ABSTRACT
The aminoacylation status of the cellular tRNA pool regulates both general amino acid control (GAAC) and target of rapamycin (TOR) stress response pathways in yeast. Consequently, fidelity of translation at the level of aminoacyl-tRNA synthesis plays a central role in determining accuracy and sensitivity of stress responses. To investigate effects of translational quality control (QC) on cell physiology under stress conditions, phenotypic microarray analyses were used to identify changes in QC deficient cells. Nitrogen source growth assays showed QC deficient yeast grew differently compared to WT. The QC deficient strain was more tolerant to caffeine treatment than wild type through altered interactions with the TOR and GAAC pathways. Increased caffeine tolerance of the QC deficient strain was consistent with the observation that the activity of Gln3p, a transcription factor controlled by the TOR pathway, is decreased in the QC deficient strain compared to WT. GCN4 translation, which is typically repressed in the absence of nutritional stress, was enhanced in the QC deficient strain through TOR inhibition. QC did not impact cell cycle regulation; however, the chronological lifespan of QC deficient yeast strains decreased compared to wild type, likely due to translational errors and alteration of the TOR-associated regulon. These findings support the idea that changes in translational fidelity provide a mechanism of cellular adaptation by modulating TOR activity. This, in turn, supports a central role for aminoacyl-tRNA synthesis QC in the integrated stress response by maintaining the proper aa-tRNA pools necessary to coordinate the GAAC and TOR.
KEYWORDS: Aminoacyl-tRNA, GAAC, nutritional stress, TOR, tRNA editing
Introduction
Direct interactions between stress signaling pathways and the translation machinery allow for cellular responses to environmental cues via changes in the intracellular tRNA pool. As environmental nutrient availability shifts, so too does the internal pool of amino acids and other cellular metabolites. Metabolic reprogramming in response to nutritional stress involves both biosynthesis and catabolism of amino acids which ultimately effect the composition of the aminoacyl-tRNA (aa-tRNA) pool. Cellular stress response networks, such as the general amino acid control (GAAC) and target of rapamycin (TOR) pathways, rely heavily on their ability to monitor changes in aa-tRNA pools, making aa-tRNA pool fidelity central to the appropriate regulation of nutrient sensing mechanisms which direct the transcriptional regulation of gene expression in response to nutritional stresses.1-5
During normal growth conditions, the TOR pathway positively regulates cellular growth by stimulating ribosome biogenesis and by controlling the generation and utilization of precursors for the synthesis of amino acids and other nitrogenous macromolecules. As nutrient availability changes, yeast cells initiate metabolic reprogramming via signal transduction pathways involving two homologous protein kinases, Tor1p and Tor2p, found in two distinct complexes, TORC1 and TORC2, respectively.6 TORC1 specifically regulates a number of signal transduction pathways which monitor and adjust metabolic pathways in response to changes in nitrogen availability. Yeast, like many bacteria, have a hierarchy of preferred nitrogen sources governed by a regulatory scheme known as nitrogen catabolite repression (NCR). In the presence of a primary (preferred) nitrogen source, GATA family transcriptional activators, Gln3 and Gat1, form complexes with Ure2 and are localized to the cytoplasm, which decreases NCR-sensitive gene expression. During nitrogen limitation or in the presence of secondary (non-preferred) nitrogen sources, Gln3 and Gat1 are dephosphorylated and moved from the cytoplasm to the nucleus, resulting in increased expression of NCR-sensitive genes.7 While the functional role of TORC1 mediated nutrient sensing in NCR-mediated metabolic programming has been established, many questions remain regarding coordination of nutrient sensing with other stress response pathways and the mechanisms by which this process occurs.
Regulatory checkpoints and cross-talk with other stress response pathways provide NCR-mediated metabolic reprogramming with additional contextual information which provides the flexibility required to efficiently and accurately coordinate a response to nitrogen stress. One of the most well-defined example of stress response pathway cross-talk in yeast involves the integration of the GAAC (amino acid starvation response) and TOR pathways, which together allow the cell to mount a comprehensive transcriptional response to changes in the environment or nutrient availability. Activation of the protein kinase Gcn2p, which monitors amino acid limitation through interaction with deacylated tRNA, is repressed in the presence of preferred nitrogen sources by phosphorylation of S577 by unknown TOR pathway components as a mechanism to limit GAAC activation during nutrient rich growth conditions.8 Conversely, during amino acid starvation, the protein kinase Gcn2p represses TOR activity by phosphorylating TOR pathway components, thereby preventing further depletion of internal amino acid pools by TOR-initiated degradation of amino acids. This complex regulatory crosstalk ultimately coordinates the transcriptional response to nutrient stress which, in many cases, requires both Gcn4p and Gln3p to mount an appropriate response.9-12 These observations suggest a shared stress sensing mechanism mediated by Gcn2p and its interactions with the intracellular aa-tRNA pool.
Aside from their role as substrates for protein synthesis, aa-tRNAs have recently been shown to play critical roles in the maintenance of cellular stress response and viability. The composition and fidelity of the aa-tRNA pool is maintained by aminoacyl-tRNA synthetases (aaRSs), enzymes which pair amino acids with their cognate tRNAs. For example, phenylalanyl-tRNA synthetase (PheRS) is responsible for pairing phenylalanine with its cognate tRNAPhe isoacceptors.13 Mispaired aminoacyl-tRNA species are occasionally synthesized (at a rate of ∼1%) due to incomplete discrimination against non-cognate amino acids within the PheRS active site, in this case resulting in the misacylation of tyrosine onto tRNAPhe (Tyr-tRNAPhe).14 Inherent proofreading mechanisms prevent accumulation of misacylated tRNAPhe through hydrolysis of misactivated aminoacyl adenylates (pre-transfer editing) and hydrolysis of misacylated aa-tRNA (post-transfer editing) resulting in ∼0.02% and ∼0.09% Phe misincorporation at Tyr codons in WT and proofreading deficient PheRS strains, respectively.15 While aa-tRNA quality control has recently been ascribed a role in maintaining the sensitivity of GAAC pathway activation, little is known about how this conserved step in translation quality control might regulate other related cellular processes such as cross talk with the TOR pathway.15-17
Results
Defects in translation quality control perturb regulation of multiple stress response networks
To effectively manage fluctuations in environmental and nutritional status, the cell must continually monitor and integrate stress signals to mount an efficient and coordinated adaptive response. As a result, most cellular stress response pathways are connected in a series of cross-talk pathways to ensure an appropriate and proportional response to stress. Although aaRS-mediated QC has recently been implicated in the regulation of specific cellular stress responses, a comprehensive phenotypic survey of stress responses has yet to be conducted. To identify other stress response pathways affected by changes in aaRS-mediated QC, we used a WT and isogenic strain of yeast with a proofreading deficient PheRS in a series of growth experiments with phenotypic microarrays (PM). Panels designed to assay nitrogen, carbon, and amino acid utilization, as well as chemical sensitivity were conducted with both WT and QC deficient strains. Conditions resulting in differential growth were compiled and assigned an associated allelic designation based on known chemicogenomic interactions mined from the publicly available data. The interaction of associated alleles was examined and mapped onto an interaction network (expanded shell by 10 interactions) to identify context within the larger stress response network. As expected, we found significant enrichment for GAAC pathway associated genes. Additionally, we observed enrichment for genes associated with the TOR pathway (Fig. 1A, Supplemental Table 1). From the chemical sensitivity PM, we observed differential growth in response to caffeine. Caffeine is a low affinity ATP analog which interacts with a number of cellular processes, including direct interaction with components of the TOR pathway.18 Recent work has established mechanistic links between caffeine tolerance, Tor1p, and several TOR complex proteins. In the absence of Tor1p, sensitivity to caffeine is increased, while hyperactive mutations of Tor1p kinase activity generally increase caffeine tolerance.19,20 In response to caffeine, the QC deficient strain displayed increased tolerance compared to WT in both the PM and additional more detailed analyses of this phenotype (Fig. 1B, C). The magnitude of increase in caffeine tolerance for the QC deficient strain was less than previous observations using TOR null or mutant strains. As a whole, the PM data and specifically the differential response to caffeine treatment indicates that TOR activity or the regulation of TOR pathway components is sensitive to defects in aaRS-mediated QC. Furthermore, the results of the PM reiterate known associations between translation QC and regulation of the GAAC and highlight potential overlap of GAAC and TOR pathway components whose regulation may be affected by aa-tRNA pool fidelity.
Figure 1.
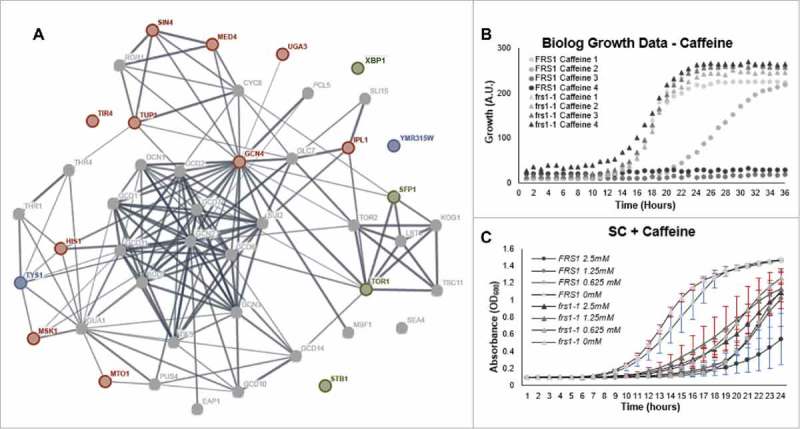
Phenotypic analysis of WT and PheRS QC deficient yeast. (A) WT and QC deficient strains of yeast were used for phenotypic screening by Biolog Phenotypic Microarray (PM). PM conditions which facilitated growth different from wild type were mapped onto a gene interaction network (expanded shell by 10 connections) using stringDB. Genes linked to growth conditions conferring growth advantage are indicated by blue and red colored nodes for the WT (FRS1) and proofreading deficient (frs1-1) strains, respectively. See supplement for gene table. Green nodes indicate gene association with both WT and frs1-1 depending on growth condition. (B) PM data for growth in response to caffeine (included within the chemical sensitivity PM plate) was extracted from cumulative data set and plotted as a function of time. (C) Caffeine growth phenotypes observed from the PM were validated through independent growth experiments. WT and QC deficient yeast were grown in synthetic complete (SC) medium with varying amounts of caffeine supplementation. Growth was followed by monitoring the OD600 over time. Results are representative of three independent replicates. Error bars colored blue for FRS1 and red for frs1-1 and indicate 1 S.D.
PheRS QC impacts nitrogen source dependent growth
When faced with nitrogen limitation, yeast cells are able to maintain viability by altering their metabolism to utilize other sources of nitrogen. This process involves crosstalk between the TOR and GAAC pathways to coordinate the efficient use of secondary nitrogen sources, including amino acids.8 As evidenced by the results of the PM analysis, lack of PheRS QC can differentially impact cellular growth during nutrient stress. To examine whether aaRS proofreading and the TOR mediated NCR regulatory pathway interact to alter cellular growth, we conducted growth assays of WT and PheRS QC deficient strains of yeast in the presence of varying nitrogen sources. Growth of both WT and QC deficient strains on media containing ammonium (YNB-NH4) supplemented individually with 2mM of each amino showed no significant differences in growth (Fig. 2A), with similar results observed on solid YNB-NH4 medium with and without amino acid supplementation (Fig. 2D). When grown in liquid medium containing gamma-aminobutyric acid (YNB-GABA) or amino acids (YNB) as the sole nitrogen source, cellular growth was moderately repressed for the QC deficient strain compared to WT (Fig. 2B, C). On solid medium of similar composition, however, the observed growth deficiency was less pronounced (Fig. 2E, F). Taken together with the results of the PM, these results suggest that under certain conditions, lack of aaRS QC may alter the growth of yeast in a nitrogen source dependent manner.
Figure 2.
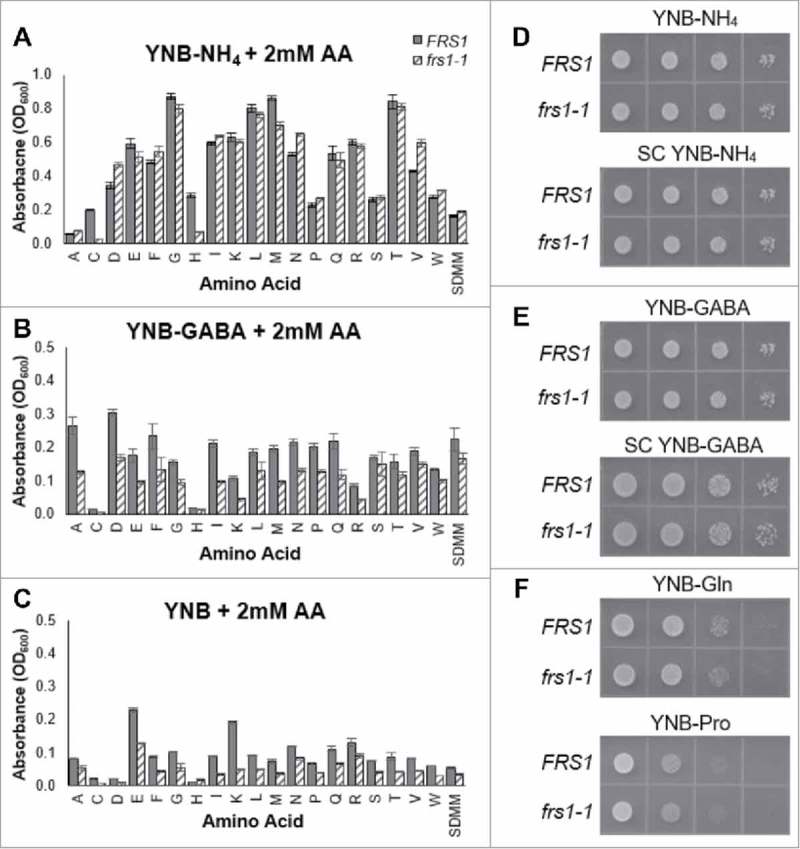
Growth of WT and QC deficient strains with different nitrogen sources. WT and PheRS QC deficient strains of yeast were inoculated to an OD600 of 0.01 and grown for 24 or 48 hours (A-C indicate maximum OD600 values after incubation) on synthetic defined minimal media (SDMM) with varying sources of nitrogen. (A) Multiplexed growth assays were conducted in 96 well plates with yeast nitrogen base containing ammonium sulfate (YNB-NH4) as the primary nitrogen source and supplemented with individual amino acids (2 mM). (B) Multiplexed growth assays were conducted in 96 well plates with yeast nitrogen base containing GABA (YNB-GABA) as the primary nitrogen source and supplemented with individual amino acids (2mM). (C) Multiplexed growth assays were conducted in 96 well plates with yeast nitrogen base supplemented with individual amino acids (YNB-2mM AA) as the primary nitrogen source. (D-F) WT and QC deficient cells were grown in YPD and serial dilutions (1 – 1 × 10−4) were spotted on solid SDMM with varying sources of nitrogen and grown for 24–48 hours. Growth assays were conducted in triplicate. Error bars are representative of 1 S.D.
aa-tRNA editing is required for proper regulation of GATA-mediated response to nitrogen stress
The ability to scavenge nitrogen from a variety of environmental sources is of particular importance as nitrogen availability is central to the synthesis of amino acids. Utilization of secondary nitrogen sources is mediated through the TOR pathway, with downstream activation of the transcription factor Gln3p resulting in corresponding reprogramming of gene expression.11,21 We have previously established that reduction in PheRS QC impacts regulation of the GAAC during amino acid stress.15 To assess whether QC-dependent perturbations of growth on various nitrogen sources involve changes in the TOR pathway, we monitored activity of the transcription factor Gln3p. Activation of Gln3p targets was examined using a set of β-galactosidase based reporters described previously,8 which provide information regarding Gln3p dependent transcription of consensus binding GATA sequence as well as the downstream target UGA1. Both WT and QC deficient strains harboring GATA: LacZ or UGA1:LacZ reporter plasmids were grown in YNB-NH4 and YNB-GABA. Surprisingly, the activity of Gln3p was found to be significantly repressed in the presence of ammonium (YNB-NH4), where no obvious growth defects were observed (Fig. 3). Gln3p activity of WT and QC deficient strains during growth in YNB-GABA was elevated compared to activity in YNB-NH4, yet the decrease in activity between strains was less significant, likely due to Gln3p-independent regulation of GABA catabolism22 (Fig. 3A). In accordance with previously published results, treatment of cells with 3-aminotriazole (3-AT), a chemical activator of the GAAC, resulted in repression of TOR-mediated Gln3p activity 8 (Fig. 3A).
Figure 3.
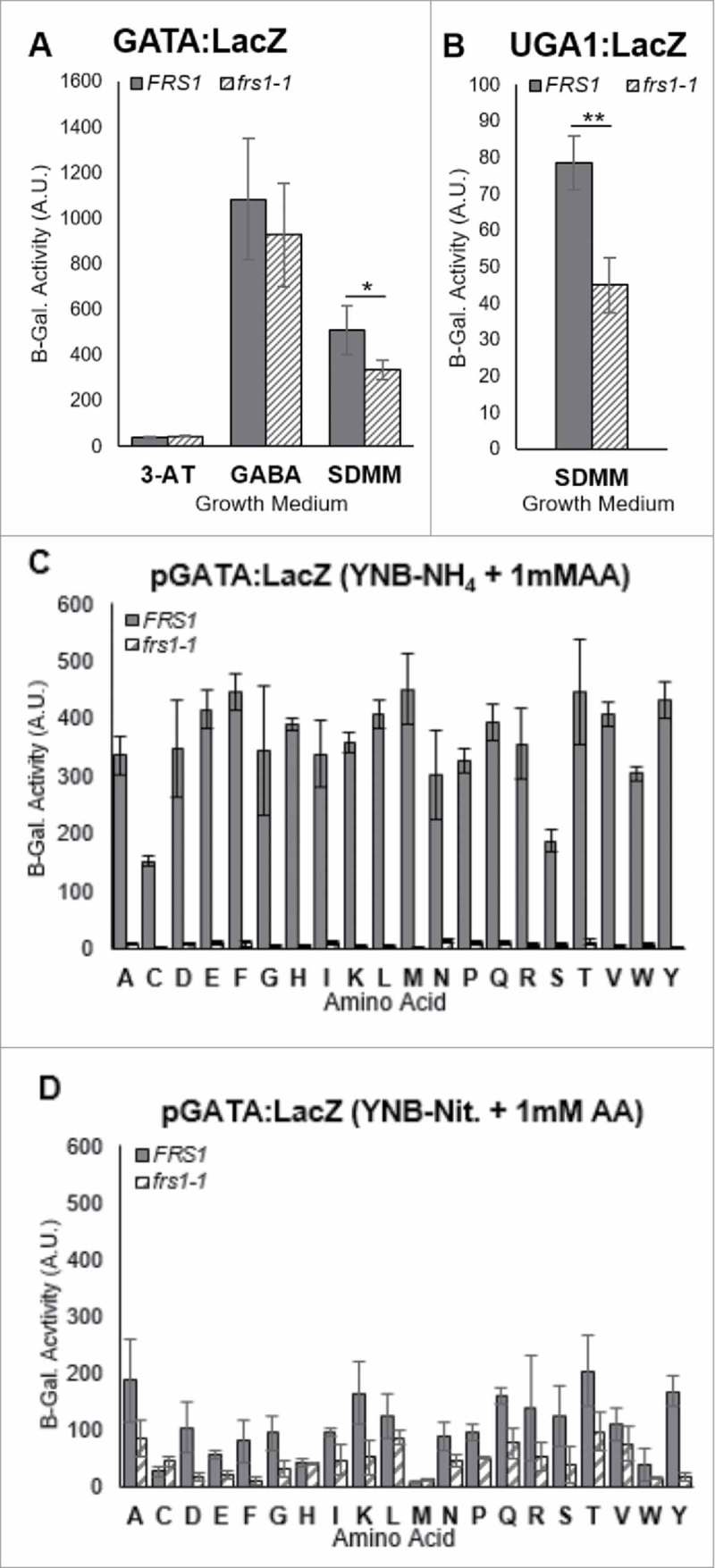
Regulation of Gln3p targets in response to diverse amino acid nitrogen sources. (A-B) WT (FRS1) and QC deficient (frs1-1) cells harboring plasmid pGATA:LacZ or pUGA1:LacZ were grown in the indicated medium. GABA was used to induce Gln3p expression. Significance was assessed using paired sample t-test where “*” represents p < 0.05, “**” represents p < 0.005 (C) WT and QC deficient cells harboring plasmid pGATA:LacZ were grown in synthetic defined minimal medium (SDMM) with ammonium sulfate as the primary nitrogen source and supplemented individually with each amino acid (1 mM). (D) WT and QC deficient cells harboring plasmid pGATA:LacZ were grown in synthetic defined minimal medium (SDMM) with each amino acid (1 mM) as the primary nitrogen source. Cells were analyzed for betagalactosidase activity using CPRG chromogenic substrate while monitoring the OD580 over time. Rate was determined in triplicate and normalized to OD600 values. Error bars reflect S.D. determined from at least three independent experiments.
To further investigate Gln3p activity during growth in ammonium containing medium, the GATA:LacZ reporter was used to monitor Gln3p activity in both WT and QC deficient strains grown under similar conditions as the high throughput growth assay (Fig. 2). The activity of Gln3p was strikingly reduced in QC deficient strains compared to WT during growth in medium with ammonium (Fig. 3C). Reduction in Gln3p activity was also observed in QC deficient strains grown in media with amino acids as the sole nitrogen source (Fig. 3D). These results indicate that PheRS QC is required for proper regulation of the TOR pathway component, Gln3p. Differences in magnitude of Gln3p activity depending on nitrogen source further suggest that the impact of aaRS QC may vary depending on the source of available nitrogen.
QC dependent TOR inhibition de-represses GAAC pathway activation
The GAAC mediates cellular reprogramming of gene expression in response to fluctuations in amino acid availability, with deacylated tRNA serving as the primary sensor. Monitoring the aminoacylation status of tRNA is an efficient means to accurately assess global amino acid availability and bypasses the requirement for sensing of individual amino acid levels. We have recently shown that aa-tRNA proofreading plays a key role in the selectivity and sensitivity of the GAAC response during amino acid stress. The ability of yeast to scavenge nitrogen from multiple secondary sources is a vital component of the response to environmental stressors, and nitrogen starvation has been shown to prevent the translation of GCN4 during periods of amino acid limitation23 and, conversely, inhibition of TOR activates the GAAC through de-phosphorylation of Ser577.24 To investigate how the GAAC participates in PheRS QC mediated TOR repression, we monitored the translation of the GAAC effector, Gcn4p. Translation of GCN4 mRNA was assessed using a GCN4:GFP reporter plasmid harboring the complete upstream open reading frame (uORF) regulatory region found in the 5’ UTR of the native GCN4 mRNA.25,26 Basal levels of GCN4 translation (∼10-20,000 A.U.) for both WT and QC deficient strains were decreased in synthetic complete ammonium medium (SC YNB-NH4) compared to medium lacking amino acids, with QC deficient cells exhibiting decreased GCN4 translation (Fig. 4A). GCN4 expression, ∼30-50,000 A.U., was general increased in amino acid free ammonium medium (YNB-NH4). Consistent with previous observations of TOR mediated GCN4 de-repression, WT cells maintained consistent translation of GCN4 across all time points, but QC deficient cells had significantly higher levels of GCN4 translation (Fig. 4B). These observations indicate that GAAC pathway activation and the ability to sense amino acid limitation is maintained in both WT and QC deficient strains grown in medium with ammonium as the nitrogen source, but during amino acid limitation, GCN4 translation is further de-repressed in QC-deficient compared to WT cells.
Figure 4.
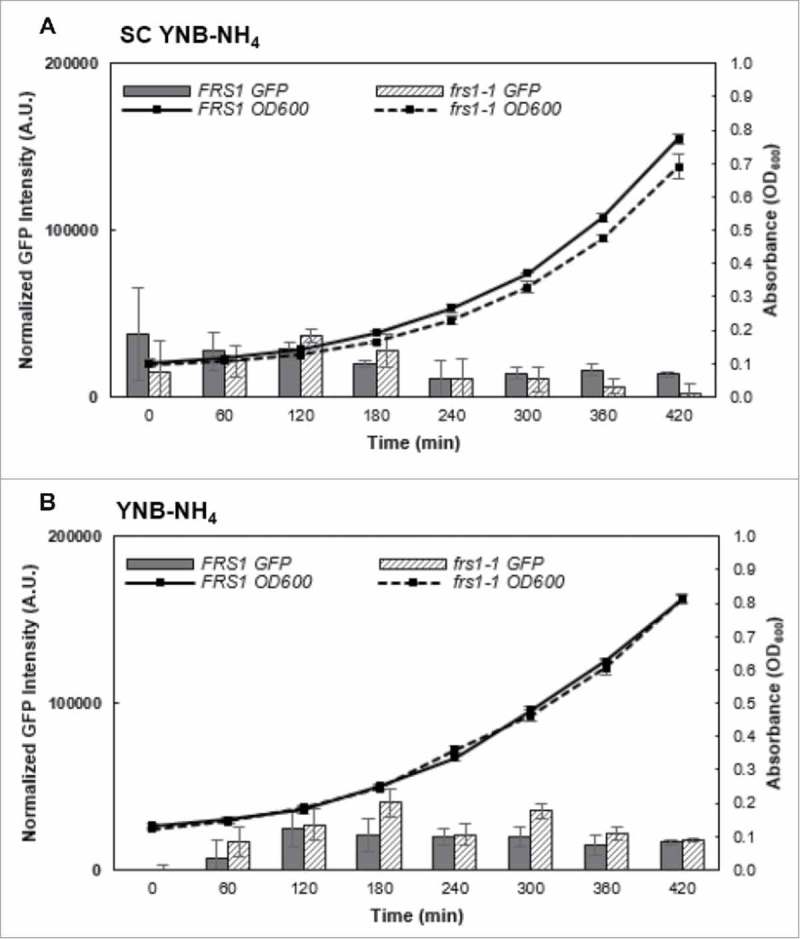
Translation of GCN4 during growth on ammonium. WT and QC deficient strains harboring a GCN4-GFP translation reporter plasmid (pGCN4Rep) were grown in either (A) synthetic complete (SC) or (B) synthetic defined minimal medium (SDMM) with ammonium as the primary nitrogen source (SC YNB-NH4 and YNB-NH4, respectively). GCN4 translation was followed by measuring GFP fluorescence over time and normalized to cellular concentration using OD600 values. Results are the average of three independent replicates. Error bars represent 1 S.D.
Impact of QC on TOR-mediated cellular physiology
Positioned as an integral sensor of nutritional stress within the cell, the TOR pathway is able to couple environmental cues to the cell cycle machinery, rapidly modulating cellular growth in response to nutrient limitation.27 Multiple TOR pathway components have been directly linked to cell cycle regulation, and acute TORC1 inhibition through treatment with rapamycin has been shown to cause G1 arrest.28 To determine whether the QC-mediated alteration of TOR pathway activity caused defects in cell cycle regulation, the DNA content of asynchronous cell populations of WT and PheRS QC deficient strains was measured during growth in medium containing ammonium. No differences were observed in the fraction of cells with G1 or non-G1 DNA content in the QC deficient strain when compared to WT (Fig. 5A). When translation is decreased due to nutritional stress, translation of the G1 cyclin CLN3 is also reduced via a uORF in the 5’ UTR, delaying cell cycle progression.29 A reduction in the expression of regulatory proteins sensitive to translational status, e.g. CLN3, could account for delayed cell cycle progression. The translational status of CLN3 was measured using a CLN3:LacZ reporter plasmid containing the regulatory uORF element fused to beta-galactosidase. As expected, expression of CLN3 in wild type cells was higher in rich medium (SC), than in the same medium containing 3-AT, or in minimal medium (SDMM-NH4). While the same general trend was observed in WT and QC deficient strains, expression of CLN3 was significantly higher in the QC deficient strain in the presence of ammonium (Fig. 5B), reinforcing the observation that aaRS-mediated QC is unlikely to directly modulate TOR-regulated cell cycle progression. Taken together, these results indicate that the QC-dependent decrease in Gln3p activity is not likely the result of general TORC1 inhibition, but rather through NCR specific TOR pathway components.
Figure 5.
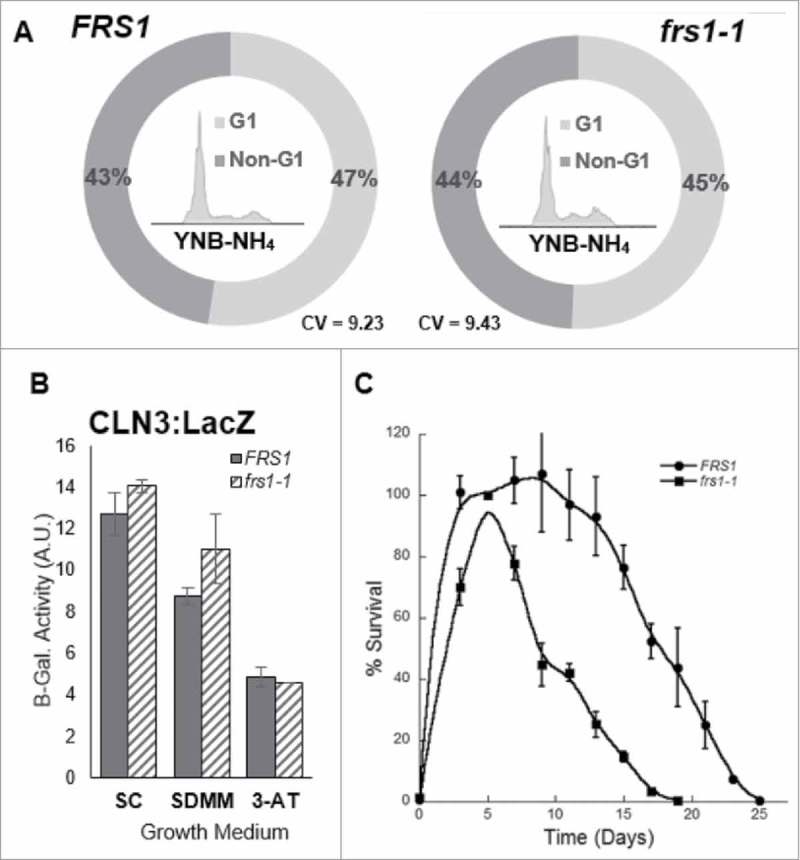
Impact of QC on TOR-dependent cellular physiology. (A) WT (FRS1) and QC deficient (frs1-1) cells were grown in minimal medium (SDMM) or minimal medium supplemented with 1.2 mM Tyrosine (SDMM + Tyr) to an OD600 of 0.8-1.0. The DNA content of cells stained with SYBR Green I was analyzed by flow cytometry. Univariate cell cycle analysis was conducted using a modified Watson Pragmatic algorithm (FlowJo). Coefficient of variation (CV) was determined using width at ½ G1 peak height from at least three independent experiments. (B) WT (FRS1) and QC deficient (frs1-1) cells harboring plasmid uORF-CLN3:LacZ were grown in the indicated medium. 3-aminotriazole (3-AT) was used to induce amino acid starvation. Cells were analyzed for betagalactosidase activity using CPRG chromogenic substrate while monitoring the OD580 over time. Rate was determined in triplicate and normalized to OD600 values. (C) WT (FRS1) and QC deficient (frs1-1) cells were grown in 50 mL of synthetic complete medium with ammonium as the primary nitrogen source. Percent survival was normalized for each culture to CFU/mL at day 5. Error bars reflect 1 S.D. determined from at least three independent experiments.
Stationary phase cells are characterized by an increased resistance to a variety of environmental and nutrient stressors. Stationary phase survival can be determined by monitoring cell viability over time, forming a chronological lifespan. Inhibition of TOR and deletion of TOR pathway components, including GLN3, increase the chronological lifespan of yeast.30 To identify potential defects in the chronological lifespan of QC deficient cells, cultures were grown to stationary phase, with samples collected every two days, thereafter. Initially, WT cells entered stationary phase earlier than QC deficient cells and maintained stationary phase for a longer period of time before a significant loss of viability was observed (Fig. 5C). At day 7, WT cells maintained in YNB-NH4 showed ∼100% survival, whereas the QC deficient cells showed 77–80% survival. WT cells maintained stationary phase for 8–10 days with more than 93% survival. The QC deficient cells, on the other hand, maintained stationary phase for less than 4 days, with only the day 5 time point showing survival above 81%. By day 19, the survival of QC deficient cells was between 0.25% and 0.4%, while the WT cells showed between 20% and 40% survival (Fig. 5C). The significant decrease in the chronological lifespan of the QC deficient cells is consistent with a connection between aa-tRNA pool fidelity and the maintenance of cellular longevity through the TOR pathway.
Discussion
Abolishment of PheRS QC decreases Gln3p activity via TORC1
Translation quality control is normally essential for the maintenance of proteome fidelity and cellular viability. There is, however, an emerging body of work which shows an expanding role for changes in translation fidelity as a mechanism of cellular adaptation.31 Examples of adaptive mistranslation, a term associated with enhanced fitness outcomes, are described with increasing frequency, yet the full scope and mechanisms by which this process occurs remains unexplored. At the proteome level, mistranslation provides an opportunity to increase proteome diversity, thereby enhancing antigenic diversity, alternative protein function, and indirectly affecting mutation rate by enhancing replication error. Organisms that leverage these mechanisms to enhance protein mistranslation and survival during stress have been well documented, but the effects on the cell of a reduction in quality control prior to amino acid misincorporation are less well documented. A number of prokaryotic and eukaryotic cellular stress responses rely on surveillance of the aa-tRNA pool as a primary signal for nutritional stress. Reducing aaRS-mediated quality control decreases aa-tRNA pool fidelity, ultimately altering aa-tRNA pool composition. In both prokaryotes and eukaryotes, this leads to substantial reductions to the cells ability to accurately detect and respond to amino acid starvation.15-17
Through phenotypic analysis, we identified additional stress response pathways which are directly affected by a reduction in aaRS-mediated QC. The TOR pathway is involved in monitoring nutrient availability and modulating cellular growth factors in response to nutritional stress. In yeast, the TOR pathway contributes to the regulation of cell growth and is the primary regulator of nitrogenous macromolecule precursor catabolism and utilization. The GATA family transcription factor, Gln3p, acts as the penultimate effector of the signal transduction pathway responsible for NCR (Fig. 6). Although interactions between the GAAC and the TOR pathway have been characterized, no direct connection between aa-tRNA pool fidelity and the regulation of pathways outside of the GAAC has been described. Our results demonstrate a clear reduction in the activity of the TOR pathway component, Gln3p, in an aaRS-mediated QC dependent manner. Under the same growth conditions, translation of the GAAC effector, Gcn4p, was derepressed, further indicating TOR pathway inhibition. While the exact nature of the interaction is undetermined, the above results implicate aa-tRNA pool fidelity in the regulation of TOR pathway components and serves as an example of aaRS-mediated QC participating in cellular stress response outside of the GAAC.
Figure 6.
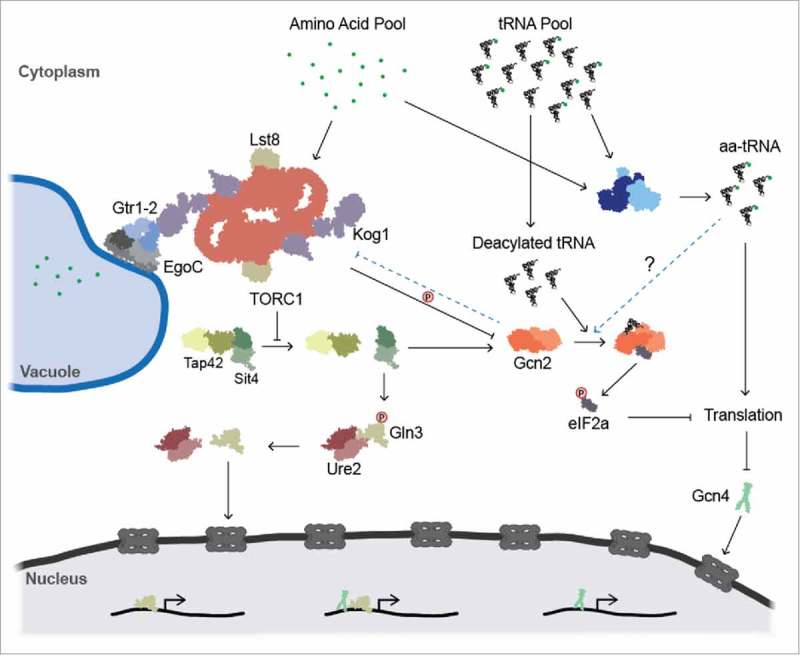
Activation and cross-regulation of the GAAC and TOR pathways. Both the GAAC and TOR pathways respond to changes in amino acid availability. The TORC1 complex monitors the availability of nitrogenous compounds and in response to nitrogen limitation, mounts a stress response by freeing Sit4p from its complex. Sit4p then dephosphorylates Gln3p, facilitating dissociation from Ure2p and allowing for translocation of Gln3p to the nucleus to activate NCR gene transcription. Sit4p also dephosphorylates S577 on Gcn2p. Gcn2p is subsequently able to efficiently bind deacylated tRNA during periods of amino acid starvation. Activated Gcn2p phosphorylates eIF2α, decreasing levels of translation while increasing translation of Gcn4p. Gcn4p enters the nucleus and facilitates transcription of amino acid biosynthesis genes. Gcn4p and Gln3p co-regulate a subset of stress response genes which are common to both stress response pathways. The intersection of the aa-tRNA pool with the TOR pathway (blue arrows) occurs through Gcn2p, yet the exact mechanism of the interaction is unclear.
GAAC and TOR pathway interactions coordinated by aminoacyl-tRNA QC
To mount an effective response to environmental or nutritional stress, the cell must monitor multiple extra- and intracellular inputs and coordinate signal transduction and response to efficiently abrogate stress and adapt to the changing environment. Hierarchical structuring of stress response pathways is essential to a coordinated stress response. Recent work has provided examples of crosstalk between the GAAC and the TOR pathways.8,32,33 Together, these pathways facilitate a “checks and balances” mechanism of nutritional stress response that prioritizes conservation of resources and modulates cellular growth. Many of the known interactions between the two pathways occur through the deacylated tRNA monitoring protein, Gcn2p. Gcn2p is able to indirectly monitor amino acid availability by directly interacting with the cellular tRNA pool via a histidyl-tRNA synthetase like domain. Deacylated tRNA activates the kinase domain of Gcn2p, with downstream activity causing GAAC activation. As nitrogenous compounds, amino acids within the vacuole are also monitored by TOR pathway components. When amino acids are abundant, TORC1 components phosphorylate Serine 557 on Gcn2p, preventing GAAC activation. When amino acids are limiting, TOR pathway component Sit4p phosphatase dephosphorylates Gcn2p, thus allowing for activation of the GAAC. At the same time, Gcn2p has recently been shown to directly inhibit the activity of TORC1 by phosphorylating components of the TOR complex (Fig. 6). While less is known about this process, it remains clear that the interaction of Gcn2p with the tRNA pool is central to the coordinated regulation of the GAAC and TOR pathways.
In the absence of PheRS QC, several phenotypes were identified which revealed direct connections to TORC1. The inhibitory effect of caffeine on TORC1 is well documented. While our results suggest inhibition of TOR activation, the increase in caffeine tolerance in the QC defieicent strain compared to WT is suggestive of an increase in TOR activation. Similarly, the decrease in chronological lifespan for the QC deficient strain appears to be opposite of the previously reported effects of TOR inhibition. These observations highlight the complexities of stress response pathway coordination. On preferred nitrogen sources (e.g., ammonium containing medium), careful regulation of aa-tRNA pool fidelity may be more important to the cell, thus the activity of the GAAC is prioritized over the TOR pathway in the hierarchy of cellular stress response. During growth on a secondary nitrogen source, activation of the TOR pathway takes priority as the cell adjusts its metabolic profile to maintain levels of nitrogenous metabolites. Given the available data, it is clear that the interactions between the GAAC and TOR pathways are connected by amino acid and aa-tRNA pool composition and that the intricacies of the cross-regulatory mechanisms for these pathways warrant further investigation.
Materials and methods
Genetic techniques
Construction of strains NR1 and NR2 was described previously.34 Plasmid pGCN4RepDeg was constructed using the GCN4 promoter and yeGFP, amplified from plasmids p180 and pYM25, respectively, and the c-terminal Gcn4p degradation signal amplified from yeast strain s288c. Fragments were assembled into the MCS of plasmid pXRU3 using Gibson assembly.
Growth analysis and complementation
Growth of all S. cerevisiae strains was conducted as described previously.35 Synthetic defined minimal media (SDMM) was comprised of Difco yeast nitrogen base without amino acids, 2% glucose, 0.002% adenine, 0.002% uracil, 0.002% L-histidine, and 0.01% L-leucine. Tyrosine stress media (SDMM + Phe:Tyr) was made with SDMM and varying concentrations of Phe:Tyr where [Phe] was kept at 0.003 mM and [Tyr] varied from 0.003-1.2 mM. Synthetic complete media (SC) was made using drop out base powder (DOB) and SC supplement (Sunrise Science). For all growth assays, S. cerevisiae strains FRS1, frs1-1 and derivatives were streaked on YPDA and grown at 30˚C for 72 hours. Single colonies were suspended in sterile water and used to inoculate 250 µl of media in a microtiter plate to a final OD600 of 0.01. Growth was monitored using an xMark microplate absorbance spectrophotometer (Bio-Rad Laboratories, Hercules, CA) by measuring the absorbance at 600 nm every five minutes for 24–48 hours.
Spot growth assays
Yeast cultures were prepared by streaking strains from frozen stocks onto YPD agar plates. After incubating the cells at 30 °C for 48 hours, single colonies were resuspended in autoclaved H2O to a final OD600 of 1.0. Serial dilutions (10−1 to 10−4) were prepared by mixing 100 μL of culture into 900 μL sterile H2O. From each dilution, 8 μL was spotted in sequence onto solid agar plates with varying composition depending upon the strain and experimental design. Plates were incubated at 30 °C for 24–48 hours and scanned for growth comparison.
β-galactosidase assays
Cultures were inoculated to an OD600 of 0.01 in various media. Culture (1 ml) at an OD600 of 0.8-1.0 was harvested by centrifugation and resuspended in 1 ml of Z-Buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, and 0.27% mercaptoethanol). Cells were permeabilized using 20 µl 0.1% SDS and 40 µl CHCl3, vortexed for 15 seconds and incubated at 30˚C for 10 minutes. Permeabilized cell suspension (125 µl) was removed from the aqueous layer into a 96 well plate, diluted with an equal volume of Z-buffer, and the absorbance at 600 nm was determined. Subsequently, 200 µl of the same cell suspension was removed from the aqueous layer into a 96 well plate and mixed with 50 µl of 5 mg/ml CPRG substrate to a final concentration of 1 mg/ml. The absorbance at 580 nm was recorded at regular intervals and the linear rate was determined. Enzyme activity was determined from linear rates normalized to absorbance at 600 nm. All absorbance measurements were recorded using an xMark microplate absorbance spectrophotometer (Bio-Rad Laboratories, Hercules, CA).
GCN4 activation
GCN4 translation was monitored in WT (FRS1) and PheRS QC deficient (frs1-1) strains harboring GCN4 uORF:GFP reporter plasmid pGCN4Rep. Cells were grown in 5 mL tubes of SC medium with ammonium as the sole nitrogen source overnight at 30C. Overnight cultures were used to inoculate 50 mL cultures to an OD600 of 0.1 in either minimal (SDMM) or synthetic complete (SC) medium with ammonium as the sole nitrogen source. Translation of GCN4 was monitored by measuring GFP fluorescence (ex: 487, em: 501) over time using a flourolog-3 fluorimeter (Horiba Group, Edison, NJ). Cells fluorescence was normalized to cell density by simultaneous measurement of OD600.
Biolog phenotypic microarray analysis
Phenotype Microarray (PM) panels 1, 2A, 3B, 4A, 5, 6, 7, 8, 9, 21D, 22D, 23A, 24C, 25D, 100x Redox Dye Mix D (PM 1, 2A, 3B, 4A, 5–9) and 100x Redox Dye Mix E (PM 21D, 22D, 23A, 24C and 25D), and 1.2 × IFY-0 Base were all obtained from Biolog. Chemicals used to prepare the PM additives were obtained from Sigma. Growth phenotypes of the WT and PheRS QC deficient strains were determined for Biolog PM 1, 2A, 3B, 4A, 5–9, 21D, 22D, 23A, 24C, 25D plates when grown at 30°C for 48–72 h by following the S. cerevisiae protocol supplied by Biolog. Strains were compared in a pairwise manner. Phenotypes exhibiting growth of one strain 2-fold over that of the other strain in one concentration and demonstrating a consistent trend across all four concentrations of particular substances, when applicable, were considered significant.
Confirmation of selected biolog phenotypes
The nitrogenous compound caffeine was chosen to confirm Biolog results and characterize defects in nitrogen catabolism for the PheRS QC deficient strain over a larger range of concentrations. A 100 mM stock of caffeine was filter sterilized, and stored at 4°C. Serial dilutions of the 100 mM caffeine stock were made with SC media to create a range of concentrations from 0.625-80 mM. Cultures of the WT and PheRS QC deficient strains were diluted to a final OD600 of 0.1 in SC media. To inoculate the wells of a greiner bio-one Cellstar 96-well suspension culture plate, 225 μL of the various caffeine growth conditions were added with 25 μL of yeast cell suspension. Growth of the strains at 30°C in the presence of varying concentrations of caffeine over a period of 24 h was monitored using a Labsystems iEMS Reader MF plate reader. The OD340 and OD600 of the wells were monitored, and readings were taken every hour during the 24 h period.
DNA content analysis
Yeast cell cycle analysis was conducted using SYBR Green I fluorescent dye. Briefly, cells were grown under various conditions to an OD600 between 0.5 and 1.0. 1 mL of culture was harvested by centrifugation at 14,000 rpm for 1 minute, resuspended in 70% EtOH, and incubated at RT for at least one hour. 0.5 ml of the fixed cells were collected by centrifugation and washed in the same volume of ddH2O. The cells were resuspended in 200 µl RNase A solution (4 µl of 10 mg/ml RNase A in 196 µl 50 mM Tris-HCl pH 8.0) and incubated at 37˚C for 4 hours. Cells were harvested by centrifugation, resuspended in 200 µl proteinase K solution (2 mg/ml proteinase K in 50 mM Tris-HCl pH 7.5) and incubated at 50˚C for one hour. Following incubation, the cells were spun down and resuspended in 200–400 µl FACS buffer (200 mM Tris-HCl pH 7.5, 200 mM NaCl, 78mM MgCl2). 10 µl of treated cells were mixed with 200 µl of SYBR Green solution (2X in 50 mM Tris-HCl pH 7.5), sonicated for 3 seconds (5-10 watt output), and analyzed using a Guava EasyCyte HT5 flow cytometer (EMD Millipore Corp., Bedford, MA). Data analysis was conducted using univariate statistical analysis (FloJo v10, TreeStar Inc.).
Chronological lifespan assay
Aging assays were completed using the methods of Fabrizio and Longo (115). The S. cerevisiae strains W303 wild type (FRS1) and W303 frs1-1 were streaked on YPDA and incubated at 30°C. After approximately 72 h several colonies were picked and resuspended in sterile water and used to inoculate 50 mL of synthetic complete medium (SDC) with variable Phe:Tyr ratios in a 250 mL flask to an initial OD600 of 0.1. Cultures were grown in SDC + Phe:Tyr 1:1 or SDC + 1:50, where Phe was at a concentration of 0.003 mM. SDC media consisted of 0.18% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 0.14% NaH2PO4, 80 mg/L adenine, 80 mg/L uracil, 80 mg/L tryptophan, 80 mg/mL histidine-HCl, 40 mg/L 53 mg/ml arginine-HCl, 80 mg/L methionine, 1200 mg/L leucine, 60 mg/L isoleucine, 60 mg/L lysine-HCl, 100 mg/L glutamic acid, 100 mg/L aspartic acid, 150 mg/L valine, 200 mg/L threonine, and 400 mg/L serine. Cultures were shaken at 220 rpm at 30°C. Cultures were allowed to grow for 72 h after which time samples of 1 mL were taken every 48 h, serial dilution performed in sterile water, and plated on YPDA. Plates were allowed to grow for 2–3 d at 30°C, colonies counted, and CFU/mL calculated for each culture. Assays were performed in triplicate.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Science Foundation (MCB 1412611), an Ohio State University STEP fellowship (to A.K.), Ohio State University Center for RNA Biology Fellowships (to K.M. and R.M.) and NIH Training Grant T32 GM086252 (to K.M.).
References
- 1.Zaborske JM, Wu X, Wek RC, Pan T. Selective control of amino acid metabolism by the GCN2 eIF2 kinase pathway in Saccharomyces cerevisiae. BMC Biochem. 2010;11:29. doi: 10.1186/1471-2091-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joo YJ, Kim JH, Kang UB, Yu MH, Kim J. Gcn4p-mediated transcriptional repression of ribosomal protein genes under amino-acid starvation. EMBO J. 2011;30:859–72. doi: 10.1038/emboj.2010.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goossens A, Dever TE, Pascual-Ahuir A, Serrano R. The protein kinase Gcn2p mediates sodium toxicity in yeast. J. Biol. Chem. 2001;276:30753–60. doi: 10.1074/jbc.M102960200. [DOI] [PubMed] [Google Scholar]
- 4.Yang R, Wek SA, Wek RC. Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol. Cell. Biol. 2000;20:2706–17. doi: 10.1128/MCB.20.8.2706-2717.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marbach I, Licht R, Frohnmeyer H, Engelberg D. Gcn2 mediates Gcn4 activation in response to glucose stimulation or UV radiation not via GCN4 translation. J. Biol. Chem. 2001;276:16944–51. doi: 10.1074/jbc.M100383200. [DOI] [PubMed] [Google Scholar]
- 6.De Virgilio C Loewith R. The TOR signalling network from yeast to man. Int J Biochem Cell Biol. 2006;38:1476–81. doi: 10.1016/j.biocel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Soulard A, Cohen A, N. HM. TOR signaling in invertebrates. Curr Opin Cell Biol. 2009;21:825–36. doi: 10.1016/j.ceb.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Staschke KA, Dey S, Zaborske JM, Palam LR, McClintick JN, Pan T, Edenberg HJ, Wek RC. Integration of General Amino Acid Control and Target of Rapamycin (TOR) Regulatory Pathways in Nitrogen Assimilation in Yeast. J Biol Chem. 2010;285:16893–911. doi: 10.1074/jbc.M110.121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubota H, Obata T, Ota K, Sasaki T, Ito T. Rapamycin-induced translational derepression of GCN4 mRNA involves a novel mechanism for activation of the eIF2 alpha kinase GCN2. J Biol Chem. 2003;278:20457–60. doi: 10.1074/jbc.C300133200. [DOI] [PubMed] [Google Scholar]
- 10.Neklesa TK, Davis RW. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genetics. 2009;5. doi: 10.1371/journal.pgen.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valenzuela L, Aranda C, Gonzalez A. TOR Modulates GCN4-Dependent Expression of Genes Turned on by Nitrogen Limitation. J Bacteriol. 2001;183. doi: 10.1128/JB.183.7.2331-2334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardwick JS, Tong JK, Schreiber SL. The transcriptional profile of Saccharomyces cerevisiae exposed to rapamycin mimics the profile induced by amino acid starvation. Nature Genetics. 1999;23:49–49. doi: 10.1038/14319. [DOI] [Google Scholar]
- 13.Kotik-Kogan O, Moor N, Tworowski D, M. S. Structural basis for discrimination of L-phenylalanine from L-tyrosine by phenylalanyl-tRNA synthetase. Structure. 2005;13:1799–807. doi: 10.1016/j.str.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 14.Ling J, Yadavalli SS, Ibba M. Phenylalanyl-tRNA synthetase editing defects result in efficient mistranslation of phenylalanine codons as tyrosine. RNA. 2007;13:1881–6. doi: 10.1261/rna.684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohler K, Mann R, Bullwinkle TJ, Hopkins K, Hwang L, Reynolds NM, Gassaway B, Aerni HR, Rinehart J, Polymenis M, et al. Editing of misaminoacylated tRNA controls the sensitivity of amino acid stress responses in Saccharomyces cerevisiae. Nucleic Acids Res. 2017;45:3985–96. doi: 10.1093/nar/gkx077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy H, Ling J, Irnov M, Ibba M. Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase. EMBO J. 2004;23:4639–48. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bullwinkle TJ, Ibba M. Translation quality control is critical for bacterial responses to amino acid stress. Proc Natl Acad Sci USA. 2016;113:2252–7. doi: 10.1073/pnas.1525206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinke A, Chen JC, Aronova S, Powers T. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J Biol Chem. 2006;281:31616–26. doi: 10.1074/jbc.M603107200. [DOI] [PubMed] [Google Scholar]
- 19.Kuranda K, Leberre V, Sokol S, Palamarczyk G, Francois J. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol Microbiol. 2006;61:1147–66. doi: 10.1111/j.1365-2958.2006.05300.x. [DOI] [PubMed] [Google Scholar]
- 20.Rallis C, Codlin S, Bahler J. TORC1 signaling inhibition by rapamycin and caffeine affect lifespan, global gene expression, and cell proliferation of fission yeast. Aging Cell. 2013;12:563–73. doi: 10.1111/acel.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godard P, Urrestarazu A, Vissers S, Kontos K, Bontempi G, van Helden J, André B. Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:3065–86. doi: 10.1128/MCB.01084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coffman JA, Rai R, Cooper TG. Genetic evidence for Gln3p-independent, nitrogen catabolite repression-sensitive gene expression in Saccharomyces cerevisiae. J Bacteriol. 1995;177:6910–8. doi: 10.1128/jb.177.23.6910-6918.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grundmann O, Mösch HU, Braus GH. Repression of GCN4 mRNA translation by nitrogen starvation in Saccharomyces cerevisiae. J Biol Chem. 2001;276:25661–71. doi: 10.1074/jbc.M101068200. [DOI] [PubMed] [Google Scholar]
- 24.Cherkasova VA, Hinnebusch AG. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 2003;17:859–72. doi: 10.1101/gad.1069003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–50. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 26.Hinnebusch A. Novel mechanisms of translational control in Saccharomyces cerevisiae. Trends Genet. 1988;4:169–74. doi: 10.1016/0168-9525(88)90023-6. [DOI] [PubMed] [Google Scholar]
- 27.Thomas G, Hall MN. TOR signalling and control of cell growth. Curr Opin Cell Biol. 1997;9:782–7. doi: 10.1016/S0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- 28.Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polymenis M, V. SE. Coupling of cell division to cell growth by translational control of the G1∼ cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–31. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powers RW 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–84. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohler K, Ibba M. Translational fidelity and mistranslation in the cellular response to stress. Nat Microbiol. 2017;2:17117. doi: 10.1038/nmicrobiol.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen R, Zou Y, Mao D, Sun D, Gao G, Shi J, Liu X, Zhu C, Yang M, Ye W, et al. The general amino acid control pathway regulates mTOR and autophagy during serum/glutamine starvation. J Cell Biol. 2014;206:173–82. doi: 10.1083/jcb.201403009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tate JJ, Buford D, Rai R, Cooper TG. General Amino Acid Control and 14-3-3 Proteins Bmh1/2 Are Required for Nitrogen Catabolite Repression-Sensitive Regulation of Gln3 and Gat1 Localization. Genetics. 2017;205:633–55. doi: 10.1534/genetics.116.195800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bullwinkle T, Reynolds NM, Raina M, Moghal AB, Matsa E, Rajkovic A, Kayadibi H, Fazlollahi F, Ryan C, Howitz N, et al. Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code. eLife. 2014;e02501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds NM, Ling J, Roy H, Banerjee R, Repasky SE, Hamel P, Ibba M. Cell-specific differences in the requirements for translation quality control. Proc. Natl. Acad. Sci. USA. 2010;107:4063–8. doi: 10.1073/pnas.0909640107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


