ABSTRACT
Posttranslational modification (PTM) is a key mechanism for regulating diverse protein functions, and thus critically affects many essential biological processes. Critical for systematic study of the effects of PTMs is the ability to obtain recombinant proteins with defined and homogenous modifications. To this end, various synthetic and chemical biology approaches, including genetic code expansion and protein chemical modification methods, have been developed. These methods have proven effective for generating site-specific authentic modifications or structural mimics, and have demonstrated their value for in vitro and in vivo functional studies of diverse PTMs. This review will discuss recent advances in chemical biology strategies and their application to various PTM studies.
KEYWORDS: Chemical biology approach, genetic code expansion, posttranslational modification (PTM), protein chemical modification
Introduction
Posttranslational modification (PTM) generally refers to the covalent attachment of functional groups to the amino acid side chains or C- or N- terminus of a protein.1 Such functional groups are highly diverse, comprising more than 200 different types of modifications ranging from small chemical groups (phosphorylation and acetylation) to different types of carbohydrate chains (glycosylation), lipids (lipidation), and even small proteins (ubiquitination).2 These modifications are reversibly modulated primarily through the action of a specific set of modifying and demodifying enzymes (e.g., kinases and phosphatases) and occasionally by certain reactive chemical species. PTM is a widespread mechanism for regulating most eukaryotic proteins. For example, more than half of all human proteins are known to be modified by phosphorylation, one-third are modified by acetylation, and about a quarter are modified by ubiquitination.3 Since PTMs critically influence numerous biological processes, including cell signaling and metabolism, abnormal PTMs, such as aberrant phosphorylation and acetylation, are associated with the genesis and development of many human diseases.4 Therefore, achieving selective and homogenous PTMs in proteins is essential for deciphering cellular function of PTMs and understanding fundamental mechanisms of PTM-mediated biological processes. In this review, we will focus on recent progress in synthetic and chemical biology approaches—including newly emerging technologies that enable selective protein chemical modifications—and their applications in studies of various PTMs that are listed in Fig. 1. For further information, readers are referred to earlier excellent reviews.5-12
Figure 1.
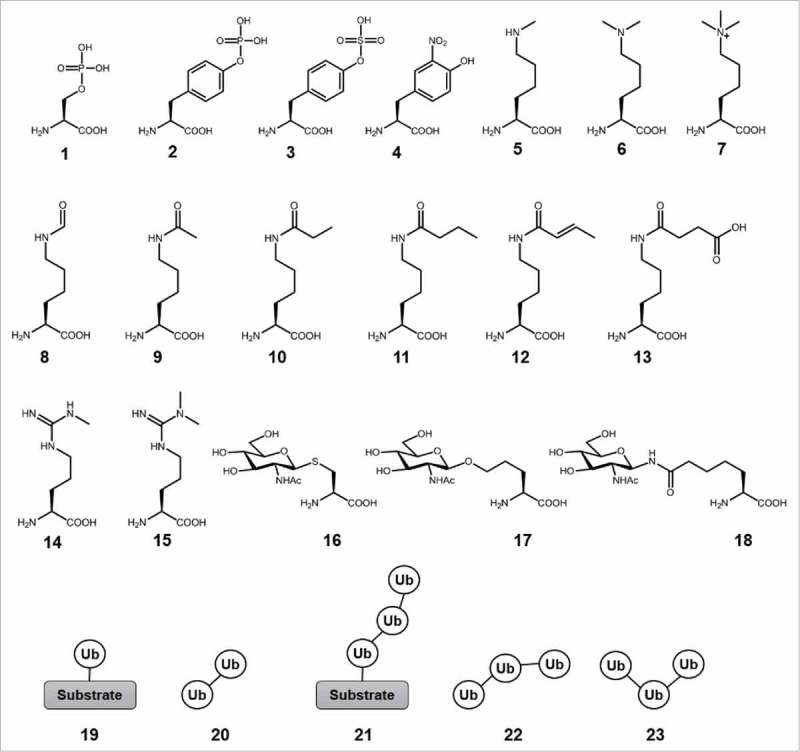
Types of PTMs accessed by chemical biology approaches. Chemical structures of the modified amino acids are shown. 1, phospho-Ser; 2, phospho-Tyr; 3, sulfo-Tyr; 4, nitro-Tyr; 5, monomethyl-Lys; 6, dimethyl-Lys; 7, trimethyl-Lys; 8, formyl-Lys; 9, acetyl-Lys; 10, propionyl-Lys; 11, butyryl-Lys; 12, crotonyl-Lys; 13, succinyl-Lys; 14, monomethyl-Arg; 15, dimethyl-Arg; 16, O-GlcNAc mimic; 17, O-GlcNAc homo-Ser; 18, N-GlcNAc mimic; 19, mono Ub; 20, di Ub; 21, homotypic Ub chain; 22, mixed Ub chain; 23, branched Ub chain.
Synthetic and chemical biology methodologies for PTMs
Chemical ligation methods
Native chemical ligation (NCL) is carried out by nucleophilic attack of the thiol group of an N-terminal Cys residue of a polypeptide on the C-terminal thioester of a synthetic peptide through transthioesterification and then a sulfur-to-nitrogen acyl-transfer reaction to form a native peptide bond, generating full-size recombinant protein (Fig. 2A).13 Because any modifiable residue can be easily inserted into a specific position of a synthetic peptide by solid-phase peptide synthesis (SPPS), recombinant proteins with specific modifications at the N-terminal region can be easily generated. A polypeptide carrying a C-terminal thioester can also be prepared using the self-splicing protein intein, and can be ligated to a synthetic peptide through expressed protein ligation (EPL) to generate recombinant proteins with specific modifications in the C-terminal region.14 Both chemical ligation methods are efficient and practical, although their applications are largely limited to terminal regions of a target protein and occasionally require refolding and oxidation of ligation products.
Figure 2.
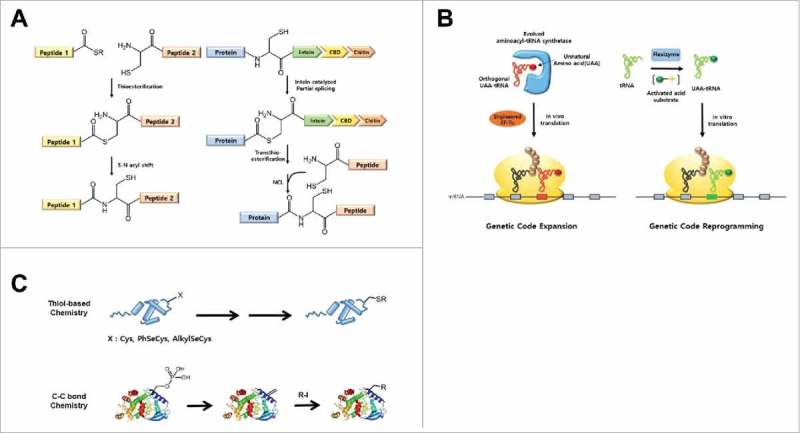
Schematics of chemical biology methodologies for site-specific installation of PTMs in proteins. (A) Native chemical ligation (right) and Expressed protein ligation (left) methods. (B) Genetic code expansion (right) and genetic code reprogramming (left) methods. (C) Chemical modification methods; thiol-based chemistry (top) and a three-step chemoselective carbon-carbon bond conjugation approach (bottom).
Genetic code expansion and reprogramming
In addition to the 20 standard amino acids, nature has expanded the genetic code to include two atypical amino acids, selenocysteine (Sec) and pyrrolysine (Pyl), which are considered the 21st and 22nd amino acids in the genetic code.15 Sec and Pyl are co-translationally inserted into nascent polypeptide chains at specific opal (UGA) and amber (UAG) codons, respectively, through specialized translation machineries.16,17 In addition, reassignment of sense and nonsense codons that are mediated by naturally occurring modified tRNAs and suppressor tRNAs, respectively, has been a vital mechanism for the generation of genetic diversity.18 Genetic code expansion techniques that mimic such natural mechanisms have been developed to more efficiently expedite expansion of the functional diversity of the proteome in the laboratory19 (Fig. 2B). This approach allows site-specific incorporation of a wide range of unnatural amino acids with diverse chemical functionalities into proteins using evolved and engineered orthogonal aminoacyl-tRNA synthetase/tRNA pairs. In particular, because this method enables cotranslational insertion of unnatural amino acids carrying specific functional groups, it is very effective for generating recombinant proteins with selective and homogenous modifications, thereby facilitating in vitro and in vivo functional studies of PTMs, including phosphorylation and acetylation. But this approach relies on the ability to acquire customized orthogonal AARS/tRNA pairs, which inevitably requires extensive, but unguaranteed, molecular evolution efforts. As an alternative, genetic code reprogramming using a flexible tRNA acylation ribozyme (flexizyme) combined with a tailor-made cell-free protein synthesis system has been developed20 (Fig. 2B). Before the era of flexizyme, chemical acylation of tRNA by using T4 RNA ligase was attempted for in vitro amber codon suppression.21 Although this approach is limited to in vitro functional studies of PTMs, it is useful for generating recombinant proteins with diverse modifications without the need for tailored AARS/tRNA pairs, because flexizyme is used for the preparation of aminoacyl-tRNAs,
Selective chemical modification methods
Unlike chemical ligation methods, chemical modification methods directly transform the chemical structure of specific residues in proteins, albeit allowing only for in vitro installation of PTMs. Such chemical modifications have been challenging owing to the presence of various reactive groups, such as amines, carboxylic acids, alcohols and thiols, in target proteins. Recently, thiol-based chemoselective conjugation approaches have been developed for various purposes. In particular, cysteine-based chemistry has been utilized to generate mimics of natural protein modifications22-25 (Fig. 2C). Similarly, a thiol-ene coupling reaction using a dehydroalanine residue converted from either a Cys or Sec derivative (PhSeCys and AlkylSeCys) has been proposed26-28 (Fig. 2C). These methods can be useful for producing various forms of thioether-linked modification mimics, such as methylation, glycosylation and PEGylation, among others.29 Very recently, two research groups, led by Park and Davis, separately demonstrated the first authentic PTM of protein molecules through radical-mediated carbon-carbon bond formation.30,31 Specifically, a three-step versatile strategy (also named the Marking-Activation-Coupling, or MAC strategy) for site-specific installation of authentic PTMs in proteins was established30 (Fig. 2C). First, a phosphoserine (Sep)-containing recombinant protein is made using the phosphoserine orthogonal translation system. The Sep residue is then dephosphorylated to dehydroalanine. Finally, addition of alkyl iodides by Zn-Cu–mediated conjugation enables chemo-selective carbon-carbon bond formation, leading to authentic PTMs in proteins. Alkyl iodide conjugation can also be achieved by a hydride-mediated radical addition reaction.31 These approaches have proved effective in producing recombinant proteins with diverse, authentic modifications or mimics, including several challenging modifications (e.g., lysine tri-methylation; arginine mono- and di-methylation; methyl-glutamine) and O- and N-GlcNac mimics, most of which were not previously accessible.
Selective protein modifications and functional studies
Protein phosphorylation
Phosphorylation, regulated by over 500 kinases and ∼100 phosphatases, is the most pervasive posttranslational modification in the mammalian proteome.1 Nearly 300,000 phosphorylation sites in human and mouse genomes have been identified, corresponding to more than half of all known posttranslational modification sites. Phosphorylation plays a key role in many essential biological processes, including cell signaling and metabolism. Because aberrant regulation of phosphorylation can cause various human diseases, protein kinases have been pursued as major therapeutic targets for decades.32 However, detailed regulatory mechanisms of many kinases and phosphatases have remained poorly understood. Facile strategies that enable homogeneous and selective phosphorylation of proteins would lead to a more detailed understanding of catalytic and regulatory features of many phosphoproteins, including enzymatic activity, stability, structure, and protein-protein interactions. Early attempts to mimic phosphorylation in proteins generally used conventional mutagenesis strategies to replace Ser or Thr with a negatively charged amino acid (Asp or Glu). Although this method is still in use, recent findings suggest that Asp or Glu substitution cannot genuinely mimic Ser or Thr phosphorylation.33-35 Several research groups have attempted semi-synthetic approaches for site-specific protein phosphorylation.36 For example, it has been shown that a synthetic peptide carrying a phospho-amino acid, non-hydrolysable mimic, or photocaged group can be ligated to fragments of recombinant target proteins by NCL or EPL, to generate a full-length protein with a phosphorylation in terminal regions. Dehydroalanine-based chemical conjugation with phospho-thiol or phospho-iodide has also been attempted as a strategy for generating phosphorylation mimics.29,31 For temporal control of phosphorylation, genetic incorporation of photocaged-amino acids has also been tested.37
Serine phosphorylation
The genetic incorporation of genuine phosphoserine, which enables the selective introduction of authentic serine phosphorylation in proteins, was established in 2011.33 The cotranslational installation of phosphoserine in Escherichia coli unlike other unnatural amino acids, essentially required engineering of elongation factor Tu to generate phosphoserine-specific elongation factor (EF-Sep) together with an orthogonal aminoacyl-tRNA synthetase/tRNA pair (SepRS/tRNASep) derived from Methanococcus maripaludis. The negative charge of the phosphate group in phosphoserine proved to be a critical barrier for endogenous EF-Tu in the bacterial translation machinery. Engineered EF-Tu was shown to be effective in incorporating other bulky unnatural amino acids as well.38 The site-specific phosphoserine-incorporation system was further improved by fine-tuning of SepRS and EF-Sep, evolution of the SepRS/tRNASep pair, deletion of release factor 1, and genomic recoding of the E. coli strain.39-42 The phosphoserine-incorporation system has also been applied to cell-free protein synthesis.43 However, this system has not been efficiently applied to mammalian cells yet. It is likely that engineering or evolution of a eukaryotic version of elongation factor (EF1A) will be essentially required for the system to work in mammalian cells.
The genetic phosphoserine-incorporation system has proved to be invaluable in deciphering diverse roles of phosphorylation in many biological processes, including gene expression and cell signaling, which are otherwise virtually inaccessible using conventional methods. For example, histone phosphorylation has been extensively studied, but its effects were considered ambiguous since it was found to be involved in opposite processes: gene activation during interphase and gene silencing through condensation of chromatin during mitosis.44 The genetic incorporation of phosphoserine allowed different levels of chromatin substrates carrying specific phosphorylation, from histone monomers to nucleosomal arrays, to be generated and tested, enabling a clear examination of the effect of phosphorylation of H3S10.39 This analysis revealed a direct correlation between phosphorylation of H3S10 and H3 acetylation (Fig. 3A). In particular, only the most physiologically relevant substrate, a nucleosomal array with an H3S10 phosphorylation, showed highly enhanced acetylation of H3 mediated by the Spt-Ada-Gcn5-Acetyl transferase (SAGA) complex. In addition, genetic installation of a phosphoserine in histone revealed that phosphorylation of H3S28 facilitates p300/CBP-mediated K27 acetylation and transcription in cell-free transcription assays (Fig. 3B).45 Taken together with other pharmacological results, these findings suggest that mitogen- and stress-activated protein kinases (MSKs) can regulate inducible transcription upon bacterial stimulation through phosphorylation of H3S28.
Figure 3.
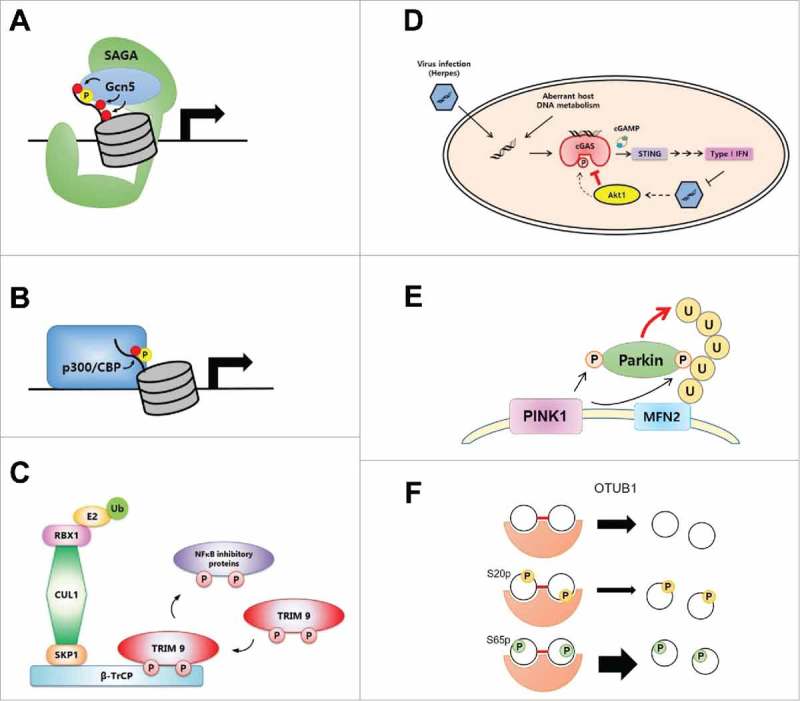
Functional studies of serine phosphorylation using the genetic phosphoserine-incorporation system. (A) Phosphorylation of H3S10 induces enhanced H3 acetylation by Gcn5-SAGA complex in yeast. (B) H3S28 phosphorylation facilitates p300/CBP-mediated H3K27 acetylation and activates transcription. (C) Phosphorylated TRIM9 negatively regulates NF-kB pathway by stabilizing NF-kB inhibitory proteins. (D) S291 phosphorylation in cGAS reduces its enzymatic activity to regulate innate immune DNA-sensing pathway. (E) PINK1-mediated S65 phosphorylation of Ub stimulates E3 ligase activity of PARKIN. (F) Phosphorylation in Ub controls the specificity of E3 ligase and DUB activities.
Protein phosphorylation also regulates essential cell signaling pathways by modulating enzymatic activity or interactions with binding partners. The genetic phosphoserine-incorporation system has proved useful for elucidating biological functions of phosphorylation in cell signaling. For example, the NF-κB signaling pathway is activated by cytokine stimulation and regulates cell proliferation and survival. The β-transducin repeat-containing protein (β-TrCP) is known to regulate the NF-κB pathway by recognizing phosphorylated forms of the NF-κB inhibitory proteins, IκBα and p100, and facilitating their degradation and processing. Brain-specific TRIpartite Motif protein 9 (TRIM9) was identified as a potential negative regulator of the NF-κB pathway, based on the presence of a degron motif in its RING domain that possesses homology with NF-κB inhibitory proteins. However, its detailed regulatory mechanism was not elucidated. Genetic installation of phosphoserine at potential serine phosphorylation sites in recombinant TRIMP9 revealed that β-TrCP binds specifically to TRIM9 with a bone fide phosphorylation at Ser76, while failing to bind to TRIM9 carrying Asp (phospho-mimic) at Ser76 (Fig. 3C).34 These results illustrate that the action of β-TrCP is controlled by binding of phosphorylated TRIMP9, which outcompetes phosphorylated NF-κB inhibitory proteins and directly affects their stability, demonstrating that TRIM9 is a negative regulator of NF-κB signaling.
Cyclic GMP-AMP synthase (cGAS), a cytosolic DNA sensor, can sense microbial and host nucleic acids in the cytoplasm and trigger an innate immune response. Upon infection, cGAS is activated and synthesizes the second messenger, cyclic GMP-AMP (cGAMP), which then binds to the stimulator of interferon genes (STING) to induce production of type I interferon.46 Phosphorylation of cGAS at S291, which is mediated by the kinase Akt, was found to abolish cGAS enzymatic activity toward the second messenger, thereby shutting down the interferon production pathway and preventing an excessive immune response (Fig. 3D).47 Thus, the genetic phosphoserine-incorporation system revealed that the DNA-sensing pathway is regulated by selective phosphorylation of cGAS.
A recent proteomics analysis has suggested that phosphorylation of ubiquitin (Ub) may play a significant role in a wide spectrum of cellular events. A single phosphorylation in Ub can induce drastic structural changes and influence Ub chain assembly and hydrolysis processes.48 Nine positions containing Ser, Thr, or Tyr are phosphorylated in Ub, but their biological roles and modulating enzymes remain largely unknown. For example, PINK1 (PTEN-induced putative kinase protein 1) and the ubiquitin E3 ligase PARKIN were found to take part in the selective degradation of mitochondria by autophagy, a process known as mitophagy. However, the precise functions of PINK-mediated PARKIN and Ub phosphorylation are poorly understood. The phosphoserine-incorporated Ub variants revealed clear stimulation of PARKIN activity by S65-phosphorylated Ub and further demonstrated the existence of a feed-forward mechanism in the mitochondrial ubiquitination pathway (Fig. 3E).35,49 Ub phosphorylation was also found to alter the specificity of E3 ligase and deubiquitinase (DUB) activities. Twenty different phosphorylated isomeric Ub dimers generated using phosphoserine-incorporated Ub variants and E3 ligase were used to profile activities of 31 DUBs toward diverse Ub substrates (Fig. 3F).50 Interestingly, phosphorylation of Ub was found to control DUB activity and specificity. In particular, phosphorylation at S20 in Ub was shown to convert UBE3C from a dual-specificity E3 ligase to a K48-specific ligase.
Threonine phosphorylation
The genetic incorporation of phosphothreonine using SepRS/tRNASep pair was challenging due to structural similarity to phosphoserine and its low intracellular concentration in E. coli. Recently, multi-directional efforts for biosynthesis of phosphothreonine and evolution of a pThrRS/tRNA pair via parallel selections with deep sequencing led to an efficient genetic encoding of phosphothreonine.51 The utility of this system was demonstrated by producing catalytically active protein kinase Cdk2 installed with phosphothreonine at position 160.
Tyrosine phosphorylation
For selective tyrosine phosphorylation in proteins, genetic incorporation of phospho-Tyr analog, p-carboxymethyl-L-phenylalanine (pCMF), was initially attempted in E. coli using an evolved Methanocaldococcus jannaschii tyrosyl-tRNA synthetase/tRNA pair.52 Tyrosine phosphorylation has also been mimicked by converting the azide group of a genetically installed azidophenylalanine to phosphoramidate by a chemoselective reaction.53 For in vivo study of tyrosine phosphorylation, genetic incorporation of photocaged-Tyr or a phospho-Tyr mimetic has also been tested.54 In particular, genetically installed, photocaged-Tyr was found to facilitate photo-control of phosphorylation in the signal transducer and activator of transcription 1 (STAT1), which would be useful for controlling tyrosine phosphorylation in many key signaling molecules. The genetic incorporation of authentic phosphotyrosine was first achieved in E. coli through deletion of five endogenous phosphatases together with introduction of an engineered EF-Tu and MjTyrRS-tRNA pair.55 Very recently, two other reports also demonstrated the genetic incorporation of phosphotyrosine and its analogs via a propeptide strategy and evolution of orthogonal aminoacyl-tRNA synthetase/tRNA pair.56,57 These strategies will be widely applied for functional analysis of tyrosine phosphorylation
Lysine modifications
Lysine acetylation
Early studies on protein acetylation have focused extensively on histone acetylation, revealing that this modification is a key epigenetic mechanism for regulating gene expression. A recent high-resolution mass spectrometry-based acetylome analysis identified numerous acetylation sites in non-nuclear and mitochondrial proteins.58 In addition, recent findings have uncovered links between lysine acetylation and metabolism, and demonstrated a regulatory role of lysine acetylation in enzymatic activity or protein subcellular localization, showing that lysine acetylation is a ubiquitous modification that affects many biological processes.59 For biochemical studies of acetylation, researchers have attempted enrichment of acetylated proteins using acetyl group-specific antibodies or modification of the target proteins by specific acetyltransferase(s), but these modifications are frequently not selective or homogeneous. For in vivo functional analyses of acetylation, replacement of a lysine residue with glutamine or arginine has been frequently used as a strategy for mimicking acetylated lysine or unacetylated lysine, respectively. However, there is growing evidence that such mimics cannot functionally substitute for acetylated and unacetylated lysine residues.60 To overcome problems of such conventional methods, researchers have made extensive use of various approaches to selectively acetylate proteins. In particular, the genetic incorporation of acetyllysine in E. coli has been achieved using evolved orthogonal MbPylRS/tRNAPyl or MmPylRS/tRNAPyl pairs.61,62 The acetyllysine-incorporation system has been expanded to eukaryotic cells, including both yeast and mammalian cells.63,64 In addition, installation of acetyllysine-incorporation machinery at the genome level has been achieved in stem cells and mice.65,66
This approach has been broadly employed to produce site-selectively acetylated proteins for in vitro assays to elucidate biological roles of specific acetylations in various proteins. For example, recombinant histones and reconstituted nucleosomes with site-specific acetylation generated using this method have facilitated investigation of the cellular function of acetylation of histones and transcription factors and their regulatory mechanisms. Nucleosomes reconstituted using K56-acetylated H3 have assisted in the study of chromatin remodeling and nucleosome stability.67,68 The specificity of NAD+-dependent histone deacetylases (sirtuins) has also been investigated using various nucleosomes carrying diversely acetylated H3 histones.69,70 Interestingly, three SIRT enzymes—SIRT1, −2 and −6—were shown to display noticeably higher activities toward nucleosome substrates than their peptide counterparts (Fig. 4A). This method has also proved useful for functional studies of acetylation of non-histone proteins, such as metabolic enzymes and cell signaling regulators. For example, in a study of cellular NADPH homeostasis during oxidative stress, site-specific acetylation of glycolytic enzymes, such as glucose-6-phosphate dehydrogenase and phosphoglycerate mutase, was found to reduce enzymatic activity by interfering with dimerization or interactions with the deacetylase SIRT2 (Fig. 4B).71,72
Figure 4.
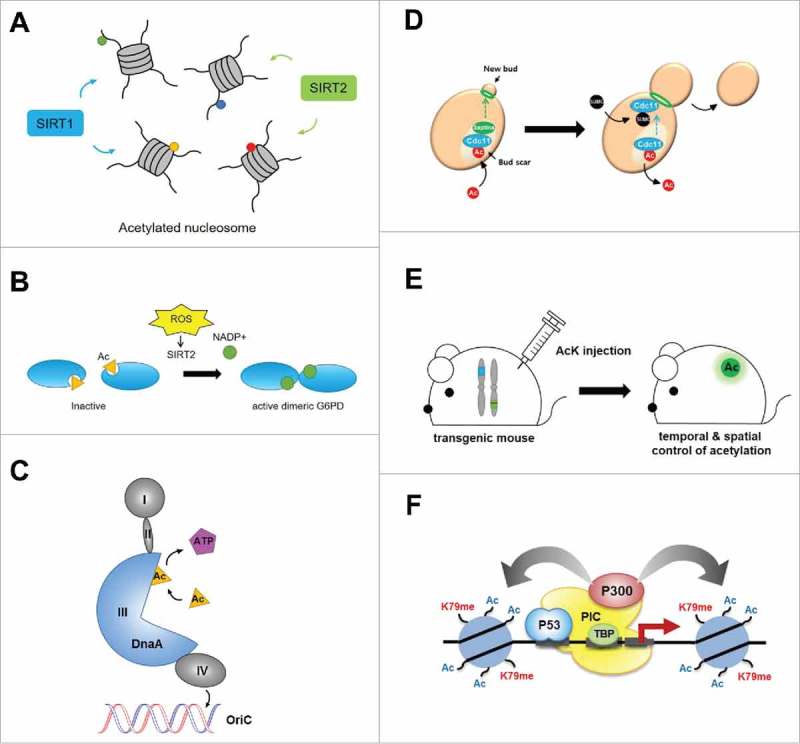
Functional studies of lysine modifications using the site-specific lysine acetylation and methylation systems. (A) Nucleosomes carrying acetylated H3 histones can be used to determine the specificity of NAD+-dependent histone deacetylases (SIRTs). (B) K403 acetylation in glucose-6-phosphate dehydrogenase negatively regulates its enzymatic activity. (C) K178 acetylation in DnaA blocks DNA replication initiation by blocking its binding to ATP or oriC in E. coli. (D) K412 acetylation in Cdc11 controls its function and localization via acetylation-SUMOylation switch during the cell cycle in yeast. (E) Transgenic mouse with expanded genetic code with acetyllysine allows temporal and spatial control of protein acetylation and facilitates systematic in vivo acetylome studies. (F) K79 methylation in histone H3 induces chromatin transcription through enhanced p300-mediated histone acetylation.
Proteome-wide mapping of the Drosophila acetylome has allowed identification of evolutionarily conserved acetylation positions in Ub-conjugating E2 enzymes. The site-specific acetylation approach revealed that acetylation of UBE2D3 at position Lys8 drastically reduces its enzymatic activity, whereas a glutamine substitution produces a much lesser effect, demonstrating that this substitution is an insufficient acetylation mimic.73 The site-specific acetylation approach has been utilized to demonstrate how K5 acetylation on lactate dehydrogenase A (LDH-A) negatively regulates its enzymatic activity and to elucidate mechanisms underlying LDH-A upregulation in pancreatic cancer.74 This approach also revealed that the Tat-interactive protein 60 (TIP60)-mediated selective acetylation of Aurora B kinase, involved in attaching mitotic spindles to the centromere, protects Aurora B from dephosphorylation by phosphatase PP2A, ensuring robust activity during the mitotic process.75 In bacterial systems, acetylation of DnaA was found to block binding of DnaA to ATP or oriC in E. coli, thereby inducing inhibition of DNA replication initiation (Fig. 4C).76 Lastly, specific acetylation of a key regulator of bacterial virulence, PhoP, was found to inhibit the DNA-binding ability of PhoP and affect the transcription of its target genes, which are involved in regulation of virulence.77
The amber suppression method has also been found useful for in vivo functional studies of lysine acetylation in eukaryotes. Genetic incorporation of acetyllysine into the yeast septin protein Cdc11 at K412 led to the finding that the cellular function and localization of Cdc11 is precisely controlled by a newly identified acetylation-SUMOylation switch in Cdc11 during the cell cycle (Fig. 4D).78 Again, Gln, used as an acetylated lysine mimic, failed to functionally substitute for acetylation in Cdc11. Notably, genetic incorporation of acetyllysine was recently extended to the mouse, the most prevalent animal model of human physiology and disease (Fig. 4E).66 In this application, an acetyllysine-incorporation system was first constructed in mammalian cells using green fluorescent protein (GFP) as a model protein. Then, the corresponding gene constructs were separately microinjected into fertilized eggs of the mouse to generate AcK and GFPamber transgenic mice. The AcK mouse and GFPamber mouse were crossed to produce a double-heterozygous, transgenic AcK-GFPamber mouse. The resulting AcK-GFPamber mouse was found to exhibit rapid onset of acetylation of a specific lysine residue of a model protein at any developmental stage or in any specific tissue upon administration of acetyllysine. Such a mouse model that enables selective and controlled acetylation is expected to facilitate systematic in vivo acetylome studies, providing insight into many essential acetylation-related biological processes and human diseases at the tissue and organism level.
Lysine methylation
Reversible lysine methylation controlled by lysine methyltransferases (KMTs) and lysine demethylases (KDMs) occurs in three different states: monomethylated (me1), dimethylated (me2), and trimethylated (me3). Various proteins, including histones, are known to be methylated, and their roles in many cellular processes, including gene expression, DNA repair, X chromatin inactivation, cell cycle progression and tumorigenesis, are beginning to be understood.79 Since lysine methylation presents one of the most complex and dynamic modifications, production of recombinant proteins with homogeneous and site-specific methylation is essential for the systematic study of methylation-dependent processes. Direct genetic incorporation of methylated lysine(s) has proved to be highly challenging owing to similarities in the chemical structure of methylated lysines and the canonical amino acid lysine. Thus, several indirect amber suppression methods have been proposed by several different groups. For example, for lysine monomethylation in histones, protected methyllysines are genetically incorporated and then converted into monomethyllysine after cleavage of the protecting groups.80-83 For lysine dimethylation, genetic installation of a dimethyllysine precursor requiring chemical transformation has also been proposed.84 For lysine trimethylation, however, no method is yet available. Thiol-based conjugation methods have been developed to generate thioether-linked analogs at the target lysine residue.22,28 But the resulting methyllysine analogs exhibit different properties and binding abilities toward reader molecules; thus, their biological relevance is often questionable.85
Very recently, a facile chemical biology route that enables attachment of PTM moieties at a selected position of a target protein through chemoselective C-C bond formation was developed.30 Using this method, which applies a clever combination of genetic code expansion and selective chemical conjugation, these researchers generated recombinant histones containing selective methylations in three modification states. In vitro transcription assays using modified nucleosomes generated from the differentially methylated histones H3K79me1, H3K79me2, and H3K79me3 clearly demonstrated the role of these modifications in the regulation of gene expression, showing that methylation at K79 in histone H3 directly induces chromatin transcription through enhanced histone acetylation mediated by p300 (Fig. 4F).30 Interestingly, the level of H3K79 methylation differentially affects histone acetylation; in particular, H3K79me2 most notably enhances histone acetylation, followed by H3K79me1 and H3K79me3, demonstrating the dependence of different biological functions of lysine methylation on the modification state.
Other lysine modifications
It has recently been shown that lysine also undergoes numerous acylations, such as formylation, propionylation, butyrylation, crotonylation, succinylation and acylation by longer fatty acids, but our understanding of these processes is still fragmentary.86 For example, propionylation and butyrylation of histones are effectively catalyzed by the acetyltransferase p300/CBP, and deacylation of various other acyl groups, such as succinyl, malonyl, glutaryl, and fatty acyl (myristoyl and palmitoyl) on lysine residues is catalyzed by members of the NAD-dependent sirtuin family deacetylases.87,88 On the other hand, crotonylation of histones, considered an important marker for male germ cell differentiation, was found to be marginally catalyzed by any known acetyltransferases.89 Thus, in-depth studies of these modifications and their regulatory mechanisms are hampered by difficulties in generating homogeneously modified proteins. The amber suppression method has proved useful for genetic installation of several short-chain acylated lysines, such as propionylated lysine (PrK), butyrylated lysine (BuK) and crotonylated lysine (CrK), using wild-type PylRS, and efficiency was further improved using evolved PylRS mutants.90-93 Recently, a more versatile strategy that enables site-specific modification of lysines with various acyl groups was developed.94 In this approach, azidonorleucine is first incorporated into a specific position of a target protein by evolved PylRS, after which acyl derivatives carrying a phosphinothioester can be conjugated to the azide group via traceless Staudinger ligation. This scheme should be applicable to the study of cellular and molecular mechanisms of lysine acylation and the detailed functions of modulating enzymes and reader proteins associated with acylation.
Tyrosine modifications
Tyrosine sulfation
Protein tyrosine sulfation (PTS) is catalyzed by tyrosylprotein transferase (TPST) using 3′-phosphoadenosine-5′-phosphosulfate (PAPS) as a sulfate donor. TPST, an enzyme of the sulfotransferase family, is known to catalyze sulfation of various biomolecules, including proteins, neurotransmitters, hormones, and carbohydrates.95 Although PTS was identified decades ago, its biological role is just beginning to come into focus. PTS is thought to be associated with cellular processes such as homeostasis, pathogen recognition, viral infection, leukocyte adhesion, and trafficking.96 However, the details of these functions remain largely unknown because of difficulties in controlling sulfation by TPST. Genetic incorporation of sulfated tyrosine (sulfoTyr), established in 2006, demonstrated its utility in the study of biological functions of PTS.97 PTS of the chemokine receptor CCR5 is known to induce HIV-1 entry through recognition by the envelope glycoprotein, gp120.98 Human antibodies directed against gp120 (e.g., doubly sulfated anti-gp120 antibody 412d) also carry sulfated tyrosine residues, whose contribution to antigen binding was elegantly verified using a recombinant Fab fragment bearing site-specific sulfoTyr.99 The receptor kinase-like protein XA21, found in rice, is known to recognize Y41 sulfation in the secreted plant bacterial pathogen protein RaxX, leading to activation of XA21-mediated immunity.100 Recombinant RaxX carrying site-specific sulfoTyr (at Y41) was found to be effective in reducing disease progression in rice infected with the pathogen.101 Recombinant hirudin, utilized as a commercial anticoagulant, is also modified by PTS. Sulfated hirudin carrying site-specific sulfoTyr was found to exhibit a 10-fold higher affinity toward thrombin compared with hirudin, demonstrating its potential therapeutic applications as an effective thrombin inhibitor (Fig. 5A).97
Figure 5.
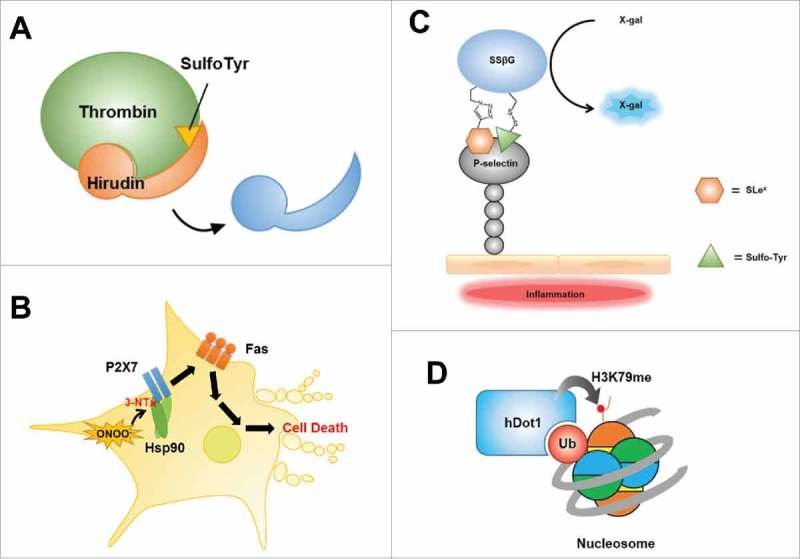
Functional studies of other modifications. (A) Site-specific sulfation in hirudin results in highly enhanced binding toward thrombin. (B) Nitration in Hsp90 induces motor neuron death through the Fas pathway. (C) LacZ-type reporter enzyme SSβG carrying mimics of glycosylation and sulfation can be used to probe brain inflammation in vivo. (D) K120 mono-ubiquitination in H2B enhances hDot1L-mediated H3K79 methylation.
Tyrosine nitration
Protein oxidation is an important modulator of redox-dependent signaling and related biological processes.102 In particular, protein tyrosine nitration (PTN) is known to be involved in aging and various pathological processes, including autoimmune disorders, cancers, and neurodegenerative diseases. Unlike other PTMs, which are mediated by specific enzymes, PTN is caused by various reactive nitrogen species (RNS), especially secondary radical species (·NO2, CO3·−) generated from peroxynitrite (ONOO-). Nonetheless, PTN is not random, but is instead a highly selective process. Accordingly, PTN can affect the structure and enzymatic activity of target proteins, thereby affecting cell viability; it can also function as a marker for oxidative damage. In addition, 3-nitrotyrosine (3-NT) can be further converted to 3-aminotyrosine or back to tyrosine through denitration, implying other possible regulatory roles in different cellular environments. 3-NT can also function as a neoantigen, inducing formation of autoantibodies in various autoimmune diseases.103 Pretreatment with RNS is not an effective approach for studying PTN, because this method is not capable of generating site-specific tyrosine nitration and can lead to unwanted side reactions, causing dinitrotyrosine cross-linked proteins.
The amber suppression method, which enables site-specific genetic incorporation of 3-NT, has aided in detailed functional analyses of PTN in many cellular processes.104,105 Nitration of manganese superoxide dismutase (MnSOD) (at Y3) caused by peroxynitrite was found to be associated with cardiovascular disease and inflammatory responses.106 Genetic incorporation of 3-NT at Y34 in MnSOD dramatically reduced the catalytic activity of MnSOD, demonstrating the negative regulatory role of 3-NT in MnSOD.104 Free cholesterol is exported in the form of ApoA-1 via the ABCA1 transporter, a process that is important for homeostasis since most cells cannot catabolize cholesterol.107 PTN is found at high levels in ApoA-1 in atherosclerotic human coronary arteries. Experiments with reconstituted nascent high density lipoprotein (HDL) particles using nitrated ApoA-1 containing a site-specific 3-NT at Y166 revealed that PTN at 166Y does not affect ABCP1-dependent cholesterol efflux activity, but drastically reduces lecithin:cholesterol acyltransferase (LCAT) activity, demonstrating the importance of nitration of ApoA-1 in the diseased artery wall.108 The heat-shock protein Hsp90 is a well-known chaperone that serves to ensure proper folding of many cellular proteins under stress conditions. Nitration of Hsp90 was found to be involved in peroxynitrite-induced apoptosis of motor neurons and PC12 cells. Genetic incorporation of 3-NT into five possible nitration positions revealed that nitration at Y33 or Y56 of Hsp90 is directly associated with Fas-mediated cell death in motor neurons (Fig. 5B).109
Protein glycosylation
Protein glycosylation is characterized by the attachment of carbohydrates to proteins through various glycosidic linkages, including N-linked (on Asn residues) and O-linked (on Ser or Thr residues) glycosylation. Glycosylation of proteins that are destined for the cell surface and extracellular regions is critical for many biological processes, including cell adhesion, receptor activation, endocytosis, and cell-cell communication.110 Glycosylation also occurs in many cytoplasmic and nuclear proteins, and is critical for their functional regulation. Thus, defects in glycosylation are associated with a number of human diseases, including cancer, diabetes, and autoimmune and neurodegenerative diseases. Glycosylation is a highly complex process regulated by the interplay of various types of glycosyltransferases and glycosidases.
Initial attempts at functional analysis of glycosylation used synthetic or semi-synthetic methods, including chemoenzymatic strategies, to obtain peptides containing glycosylated amino acids.111-113 In particular, EPL and NCL employing glycosylated synthetic peptides have been employed to generate full-sized proteins containing specific glycosylations at terminal regions,114,115 and a number of different chemical ligation methods for improving the efficiency of protein glycosylation have been tried. These methods include disulfide linkages using glycomethanethiosulfonate (glyco-MTS),116 triazole linkages through Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC),117 attachment of allyl C-glycosides to Cys through thiol-ene coupling,118 and linkage of glycosyl thiols using genetically installed homoallyglycine.119 Lastly, recently developed chemical modification methods using dehydroalanine-based chemistry have shown great potential and versatility for homogeneous and selective glycosylation of proteins.31 This approach enables generation of recombinant proteins with site-specific O- or N-glycosylation mimics through C-C bonds, forming radical conjugations on specific dehydroalanine residues using N- or O-linked glycosidic iodide.
P-selectin glycoprotein ligand-1 (PSGL-1) is known to regulate leukocyte rolling by binding to the cell adhesion molecule, P-selectin, expressed on inflamed endothelial cells.120 In binding to its ligand, P-selectin recognizes two specific modifications—LewisX(SLeX)-like glycans and tyrosine sulfation—in the N-terminal region of PSGL-1.121 These two modification-mimics have been introduced at specific positions of the LacZ-type reporter enzyme SSβG using a chemical ligation method in which sialylated glycan was conjugated through CuAAC and SulfoTyr-methanethiosulphonates (MTS) was ligated onto a Cys residue via a disulfide bond (Fig. 5C).117 The resulting recombinant PSGL-1 mimic containing specific double modifications exhibited P-selectin binding and galactosidase activity, enabling X-gal visualization, which could prove useful for probing brain inflammation in vivo. GlcNAcylation of histones was recently detected, and its detailed molecular mechanism in transcriptional regulation has come under scrutiny. Glycosylated H2A (at T101) was generated via dehydroalanine-based conjugation with GlcNAc-thiol and used for nucleosome reconstitution experiments, which showed that GlcNAcylation of H2A at T101 affected H2A-H2B dimer and tetramer interactions such that they destabilized nucleosomal structure.122 H2B GlcNAcylated at S112 was similarly produced and shown by a MS-based proteomics analysis using the reconstituted GlcNAcylated nucleosome to interact with the facilitated chromatin transcription (FACT) complex.123
Ubiquitination
Ub is a highly conserved, small regulatory protein (76 amino acids, 8.5 kDa) that is found in all eukaryotes. Ubiquitination, the enzymatic process by which Ub is added to a substrate protein, is carried out by a cascade of three enzymes: an E1 Ub-activating enzyme, an E2 Ub-conjugating enzyme, and an E3 Ub ligase. Ub conjugation occurs between the C-terminus of Ub and specific Lys residues in a substrate protein, forming a covalent isopeptide linkage. A single Ub can be added to a protein, a process known as mono-ubiquitination. Alternatively, multiple Ubs can be added to form different Ub chain topologies (linear or branched Ub chain), reflecting the availability of multiple Lys residues (K6, K11, K27, K29, K33, K48, and K63) or N-terminal methionine (M1) in Ub for conjugation, a process known as poly-ubiquitination. Poly-ubiquitination via elongation of single linkages generates linear homotypic Ub chains, of which K48- and K64-linked poly-Ub, referred to as typical chains, are the most abundant; others chains are called atypical chains. In contrast, poly-ubiquitination via elongation of mixed linkages or using the distinct small Ub-like modifier (SUMO) pathway, forms mixed and branched chains, also known as heterotypic chains.124 In addition, various PTMs, including phosphorylation and acetylation, occur on Ub molecules in chain topologies, adding further complexity to the Ub code.125 Ubiquitination regulates various properties of target proteins, including their activity, stability and cellular location, thereby affecting many cellular processes.
Simple and conventional approaches, including expression of Ub mutants, detection of ubiquitination using MS spectrometry and linkage-specific antibodies, and Ub chain synthesis using Ub ligase/DUBs, have been widely applied for functional analysis of ubiquitination.124 Synthetic conjugation approaches employing NCL and EPL, which enable peptide ligation or chemoselective coupling, have proven efficient for producing proteins with position-specific ubiquitination. Mono- or di-ubiquitination can be generated by attaching a Ub thioester, generated from intein-fused recombinant Ub via thiolysis, to a substrate protein or another Ub carrying a thiol-bearing branch synthesized by SPPS. For example, an SPPS-synthesized Ub peptide fragment carrying δ-mercaptolysine at a linkage lysine position can be linked to Ub thioester via NCL to generate di-Ub.126 An auxiliary-linked, synthetic H2B C-terminal peptide bearing thiol, generated by SPPS, can be linked with Ub thioester and H2B N-terminal fragment thioester produced by thiolysis of intein fusion proteins through two successive traceless orthogonal EPL reactions to produce full-length mono-ubiquitinated histone H2B-K120Ub. Mono-ubiquitination of H2B at K120 was shown to strongly induce hDot1L-mediated methylation of H3 at K79, demonstrating direct crosstalk between ubiquitination and methylation in the context of the nucleosome (Fig. 5D).127
The amber suppression method has proven effective in generating a thiol-bearing branch in target proteins. For example, a thiol-bearing Lys derivative (Cys-Lys) can be directly incorporated into proteins.128 Moreover, protected δ-thiol-L-lysine can be incorporated into a protein and ligated with Ub thioester using EPL, and then further converted into a native isopeptide bond by desulfurization.129 Ub containing a genetically installed Boc-Lys can be coupled with a Ub thioester generated from thiolysis of intein-fused Ub via silver-mediated condensation after a series of protection (with Cbz group) and deprotection (using TFA) steps applied to lysine residues of both Ub molecules to produce isopeptide-coupled di-Ub.130 Ub containing Boc-Lys can also be used to make mixed and branched Ub trimer chains through conjugation with a Ub thioester generated from E1 and sodium 2-mercaptoethane sulfate (MESNa).131 Application of CuAAC to genetically encoded unnatural amino acids carrying azide and alkyne is an effective approach for making non-isopeptide triazole-linked di-Ub, which is uncleavable by DUBs.132 Lastly, Ub containing a genetically installed allyl amine at the C-terminus has been coupled with Ub carrying Cys at the linkage site through thiol-ene coupling to generate homotypic chains and mixed or branched Ub trimers.133,134 This approach was utilized to create synthetic Ub chains from Ub functionalized with fluorescent dye or biotin for functional analyses of DUB specificity.135 A better understanding of detailed molecular features of the puzzling ubiquitin code and its cellular effects will ultimately require the development of more versatile methods for Ub modifications so as to enable the creation of diverse forms of poly Ubs with various chain lengths and topology.
Concluding remarks
Recent MS-based proteome analyses have revealed that PTMs occur on most eukaryotic proteins.136 Over 400,000 modification sites have been identified and certainly more will be discovered. Considering the overwhelming number of PTMs, a full array of efficient experimental tools and versatile strategies will be needed to explore fundamental mechanisms of such complicated PTMs. There have been remarkable advances in synthetic and chemical biology approaches for in vitro and in vivo PTM studies. The genetic code expansion strategy, which has already proven useful for several important PTMs, such as phosphorylation, acetylation and sulfation, will continue to broaden its applications to achieve many challenging modifications in the future. In particular, animal models with an expanded genetic code will facilitate temporal and spatial control of specific modifications in target proteins, providing a powerful tool for studies of human physiology and disease. Notably, newly emerging chemical modification methods that enable direct installation of specific modifications will further expand their utility, allowing access to numerous major and minor PTMs considered untouchable by previous methods. The combination of individual powerful approaches may lead to installation of distinct forms of PTMs at multiple positions of a target protein, which is still challenging but essentially required to explore complicated crosstalk and interaction between PTMs. Such efforts will dramatically expand our ability to decipher molecular and cellular functions of PTMs.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We apologize to all scientists in the field whose important work could not be cited in this report due to space limitations.
Funding
This work was supported by grants from the National Research Foundation of Korea (2014M3A6A4075060 and 2017R1A2B3011543) and Systems Healthcare from KAIST.
References
- 1.Walsh CT, Garneau-Tsodikova S, Gatto GJ. Protein posttranslational modifications: The chemistry of proteome diversifications. Angew Chem Int Ed. 2005;44:7342–72. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 2.Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17:666–72. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 3.Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al.. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582-+. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 4.Harper JW, Bennett EJ. Proteome complexity and the forces that drive proteome imbalance. Nature. 2016;537:328–38. doi: 10.1038/nature19947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–44. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 6.Spicer CD, Davis BG. Selective chemical protein modification. Nat Commun. 2014;5:4740. doi: 10.1038/ncomms5740. [DOI] [PubMed] [Google Scholar]
- 7.Chin JW. Expanding and reprogramming the genetic code of cells and animals In: Kornberg RD, editors. Annual Review of Biochemistry. Vol. 83 Palo Alto: Annual Reviews; 2014. p. 379–408. [DOI] [PubMed] [Google Scholar]
- 8.Wan W, Tharp JM, Liu WR. Pyrrolysyl-tRNA synthetase: An ordinary enzyme but an outstanding genetic code expansion tool. Biochim Biophys Acta. 2014;1844:1059–70. doi: 10.1016/j.bbapap.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holt M, Muir T. Application of the protein semisynthesis strategy to the generation of modified chromatin. Annu Rev Biochem. 2015;84:265–90. doi: 10.1146/annurev-biochem-060614-034429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terasaka N, Iwane Y, Geiermann AS, Goto Y, Suga H. Recent developments of engineered translational machineries for the incorporation of non-canonical amino acids into polypeptides. Int J Mol Sci. 2015;16:6513–31. doi: 10.3390/ijms16036513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuh KN, Batt AR, Pratt MR. Chemical methods for encoding and decoding of posttranslational modifications. Cell Chem Biol. 2016;23:86–107. doi: 10.1016/j.chembiol.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krall N, da Cruz FP, Boutureira O, Bernardes GJL. Site-selective protein-modification chemistry for basic biology and drug development. Nat Chem. 2016;8:102–12. [DOI] [PubMed] [Google Scholar]
- 13.Dawson PE, Muir TW, Clarklewis I, Kent SBH. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–9. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 14.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: A general method for protein engineering. Proc Natl Acad Sci U S A. 1998;95:6705–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambrogelly A, Palioura S, Soll D. Natural expansion of the genetic code. Nat Chem Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 16.Zinoni F, Birkmann A, Leinfelder W, Bock A. Cotranslational insertion of selenocysteine into formate dehydrogenase from Escherichia-coli directed by a UGA codon. Proc Natl Acad Sci U S A. 1987;84:3156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao B, Gong WM, Ferguson TK, James CM, Krzycki JA, Chan MK. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–6. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 18.Beier H, Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001;29:4767–82. doi: 10.1093/nar/29.23.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 20.Goto Y, Katoh T, Suga H. Flexizymes for genetic code reprogramming. Nat Protoc. 2011;6:779–90. doi: 10.1038/nprot.2011.331. [DOI] [PubMed] [Google Scholar]
- 21.Noren CJ, Anthonycahill SJ, Griffith MC, Schultz PG. A general-method for site-specific incorporation of unnatural amino-acids into proteins. Science. 1989;244:182–8. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 22.Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–12. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang R, Holbert MA, Tarrant MK, Curtet S, Colquhoun DR, Dancy BM, Dancy BC, Hwang Y, Tang Y, Meeth K, et al.. Site-specific introduction of an acetyl-lysine mimic into peptides and proteins by cysteine alkylation. J Am Chem Soc. 2010;132:9986–7. doi: 10.1021/ja103954u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li FP, Allahverdi A, Yang RL, Lua GBJ, Zhang XH, Cao Y, Korolev N, Nordenskiöld L, Liu CF. A direct method for site-specific protein acetylation. Angew Chem Int Ed. 2011;50:9611–4. doi: 10.1002/anie.201103754. [DOI] [PubMed] [Google Scholar]
- 25.Le DD, Cortesi AT, Myers SA, Burlingame AL, Fujimori DG. Site-specific and regiospecific installation of methylarginine analogues into recombinant histones and insights into effector protein binding. J Am Chem Soc. 2013;135:2879–82. doi: 10.1021/ja3108214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chalker JM, Gunnoo SB, Boutureira O, Gerstberger SC, Fernandez-Gonzalez M, Bernardes GJL, Griffin L, Hailu H, Schofield CJ, Davis.. Methods for converting cysteine to dehydroalanine on peptides and proteins. Chem Sci. 2011;2:1666–76. doi: 10.1039/c1sc00185j. [DOI] [Google Scholar]
- 27.Guo J, Wang J, Lee JS, Schultz PG. Site-specific incorporation of methyl- and acetyl-lysine analogues into recombinant proteins. Angew Chem Int Ed. 2008;47:6399–401. doi: 10.1002/anie.200802336. [DOI] [PubMed] [Google Scholar]
- 28.Wang ZU, Wang Y-S, Pai P-J, Russell WK, Russell DH, Liu WR. A facile method to synthesize histones with posttranslational modification mimics. Biochemistry. 2012;51:5232–4. doi: 10.1021/bi300535a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chalker JM, Lercher L, Rose NR, Schofield CJ, Davis BG. Conversion of cysteine into dehydroalanine enables access to synthetic histones bearing diverse post-translational modifications. Angew Chem Int Ed. 2012;51:1835–9. doi: 10.1002/anie.201106432. [DOI] [PubMed] [Google Scholar]
- 30.Yang A, Ha S, Ahn J, Kim R, Kim S, Lee Y, Kim J, Söll D, Lee HY, Park HS. A chemical biology route to site-specific authentic protein modifications. Science. 2016;354:623–6. doi: 10.1126/science.aah4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright TH, Bower BJ, Chalker JM, Bernardes GJL, Wiewiora R, Ng WL, Raj R, Faulkner S, Vallee MRJ, Phanumartwiwath A, et al.. Posttranslational mutagenesis: A chemical strategy for exploring protein side-chain diversity. Science. 2016;354:11. doi: 10.1126/science.aag1465. [DOI] [PubMed] [Google Scholar]
- 32.Roskoski R. A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol Res. 2015;100:1–23. doi: 10.1016/j.phrs.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Park H-S, Hohn MJ, Umehara T, Guo L-T, Osborne EM, Benner J, Noren CJ, Rinehart J, Söll D. Expanding the genetic code of Escherichia coli with phosphoserine. Science. 2011;333:1151–4. doi: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi MD, Cho H, Inn KS, Yang AR, Zhao Z, Liang QM, Versteeg GA, Amini-Bavil-Olyaee S, Wong LY, Zlokovic BV, et al.. Negative regulation of NF-kappa B activity by brain-specific TRIpartite Motif protein 9. Nat Commun. 2014;5:11. doi: 10.1038/ncomms5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ordureau A, Heo JM, Duda DM, Paulo JA, Olszewski JL, Yanishevski D, Rinehart J, Schulman BA, Harper JW. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc Natl Acad Sci U S A. 2015;112:6637–42. doi: 10.1073/pnas.1506593112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Z, Cole PA. Synthetic approaches to protein phosphorylation. Curr Opin Chem Biol. 2015;28:115–22. doi: 10.1016/j.cbpa.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemke EA, Summerer D, Geierstanger BH, Brittain SM, Schultz PG. Control of protein phosphorylation with a genetically encoded photocaged amino acid. Nat Chem Biol. 2007;3:769–72. doi: 10.1038/nchembio.2007.44. [DOI] [PubMed] [Google Scholar]
- 38.Mittelstaet J, Konevega AL, Rodnina MV. A kinetic safety gate controlling the delivery of unnatural amino acids to the ribosome. J Am Chem Soc. 2013;135:17031–8. doi: 10.1021/ja407511q. [DOI] [PubMed] [Google Scholar]
- 39.Lee S, Oh S, Yang A, Kim J, Soll D, Lee D, Park HS. A facile strategy for selective incorporation of phosphoserine into histones. Angew Chem Int Ed. 2013;52:5771–5. doi: 10.1002/anie.201300531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogerson DT, Sachdeva A, Wang KH, Haq T, Kazlauskaite A, Hancock SM, Huguenin-Dezot N, Muqit MM, Fry AM, Bayliss R, et al.. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat Chem Biol. 2015;11:496-+. doi: 10.1038/nchembio.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heinemann IU, Rovner AJ, Aerni HR, Rogulina S, Cheng L, Olds W, Fischer JT, Söll D, Isaacs FJ, Rinehart J. Enhanced phosphoserine insertion during Escherichia coli protein synthesis via partial UAG codon reassignment and release factor 1 deletion. FEBS Lett. 2012;586:3716–22. doi: 10.1016/j.febslet.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pirman NL, Barber KW, Aerni HR, Ma NJ, Haimovich AD, Rogulina S, Isaacs FJ, Rinehart J.. A flexible codon in genomically recoded Escherichia coli permits programmable protein phosphorylation. Nat Commun. 2015;6:6. doi: 10.1038/ncomms9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oza JP, Aerni HR, Pirman NL, Barber KW, ter Haar CM, Rogulina S, Amrofell MB, Isaacs FJ, Rinehart J, Jewett MC.. Robust production of recombinant phosphoproteins using cell-free protein synthesis. Nat Commun. 2015;6:7. doi: 10.1038/ncomms9168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sawicka A, Seiser C. Histone H3 phosphorylation–a versatile chromatin modification for different occasions. Biochimie. 2012;94:2193–201. doi: 10.1016/j.biochi.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Josefowicz SZ, Shimada M, Armache A, Li CH, Miller RM, Lin S, Yang A, Dill BD, Molina H, Park HS, et al.. Chromatin kinases act on transcription factors and histone tails in regulation of inducible transcription. Mol Cell. 2016;64:347–61. doi: 10.1016/j.molcel.2016.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–80. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seo GJ, Yang A, Tan B, Kim S, Liang QM, Choi Y, Yuan W, Feng P, Park HS, Jung JU. Akt kinase-mediated checkpoint of cGAS DNA sensing pathway. Cell Rep. 2015;13:440–9. doi: 10.1016/j.celrep.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wauer T, Swatek KN, Wagstaff JL, Gladkova C, Pruneda JN, Michel MA, Gersch M, Johnson CM, Freund SM, Komander D. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 2015;34:307–25. doi: 10.15252/embj.201489847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.George S, Aguirre JD, Spratt DE, Bi YM, Jeffery M, Shaw GS, O'Donoghue P. Generation of phospho-ubiquitin variants by orthogonal translation reveals codon skipping. FEBS Lett. 2016;590:1530–42. doi: 10.1002/1873-3468.12182. [DOI] [PubMed] [Google Scholar]
- 50.Huguenin-Dezot N, De Cesare V Peltier J, Knebel A, Kristaryianto YA, Rogerson DT, Kulathu Y, Trost M, Chin JW. Synthesis of Isomeric Phosphoubiquitin Chains reveals that phosphorylation controls deubiquitinase activity and specificity. Cell Rep. 2016;16:1180–93. doi: 10.1016/j.celrep.2016.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang MS, Brunner SF, Huguenin-Dezot N, Liangl AD, Schmied WH, Rogerson DT, Chin JW. Biosynthesis and genetic encoding of phosphothreonine through parallel setection and deep sequencing. Nat Methods. 2017;14:729-+. doi: 10.1038/nmeth.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie J, Supekova L, Schultz PG. A genetically encoded metabolically stable analogue of phosphotyrosine in Escherichia coli. ACS Chem Biol. 2007;2:474–8. doi: 10.1021/cb700083w. [DOI] [PubMed] [Google Scholar]
- 53.Serwa R, Wilkening I, Del Signore G, Muehlberg M, Claussnitzer I, Weise C, Gerrits M, Hackenberger CP. Chemoselective Staudinger-phosphite reaction of azides for the phosphorylation of proteins. Angew Chem Int Ed. 2009;48:8234–9. doi: 10.1002/anie.200902118. [DOI] [PubMed] [Google Scholar]
- 54.Arbely E, Torres-Kolbus J, Deiters A, Chin JW. Photocontrol of tyrosine phosphorylation in mammalian cells via genetic encoding of photocaged tyrosine. J Am Chem Soc. 2012;134:11912–5. doi: 10.1021/ja3046958. [DOI] [PubMed] [Google Scholar]
- 55.Fan CG, Ip K, Soll D. Expanding the genetic code of Escherichia coli with phosphotyrosine. FEBS Lett. 2016;590:3040–7. doi: 10.1002/1873-3468.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo XZ, Fu GS, Wang RSE, Zhu XY, Zambaldo C, Liu RH, Liu T, Lyu XX, Du JT, Xuan WM, et al.. Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria. Nat Chem Biol. 2017;13:845-+. doi: 10.1038/nchembio.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoppmann C, Wong A, Yang B, Li S, Hunter T, Shokat K M, Wang L. Site-specific incorporation of phosphotyrosine using an expanded genetic code. Nat Chem Biol. 2017;13:842-+. doi: 10.1038/nchembio.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baeza J, Smallegan MJ, Denu JM. Mechanisms and dynamics of protein acetylation in mitochondria. Trends Biochem Sci. 2016;41:231–44. doi: 10.1016/j.tibs.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–50. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 60.Albaugh BN, Arnold KM, Lee S, Denu JM. Autoacetylation of the histone acetyltransferase Rtt109. J Biol Chem. 2011;286:24694–701. doi: 10.1074/jbc.M111.251579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N-epsilon-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–4. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 62.Umehara T, Kim J, Lee S, Guo LT, Soll D, Park HS. N-Acetyl lysyl-tRNA synthetases evolved by a CcdB-based selection possess N-acetyl lysine specificity in vitro and in vivo. FEBS Lett. 2012;586:729–33. doi: 10.1016/j.febslet.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 63.Hancock SM, Uprety R, Deiters A, Chin JW. Expanding the genetic code of yeast for incorporation of diverse unnatural amino acids via a pyrrolysyl-tRNA synthetase/tRNA pair. J Am Chem Soc. 2010;132:14819–24. doi: 10.1021/ja104609m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, Yokoyama S. Adding L-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem Biophys Res Commun. 2008;371:818–22. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- 65.Elsasser SJ, Ernst RJ, Walker OS, Chin JW. Genetic code expansion in stable cell lines enables encoded chromatin modification. Nat Methods. 2016;13:158-+. doi: 10.1038/nmeth.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han S, Yang A, Lee S, Lee HW, Park CB, Park HS. Expanding the genetic code of Mus musculus. Nat Commun. 2017;8:14568. doi: 10.1038/ncomms14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histories defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–63. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell. 2010;37:834–42. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hsu WW, Wu B, Liu WSR. Sirtuins 1 and 2 are universal histone deacetylases. ACS Chem Biol. 2016;11:792–9. doi: 10.1021/acschembio.5b00886. [DOI] [PubMed] [Google Scholar]
- 70.Wang WW, Zeng Y, Wu B, Deiters A, Liu WR. A chemical biology approach to reveal Sirt6-targeted histone H3 sites in nucleosomes. ACS Chem Biol. 2016;11:1973–81. doi: 10.1021/acschembio.6b00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang YP, Zhou LS, Zhao YZ, Wang SW, Chen LL, Liu LX, Ling ZQ, Hu FJ, Sun YP, Zhang JY, et al.. Regulation of G6PD acetylation by SIRT2 and KAT9 modulates NADPH homeostasis and cell survival during oxidative stress. EMBO J. 2014;33:1304–20. doi: 10.1002/embj.201387224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu YP, Li FL, Lv L, Li TT, Zhou X, Deng CX, Guan KL, Lei QY, Xiong Y. Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase. Cancer Res. 2014;74:3630–42. doi: 10.1158/0008-5472.CAN-13-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weinert BT, Wagner SA, Horn H, Henriksen P, Liu WSR, Olsen JV, Choudhary C.. Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci Signal. 2011;4:11. doi: 10.1126/scisignal.2001902. [DOI] [PubMed] [Google Scholar]
- 74.Zhao D, Zou SW, Liu Y, Zhou X, Mo Y, Wang P, Xu YH, Dong B, Xiong Y, Lei QY, et al.. Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer Cell. 2013;23:464–76. doi: 10.1016/j.ccr.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mo F, Zhuang XX, Liu X, Yao PY, Qin B, Su ZQ, Zang J, Wang Z, Zhang J, Dou Z, et al.. Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat Chem Biol. 2016;12:226-+. doi: 10.1038/nchembio.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang QF, Zhou AP, Li SX, Ni JJ, Tao J, Lu J, Wan BS, Li S, Zhang J, Zhao SM, et al.. Reversible lysine acetylation is involved in DNA replication initiation by regulating activities of initiator DnaA in Escherichia coli. Sci Rep. 2016;6:13. doi: 10.1038/srep30837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ren J, Sang Y, Tan YC, Tao J, Ni JJ, Liu ST, Fan X, Zhao W, Lu J, Wu WJ, et al.. Acetylation of Lysine 201 inhibits the DNA-binding ability of PhoP to regulate salmonella virulence. PLoS Pathog. 2016;12:29. doi: 10.1371/journal.ppat.1005458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim S-W, Lee KJ, Kim S, Kim J, Cho K, Ro H-S, Park H-S, et al.. Genetic incorporation of Nε-acetyllysine reveals a novel acetylation-sumoylation switch in yeast. Biochim Biophys Acta. 2017. doi: 10.1016/j.bbagen.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 79.Zhang X, Huang YL, Shi XB. Emerging roles of lysine methylation on non-histone proteins. Cell Mol Life Sci. 2015;72:4257–72. doi: 10.1007/s00018-015-2001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nguyen DP, Alai MMG, Kapadnis PB, Neumann H, Chin JW. Genetically encoding N-epsilon-methyl-L-lysine in recombinant histones. J Am Chem Soc. 2009;131:14194-+. doi: 10.1021/ja906603s. [DOI] [PubMed] [Google Scholar]
- 81.Wang Y-S, Wu B, Wang Z, Huang Y, Wan W, Russell WK, Pai PJ, Moe YN, Russell DH, Liu WR.. A genetically encoded photocaged N-epsilon-methyl-L-lysinewz. Mol Biosyst. 2010;6:1575–8. doi: 10.1039/c002155e. [DOI] [PubMed] [Google Scholar]
- 82.Groff D, Chen PR, Peters FB, Schultz PG. A genetically encoded epsilon-N-methyl lysine in mammalian cells. Chembiochem. 2010;11:1066–8. doi: 10.1002/cbic.200900690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ai H-w, JW Lee, Schultz PG. A method to site-specifically introduce methyllysine into proteins in E. coli. Chem Commun. 2010;46:5506–8. doi: 10.1039/c0cc00108b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nguyen DP, Alai MMG, Virdee S, Chin JW. Genetically directing epsilon-N, N-dimethyl-L-lysine in recombinant histones. Chem Biol. 2010;17:1072–6. doi: 10.1016/j.chembiol.2010.07.013 [DOI] [PubMed] [Google Scholar]
- 85.Yanagisawa T, Takahashi M, Mukai T, Sato S, Wakamori M, Shirouzu M, Sakamoto K, Umehara T, Yokoyama S. Multiple site-specific installations of N-epsilon-monomethyl-L-lysine into histone proteins by cell-based and cell-free protein synthesis. Chembiochem. 2014;15:1830–8. doi: 10.1002/cbic.201402291. [DOI] [PubMed] [Google Scholar]
- 86.Lin HN, Su XY, He B. Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem Biol. 2012;7:947–60. doi: 10.1021/cb3001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6:812–9. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Resh MD. Fatty acylation of proteins: The long and the short of it. Prog Lipid Res. 2016;63:120–31. doi: 10.1016/j.plipres.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng ZY, Rousseaux S, Rajagopal N, et al.. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1015–27. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gattner MJ, Vrabel M, Carell T. Synthesis of epsilon-N-propionyl-, epsilon-N-butyryl-, and epsilon-N-crotonyl-lysine containing histone H3 using the pyrrolysine system. Chem Commun. 2013;49:379–81. doi: 10.1039/C2CC37836A [DOI] [PubMed] [Google Scholar]
- 91.Kim CH, Kang M, Kim HJ, Chatterjee A, Schultz PG. Site-specific incorporation of epsilon-N-crotonyllysine into histones. Angew Chem Int Ed. 2012;51:7246–9. doi: 10.1002/anie.201203349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee YJ, Wu B, Raymond JE, Zeng Y, Fang XQ, Wooley KL, Liu WR. A genetically encoded acrylamide functionality. ACS Chem Biol. 2013;8:1664–70. doi: 10.1021/cb400267m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wilkins BJ, Hahn LE, Heitmuller S, Frauendorf H, Valerius O, Braus GH, Neumann H. Genetically encoding lysine modifications on histone H4. ACS Chem Biol. 2015;10:939–44. doi: 10.1021/cb501011v. [DOI] [PubMed] [Google Scholar]
- 94.Wang ZPA, Kurra Y, Wang X, Zeng Y, Lee YJ, Sharma V, Lin H, Dai SY, Liu WR. A versatile approach for site-specific lysine acylation in proteins. Angew Chem Int Ed. 2017;56:1643–7. doi: 10.1002/anie.201611415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stone MJ, Chuang S, Hou X, Shoham M, Zhu JZ. Tyrosine sulfation: An increasingly recognised post-translational modification of secreted proteins. N Biotechnol. 2009;25:299–317. doi: 10.1016/j.nbt.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 96.Yang YS, Wang CC, Chen BH, Hou YH, Hung KS, Mao YC. Tyrosine sulfation as a protein post-translational modification. Molecules. 2015;20:2138–64. doi: 10.3390/molecules20022138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu CC, Schultz PG. Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat Biotechnol. 2006;24:1436–40. doi: 10.1038/nbt1254. [DOI] [PubMed] [Google Scholar]
- 98.Choe H, Li WH, Wright PL, Vasilieva N, Venturi M, Huang CC, Grundner C, Dorfman T, Zwick MB, Wang L, et al.. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell. 2003;114:161–70. doi: 10.1016/S0092-8674(03)00508-7. [DOI] [PubMed] [Google Scholar]
- 99.Liu CC, Choe H, Farzan M, Smider VV, Schultz PG. Mutagenesis and evolution of sulfated antibodies using an expanded genetic code. Biochemistry. 2009;48:8891–8. doi: 10.1021/bi9011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pruitt RN, Schwessinger B, Joe A, Thomas N, Liu F, Albert M, Robinson MR, Chan LJ, Luu DD, Chen H, et al.. The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci Adv. 2015;1:e1500245. doi: 10.1126/sciadv.1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schwessinger B, Li X, Ellinghaus TL, Chan LJG, Wei T, Joe A, Thomas N, Pruitt R, Adams PD, Chern MS, et al.. A second-generation expression system for tyrosine-sulfated proteins and its application in crop protection. Integr Biol. 2016;8:542–5. doi: 10.1039/C5IB00232J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Houee-Levin C, Bobrowski K, Horakova L, Karademir B, Schoneich C, Davies MJ, Spickett CM. Exploring oxidative modifications of tyrosine: An update on mechanisms of formation, advances in analysis and biological consequences. Free Radic Res. 2015;49:347–73. doi: 10.3109/10715762.2015.1007968. [DOI] [PubMed] [Google Scholar]
- 103.Ahsan H. 3-Nitrotyrosine: A biomarker of nitrogen free radical species modified proteins in systemic autoimmunogenic conditions. Hum Immunol. 2013;74:1392–9. doi: 10.1016/j.humimm.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 104.Neumann H, Hazen JL, Weinstein J, Mehl RA, Chin JW. Genetically encoding protein oxidative damage. J Am Chem Soc. 2008;130:4028–33. doi: 10.1021/ja710100d. [DOI] [PubMed] [Google Scholar]
- 105.Cooley RB, Feldman JL, Driggers CM, Bundy TA, Stokes AL, Karplus PA, Mehl RA. Structural basis of improved second-generation 3-nitro-tyrosine tRNA synthetases. Biochemistry. 2014;53:1916–24. doi: 10.1021/bi5001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yamakura F, Taka H, Fujimura T, Murayama K. Inactivation of human manganese-superoxide dismutase by peroxynitrite is caused by exclusive nitration of tyrosine 34 to 3-nitrotyrosine. J Biol Chem. 1998;273:14085–9. doi: 10.1074/jbc.273.23.14085. [DOI] [PubMed] [Google Scholar]
- 107.Phillips MC. Molecular mechanisms of cellular cholesterol efflux. J Biol Chem. 2014;289:24020–9. doi: 10.1074/jbc.R114.583658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.DiDonato JA, Aulak K, Huang Y, Wagner M, Gerstenecker G, Topbas C, Gogonea V, DiDonato AJ, Tang WH, Mehl RA, et al.. Site-specific nitration of apolipoprotein A-I at tyrosine 166 is both abundant within human atherosclerotic plaque and dysfunctional. J Biol Chem. 2014;289:10276–92. doi: 10.1074/jbc.M114.556506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Franco MC, Ye YZ, Refakis CA, Feldman JL, Stokes AL, Basso M, Melero Fernández de Mera RM, Sparrow NA, Calingasan NY, Kiaei M, et al.. Nitration of Hsp90 induces cell death. Proc Natl Acad Sci U S A. 2013;110:E1102–11. doi: 10.1073/pnas.1215177110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–67. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 111.Doores KJ, Mimura Y, Dwek RA, Rudd PM, Elliott T, Davis BG. Direct deprotected glycosyl-asparagine ligation. Chem Commun. 2006;13:1401–3. doi: 10.1039/b515472c. [DOI] [PubMed] [Google Scholar]
- 112.Baumann K, Kowalczyk D, Kunz H. Total synthesis of the glycopeptide recognition domain of the P-selectin glycoprotein ligand 1. Angew Chem Int Ed. 2008;47:3445–9. doi: 10.1002/anie.200705762. [DOI] [PubMed] [Google Scholar]
- 113.Rodriguez EC, Winans KA, King DS, Bertozzi CR. A strategy for the chemoselective synthesis of O-linked glycopeptides with native sugar-peptide linkages. J Am Chem Soc. 1997;119:9905–6. doi: 10.1021/ja971633p [DOI] [Google Scholar]
- 114.Marotta NP, Lin YH, Lewis YE, Ambroso MR, Zaro BW, Roth MT, Arnold DB, Langen R, Pratt MR. O-GlcNAc modification blocks the aggregation and toxicity of the protein alpha-synuclein associated with Parkinson's disease. Nat Chem. 2015;7:913–20. doi: 10.1038/nchem.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Izumi M, Makimura Y, Dedola S, Seko A, Kanamori A, Sakono M, Ito Y, Kajihara Y. Chemical synthesis of intentionally misfolded homogeneous glycoprotein: A unique approach for the study of glycoprotein quality control. J Am Chem Soc. 2012;134:7238–41. doi: 10.1021/ja3013177. [DOI] [PubMed] [Google Scholar]
- 116.Davis BG, Lloyd RC, Jones JB. Controlled site-selective glycosylation of proteins by a combined site-directed mutagenesis and chemical modification approach. J Org Chem. 1998;63:9614–5. doi: 10.1021/jo9816461 [DOI] [Google Scholar]
- 117.van Kasteren SI, Kramer HB, Jensen HH, Campbell SJ, Kirkpatrick J, Oldham NJ, Anthony DC, Davis BG. Expanding the diversity of chemical protein modification allows post-translational mimicry. Nature. 2007;446:1105–9. doi: 10.1038/nature05757. [DOI] [PubMed] [Google Scholar]
- 118.Dondoni A, Massi A, Nanni P, Roda A. A new ligation strategy for peptide and protein glycosylation: Photoinduced thiol-ene coupling. Chemistry. 2009;15:11444–9. doi: 10.1002/chem.200901746. [DOI] [PubMed] [Google Scholar]
- 119.Floyd N, Vijayakrishnan B, Koeppe AR, Davis BG. Thiyl glycosylation of olefinic proteins: S-linked glycoconjugate synthesis. Angew Chem Int Ed. 2009;48:7798–802. doi: 10.1002/anie.200903135. [DOI] [PubMed] [Google Scholar]
- 120.Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: The leukocyte adhesion cascade updated. Nat Rev Immunol. 2007;7:678–89. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- 121.Somers WS, Tang J, Shaw GD, Camphausen RT. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell. 2000;103:467–79. doi: 10.1016/S0092-8674(00)00138-0. [DOI] [PubMed] [Google Scholar]
- 122.Lercher L, Raj R, Patel NA, Price J, Mohammed S, Robinson CV, Schofield CJ, Davis BG.. Generation of a synthetic GlcNAcylated nucleosome reveals regulation of stability by H2A-Thr101 GlcNAcylation. Nat Commun. 2015;6:9. doi: 10.1038/ncomms8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Raj R, Lercher L, Mohammed S, Davis BG. Synthetic nucleosomes reveal that GlcNAcylation modulates direct interaction with the FACT complex. Angew Chem Int Ed. 2016;55:8918–22. doi: 10.1002/anie.201603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18:579–86. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 125.Herhaus L, Dikic I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 2015;16:1071–83. doi: 10.15252/embr.201540891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kumar KSA, Spasser L, Erlich LA, Bavikar SN, Brik A. Total chemical synthesis of Di-ubiquitin chains. Angew Chem Int Ed. 2010;49:9126–31. doi: 10.1002/anie.201003763. [DOI] [PubMed] [Google Scholar]
- 127.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–U12. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li X, Fekner T, Ottesen JJ, Chan MK. A pyrrolysine analogue for site-specific protein ubiquitination. Angew Chem Int Ed. 2009;48:9184–7. doi: 10.1002/anie.200904472. [DOI] [PubMed] [Google Scholar]
- 129.Virdee S, Kapadnis PB, Elliott T, Lang K, Madrzak J, Nguyen DP, Riechmann L, Chin JW. Traceless and site-specific ubiquitination of recombinant proteins. J Am Chem Soc. 2011;133:10708–11. doi: 10.1021/ja202799r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat Chem Biol. 2010;6:750–7. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- 131.Dixon EK, Castaneda CA, Kashyap TR, Wang Y, Fushman D. Nonenzymatic assembly of branched polyubiquitin chains for structural and biochemical studies. Bioorg Med Chem. 2013;21:3421–9. doi: 10.1016/j.bmc.2013.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rosner D, Schneider T, Schneider D, Scheffner M, Marx A. Click chemistry for targeted protein ubiquitylation and ubiquitin chain formation. Nat Protoc. 2015;10:1594–611. doi: 10.1038/nprot.2015.106. [DOI] [PubMed] [Google Scholar]
- 133.Valkevich EM, Guenette RG, Sanchez NA, Chen Y-c, Ge Y, Strieter ER. Forging isopeptide bonds using thiol-ene chemistry: Site-specific coupling of ubiquitin molecules for studying the activity of isopeptidases. J Am Chem Soc. 2012;134:6916–9. doi: 10.1021/ja300500a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Trang VH, Valkevich EM, Minami S, Chen Y-C, Ge Y, Strieter ER. Nonenzymatic polymerization of ubiquitin: Single-step synthesis and isolation of discrete ubiquitin oligomers. Angew Chem Int Ed. 2012;51:13085–8. doi: 10.1002/anie.201207171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Trang VH, Rodgers ML, Boyle KJ, Hoskins AA, Strieter ER. Chemoenzymatic synthesis of bifunctional polyubiquitin substrates for monitoring ubiquitin chain remodeling. Chembiochem. 2014;15:1563–8. doi: 10.1002/cbic.201402059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Swaney DL, Villén J. Proteomic analysis of protein posttranslational modifications by mass spectrometry. Cold Spring Harb Protoc. 2016;2016:pdb.top077743. doi: 10.1101/pdb.top077743. [DOI] [PMC free article] [PubMed] [Google Scholar]


