ABSTRACT
The modification of adenosine to inosine at position 34 of tRNA anticodons has a profound impact upon codon-anticodon recognition. In bacteria, I34 is thought to exist only in tRNAArg, while in eukaryotes the modification is present in eight different tRNAs. In eukaryotes, the widespread use of I34 strongly influenced the evolution of genomes in terms of tRNA gene abundance and codon usage. In humans, codon usage indicates that I34 modified tRNAs are preferred for the translation of highly repetitive coding sequences, suggesting that I34 is an important modification for the synthesis of proteins of highly skewed amino acid composition. Here we extend the analysis of distribution of codons that are recognized by I34 containing tRNAs to all phyla known to use this modification. We find that the preference for codons recognized by such tRNAs in genes with highly biased codon compositions is universal among eukaryotes, and we report that, unexpectedly, some bacterial phyla show a similar preference. We demonstrate that the genomes of these bacterial species contain previously undescribed tRNA genes that are potential substrates for deamination at position 34.
KEYWORDS: Translation, Evolution, Speciation, tRNA, Transcriptome, mRNA, ADAT, TadA, CDS
Introduction
Transfer RNAs (tRNA) are the universal adaptors of the genetic code,1 linking codons to their cognate amino acids during protein synthesis. While the structure of the genetic code is mostly conserved across all domains of life, the genomic composition of tRNA genes varies widely between species.2 Some archaeal species have only 20–25 different tRNAs, while some Eukaryotes can present up to 40–45 different tRNAs in their genomes3,4 Importantly, the number of different tRNAs is always lower than the number of codons used and, therefore, some tRNAs need to recognize more than one codon. This is achieved by a higher pairing permissiveness between the third position of the mRNA codons and the first position of the tRNA anticodon in what is known as “wobble” or degenerate pairing.5 Due to codon degeneracy those amino acids coded by at least four codons (threonine, alanine, proline, serine, glycine, leucine, valine and arginine) can, in principle, be specifically incorporated with just two specific pairings (Fig. 1a).6
Figure 1.
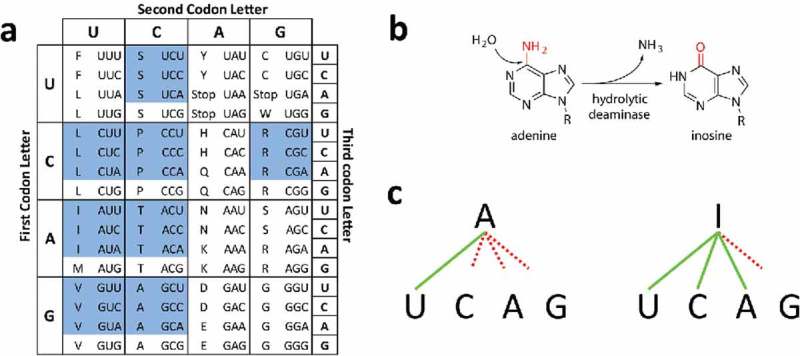
Impact of I34 in translation. (a) Standard genetic code. Blue cells correspond to ADAT-sensitive codons. (b) Inosine modification by hydrolytic deamination. (c) Schematic representation of possible base pairings between Adenine and Inosine. Continuous green lines indicate preferred pairings and dashed red line indicates poor or no pairing.
Posttranscriptional tRNA modifications are essential for tRNA function. While certain tRNA modifications are present in all living organisms, others are kingdom specific.4 In general, modifications in the acceptor stem of tRNAs are important for amino acid charging, modifications in the main body of the tRNA can affect tRNA structure and stability, and modifications in the anticodon loop of tRNAs modulate codon recognition.7–9 Position 34 in the anticodon pairs with the third base of codons, and harbors the widest variety of known modifications.10,11 Modifications at position 34 fine-tune wobble pairing and extend or restrict the number of codons that anticodons recognize. We have shown that an improved correlation between codon usage and tRNA gene copy number in Bacteria and Eukaryotes can be observed when the effects that modified bases at position 34 have upon codon recognition are taken into account.6
In bacterial genomes, tRNA genes complementary to codons for four-box and six-box amino acids predominantly have guanine at position 34 (G34) over adenosine at this position (A34), with the exception of genes for tRNAArg. In eukaryotic genomes, on the other hand, tRNA genes for the same codons almost exclusively use A34.6 These differences in decoding strategies between Bacteria and Eukarya are explained by the presence or absence of a modified base (inosine) at position 34 (I34) in these tRNAs. I34 is absent in archaeal tRNAs, occurs in bacterial tRNAArg (ACG), and in eukaryotic tRNAAla (AGC), tRNAPro (AGG), tRNAThr (AGT), tRNAVal (AAC), tRNASer (AGA), tRNAArg (ACG), tRNALeu (AAG) and tRNAIle (AAT).12,13
The conversion by hydrolytic deamination of adenosine (A) to inosine (I) at position 34 of tRNAs is catalyzed by adenosine deaminases (Fig. 1b).12–18 I34-modified tRNAs can wobble pair with codons ending in A3, C3 or U3, but not G3, expanding to 3 the number of codons that genetically encoded A34 tRNAs can recognize (Fig. 1c).5,13 However, the selective pressure that drove the increase from one I34-containing tRNA in bacteria to eight modified tRNAs in eukaryotic translation is still unclear.13,19 We have recently shown that tRNAGly escaped this process because its anticodon structure is incompatible with the presence of an adenosine at position 34.20
In Bacteria, an essential homodimer called tRNA adenosine deaminase A (TadA) modifies tRNAArg (ACG) to tRNAArg (ICG).21 tRNAArg (ACG) is, to date, the only A34-containing tRNA known to be deaminated to I34 in Bacteria, and this explains why bacterial arginine codons are preferentially translated by tRNAs initially transcribed with A34. A bacterial tadA gene was transferred to eukaryotes (possibly during the mitochondrial endosymbiotic event) where, through duplication and divergence, evolved into the heterodimeric enzyme adenosine deaminase acting on tRNA (ADAT, formed by the subunits ADAT2 and ADAT3). This acquisition increased the substrate repertoire of ADAT to eight different tRNAs,13,22 which are preferentially used by eukaryotes to translate the codons for threonine, alanine, proline, serine, leucine, isoleucine, valine and arginine (hereinafter “ADAT amino acids”).
We have shown that the impact of ADAT activity in the translation of the human genome is particularly relevant to the synthesis of proteins highly enriched in ADAT amino acids.23 We found that human ORFs enriched in ADAT amino acids are also significantly enriched in codons that are translated by I34-containg tRNAs (hereinafter “ADAT-sensitive codons”)(23). Interestingly, although eight amino acids are translated by I34-containing tRNAs, only four amino acids (T, A, P, and S; hereinafter “TAPS”) are found to accumulate to high levels (over 84% of positions) in human proteins.23
Here we have extended our initial analysis of the human proteome to 64 eukaryotic and 980 bacterial species spanning the whole tree of life. Again we find that, for both Eukarya and Bacteria, proteins enriched in ADAT amino acids (>84% threshold) show an enrichment limited to TAPS. When all eukaryotic sequences are considered, a bias toward the use of ADAT-sensitive codons in genes coding for proteins enriched in ADAT amino acids is apparent. Similarly, a positive correlation exists between length of the stretches enriched in ADAT amino acids and enrichment in ADAT-sensitive codons. Bacterial sequences as a whole do not display such correlations.
We have continued this analysis organizing the organisms into different groups (Table S1), to check whether our findings within eukaryotes and bacteria are applicable to all the groups. Surprisingly, this additional analysis revealed that neither eukaryotes nor bacteria behave uniformly with regards to the frequency of ADAT-sensitive codons in their genomes. Instead, our data indicates that the evolution of I34 as an adaptation for the synthesis of TAPS-enriched proteins is different between bacterial and eukaryotic phyla. In support for this idea we present new evidence that bacterial species showing an unexpected enrichment in ADAT-sensitive codons contain in their genomes tRNA genes with A34 that are cognate for amino acids other than arginine and could possibly also be deaminated by TadA.
We postulate that this situation is an adaptation of these species that allows them to translate a larger number of transcripts enriched in ADAT-sensitive codons. We propose that I34 was selected in eukaryotes because of its contribution to the potential diversity of the eukaryotic proteome.24,25 It is possible that some bacterial phyla have undergone a similar evolution of their translation apparatus.
Results
Stretches of ADAT amino acids are more abundant, and more enriched in ADAT-sensitive codons, in eukarya than in bacteria
Using previously described methods,23 we have analyzed 1044 genomes in search of transcript coding sequences (CDSs) coding for protein stretches enriched in ADAT amino acids (see Materials and Methods). Figure 2 shows the number of such stretches obtained for our set of 64 eukaryotic and 980 bacterial genomes (Table 1). As previously reported for the human transcriptome23, the observed stretches both in Eukarya and Bacteria are mainly composed of TAPS amino acids (p-value = 6.3e-13) (Fig. 3), indicating that the enrichment in TAPS within stretches probably is a consequence of the physicochemical characteristics of these amino acids and not of a phylogenetic parameter.
Figure 2.
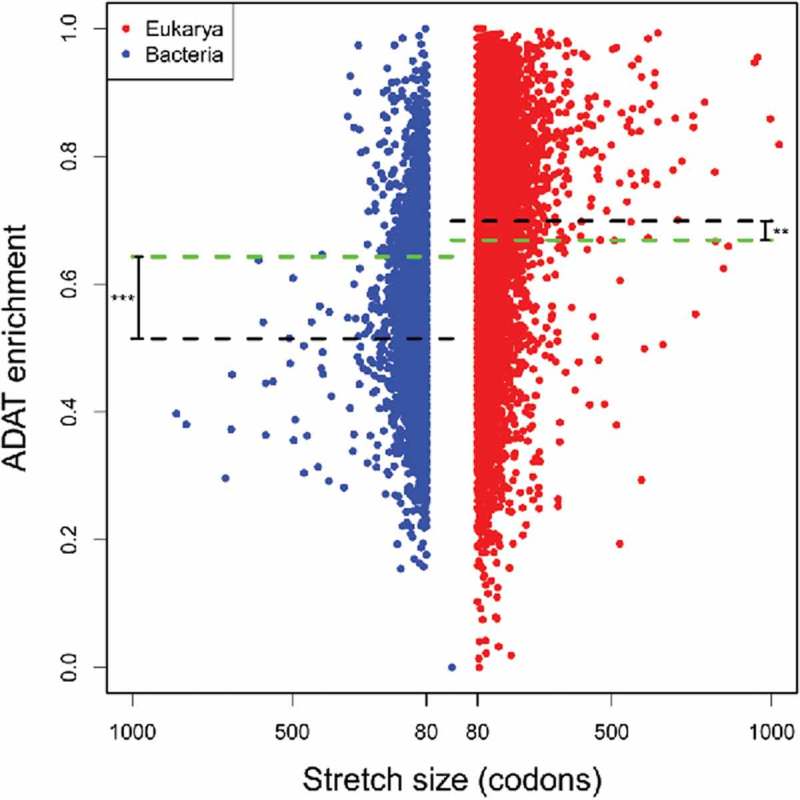
Characterization of proteins enriched in ADAT amino acids in Bacteria (blue dots) and Eukarya (red dots). Each dot represents a stretch of amino acids of length >80, with a composition of ADAT amino acids higher than 83%. Black dashed lines denote the mean enrichment in ADAT sensitive codons in stretches of ADAT amino acids. The green dashed lines denote the mean enrichment in ADAT-sensitive codons for all CDSs analyzed. There are significant differences between black and green dashed lines (p-value = 0.002 for Eukarya, p-value <2.2e-16 for Bacteria). Stretches higher than 1000 codons were not considered.
Table 1.
Statistics from the analysis in Figure 1.
| Bacteria | Eukarya | p-val | |
|---|---|---|---|
| Num. organisms | 980 | 64 | n.a. |
| with stretches | 531 (54%) | 64 (100%) | n.a. |
| Num. Stretches | 7047 | 11769 | n.a. |
| Mean stretch size | 107 ± 42 | 118 ± 58 | < 2.2e-16 |
| Mean stretch enrich. | 0.546 ± 0.12 | 0.699 ± 0.15 | < 2.2e-16 |
| Mean CDSs enrich. | 0.643 ± 0.08 | 0.668 ± 0.06 | 0.0159 |
| Num. CDSs | 3.39E+06 | 1.36E+06 | n.a. |
| Stretch/CDS | 2.08E-03 | 8.64E-03 | n.a. |
“enrich” = enrichment.
Figure 3.
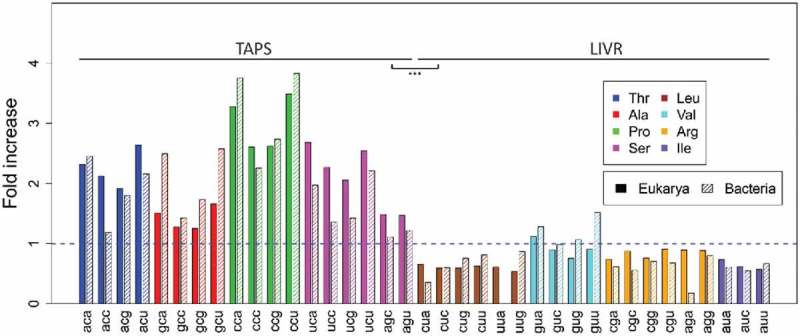
Fold increase observed for each amino acid in the stretches identified in Fig. 2 for Eukarya (uniform color) and Bacteria (shading lines). Synonymous codons for ADAT amino acids are colored (legend).
We find that the total number of stretches rich in ADAT amino acids in Eukarya is higher (11769) than in Bacteria (7747) (Table 1) (a 4-fold increase when corrected for the total number of CDSs analyzed). Similarly, the mean stretch length is also significantly longer in eukaryotic proteomes (Table 1). The number of eukaryotic stretches is higher than the number of bacterial stretches at any length interval (Fig. 4), and this difference reaches a maximum of eight fold at the interval 227–236 codons (Fig. 4, black line). However, this differences in stretch length are likely not due to general differences in gene length between eukaryotes and bacteria, as no correlation was apparent between the length of ADAT stretches and the length of their corresponding CDSs for any species (data not shown).
Figure 4.
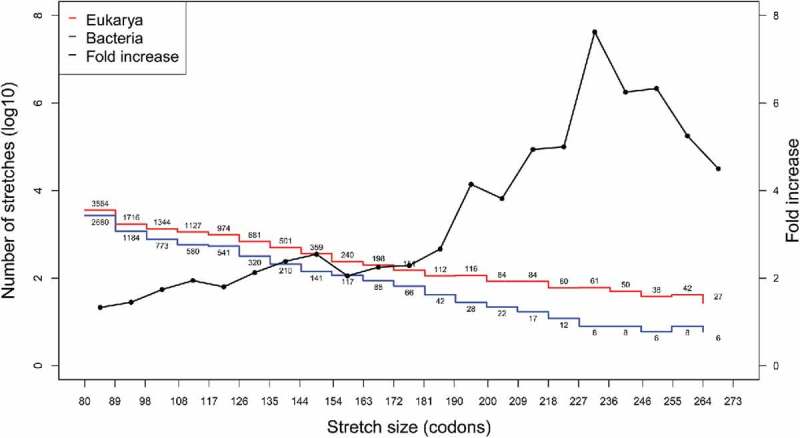
Histogram of number of ADAT stretches (in log10) for Eukarya (red) and Bacteria (blue) according to their length. ADAT stretches were calculated as in23 (See Materials and Methods) and their lengths (in codons) were clustered in equidistant intervals. Black line shows the fold increase in number of stretches of Eukarya compared with Bacteria (right axis). Numbers close to red and blue lines corresponds to the number of samples for each interval. Intervals with less than five samples were omitted.
When considering all coding sequences, eukaryotic genes display a significantly higher frequency of ADAT-sensitive codons with respect to bacterial genes (Table 1, p-value = 0.016). This difference is further increased when only genes coding for proteins enriched in ADAT amino acids are considered (Table 1, p-value < 2.2e-16). In fact, in Eukaryotes, a significant increase of ADAT-sensitive codons is seen in genes coding for proteins rich in ADAT amino acids (p-value < 2.2e-16), while the opposite is true for Bacteria (p-value = 0.002) (Fig. 2), suggesting a domain-specific bias in codon composition within ADAT stretches. Importantly, there is no correlation between the number of CDSs (or the GC content) and the number of stretches present in any given organism, either when comparing Eukaryotes versus Bacteria or when comparing individual phyla (see below), meaning that our observations are not biased by the size of the genomes, nor by the CG content of the genomes used in our analyses (Figure S2, S7 and S8).
We next compared the median enrichment in ADAT-sensitive codons in stretches of different sizes in Bacteria and Eukarya (Fig. 5). In general, no correlation was apparent between the length of ADAT stretches and the length of their corresponding CDSs (data not shown). Enrichment in ADAT-sensitive codons in Eukarya was higher than in Bacteria for all the intervals. Additionally, we found a strong positive correlation of (See Materials and Methods for definition) between the stretch size and enrichment in ADAT-sensitive codons in Eukarya, while in Bacteria this correlation was not present (Fig. 5).
Figure 5.
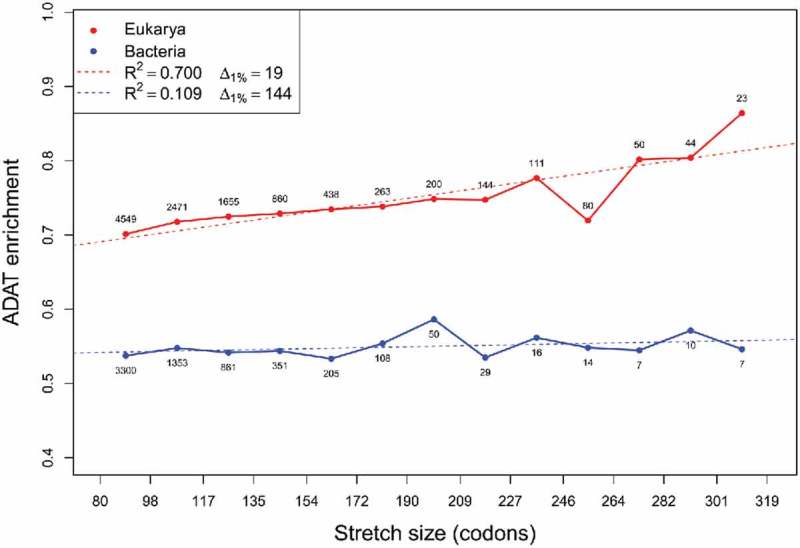
Median values of ADAT-sensitive codon frequency by length of ADAT amino acid stretches plotted for Eukarya (red) and bacteria (blue). Numbers close to the dots correspond to the number of samples for each interval. Linear regression was calculated for Eukarya (dashed red line and legend) and Bacteria (dashed blue line and legend). Intervals with less than five samples were omitted.
The enrichment in ADAT-sensitive codons in stretches behaves differently across eukaryotic and bacterial phyla
To further study ADAT stretches in Eukarya, we divided the data into the four eukaryotic groups: Metazoa, Fungi, Plantae and the rest of Eukarya (hereinafter “Protists”) (Table S1). Figure 6 shows a histogram analysis of enrichment in ADAT-sensitive codons for these kingdoms according to stretch size. The highest overall enrichment in ADAT-sensitive codons within stretches is observed for Metazoa, followed by Fungi. Note that Metazoa have the longest stretches and is the only kingdom with ADAT stretches higher than 356 codons. Significant differences in enrichment in ADAT-sensitive codons can be seen for stretch sizes higher than 190 codons, where Metazoan sequences are clearly more ADAT enriched compared to Fungi (Figure S9). Both Metazoa and Plantae show a positive correlation between enrichment in ADAT-sensitive codons and stretch length ( and respectively); while Fungi (R2 = 0.27) and Protists (R2 = 0.01) present no correlation (Fig. 6).
Figure 6.
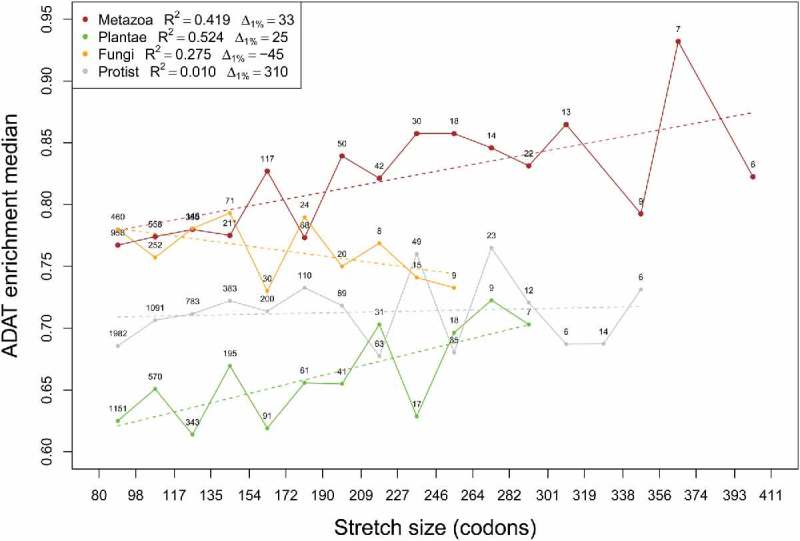
Median values of ADAT-sensitive codon enrichment by length of ADAT amino acid stretches plotted for different eukaryotic kingdoms (see legend). Linear regression was calculated for each kingdom (colored dashed lines and legend). Numbers close to the dots correspond to the number of samples for each interval. Intervals with less than five samples were omitted.
We performed a similar analysis by dividing the 980 Bacteria species into 20 phyla (Table S1). Figure 7 shows a histogram analysis of enrichment in ADAT-sensitive codons for these phyla according to their stretch size. Unexpectedly, cyanobacteria (cyan) and Firmicutes (yellow) show a high degree of enrichment in ADAT-sensitive codons throughout all the intervals, consistently higher than 0.65 ( Fig. 7). Moreover, Firmicutes present a strong positive correlation of , indicating that longer stretches are coded by sequences richer in ADAT-sensitive codons (Fig. 7). The rest of bacterial phyla (Fig. 7, black) have a depletion in ADAT-sensitive codons with most values below the mean when considering all the CDSs (Table 1, 0.643 ± 0.08) for all intervals.
Figure 7.
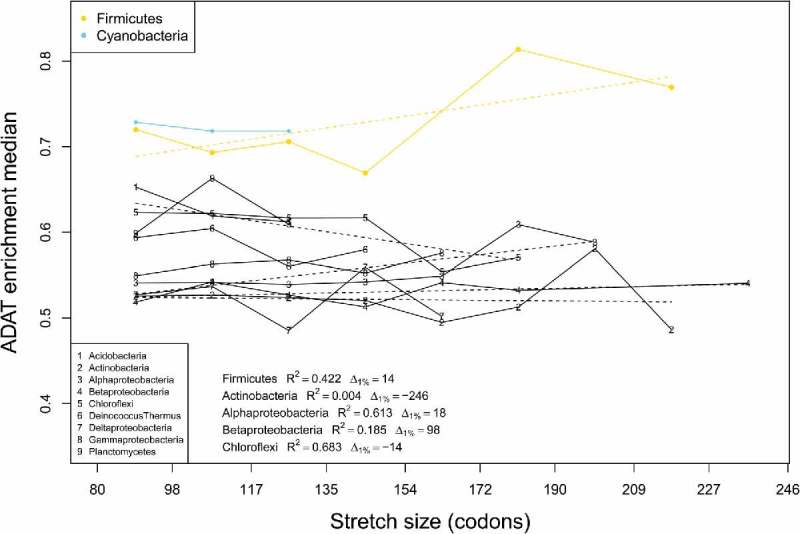
Median values of ADAT-sensitive codon enrichment by length of ADAT amino acid stretches plotted for different bacterial phyla (see legend). Phyla with less than 50 stretches were not considered. Linear regression was calculated for each phylum with more than 5 dots (dashed lines and legend). Intervals with less than five samples were omitted.
Firmicutes and Cyanobacteria are the two bacterial phyla with the highest overall enrichment in ADAT-sensitive codons compared with the rest of bacterial phyla (Fig. 8), and are comparable to Eukarya in terms of enrichment in ADAT-sensitive codons within stretches, with values not statistically different from those found for Protists (Fig. 8). Alphaproteobacteria (868 stretches), Actinobacteria (3360 stretches) and Betaproteobacteria (1115 stretches) have the lowest median enrichment in ADAT-sensitive codons despite being the bacterial phyla with most stretches, representing 57% of all the ADAT stretches (Fig. 8).
Figure 8.
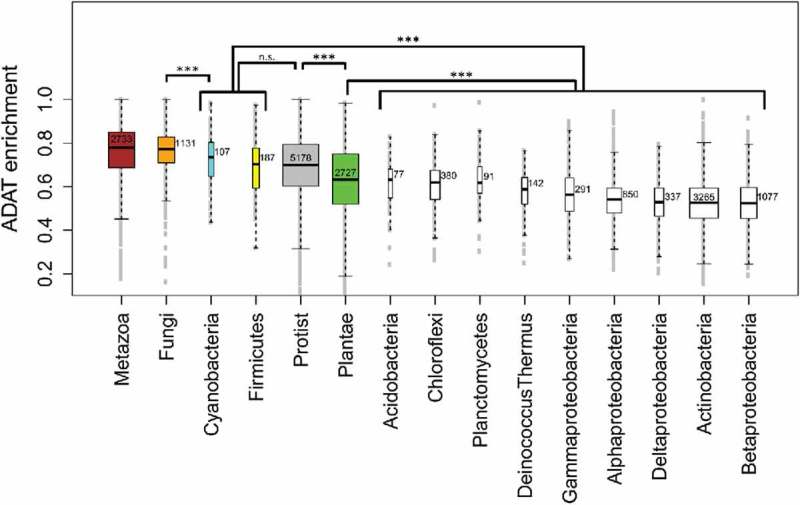
Boxplot of the enrichment in ADAT-sensitive codons for four different eukaryotic kingdoms and eleven different bacterial phyla. Colored boxes correspond to eukaryotic kingdoms except two bacterial phyla, Cyanobacteria (cyan) and Firmicutes (yellow). Numbers close to the median correspond to the number of stretches for each interval. Phyla with less than 50 stretches were not considered.
Although the discovery of proteins highly enriched in ADAT amino acids in bacteria was not unexpected given our initial analysis (Fig. 2), it was surprising to discover that the coding sequences for these stretches in Cyanobacteria and Firmicutes are also enriched in ADAT-sensitive codons and resemble Eukaryotes in this regard. Given that this observation suggests that I34 may also be playing a role in the translation of genes from these bacterial species, we searched their genomes for tRNA genes cognate for ADAT amino acids and with A34 containing anticodons. This analysis revealed that several species of Firmicutes contain this type of tRNA genes (Table S2). Possibly, these tRNAs are deaminated by TadA and influence the translation of ADAT-sensitive codons in these species.
Materials and methods
Definitions
ADAT amino acids are defined as those amino acids that are charged to tRNAs that can be modified by ADAT in most Eukarya: Thr, Ala, Pro, Thr, Ser (TAPS), and Leu, Ile, Val, and Arg. ADAT codons are defined as the set of 37 codons that code for any of the ADAT amino acids (Fig. 1a). ADAT-sensitive codons are defined as the set of 24 codons that are recognized by I34 tRNAs (Fig. 1a, blue codons). ADAT stretches are defined as those regions in the CDSs that have an enrichment in ADAT amino acids and are found using the “Running Windows” Method.26 For each ADAT stretch we define ADAT enrichment as the fraction ADAT-sensitive codons/ADAT codons. Fold increase is calculated by normalizing ADAT stretch codon composition to total CDSs codon usage, for each organism. represents the number of codons needed to increase the ADAT enrichment by 1% based on the slope of the linear model.
Eukaryotic and bacterial transcriptome retrieval
We have analyzed the transcriptomes of 64 eukaryotic species and 980 bacterial species. Eukaryotic species were downloaded from Ensembl and bacterial sequences were downloaded from NCBI. We analyzed a total of 1.13 million eukaryotic CDSs and 3.58 million bacterial CDSs. Only CDSs with a start codon, a stop codon and a number of nucleotides multiple of 3 were used for our analysis. 26% (27% in Bacteria and 19% in Eukarya) of the initial data were discarded because it did not fulfill these three requisites.
Identification of stretches by the “running windows” method
To study in more detail the presence of stretches of ADAT codons, we applied the “Running Windows” method based on software that applies similar methodologies.26–28 For each CDS of the human transcriptome a window (fragment of the sequence with a fixed length in codons) slides codon by codon from the beginning to the end of the sequence. For each position, the percentage of ADAT codons is calculated and represented with respect to its location.23 We fix the window size to 80 codons, and the threshold to be considered an enriched window in ADAT codons was set at 83% based on our previous analysis of the human proteome23 in order to have comparable datasets. We define an ADAT stretch as those regions corresponding to a window, or a set of consecutive windows, enriched in ADAT codons. Two (or more) windows are considered consecutive if the intersection between them in the cognate CDS is not void.
Statistical analyses
In order to perform robust statistics we discarded intervals of stretch length represented by less than 5 ADAT stretches in either Bacteria or Eukarya in Figs. 3–6 (full data can be observed in Figures S3-S6 respectively). In Figs. 7 and 8 we only used for the analyses bacterial phyla with more than 50 stretches in total. In Fig. 7, linear regression was made for phyla with at least 5 points of approximation. Linear regression in Figs. 4–6 were fitted using the lm() function in R software. Significant differences in all the analysis were obtained using two-samples Wilcoxon test with wilcox.test() function in R software.29 In Figure S1 we have considered as outliers those sequences longer than 1000 codons since they represent only 0.13% of the data (24 stretches out of 18840 stretches in total).
Identification of tRNA genes
tRNA genes were predicted with tRNAscan-SE software.30,31 We used the version 1.3.1 with the options -B for bacterial genomes and -G for eukaryotic genomes. Pseudogenes and undetermined tRNAs from standard output were discarded from the analysis.
Discussion
I34 tRNAs modified by ADAT are essential for cell survival.12,16,18,21,32 They regulate the process of translation by increasing tRNA pairing ability to synonymous codons ended in C, U or A, and avoiding those ended in G. It is clear that inosine is important to balance codon usage and tRNA gene copy number in Eukaryotes, and that highly translated genes in these species tend to be enriched in ADAT-sensitive codons.19 Consistent with these observations, an initial analysis of the human genome23 showed: (1) that CDSs coding for proteins rich in ADAT amino acids are overrepresented in the human genome, (2) that ADAT stretches in the human proteome are specifically enriched in TAPS but not in LIVR, and (3) that the more enriched in ADAT amino acids an ORF is, the higher its tendency to use ADAT-sensitive codons instead of G-ended codons.
To explore the impact of I34 in evolution we extended our initial analysis to proteomes from a number of Eukarya and Bacteria. This analysis was designed to identify sections of proteins potentially more dependent on I34 for translation. Our approach was to screen the complete CDSs of different organisms and classify their genes on the basis of ADAT amino acid abundance. Using this initial curation, we identified those transcripts whose proportion of codons for ADAT amino acids is significantly enriched, and we used this subset of sequences to determine their relative enrichment in ADAT amino acids and ADAT-sensitive codons.
First, we found that the majority of eukaryotic and bacterial proteins rich in ADAT amino acids are specifically enriched in TAPS (Fig. 3). Physicochemical parameters probably explain why, among all ADAT amino acids, only TAPS can reach higher relative frequencies. Functional features of proteins rich in TAPS must have driven the selection of these extremely biased protein sequences.
Our data shows that in Eukarya ADAT stretches are longer and more frequent than in Bacteria (Figs. 2, 3 and Table 1). Eukaryotic genes coding for stretches of ADAT amino acids are generally biased towards ADAT-sensitive codons, with a positive correlation with respect to their length. Conversely, in Bacteria, the presence of stretches rich in ADAT amino acids is 4-fold lower, and the corresponding genes are generally depleted of ADAT-sensitive codons (Fig. 5 and Table 1). These results likely reflect the differences in I34 dependence between Eukaryotes and Bacteria.
Among Eukarya, only animals and plants have a positive correlation between lengths of stretches rich in ADAT amino acids with enrichment in ADAT-sensitive codons. Fungi and Protists show no such correlation. Moreover, Metazoa contain the longest ADAT stretches and the highest enrichment in ADAT-sensitive codons among Eukarya (Fig. 6). These results indicate a preference for ADAT-sensitive codons in longer stretches of ADAT amino acids in kingdoms with embryonic development where multicellularity is prevalent. In this regard, a connection between variation in codon usage and the establishment of multicellularity was already proposed by Ikemura.33
Surprisingly we have discovered that in the bacterial phyla of Firmicutes and Cyanobacteria genes for proteins rich in ADAT amino acids are also enriched in ADAT-sensitive codons (Fig. 7). This is initially surprising because, in bacteria, only tRNAArg (ACG) is believed to be deaminated by TadA, and arginine is not significantly enriched in the proteins that we have identified. This apparent contradiction may be resolved by our discovery that the genomes of several Firmicutes code for previously unnoticed tRNAs with A34 for amino acids Threonine, Proline, Serine, Leucine and Isoleucine (Table S2). These tRNAs could potentially be deaminated to I34. Thus, it is possible that the enrichment in ADAT-sensitive codons in these species is due, at least partially, to their use of additional I34-modified tRNAs. Cyanobacteria genomes do not contain this type of tRNAs, but their stretch length is shorter than in Firmicutes, and the frequency of ADAT-sensitive codons does not increase with stretch length (Fig. 7).
Our results suggest that a dependence on ADAT activity for the synthesis of TAPS-enriched proteins is an eukaryote-wide phenomenon that possibly extends to some bacterial species. It has previously been reported that some bacterial groups have secondarily lost A34-containing tRNAs and the ability to deaminate A34 to I34.34,35 Here, we show that other bacterial groups display the opposite behavior, and possibly have gained the ability to modify the A34 to I34 in tRNAs other than tRNAArg (AGC). We would like to offer the hypothesis that the enrichment in ADAT-sensitive codons is an adaptation of the translational apparatus to improve the synthesis of proteins extremely rich in threonine, alanine, proline, and serine. As this type of polypeptides is particularly prevalent in extracellular matrices it is possible that ADAT, I34, and ADAT-sensitive codons were important in the evolution of multicellularity.24
Supplementary Material
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This work was supported by the Spanish Ministry of Economy and Competitiveness under Grant [BES2013-064551] to AR-Y; and [BIO2015-64572] to LRdP. IR-T acknowledges financial support from an ERC Consolidator (ERC-2012-Co -616960) grant and a grant (BFU2014-57779-P) from Ministerio de Economía y Competitividad (MINECO), the latest with help from the Fondo Europeo de Desarrollo regional (FEDER).
References
- 1.Crick FH. On protein synthesis. Symp Soc Exp Biol. 1958;12:138–63. [PubMed] [Google Scholar]
- 2.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8(10):1189–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37(Database issue):D93–7. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grosjean H, de Crecy-Lagard V, Marck C. Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010;584(2):252–64. doi: 10.1016/j.febslet.2009.11.052. [DOI] [PubMed] [Google Scholar]
- 5.Crick FH. Codon–anticodon pairing: the wobble hypothesis. J Mol Biol. 1966;19(2):548–55. doi: 10.1016/S0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 6.Novoa EM, Pavon-Eternod M, Pan T, Ribas de Pouplana L. A role for tRNA modifications in genome structure and codon usage. Cell. 2012;149(1):202–13. doi: 10.1016/j.cell.2012.01.050. [DOI] [PubMed] [Google Scholar]
- 7.El Yacoubi B, Bailly M, de Crecy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 8.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24(17):1832–60. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Towns WL, Begley TJ. Transfer RNA methytransferases and their corresponding modifications in budding yeast and humans: activities, predications, and potential roles in human health. DNA Cell Biol. 2012;31(4):434–54. doi:10.1089%2Fdna.2011.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med. 2014;20(6):306–14. doi: 10.1016/j.molmed.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Jackman JE, Alfonzo JD. Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip Rev RNA. 2013;4(1):35–48. doi:10.1002%2Fwrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber AP, Keller W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Sci. 1999;286(5442):1146–9. doi: 10.1126/science.286.5442.1146. [DOI] [PubMed] [Google Scholar]
- 13.Torres AG, Pineyro D, Filonava L, Stracker TH, Batlle E, Ribas de Pouplana L. A-to-I editing on tRNAs: biochemical, biological and evolutionary implications. FEBS lett. 2014;588(23):4279–86. doi: 10.1016/j.febslet.2014.09.025. [DOI] [PubMed] [Google Scholar]
- 14.Auxilien S, Crain PF, Trewyn RW, Grosjean H. Mechanism, specificity and general properties of the yeast enzyme catalysing the formation of inosine 34 in the anticodon of transfer RNA. J Mol Biol. 1996;262(4):437–58. doi: 10.1006/jmbi.1996.0527. [DOI] [PubMed] [Google Scholar]
- 15.Zhou W, Karcher D, Bock R. Identification of enzymes for adenosine-to-inosine editing and discovery of cytidine-to-uridine editing in nucleus-encoded transfer RNAs of Arabidopsis. Plant Physiol. 2014;166(4):1985–97. doi: 10.1104/pp.114.250498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubio MA, Pastar I, Gaston KW, Ragone FL, Janzen CJ, Cross GA, Papavasiliou FN, Alfonzo JD.. An adenosine-to-inosine tRNA-editing enzyme that can perform C-to-U deamination of DNA. Proc Natl Acad Sci U S A. 2007;104(19):7821–6. doi: 10.1073/pnas.0702394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elias Y, Huang RH. Biochemical and structural studies of A-to-I editing by tRNA:A34 deaminases at the wobble position of transfer RNA. Biochemistry. 2005;44(36):12057–65. doi: 10.1021/bi050499f. [DOI] [PubMed] [Google Scholar]
- 18.Torres AG, Pineyro D, Rodriguez-Escriba M, Camacho N, Reina O, Saint-Leger A, Filonava L, Batlle E, Ribas de Pouplana L.. Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res. 2015;43(10):5145–57. doi: 10.1093/nar/gkv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novoa EM, Ribas de Pouplana L. Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet. 2012;28(11):574–81. doi: 10.1016/j.tig.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Saint-Leger A, Bello C, Dans PD, Torres AG, Novoa EM, Camacho N, Orozco M, A Kondrashov F, de Pouplana L.. Saturation of recognition elements blocks evolution of new tRNA identities. Sci Adv. 2016;2(4):e1501860. doi: 10.1126/sciadv.1501860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf J, Gerber AP, Keller W. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J. 2002;21(14):3841–51. doi:10.1093%2Femboj%2Fcdf362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyer LM, Zhang D, Rogozin IB, Aravind L. Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res. 2011;39(22):9473–97. doi:10.1093%2Fnar%2Fgkr691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rafels-Ybern A, Attolini CS, Ribas de Pouplana L. Distribution of ADAT-Dependent Codons in the Human Transcriptome. Int J Mol Sci. 2015;16(8):17303–14. doi: 10.3390/ijms160817303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribas de Pouplana L, Torres AG, Rafels-Ybern A. What Froze the Genetic Code? Life. 2017;7(2):14. doi: 10.3390/life7020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaub M, Keller W. RNA editing by adenosine deaminases generates RNA and protein diversity. Biochimie. 2002;84(8):791–803. doi: 10.1016/S0300-9084(02)01446-3. [DOI] [PubMed] [Google Scholar]
- 26.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–2. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 27.Hutter S, Vilella AJ, Rozas J. Genome-wide DNA polymorphism analyses using VariScan. BMC bioinformatics. 2006;7:409. doi: 10.1186/1471-2105-7-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald JH. Detecting non-neutral heterogeneity across a region of DNA sequence in the ratio of polymorphism to divergence. Mol Biol Evol. 1996;13(1):253–60. [DOI] [PubMed] [Google Scholar]
- 29.The R project for statistical computing. https://www.r-project.org/.
- 30.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29(22):2933–5. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fichant GA, Burks C. Identifying potential tRNA genes in genomic DNA sequences. J Mol Biol. 1991;220(3):659–71. doi: 10.1016/0022-2836(91)90108-I. [DOI] [PubMed] [Google Scholar]
- 32.Tsutsumi S, Sugiura R, Ma Y, Tokuoka H, Ohta K, Ohte R, Noma A, Suzuki T, Kuno T.. Wobble inosine tRNA modification is essential to cell cycle progression in G(1)/S and G(2)/M transitions in fission yeast. J Biol Chem. 2007;282(46):33459–65. doi: 10.1074/jbc.M706869200. [DOI] [PubMed] [Google Scholar]
- 33.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2(1):13–34. [DOI] [PubMed] [Google Scholar]
- 34.Yokobori S, Kitamura A, Grosjean H, Bessho Y. Life without tRNAArg-adenosine deaminase TadA: evolutionary consequences of decoding the four CGN codons as arginine in Mycoplasmas and other Mollicutes. Nucleic Acids Res. 2013;41(13):6531–43. doi:10.1093%2Fnar%2Fgkt356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Crecy-Lagard V, Marck C, Brochier-Armanet C, Grosjean H. Comparative RNomics and modomics in Mollicutes: prediction of gene function and evolutionary implications. IUBMB life. 2007;59(10):634–58. doi: 10.1080/15216540701604632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


