ABSTRACT
Translation begins at AUG, GUG, or UUG codons in bacteria. Start codon recognition occurs in the P site, which may help explain this first-position degeneracy. However, the molecular basis of start codon specificity remains unclear. In this study, we measured the codon dependence of 30S•mRNA•tRNAfMet and 30S•mRNA•tRNAMet complex formation. We found that complex stability varies over a large range with initiator tRNAfMet, following the same trend as reported previously for initiation rate in vivo (AUG > GUG, UUG > CUG, AUC, AUA > ACG). With elongator tRNAMet, the codon dependence of binding differs qualitatively, with virtually no discrimination between GUG and CUG. A unique feature of initiator tRNAfMet is a series of three G-C basepairs in the anticodon stem, which are known to be important for efficient initiation in vivo. A mutation targeting the central of these G-C basepairs causes the mRNA binding specificity pattern to change in a way reminiscent of elongator tRNAMet. Unexpectedly, for certain complexes containing fMet-tRNAfMet, we observed mispositioning of mRNA, such that codon 2 is no longer programmed in the A site. This mRNA mispositioning is exacerbated by the anticodon stem mutation and suppressed by IF2. These findings suggest that both IF2 and the unique anticodon stem of fMet-tRNAfMet help constrain mRNA positioning to set the correct reading frame during initiation.
KEYWORDS: IF3, IF1, initiation, ribosome, start codon selection, P site
Introduction
Initiation of translation entails the assembly of a ribosome complex at the start codon of mRNA. In bacteria, this process is facilitated by three initiation factors (IF1, IF2, and IF3) and occurs in two major steps (reviewed in1,2). The first step is formation of the 30S initiation complex (IC), which contains all three factors and initiator tRNA [N-formyl-methionyl-tRNAfMet (fMet-tRNAfMet)] bound to the peptidyl (P) site and paired to the start codon of mRNA. There is no obligate order of ligand binding during 30S IC formation, although the factors bind before fMet-tRNAfMet in the predominant pathway.3 The second step is docking of the 50S subunit and concomitant release of the initiation factors, resulting in the 70S IC, which is competent to enter the elongation phase of translation. The factors kinetically control the process to increase speed and accuracy.4-11 The largest factor, IF2, is a GTPase that binds the 30S shoulder and stimulates both steps of the process. IF2 also plays a key role in tRNA selection, by forming specific contacts with the acceptor end of fMet-tRNAfMet. IF3 binds the 30S platform and negatively regulates both steps of initiation to ensure accurate start codon selection. IF1 binds the 30S aminoacyl (A) site and enhances the activities of the other two factors.
In all cells, a dedicated tRNA—initiator tRNA—is responsible for decoding the start codon during translation initiation. There are several unique features of initiator tRNA that ensure this specific role (reviewed in12). In bacteria, the initiator tRNAfMet is charged with methionine and then formylated, yielding fMet-tRNAfMet. Critical for formylation are conserved identity elements in the acceptor end of tRNAfMet, particularly a mismatch between nucleotide (nt) 1 and nt 72 (C1×A72 in E. coli) (Fig. 1). The formyl group is a positive determinant for IF2 binding and a negative determinant for EF-Tu binding,11,13 shunting fMet-tRNAfMet to the initiation pathway. Another unique feature of initiator tRNAfMet is a series of three G-C basepairs in the anticodon stem (Fig. 1). These basepairs are important for efficient initiation in the cell,14 presumably by contributing to P-site binding.
Figure 1.
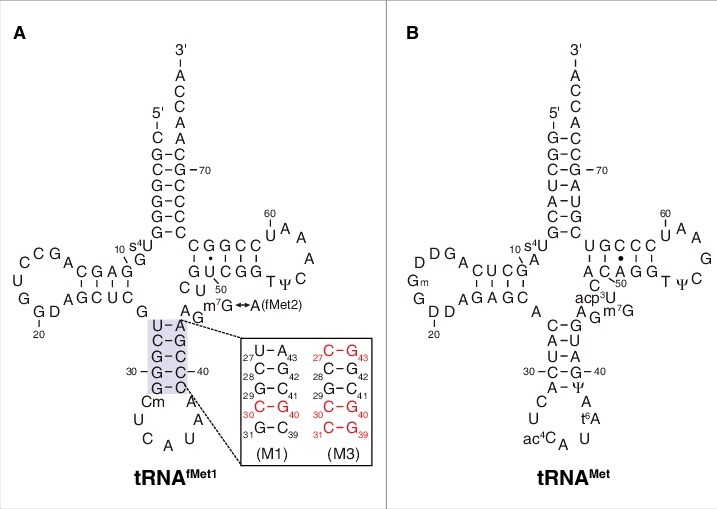
Transfer RNA molecules used in this study. Secondary structures of E. coli tRNAfMet1 (A) and tRNAMet (B) are shown, with mutations M1 and M3 of the anticodon stem of tRNAfMet indicated. Isoacceptor tRNAfMet2 is identical to tRNAfMet1 except that nucleotide 46 is A rather than m7G. D, dihydrouridine; Ψ, pseudouridine; s4U, 4-thiouridine; Cm, 2′-O-methylcytidine; Gm, 2′-O-methylguanosine; m7G, 7-methylguanosine; ac4C, N4-acetylcytidine; acp3U, 3-(3-amino-3-carboxypropyl)uridine; t6A, N6-threonylcarbamoyladenosine.
Natural start codons in bacteria include AUG, GUG, and UUG. This first-position degeneracy presumably stems from the fact that the start codon is decoded in the ribosomal P site, whereas all other codons are decoded in the A site. The 30S P site is formed by a number of 16S rRNA nucleotides, which occupy similar relative positions in the absence and presence of tRNA.15-17 Nucleotides m2G966 and C1400 interact with nt 34 of tRNA, the latter stacking “beneath” codon-anticodon basepair 3:34. A790 interacts with the backbone of P-site tRNA (P-tRNA) at nt 38, and G926 contacts the phosphate of nt 1 of the P-site codon (P codon). A1339 and G1338 dock into the minor groove of the anticodon stem of P-tRNA, forming Type I and II interactions, respectively. Recent structural studies suggest that, during initiation, A1339 and G1338 interact initially with G29-C41 and G42 of fMet-tRNAfMet, and then “slide down” by one basepair to interact with G30-C40 and C41 at later stages of the process.18 The importance of G29-C41 and G30-C40 in initiation is well established, as basepair substitutions at these positions reduce translation in the cell by 7- and 20-fold, respectively.14
The molecular basis of start codon recognition remains unclear. In this work, we measure the stabilities of 30S•mRNA•tRNAfMet complexes with various cognate and near-cognate start codons. We find that the codon dependence of tRNAfMet binding mirrors that of translation initiation in vivo and differs from that of elongator tRNAMet. For certain complexes containing fMet-tRNAfMet, we observe mispositioning of mRNA, such that codon 2 is no longer programmed in the A site. This mispositioning is exacerbated by mutations in the anticodon stem and suppressed by IF2. These findings suggest that both IF2 and the unique anticodon stem of tRNAfMet help set the correct reading frame during initiation.
Results
Codon dependence of initiator tRNAfMet binding to the 30S P site
Using toeprinting, we measured the effect of the start codon sequence on the overall equilibrium association constant for 30S•mRNA•tRNAfMet1 ternary complex formation (K TC) in the absence of initiation factors, as described previously.19 Complex stability varied over a wide range (>3 orders of magnitude), following the trend AUG > GUG, UUG > CUG, AUC, AUA > ACG (Fig. 2A, Table 1). Previously, Simons and coworkers measured rates of translation initiation from these codons in E. coli cells and observed the same trend.20 This correlation suggests that fMet-tRNAfMet is fundamentally responsible for the codon dependence of initiation observed in vivo.
Figure 2.
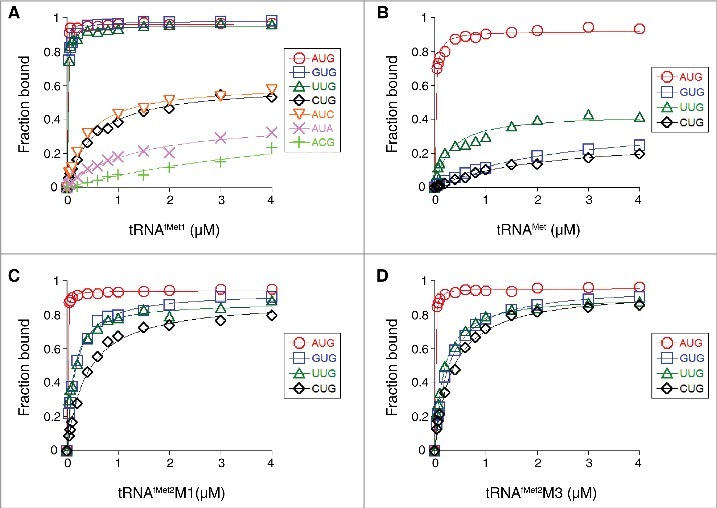
Effects of the start codon and tRNA sequence on the thermodynamic stability of the 30S•mRNA•tRNA complex. 30S subunits (1 µM) were incubated with mRNA (0.01 µM; with indicated start codon and preannealed radiolabeled primer) and various concentrations of tRNAfMet1 (A), tRNAMet (B), tRNAfMet2M1 (C), or tRNAfMet2M3 (D) for 2 h at 37°C, and then complexes were analyzed by toeprinting. Fraction of bound mRNA (F) was quantified as [toeprint signal / (toeprint + run-off signal)] and plotted versus input tRNA concentration. Data were fit to the equation F = Fmax[bc/(bc+1/K TC)], where b is the input tRNA concentration, c is the input 30S concentration, K TC is the equilibrium association constant, and Fmax is the maximal level of detected complex. For the ACG case of panel A, the Fmax parameter was set arbitrarily at 1.0 prior to fitting the curve shown. All K TC and Fmax values are listed in Table 1.
Table 1.
Binding parameters for various 30S complexes.
| tRNA | Start codon | KTC | Fmax | koff |
|---|---|---|---|---|
| tRNAfMet1 | AUG | 390 ± 60 | 0.96 ± 0.01 | 0.0010 ± 0.0002 |
| GUG | 67 ± 2 | 0.98 ± 0.01 | 0.014 ± 0.002 | |
| UUG | 80 ± 7 | 0.95 ± 0.01 | 0.032 ± 0.003 | |
| CUG | 1.9 ± 0.2 | 0.61 ± 0.02 | 0.074 ± 0.006 | |
| AUC | 2.6 ± 0.2 | 0.61 ± 0.01 | ND | |
| AUA | 0.77 ± 0.13 | 0.41 ± 0.03 | ND | |
| ACG | ∼ 0.1 | set | ND | |
| tRNAMet | AUG | 55 ± 5 | 0.92 ± 0.01 | 0.014 ± 0.003 |
| GUG | 0.45 ± 0.04 | 0.39 ± 0.02 | 0.13 ± 0.03 | |
| UUG | 3.4 ± 0.7 | 0.43 ± 0.03 | 0.11 ± 0.02 | |
| CUG | 0.49 ± 0.05 | 0.30 ± 0.02 | 0.093 ± 0.018 | |
| tRNAfMet2M1 | AUG | 230 ± 20 | 0.94 ± 0.01 | 0.005 ± 0.001 |
| GUG | 6.4 ± 0.3 | 0.93 ± 0.01 | 0.025 ± 0.003 | |
| UUG | 8.5 ± 0.6 | 0.87 ± 0.02 | 0.061 ± 0.007 | |
| CUG | 2.7 ± 0.2 | 0.89 ± 0.02 | 0.040 ± 0.003 | |
| tRNAfMet2M3 | AUG | 150 ± 10 | 0.95 ± 0.01 | 0.004 ± 0.002 |
| GUG | 3.9 ± 0.1 | 0.97 ± 0.01 | 0.026 ± 0.002 | |
| UUG | 5.9 ± 0.2 | 0.91 ± 0.01 | 0.038 ± 0.004 | |
| CUG | 2.9 ± 0.1 | 0.95 ± 0.01 | 0.025 ± 0.003 |
Reported values and their standard errors derive from the curve fits shown in Figs. 2 and 3 . ND, not determined.
Overall equilibrium association constant in units of µM−2.
Maximal fraction of toeprint signal, defined by the horizontal asymptote of the tRNA titration curve (Fig. 2). Fmax presumably reflects the probability that the complex resists disruption by reverse transcriptase.
Messenger RNA dissociation rate in units of min−1.
In this case, the best-fitting curve returned an unreasonable Fmax value of 2.6. Arbitrarily setting the Fmax parameter to 1.0 and 0.5 yielded K TC values of 0.07 and 0.17 µM−2, respectively.
In the NUG cases, we also assessed complex stability by measuring the mRNA dissociation rate (k off) (Fig. 3A, Table 1). In these experiments, the ternary complex (carrying radiolabeled primer annealed to the 3′ end of the mRNA) was formed, an excess of competitor mRNA lacking the primer-binding site was added at time t = 0, and portions of the reaction were removed at various times and subjected to primer extension analysis. The toeprint signal decreased as a function of time, and the data were fit to a single-exponential function to obtain k off. Values for k off varied over a 70-fold range, with mRNA binding tightest in the presence of AUG (k off = 0.001 min−1), intermediate in the presence of GUG (k off = 0.014 min−1) and UUG (k off = 0.032 min−1), and loosest in the presence of CUG (k off = 0.074 min−1). These data are in line with the equilibrium binding data described above.
Figure 3.
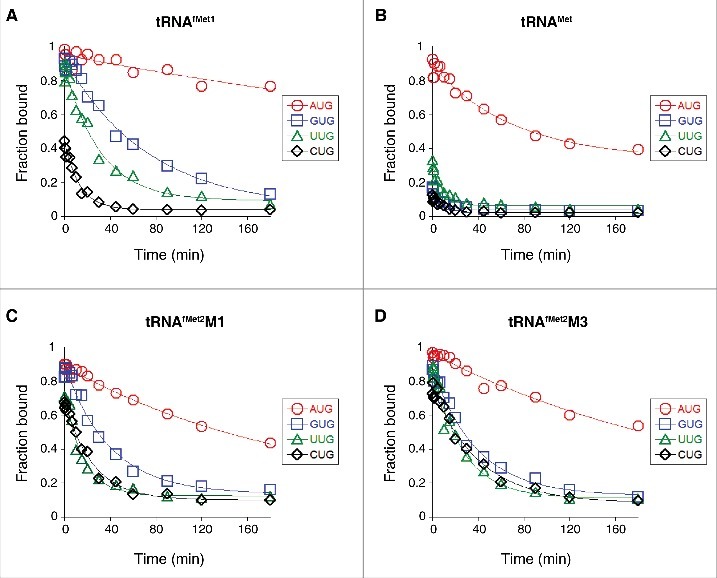
Effects of the start codon and tRNA sequence on the kinetic stability of the 30S•mRNA•tRNA complex. Complexes were pre-formed by incubating 30S subunits (1 µM), mRNA (0.05 µM; with indicated start codon and preannealed radiolabeled primer), and 1.5 µM tRNAfMet1 (A), tRNAMet (B), tRNAfMet2M1 (C), or tRNAfMet2M3 (D) for 2 h at 37°C. At time t = 0, an excess of chase mRNA (containing start codon AUG and lacking the primer binding site) was added, and aliquots were removed at various time points and subjected to primer extension analysis. Fraction of toeprint signal (F) was quantified and plotted as a function of time. Data were fit to a single exponential function to obtain the mRNA dissociation rate, k off, for each complex (listed in Table 1).
Codon dependence of elongator tRNAMet binding to the 30S P site
The anticodon loops of tRNAfMet and tRNAMet have identical nucleotide sequences and differ only in their posttranscriptional modifications (Fig. 1). Elongator tRNAMet harbors N4-acetylcytidine (ac4C) and N6-threonylcarbamoyladenosine (t6A) at positions 34 and 37, respectively, modified nucleotides which prevent misreading and frameshifting during translation.21-23 We measured the codon dependence of 30S•mRNA•tRNAMet formation, as described above for tRNAfMet1. Complexes containing tRNAMet were generally less stable than those containing tRNAfMet1, as expected from earlier studies.24 30S•mRNA•tRNAMet was most stable with AUG (K TC = 55 µM−2), less stable with UUG (K TC = 3.4 µM−2), and even less stable with GUG or CUG (K TC ≈ 0.5 µM−2) (Fig. 2B, Table 1). Messenger RNA dissociation rate (k off) measurements gave congruent results (Fig. 3B, Table 1). These data show that the codon dependence of tRNAMet and tRNAfMet1 binding differs qualitatively. Favorable pairing to GUG appears to be a unique property of tRNAfMet, as tRNAMet fails to distinguish GUG from CUG.
Effects of mutations in the anticodon stem of tRNAfMet on codon discrimination
A unique feature of initiator tRNA is a series of three G-C pairs in the anticodon stem, which are important for efficient initiation.14 To investigate the role of these G-C pairs in P codon recognition, we generated and purified variants of tRNAfMet2 with either one (M1) or three (M3) basepair changes in the stem (Fig. 1). Isoacceptor fMet2 is nearly identical to fMet1 (differing by one nucleotide) and was chosen for the practical reason that it can be overexpressed in E. coli B cells and purified using a one-step electrophoretic method.25,26 The three-basepair change of M3 was originally designed for another study24 and converts the anticodon stem sequence to that of tRNAGlu. The basepair substitution G30C / C40G, common to both tRNAfMet2 variants, was shown previously to reduce translation by 20-fold.14 When tested in 30S•mRNA•tRNA formation / stability assays, the two mutant tRNAs behaved similarly to one another and differently than wild-type tRNAfMet1 (Fig. 2C, D, Fig. 3C, D, Table 1). Complex stability varied over a narrower range, suggesting reduced codon specificity. Discrimination against CUG was diminished, as reflected by larger K TC (and Fmax) values, and smaller k off values. Moreover, recognition of the alternative cognate start codons UUG and GUG was compromised, based on the ∼10-fold smaller K TC values and ∼2-fold larger k off values. Indeed, the codon dependence of the mutant tRNAfMet2 resembled that of tRNAMet, in that little distinction between UUG, GUG, and CUG was seen.
Effects of the anticodon stem sequence on positioning of mRNA in the ribosome
The toeprinting technique can accurately map the position of mRNA on the 30S subunit, with the first nucleotide of the P codon (+1) fifteen nucleotides upstream of the toeprint position (+16) (Fig. 4A). In the course of these experiments, we noticed an altered toeprint pattern in the presence of CUG, which was strongly exacerbated by the anticodon stem mutations (Fig. 4). In 30S complexes containing tRNAfMet1 paired to AUG, UUG, or GUG, toeprint bands at +16 and +17 were observed, the former being ∼2-fold stronger. In the presence of CUG, a shift in the toeprint pattern was observed, which was more evident with tRNAfMet2 than tRNAfMet1. Higher signal was seen at downstream positions (+17, +18) concomitant with reduced signal at +16. With the mutant forms of tRNAfMet2, a further shift in the toeprint pattern was seen, with loss of signal at +16 and highest signal at +18 (Fig. 4B, C). The most straightforward interpretation of these data is that the mRNA at the entrance channel of the subunit is being drawn inward toward the P site, by 1–2 nt.
Figure 4.
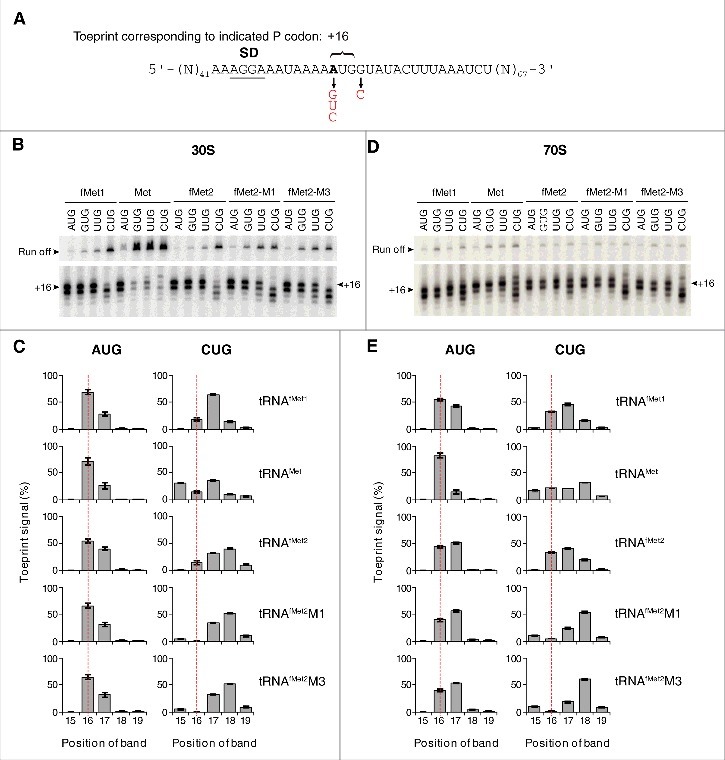
Positioning of mRNA in various ribosomal complexes. (A) Model mRNAs used in this study. The Shine-Dalgarno element (SD) is underscored and position +1 of the start codon is highlighted in bold text. Various base substitutions made at positions +1 and +4 are shown. 30S (B-C) or 70S (D-E) complexes containing P-site tRNA (as indicated) paired to start codon (as indicated) were analyzed by toeprinting. Complexes were formed in the absence of factors by incubating ribosomes or subunits (1 µM), mRNA (0.01 µM, with preannealed radiolabeled primer), and tRNA (1.5 µM) for 2 h at 37°C, prior to primer extension analysis. Top gel panels show the relative intensities of the full-length cDNA products (Run off). Bottom gel panels show the toeprint bands, with +16 indicated. Histograms show the distribution of toeprint signal versus toeprint position for each 30S (C) and 70S (E) complex. Data were quantified for each complex as (specific toeprint / all toeprints) ×100% and correspond to the mean ± SEM from ≥ 3 independent experiments. The dotted red line benchmarks position +16.
For 30S complexes containing tRNAMet paired to AUG, UUG, or GUG, the predominant toeprint observed was +16, as expected (Fig. 4B, C). In the presence of CUG, a broader distribution of signal was seen, although no obvious directional shift in the overall signal (upstream or downstream) was observed.
We next analyzed analogous 70S complexes by toeprinting and obtained highly similar results (Fig. 4D, E). Again, complexes containing tRNAfMet2M1 or tRNAfMet2M3 paired to CUG gave the shifted toeprint pattern, with the strongest band at +18 and additional bands at +15, +17, and +19. These data indicate that the interactions responsible for the anomalous toeprint pattern occur in the context of the 30S subunit or 70S ribosome.
The 3′ shift of toeprint pattern raised the possibility that the mutant tRNAfMet2 interacts in some way with nucleotide following CUG, which is G (+4) in these model mRNAs. Hence we changed this nucleotide to C and repeated the toeprinting analysis. This substitution largely eliminated the 3′ shift, substantially reducing the signal intensity at +18 in both 30S and 70S contexts (Fig. 5A, B). These data are consistent with an unconventional interaction between mutant tRNAfMet2 and nt +4 of mRNA, which is more favorable in the G (+4) case.
Figure 5.
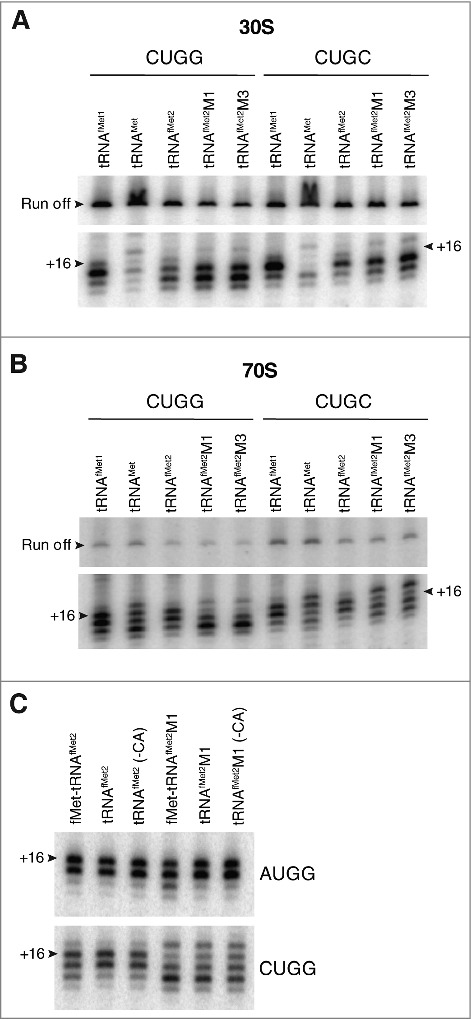
Contribution of nucleotide +4 of mRNA to anomalous toeprint patterns. Toeprints of 30S (A) or 70S (B) complexes containing P-site tRNA (as indicated) paired to mRNA with CUGG or CUGC (as indicated). (C) Comparison of toeprints of 70S complexes carrying acylated, deacylated, or 3′ truncated tRNAfMet2 (as indicated) bound to the P site and paired with AUGG or CUGG (as indicated). Experimental conditions as in Fig. 4, except that incubation time for complex formation was 30 min for the experiments of panel C.
In the 70S ribosome, deacylated tRNA spontaneously fluctuates between the P/P and P/E sites.27,28 To test whether P/E occupancy had any bearing on mRNA positioning, we made charged (N-formyl-methionyl) and 3′-truncated (lacking CCA-3′ or CA-3′) tRNAs, which are unable to bind the P/E site,29,30 and repeated the experiment. Similar toeprint patterns were seen for all forms of the tRNA (Fig. 5C), suggesting that the interactions responsible for the aberrant +18 toeprint can take place with P/P-bound tRNAfMet.
It is possible that the anomalous +18 toeprint corresponds to a frameshifted complex—e.g., with tRNAfMet2M1 paired to GGU in the P site (+2 frameshift). An alternative possibility is that codon 2 lies in the A site but adopts a distorted conformation in the presence of P-site tRNAfMet2M1, resulting in the shortened (+18) toeprint. We reasoned that, in either case, the A site would not be functionally programmed with codon 2 (GUA). To test this, we non-enzymatically formed 70S initiation complexes (70S ICs) with f[35S]Met-tRNAfMet2 in the P site, and then measured dipeptide formation upon addition of EF-Tu•GTP•Val-tRNA (Fig. 6, Table 2). In the presence of AUG, fMet-Val formation was rapid, regardless of the P-site tRNA (Fig. 6B). In the presence of CUG and wild-type fMet-tRNAfMet2, fMet-Val formation was rapid but the reaction amplitude was reduced by >2-fold. In the presence of CUG and fMet-tRNAfMet2M1, dipeptide formation was barely detected and its apparent rate was small (Fig. 6B). Thus, as predicted from the toeprinting data, this complex does not have codon 2 (GUA) functionally positioned in the A site.
Figure 6.
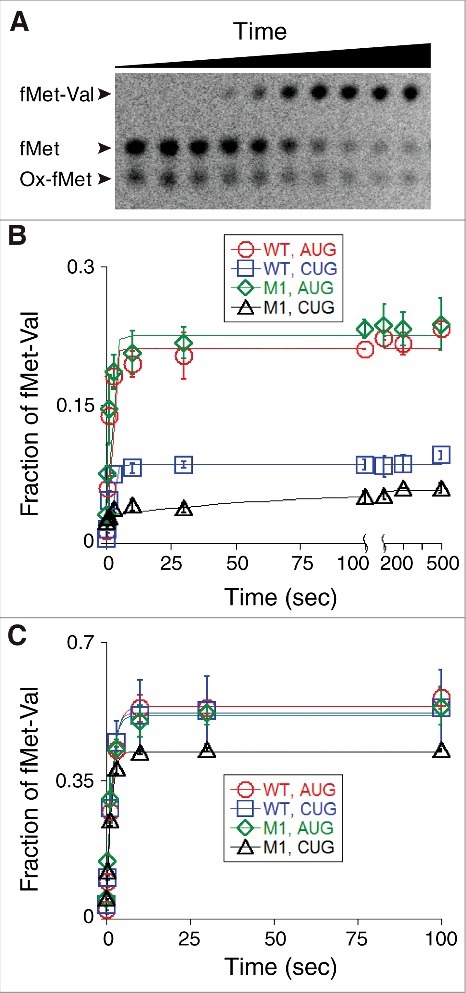
Functional assessment of various 70S ICs with respect to decoding of codon 2. (A) Example of an experiment measuring dipeptide formation. A 70S IC containing f[35S]Met-tRNAfMet2 paired to AUG in the P site and codon GUA in the A site was rapidly mixed with EF-Tu•GTP•Val-tRNA at time t = 0, and samples quenched at various time points were analyzed by electrophoretic TLC. Ox-fMet, oxidized fMet. (B) 70S complexes containing P-site fMet-tRNAfMet2 (WT) or fMet-tRNAfMet2M1 (M1) paired to AUG or CUG mRNA (as indicated) were formed non-enzymatically. Each was rapidly mixed with EF-Tu•GTP•Val-tRNA, and the rate of fMet-Val formation was quantified as a function of time. Data points represent mean ± range values (n = 2), which were used to fit to a signal exponential equation, generating the curves shown. (C) Experimental setup as in panel B except that the 70S ICs were formed enzymatically, in the presence of initiation factors and GTP.
Table 2.
Apparent rates and amplitudes of fMet-Val formation in various 70S complexes.
| Factors | P-site tRNA | Start codon | kapp, s−1 | A |
|---|---|---|---|---|
| Absent | fMet-tRNAfMet2 | AUG | 0.99 ± 0.21 | 0.21 ± 0.02 |
| fMet-tRNAfMet2 | CUG | 0.74 ± 0.10 | 0.087 ± 0.004 | |
| fMet-tRNAfMet2M1 | AUG | 0.78 ± 0.17 | 0.20 ± 0.02 | |
| fMet-tRNAfMet2M1 | CUG | 0.014 ± 0.008 | 0.028 ± 0.004 | |
| Present | fMet-tRNAfMet2 | AUG | 0.61 ± 0.07 | 0.54 ± 0.02 |
| fMet-tRNAfMet2 | CUG | 0.71 ± 0.05 | 0.52 ± 0.01 | |
| fMet-tRNAfMet2M1 | AUG | 0.74 ± 0.12 | 0.47 ± 0.03 | |
| fMet-tRNAfMet2M1 | CUG | 0.80 ± 0.05 | 0.40 ± 0.01 |
Reported values and their standard errors derive from the curve fits shown in Fig. 6.
We repeated the decoding experiments using initiation factors (IFs) to increase the efficiency of 70S IC formation prior to EF-Tu•GTP•Val-tRNA addition (Fig. 6C, Table 2). Remarkably, the IFs largely rescued the functional defects caused by the start codon substitution and anticodon stem mutation. Dipeptide formation was rapid and efficient in all cases, although the amplitude in the fMet-tRNAfMet2M1-CUG case was ∼20% lower than the others.
Suppression of mRNA mispositioning by initiation factors
To directly test the impact of IFs on mRNA positioning, we used fMet-tRNAfMet2M1 to form 30S complexes in the presence of IF1, IF2•GTP, and IF3 (alone and in combination), and analyzed the complexes by toeprinting (Fig. 7A, B). On its own, the fMet group had no bearing on mRNA positioning, as 30S complexes containing fMet-tRNAfMet2M1 and CUGG exhibited the same altered toeprint pattern (with strong +18 band) as described above. However, in the presence of all three factors, or IF2 and IF1, one predominant toeprint at +16 was observed. Thus, these factors can suppress the effects of the anticodon stem / start codon substitutions and restore normal mRNA positioning. In the presence of IF2 alone, or IF2 and IF3, normal mRNA positioning is largely restored, whereas reactions lacking IF2 show no such activity. Hence we conclude that IF2 is primarily responsible for this activity and aided by IF1. When the experiment was repeated with deacylated tRNA, the ability of IF2 to influence mRNA positioning was lost (Fig. 7C), indicating that interaction between IF2 and the acceptor end of fMet-tRNAfMet2M1 is required for this activity. IF3 had no appreciable effect on the toeprint pattern but uniformly reduced the toeprint intensities (Fig. 7A, C), consistent with earlier evidence that the factor destabilizes near-cognate complexes.5,11,19
Figure 7.
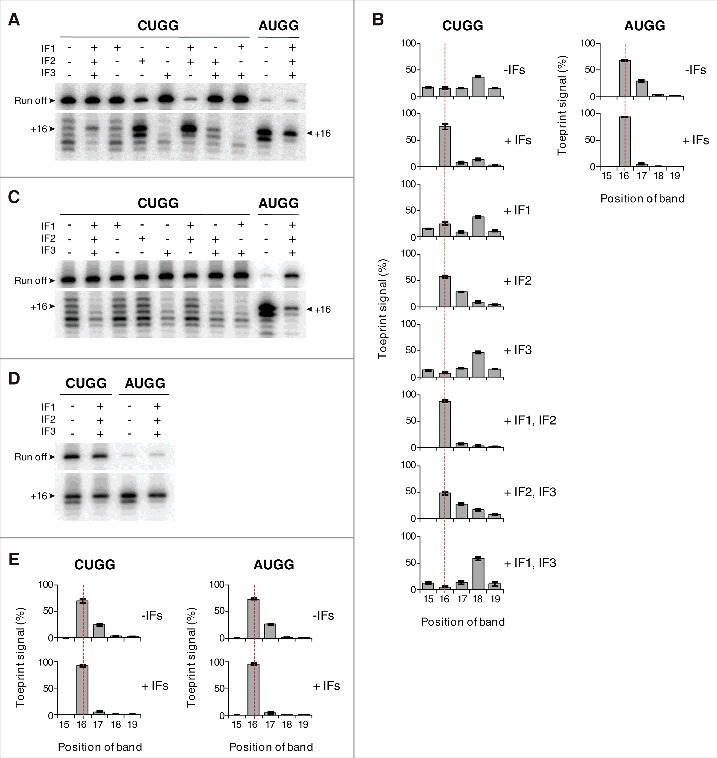
Initiation factors rectify mRNA positioning in various 30S complexes. 30S subunits (2 µM, CUGG; 1 µM, AUGG) were incubated with fMet-tRNAfMet2M1 (A-B, 2 µM), tRNAfMet2M1 (C, 2 µM), or fMet-tRNAfMet2 (D-E, 2 µM) in the presence of mRNA (CUGG, 1 µM; AUGG, 0.1 µM), GTP (100 µM), and in the absence or presence of initiation factors (3 µM each, as indicated) at 37°C for 5 min, and complexes were analyzed by toeprinting. Histograms show the distribution of toeprint signal versus toeprint position for complexes containing mutant (B) or control (E) tRNA. Data represent the mean ± SEM from ≥ 3 independent experiments. The dotted red line benchmarks position +16.
Effects of the IFs were also seen in 30S complexes containing fMet-tRNAfMet2M1 paired to AUGG (Fig. 7A, B) and in complexes with wild-type fMet-tRNAfMet2 paired to AUGG or CUGG (Fig. 7D, E). In the absence of factors, these complexes gave toeprint bands at both +16 and +17. With IFs present, the +17 band was specifically reduced, yielding one predominant toeprint at +16 (Fig. 7). These data suggest that the ability of IFs to constrain mRNA positioning in the 30S IC also applies to cognate and/or wild-type contexts.
We also compared the effects of IFs on mRNA positioning in 70S complexes (Fig. 8). 70S•CUGG•fMet-tRNAfMet2M1 formed in the absence of factors showed a 3′-shifted toeprint pattern, reminiscent of that seen for the analogous 30S complex under identical conditions. In the presence of all IFs, or IF2 and IF3, normal mRNA positioning is largely restored (as indicated by +16/17 toeprints) (Fig. 8), consistent with the factor dependence of fMet-Val formation in analogous complexes (Fig. 6). Notably, little-to-no enhancement of mRNA positioning is seen in the absence of either IF2 or IF3 (Fig. 8). It is well known that IF3 promotes dissociation of the ribosomal subunits.31,32 By shifting the 30S + 50S ⇌ 70S equilibrium leftward, IF3 may give IF2 access to the subunit interface and fMet-tRNAfMet2M1. This would explain why both IF3 and IF2 are needed in the context of the 70S ribosome to facilitate mRNA repositioning.
Figure 8.
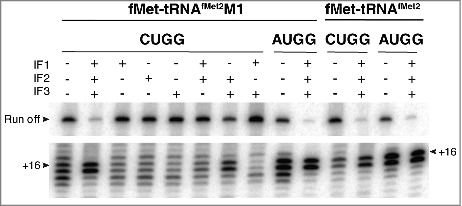
Initiation factors rectify mRNA positioning in 70S complexes. Ribosomes (2 µM, CUGG; 1 µM, AUGG) were incubated with fMet-tRNAfMet2M1 or fMet-tRNAfMet2 (2 µM, as indicated) in the presence of mRNA (CUGG, 1 µM; AUGG, 0.1 µM), GTP (100 µM), and in the absence or presence of initiation factors (3 µM each, as indicated) at 37°C for 5 min, and complexes were analyzed by toeprinting.
Discussion
Here, we provide evidence that both IF2 and the unique anticodon stem of fMet-tRNAfMet play a role in the positioning of mRNA during initiation. Ribosome complexes (30S or 70S) containing tRNAfMet2M1 and CUGG exhibit an irregular toeprint pattern, with the prominent bands shifted downstream (to +18). We suspect that this stems from some type of noncanonical interaction between tRNAfMet2M1 and mRNA, because the +18 toeprint depends on G(+4) of mRNA and 30S•CUGG•tRNAfMet2M1 is more stable than 30S•CUGG•tRNAfMet1. The anomalous toeprints might reflect a frameshifted complex—i.e., positioning of UGG or GGU in the P site, which would entail codon-anticodon mismatches. Alternatively, the conformation of the A-codon nucleotides may be altered, such that the 3′ end of the mRNA is drawn toward the P site by 1–2 nt. In any case, codon 2 (GUA) is not appropriately positioned in the A site, based on the reduced reactivity of corresponding 70S complexes toward EF-Tu•GTP•Val-tRNA. Remarkably, initiation factors IF2 and IF1 completely restore normal mRNA positioning in 30S complexes containing fMet-tRNAfMet2M1 and CUGG. IF2 is primarily responsible for this activity, since IF1 alone or in combination with IF3 has virtually no effect. IFs enhance mRNA positioning in 30S complexes containing fMet-tRNAfMet2 (WT) and/or AUG as well. Complexes containing fMet-tRNAfMet2 and AUGG give toeprint bands at +16 and +17 in the absence of IFs and the signal shifts almost exclusively to +16 in the presence of IFs. Together these findings suggest that IF2 and the unique anticodon stem of fMet-tRNAfMet help constrain mRNA in the 30S IC to set the correct reading frame.
Structural studies of initiation complexes indicate that IF2 and basepair G30-C40 of fMet-tRNAfMet lie some distance from the mRNA,18,33 so the observed effects on mRNA positioning are presumably indirect. Domain II of IF2 binds the shoulder domain of the 30S subunit while domain IV of the factor extends out to interact with the fMet group and 3′ end of fMet-tRNAfMet. These interactions constrain fMet-tRNAfMet in the P/I orientation, which resembles the P/P position except that the elbow region is displaced toward the E site. Notably, the 50S subunit, which constrains fMet-tRNAfMet in the P/P site, fails to restore normal mRNA positioning in various complexes (Fig. 4). This raises the possibility that the P/I configuration imposes constraints on tRNA-mRNA interactions in the complex, thereby increasing the stringency of mRNA positioning. Consistent with this idea, the ability of IF2 to influence mRNA positioning depends on the presence of the fMet group (Fig. 7C). G30-C40 represents the middle of three G-C pairs characteristic of the anticodon stem of initiator tRNA, and 16S rRNA nucleotides A1339 and G1338 dock into the minor groove of fMet-tRNAfMet precisely in this region.18 Mutation M1 (G30C / C40G) alters these contacts, which may enable formation of an aberrant tRNA-mRNA interaction that causes mRNA mispositioning. Alternatively, this basepair substitution may act intra-molecularly to alter the structure and/or dynamics of the anticodon loop. Support for this possibility comes from earlier evidence that the three G-C pairs of tRNAfMet can allosterically influence the anticodon loop.26,34,35 Future experiments will be needed to better understand how IF2 and the unique stem of fMet-tRNAfMet restrict mRNA positioning in the ribosome.
Comparison of complexes containing tRNAfMet1 versus tRNAfMet2 revealed some toeprint-pattern differences, even though these two isoacceptors differ by only one nucleotide (m7G vs. A at position 46).25 Previous studies comparing the two isoacceptors have shown that m7G46 helps stabilize the tertiary structure of tRNAfMet through interactions with the D stem.36,37 It is possible that the less rigid fold of A46-containing tRNAfMet2 reduces its mRNA pairing constraints in some way, causing the more anomalous toeprint pattern observed. Another possibility is that the functional difference stems from hypo-modification of tRNAfMet2, as only this tRNA was overexpressed prior to purification. The only modified nucleotide in the anticodon step loop is 2′-O-methylcytidine at position 32. This modification is predicted to stabilize the C3′-endo conformation of the nucleotide,23 hence loss of this modification would likely increase the flexibility of the anticodon loop.
Mutations of the anticodon stem of tRNAfMet reduce the codon dependence of P-site binding (Fig. 2, Fig. 3, Table 1). Based on this observation alone, one might predict such mutations would decrease the accuracy of start codon selection (e.g., increase spurious initiation from CUG). However, it is important to keep in mind that the complexes containing fMet-tRNAfMet2M1 paired to CUG are incompetent for in-frame elongation, due to mispositioning of codon 2 (Fig. 6). Although IFs can restore codon 2 positioning, how mutation M1 affects the rate of initiation from CUG versus AUG in the cell remains to be determined. Notably, the stringency of start codon selection is increased by mutations of the 30S P site that destabilize tRNAfMet and decreased by the one mutation (G1338A) that stabilizes tRNAfMet.38,39 By analogy, mutation M1 probably reduces the relative rate of in-frame initiation from near-cognate start codons (e.g., CUG versus AUG), although may increase other types of errors (e.g., frame establishment errors).
Finally, our data suggests that the P site exhibits considerable plasticity. For several complexes, multiple toeprints are observed, which may reflect dynamic equilibria between alternative tRNA-mRNA pairing states. This view seems at odds with that of Yusupova and coworkers.40 They have proposed that the P site is highly restrictive, constraining the codon-anticodon basepairs in Watson-Crick geometry, even though no ribosomal residues are positioned to “monitor” this geometry (as in the A site). Future comparative studies of these various near-cognate complexes, using both structural and biochemical methods, should resolve this apparent discrepancy and lead to a deeper understanding of the ribosomal P site.
Materials and methods
Ribosomes and translation factors
70S ribosomes and 30S subunits were purified from E. coli strain MRE600 as described previously.38,41 Untagged IF1 and His6-tagged factors IF2 (α form), IF3, and EF-Tu were overexpressed and purified as described.39,42,43
tRNA and mRNA
Native tRNAfMet1, tRNAMet, and tRNAVal of E. coli were purchased from Chemical Block Ltd. (Moscow, Russia). Wild-type tRNAfMet2 and its mutant derivatives were overexpressed in E. coli strain B105 and purified using native gel electrophoresis.25,26 Mutations M1 (G30C, C40G; Fig. 1) and M3 (U27C, G30C, G31C, C39G, C40G, A43G; Fig. 1) were introduced into pUC13-trnfM, using QuikChange™ mutagenesis, yielding plasmids to overexpress tRNAfMet2, tRNAfMet2M1, and tRNAfMet2M3. E. coli B105 was transformed with each of these plasmids, and transformants were grown in LB with ampicillin (100 µg/mL). Cells from 1L culture were pelleted, washed with PBS [10 mM phosphate (pH 7.4), 137 mM NaCl, 3 mM KCl], resuspended in 10 mL TM buffer [20 mM Tris-HCl (pH 7.6), 20 mM MgCl2], and extracted several times with phenol [saturated with 25 mM NaOAc (pH 5.2), 50 mM NaCl]. Crude tRNA was precipitated from the aqueous phase by adding one-tenth volume 3M NaOAc (pH 5.2) and one volume isopropanol. The RNA pellet was washed with 70% ethanol and dissolved in 4 mL 200 mM Tris-acetate (pH 9.0), and the solution was incubated at 37°C for 30 min to deacylate tRNA. The RNA was re-precipitated with ethanol, pelleted, washed with 70% ethanol, and dissolved in water. Approximately 60 A260 units of crude tRNA was loaded per 12% polyacrylamide gel containing 1X TBE buffer, and subjected to electrophoresis in the same buffer for 12 h at 400V and 4°C. The band corresponding to tRNAfMet2 was identified by UV shadowing and excised. The gel slice was crushed and added to 4 mL of elution buffer [300 mM NaOAc (pH 5.2), 0.1% SDS, 1 mM EDTA] in a capped tube, which was rotated overnight. The eluate was extracted with water-saturated phenol and then with CHCl3 / isoamyl alcohol (24:1), and finally the purified tRNA was ethanol precipitated, pelleted, washed with 70% ethanol, and dissolved in water. Purified tRNAs were charged and formylated as described.44 3′-truncated tRNAs were prepared using snake venom phosphodiesterase as described.45
The mRNAs used are variants of m292,46 a model mRNA based on gene 32 of phage T4. All mRNAs were made by in vitro transcription and gel purified.
Analysis of ribosomal complexes by toeprinting
30S•mRNA•tRNA and 70S•mRNA•tRNA complexes were detected by toeprinting.47 Typically, 5′[32P]-labeled primer #132 (5′-CTTTATCTTCAGAAGAAAAACC-3′) was annealed to mRNA (variable concentration, as indicated in legends) in 50 mM Tris-HCl (pH 7.6), 150 mM NH4Cl, and 30 mM KCl. MgCl2 (7 mM), DTT (1 mM), GTP (0.1 mM), ribosomes or heat-activated 30S subunits (1 µM), and tRNA (1.5 µM) were then added, and the reaction was incubated at 37° C (for 5–120 min, see legends). Reverse transcriptase (2 U, Life Sciences Advanced Technologies, Inc.) and dNTPs (0.2 mM each) were then added to extend the primer, and the cDNA products were resolved using 7% PAGE under denaturing (8M urea) conditions. Gel imaging and quantification were performed using a Typhoon FLA 9000 phophorimager (GE Healthcare) and associated software (ImageQuant 5.2).
Overall equilibrium association constants for 30S•mRNA•tRNA formation (K TC) were measured using toeprinting as described previously.19 In these experiments, the concentration of subunits (1 µM) and tRNA (variable, 0.05–4 µM) always exceeded that of mRNA (0.01 µM) by >5-fold. Hence the data were fit to the equation F = Fmax [bc/(bc+1/ K TC)], where b is the input tRNA concentration, c is the input 30S concentration, and K TC is the equilibrium association constant. Fmax corresponds to the maximal level of detected complex and presumably reflects the probability that the complex resists disruption by reverse transcriptase.
The rate of mRNA dissociation (k off) from a given complex was measured as follows. The 30S complex was formed by incubating heat-activated 30S subunits (1 µM) with mRNA (0.05 µM, containing preannealed radiolabeled primer) and tRNA (1.5 µM) for 2 h at 37° C. Then, at time t = 0, the reaction was diluted by 5-fold in the presence of an excess of chase mRNA (containing start codon AUG and lacking the primer binding site; 5 µM final concentration), and aliquots were removed at various times and subjected to primer extension analysis. Loss of the toeprint signal as a function of time was quantified, and the data were fit to a single-exponential function to obtain k off.
Decoding assay
To assess the ability of various 70S complexes to enter the elongation phase, a single-turnover decoding assay was employed as described.10
Acknowledgements
We thank U. RajBhandary for providing plasmid pUC13-trnfM and E. coli strain B105, B. P. Roy for contributing to Figure preparation, and Z. McNutt, D. Watkins, and M. Chen for comments on the manuscript. This work was supported by the National Science Foundation under Grant MCB-1614990.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Milon P, Rodnina MV. Kinetic control of translation initiation in bacteria. Critical Reviews in Biochemistry and Molecular Biology. 2012;47:334–48. doi: 10.3109/10409238.2012.678284. [DOI] [PubMed] [Google Scholar]
- 2.Caban K, Gonzalez RL Jr.. The emerging role of rectified thermal fluctuations in initiator aa-tRNA- and start codon selection during translation initiation. Biochimie. 2015;114:30–8. doi: 10.1016/j.biochi.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milon P, Maracci C, Filonava L, Gualerzi CO, Rodnina MV. Real-time assembly landscape of bacterial 30S translation initiation complex. Nature Structural & Molecular Biology. 2012;19:609–15. doi: 10.1038/nsmb.2285. [DOI] [PubMed] [Google Scholar]
- 4.Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Molecular Cell. 2006;23:183–93. doi: 10.1016/j.molcel.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 5.Milon P, Konevega AL, Gualerzi CO, Rodnina MV. Kinetic checkpoint at a late step in translation initiation. Molecular Cell. 2008;30:712–20. doi: 10.1016/j.molcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Grigoriadou C, Marzi S, Pan D, Gualerzi CO, Cooperman BS. The translational fidelity function of IF3 during transition from the 30 S initiation complex to the 70 S initiation complex. Journal of Molecular Biology. 2007;373:551–61. doi: 10.1016/j.jmb.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elvekrog MM, Gonzalez RL Jr.. Conformational selection of translation initiation factor 3 signals proper substrate selection. Nature Structural & Molecular Biology. 2013;20:628–33. doi: 10.1038/nsmb.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacDougall DD, Gonzalez RL Jr.. Translation initiation factor 3 regulates switching between different modes of ribosomal subunit joining. Journal of Molecular Biology. 2015;427:1801–18. doi: 10.1016/j.jmb.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavlov MY, Zorzet A, Andersson DI, Ehrenberg M. Activation of initiation factor 2 by ligands and mutations for rapid docking of ribosomal subunits. The EMBO Journal. 2011;30:289–301. doi: 10.1038/emboj.2010.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Q, Fredrick K. Roles of helix H69 of 23S rRNA in translation initiation. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:11559–64. doi: 10.1073/pnas.1507703112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Caban K, Gonzalez RL Jr.. Ribosomal initiation complex-driven changes in the stability and dynamics of initiation factor 2 regulate the fidelity of translation initiation. Journal of Molecular Biology. 2015;427:1819–34. doi: 10.1016/j.jmb.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayer C, Stortchevoi A, Kohrer C, Varshney U, RajBhandary UL. Initiator tRNA and its role in initiation of protein synthesis. Cold Spring Harb Sym. 2001;66:195–206. doi: 10.1101/sqb.2001.66.195. [DOI] [PubMed] [Google Scholar]
- 13.Faulhammer HG, Joshi RL. Structural features in aminoacyl-tRNAs required for recognition by elongation factor Tu. FEBS Letters. 1987;217:203–11. doi: 10.1016/0014-5793(87)80664-6. [DOI] [PubMed] [Google Scholar]
- 14.Mandal N, Mangroo D, Dalluge JJ, McCloskey JA, Rajbhandary UL. Role of the three consecutive G:C base pairs conserved in the anticodon stem of initiator tRNAs in initiation of protein synthesis in Escherichia coli. RNA. 1996;2:473–82. [PMC free article] [PubMed] [Google Scholar]
- 15.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, et al.. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–34. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 16.Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15830–4. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, et al.. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 18.Hussain T, Llacer JL, Wimberly BT, Kieft JS, Ramakrishnan V. Large-Scale movements of IF3 and tRNA during bacterial translation initiation. Cell. 2016;167:133–44 e13. doi: 10.1016/j.cell.2016.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin D, Liu Q, Devaraj A, Fredrick K. Role of helix 44 of 16S rRNA in the fidelity of translation initiation. RNA. 2012;18:485–95. doi: 10.1261/rna.031203.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sussman JK, Simons EL, Simons RW. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Molecular Microbiology. 1996;21:347–60. doi: 10.1046/j.1365-2958.1996.6371354.x. [DOI] [PubMed] [Google Scholar]
- 21.Kumbhar BV, Kamble AD, Sonawane KD. Conformational preferences of modified nucleoside N(4)-acetylcytidine, ac4C occur at “wobble” 34th position in the anticodon loop of tRNA. Cell Biochemistry and Biophysics. 2013;66:797–816. doi: 10.1007/s12013-013-9525-8. [DOI] [PubMed] [Google Scholar]
- 22.Stern L, Schulman LH. The role of the minor base N4-acetylcytidine in the function of the Escherichia coli noninitiator methionine transfer RNA. The Journal of Biological Chemistry. 1978;253:6132–9. [PubMed] [Google Scholar]
- 23.Yokoyama S, Nishimura S. Modified nucleosides and codon recognition In:Soll D, RajBhandary UL, editors. tRNA: Structure, Biosynthesis, and Function. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 24.Shoji S, Abdi NM, Bundschuh R, Fredrick K. Contribution of ribosomal residues to P-site tRNA binding. Nucleic Acids Research. 2009;37:4033–42. doi: 10.1093/nar/gkp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandal N, RajBhandary UL. Escherichia coli B lacks one of the two initiator tRNA species present in E. coli K-12. Journal of Bacteriology. 1992;174:7827–30. doi: 10.1128/jb.174.23.7827-7830.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seong BL, RajBhandary UL. Escherichia coli formylmethionine tRNA: mutations in GGGCCC sequence conserved in anticodon stem of initiator tRNAs affect initiation of protein synthesis and conformation of anticodon loop. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:334–8. doi: 10.1073/pnas.84.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fei J, Kosuri P, MacDougall DD, Gonzalez RL Jr.. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Molecular Cell. 2008;30:348–59. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Molecular Cell. 2007;25:505–17. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lill R, Robertson JM, Wintermeyer W. Binding of the 3′ terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. The EMBO Journal. 1989;8:3933–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGarry KG, Walker SE, Wang H, Fredrick K. Destabilization of the P site codon-anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome. Molecular Cell. 2005;20:613–22. doi: 10.1016/j.molcel.2005.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grunberg-Manago M, Dessen P, Pantaloni D, Godefroy-Colburn T, Wolfe AD, Dondon J. Light-scattering studies showing the effect of initiation factors on the reversible dissociation of Escherichia coli ribosomes. Journal of Molecular Biology. 1975;94:461–78. doi: 10.1016/0022-2836(75)90215-6. [DOI] [PubMed] [Google Scholar]
- 32.Sabol S, Ochoa S. Ribosomal binding of labelled initiation factor F3. Nature: New Biology. 1971;234:233–6. [DOI] [PubMed] [Google Scholar]
- 33.Simonetti A, Marzi S, Myasnikov AG, Fabbretti A, Yusupov M, Gualerzi CO, et al.. Structure of the 30S translation initiation complex. Nature. 2008;455:416–20. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 34.Wrede P, Rich A. Stability of the unique anticodon loop conformation of E.coli tRNAfMet. Nucleic Acids Research. 1979;7:1457–67. doi: 10.1093/nar/7.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barraud P, Schmitt E, Mechulam Y, Dardel F, Tisne C. A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis. Nucleic Acids Research. 2008;36:4894–901. doi: 10.1093/nar/gkn462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daniel WE Jr., Cohn M. Changes in tertiary structure accompanying a single base change in transfer RNA. Proton magnetic resonance and aminoacylation studies of Escherichia coli tRNAMet f1 and tRNAMet f3 and their spin-labeled (s4U8) derivatives. Biochemistry. 1976;15:3917–24. doi: 10.1021/bi00663a003. [DOI] [PubMed] [Google Scholar]
- 37.Crothers DM, Cole PE, Hilbers CW, Shulman RG. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. Journal of Molecular Biology. 1974;87:63–88. doi: 10.1016/0022-2836(74)90560-9. [DOI] [PubMed] [Google Scholar]
- 38.Qin D, Abdi NM, Fredrick K. Characterization of 16S rRNA mutations that decrease the fidelity of translation initiation. RNA. 2007;13:2348–55. doi: 10.1261/rna.715307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin D, Fredrick K. Control of translation initiation involves a factor-induced rearrangement of helix 44 of 16S ribosomal RNA. Molecular Microbiology. 2009;71:1239–49. doi: 10.1111/j.1365-2958.2009.06598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. Structural insights into the translational infidelity mechanism. Nature Communications. 2015;6:7251. doi: 10.1038/ncomms8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lancaster L, Kiel MC, Kaji A, Noller HF. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell. 2002;111:129–40. doi: 10.1016/S0092-8674(02)00938-8. [DOI] [PubMed] [Google Scholar]
- 42.Boon K, Vijgenboom E, Madsen LV, Talens A, Kraal B, Bosch L. Isolation and functional analysis of histidine-tagged elongation factor Tu. European Journal of Biochemistry. 1992;210:177–83. doi: 10.1111/j.1432-1033.1992.tb17406.x. [DOI] [PubMed] [Google Scholar]
- 43.Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Molecular Cell. 2001;8:855–64. doi: 10.1016/S1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 44.Walker SE, Fredrick K. Preparation and evaluation of acylated tRNAs. Methods. 2008;44:81–6. doi: 10.1016/j.ymeth.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–62. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 46.Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Molecular Cell. 2006;24:931–42. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartz D, McPheeters DS, Gold L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes & Development. 1989;3:1899–912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]


