tissues; gain-and-loss-of-function analyses in vitro showed that LINC00152 facilitates HCC cell cycle progression through regulating the expression of CCND1. LINC00152 knockdown inhibits tumorigenesis in vivo. MS2-RIP analysis indicated that LINC00152 binds directly to miR-193a/b-3p, as confirmed by luciferase reporter assays. Furthermore, ectopic expression of LINC00152 partially halted the decrease in CCND1 expression and cell proliferation capacity induced by miR-193a/b-3p overexpression. Thus, LINC00152 acts as a competing endogenous RNA (ceRNA) by sponging miR-193a/b-3p to modulate its target gene, CCND1. Our findings establish a ceRNA mechanism regulating cell proliferation in HCC via the LINC00152/miR-193a/b-3p/CCND1 signalling axis, and identify LINC00152 as a potential therapeutic target for HCC.
KEYWORDS: Hepatocellular carcinoma, LINC00152, miR-193a/b-3p, CCND1, cell cycle
Introduction
As one of the most aggressive human malignancies, hepatocellular carcinoma (HCC) has a high mortality rate. The survival rate is modest because of rapid proliferation and early intrahepatic or extrahepatic metastasis; therefore, understanding the molecular mechanisms of malignant progression in HCC may enable the development of new diagnostic techniques and improve prognosis.
Long non-coding RNAs (lncRNAs) have attracted attention as potential biological regulators in a wide range of activities, including chromatin organization, genome defense, protein-coding gene translation, gene transcription, epigenetic regulation and RNA turnover. Notably, most of the functions involve gene expression regulation, including both of protein-coding genes and non-coding RNAs [1]. Recently, long intergenic non-coding RNA 00152 (LINC00152) has been reported to exert a ubiquitous oncogenic influence in various human cancers. For example, LINC00152 positively correlated with hepatitis B virus (HBV) infection, advanced tumour-node-metastasis (TNM) stage and poor overall survival, is supposed to be a novel biomarker in HBV-associated HCC [2,3]. And upregulation of LINC00152 is also associated with tumour invasion, lymph node metastasis and TNM stage in gastric cancer, lung cancer and tongue squamous cell carcinoma [4–6]. Further study showed that LINC00152 involves in glioma stem cell, gastric cancer and HCC cell proliferation, migration and invasion [7,8]. Collectively, these findings strongly imply that LINC00152 may be a ubiquitous oncogene in cancers. Latest studies have shown that LINC00152 interacts with a network of proteins associated with M phase of the cell cycle progression [9]. CCND1 a key member of the cyclins, and its associated kinases are responsive to mitogenic signals and promote cell cycle progression from G1 to S phase by binding and activating the cyclin dependent kinases CDK4 and CDK6. We hypothesize whether LINC00152 regulates HCC cell proliferation via regulating CCND1.
Previous evidences indicated that LINC00152 localizes in both cytoplasm and nuclear, preferentially to cytoplasm [5,10]. And the portion of LINC00152 localized in nuclear, functions in a series of mechanisms, such as binding to the promoter of EpCAM through a cis-regulation in HCC [11], and recruiting EZH2 to the IL24 promoter in lung adenocarcinoma [5]. While most of the well-studied lncRNAs regulate cellular activities at the nuclear level, others are localized within the cytoplasm and mediate post-transcriptional control via RNA-RNA interactions [12]. They can act as sponges and interact with short non-coding RNAs, such as miRNAs to simultaneously influence the expression of several thousands of miRNA-target genes leading to an overall change in cellular physiology [13]. Although ceRNA research is still in its initial stage, we suppose LINC00152 in cytoplasm can also participate in ceRNAs crosstalks in HCC.
In this study, we identified LINC00152 upregulation in bioinformatics and a cohort of HCC tissues and discovered an association between LINC00152 and HCC cell proliferation potentials. Our data demonstrates LINC00152-mediated miR-193a/b-3p crosstalk via a ceRNA mechanism that, modulates repression of CCND1, thereby facilitating HCC progression.
Results
LINC00152 is strongly upregulated in HCC tissues, and LINC00152 is required for efficient proliferation of HCC cells in vitro
Analysis of the expression levels of 441 recurrently dysregulated tumourigenesis-associated lncRNAs in 20 HCC patients using the bioinformatics tool NCBI Gene Expression Omnibus (GEO Series Accession Number GSE77509) revealed that LINC00152 expression was significantly higher (~2.24-fold) in HCC tumour tissues than in adjacent non-neoplastic liver tissues (Figure 1(a,b)). To validate this finding, we measured the expression level of LINC00152 in 80 pairs of HCC/non-tumour tissues by qRT-PCR and found, significant upregulation of LINC00152 in the HCC tumour compared to the normal tissues (Figure 1(c)).
Figure 1.
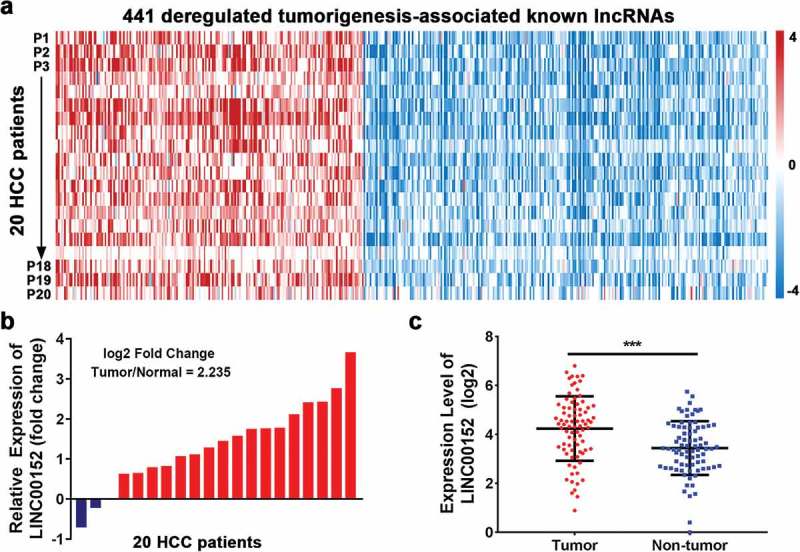
Recurrently dysregulated lncRNAs and LINC00152 expression in HCC tissues. (a) Analysis of 441 recurrently dysregulated tumourigenesis-associated lncRNAs according to the NCBI Gene Expression Omnibus bioinformatics tool (GEO Series Accession Number GSE77509). lncRNAs were rank-ordered by differential expression between adjacent normal tissue and primary tumour samples (n = 20). (b) Relative expression of LINC00152 in HCC compared with adjacent non-neoplastic liver tissues according to NCBI Gene Expression Omnibus analysis. The fold change was ~2.24 higher in tumour than in normal tissues (n = 20). (c) Upregulation of LINC00152 in HCC tissues compared with non-tumour tissues as analysed by qRT-PCR. LINC00152 expression was normalized to that of U6 (n = 80) (*** p < 0.001).
To investigate the potential biological function of LINC00152 in HCC cell lines, we first performed RNAi-based loss-of-function assays using Huh7 and HCCLM3 cells, which showed both shRNAs against LINC00152 reduced endogenous LINC00152 expression (Figure 2(a)). Moreover, LINC00152 silencing significantly decreased cell proliferation in Huh7 and HCCLM3 cells, as determined by CCK-8 assay (Figure 2(b)). Additionally, EdU incorporation assays revealed that LINC00152 knockdown decreased Huh7 and HCCLM3 cell proliferation capacity (Figure 2(c)); colony formation assays yielded similar results (Figure 2(d)). Furthermore, we overexpressed LINC00152 to investigate the effect of ectopic LINC00152 on Hep3B cell proliferation. The results showed that p-LINC00152 effectively induced the increased expression (~9.0 fold) of endogenous LINC00152 (Fig. S1A); and a sharp decrease in G0/G1 proportion, a corresponding increase in S and G2/M phase according to the cell cycle distribution (Fig. S1B); and increased proliferation of HCC cancer cell in vitro (Fig. S1C). Consistent with the fluorescence-activated cell sorting (FACS) analysis, the mRNA and protein level of CCND1 was also significantly increased in LINC00152 overexpressed Hep3B cell (Fig S1D).
Figure 2.
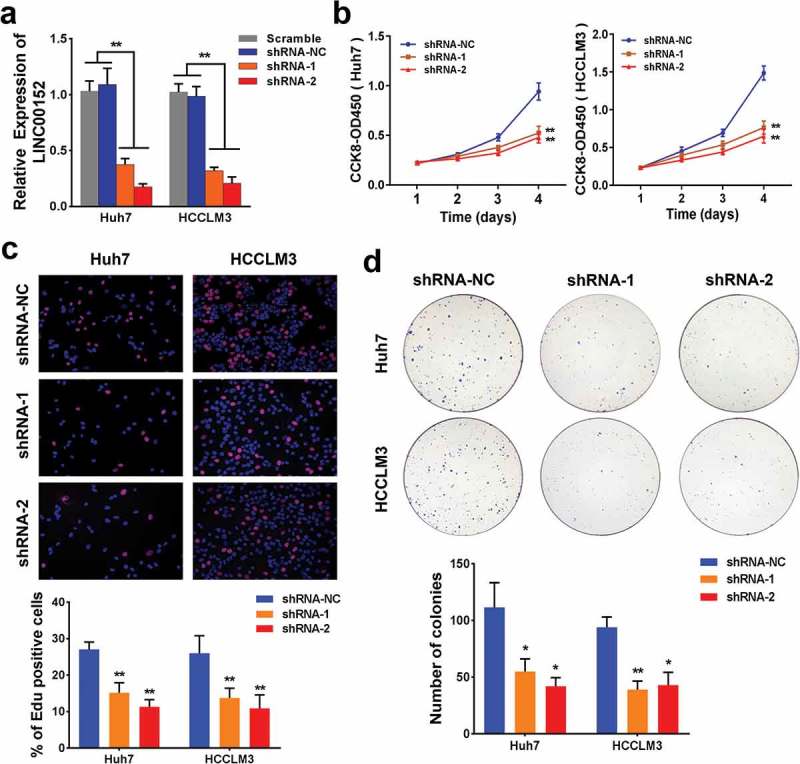
Silencing of LINC00152 inhibits HCC cellular proliferation in vitro. (a) LINC00152 shRNAs reduced endogenous LINC00152 expression compared with either scrambled shRNAs or shRNA-NC in both Huh7 and HCCLM3 cells (** p < 0.01). (b) Huh7 and HCCLM3 cell proliferation was decreased 4 days after LINC00152-shRNAs transfection as evaluated by CCK-8 assay (** p < 0.01). (c) EdU (5-Ethynyl-2ʹ-deoxyuridine) incorporation assays showed that Huh7 and HCCLM3 cell growth was suppressed 48 h after LINC00152-shRNAs transfection (** p < 0.01). (d) The colony formation ability of HCC cells was greatly repressed following LINC00152 knockdown (* p < 0.05, ** p < 0.01).
LINC00152 knockdown inhibits tumorigenesis of HCC cells in vivo
For our in vivo studies, either HCCLM3 cells stably transfected with shRNA-LINC00152 or control cells transfected with shRNA-NC were injected into the armpits of male nude mice. We found that LINC00152-shRNA #2 dramatically inhibited tumour growth compared to the control shRNA-NC according to the mean volumes of the xenograft tumours and the tumour growth curves (Figure 3(a)). Ki-67, a proliferation index, was dramatically reduced in xenograft tumours from LINC00152 knockdown cells than in tumours from the control cells (Figure 3(b,c)). Collectively, these results demonstrated that LINC00152 is required for HCC cell proliferation both in vitro and in vivo.
Figure 3.
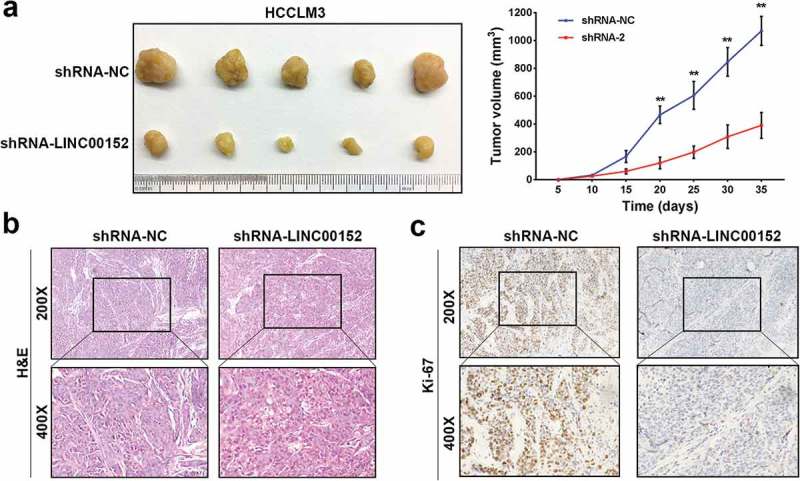
LINC00152 knockdown inhibits HCC tumour growth in vivo. (a) Representative images of tumour formation in nude mice injected subcutaneously with HCCLM3 cells downexpressing either LINC00152 (lower panel) (n = 5) or shRNA-NC (upper panel) (n = 5). Tumour growth curve of xenograft tumours from nude mice in the LINC00152-knockdown group and the control group. The data are shown as the mean ± s.d. (** p < 0.01). All mice were sacrificed 5 weeks after injection. (b, c) Representative images of xenograft tumours stained with haematoxylin and eosin (H&E) and with immunohistochemical stain for Ki-67. Expression of the proliferative indicator, Ki-67, was decreased in xenograft tumours from LINC00152-knockdown cells compared with control cells.
LINC00152 retards the progression of the G0/G1 transition by regulating CCND1
We speculated that LINC00152 might have a critical role in the cell cycle in HCC cells. Therefore, we analysed the cell cycle distribution after LINC00152 knockdown using FACS and found a corresponding increase in the G0/G1 proportion cells in LINC00152-knockdown Huh7 and HCCLM3 cells (Figure 4(a)). Consistent with the FACS analysis, the mRNA level of CCND1, one of the most important proteins of G1/S checkpoint, was significantly reduced in LINC00152-knockdown Huh7 and HCCLM3 cells. Likewise, western blot analysis showed that CCND1 protein expression was also reduced in LINC00152-knockdown Huh7 and HCCLM3 cells (Figure 4(b)). To confirm the association between LINC00152 and CCND1, we studied the correlation between LINC00152 and CCND1 mRNAs in HCC tissues by qRT-PCR, and found a positive relationship between LINC00152 and CCND1 mRNA expression in 32 HCC tissues (R2 = 0.354, p < 0.001) (Figure 4(c)). Taken together, these data indicate that CCND1 is a downstream effecter of LINC00152.
Figure 4.
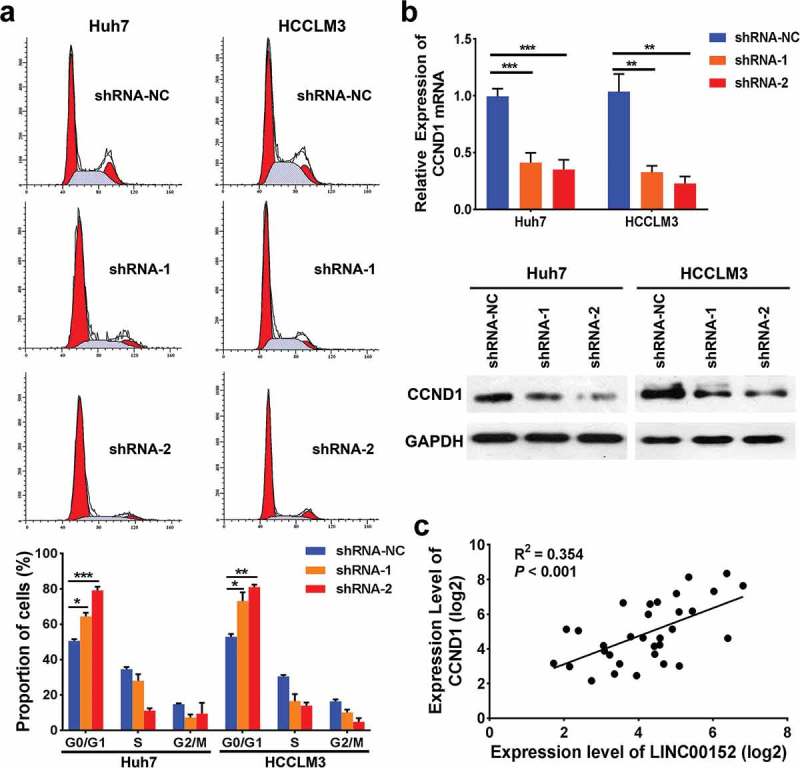
LINC00152 inhibits cell cycle progression by controlling CCND1 in HCC cells. (a) Fluorescence-activated cell sorting (FACS) analysis showed a marked increase in the proportion of cells in G0/G1 in the LINC00152-knockdown Huh7 and HCCLM3 cells (* p < 0.05, ** p < 0.01, *** p < 0.001). (b) The CCND1 mRNA and protein expression levels were reduced in qPCR and western blot analyses following LINC00152 knockdown (** p < 0.01, *** p < 0.001). (c) LINC00152 mRNA expression was positively correlated with CCND1 mRNA expression in 32 HCC specimens as detected by qRT–PCR (R2 = 0.354, p < 0.001).
Moreover, rescue experiments showed that, CCND1 overexpression partially abrogated G0/G1 arrest induced by LINC00152 silencing in Huh7 and HCCLM3 cells (Figure 5(a)). In contrast, CCND1 silencing abolished the decrease in G0/G1 arrest observed in Hep3B overexpressing LINC00152 (Figure 5(b)). Consistent with the cell cycle analysis, CCND1 overexpression reversed the effect of reduced cell proliferation capacity in LINC00152-knockdown HCC cells, while CCND1 silencing reversed the effect of increased cell proliferation capacity caused by LINC00152 overexpression in HCC cells (Figure 5(c,d)), suggesting that the function of LINC00152 depended on CCND1. Collectively, these data indicated that LINC00152 retards the progression of the G1/S transition by controlling CCND1.
Figure 5.
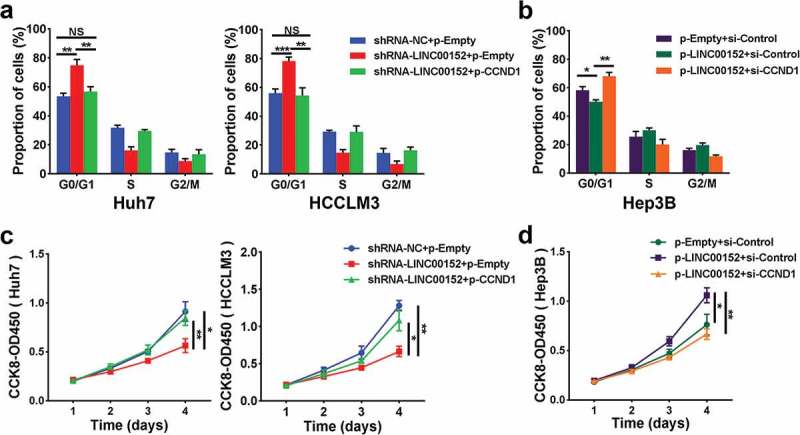
The function of LINC00152 depends on CCND1. (a) Knockdown LINC00152 induced a marked increase in the proportion of cells in G0/G1, and ectopic expression of CCND1 partially abrogated the LINC00152 knockdown inducing G0/G1 arrest in both Huh7 and HCCLM3 cells (** p < 0.01, *** p < 0.001). (b) Ectopic expression of LINC00152 reduced the proportion of cells in G0/G1, and CCND1 knockdown halted the decreased in G0/G1 arrest seen in Hep3B cells with overexpressubg LINC00152 (* p < 0.05, ** p < 0.01). (c, d) CCND1 overexpression reversed the effect of reduced cell proliferation capacity in LINC00152-knockdown HCC cells, while CCND1 underexpression reversed the effect of increased cell proliferation capacity caused by overexpressing LINC00152 in HCC cells (* p < 0.05, ** p < 0.01).
LINC00152 functions as a competing endogenous RNA (cerna) by sponging mir-193a/b-3p
Since LINC00152 regulates the CCND1 transcript, we speculated that LINC00152 might be involved in the transcriptional control of CCND1. According to previous studies, LINC00152 localizes preferentially to the cytoplasm, and cytoplasmic lncRNAs harbouring miRNA-binding sites can communicate and regulate each other’s expression levels by competing specifically with mRNAs for binding to shared miRNAs, thus acting as ceRNAs. Bioinformatics analyses using StarBase v2.0 (http://starbase.sysu.edu.cn/mirLncRNA.php) and the miRcode database (http://www.mircode.org/) predicted that all the LINC00152 transcripts contain a potential binding site for Argonaute 2 (AGO2), which combines miR-193a-3p and miR-193b-3p (Figure 6(a,b), Supplementary Table 2a, 2b).
Figure 6.
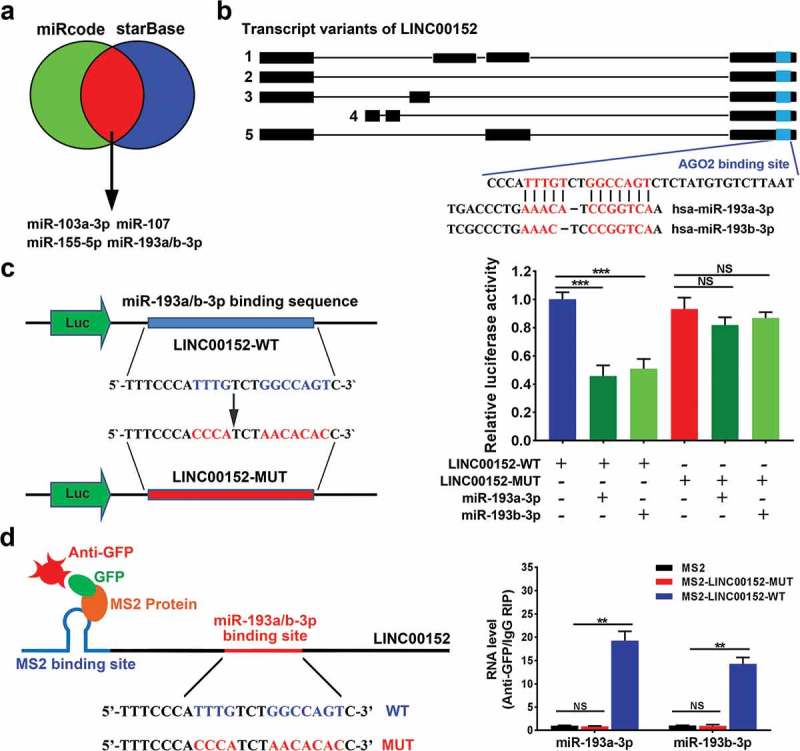
LINC00152 functions as a competing endogenous RNA (ceRNA) by sponging miR-193a/b-3p. (a, b) Bioinformatics analyses using miRcode (http://www.mircode.org/) and StarBase v2.0 (http://starbase.sysu.edu.cn/mirLncRNA.php) database predicted that all LINC00152 transcript variants contained the same potential binding sites for both AGO2 and miR-193a/b-3p (shown in blue). (c) Wild type and mutant LINC00152 sequences were cloned into pmirGLO vectors and co-transfected with either miR-193a-3p or miR-193b-3p into Hep3B cells. Luciferase activities were measured using the dual-luciferase assay. The results showed that co-transfection of miR-193a/b-3p and the wild type LINC00152, but not the mutant LINC00152, significantly decreased luciferase activity (***p < 0.001, NS, not significant). (d) MS2-RNA immunoprecipitation (MS2-RIP) assays followed by qRT–PCR to detect miR-193a/b-3p (**p < 0.01, NS, not significant).
Based on these analyses, we speculated that LINC00152 might act as a ceRNA by sponging miR-193a/b-3p in HCC. To determine whether miR-193a/b-3p recognize the predicted target sites within the LINC00152 sequence, we constructed luciferase vectors containing either wild type or mutant LINC00152. We performed luciferase assays using Hep3B cells transfected with the different vectors and, found that co-transfection of either miR-193a-3p mimic or miR-193b-3p mimic together with wild-type pmirGLO-LINC00152, but not with the mutant LINC00152, significantly decreased luciferase activities (Figure 6(c)).
We also performed MS2-RNA immunoprecipitation (MS2-RIP) assays to demonstrate a direct interaction between LINC00152 and miR-193a/b-3p. The results showed that wild-type LINC00152 could be immunoprecipitated with miR-193a/b-3p; however, LINC00152 containing mutant miR-193a/b-3p-binding sites showed no interaction with miR-193a/b-3p in Hep3B cells (Figure 6(d)), indicating that miR-193a/b-3p can bind directly to LINC00152.
LINC00152 regulates CCND1 by binding to miR-193a/b-3p
To determine whether LINC00152 regulates CCND1 transcription by binding to miR-193a/b-3p, we first performed bioinformatic analysis using TargetScan, which identified CCND1 as a candidate target gene of miR-193a/b-3p (Figure 7(a), Supplementary Table 2c). Subsequently, the pmirGLO-CCND1-3ʹ-UTR plasmid was co-transfected with the miR-193a-3p mimic, the miR-193b-3p mimic or the pcDNA3.1-LINC00152 plasmid in Hep3B. Co-transfection with either the miR-193a-3p mimic or the miR-193b-3p mimic reduced luciferase activity, confirming that CCND1 is a target of miR-193a/b-3p. In contrast, cells co-transfected with LINC00152 alone showed increased luciferase activity, while co-transfected with the miR-193a-3p or miR-193b-3p mimics together with LINC00152 partially abrogated the increase in luciferase activity induced by the LINC00152 plasmid (Figure 7(b)). These data demonstrated LINC00152-CCND1 crosstalk through competition for binding to miR-193a/b-3p. As expected, the mRNA expression of CCND1 was increased with ectopically expressed LINC00152, decreased with miR-193a/b-3p overexpression, and moderately changed with LINC00152 together with miR-193a/b-3p (Figure 7(c)). Moreover, the protein level of CCND1 was consistent with the mRNA level (Figure 7(d)). Finally, we suspected that LINC00152 may promote cell proliferation through the regulation of CCND1 in HCC. EdU incorporation assays demonstrated that overexpression of LINC00152 together with miR-193a/b-3p moderately promoted cell proliferation compared with LINC00152 overexpression alone (Figure 7(e)). Collectively, these data indicated that LINC00152 regulates CCND1 by binding to miR-193a/b-3p.
Figure 7.
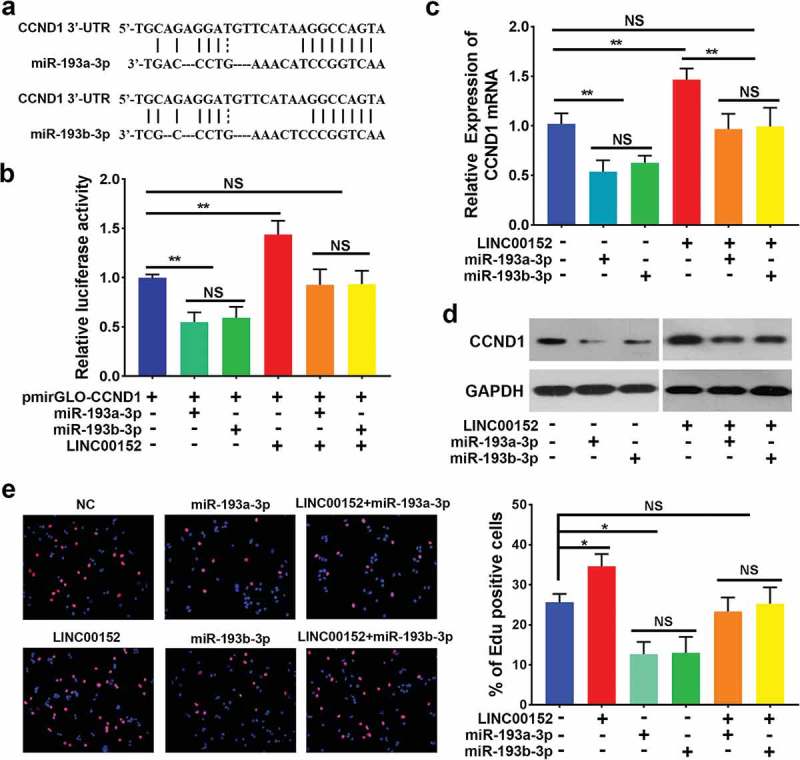
LINC00152 regulates the expression of the miR-193a/b-3p target gene, CCND1. (a) Bioinformatic analysis using TargetScan identified CCND1 as a candidate target gene of miR-193a/b-3p. (b) The 3ʹ-UTR of CCND1 was cloned into the pmirGLO vector and co-transfected into Hep3B cells along with either the miR-193a-3p, or miR-193b-3p mimic, or with pcDNA3.1-LINC00152. Luciferase activities were detected using the dual-luciferase assay. Cells co-transfected with pmirGLO-CCND1 and either miR-193a-3p or miR-193b-3p showed a decrease in luciferase activity, while cells co-transfected with pcDNA3.1-LINC00152 and pmirGLO-CCND1 showed an increase in luciferase activity. Cells co-transfected with pcDNA3.1-LINC00152, pmirGLO-CCND1 and either miR-193a-3p or miR-193b-3p showed no marked change (**p < 0.01, NS, not significant). (c) The expression of CCND1 mRNA was analysed in HCC cells following the overexpression of LINC00152 and either miR-193a-3p or miR-193b-3p. CCND1 mRNA expression decreased in cells transfected with either miR-193a-3p or miR-193b-3p compared with negative control cells, and CCND1 mRNA expression increased in cells transfected with pcDNA3.1-LINC00152. Cells co-transfected with pcDNA3.1-LINC00152 and either miR-193a-3p or miR-193b-3p showed a marked decrease in CCND1 mRNA expression compared with cells pcDNA3.1-LINC00152 alone (**p < 0.01, NS, not significant). (d) The expression of CCND1 protein was analysed in HCC cells following the overexpression of LINC00152 and either miR-193a-3p or miR-193b-3p. The expression level of the CCND1 protein was consistent with that of the CCND1 mRNA. (e) MiR-193a/b-3p abrogated the ability of LINC00152 to induce cell proliferation in Hep3B cells (*p < 0.05, NS, not significant).
Discussion
Recently, many studies have revealed crucial roles for lncRNAs in the development and progression of human cancers. In the case of HCC, several lncRNAs, such as PCNA-AS1[14], lncRNA-ATB [15], HULC [16] and Dreh [17], have been characterized, and the underlying mechanisms have been explored. In the present study, we identified another non-coding RNA, LINC00152 is upregulated in HCC tissues; moreover, LINC00152 knockdown inhibits HCC cell cycle progression by controlling CCND1 and thus retarding cell proliferation. These findings suggest that LINC00152 may exert an oncogenic function and play a key role in HCC development.
Recently, recurrent tumourigenesis-associated lncRNAs were identified via an analysis of RNA-sequence data from 60 clinical HCC samples [18], which was correlated with clinical data in a TCGA cohort and published liver cancer data; moreover, LINC00152 was considered to represent a recurrently deregulated lncRNA potentially associated with tumourigenesis. Further research showed that LINC00152 overexpression leads to increased proliferation in clear cell renal cell carcinoma [19], glioma stem cell [7], gastric cancer [8] and HCC [3]. Consistent with these findings, the present study found that LINC00152 promotes HCC cell proliferation by inducing cell cycle progression via regulating the cell cycle-related protein, CCND1. CCND1 functions as a regulatory subunit of CDK4 or CDK6, whose activity is required for cell cycle progression [20]. Deregulated cell cycle contributes to the unscheduled proliferation in cancer cells. Mutant CCND1 is one of the most common molecular anomalies in HCC, besides mutations in the TERT promoter, TP53, CTNNB1, AXIN1, ARID1A, CDKN2A genes. On the other hand, CCND1 alterations (focal amplifications or deletions) are also associated with poor prognosis in HCC patients [21]. Previous results showed that CCND1 amplification and overexpression is also associated with tumour prognosis in hairy cell leukemia [22] and human malignant gliomas [23]. Our data indicated that LINC00152 silencing leads to decreased expression of CCND1 at both the mRNA and protein levels; LINC00152 is positively correlated with CCND1 expression in a group of HCC tissues. And moreover LINC00152 regulates CCND1 by binding to miR-193a/b-3p. The coordinated regulation of LINC00152 and CCND1 requires future investigation in other cancers.
LINC00152, belongs to a major class of intergenic lncRNAs, which originate from the region between two protein-coding genes; and the structural complexity of intergenic lncRNAs offer multiple possibilities for interactions with DNA, RNA and proteins and enable the formation of multicomponent complexes such as those involved in epigenetic regulation of transcription. Likewise, LINC00152 can bind to the promoter of EpCAM in HCC [11], the enhancer of EZH2 in lung adenocarcinoma [5], renal cell cancer [19] and the promoter of EGFR in gastric cancer [24], which reflects the function of cis-acting lncRNAs in regulating gene expression in a locus and allele-specific manner because of their genomic proximity to their target genes. The modulation of neighboring gene expression can occur via directly binding or through the recruitment of chromatin-binding proteins, which can modify the chromatin structure in the vicinity [1]. Previous evidences indicated that LINC00152 localizes in both cytoplasm and nuclear, preferential to cytoplasm. And trans-acting nuclear lncRNAs can act in together with chromatin modifiers, such as histone modifying complexes and DNA methyltransferases, to epigenetically regulate transcription. In contract, cis-acting nuclear lncRNAs may be able to regulate gene expression via directly binding or through the recruitment of chromatin-binding proteins [1]. Recently, a novel regulatory mechanism has been identified in which crosstalk between lncRNAs in cytoplasm and mRNAs occurs due to competition for shared miRNAs. These emerging links with miRNAs provided an attractive potential mechanism for the regulation of lncRNAs. For instance, HOXD-AS1 competitively binds to miR-130a-3p, and prevents SOX4 from miRNA-mediated degradation, thus activates the expression of EZH2 and MMP2, facilitating HCC metastasis [25]. LncRNA nuclear paraspeckle assembly transcript 1 regulates CDK6 expression in laryngeal squamous cell cancer by sponging miR-107, then controls cell proliferation and apoptosis [26]. And likewise, LINC00152 can directly bind to miR-103a-3p and regulate its direct target, forebrain embryonic zinc finger protein 1 (FEZF1), which plays an oncogenic role in GSCs [7]. Our current study revealed that LINC00152 functions as a ceRNA to modulate CCND1 by competitively binding to the same major response elements on miR-193a/b-3p. First, we found that LINC00152 binds directly to miR-193a/b-3p and confirmed, an endogenous interaction between LINC00152 and miR-193a/b-3p using RIP. Second, LINC00152 partially abolished miR-193a-3p and miR-193b-3p mediated biological effects. Third, LINC00152 indirectly regulated the expression of the miR-193a/b-3p target gene, CCND1. Currently, LINC00152 has been considered to mediate acquired resistance to oxaliplatin in colon cancer; mechanistically, LINC00152 confers resistance to oxaliplatin by competitively binding miR-193a-3p, upregulating ERBB4, and then inducing the activation of AKT signaling pathway [27]. Consistently, our data strongly supported crosstalk between LINC00152 and CCND1 due to competition for shared miRNAs, miR-193a/b-3p.
Based on our work, we propose a ceRNA mechanism regulating proliferation via a LINC00152/miR-193a/b-3p/CCND1 signalling axis in HCC. Mir-193a/b-3p is part of the miR-193 family, together with miR-193a-5p and miR-193b-5p. Previous findings suggest that miR-193a-3p acts as a tumour suppressor in human malignancies. Exogenous miR-193a-3p/5p inhibits NSCLC cell migration, invasion and EMT in vitro and lung metastasis formation in vivo, by inactivating the AKT/mTOR signalling pathway [28].
In summary, our study illustrates that LINC00152 functions as a ceRNA and, regulates CCND1 expression by competitively binding to miR-193a/b-3p, thus promoting cell proliferation. These findings provided mechanistic insights into the role of LINC00152 in promoting HCC proliferation and suggest that LINC00152 may be an important prognostic factor and therapeutic target for HCC.
Conclusions
The current study reveals that LINC00152 controls cell cycle progression by regulating CCND1 and promotes HCC proliferation; LINC00152 acts as a ceRNA by binding to miR-193a/b-3p and thereby modulating CCND1. Thus, our findings establish a ceRNA mechanism regulating proliferation via the LINC00152/miR-193a/b-3p/CCND1 signalling axis and identify LINC00152 as a potential therapeutic target for HCC.
Materials and methods
Tissue samples and cell lines
All human tissue samples were obtained with written informed consent from all subjects, and this study was approved by the medical ethical committee of Huazhong University of Science and Technology. A total of 80 paired HCC and non-neoplastic liver tissues were obtained from patients who underwent surgery at Wuhan Central Hospital of Tongji Medical College, Huazhong University of Science and Technology from Jan., 2015 to Dec., 2016. All patients were diagnosed with HCC according to histopathological evaluation. None of the patients had received preoperative adjuvant transcatheter arterial chemoembolization or percutaneous ethanol injection before hepatectomy. Samples were immediately snap-frozen in liquid nitrogen and then stored at −80°C until use.
Huh7, HCCLM3 and Hep3B were purchased from the Cell Bank of Type Culture Collection (Chinese Academy of Science, Shanghai, China) and were authenticated. The cell lines were incubated in DMEM (Gibco, USA) with 10% fetal bovine serum (Gibco, USA) and maintained at 37°C in a humidified incubator containing 5% CO2. All cell lines have been passaged for less than 6 months.
RNA extraction, quantitative real-time PCR, protein extraction and western blot
Detailed descriptions of RNA isolation, cDNA synthesis, quantitative real-time PCR (qRT-PCR) and western blot are provided in the Supplementary Methods. The primers used are presented in Supplementary Table 1.
Shrna interference, plasmid construction
For the RNA interference (RNAi)-mediated knockdown of LINC00152 and CCND1, two different shRNAs for LINC00152, one siRNA for CCND1 and one negative control shRNA were generated by GenePharma (Suzhou, China), as shown in Supplementary Table 1. The cDNAs encoding LINC00152 and the 3ʹUTR/CDS of CCND1 were amplified and subcloned into the pcDNA3.1 and pmirGLO vectors, respectively. Hsa-miR-193a-3p and Hsa-miR-193b-3p mimics were purchased from GenePharma (Suzhou, China). A LINC00152 construct, designated LINC00152-mut, containing mutations at the putative miR-193a/b-3p response elements was created using the QuikChange Site-Directed Mutagenesis kit (Stratagene, USA).
Tumour formation assays
Male athymic BALB/c nude mice (4-weeks-old) were purchased from the Central Laboratory of Animal Science, Wuhan University (Wuhan, China) and were maintained under specific pathogen-free conditions. HCCLM3 cells stably transfected with either shRNA-LINC00152 or shRNA-NC were harvested from 6-well plates and suspended at 1 × 108 cells/ml. The suspended cells (100 μl) were subcutaneously injected into the armpits of ten mice (2 groups), and the mice were sacrificed 5 weeks after injection. The visible tumours on the armpits were counted and used for further analysis.
Cell viability, EdU, colony formation, cell cycle assay and luciferase assay
Detailed descriptions of cell viability, EdU, colony formation, cell cycle distribution and luciferase assay are provided in the Supplementary Methods.
lncRNA-miRNA MS2-RNA immunoprecipitation (MS2-RIP) assay
Hep3B cells transfected with pMS2-GFP were co-transfected with pcDNA3.1-MS2, pcDNA3.1-MS2-LINC00152 or pcDNA3.1-MS2-LINC00152-mut (miR-193a/b-3p). After 48 hours, the cells were subjected to RIP experiments using a GFP antibody (Roche, Germany) and the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) according to the manufacturer’s instructions (schematic diagram is shown in Figure 6(d)). After the cells were harvested and lysed, the cell suspensions and magnetic beads were incubated in microcentrifuge tubes and vortex-oscillated to prepare the suspensions and beads for immunoprecipitation. The cell lysate supernatants and magnetic bead plus antibody complexes were incubated overnight. Salt solutions and precipitate enhancer were used for RNA purification. The RNA fraction isolated by RIP was quantified with the NanoDrop spectrophotometer (Thermo Fisher Scientific), and the RNA quality was assessed with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). After reverse transcription, qRT-PCR of miR-193a/b-3p was used to confirm miR-193a/b-3p adsorption by LINC00152.
Statistical analysis
All analyses were performed using PASW Statistics, version 13.0 (SPSS). Variables were compared using t test or one-way ANOVA in univariate analysis. Spearman correlation coefficient was used to assess the expression correlation assay. All p values shown were two sided and p < 0.05 was considered statistically significant.
Funding Statement
This study was funded by the Natural Science Foundation of Hubei Province [No. 2018CFB120], and National Basic Research Program of China (973 Program) [No. 2012CB720600, 2012CB720605].
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Parasramka MA, Maji S, Matsuda A, et al. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol Ther. 2016;161:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Li J, Wang X, Tang J, et al. HULC and Linc00152 Act as Novel biomarkers in predicting diagnosis of hepatocellular Carcinoma. Cell Physiol Biochem. 2015;37:687–696. [DOI] [PubMed] [Google Scholar]
- [3].Deng X, Zhao XF, Liang XQ, et al. Linc00152 promotes cancer progression in hepatitis B virus-associated hepatocellular carcinoma. Biomed Pharmacother. 2017;90:100–108. [DOI] [PubMed] [Google Scholar]
- [4].Cai Q, Wang Z, Wang S, et al. Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial-mesenchymal transition by regulating HIF-1α via miR-138. Open Biol. 2017;7:160247. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [5].Chen QN, Chen X, Chen ZY, et al. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression. Mol Cancer. 2017;16:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yu J, Liu Y, Guo C, et al. Upregulated long non-coding RNA LINC00152 expression is associated with progression and poor prognosis of tongue squamous cell carcinoma. J Cancer. 2017;8:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yu M, Xue Y, Zheng J, et al. Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway. Mol Cancer. 2017;16:110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [8].Zhao J, Liu Y, Zhang W, et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nötzold L, Frank L, Gandhi M, et al. The long non-coding RNA LINC00152 is essential for cell cycle progression through mitosis in HeLa cells. Sci Rep. 2017;7:2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Feng S, Zhang J, Su W, et al. Overexpression of LINC00152 correlates with poor patient survival and knockdown impairs cell proliferation in lung cancer. Sci Rep. 2017;7:2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ji J, Tang J, Deng L, et al. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6:42813–42824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chapman EG, Costantino DA, Rabe JL, et al. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science. 2014;344:307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tay Y, Rinn J, Pandolfi PP.. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yuan SX, Tao QF, Wang J, et al. Antisense long non-coding RNA PCNA-AS1 promotes tumour growth by regulating proliferating cell nuclear antigen in hepatocellular carcinoma. Cancer Lett. 2014;349:87–94. [DOI] [PubMed] [Google Scholar]
- [15].Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. [DOI] [PubMed] [Google Scholar]
- [16].Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. [DOI] [PubMed] [Google Scholar]
- [17].Huang JF, Guo YJ, Zhao CX, et al. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;57:1882–1892. [DOI] [PubMed] [Google Scholar]
- [18].Yang Y, Chen L, Gu J, et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun. 2017;8:14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang Y, Liu J, Bai H, et al. Long intergenic non-coding RNA 00152 promotes renal cell carcinoma progression by epigenetically suppressing P16 and negatively regulates miR-205. Am J Cancer Res. 2017;7:312–322. [PMC free article] [PubMed] [Google Scholar]
- [20].Belinky F, Nativ N, Stelzer G, et al. PathCards: multi-source consolidation of human biological pathways. Database (Oxford). 2015;2015:bav006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].de Boer CJ, Kluin-Nelemans JC, Dreef E, et al. Involvement of the CCND1 gene in hairy cell leukemia. Ann Oncol. 1996;7:251–256. [DOI] [PubMed] [Google Scholar]
- [23].Buschges R, Weber RG, Actor B, et al. Amplification and expression of cyclin D genes (CCND1, CCND2 and CCND3) in human malignant gliomas. Brain Pathol. 1999;9:435–442, 432–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhou J, Zhi X, Wang L, et al. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J Exp Clin Cancer Res. 2016;34(9):135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang H, Huo XS, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang P, Wu T, Zhou H, et al. Long noncoding RNA NEAT1 promotes laryngeal squamous cell cancer through regulating miR-107/CDK6 pathway. J Exp Clin Cancer Res. 2016;35:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yue B, Cai D, Liu C, et al. Linc00152 functions as a competing endogenous RNA to confer oxaliplatin resistance and holds prognostic values in colon cancer. Mol Ther. 2016;24:2064–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yu T, Li J, Yan M, et al. MicroRNA-193a-3p and −5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene. 2015;34:413–423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


