ABSTRACT
A simple post-transcriptional modification of tRNA, deamination of adenosine to inosine at the first, or wobble, position of the anticodon, inspired Francis Crick's Wobble Hypothesis 50 years ago. Many more naturally-occurring modifications have been elucidated and continue to be discovered. The post-transcriptional modifications of tRNA's anticodon domain are the most diverse and chemically complex of any RNA modifications. Their contribution with regards to chemistry, structure and dynamics reveal individual and combined effects on tRNA function in recognition of cognate and wobble codons. As forecast by the Modified Wobble Hypothesis 25 years ago, some individual modifications at tRNA's wobble position have evolved to restrict codon recognition whereas others expand the tRNA's ability to read as many as four synonymous codons. Here, we review tRNA wobble codon recognition using specific examples of simple and complex modification chemistries that alter tRNA function. Understanding natural modifications has inspired evolutionary insights and possible innovation in protein synthesis.
KEYWORDS: Wobble Hypothesis, Modified Wobble Hypothesis, wobble decoding, tRNA, wobble nucleoside, modified nucleosides, nucleoside tautomers, cognate and wobble codon recognition, translation
Introduction
Translation of the Universal Genetic Code (Fig. 1) into the amino acid sequence of proteins requires accurate and efficient decoding of mRNA (mRNA) on the ribosome by tRNA (tRNA). Sixty-one of 64 3-nucleoside codons of the mRNA, N1N2N3, are decoded in frame with a complementary sequence of the tRNA anticodon, N34N35N36. Three codons are recognized by protein factors and correspond to translation termination signals. Complementariness of base pairing between tRNA's anticodon and the mRNA codon, A•U, U•A, G•C and C•G where • denotes canonical Watson-Crick hydrogen bonding, does not explain how the 61 amino acid codons are decoded by far fewer tRNAs. Fifty years ago, Francis Crick published the Wobble Hypothesis.1 At the time there was evidence to suggest that the first two positions, N1N2, of mRNA's 3-nucleoside codons were uniquely identified by the tRNA with some ambiguity in the third position. Crick offered the idea that uridine (U) at the first position of the anticodon, position 34 in tRNA (Fig. 2), would base pair with guanosine (G) and that inosine (I), which at the time had only recently been found at position 34 in yeast tRNAAla,2 would base pair with uridine, cytidine (C) and adenosine (A). Thus, canonical, Watson-Crick base pairing with tRNA and mRNA on the ribosome was supplemented with non-canonical hydrogen bonding, ‘wobble’ base pairing, including that of a purine with another purine. Though conceding that the wobble position base pairing of two purines would widen the anticodon-codon double helix at the point of an I34○A3 base pair where A3 is the third nucleoside of the codon and ○ denotes non-canonical hydrogen bonding, Crick excluded the possibility of a wobble position pyrimidine-pyrimidine base pairing. He argued that the narrowing of the helix would be too dramatic in comparison to the neighboring canonical purine-pyrimidine distances at the first and second positions.
Figure 1.
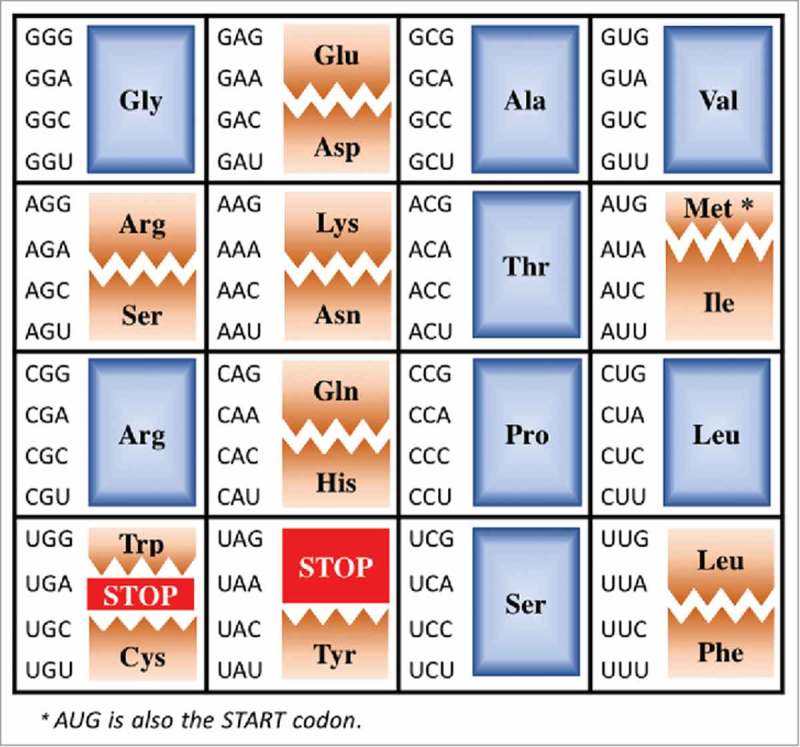
The Universal Genetic Code. The Universal Genetic Code is shown in an atypical array to highlight those codons and their decoding by tRNAs discussed here. Fully degenerate codon boxes are shown in blue, split codon boxes in brown and stop codons in red.
Figure 2.
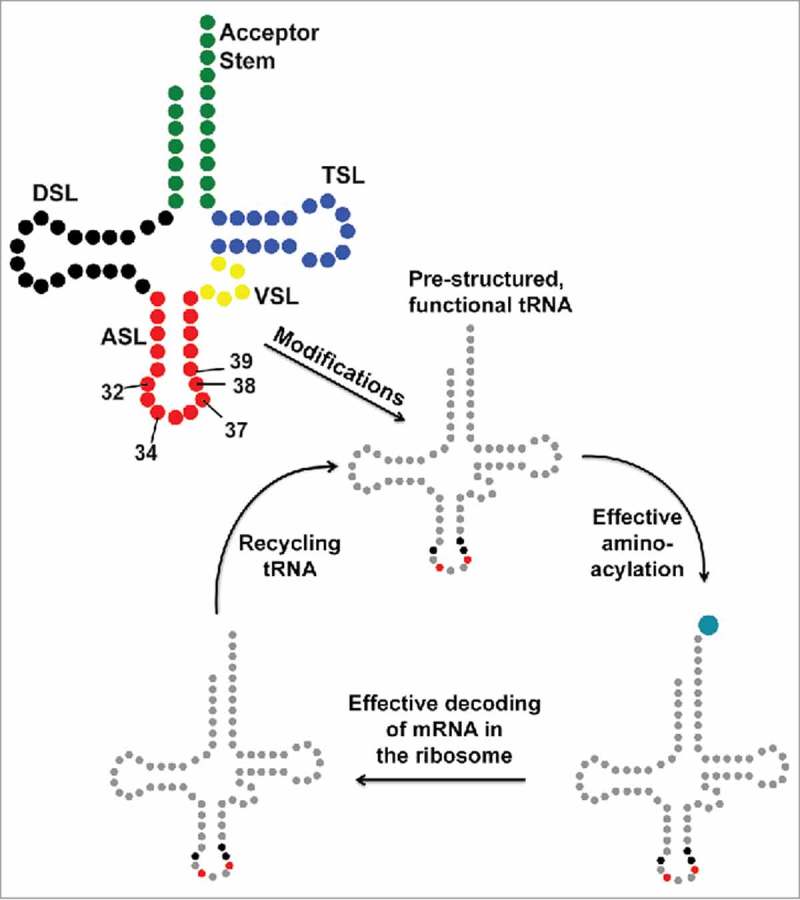
The tRNA journey. The secondary structure of tRNA with its constituent domains marked in different colors (top left): Acceptor Stem (green); Dihydrouridine Stem and Loop, DSL (black); Anticodon Stem and Loop, ASL (red); Variable Stem and Loop, VSL (yellow); Thymidine Stem and Loop, TSL (blue). tRNA transcripts are processed by sizing and modification, some are spliced, before functioning in translation. Modification of tRNAs, particularly the anticodon stem and loop (ASL) domain at positions 32, 34, 37, 38 and 39, is an important step toward achieving functional chemistry and architecture. The wobble nucleoside, first of the anticodon, is position 34. Red and black highlights of mature tRNA after modification indicate the locations in the ASL where it is heavily modified.
The post-transcriptional modification of tRNA, the original soluble RNA, has been known for over 50 years pioneered by the extraction and characterization methods of Ross Hall in the 1960s.3 At the time the Universal Genetic Code was unveiled,4 there were several RNAs known to contain modified nucleosides, but their sequence locations and functions were a mystery before the evolution of RNA sequencing methods. By 1991, 25 years later, a sufficient number of modified nucleosides had been found to occupy the wobble position 34 of tRNA's anticodon that a Modified Wobble Hypothesis was advanced.5 Biophysical and biochemical experiments had suggested and continue to support the principle that some position 34 modifications structure the architecture of the anticodon stem and loop (ASL) to counter wobble whereas other modifications shape the ASL to enable a tRNA to decode three or even four synonymous codons (codons differing only in the wobble position N3).6-10
Of what importance are these ubiquitous and highly conserved anticodon modified nucleosides to the decoding of mRNA codons? In general, modified nucleosides of tRNA's anticodon stem and loop (ASL) domain are found within the stem at positions 27, 28, 31, 39, and 40 and at loop positions 32, 34, 35, 37, and 38 (Fig. 2).11-13 These ten nucleoside positions of the ASL are not simultaneously modified in any one tRNA, rather a set of 3–5 specific nucleosides are found to be modified in an individual tRNA. Many times in the sequencing of a tRNA species one finds that as many as three nucleoside positions of the seven residue loop are modified. Considering that the ASL of tRNA constitutes 17 of the molecule's ∼76 nucleosides and that the stem constitutes 5 base pairs, with 10 nucleosides, the modifications within the ASL loop represent a dense population of altered nucleoside chemistries. Often the modifications at wobble position 34 and 3′-adjacent to the anticodon at position 37 are the most chemically complex of all RNA modifications and are composed of hydrophobic aliphatic chains or aromatic substituents, or of highly hydrophilic or even charged functional groups participating in translational efficiency and fidelity.11 Thus, posttranscriptional modifications of the ASL nucleosides emulate the chemistries of amino acid side chains.14 Even what appears to be one of the simplest of modifications, such as the substitution of a sulfur atom for an oxygen, a thioketone for a carbonyl group, or the deamination of adenosine to inosine (I), takes on significance in the decoding of mRNA. The modification of wobble position U34 to s2U34 alters tRNA's ability to wobble to G3;5 the modification of C32 to s2C32 negates the ability of tRNAs with I34 to decode A3 of the codon.6
The importance of being modified
The high percentage of anticodon modification and their composite chemistries provides different functionalities to tRNA in its role in decoding mRNA (Fig. 3). Individual anticodon domain modified nucleosides are identity determinants for protein recognition, particularly aminoacylation,15-17 increase accuracy and efficiency in codon recognition,18-24 and pre-structure the ASL for translation.9,25-30 Each of the modified nucleosides contribute distinct chemistries, nucleoside conformations and dynamics, and their contributions to decoding have been studied extensively over decades and most recently reviewed.20,22,24,28,31-38 However, there is significant evidence that a combination of two or three anticodon domain modifications play a synergistic role in tRNA function where modification of a wobble position U is crucial.9,20,24,39-46 Today, we know that certain modifications of U34 enable expansion of codon from NNA/G recognition to synonymous codons ending in pyrimidines, N1-N2-Pyr, where N is any of the 4 nucleosides and Pyr is either U or C.29 Yet, the anticodon domain of some tRNA species lack modification and can be totally devoid of modified nucleosides. Bacteriophage T4 tRNAGly is an early example of a tRNA lacking modification.47 The unmodified U34 nucleosides of native mitochondrial alanine, leucine, threonine and valine tRNA species in vivo, and that of a totally unmodified tRNA transcript in vitro were shown to read codons ending in U3 and C3, as well as A3 and G3.48,49 To understand the forces that maintained and propagated tRNAs' anticodon domain modifications throughout all life, first we will discuss the limited number of examples revealed over many years to have unmodified Us and As at wobble position 34, and the influence of position 32 nucleosides on decoding.
Figure 3.
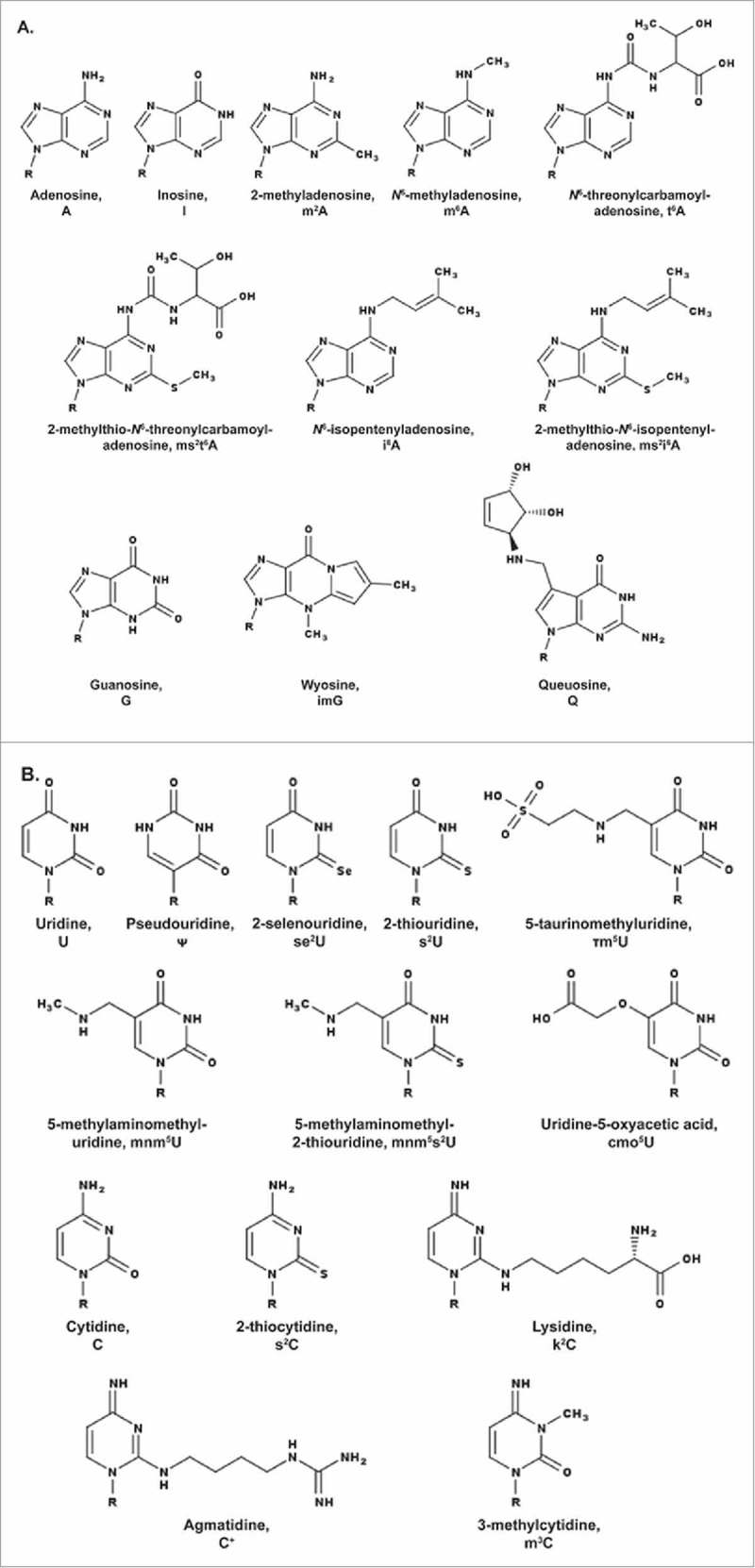
Modifications present in tRNA's anticodon stem and loop domain (ASL) featured in the text and displayed in their neutral state. A. Modified purines, adenosine (A) and guanosine (G). B. Modified pyrimidines, uridine (U) and cytidine (C). R = ribose.
Unmodified wobble position 34 and the importance of position 32
Bacteria, archaea and eukaryotes have some 40 tRNA species decoding the Universal Genetic Code. However, many times we find that in mammalian and yeast mitochondria, in chloroplasts, and in Mycoplasma ssp only a single tRNA species decodes all 4 codons of a fully degenerate codon box of the Universal codes (Fig. 1).49-57 Wobble codon recognition is at its extreme in these circumstances and has been referred to as ‘superwobble’.58 An unmodified U in tRNA's wobble position 34 facilitates the use of far fewer tRNAs in organelles than the over 40 cytoplasmic species typically reading the 61 amino acid codes. Fewer tRNAs is certainly an advantage for the small genomes of the organelles and the minimal single cell organism, mycoplasma. An unmodified U34 also abrogates the need for the extensive array of genes encoding the modified nucleoside pathways of enzymes and substrates required of the simplest to the most complex modifications of U34.11 However, the superwobble reading of codons compromises translational efficiency.58
An unmodified U34 seems to be particularly efficient for the organellar tRNAGly species. The codons for the amino acid glycine, GGA, GGG, GGU or GGC, are found in a 4-fold degenerate codon box, whose first two nucleosides are the same (Fig. 1). They are read by as many as three different tRNA isoacceptors in bacteria with cognate and wobble anticodons. Early in the study of glycine tRNAs, the Escherichia coli tRNAGly isoacceptors were grouped into 3 subspecies based on a chromatographic separation: tRNAGly1 CCC, tRNAGly2 UCC, and tRNAGly3 GCC.59 In an E. coli in which there were multiple copies of suppressors increasing the levels of wild-type tRNAGly1 CCC, −1 translational frameshifting occurred at the 5′-GGG-3′ codon allowing the near-cognate tRNA to read GGA codons.60 Surprisingly, experiments with E. coli tRNAGly2 UCC demonstrated that the unmodified UCC anticodon discriminates among the four glycine codons depending on the nucleoside in position 32, an unmodified U32 or C32 (Fig. 4A). Thus, the unmodified UCC anticodon reads GGA and wobbles to GGG, but does not recognize GGU and GGC.61
Figure 4.
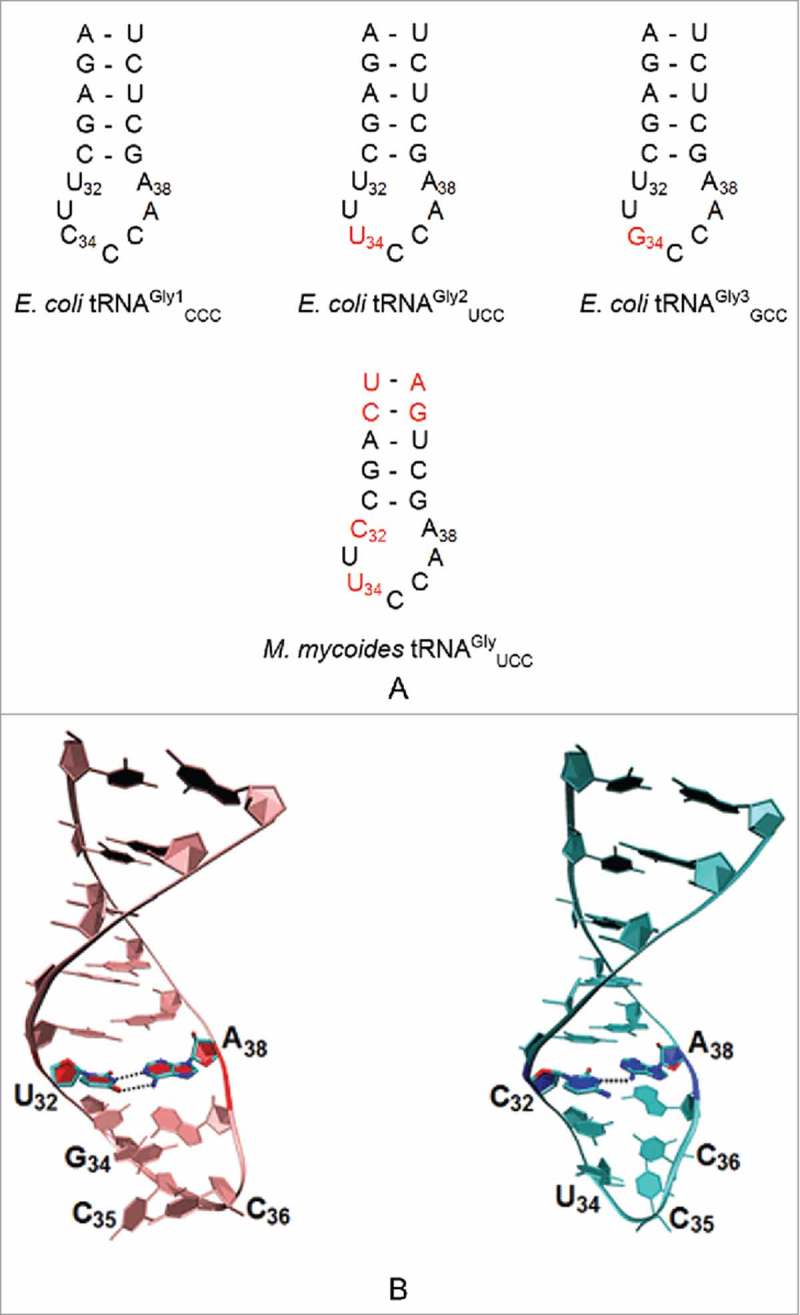
Sequences of unmodified ASL of tRNAGly. A. ASLs of E. coli tRNAGly1 CCC, tRNAGly2 UCC, and tRNAGly3 GCC indicating the changes in nucleoside position 32 and 34 (marked in red) from the tRNAGly1 sequence. All 3 E. coli tRNAGly have the same nucleoside at position 32 (U32). The Mycoplasma mycoides tRNAGly UCC differs from the E. coli tRNAGly as a result of a cytidine at position 32 as well as the presence of inverted base pairs in the anticodon stem at positions 27•43 and 28•42. B. Three Dimensional Structure of the tRNAGly GCC and tRNAGly UCC ASLs of B. subtilis. The interactions between the U32•A38 and C32○A38 nucleosides (PDB ID: 2LBJ and 2LBL) are shown. Both ASLs lack the sharp U-turn characteristic of ASLs of other tRNAs. Nucleosides of the anticodon are labeled 34 to 36.
Although the anticodon UCC could discriminate efficiently with a U in position 32, it loses its ability to differentiate when substituted with a C32 as was also true for M. mycoides glycine tRNA. In wild type M. mycoides tRNAGly, the anticodon UCC failed to discriminate between the glycine codons with a position 32 cytidine, but when changed to a U it acted like E. coli tRNAs and discriminated between the four glycine codons.62 In M. mycoides, the single tRNAGly UCC that decodes all four glycine codons is devoid of anticodon modifications and has a C32○A38 mismatch which leads to decreased fidelity in vivo (Fig. 4).63 The tRNAGly UCC transcript without any modifications reads all 4 codons in vitro.48 Ribosome binding experiments with U32/C32 mutants of tRNAGly showed an increased affinity of the C32 mutant to the cognate codon and to codons with third position mismatches in the ribosome's A-site.64 The rate of dissociation of the U32-containing tRNAGly from the near-cognate GGC, GGA and GGU codons was much more rapid - 12-fold faster than from the GGG cognate codon - and stabilized the binding of tRNA to codons with third position mismatches.64 In contrast, the mutated tRNAGly1 with C32 dissociated much more slowly from near-cognate codons. Analysis of the UV-thermal denaturation (melting) curves of the anticodon stem and loop domains demonstrated that tRNAGly UCC with a protonated C32○A+ 38 non-canonical base pair melted at a temperature 10 °C lower than tRNAGly GCC or np-tRNAGly UCC, used exclusively for non-protein cell wall synthesis in Staphylococcus species, which exhibited a Tm of 70 °C.65 Although the sequences of these tRNA differed from one another, none of their solution structures formed the classical U-turn motif seen in other tRNA anticodon loops (Fig. 4B). tRNA molecules fulfill different functional roles by interacting with other cellular molecules, and the structural variations of the glycine ASL of S. aureus could contribute to its functional diversity. tRNAGly GCC, without any base modification participates in transcriptional regulation and transcription. The tRNAGly UCC which more often contains a U34 modification except in some organisms including M. mycoides, participates in translation. The np-tRNAGly UCC contains no base modification and participates in cell wall synthesis.65 The presence of pyrimidine at position 37, along with a reduced affinity for EF-Tu, could limit their involvement in transcription and translation. Modifications of U34 could increase or decrease the ability to wobble, thus enhancing discrimination. A more dynamic ASL tRNAGly GCC was observed in the presence of multivalent cations, whereas tRNAGly UCC and np-tRNAGly UCC were more structurally ordered in their presence. A more dynamic loop structure would therefore, better accommodate the different functional roles of an unmodified tRNAGly in protein translation, in tRNA-dependent gene regulation, and cell wall biosynthesis.65
The nucleoside at position 32 of the tRNA's anticodon loop is recognized as important for translation, though the position 32 nucleoside is remote from the anticodon. The intraloop hydrogen bonding between the nucleosides at 32 and 38 strongly influences the affinity of tRNA to the A-site.64 tRNA species other than those for glycine also have an unmodified wobble position U34, and the nature of the N32○N38 intraloop hydrogen bonding in these tRNAs does appear to regulate expansion/discrimination of codon reading. The A32•U38 interaction, highly conserved in tRNAAla and also seen in tRNAPro, decreases the tRNA affinity to the ribosomal A-site compared with other N32○N38 intraloop hydrogen bonding nucleosides.66 Five Mycoplasma capricolum tRNA species with the unmodified anticodon UNN and decoding for five different amino acids have anticodon loop sequences with C32○A38. In addition, tRNAAla UGC has an intraloop interaction of C32○C38. Because these 6 tRNA species have a C32 and an unmodified U34, in theory they could efficiently read all four synonymous codons for leucine, valine, proline, alanine, glycine and serine.67 Although tRNAThr UGU was shown to translate the codons ACU, ACA and ACG, it was inefficient in reading the codon ACC.
Interestingly, the sufD42 mutant of E. coli tRNAGly1, encoded by glyU, is a derivative of tRNAGly CCC with an extra C in the anticodon loop and contains no modification in the anticodon loop.68 This mutant is considered dominant and contains four bases, 5′-CCCC-3′ that make up the anticodon and suppress +1 frameshift mutants with an extra G inserted into a GGN codon (GGGN). The quadruplet translocation theory is used to deduce the pairing of these four cytidines with four bases of the codon in the A site. The tRNA anticodon sequence has been suggested to act as a molecular ruler which determines the codon size during translation, within certain limits,69 thereby restoring some ribosomes to the wild type frame. Thus, the nature of the N32○N38 base interaction affects the binding of the anticodon to the codon suggesting that the intraloop hydrogen bonding alters the conformation and dynamics of the anticodon stem and loop domain within the ribosomal complex.62,70
Very few unmodified, wobble position A34 have been found in tRNA sequences: a yeast mitochondrial tRNAArg ACG71 and Mycoplasma tRNAThr AGU.50,56,72 In all domains of life, A34 of the tRNA transcript is almost always deaminated to form inosine at the wobble position (I34). The modification to I34 expands the coding capacity to read the bases U, C and A. In Mycoplasma the unmodified A34 of tRNAThr AGU efficiently translates the ACC codon,67 but mutants of E. coli tRNASer and tRNAGly with A34 could only weakly read UCC and GCC in vitro, respectively.73,74 In Salmonella typhimurium, tRNAPro GGG is the only tRNA that reads the CCC codon. With G34 replaced by an unmodified A, a mutant with no cognate codon for CCC, grew normally.75 The mutant tRNAPro AGG efficiently read the CCC codon similarly to its wild-type counterpart with a GGG. It formed a wobble base pair using a protonated A with the third position C in mRNA. Similarly, a mutant tRNAGly1 with a UCC to ACC mutation containing an unmodified A34, lost its ability to discriminate between the third position nucleosides of the glycine codons.73 The possibility of a purine○purine base pair being more stable than a pyrimidine○pyrimidine pair,76 as well as the two out of three model77 was suggested to explain the non-discrimination by A in the wobble position.73 The presence of A in the wobble position could change the conformation of the anticodon by preventing hydrogen bonding between U33 and the phosphate of nucleoside 36,78 which stabilizes the U-turn conformation of the ASL. The rare occurrence of an unmodified A34 is supported by the hypothesis that a wobble position A cannot discriminate and ensure translational fidelity of tRNAs reading split box codons. In contrast, the presence of A34 would be advantageous in the reading of fully degenerate synonymous codons where there is a lack of discrimination and in cases where only a single tRNA exists for the reading all four codons.73
In addition to their primary role of translation, some tRNAs have other functions including regulation of gene expression, bacterial cell wall synthesis, viral replication, antibiotic biosynthesis and suppression of alternative splicing.64,65 In Bacillus subtilis and many other Gram-positive bacteria, tRNA molecules regulate gene expression by the tRNA dependent, ‘T-box’, mechanism of transcription attenuation to maintain a balanced pool of aminoacyl-tRNAs that is essential for cell viability.79,80 The tRNA ligand for the T-box mechanism regulating the expression of glycyl-tRNA synthetase is tRNAGly UCC with an unmodified U34. A Rho-independent, terminator helix in the 5′UTR of the leader mRNA of the glyQS operon for glycyl-tRNA synthetase prevents the operational binding of an aminoacylated glycyl-tRNAGly UCC.81 Conversely, the anticodon of an uncharged tRNA interacts with a loop, the Specifier Loop, containing the complementary codon and the tRNA's 3′-terminal CCA hydrogen bonds to an anti-terminator helix, re-conformed from the terminator helix. These interactions as well as others between the tRNA and the mRNA stabilize the anti-terminator conformation of the 5′UTR and allow transcription to proceed downstream through the coding sequence. Thus, the unacylated tRNAGly UCC is similar to the much smaller metabolic products that affect the riboswitch mechanisms controlling gene expression; the 5′UTR undergoes a conformational change with the binding of the ligand.82 The tRNAGly UCC is also predicted to bind to the 5′-GGA-3′ Specifier codon in Bacillus and Staphylococcus species glycyl T-box riboswitches.83
A third glycine tRNA (UCC) without a modified U34 has been identified in Staphylococcus species as participating in cell wall biosynthesis, but not in protein translation, and was termed non-proteinogenic (np-tRNAGly).84-87 These np-tRNA have an unmodified U34 nucleoside and a cytidine rather than a purine at position 37.88 In Thermus thermophiles, the np-tRNAGly species are found to have reduced affinity for the elongation factor Tu (EF-Tu) due to base substitutions of A51-U63 for G51-C63 in the base of the T-stem, thus decreasing their involvement in ribosomal protein synthesis.65 These weak EF-Tu binders could act as glycine donors in forming essential pentaglycine bridges which stabilize the staphylococcal cell wall.85,89-91
The biosynthesis of peptidoglycan in S. aureus involves two uridine nucleotide substrates, UDP-MurNAc-pentapeptide and UDP-GlcNAc (N-acetyl glucosamine), which combine to form a lipid intermediate GlcNAc-MurNAC(pentapeptide)-P-P-lipid. The lipid intermediate gets further modified by amidation of the α-carboxylic group of glutamic acid and the addition of a pentaglycine chain to the ε-amino group of lysine. The weak EF-Tu binding glycyl tRNAs serve as intermediates in these reactions to form the pentaglycine bridges, that stabilize the peptidoglycan chains and are essential for cell viability. These short peptide bridges are synthesized in a non-ribosome catalyzed peptidyl transferase reaction, which uses the charged glycyl-tRNA,1 np-tRNAGly and a ‘pseudo’-tRNAGly UCC as substrates.65,85,92 Glycine and serine act as substrates that are successively added to form small peptide bridges which are catalyzed by a family of non-ribosomal peptidyl transferases known as FEM-XAB (Factors Essential for Methicillin Resistance) - mediated cell wall synthesis.93 The incomplete formation of these interpeptide bridges can lead to increased antibiotic susceptibility or lethality.85
The wobble hypothesis and the modulation of inosine wobbling
Though there are instances of unmodified nucleosides at tRNA's wobble position 34 as described, more often than not U34 is post-transcriptionally modified and A34 is deaminated to inosine. Using specific modifications of U and the modulation of I reading A, U and C, we illustrate here the importance of wobble position 34 modifications to tRNAs' accuracy and efficiency of translation. The modification of adenosine to inosine was first recognized by Francis Crick for enabling tRNA recognition of synonymous codons.1 Inosine (Fig. 3) results from the deamination of adenosine, a transformation that is facilitated by the adenosine deaminase (ADAR) family of enzymes that act on RNA.94 Although inosine is a marker of damage or mutation in DNA, the presence of this very same modified nucleoside is considered to be essential in various RNAs.95 Inosine plays a vital role in the function of tRNA, in particular. As the first recorded nucleoside modification within the sequence of an anticodon,2 Crick introduced inosine in his 1966 Wobble Hypothesis.1 While the first two bases of the codon undergo traditional base-pairing without exception,96 Crick proposed the potential for non-canonical base pairs between the first base of the anticodon (“wobble” position 34) and the third base of the codon, U34○G3, or I34○A3/U3/C3 (Fig. 5).1 This flexibility of the genetic code is not without limitations; however, in accordance with this hypothesis, a given tRNA isoacceptor may recognize multiple codons, thus explaining the degeneracy of the genetic code.
Figure 5.
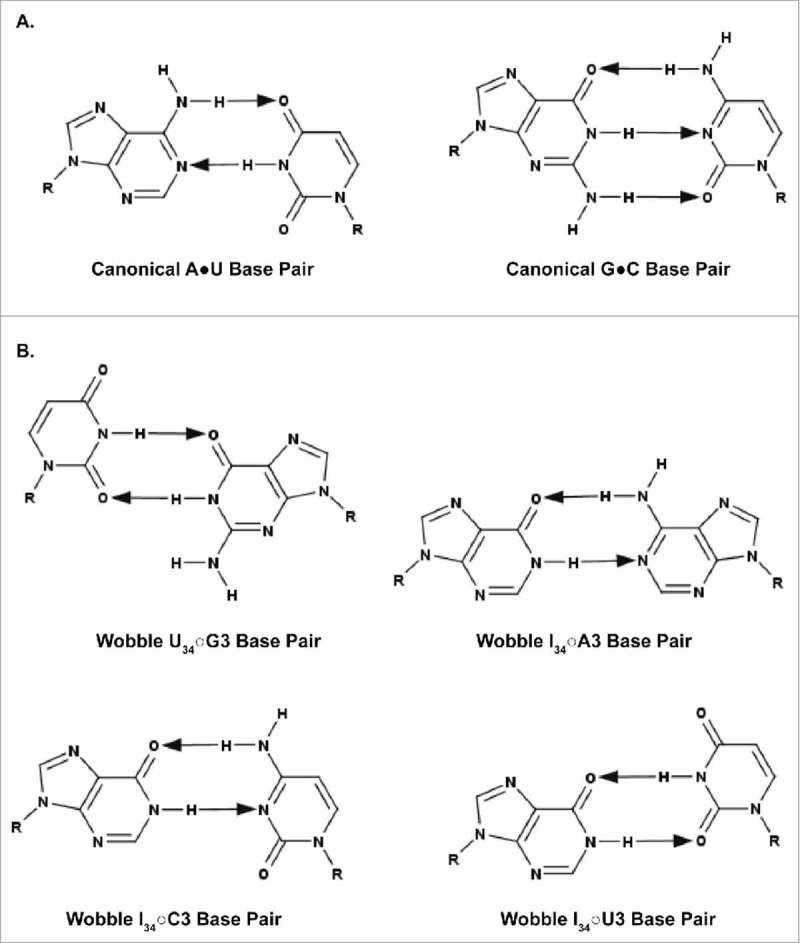
Canonical and wobble base pairing of tRNA to mRNA. A. Canonical A•U and G•C base pairs. B. Wobble U34○G3, I34○A3, I34○C3, and I34○U3 base pairs. G34○U3 pairings are virtually nonexistent; therefore, the pairing is not shown. The arrows point away from the hydrogen bond donor and toward the hydrogen bond acceptor.
The Wobble Hypothesis states that position 34 inosine may base pair with uridine, cytidine, and adenosine. The ability of inosine at the wobble position to promote the reading of multiple codons, in some cases, proves essential to survival. The heterodimeric enzyme consisting of the Tad2p and Tad3p subunits of Saccharomyces cerevisiae catalyzes the deamination of adenosine to inosine on tRNA.97 A strain of Schizosaccharomyces pombe containing a mutant tad3–1, the homolog of TAD3, experienced temperature-dependent arrested growth at Gap 1 and Gap 2 of the cell cycle.98 The S. pombe genome utilizes only 3 tRNAAla isoacceptors for the 4 alanine codons GCU, GCC, GCG, and GCA: tRNAAla IGC, tRNAAla CGC, and tRNAAla UGC. According to the wobble rules, tRNAAla IGC must be responsible for decoding of the GCC codon. The otherwise unmodified tRNAAla AGC would be able to decode its complementary GCU codon but could not wobble to GCC, inhibiting the translation of gene products vital to the G1/S and G2/M transitions of the cell cycle.98
A tRNAArg IGC may be modified from A34 to I34 yet still be unable to decode A3. The E. coli tRNAArg1 ICGand tRNAArg2 ICG isoacceptors should bind and effectively decode the CGU and CGC codons, and even the CGA codon. These wobble pairings were confirmed experimentally, as the singly modified anticodon stem and loop of tRNAArg ICG, ASLArg ICG, is able to bind the CGU, CGC, and CGA codons within the ribosomal A-site.39 In fact, tRNAArg1 ICG and tRNAArg2 ICG are the only isoacceptors available to recognize the 3 aforementioned codons. Furthermore, adenosine must be modified to inosine at position 34 of the tRNAArg1,2 ICG isoacceptors in order for wobble pairing to occur. Unmodified ASLArg1,2 ACG is able to bind its cognate codon, CGU, but unable to bind CGC or CGA as expected, as Crick did not explicitly delineate any wobble capabilities of position-34 adenosine.1,39
The tRNAArg1,2 ICG decoding of the CGA codon is relatively inefficient compared with translation of the CGU.99,100 Although the energy barrier for I○A base-pair formation is greater than those of I○C and I○U, the increased distance between the N-glycosyl bonds of I34 and A3 required to accommodate the purine○purine wobble pair can be achieved when the nucleosides adopt an Ianti○Aanti conformation.1,44,101 Additional modifications at position 32 and 37 of the ASLArg ICG may further contribute to the difficulties of I○A wobble pairing. The E. coli tRNAArg1 ICG species contains the naturally-occurring 2-thiocytidine at position 32 (s2C32) and 2-methyladenosine at position 37 (m2A37). The thrice modified ASL construct could not be synthesized, but the doubly modified ASLArg ICG-s2C32 was unable to bind the CGA codon within the ribosomal A-site.39 Here, the complete wobble capabilities of inosine do not apply due to the restrictive effects of the s2C32 modification with respect to the otherwise feasible I○A wobble pair.
The tRNAArg2 ICG species lacks the s2C32 modification but does contain the m2A37 modification. As evidenced by the aforementioned ribosomal binding study, m2A37 prohibits I○A pairing.39 However, reading of the CGA codon defaults to tRNAArg2 ICG, as the inclusion of the s2C32 modification disqualifies the tRNAArg1 ICG from decoding of the CGA codon in vivo. Yet the E. coli genome still must compensate for the overall poor capacity of the tRNAArg ICG isoacceptors to wobble to the CGA codon by biasing codon usage. The inclusion of CGA codons in mRNA transcripts increases energetic costs and decreases the efficiency of translation. The prevalence of the CGU, CGC, and CGA codons is heavily biased against the CGA codon and in favor of the CGU and CGC codons.102,103
The modified wobble hypothesis and decoding at position 34
For many years, Crick's Wobble Hypothesis appeared to sufficiently explain the function of modified nucleosides at tRNA's wobble position without the need for alteration. However, the discovery of numerous new modifications, a large number of which are found exclusively at the wobble position, led to the development of a modified wobble hypothesis.5 The first base of the anticodon is so often modified to either expand or restrict the binding abilities of the wobble nucleoside, therefore enabling the specific recognition of cognate and synonymous codons. As such, near-cognate codons can be selected against, or the recognition of multiple codons can be made feasible with various chemical moieties introduced onto the wobble base.5 There are several examples of both expansion as well as restriction of codon recognition which are presented with a focus on the mechanistic details of both expansion and restriction of recognition, and the many factors that must come to play to make either possible.
Expansion of codon recognition through modified nucleosides pre-structuring of the ASL
The high freedom of rotation coupled with the limited chemical variation of the four major nucleosides enables RNAs to adopt multiple conformations with comparable stability yet limited chemistry. Modifications add to the chemistry and can either limit or expand the number of available conformations, thereby influencing the structure toward a more specific architecture or provide dynamics important to translational effectiveness. There are six amino acids represented by 4-fold degenerate codon boxes which include alanine, glycine, proline, serine, threonine, and valine, and are of particular interest in U34 modification. As with S. pombe and the GCN alanine codon box, Salmonella enterica utilizes 3 distinct species of tRNAPro for recognition of the entire CCN codon box: tRNAPro CGG, tRNAPro GGG, and tRNAPro UGG. Deletion of the genes for tRNAPro CGG and tRNAPro GGG simultaneously is not lethal to S. enterica.104 Only tRNAPro UGG proves necessary for viability of the organism by surpassing its expected decoding capabilities due to the presence of the 5-oxyacetic acid modification (cmo5) on position-34 uridine in the ASL. The cmo5U34 modification is present in one species of each of the tRNA isoacceptors for alanine, proline, serine, threonine, and valine. Similarly, tRNAVal UAC-cmo5 and tRNAAla UGC-cmo5 partially rescue the E. coli growth phenotype caused by knockout of the 2 tRNAVal GAC isoacceptors and the tRNAAla GGC isoacceptor, respectively, indicating the ability of tRNA containing the cmo5U34 modification to decode an entire codon box, albeit somewhat inefficiently.105 The presence of the cmo5U34 modification facilitates pre-structuring of the ASLVal UAC that enhances its ability to bind near cognate codons. When binding each of the GUN valine codons, the ribose of cmo5U34 of the modified ASLVal UAC assumes the C3′-endo conformation.29 Prevalent within A-type helical regions of tRNA, the C3′-endo sugar pucker is synonymous with stability and rigidity.106-108 The existence of a hydrogen bond between the 2′-OH of the almost invariant U33 of the ASL and O5 of the cmo5 modification further constrains cmo5U34.29 This intramolecular hydrogen bond in particular expands the ability of ASLVal UAC-cmo5U34 to decode codons ending in U and C, pre-structuring the ASL such that the entropic cost of a pyrimidine○pyrimidine base pair, initially believed to be unfavorably short, is surpassed.1,29
The cmo5 modification has surprising implications for the U34•G3 wobble pair. Although this pairing was originally predicted without regard for the potential of modified uridines, the uridine at position 34 of the ASL must be modified in order for the U34•G3 pairing to occur.18,105 Rather than enhancing the ability of U34 to wobble to G3, cmo5 merely enables the interaction. In a second divergence from the original Wobble Hypothesis, the modified cmo5U34•G3 does not adopt the predicted wobble geometry with two hydrogen bonds between the two bases. The cmo5U34 instead forms three hydrogen bonds with G3 as in the traditional Watson-Crick geometry (Fig. 6).29 The steric and electronic properties of the cmo5 modification are thought to promote the enol form of cmo5U34, thus allowing for the Watson-Crick geometry of the cmo5U34•G3 base pair reminiscent of a C•G base pair.29,109
Figure 6.
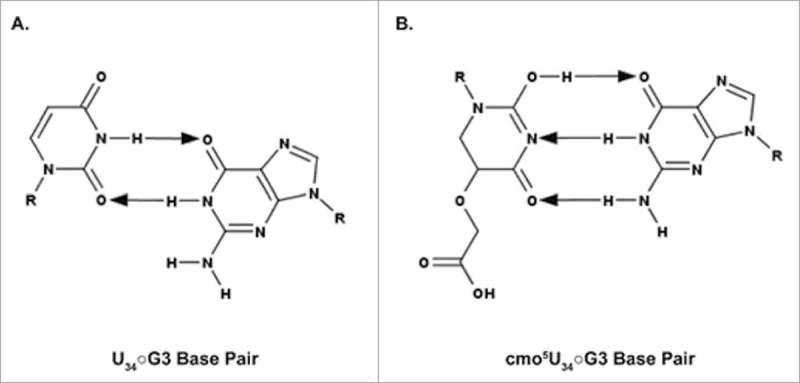
Watson-Crick geometry of the cmo5U34○G3 base pair. A. The predicted geometry of the unmodified U34○G3 base pair containing two hydrogen bonds. B. The observed geometry of cmo5U34○G3 base pair containing three hydrogen bonds. For the cmo5U34○G3 to resemble a C•G base pair, the cmo5 modification is proposed to facilitate formation of the enol tautomer. The arrows point away from the hydrogen bond donor and toward the hydrogen bond acceptor.
Additionally, E. coli tRNAVal UAC containing the cmo5U34 modification is also methylated at the position-37 adenosine to yield N6-methyladenosine (m6A). As seen with tRNAArg1 ICG-s2C32;m2A37 and tRNAArg2 ICG-m2A37, it is not uncommon for the ASL of a given tRNA to contain multiple modified nucleosides. Modifications written onto N6 of the universal purine located at position 37 of the ASL are proposed to interfere with intraloop hydrogen bonding and to enhance base-stacking, thereby promoting an open loop structure within the ASL.18 The open loop structure of the ASL is a prerequisite for anticodon-codon binding within the ribosomal complex.110 ASLVal UAC lacking both the cmo5U34 and m6A modifications does not appreciably bind the GUU, GUC, or GUG codons within the 30S ribosomal subunit.111
While G is able to base pair with both C as well as U, the interaction with the latter is thermodynamically less stable. As such, tRNAs coding for aspartate, asparagine, histidine, and tyrosine commonly contain 7-(((4,5-cis-dihydroxy-2-cyclopenten-1-yl)amino)methyl)-7-deazaguanine, queuosine or Q (Fig. 3A), at the wobble position, which enables recognition of codons ending in either C or U without favoring one or the other.112 This is true even for the more simplistic and somewhat less commonly modified mitochondrial tRNAs, implying an evolutionary necessity for enhanced codon recognition that Q provides.113 Q is synthesized by bacteria, and eukaryotes acquire Q either as a nutrient or from the intestinal flora as quenine, the free base form of Q, which is then directly inserted into the pertinent tRNAs through substitution of the base quenine.114 Interestingly, the E. coli tRNATyr contains pseudouridine at position 35, the second position of the anticodon and adjacent to Q, ms2i6A at position 37, and Ψ at position 39. Loss of Ψ at the rarely modified position 35 compromises translational fidelity in vitro, implying that some ASLs need even more than three modified nucleosides for accurate function.115 Like many other large modifications, Q causes a steric effect in the ASL, influencing the structure of the loop. The effect is largely mediated by hydrogen bonding within the ASL, which has been shown to maintain the structure of the ASL in silico, thus pre-structuring the ASL for codon recognition in the ribosome. The electrostatic potential of Q compared with G is important. Due to the intramolecular hydrogen bond between O6 and the quaternary amine of the aminomethyl side chain, the electronegativity of O6 in Q is reduced compared with O6 in G. In G, O6 acts as a hydrogen bond acceptor, however, this is not the case for Q. The two amines act as hydrogen bond donors in both species. The side-chain of Q can also engage in hydrogen bonding with the backbone of U33, which in turn enhances the U33○C36 interaction, stabilizing the ASL conformation. This same effect is absent or reduced if Q is replaced with G.112 The pre-structuring is further enhanced by the large modification wyosine, (3,4-dihydro-4,6-dimethyl-3-β-D-ribofuranosyl-9H-imidazo[1,2-α]purin-9-one), imG, also derived from G and found at position 37 along with other derivatives often paramount to tRNA structure and function.112
The high degree of hydrophobicity of wyosine (Fig. 3A) and its derivatives enhances the nucleoside's ability to form hydrophobic interactions such as base stacking, which enhances codon binding stability. The bulky residue prevents the intraloop hydrogen bond between residues 32 and 37, thus promoting proper pre-structuring of the ASL.116,117 In mitochondrial tRNAs, imG37 or its derivatives are commonly replaced by i6A37 or a derivative thereof, or by a methylated purine, therefore suggesting the evolution of an alternative strategy for establishing the same functional properties by using different chemical moieties in RNA modifications.113,116
The nearly universal modification at position 37 that is required for decoding codons beginning with A, N6-threonylcarbamoyladenosine, t6A37, may be further modified to 2-methylthio-N6-threonylcarbamoyladenosine, ms2t6A37.118 Interestingly, a cyclic derivative of t6A, ct6A, has also been shown to exist, functioning in a manner analogous to the more studied t6A.118,119 Hydrogen bonding between N1 and N11 of t6A lead to a pseudocyclic conformation that enhances the rigidity of the conformation.120 The same hydrogen bonding exists in ct6A, but a further isomerization occurs as well, where C10 and C13 are linked via an ether, thus forming a oxazolidine.118-120 The C14 alcohol of ct6A can hydrogen bond with the N7 of the first codon, an A, increasing the stabilization that enhanced stacking alone provides.118,120,121
Restriction of codon recognition and pre-structuring of the ASL
RNAs are highly flexible molecules with each base containing several bonds about which free rotation can take place. The flexibility and the possibility of not only Watson-Crick base pairing but also wobble or Hoogsteen interactions provide suitable conditions for multiple structures to form. Certain modifications restrict intraloop base pairing interactions, thus conforming the architecture to one suitable for ribosomal decoding. Restriction of codon recognition may become necessary in cases where an unmodified nucleoside could base pair with a near-cognate codon, therefore causing misreading of the codon. U, for instance, can base pair with all of the 4 major nucleosides with a strong preference to purines.18 As such, modification can provide an enhanced specificity. As an example, isoleucine shares a codon box with methionine where AUA, AUC, and AUU code for isoleucine, and AUG codes for methionine, with the exception of mitochondria where AUA also codes for methionine.6,113,122 Two codons can be read by tRNAIle GAU, but the AUA codon requires a different isoacceptor that would also not read AUG as isoleucine. Eukaryotes commonly have a tRNAIle IAU which can read all three codons, but not AUG.122 The genomes of bacteria and some eukaryotic organelles encode a tRNAIle GAU and tRNAIle LAU where L is lysidine (k2C).123 Modification of C to k2C (Fig. 3B) effectively changes the base pairing capabilities of the nucleoside, causing a switch in preference from G to A for tRNAIle. The modification is required to prevent the recognition of the near-cognate AUG codon (Fig. 7).7 Lysidine is a modified cytidine containing a lysine residue in lieu of O2 at the wobble position. The modification lysidine is the recognition determinant for the amino acid specificity in isoleucyl-tRNA synthetase (IleRS) aminoacylation of tRNAIle LAU and not tRNAMet CAU. An intriguing minor tRNAIle ΨAΨ in yeast contains pseudouridine at position 34 instead of lysidine,121 and many archeal species utilize agmatidine [N-(4-carbamimidamidobutyl)-4-imino-1-(b-D-ribofuranosyl)-1,4-dihydro-2-pyrimidinamine] (Fig. 3B) for decoding of the AUA isoleucine codon. Thus, there have been multiple and convergent evolutionary paths to accomplish specificity in decoding through restriction of codon recognition with the help of modified nucleosides.122
Figure 7.
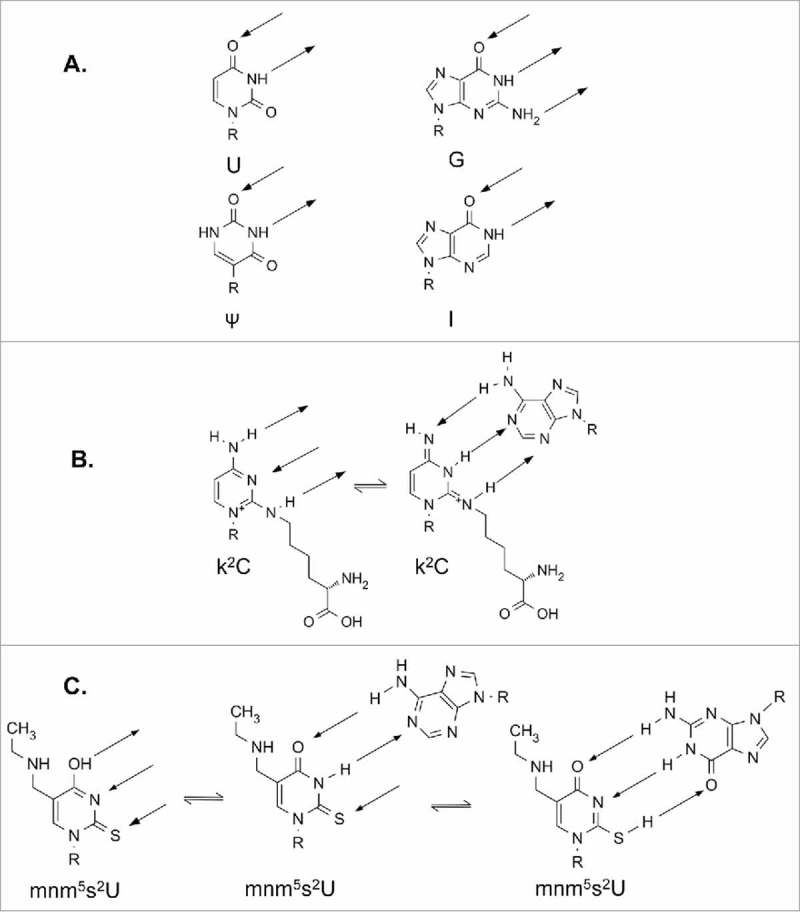
Modifications influence protein and codon recognition of tRNA through altered hydrogen bonding. R represents the ribose sugar. Hydrogen bonding is indicated by arrows. A. Nucleosides commonly found at position 34 of tRNAIle. All 4 bases are capable of recognition by isoleucyl-tRNA synthetase, IleRS. B. Two possible tautomers of lysidine. Structure on the right can be recognized by IleRS, and could putatively recognize A as shown. C. Possible tautomers of mnm5s2U34 of E. coli tRNALys UUU. Likely interactions with A and G are shown.
Tautomerism: A chemical mechanism by which modified nucleosides restrict codon recognition
Tautomers and rare ionic forms of nucleosides have been shown to exist at low populations in vitro;124 it is commonly postulated that the existence of tautomers of modified nucleosides at the ribosome's decoding site has a strong effect on codon recognition, either preventing or enabling it.28 The nature of the C5 modification affects the tautomerism of the uridine and 2-thio modified uridine125 producing either an expanded codon recognition, or one which is restricted. Restriction of codon recognition is a common mechanism for fidelity when a tRNA anticodon can mistakably base pair with a near-cognate codon in a shared codon box. In contrast, expansion of codon recognition by tRNA is often required when an interaction of an unmodified nucleoside would either lack the base pairing ability, thermodynamic stability, or the required structural orientation to achieve accuracy and efficiency of translation.18
Both lysidine and agmatidine can exist in several tautomeric forms. The exact tautomeric form through which cognate codon recognition can occur still needs to be elucidated.122 However, it is a tautomer of lysidine that disallows recognition of the near-cognate AUG codon. The modification contains a secondary amine in place of oxygen at the two position of C, thus switching a hydrogen bond acceptor into a donor. The two tautomers (Fig. 7B) have different base pairing capabilities due to distinct local electron densities. Interestingly, neither tautomer is ideal for base pairing with G, but the non-aromatic tautomer is capable of recognizing A; the primary amine at position 4 on the ring is switched into an imine, thereby enabling it to function as a hydrogen bond acceptor. Similarly, N3 of the base becomes a hydrogen bond donor, thus enabling base pairing with A (Fig. 7B).126 Agmatidine forms a similar tautomer as lysidine.122,126 The hydrogen bonding capability of modified nucleosides commonly found at the wobble position of tRNAIle illustrates how small changes in the chemistry of a single nucleoside can have broad and highly specific effects, in this case altering codon recognition and specificity while simultaneously preserving aaRS recognition.121
Tautomers of modified nucleosides expand reading of synonymous codons
U•A base pairs are thermodynamically much weaker than C•G base pairs. In tRNAs such as tRNALys UUU this problem is exacerbated, as all anticodon to codon base pairs are U•A pairs. Furthermore, in the absence of modifications, the U-rich ASL cannot effectively form stacking interactions to stabilize its structure nor negate intraloop hydrogen bonding that effectively condenses the size of the ASL.117 The nearly ubiquitously modified, invariable purine at position 37 can alleviate the thermodynamic penalty of a pyrimidine rich anticodon. In tRNALys UUU the purine at position 37 is modified to N6-threonylcarbamoyladenosine, t6A or a derivative thereof such as 2-methylthio-N6-threonylcarbamoyladenosine, ms2t6A37 found in mammalian tRNALys3 UUU. Additionally, Ψ at position 39 is important for function.127 The wobble position is extensively modified with an xm5s2U type of modification, illustrating the importance of having three modified nucleosides in the anticodon loop for at least this tRNA.28,128
Lysine is coded by AAA and AAG codons. In many organisms, as well as in organelles, a single tRNALys UUU reads both codons. In mammals, two isoacceptors, tRNALys1,2 CUU, decode the AAG codon, and a third, tRNALys UUU, translates both codons. The wobble xm5s2U modification is required for the efficient and specific recognition of both lysine codons, and for translocation on the ribosome.129 The xm5s2U34 modifications, such as bacterial tRNA modification mnm5s2U34, can undergo keto-enol tautomerism (Fig. 7C), which allows recognition of both AAA and AAG by expanding the hydrogen bonding capabilities of the nucleoside. Similarly, expanded codon recognition could be achieved via ionization of the nucleoside. In the case of mnm5s2U, the secondary amine in the sidechain could be positively charged and the thio-group negatively charged. This zwitterion would have the same hydrogen bonding qualities as the tautomer (Fig. 7C).130
Group (VI) elements at the 2 position of U – the influence of atom size
When U is found at tRNA's position 34 it is nearly always modified. Modifications of U at the wobble position include 2-thiouridine (s2U) and 2-selenouridine (se2U) (Fig. 8)131 that affect both ribose sugar pucker as well as the glycosidic χ angle resulting in a restrictive reading of codons. Both sulfur as well as selenium have similar chemical properties to the carbonyl oxygen at the C2 position of U. However, the larger size of sulfur and selenium elicit different steric properties on their environment, being approximately 40% and 100% larger than oxygen, respectively (Fig. 8). Also the electronegativity of the substituent atoms (sulfur and selenium) is smaller than that of oxygen. As would be expected, the modified U exhibits a dominant anti conformation. The larger atomic radii of these modifications also affect the nucleoside's sugar pucker particularly when the glycosidic bond angle χ is in the anti conformation. While the ribose of unmodified uridines populate equally the C3′ and C2′ endo conformations, the ribose sugar pucker of s2U and se2U exist preferentially in the C3′ endo conformation only occasionally switching to the C2′ endo pucker. The large size difference in the Van der Waal radius of the group IV elements bonded to C2 of U forces the sugar to adopt the C’3 endo conformation; a conformation that has been principally associated with increased stability through enhanced stacking.28 In the case of the unmodified uridine, the carbonyl oxygen at C2 can form a hydrogen bond with the 2′OH group on the ribose thus stabilizing the C2′ endo conformation of the ribose (Fig. 8A). The less electronegative S or Se have a reduced propensity for such hydrogen bond interactions, and combined with the steric effects introduced by the larger size of the atoms, these modifications push the sugar pucker distribution toward C3′endo (Fig. 8B and 8C). Thus, the conformation of uridines at wobble position 34 and modified with s2 and se2 is anti, C3′ endo, and the base pairing is preferentially to A, rather than wobbling to G.
Figure 8.
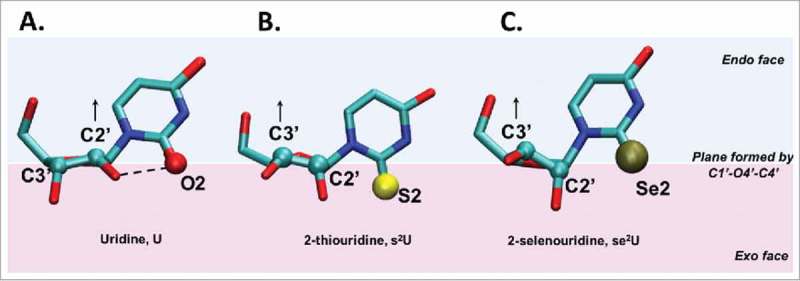
Sulfur and selenium atomic radii affect sugar pucker when modifying the C2 position of uridine. A. Uridine, B. 2-thiouridine and C. 2-selenouridine nucleosides are shown. All 3 nucleosides are depicted in their anti conformation. The plane defined by atoms C1′-O4′-C4′ of the sugar separates the endo face (the side in which C5′ projects from the plane) from the exo face. In the case of uridine, the sugar pucker is C2′ endo as indicated by atom C2′ in the endo face (above the C1′-O4′-C4′ plane), while in 2-thiouridine and 2-selenouridine, the relative increase in atomic radius of substituent atom X at position C2 and the weakening of the hydrogen bonding interaction between 2′OH and X (where X – O2, S2 or Se2) shifts the equilibrium toward a C3′ endo conformation.
Discussion
We and others have extolled the virtues of modifications at wobble position 34 and the invariant purine 3′-adjacent to the anticodon at position 37. Wobble base pairing does not appear to occur at positions other than tRNA position 34 with rare exception such as arthropod mitochondrial translation of the codon AGG as lysine instead of serine (invertebrates) or arginine (standard Universal Genetic Code) in which there is position 35 wobble base pairing, U35○G2.132 Modifications at tRNA's nucleosides 34 and 37 frame the anticodon and pre-structure the ASL for decoding. In doing so, they reduce energy barriers to conformational change required for ribosomal A-site binding, maintain the translational reading frame and either expand or restrict cognate and wobble codon recognition. In S cerevisiae, loss of either the xm5 or s2 modifications of mcm5s2U34 increases observation of +1 translational frameshifts,133 probably caused by a decrease in the affinity of the tRNA for the ribosomal A-site and an impaired translocation.9,129 The nearly ubiquitous t6A37 modification at position 37 in tRNAs responding to codons beginning with A, ANN, enhances base stacking of U36 with A1 of the codon, as well as prevents intraloop base pairing within the ASL.115 Thereby, t6A37 facilitates the U-turn and presentation of the anticodon to the codon on the ribosome. t6A37 also facilitates ribosomal codon binding and maintains the translational frame.26,126 Other modifications at position 37, particularly in tRNAs responding to codons beginning with U, UNN, such as wyosine, contribute similarly to base stacking, maintaining the open loop structure and by doing so maintain the translational reading frame.26,126 The fact that the ASL can be stabilized by various purine 37 modifications implies convergent evolutionary pathways that relate not only to the identity of the codon, but also to that of the nucleoside at position 34 and perhaps additionally either 32 or 38/39.
Yet, there appears to be a need for some tRNAs to further modify the anticodon loop for acceptance on the ribosome and codon recognition. For instance, the nucleoside N3-methylcytidine (m3C) was first found in tRNA over 50 years ago134 and appears to be located exclusively at position 32 of the anticodon loop in specific eukaryotic tRNAs.20 In addition, 2-thiocytidine is found at position 32 of specific prokaryotic tRNAs. The s2C32 modification is present in E. coli and Salmonella enterica tRNAArg ICG which decodes CGU/C and CGA in the absence of s2C32, tRNAArg CCG that decodes CGG, tRNAArg with the modified anticodon mnm5UCU that decodes AGA/G, and tRNASer GCU decoding AGC/U.19,39 The presence of s2C32 negates I34 wobbling to A3.39 It may affect translational efficiency of rare codons that are intrinsically inefficient in decoding.19,135 The s2C32 modification has also been found in archaeal tRNA.136 Pseudouridine, Ψ, although present at the anticodon wobble position 34, is another modification found predominantly at positions 32, 38 and 39.137 Pseudouridine in the anticodon loop apparently increases thermal stability but does not affect ribosome mediated codon binding, nor is its N1 position used for hydrogen bonding.127
Multiple modifications within the ASL may provide a redundancy to the structure and conformational dynamics not otherwise achievable by 40+ individual tRNAs meeting ribosome acceptance and unique codon recognition.43 Modification of tRNAs anticodon loop positions 32, 38 or 39, along with positions 34 and 37, in effect complete a triangle of modified nucleosides (Fig. 9). It is interesting that the three point presentation of ASL modifications has modified nucleosides spaced almost equally around the anticodon loop at approximately every other position 32, 34, and 37, or 34, 37 and 39. Looking back at tRNAGly NCC, the strong anticodon-codon, C•G interactions require no modifications, whereas tRNALys UUU, tRNATyr NUA, and tRNAIle NAU have at least three functionally and/or structurally essential modified nucleosides in the ASL. Weak interactions with the codon, such as multiple U•A base pairs, may require far more extensive modifications, where chemically diverse modifications affect similar biologic properties, often through structure. The ribosome bound structures of the tRNA ASLs containing post-transcriptional modifications offer molecular insights into their role in modulating structure, stability and interactions of the tRNA. While the uridine at wobble position 34 is the most heavily modified, positions 32, 37 and 38 also show significant levels of modifications. We have gathered the structures of five tRNAs bound to the ribosome at the A-site, and highlighted the interactions of their modifications (Fig. 10). It is interesting to note that U34 is primarily modified at C5 adjacent to the base-pairing face and hence the modifications do not interfere with its hydrogen bonding to the codon except in cases where the modifications introduce tautomerism. However, the modifications are either polar (as is true in case of tRNALeu 138 and tRNAVal 11) or charged, (tRNALys),44 and therefore participate in hydrogen bonding with neighboring bases or charge-charge interactions with the backbone phosphate group. Modifications on the position 37 purine nucleobase are primarily involved in stacking with the base at position 36 and the base to which it is paired in codon recognition on the ribosome. These modifications are either methylations (tRNAVal) or groups with complex chemistries, like t6A37, which significantly increases its stacking propensity. Enhanced base stacking of position 37 modified purines provides additional stability to the U-turn structure, and increases the binding strength of the ASL when U34 pairs with a near-cognate or non-cognate codon.
Figure 9.
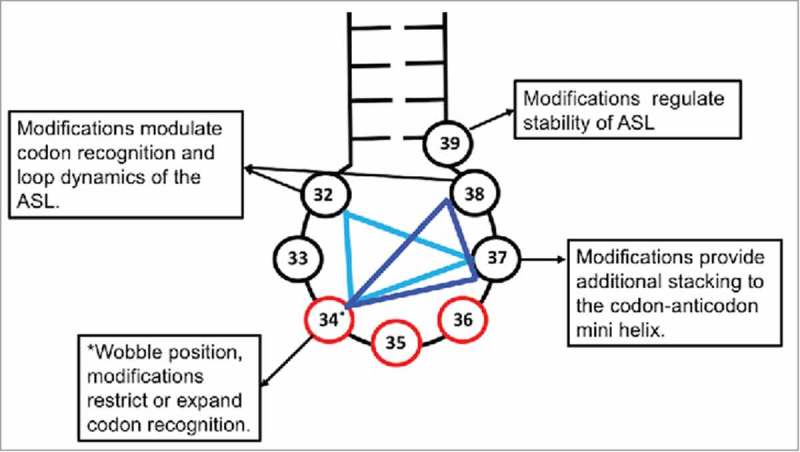
ASL modification triangle. A combination of 3 positions in the ASL (nucleotides 32 through 38) of tRNAs are commonly modified to expand or restrict codon recognition. Position 32, 34 and 37 (light blue) or position 34, 37 and 38 (dark blue) form vertices of the triangle in which modifications work in a co-surgical manner to achieve desired ribosomal binding affinity of the tRNA to different codons. The anticodon is represented in red and the nucleosides in the ASL in black.
Figure 10.
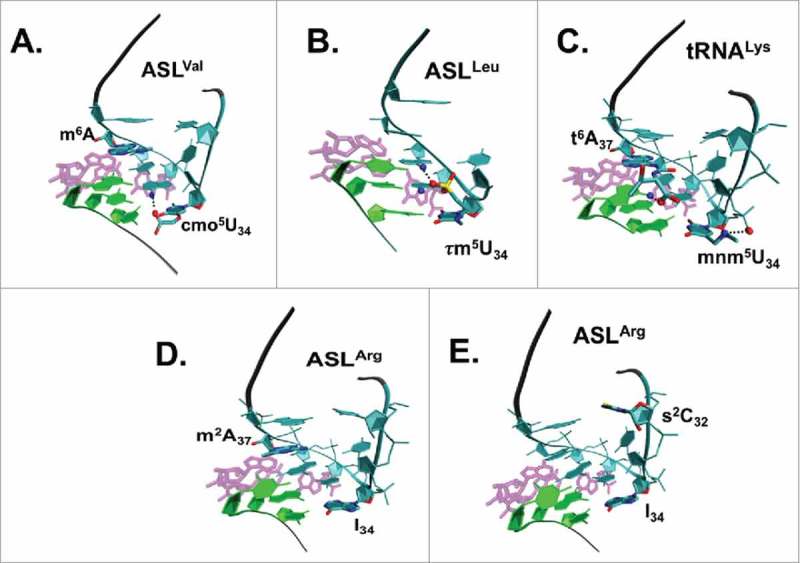
Ribosome-bound structures of the ASLs of 5 tRNA species with modifications. tRNA modified nucleosides at positions 32, 34 and 37 are influential in creating the architecture of the anticodon domain accepted into the ribosome's A-site for cognate and wobble codon binding. The codon on the mRNA is shown in green, the ASL in cyan with the modifications labeled and the rRNA interactions shown in magenta. Possible interactions of the modified groups are represented using a dashed line. A. In ASLVal UAC bound to codon GUU, (PDB ID: 2UUB), the carboxyl oxygen of cmo5U34 is within hydrogen bonding distance of the amine group on the neighboring A35. B. In ASLLeu UAA bound to codon UUG (PDB ID: 2VQF), two of the three oxygens bonded to the sulfur atom in the τm5U34 can form hydrogen bonding interactions with A35 and A36. C. In ASLLys UUU bound to codon AAA (PDB ID: 1XMQ), there are a salt-bridge between the 5-methylaminomethyl group on U34 and its phosphate group, an enhanced stacking interaction provided by t6A37, and a possible hydrogen bond between the threonyl group of t6A37 and A1 on the mRNA. D. ASLArg2 ICG bound to the codon CGC. The ASL has the two modifications I34, m2A37. E. ASLArg1 ICG bound to the codon CGC. The ASL has the modifications s2C32, I34. When either of the modifications m2A37 or s2C32 are present, then the tRNA is unable to recognize the rare CGA codon.39
The cross-loop interaction between the nucleosides at position 32 and 38 at the beginning of the ASL loop (Fig. 4B) is characterized by a single or bifurcated hydrogen bond.139 The strength of this interaction appears to be carefully modulated. For instance, pseudouridine at position 32 uses a water mediated base-backbone interaction to stabilize the interaction between nucleosides 32 and 38.138 While a stronger interaction, like that of a canonical base pair at this position could lead to a loss in flexibility of the loop domain, a weaker interaction could result in loss of stacking in the ASL resulting in a disruption of the functional U-turn conformation of the ASL. Modifications at the 32nd and 38th positions of the ASL may be important as a possible means of modulating the interaction between the two nucleosides and base stacking in the loop.
We hypothesize that the combined chemistries and conformational dynamics of modified nucleosides located at three positions, positions 34, 37 and one other within the ASL loop (Fig. 9) transform the loop architecture and dynamics to that consistent with the constraints that the ribosome places on all tRNAs. It is important that a stable, yet adaptable, U-turn is maintained for presentation of the anticodon resulting in accurate and efficient codon recognition. Multiple modifications mold the anticodon loop architecture of specific tRNAs into thermally stable, malleable triangles of strength recognizable by mRNA-programmed ribosomes (Fig. 9). In order for the architecture of the anticodon's loop to change, an edge of the triangle must collapse, as in an alteration of the 5′-side and U-turn. This is apparent in the three different anticodon loop conformers of tRNAArg1,2 that are recognized by argininyl-tRNA synthetase with a disrupted U-turn, a solution structure that does not exhibit a U-turn and a conformation on the ribosome in the A-site that has the canonical U-turn.39
Conjecture about the evolution of site- and chemically specific modifications within tRNA's ASL domain relative to their functions can yield insights into their future, investigator-designed applications. RNA polymerases, with few exceptions, in vivo and in vitro, do not accept modified nucleoside triphosphates as substrates for transcription, and if they did the modification would be randomly placed throughout the transcript in response to the template. Therefore, it is feasible to hypothesize that the evolution of post-transcriptionally modified nucleosides occurred after the appearance of proteins that could catalyze their limited existence at specific sites to which each modification would contribute chemistry and conformation to that tRNA's function in translation. In acknowledging today's understanding of the manner in which anticodon domain modifications both expand and restrict recognition of cognate and wobble codons, but not near-cognate codons, it is difficult to comprehend life in which new amino acids would be introduced with limited numbers of tRNAs and necessitating 2-fold degenerate codons. For instance, without the restrictions imposed by modifications, one could conceive of asparagine and lysine, aspartic and glutamic acids, and perhaps arginine and serine being mistakenly incorporated. More likely, tRNAs with unmodified anticodon domains much like the very limited number of 22 tRNAs transcribed in mammalian mitochondria decoded 4-fold degenerate codon boxes without error, but perhaps at reduced efficiencies. In contrast, cytoplasmic and mitochondrial translation of codons from 2-fold degenerate codon boxes (Fig. 1) preceded in the absence of modification with significantly reduced translational fidelity and lack of efficiency until restrictive modifications had evolved. If tRNA anticodon domain modifications facilitated the accurate and efficient entry of new amino acids through split codon boxes in the evolution of proteins as we know them, then why couldn't the emergence of still other amino acid incorporations continue by re-appropriating sense codons?140,141 The use of 4-fold degenerate sense codons to incorporate new amino acids would require the splitting of 4-fold degenerate codons. Perhaps, this could be accomplished by removal of cytoplasmic tRNAs from redundant codon recognition (which does not occur in the mitochondria), restructuring the modification enzyme recognition of specific tRNA species and aminoacyl-tRNA synthetase recognition of tRNA and the non-natural amino acid substrates. The introduction of novel amino acids into proteins portends applications to biomaterial science and medicine. New investigator-designed, protein-based materials and potential therapeutics would become available through biomanufacturing.
In conclusion, The Wobble Hypothesis is being revised continually for as we learn more every year about modified nucleoside chemistry, structure and function and how it reflects on RNA chemistry, structure and function, we understand the subtleties of translation being able to manipulate and apply them. We have discussed the different instances in which wobble base pairs are used to expand/restrict codon recognition by tRNA's anticodon with modified and unmodified wobble position 34, and the importance of modified nucleoside positions outside of anticodon, 32, 37 and 38 (Fig. 11). Wobble and non-canonical base pairs in the ASL domain of tRNA are integral to the translation of the genetic code. They are modulated either by unique structural and dynamic features introduced in a sequence dependent manner by unmodified bases (e.g., tRNAGly with C32/U32) or by nucleoside modifications that can expand or restrict decoding capacity of tRNAs by stabilizing or disrupting native interactions at the decoding center of the ribosome. An in-depth understanding of the origins and mechanisms of the variety of ways by which wobbling is achieved will equip us with the tools to tap into this often overlooked potential of using modified nucleoside contributions to translation for therapeutics and other applications.
Figure 11.
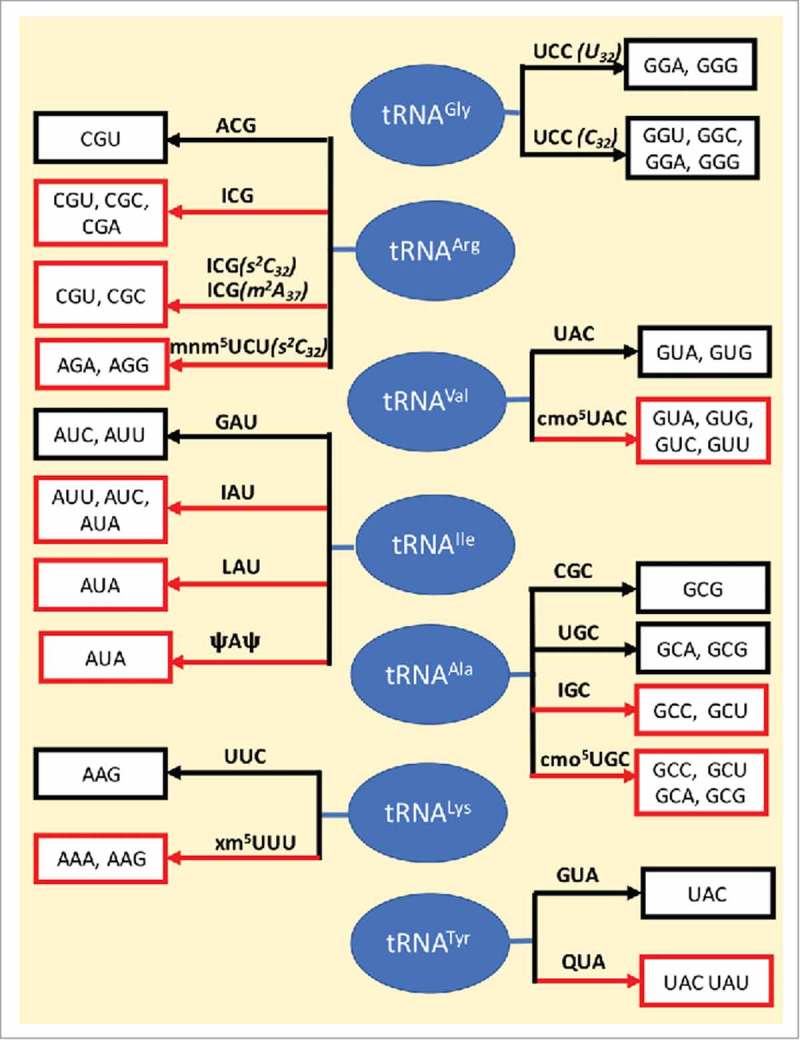
Wobble map. Anticodon domain modified nucleosides play an important role in the wobbling of tRNA's codon recognition, some expanding codon recognition to synonymous codons while others restrict recognition. The different cases of wobbling in the presence and absence of modifications as discussed in this paper, are shown. The black arrow represents no modifications, whereas a red arrow indicates the presence of one or more modifications. The tRNAs are shown in blue, the anticodons are indicated on the arrow and codons decoded by the tRNA bearing the anticodon are enclosed in white boxes.
Disclosure of potential conflicts of interest
The authors report no conflict of interest.
Funding
Research conducted by the authors is supported by grants to P.F.A. from the Department of Defense Medical Research Program under grant number W81XWH-16-1-0428, PR151248, the National Science Foundation under grant number CHE1407042 and the National Institutes of Health, National Institute of General Medical Sciences under grant number 5R01GM110588-03 to Manal Swairjo, P.F.A. co-investigator.
References
- 1.Crick FH. Codon—anticodon pairing: The wobble hypothesis. J Mol Biol. 1966; 19:548–55. doi: 10.1016/S0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 2.Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, Penswick JR, Zamir A. Structure of a ribonucleic acid. Science. 1965;147:1462–5. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- 3.Hall RH. A general procedure for the isolation of “minor” nucleosides from ribonucleic acid hydrolysates. Biochemistry. 1965;4:661–70. doi: 10.1021/bi00880a008. [DOI] [PubMed] [Google Scholar]
- 4.Crick FH. The genetic code—yesterday, today, and tomorrow. Cold Spring Harb Symp Quant Biol. 1966;31:1–9. doi: 10.1101/SQB.1966.031.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Agris PF. Wobble position modified nucleosides evolved to select transfer RNA codon recognition: A modified-wobble hypothesis. Biochimie. 1991;73:1345–9. doi: 10.1016/0300-9084(91)90163-U. [DOI] [PubMed] [Google Scholar]
- 6.Cantara WA, Murphy FV, Demirci H, Agris PF. Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc Natl Acad Sci U S A. 2013;110:10964–9. doi: 10.1073/pnas.1222641110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muramatsu T, Nishikawa K, Nemoto F, Kuchino Y, Nishimura S, Miyazawa T, Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988;336:179–81. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- 8.Takai K, Yokoyama S. Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Res. 2003;31:6383–91. doi: 10.1093/nar/gkg839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vendeix FAP, Murphy FV, Cantara WA, Leszczyńska G, Gustilo EM, Sproat B, Malkiewicz A, Agris PF. Human tRNALys3UUU is pre-structured by natural modifications for cognate and wobble codon binding through keto-enol tautomerism. J Mol Biol. 2012;416:467–85. doi: 10.1016/j.jmb.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama S, Watanabe T, Murao K, Ishikura H, Yamaizumi Z, Nishimura S, Miyazawa T. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc Natl Acad Sci U S A. 1985;82:4905–9. doi: 10.1073/pnas.82.15.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackman JE, Alfonzo JD. Transfer RNA modifications: Nature's combinatorial chemistry playground. Wiley Interdiscip Rev RNA. 2013;4:35–48. doi: 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al.. MODOMICS: A database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013;41:D262–7. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agris PF. The importance of being modified: Roles of modified nucleosides and Mg2+ in RNA structure and function. Prog Nucleic Acid Res Mol Biol. 1996;53:79–129. doi: 10.1016/j.jmb.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Hernandez A, Spears JL, Gaston KW, Limbach PA, Gamper H, Hou YM, Kaiser R, Agris PF, Perona JJ. Structural and mechanistic basis for enhanced translational efficiency by 2-thiouridine at the tRNA anticodon wobble position. J Mol Biol. 2013;425:3888–906. doi: 10.1016/j.jmb.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seno T, Agris PF, Soll D. Involvement of the anticodon region of Escherichia coli tRNAGln and tRNAGlu in the specific interaction with cognate aminoacyl-tRNA synthetase. Alteration of the 2-thiouridine derivatives located in the anticodon of the tRNAs by BrCN or sulfur deprivation. Biochim Biophys Acta. 1974;349:328–38. doi: 10.1016/0005-2787(74)90120-8. [DOI] [PubMed] [Google Scholar]
- 17.Sylvers LA, Rogers KC, Shimizu M, Ohtsuka E, Söll D. A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry. 1993;32:3836–41. doi: 10.1021/bi00066a002. [DOI] [PubMed] [Google Scholar]
- 18.Agris PF, Vendeix FA, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J Mol Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 19.Björk GR, Hagervall TG. Transfer RNA modification: Presence, synthesis, and function. EcoSal Plus. 2014;6:1–68. doi: 10.1128/ecosalplus.ESP-0007-2013. [DOI] [PubMed] [Google Scholar]
- 20.Maraia R, Arimbasseri A. Factors that shape eukaryotic tRNAomes: Processing, modification and anticodon–codon use. Biomolecules. 2017;7:26. doi: 10.3390/biom7010026. [DOI] [Google Scholar]
- 21.Numata T. Mechanisms of the tRNA wobble cytidine modification essential for AUA codon decoding in prokaryotes. Biosci Biotechnol Biochem. 2015;79:347–53. doi: 10.1080/09168451.2014.975185. [DOI] [PubMed] [Google Scholar]
- 22.Ranjan N, Rodnina MV. tRNA wobble modifications and protein homeostasis. Translation. 2016;4:e1143076. doi: 10.1080/21690731.2016.1143076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schweizer U, Bohleber S, Fradejas-Villar N. The modified base isopentenyladenosine and its derivatives in tRNA. RNA Biol. 2017;17:1–12. doi: 10.1080/15476286.2017.1294309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuorto F, Lyko F. Genome recoding by tRNA modifications. Open Biol. 2016;6:160287. doi: 10.1098/rsob.160287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agris PF. Bringing order to translation: The contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008;9:629–35. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn CD. RNA versatility governs tRNA function: Why tRNA flexibility is essential beyond the translation cycle. Bioessays. 2016;38:465–73. doi: 10.1002/bies.201500190. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt PG, Sierzputowska-Gracz H, Agris PF. Internal motions in yeast phenylalanine transfer RNA from 13C NMR relaxation rates of modified base methyl groups: A model-free approach. Biochemistry. 1987;26:8529–34. doi: 10.1021/bi00400a006. [DOI] [PubMed] [Google Scholar]
- 28.Väre V, Eruysal E, Narendran A, Sarachan K, Agris P. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules. 2017;7:29. doi: 10.3390/biom7010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weixlbaumer A, Murphy FVt, Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, Ramakrishnan V. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat Struct Mol Biol. 2007;14:498–502. doi: 10.1038/nsmb1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Walker RC, Phizicky EM, Mathews DH. Influence of sequence and covalent modifications on yeast tRNA dynamics. J Chem Theory Comput. 2014;10:3473–83. doi: 10.1021/ct500107y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Čavužić M, Liu Y. Biosynthesis of sulfur-containing tRNA modifications: A comparison of bacterial, archaeal, and eukaryotic pathways. Biomolecules. 2017;7:27. doi: 10.3390/biom7010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duechler M, Leszczyńska G, Sochacka E, Nawrot B. Nucleoside modifications in the regulation of gene expression: Focus on tRNA. Cell Mol Life Sci. 2016;73:3075–95. doi: 10.1007/s00018-016-2217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ehrenhofer-Murray A. Cross-talk between Dnmt2-dependent tRNA methylation and queuosine modification. Biomolecules. 2017;7:14. doi: 10.3390/biom7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hori H. Transfer RNA methyltransferases with a SpoU‐TrmD (SPOUT) fold and their modified nucleosides in tRNA. Biomolecules. 2017;7:23. doi: 10.3390/biom7010023. [DOI] [Google Scholar]
- 35.Nakai Y, Nakai M, Yano T. Sulfur modifications of the wobble U34 in tRNAs and their intracellular localization in eukaryotic cells. Biomolecules. 2017;7:17. doi: 10.3390/biom7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rintala-Dempsey AC, Kothe U. Eukaryotic stand-alone pseudouridine synthases - RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 2017;3:1–12. doi: 10.1080/15476286.2016.1276150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaffrath R, Leidel SA. Wobble uridine modifications-a reason to live, a reason to die?! RNA Biol. 2017;23:1–14. doi: 10.1080/15476286.2017.1295204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Haute L, Powell CA, Minczuk M. Dealing with an unconventional genetic code in mitochondria: The biogenesis and pathogenic defects of the 5-formylcytosine modification in mitochondrial tRNAMet. Biomolecules. 2017;7:24. doi: 10.3390/biom7010024. [DOI] [Google Scholar]
- 39.Cantara WA, Bilbille Y, Kim J, Kaiser R, Leszczyńska G, Malkiewicz A, Agris PF. Modifications modulate anticodon loop dynamics and codon recognition of E. coli tRNAArg1,2. J Mol Biol. 2012;416:579–97. doi: 10.1016/j.jmb.2011.12.054. [DOI] [PubMed] [Google Scholar]
- 40.Grosjean H, Westhof E. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016;44:8020–40. doi: 10.1093/nar/gkw608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou Y-M, Gamper H, Yang W. Post-transcriptional modifications to tRNA—a response to the genetic code degeneracy. RNA. 2015;21:642–4. doi: 10.1261/rna.049825.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klassen R, Ciftci A, Funk J, Bruch A, Butter F, Schaffrath R. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res. 2016;44:10946–59. doi: 10.1093/nar/gkw705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klassen R, Schaffrath R. Role of pseudouridine formation by Deg1 for functionality of two glutamine isoacceptor tRNAs. Biomolecules. 2017;7:E8. doi: 10.3390/biom7010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy FV 4th, Ramakrishnan V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat Struct Mol Biol. 2004;11:1251–2. doi: 10.1038/nsmb866. [DOI] [PubMed] [Google Scholar]
- 45.van der Gulik PT, Hoff WD. Unassigned codons, nonsense suppression, and anticodon modifications in the evolution of the genetic code. J Mol Evol. 2011;73:59–69. doi: 10.1007/s00239-011-9470-3. [DOI] [PubMed] [Google Scholar]
- 46.Yarian C, Townsend H, Czestkowski W, Sochacka E, Malkiewicz AJ, Guenther R, Miskiewicz A, Agris PF. Accurate translation of the genetic code depends on tRNA modified nucleosides. J Biol Chem. 2002;277:16391–5. doi: 10.1074/jbc.M200253200. [DOI] [PubMed] [Google Scholar]
- 47.Stahl S, Paddock GV, Abelson J. Nucleotide sequence determination of bacteriophage T4 glycine transfer ribonucleic acid. Nucleic Acids Res. 1974;1:1287–304. doi: 10.1093/nar/1.10.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Claesson C, Samuelsson T, Lustig F, Boren T. Codon reading properties of an unmodified transfer RNA. FEBS Lett. 1990;273:173–6. doi: 10.1016/0014-5793(90)81077-2. [DOI] [PubMed] [Google Scholar]
- 49.Heckman JE, Sarnoff J, Alzner-DeWeerd B, Yin S, RajBhandary UL. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980;77:3159–63. doi: 10.1073/pnas.77.6.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andachi Y, Yamao F, Muto A, Osawa S. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J Mol Biol. 1989;209:37–54. doi: 10.1016/0022-2836(89)90168-X. [DOI] [PubMed] [Google Scholar]
- 51.Barrell BG, Anderson S, Bankier AT, de Bruijn MH, Chen E, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, et al.. Different pattern of codon recognition by mammalian mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980;77:3164–6. doi: 10.1073/pnas.77.6.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonitz SG, Berlani R, Coruzzi G, Li M, Macino G, Nobrega FG, Nobrega MP, Thalenfeld BE, Tzagoloff A. Codon recognition rules in yeast mitochondria. Proc Natl Acad Sci U S A. 1980;77:3167–70. doi: 10.1073/pnas.77.6.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guindy YS, Samuelsson T, Johansen TI. Unconventional codon reading by Mycoplasma mycoides tRNAs as revealed by partial sequence analysis. Biochem J. 1989;258:869–73. doi: 10.1042/bj2580869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, Umesono K, Shiki Y, Takeuchi M, Chang Z, et al.. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322:572–4. doi: 10.1038/322572a0 [DOI] [Google Scholar]
- 55.Samuelsson T, Elias P, Lustig F, Guindy YS. Cloning and nucleotide sequence analysis of transfer RNA genes from Mycoplasma mycoides. Biochem J. 1985;232:223. doi: 10.1042/bj2320223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samuelsson T, Guindy YS, Lustig F, Boren T, Lagerkvist U. Apparent lack of discrimination in the reading of certain codons in Mycoplasma mycoides. Proc Natl Acad Sci U S A. 1987;84:3166–70. doi: 10.1073/pnas.84.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al.. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986;5:2043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rogalski M, Karcher D, Bock R. Superwobbling facilitates translation with reduced tRNA sets. Nat Struct Mol Biol. 2008;15:192–8. doi: 10.1038/nsmb.1370. [DOI] [PubMed] [Google Scholar]
- 59.Carbon J, Squires C. Studies on genetically altered transfer RNA species in Escherichia coli. Cancer Res. 1971;31:663–6. [PubMed] [Google Scholar]
- 60.O'Connor M. tRNA imbalance promotes −1 frameshifting via near-cognate decoding1. J Mol Biol. 1998;279:727–36. doi: 10.1006/jmbi.1998.1832. [DOI] [PubMed] [Google Scholar]
- 61.Lustig F, Boren T, Claesson C, Simonsson C, Barciszewska M, Lagerkvist U. The nucleotide in position 32 of the tRNA anticodon loop determines ability of anticodon UCC to discriminate among glycine codons. Proc Natl Acad Sci U S A. 1993;90:3343–7. doi: 10.1073/pnas.90.8.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Claesson C, Lustig F, Boren T, Simonsson C, Barciszewska M, Lagerkvist U. Glycine codon discrimination and the nucleotide in position 32 of the anticodon loop. J Mol Biol. 1995;247:191–6. doi: 10.1006/jmbi.1994.0132. [DOI] [PubMed] [Google Scholar]
- 63.Samuelsson T, Axberg T, Boren T, Lagerkvist U. Unconventional reading of the glycine codons. J Biol Chem. 1983;258:13178–84. [PubMed] [Google Scholar]
- 64.Olejniczak M, Uhlenbeck OC. tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie. 2006;88:943–50. doi: 10.1016/j.biochi.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 65.Chang AT, Nikonowicz EP. Solution NMR analyses of the anticodon arms of proteinogenic and non-proteinogenic tRNAGly. Biochemistry. 2012;51:3662–74. doi: 10.1021/bi201900j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Olejniczak M, Dale T, Fahlman RP, Uhlenbeck OC. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat Struct Mol Biol. 2005;12:788–93. doi: 10.1038/nsmb978. [DOI] [PubMed] [Google Scholar]
- 67.Inagaki Y, Kojima A, Bessho Y, Hori H, Ohama T, Osawa S. Translation of synonymous codons in family boxes by Mycoplasma capricolum tRNAs with unmodified uridine or adenosine at the first anticodon position. J Mol Biol. 1995;251:486–92. doi: 10.1006/jmbi.1995.0450. [DOI] [PubMed] [Google Scholar]
- 68.Riddle DL, Carbon J. Frameshift suppression: A nucleotide addition in the anticodon of a glycine transfer RNA. Nature New Biol. 1973;242:230–4. doi: 10.1038/newbio242230a0. [DOI] [PubMed] [Google Scholar]
- 69.Atkins JF, Björk GR. A gripping tale of ribosomal frameshifting: Extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol Biol Rev. 2009;73:178–210. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cabello-Villegas J, Nikonowicz EP. Solution structure of ψ32-modified anticodon stem-loop of Escherichia coli tRNAPhe. Nucleic Acids Res. 2005;33:6961–71. doi: 10.1093/nar/gki1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sibler AP, Dirheimer G, Martin RP. Codon reading patterns in Saccharomyces cerevisiae mitochondria based on sequences of mitochondrial tRNAs. FEBS Lett. 1986;194:131–8. doi: 10.1016/0014-5793(86)80064-3. [DOI] [PubMed] [Google Scholar]
- 72.Andachi Y, Yamao F, Iwami M, Muto A, Osawa S. Occurrence of unmodified adenine and uracil at the first position of anticodon in threonine tRNAs in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1987;84:7398–402. doi: 10.1073/pnas.84.21.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boren T, Elias P, Samuelsson T, Claesson C, Barciszewska M, Gehrke CW, Kuo KC, Lustig F. Undiscriminating codon reading with adenosine in the wobble position. J Mol Biol. 1993;230:739–49. doi: 10.1006/jmbi.1993.1196. [DOI] [PubMed] [Google Scholar]
- 74.Takai K, Takaku H, Yokoyama S. In vitro codon-reading specificities of unmodified tRNA molecules with different anticodons on the sequence background of Escherichia coli tRNASer1. Biochem Biophys Res Commun. 1999;257:662–7. doi: 10.1006/bbrc.1999.0538. [DOI] [PubMed] [Google Scholar]
- 75.Chen P, Qian Q, Zhang S, Isaksson LA, Björk GR. A cytosolic tRNA with an unmodified adenosine in the wobble position reads a codon ending with the non-complementary nucleoside cytidine. J Mol Biol. 2002;317:481–92. doi: 10.1006/jmbi.2002.5435. [DOI] [PubMed] [Google Scholar]
- 76.Topal MD, Fresco JR. Complementary base pairing and the origin of substitution mutations. Nature. 1976;263:285–9. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- 77.Lagerkvist U. “Two out of three:” An alternative method for codon reading. Proc Natl Acad Sci U S A. 1978;75:1759–62. doi: 10.1073/pnas.75.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Balasubramanian R, Seetharamulu P. A conformational rationale for the wobble behaviour of the first base of the anticodon triplet in tRNA. J Theor Biol. 1983;101:77–86. doi: 10.1016/0022-5193(83)90273-4. [DOI] [PubMed] [Google Scholar]
- 79.Henkin TM. Riboswitch RNAs: Using RNA to sense cellular metabolism. Genes Dev. 2008;22:3383–90. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Henkin TM, Glass BL, Grundy FJ. Analysis of the Bacillus subtilis tyrS gene: Conservation of a regulatory sequence in multiple tRNA synthetase genes. J Bacteriol. 1992;174:1299–306. doi: 10.1128/jb.174.4.1299-1306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010;584:318–24. doi: 10.1016/j.febslet.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol. 2012;4:a003566. doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA. 2008;14:717–35. doi: 10.1261/rna.819308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bumsted RM, Dahl JL, Söll D, Strominger JL. Biosynthesis of the peptidoglycan of bacterial cell walls. X. Further study of the glycyl transfer ribonucleic acids active in peptidoglycan synthesis in Staphylococcus aureus. J Biol Chem. 1968;243:779–82. [PubMed] [Google Scholar]
- 85.Giannouli S, Kyritsis A, Malissovas N, Becker HD, Stathopoulos C. On the role of an unusual tRNAGly isoacceptor in Staphylococcus aureus. Biochimie. 2009;91:344–51. doi: 10.1016/j.biochi.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 86.Roberts RJ. Staphylococcal transfer ribonucleic acids: II. Sequence analysis of isoaccepting glycine transfer ribonucleic acids IA and IB from Staphylococcus epidermidis Texas 26. J Biol Chem. 1974;249:4787–96. [PubMed] [Google Scholar]
- 87.Roberts RJ, Lovinger GG, Tamura T, Strominger JL. Staphylococcal transfer ribonucleic acids: I. Isolation and purification of the isoaccepting glycine transfer ribonucleic acids from Staphylococcus epidermidis Texas 26. J Biol Chem. 1974;249:4781–6. [PubMed] [Google Scholar]
- 88.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–53. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–72. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 90.Nissen P, Thirup S, Kjeldgaard M, Nyborg J. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure. 1999;7:143–56. doi: 10.1016/S0969-2126(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 91.Sanderson LE, Uhlenbeck OC. The 51–63 base pair of tRNA confers specificity for binding by EF-Tu. RNA. 2007;13:835–40. doi: 10.1261/rna.485307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Anderson JS, Matsuhashi M, Haskin MA, Strominger JL. Lipid-phosphoacetylmuramyl-pentapeptide and lipid-phosphodisaccharide-pentapeptide: Presumed membrane transport intermediates in cell wall synthesis. Proc Natl Acad Sci U S A. 1965;53:881–9; doi: 10.1073/pnas.53.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Berger-Bachi B. Factors affecting methicillin resistance in Staphylococcus aureus. Int J Antimicrob Agents. 1995;6:13–21; doi: 10.1016/0924-8579(95)00021-Y. [DOI] [PubMed] [Google Scholar]
- 94.Bass BL, Nishikura K, Keller W, Seeburg PH, Emeson RB, O'Connell MA, Samuel CE, Herbert A. A standardized nomenclature for adenosine deaminases that act on RNA. RNA. 1997;3:947–9. [PMC free article] [PubMed] [Google Scholar]
- 95.Alseth I, Dalhus B, Bjørås M. Inosine in DNA and RNA. Curr Opin Genet Dev 2014;26:116–23. doi: 10.1016/j.gde.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 96.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–9. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 97.Gerber AP, Keller W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science. 1999;286:1146–9. doi: 10.1126/science.286.5442.1146. [DOI] [PubMed] [Google Scholar]
- 98.Tsutsumi S, Sugiura R, Ma Y, Tokuoka H, Ohta K, Ohte R, Noma A, Suzuki T, Kuno T. Wobble inosine tRNA modification is essential to cell cycle progression in G1/S and G2/M transitions in fission yeast. J Biol Chem. 2007;282:33459–65. doi: 10.1074/jbc.M706869200. [DOI] [PubMed] [Google Scholar]
- 99.Curran JF. Decoding with the A:I wobble pair is inefficient. Nucleic Acids Res. 1995;23:683–8. doi: 10.1093/nar/23.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jager G, Leipuviene R, Pollard MG, Qian Q, Björk GR. The conserved Cys-X1-X2-Cys motif present in the TtcA protein is required for the thiolation of cytidine in position 32 of tRNA from Salmonella enterica serovar Typhimurium. J Bacteriol. 2004;186:750–7. doi: 10.1128/JB.186.3.750-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vendeix FAP, Munoz AM, Agris PF. Free energy calculation of modified base-pair formation in explicit solvent: A predictive model. RNA. 2009;15:2278–87. doi: 10.1261/rna.1734309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. [DOI] [PubMed] [Google Scholar]
- 103.Quax TEF, Claassens NJ, Söll D, van der Oost J. Codon bias as a means to fine-tune gene expression. Mol Cell. 2015;59:149–61. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Näsvall SJ, Chen P, Björk GR. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAProcmo5UGG promotes reading of all four proline codons in vivo. RNA. 2004;10:1662–73. doi: 10.1261/rna.7106404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Näsvall SJ, Chen P, Björk GR. The wobble hypothesis revisited: Uridine-5-oxyacetic acid is critical for reading of G-ending codons. RNA. 2007;13:2151–64. doi: 10.1261/rna.731007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kawai G, Ue H, Yasuda M, Sakamoto K, Hashizume T, McCloskey JA, Miyazawa T, Yokoyama S. Relation between functions and conformational characteristics of modified nucleosides found in tRNAs. Nucleic Acids Symp Ser. 1991;(25):49–50. [PubMed] [Google Scholar]
- 107.Kawai G, Yamamoto Y, Kamimura T, Masegi T, Sekine M, Hata T, Iimori T, Watanabe T, Miyazawa T, Yokoyama S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry. 1992;31:1040–6. doi: 10.1021/bi00119a012. [DOI] [PubMed] [Google Scholar]
- 108.Noon KR, Guymon R, Crain PF, McCloskey JA, Thomm M, Lim J, Cavicchioli R. Influence of temperature on tRNA modification in Archaea: Methanococcoides burtonii (Optimum Growth Temperature [Topt], 23°C) and Stetteria hydrogenophila (Topt, 95°C). J Bacteriol. 2003;185:5483–90. doi: 10.1128/JB.185.18.5483-5490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hillen W, Egert E, Lindner HJ, Gassen HG, Vorbruggen H. 5-methoxyuridine: The influence of 5-substituents on the keto-enol tautomerism of the 4-carbonyl group. J Carbohydr Nucleos Nucleot. 1978;5:23–32 [Google Scholar]
- 110.Dao V, Guenther R, Malkiewicz A, Nawrot B, Sochacka E, Kraszewski A, Jankowska J, Everett K, Agris PF. Ribosome binding of DNA analogs of tRNA requires base modifications and supports the “extended anticodon.” Proc Natl Acad Sci U S A. 1994;91:2125–9. doi: 10.1073/pnas.91.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vendeix FA, Dziergowska A, Gustilo EM, Graham WD, Sproat B, Malkiewicz A, Agris PF. Anticodon domain modifications contribute order to tRNA for ribosome-mediated codon binding. Biochemistry. 2008;47:6117–29. doi: 10.1021/bi702356j. [DOI] [PubMed] [Google Scholar]
- 112.Morris RC, Brown KG, Elliott MS. The effect of queuosine on tRNA structure and function. J Biomol Struct Dyn. 1999;16:757–74. doi: 10.1080/07391102.1999.10508291. [DOI] [PubMed] [Google Scholar]
- 113.Suzuki T, Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014;42:7346–57. doi: 10.1093/nar/gku390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Iwata-Reuyl D. Biosynthesis of the 7-deazaguanosine hypermodified nucleosides of transfer RNA. Bioorg Chem. 2003;31:24–43. doi: 10.1016/S0045-2068(02)00513-8. [DOI] [PubMed] [Google Scholar]
- 115.Addepalli B, Limbach PA. Pseudouridine in the anticodon of Escherichia coli tRNATyr(QΨA) is catalyzed by the dual specificity enzyme RluF. J Biol Chem. 2016;291:22327–37. doi: 10.1074/jbc.M116.747865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.de Crécy-Lagard V, Brochier-Armanet C, Urbonavičius J, Fernandez B, Phillips G, Lyons B, Noma A, Alvarez S, Droogmans L, Armengaud J, et al.. Biosynthesis of wyosine derivatives in tRNA: An ancient and highly diverse pathway in Archaea. Mol Biol Evol. 2010;27:2062–77. doi: 10.1093/molbev/msq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stuart JW, Gdaniec Z, Guenther R, Marszalek M, Sochacka E, Malkiewicz A, Agris PF. Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry. 2000;39:13396–404. doi: 10.1021/bi0013039. [DOI] [PubMed] [Google Scholar]
- 118.Agris PF, Narendran A, Sarachan K, Väre VYP, Eruysal E. The importance of being modified: The role of RNA modifications in translational fidelity In: Chanfreau GF, editor. The enzymes RNA modification. San Diego: (CA, United States: ): Academic Press; 2017. p. 1–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Miyauchi K, Kimura S, Suzuki T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat Chem Biol. 2013;9:105–11; doi: 10.1038/nchembio.1137. [DOI] [PubMed] [Google Scholar]
- 120.Matuszewski M, Sochacka E. Stability studies on the newly discovered cyclic form of tRNA N6-threonylcarbamoyladenosine (ct6A). Bioorg Med Chem Lett. 2014;24:2703–6; doi: 10.1016/j.bmcl.2014.04.048. [DOI] [PubMed] [Google Scholar]
- 121.Senger B, Auxilien S, Englisch U, Cramer F, Fasiolo F. The modified wobble base inosine in yeast tRNAIle is a positive determinant for aminoacylation by isoleucyl-tRNA synthetase. Biochemistry. 1997;36:8269–75. doi: 10.1021/bi970206l. [DOI] [PubMed] [Google Scholar]
- 122.Mandal D, Köhrer C, Su D, Russell SP, Krivos K, Castleberry CM, Blum P, Limbach PA, Söll D, RajBhandary UL. Agmatidine, a modified cytidine in the anticodon of archaeal tRNAIle, base pairs with adenosine but not with guanosine. Proc Natl Acad Sci U S A. 2010;107:2872–7. doi: 10.1073/pnas.0914869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grosjean H, Björk GR. Enzymatic conversion of cytidine to lysidine in anticodon of bacterial isoleucyl-tRNA—an alternative way of RNA editing. Trends Biochem Sci. 2004;29:165–8. doi: 10.1016/j.tibs.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 124.Kimsey IJ, Petzold K, Sathyamoorthy B, Stein ZW, Al-Hashimi HM. Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature. 2015;519:315–20. doi: 10.1038/nature14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sochacka E, Lodyga-Chruscinska E, Pawlak J, Cypryk M, Bartos P, Ebenryter-Olbinska K, Leszczynska G, Nawrot B. C5-substituents of uridines and 2-thiouridines present at the wobble position of tRNA determine the formation of their keto-enol or zwitterionic forms - a factor important for accuracy of reading of guanosine at the 3′-end of the mRNA codons. Nucleic Acids Res. 2017;45:4825–36. doi: 10.1093/nar/gkw1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sambhare SB, Kumbhar BV, Kamble AD, Bavi RS, Kumbhar NM, Sonawane KD. Structural significance of modified nucleosides k2C and t6A present in the anticodon loop of tRNAIle. RSC Advances. 2014;4:14176–88. doi: 10.1039/c3ra47335j [DOI] [Google Scholar]
- 127.Yarian CS, Basti MM, Cain RJ, Ansari G, Guenther RH, Sochacka E, Czerwinska G, Malkiewicz A, Agris PF. Structural and functional roles of the N1- and N3-protons of Ψ at tRNA's position 39. Nucleic Acids Res. 1999;27:3543–9. doi: 10.1093/nar/27.17.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Murphy FV 4th, Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNALysUUU. Nat Struct Mol Biol. 2004;11:1186–91. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- 129.Phelps SS, Malkiewicz A, Agris PF, Joseph S. Modified nucleotides in tRNALys and tRNAVal are important for translocation. J Mol Biol. 2004;338:439–44. doi: 10.1016/j.jmb.2004.02.070. [DOI] [PubMed] [Google Scholar]
- 130.Rozov A, Demeshkina N, Khusainov I, Westhof E, Yusupov M, Yusupova G. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat Commun. 2016;7:10457. doi: 10.1038/ncomms10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Björk GR. Chapter 11: Biosynthesis and function of modified nucleosides In: Söll D, RajBhandary UL, editors. tRNA: Structure, biosynthesis, and function. Washington, DC: (USA: ): American Society for Microbiology; 1995. p. 165–205 [Google Scholar]
- 132.Abascal F, Posada D, Knight RD, Zardoya R. Parallel evolution of the genetic code in arthropod mitochondrial genomes. PLoS Biol. 2006;4:e127; doi: 10.1371/journal.pbio.0040127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Klassen R, Bruch A, Schaffrath R. Independent suppression of ribosomal +1 frameshifts by different tRNA anticodon loop modifications. RNA Biol. 2016;12:1–8. doi: 10.1080/15476286.2016.1267098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hall RH. Isolation of 3-methyluridine and 3-methylcytidine from solubleribonucleic acid. Biochem Biophys Res Commun. 1963;12:361–4. doi: 10.1016/0006-291X(63)90105-0. [DOI] [PubMed] [Google Scholar]
- 135.Gustilo EM, Vendeix FAP, Agris PF. tRNA's modifications bring order to gene expression. Curr Opin Microbiol. 2008;11:134–40. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.McCloskey JA, Graham DE, Zhou S, Crain PF, Ibba M, Konisky J, Söll D, Olsen GJ. Post-transcriptional modification in archaeal tRNAs: Identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 2001;29:4699–706. doi: 10.1093/nar/29.22.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tworowska I, Nikonowicz EP. Base pairing within the ψ32,ψ39-modified anticodon arm of Escherichia coli tRNAPhe. J Am Chem Soc. 2006;128:15570–1. doi: 10.1021/ja0659368. [DOI] [PubMed] [Google Scholar]
- 138.Kurata S, Weixlbaumer A, Ohtsuki T, Shimazaki T, Wada T, Kirino Y, Takai K, Watanabe K, Ramakrishnan V, Suzuki T. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U.G wobble pairing during decoding. J Biol Chem. 2008;283:18801–11. doi: 10.1074/jbc.M800233200. [DOI] [PubMed] [Google Scholar]
- 139.Auffinger P, Westhof E. Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J Mol Biol. 1999;292:467–83. doi: 10.1006/jmbi.1999.3080. [DOI] [PubMed] [Google Scholar]
- 140.Cui Z, Mureev S, Polinkovsky ME, Tnimov Z, Guo Z, Durek T, Jones A, Alexandrov K. Combining sense and nonsense codon reassignment for site-selective protein modification with unnatural amino acids. ACS Synth Biol. 2017;6:535–44. doi: 10.1021/acssynbio.6b00245. [DOI] [PubMed] [Google Scholar]
- 141.Xue H, Wong JT. Future of the genetic code. Life (Basel). 2017;7:E10. doi: 10.3390/life7010010. [DOI] [PMC free article] [PubMed] [Google Scholar]


