ABSTRACT
The core macroautophagy/autophagy machinery consists of a large group of autophagy-related (ATG) proteins, that mediate highly controlled, step-wise execution of this conserved intracellular degradation process. Whereas ATG proteins have been intensely studied in terms of protein interactions, post-translational modifications and transcriptional regulation, the mechanisms ensuring efficient translation of ATG proteins are not well understood. In a recent study, we describe an evolutionarily conserved role for EIF5A (eukaryotic translation initiation factor 5A) in autophagy. We demonstrate that EIF5A mediates Atg8-family protein lipidation and autophagosome formation via translation of the E2-like ATG3 protein. Moreover, we identify a particular motif in ATG3 causing EIF5A-dependency for its efficient translation.
KEYWORDS: ATG3, autophagy, EIF5A, lipidation, ribosome, translation
Key to the autophagy machinery are the Atg8-family proteins (in humans LC3s and GABARAPs), which associate with the growing phagophore through conjugation to the lipid phosphatidylethanolamine. The E1-like ATG7, E2-like ATG3 and E3-like ATG12-ATG5-ATG16L1 complex mediate lipidation of LC3/GABARAPs, thereby anchoring them to the membrane. This ensures proper autophagosome formation and additionally drives subsequent steps of the pathway. Prompted by recent literature assigning novel functions to RNA-binding proteins (RBPs) related to metabolic signaling pathways, we performed a high-throughput screen to identify RBPs affecting autophagy [1]. By siRNA-mediated targeting of 1530 RBPs, we systematically assessed the effects of RBP depletion on GFP-LC3B puncta in MCF-7 cells. From the screen, we identified the broadly conserved EIF5A, as a novel translational requirement for autophagy. We found that depletion of EIF5A reduced the number of autophagosomes, which coincided with decreased lipidation of LC3/GABARAP proteins. Importantly, this lipidation defect was consistent across several human and mouse cell lines. To address the potential evolutionary conservation of our findings, we turned to C. elegans where we depleted the EIF5A homolog, iff-2, and observed a significant reduction in the number of GFP::LGG1 puncta in the body-wall muscle and terminal pharyngeal bulb. Taken together, these phenotypic observations across cell lines and species suggest a functionally conserved role for EIF5A in autophagy.
EIF5A has previously been shown to bind to ribosomes and prevent them from stalling during the translation of certain geometrically challenging amino acid stretches. An interesting feature of EIF5A is that it contains the unique amino acid hypusine, formed through the post-translational modification of a specific lysine residue (K50 in humans). Because hypusine is critical for EIF5A-mediated translation, we assessed its importance in relation to autophagy by creating doxycycline-inducible cell lines expressing either wild type- or a non-hypusinated K50A mutant of EIF5A. Whereas the wild type EIF5A clearly increased GFP-LC3B puncta and rescued the autophagy-related defects caused by EIF5A depletion, the K50A mutant was unable to do so. Similarly, treatment with an inhibitor of hypusination, N1-guanyl-1,7-diaminoheptane (GC7), blocked EIF5A-induced autophagy. Collectively, these data indicate a functional requirement for EIF5A hypusination in autophagy and suggest a mechanism ascribed to translation.
EIF5A function is well-described in yeast and bacteria, yet remarkably little is known concerning its translational targets in human cells. We did not observe an effect on global protein synthesis after depletion of EIF5A, suggesting that it instead affects the translation of a subset of proteins. To pinpoint these translational targets, we performed liquid chromatography-mass spectrometry (LC-MS) analysis of newly synthesized proteins using a pulse-labeling method based on incorporation of the methionine analog L-azidohomoalanine. Our analysis revealed that EIF5A affects the translation of a subset of proteins with diverse biological roles. Filtering this list for autophagy-related function yielded a small group of 9 autophagy-relevant targets of EIF5A. Among these was ATG3, which caught our attention due to its well-known E2-like enzymatic function, required for lipidation of LC3/GABARAPs. We confirmed that both total and nascent ATG3 protein levels were reduced upon knockdown of EIF5A, corroborating the findings from our MS analysis. Importantly, we found that ATG3 overexpression could rescue the lipidation phenotype caused by loss of EIF5A.
Intrigued by a recent identification of 29 motifs displaying EIF5A-hyperdependency in yeast (Schuller et al. 2017), we searched the sequence of human ATG3 and discovered 2 such motifs, namely DDG at position 104 and PPP(P) at position 254. To address their importance, we employed a reporter system using wild type or mutated versions of ATG3 fused to mCherry. By monitoring the translational sensitivity of these constructs towards EIF5A depletion, we found that mutation of the 4 consecutive prolines had little or no effect, whereas mutation of the DDG motif of ATG3 abolished its sensitivity towards EIF5A. In light of previous literature in yeast, alluding to the importance of EIF5A in translating proline-rich motifs, the lack of effect of the proline motif in ATG3 is perhaps unexpected, and emphasizes the need to obtain further insight towards EIF5A function in mammalian cells.
Previous studies have mainly addressed EIF5A function in steady-state conditions, and little is known about stress-dependent dynamics of EIF5A. We therefore questioned whether EIF5A itself responds to autophagy induction. Whereas we did not observe changes in the levels of EIF5A upon autophagy stimulation, we found that its association with the ribosome was clearly enhanced after MTORC1 inhibition. While the cause of this strengthened interaction remains elusive, it could be due to an enhanced recruitment of EIF5A to translating ribosomes under certain conditions. Alternatively, this finding could reflect an increased stabilization of EIF5A at the ribosome, required to ensure the translation of a shifted mRNA repertoire after MTORC1 inhibition. Paradoxically, MTORC1 inhibition causes a global translational shutdown, perhaps eluding to a stress-responsive role of EIF5A in ensuring efficient translation of an mRNA subset under these conditions.
Collectively, our work sheds light on novel aspects of EIF5A function and identifies, to our knowledge, the first mechanistic requirement for a mammalian protein dependent on EIF5A for its efficient synthesis. A model summarizing our data is shown in Figure 1. From a broader perspective, the data emerging from our study offers valuable resources to the field, both by providing a comprehensive overview of functional links between RBPs and autophagy, and by elucidating a cohort of novel EIF5A targets in human cells. Most importantly, our study reveals a unique translational requirement for autophagy, and emphasizes the importance of translation per se as a regulatory layer of autophagy.
Figure 1.
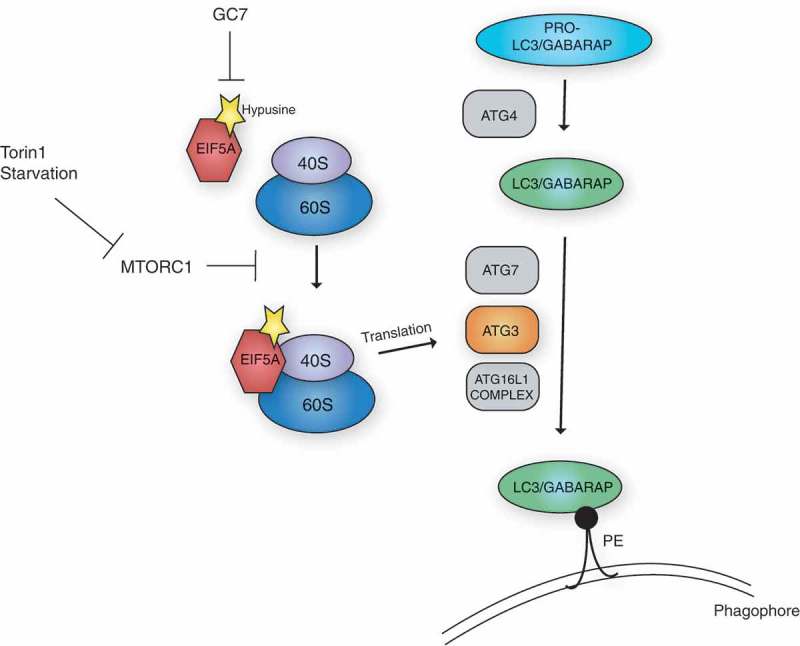
EIF5A is required for autophagy via the translation of ATG3. The LC3/GABARAP proteins are processed, first by ATG4, and subsequently by the lipidation machinery, including the E1-like ATG7, the E2-like ATG3 and the E3-like ATG16L1 complex. This results in their lipidation and anchoring to the phagophore membrane via phosphatidylethanolamine (PE), which is required for autophagy. EIF5A, via a hypusine-dependent mechanism, can associate with the ribosome and assist in the translation of ATG3. Stimulation of autophagy by MTORC1 inhibition (via starvation or Torin1 treatment), can enhance EIF5A association with the ribosome. Inhibition of hypusination by treatment with N1-guanyl-1,7-diaminoheptane (GC7), can effectively inhibit this process.
Disclosure statement
No potential conflict of interest was reported by the author.
References
- [1].Lubas M, Harder LM, Kumsta C, et al. eIF5A is required for autophagy by mediating ATG3 translation. EMBO Rep. 2018;19:e46072. [DOI] [PMC free article] [PubMed] [Google Scholar]


