ABSTRACT
Tryptophanyl-tRNA synthetase (TrpRS) in vertebrates contains a N-terminal extension in front of the catalytic core. Proteolytic removal of the N-terminal 93 amino acids gives rise to T2-TrpRS, which has potent anti-angiogenic activity mediated through its extracellular interaction with VE-cadherin. Zinc has been shown to have anti-angiogenic effects and can bind to human TrpRS. However, the connection between zinc and the anti-angiogenic function of TrpRS has not been explored. Here we report that zinc binding can induce structural relaxation in human TrpRS to facilitate the proteolytic generation of a T2-TrpRS-like fragment. The zinc-binding site is likely to be contained within T2-TrpRS, and the zinc-bound conformation of T2-TrpRS is mimicked by mutation H130R. We determined the crystal structure of H130R T2-TrpRS at 2.8 Å resolution, which reveals drastically different conformation from that of wild-type (WT) T2-TrpRS. The conformational change creates larger binding surfaces for VE-cadherin as suggested by molecular dynamic simulations. Surface plasmon resonance analysis indicates more than 50-fold increase in binding affinity of H130R T2-TrpRS for VE-cadherin, compared to WT T2-TrpRS. The enhanced interaction is also confirmed by a cell-based binding analysis. These results suggest that zinc plays an important role in activating TrpRS for angiogenesis regulation.
KEYWORDS: Anti-angiogenesis, crystal structure, H130R T2-TrpRS, Tryptophanyl-tRNA synthetase, VE-cadherin
Introduction
As a member of class I aminoacyl-tRNA synthetase (aaRS), tryptophanyl-tRNA synthetase (TrpRS) uses a Rossmann-fold catalytic domain to catalyze the attachment of tryptophan to its cognate tRNA, with the help of an anticodon binding domain.1 In vertebrates, TrpRS have an N-terminal extension of about 150 amino acids beyond the core catalytic architecture.2,3 The extension is composed of a vertebrate-specific extension (VSE), also known as WHEP domain (named after the tRNA synthetases (i.e., tryptophanyl (W), histidyl (H), and glutamyl-prolyl (EP) tRNA synthetases) first identified to contain this domain), followed by an eukaryotic specific extension (ESE)4 (Fig. 1A). The WHEP domain has a conserved helix-turn-helix conformation and is dispensable for aminoacylation but critical for regulating the non-enzymatic function of TrpRS in angiogenesis.2,3 The natural splice variant mini-TrpRS, which lacks most of the WHEP domain but contains the ESE, was found to inhibit VEGF induced angiogenesis.5,6 Further truncation by leukocyte elastase to remove the first 93 amino acid creates T2-TrpRS that is completely inactive for aminoacylation, but has enhanced anti-angiogenic activity compared to mini-TrpRS.5
Figure 1.
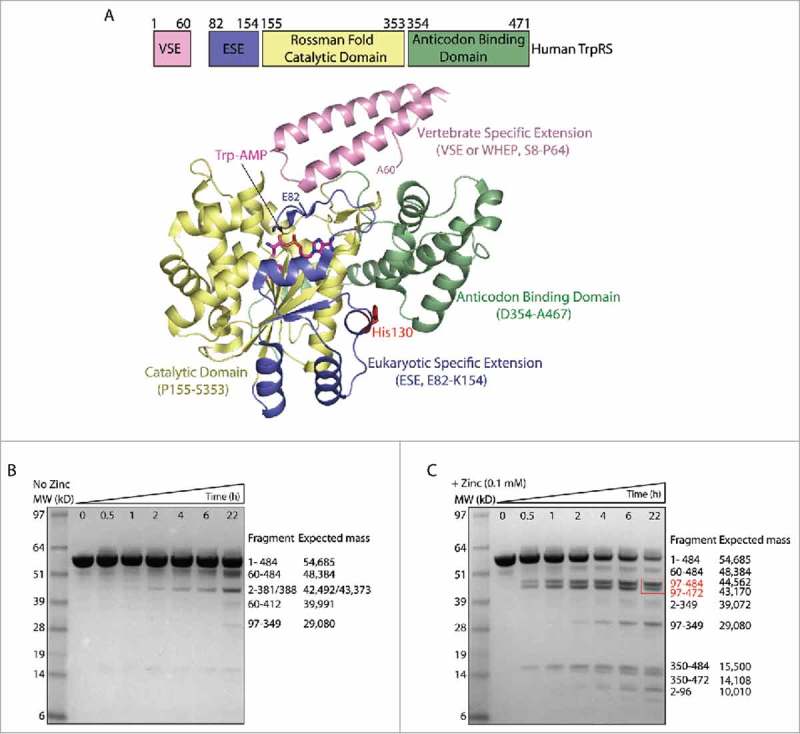
Zinc induces conformational change of human FL-TrpRS and the generation of T2-TrpRS. (A) Domain organization of human FL-TrpRS and its crystal structure in complex with Trp-AMP (PDB ID 1R6T). B, C) Time course of plasmin digestion of FL-TrpRS in the absence (B) and presence (C) of zinc (0.1 mM), followed by mass spectrometry analysis. After plasmin digestion, fragments were separated by SDS-PAGE, the bands were cut from the gel and digested with trypsin and AspN for LC-MS/MS analysis. The whole protein mass measurement was performed by MALDI_TOF MS analysis. The cleavage site of the fragments were confirmed by N-terminal sequencing, and the expected mass were calculated from the protein sequence of these fragments. A red box highlights two T2-TrpRS-like fragments generated in the presence of zinc.
T2-TrpRS was shown to inhibit angiogenesis through an interaction with VE-cadherin, which is an endothelial cell junction molecule that adheres apposed endothelial cells through a “strand-swapped” dimer formed by reciprocal swapping of two tryptophan residues of one VE-cadherin molecule into a hydrophobic pocket of the first extracellular cadherin (EC) domain of another VE-cadherin.7,8,9 Our previous study has demonstrated that T2-TpRS binds the two tryptophan residues (Trp2 and Trp4) through its tryptophan and AMP binding pockets in the active site, respectively, and thereby inhibits the VE-cadherin mediated endothelial cell-cell junctions and disrupts the formation of new blood vessels.8 Notably, human TrpRS manifests its anti-angiogenic activity only when the WHEP domain is removed. Full length (FL) TrpRS cannot inhibit angiogenesis,5,6,10 due to the steric hindrance of the WHEP domain that blocks the access of Trp2 and Trp4 of VE-cadherin to the active site pocket.8
Zinc is known to have anti-angiogenic effects, which are mediated through multiple mechanisms, including its binding to transcription factors to influence gene expression.11,12 For example, zinc can dramatically reduce the expression of pro-angiogenic growth factors such as IL-6, IL-8, VEGF and MMP-9, while promoting the production of anti-angiogenic factors such as endostatin.13,14,15 Zn2+ can also directly bind to endostatin and the binding is essential for the anti-angiogenic activity of endostatin.16 Interestingly, both bovine and human TrpRS were shown to bind zinc to enhance their aminoacylation activities.17,18,19 However, no zinc was found in the crystal structures of TrpRS,4,10,20,21,22,23 suggesting that the zinc-bound conformation may be different. Interestingly, Wakasugi has identified a single amino acid substitution in TrpRS (H130R) that eliminates zinc-induced stimulation and renders constitutively high enzymatic active,24 suggesting H130R TrpRS may mimic a zinc-bound conformation.
We sought out to investigate the effect of zinc on TrpRS. We found that zinc can induce conformational change of TrpRS and facilitate the proteolytic production of T2-TrpRS. We also found that the H130R mutation induces an alternative conformation of T2-TrpRS that may mimic the zinc-bound conformation. We determined the crystal structure of H130R T2-TrpRS and revealed conformational changes that correlate with an enhanced VE-cadherin binding. Altogether, these results suggest that zinc can promote the production and the anti-angiogenic activity of T2-TrpRS.
Results
Zinc induces conformational changes in human TrpRS and facilitates proteolytic generation of T2-TrpRS
To probe the effect of zinc on the conformation of human TrpRS, we carried out limited protease digestion with plasmin in the presence and absence of zinc, followed by mass spectrometry analysis (Fig. 1B, C). In the absence of zinc, although some proteolytic fragments were generated, the FL-TrpRS (expressed with a C-terminal His-tag to give a total of 484 residues) is relatively stable during 22 hrs' incubation with plasmin (Fig. 1B). However, when in the presence of zinc (0.1 mM), majority of the TrpRS protein were digested by plasmin within 22 hrs, producing a large number of proteolytic fragments (Fig. 1C). This increased susceptibility to protease digestion is indicative of a more relaxed structure of TrpRS induced by zinc binding. Interestingly, two new and major fragments (Gly97-His484, Gly97-Lys472) generated in the presence of zinc were similar to T2-TrpRS (Ser94-Gln471) (Fig. 1C). Therefore, zinc facilitates the generation of a T2-TrpRS-like fragment, most likely through its conformational opening effect on TrpRS.
H130R cannot induce conformational change in FL-TrpRS
Both zinc and the H130R substitution can enhance the aminoacylation activity of human TrpRS,24 suggesting H130R may mimic a zinc-bound conformation in the absence of zinc. To investigate the potential conformational change induced by H130R, we subjected WT and H130R FL-TrpRS to trypsin digestion in the absence of zinc; however, no obvious difference in the digestion pattern between the two proteins was detected (Fig. 2A). Also, no obvious difference between the two proteins was detected in the presence of zinc (Fig. 2B). Consistent with the previous result (Fig. 1B, C), TrpRS is more efficiently digested in the presence of zinc, and a T2-TrpRS-like fragment can be generated (Fig. 2B). (To rule out the possibility that the enhanced efficiency of digestion is due to enzymatic activity enhancement of trypsin in the presence of zinc, we used another tRNA synthetase as a control. No change in the digestion pattern of ProRS was observed with and without zinc (Fig. S1)). These results further support the idea that zinc induces a more relaxed conformation in FL-TrpRS; however, this conformational change cannot be induced by the H130R substitution at least under our experimental conditions.
Figure 2.
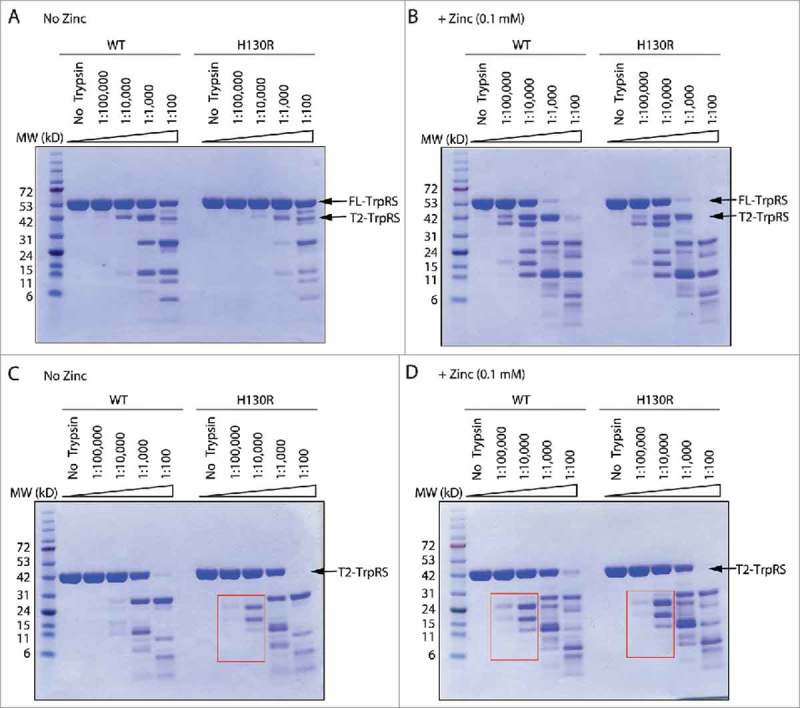
Trypsin digestion analyses suggest H130R mutation induces a zinc-bound conformation in the context of T2-TrpRS. (A, B) Trypsin digestion of wild-type (WT) and H130R FL-TrpRS with increasing concentrations of the protease in absence (A) and presence (B) of zinc (0.1 mM). The concentration of trypsin is labeled as [protease] to [TrpRS] ratio. (C, D) Trypsin digestion of wild-type (WT) and H130R T2-TrpRS in absence (C) and presence (D) of zinc (0.1 mM). Red boxes highlight the resemblance of the digestion pattern of H130R T2-TrpRS in the absence of zinc with that of WT T2-TrpRS in the presence of zinc.
We were able to obtain crystals of H130R FL-TrpRS bound with Trp-AMP but not crystals of apo H130R FL-TrpRS. The structure of the complex was solved at 2.1 Å resolution (Table S1). Consistent with the result of the trypsin digestion experiment, the complex structure resembles that of wild-type FL-TrpRS in complex with Trp-AMP (PDB ID 1R6T), with a small root-mean-square deviation (RMSD) value of 0.64 Å for the main chain Cα atoms (Fig. S2). Therefore, we conclude that H130R substitution does not induce significant conformational change in FL-TrpRS, at least when it is bound with Trp-AMP.
H130R mutation mimics zinc and induces conformational change in T2-TrpRS
Next, we tested the effect of the H130R substitution in the context of T2-TrpRS. In the presence of zinc, the trypsin digestion pattern is the same between WT T2-TrpRS and the H130R mutant (Fig. 2D). However, in the absence of zinc, there is a difference between WT and H130R T2-TrpRS in their fragmentation pattern at low trypsin concentration (trypsin to TrpRS concentration ratio as 1:100,000 and 1:10,000) (Fig. 2C). Remarkably, the pattern of H130R T2-TrpRS in the absence of zinc resembles that of WT T2-TrpRS in the presence of zinc (Fig. 2C, D), suggesting that in the context of T2-TrpRS the H130R substitution does induce conformational change, which mimics the effect of zinc.
Overall conformational changes of H130R T2-TrpRS
To visualize the conformational change, we determined the crystal structure of H130R T2-TrpRS at 2.8 Å resolution by molecular replacement using WT human T2-TrpRS structure (PDB 2QUI) as the search model.25 Like the wild-type protein (Fig. 3A), H130R T2-TrpRS is composed of a truncated ESE (Gly97-Lys154), a Rossmann fold catalytic domain (Pro155-Ser353) and an anticodon binding domain (Asp354-Gln471) (Fig. 3B). A homodimer of H130R T2-TrpRS was found in the asymmetric unit, with two virtually identical subunits A and B interacting at the catalytic domain to form a “W” form dimer (Fig. 3B). The structure is well folded overall with clear electron density, except for the disordered regions Gln344-Pro355 and Gln389-Gly392 in subunit A, Met350-Ser353 and Gly379-Gly391 in subunit B. Superposition of the structure with that of WT T2-TrpRS (PDB 2QUI) gives a large main chain RMSD value of 2.191 Å. Major conformational changes were observed at the entire ESE, KMSAS signature motif containing loop (Thr338-Thr362), and loop Ala376-Asp396 in the anticodon binding domain (Fig. 3A, B).
Figure 3.
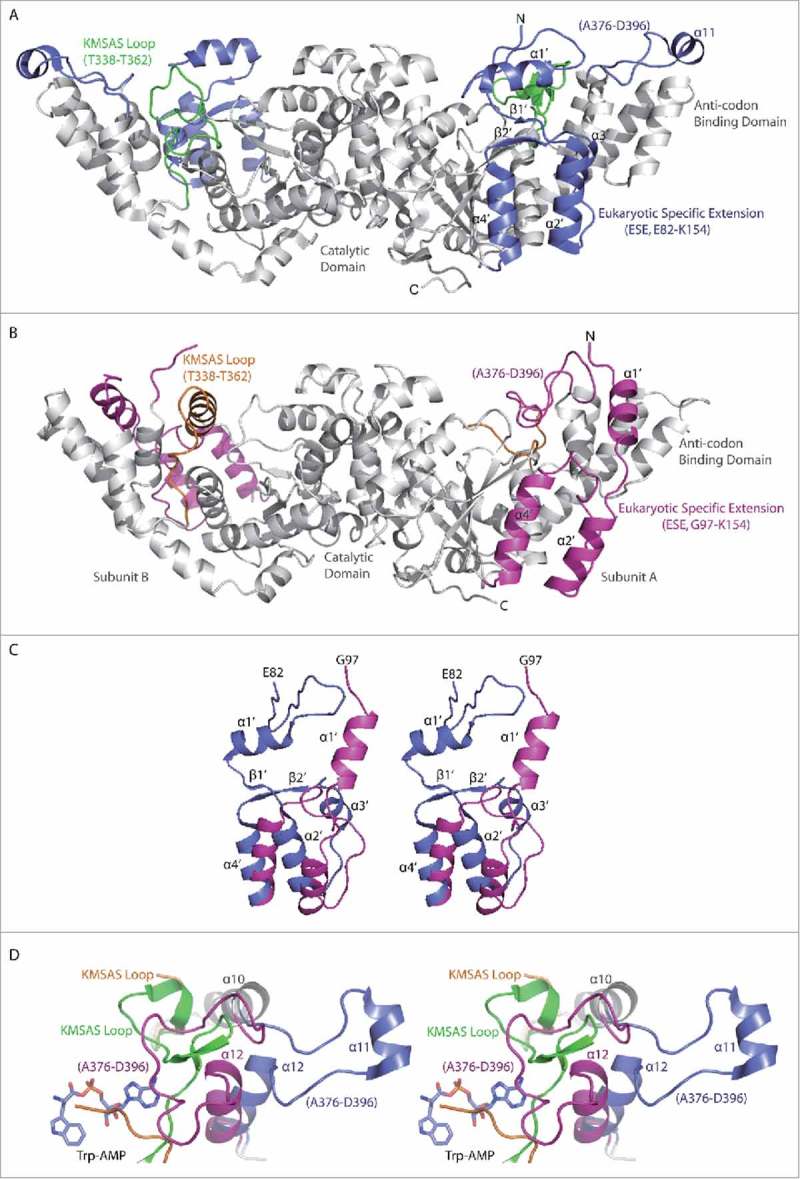
Conformational change induced by H130R substitution in T2-TrpRS. (A) Overall crystal structure of wild-type (WT) T2-TrpRS homo-dimer. The eukaryotic specific extension (ESE, Glu82-Lys154) and the Ala376-Asp397 loop in the anticodon binding domain are colored in blue, whereas the KMSAS motif containing loop (Thr338-Thr362) is colored in green. Secondary structural elements of the ESE in subunit A are labeled. (B) The crystal structure of H130R T2-TrpRS homo-dimer. The ESE (Gly97-Lys153) and the Ala376-Asp397 loop are colored in magenta, whereas the KMSAS loop (Thr338-Thr362) is in orange. (C) Stereo view of the ESE structure in H130R (magenta) versus WT (blue) T2-TrpRS. (D) Stereo view of the conformational changes surround active site. WT T2-TrpRS in complex with Trp-AMP (PDB 1R6U) is aligned with H130R mutant structure. The α11, α12 helices and the Ala376-Asp397 loop are colored in blue in WT structure and in magenta in the H130R structure. KMSAS loop (Thr338-Thr362) is colored in green in WT and in orange in the H130R mutant.
The ESE undergoes dramatic structural changes (Fig. 3A-C). Compared to the wild-type structure, the α1′ helix in the H130R mutant no longer covers the substrate binding pocket, and is rotated 90° and shifted away about 13.7 Å from the catalytic domain to the anticodon binding domain. Following the α1′ helix, α2′ helix is also rotated for 37°, while β1′, β2′ strands, and α3′ helix all become disordered. (The H130R substitution site is located in α3′; see below.) The α4′ helix is the last part of the ESE and hinged for 34° from the end of the helix (Fig. 3C).
The catalytic domain of the H130R mutant superimposes well with that of the WT T2-TrpRS, except for the loop containing the KMSAS motif. HIGH and KMSKS (170HVGH173 and 349KMSAS353 in human TrpRS) are two conserved sequence motifs of class I tRNA synthetases involved in ATP recognition and catalysis; a third and less conserved motif in the catalytic domain (310AIDQ313 in human TrpRS) also participates in ATP binding.22,23,25 The KMSAS region (Thr338-Thr362) is resolved in the WT T2-TrpRS crystal structure as flexible loops and turns;4,22,25 however, it becomes completely disordered in subunit A of H130R T2-TrpRS, while in subunit B it folds into an α helix (Thr338-Ser351) (Fig. 3A, B). As a result of these conformational changes, the Trp-AMP binding pocket is more “open” in the H130R mutant (Fig. S3).
In the structure of the WT T2-TrpRS, α11 is located at the tip of anticodon binding domain and is involved in binding the tRNA anticodon loop.4,22,26 However, in the H130R T2-TrpRS structure, it is folded as a loop (Ala376-Asp397) and flipped 180° onto the active site pocket of subunit A, while this loop is disordered from Gly379 to Gly391 in subunit B (Fig. 3B). Most interestingly, the Ala376-Asp397 loop in subunit A occupies a similar location as that of the KMSAS loop (Thr338-Thr362) in the WT T2-TrpRS, partially compensating for the disordered KMSAS loop in the mutant (Fig. 3D). (α11 is disordered in subunit B of the mutant.) Following α11, helix α12 swings ∼36° towards the active site pocket in both subunits (Fig. 3D).
Conformational changes around the H130R substitution site
In the structure of WT T2-TrpRS, His129, His130 and Phe131 are neighboring residues located on α3′ helix of the ESE (Fig. 4A). Particularly, NE2 of His130 forms a weak hydrogen bond with the main chain carbonyl oxygen of Phe405 (3.12 Å); side chain of Phe131 and His129 protrude into a hydrophobic patch formed by Ile175, Ile178, Phe179, Trp182 from α1 and Phe405, Phe406 from α12 of the anticodon binding domain. In addition, the α3′ helix is further stabilized by interactions with residues from α12 (Asp409-Ser422) and α13 (Asp397-Leu407). For example, Arg127 located on the loop between α2′ and α3′ interacts with Asp410 through a salt bridge (2.92 Å), whereas Arg134 on α3′ forms a hydrogen bond (2.56 Å) with Leu407 located on α12 (Fig. 4A).
Figure 4.
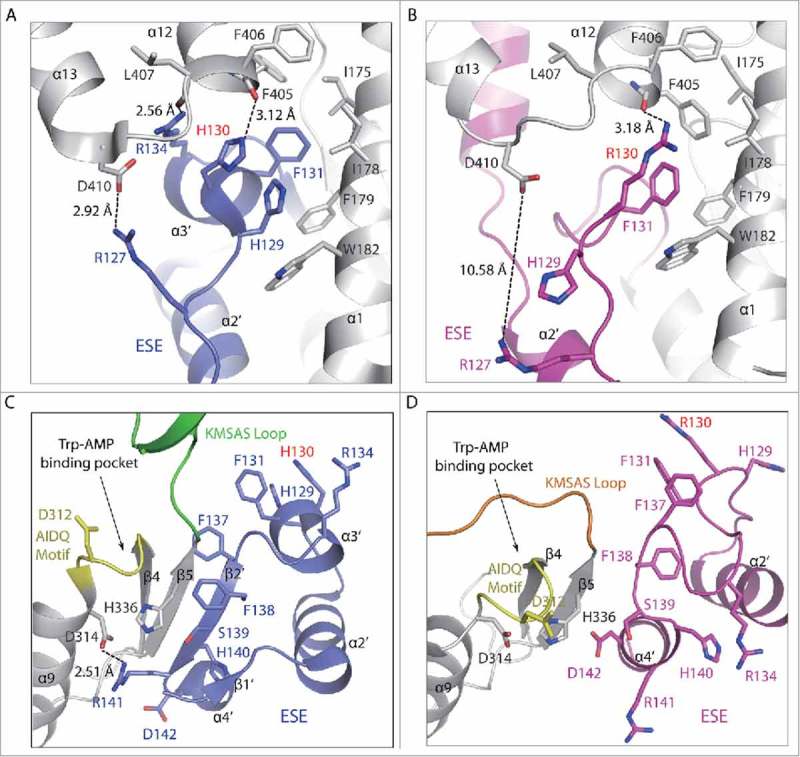
Local conformational changes around the H130R substitution site. (A, B) Conformational changes around residue 130 in WT (A) and H130R (B) T2-TrpRS. (C, D) Interactions between the ESE and catalytic domain in WT (C) and H130R (D) T2-TrpRS.
In contrast, H130R T2-TrpRS shows strikingly different local conformation around the substitution site (Fig. 4B). The α3′ helix becomes unwound. The Cα position of the 130 residue shifts 6.94 Å; the neighboring Phe131 is shifted 3.5 Å. More dramatically, the Cα position of His129 moves 5.74 Å, and its imidazole side chain flips 180° to become solvent exposed. Arg127 is no longer salt bridged with Asp410 and a large gap (10.58 Å) is opened up in between the two residues. Arg134 is also moved away from Leu407 (Fig. 4A, B).
These local conformational changes abolish the interactions of the α2′ and α3′ helices from the ESE with α1 of the catalytic domain and α12 and α13 from the anticodon binding domain (Fig. 4A, B). Many of the secondary structures such as β1′, β2′ and α3′ in the ESE become disordered. The side chains of Phe137 and Phe138 are flipped 90° and 180°, respectively, while the side chain of Arg141 is also flipped away from the catalytic domain and becomes solvent exposed (Fig. 4C, D). As a result, the entire ESE dissociates away from the catalytic domain. These and other conformational changes within the catalytic domain, make the Trp-AMP binding pocket more open in H130R T2-TrpRS (Fig. 4C, D; Fig. S3).
Molecular simulations of the interaction between T2-TrpRS and VE-cadherin
Considering the importance of the Trp and AMP binding pockets for the T2-TrpRS and VE-cadherin interaction,8 we evaluated how the H130R mutation would affect the interaction using molecular docking and dynamic simulations (MDS). We used the crystal structure of the EC1-2 domain of chicken VE-cadherin27 (PDB ID 3PPE) that share 58% sequence identity with human VE-cadherin to docked onto WT and H130R T2-TrpRS structures, respectively. Considering the expected low concentration of the extracellular T2-TrpRS, we used the monomer form of T2-TrpRS for the docking, followed by dynamic simulations to obtain final stable complexes (Fig. 5). As suggested by the small fluctuations of root-mean-square deviations (RMSDs) during the MDS process, the docking of WT and H130R T2-TrpRS are rather stable (Fig. S4A, B).
Figure 5.
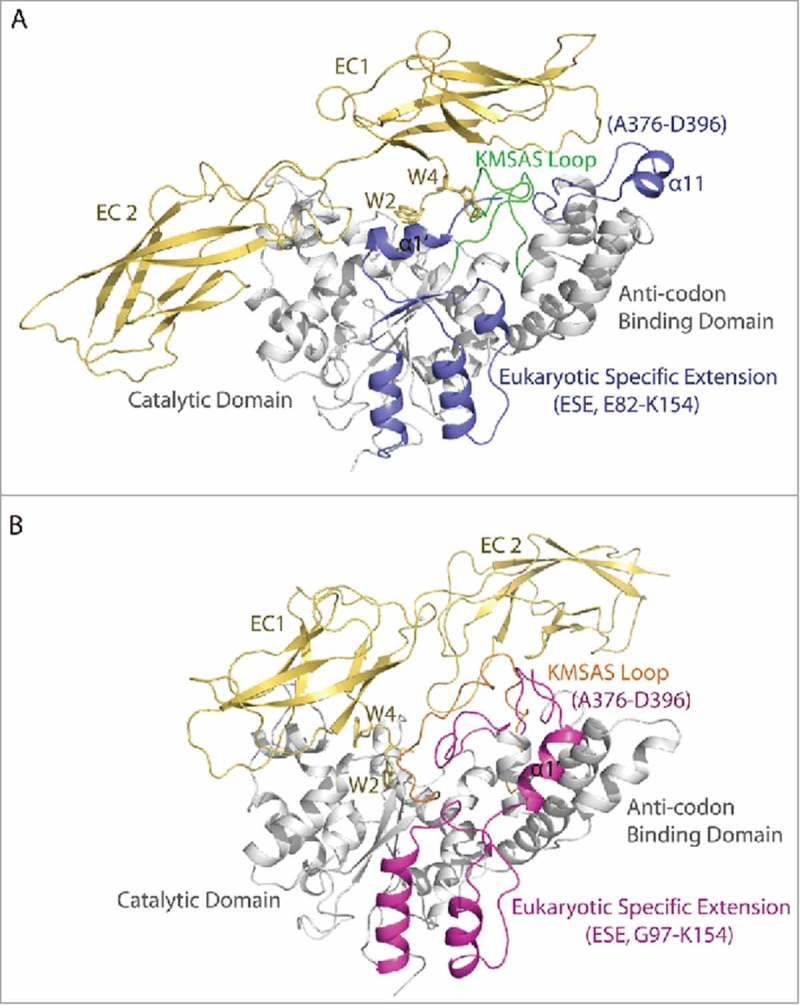
Molecular dynamic simulations (MDS) of the interaction between extracellular domain EC1-2 of VE-cadherin and the monomer form of WT (A) and H130R (B) T2-TrpRS. The eukaryotic specific extension (ESE) and the Ala376-Asp397 loop in the anticodon binding domain of T2-TrpRS are colored in blue (WT) and magenta (H130R), whereas the KMSAS loop (Thr338-Thr362) is colored in green (WT) and orange (H130R). The EC1-2 domain of VE-cadherin is colored in yellow, the Trp2 and Trp4 residues are shown in sticks.
Interestingly, the docking model of WT and H130R T2-TrpRS are dramatically different. As shown in Fig. 5A and B, residues Trp2 and Trp4 of EC1-2 are docked in the Trp and AMP binding pocket of the WT T2-TrpRS, whereas the two Trp residues are no longer inserted in the Trp and AMP binding pockets of H130R T2-TrpRS. However, the contact areas between H130R T2-TrpRS and EC1-2 (1447.45 Å) are substantially larger than that in the WT complex (1276.4 Å). The EC1-2 domain adopts an extended conformation in the WT complex, while the two domains are more compact and has a different orientation in the H130R complex.
H130R mutation enhances the interaction of T2-TrpRS with VE-cadherin
The larger contact areas in the H130R complex suggest that the mutation may enhance the T2-TrpRS and VE-cadherin interaction. We test the interaction by Surface Plasmon Resonance (Fig. 6A, B). The EC1-2 domain of VE-cadherin was expressed from baculovirus transfected insect cells. Purified EC1-2 domain of VE-cadherin was immobilized on a CM5 chip, and the E. coli expressed human WT or H130R T2-TrpRS was injected as analyte. Interestingly, the binding affinity of the H130R mutant is more than 50-fold stronger than WT T2-TrpRS for EC1-2 (Fig. 6A, B), and the binding can be detected only in the presence of 0.1 mM ZnCl2 (see discussion). The stronger affinity of H130R T2-TrpRS is mostly due to a faster association rate (ka of 2.71 × 107 M−1s−1 for H130R versus 1.76 × 105 M−1s−1 for WT), consistent with the model that the H130R structure provides a larger binding surface for VE-cadherin.
Figure 6.
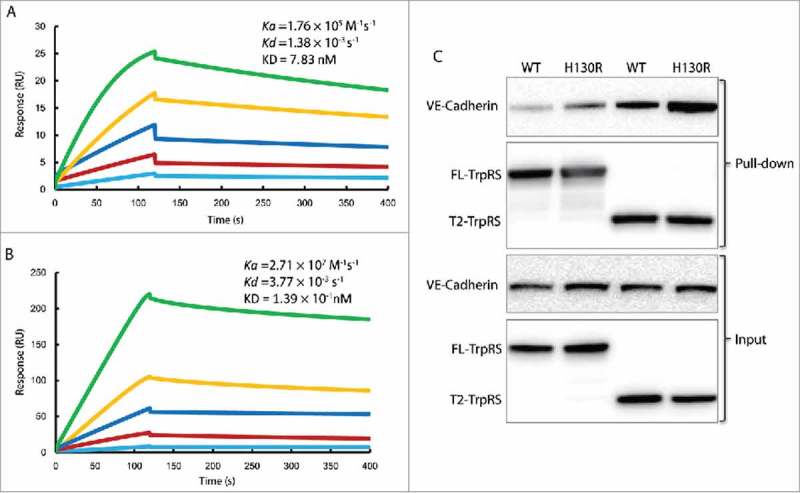
H130R substitution enhances the interaction between T2-TrpRS and VE-cadherin. (A) The fitted sensorgram of the binding between wild-type (WT) T2-TrpRS and EC1-2 domain of VE-cadherin by surface plasmon resonance. The response unit is plotted over 400 s that illustrates the association and dissociation of the complex. The concentration of WT T2-TrpRS is 2-fold increased from 12.5 nM to 200 nM. (B) The fitted sensorgram of the binding between H130R T2-TrpRS and EC1-2 domain of VE-cadherin. The response unit of H130R T2-TrpRS at concentration from 0.78 nM to 25 nM is plotted over 400 s. (C) In vitro pull down assay to test the interaction between TrpRS and endogenous VE-cadherin. The melanoma C8161 cells expressed VE-cadherin was pulled down by 6xHis tagged FL- and T2-TrpRS (WT and H130R), and immune-blotted by rabbit anti VE-cadherin. The TrpRS variants are blotted with HRP-conjugated 6xHis antibodies.
To further test the effect of H130R on VE-cadherin binding, we used a cell-based assay. Human melanoma C8161 cells express VE-cadherin and were used in a pull-down assay to detect the amount of the endogenous VE-cadherin that can be pulled down by various human TrpRS proteins (Fig. 6C). As expected from previous studies,8 T2-TrpRS pulled down substantially larger amounts of VE-cadherin than did the FL-TrpRS. Interestingly, the H130R mutation enhanced the VE-cadherin interaction, especially in the context of T2-TrpRS. Notably, the H130R mutation also enhanced the VE-cadherin interaction with FL-TrpRS. Possibly, the presence of an interaction partner helps H130R to manifest a conformational opening effect on FL-TrpRS.
Discussion
Zinc has long been implicated in the function of mammalian TrpRS.18 Bovine TrpRS was found to be zinc-bound, and elimination of zinc affected the tertiary structure and abolished the enzymatic activity.18 Later studies using human TrpRS confirmed the zinc-binding capacity and the effect of zinc binding in stimulating the synthetase activity.19 Interestingly, bovine TrpRS differs from human TrpRS at position 130 (according to the human sequence): bovine TrpRS has Arg instead of His as found in human TrpRS at this position. Remarkably, the H130R substitution in human TrpRS abolished zinc-induced stimulation and provided constitutively high enzymatic activity,24 suggesting that human H130R TrpRS mimics a zinc-bound form, while the Arg130 residue in bovine TrpRS is not sufficient to induce the zinc-bound conformation. Therefore, it seems that human and bovine TrpRS have different affinity or sensitivity towards zinc and the position 130 is a determinant for this difference.
The fact that bovine TrpRS (with Arg130) can still bind zinc18 and undergoes conformational change indicates that the H130R mutant may represent an intermediate state between zinc-free and zinc-bound conformations in human TrpRS. The conformation of this intermediate state may be shifted toward the zinc-free form by binding to Trp-AMP and towards the zinc-bound form by removal of the N-terminal residues. This speculation helps to explain the lack of conformational change in full-length human H130R TrpRS both in solution (Fig. 2) and in crystals (Fig. S2), and to interpret the observation that the enhanced VE-cadherin binding by H130R mutation in the context of T2-TrpRS is only observed in the presence of zinc (Fig. 6). It is possible that under certain conditions (such as the condition used for our SPR experiment) the H130R substitution alone is not sufficient to induce the zinc-bound conformation (like in bovine TrpRS); nevertheless, it can facilitate the conformational change induced by zinc binding. We should note that we have attempted to crystallize WT FL- and T2-TrpRS in the presence of zinc but did not yield any crystal. This suggests that the zinc-bound conformation is not as stable as the zinc-free conformation, which is consistent with our limited protease digestion analyses (Fig. 1B, C and Fig. 2).
Our previous work has demonstrated the critical role of Trp and AMP binding pockets in T2-TrpRS for VE-cadherin binding.8 However, in the H130R mutant structure, large conformational changes occur around the active site pockets. In the docking model (Fig. 5), Trp2 and Trp4 of VE-cadherin are no longer bound to the active site of H130R T2-TrpRS. It remains an open question for whether the active site of T2-TrpRS is still involved in the VE-cadherin interaction upon zinc binding.
A recent paper reported that secreted TrpRS is involved in priming innate immunity upon pathogen infection through mediating an interaction between toll-like receptor 4 (TLR4) and myeloid differentiation factor 2 (MD2).28 Interestingly, the activity of the FL-TrpRS can be recapitulated by the N-terminal 154 residues, which contain the WHEP domain and the ESE (Fig. 1A). In particular, it was proposed that the last helix of ESE is involved in the TLR4 interaction. As shown in Fig. 3, the ESE including the last helix of ESE (α4′) undergoes large conformational change in the H130R mutant that mimics a zinc-bound conformation. Therefore, it is possible that zinc binding may also influence the role of TrpRS in priming innate immunity.
Overall, our work has provided direct evidence for an alternative conformation of human TrpRS that mimics a zinc-bound conformation. This conformation may enhance the role of TrpRS in angiogenesis regulation by facilitating the proteolytic production of the anti-angiogenic fragment T2-TrpRS and by strengthening the interaction between T2-TrpRS and its receptor VE-Cadherin. For the first time, this work links zinc to regulatory functions of tRNA synthetases.
Accession numbers
The atomic coordinates and structure factors of FL-TrpRS in complex with Trp-AMP and the H130R T2-TrpRS have been deposited in the Protein Data Bank with the accession code 5UJI and 5UJJ, respectively.
Material and methods
Protein expression and purification
The genes encoding human H130R FL- and T2 (Gly97 to Gln417) TrpRS were amplified by PCR and constructed into pET20b vector (Novagen) with a C-terminal 6 × His tag. The proteins were expressed in transformed Escherichia coli BL21 (DE3) cells and induced by 0.1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at room temperature for 8 hrs. The cells were disrupted by M-110L Microfluidizer Processor (Microfluidics) in 25 mM Hepes-Na pH 7.5, 300 mM NaCl, 20 mM imidazole and 5% glycerol, the cell lysates were centrifuged at 25,000 rpm for 30 min. The soluble fraction was applied on a Ni-NTA agarose column (Qiagen) for affinity purification. T2-TrpRS was eluted by 250 mM imidazole and further purified by ion-exchange chromatography by Resouece Q column (GE Healthcare), protein was eluted by 25 mM Hepes-Na pH7.5 and sodium chloride gradient to 1 M. The fractions were concentrated and applied to HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare) for size exclusion purification, purified human H130R T2-TrpRS was concentrated to 10 mg mL−1 and set up for crystallization. H130R FL-TrpRS was purified by same procedure as H130R T2-TrpRS.
Crystallization
Crystallization was performed by micro-sitting drop vapor diffusion method using Mosquito liquid transfer robot (TTP Labtech). H130R T2-TrpRS was pre-incubated with 2 mM MgCl2 and 4 mM β-Mercaptoethanol for crystallization. Crystals were obtained by mixing 0.1 uL of protein with 0.1 uL reservoir solution (20% PEG4000, 0.1 M Na Citrate pH5.0, 0.1 M MgCl2) equilibrated against 70 uL reservoir solution at 4°C.
H130R FL-TrpRS was diluted in a buffer containing 25 mM Hepes-Na pH7.5, 100 mM KCl, 2 mM MgCl2 and 4 mM β-Mercaptoethanol, and pre-incubated with 500 uM Trp-AMP for crystallization. The crystals were obtained in 1:1 ration of the protein and reservoir solution containing 20% PEG1500, 0.1 M Bis-Tris pH6.5.
Data collection
The crystals were cryoprotected in a solution containing the reservoir solution with 15% glycerol and flash-cooled with liquid nitrogen. A 2.8 Å resolution data set of human H30R T2-TrpRS and 2.1 Å resolution data set of H30R FL-TrpRS were collected at 100 K on beamline BL7-1 at the Stanford Synchrotron Radiation Laboratory using an ADSC Q315 detector. The crystal of H130R T2-TrpRS belongs to the space group C2221, with unit cell dimensions a = 84.89 Å, b = 139.06 Å, c = 146.72Å, α = β = γ = 90° (Table 1). Whereas the crystal of H130R FL-TrpRS in complex with Trp-AMP belongs to space group C121, and with unit cell parameters a = 136.35, b = 95.28, c = 99.63, α = γ = 90°, β = 130° (Table S1). Processing of all diffraction images and scaling of the integrated intensities were performed using the HKL2000.29 There are two molecules of H130R T2-TrpRS in one asymmetric unit with Matthews coefficient VM of 2.5 Å3 Da−1, corresponding to a solvent content of 51%.30
Table 1.
X-ray crystallography data collection and refinement statistics.
| Parameters | T2-TrpRS H130R |
|---|---|
| Data collection | |
| Cell parameters (Å) | a = 84.89, b = 139.06, |
| c = 146.72 | |
| α = β = γ = 90° | |
| Space group | C2221 |
| Resolution (Å) | 50 (2.85) – 2.80 |
| No. of all reflections | 108967 |
| No. of unique reflections | 21588 |
| Completeness (%) | 98.9 (99.4) |
| Redundacy | 5.0 (5.1) |
| I/σI | 6.5 (2.5) |
| Rmerge (%) | 9.3 (47.1) |
| Refinement | |
| Resolution (Å) | 50 – 2.79 |
| Total No. of reflections | 20462 |
| No. of reflections used | 19358 |
| Rwork / Rfree (%) | 22.0 / 26.2 |
| No. of atoms (Protein) | 5762 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angle (°) | 1.15 |
| Average B-factors (Å2) | 73.0 |
| Ramachandran Plot | |
| Most favored region (%) | 96.0 |
| Allowed regions (%) | 4.0 |
| Disallowed regions (%) | 0 |
Note: Values in parentheses are for the highest resolution shell.
Structure determination
The structure of H130R T2-TrpRS was determined by molecular replacement using wild-type human T2-TrpRS structure (PDB 2QUI)25 as search model in program PHASER31 from the CCP4 package.32 One homodimer of H130R T2-TrpRS was found in one asymmetric unit. Iterative model building and refinement were performed using Coot,33and Refmac5,34 the final model with Rwork of 22.0% and Rfree of 26.2% at 2.8 Å resolution. The H130R FL-TrpRS structure was determined using the structure of WT human FL-TrpRS (PDB 1R6T) as serch model and the method molecular replacement. Final model was refined to Rwork of 21.5% and Rfree of 23.2% (Table S1).
Structure and molecular docking
To derive the binding conformations of the wild-type (PDB ID 2QUI) and H130R T2-TrpRS with EC1-2 of VE-cadherin (PDB ID 3PPE), molecular docking was performed using ZDOCK (version 3.0.2) software.35 Using a fast Fourier transform (FFT)-based initial-stage rigid-body molecular docking algorithm,36 ZDOCK performs a global search in the translational and rotational space to produce all possible binding configurations between proteins. With a composite scoring function that combines pairwise shape complementarity with desolvation and electrostatics, the performance of ZDOCK in critical assessment of prediction of interaction (CAPRI) challenge proves that it is among the best protein-protein docking algorithms.37,38 ZDOCK created 3600 and retained 2000 putative binding configurations of WT / H130R T2-TrpRS and EC1-2, the complex models with the top default ZDOCK scores were chosen to be the most possible binding configurations. To remove bad atomic contacts around the binding interface, each binding model was then subjected to 2000 steps steepest descent and 2000 steps conjugate gradient energy minimizations using SANDER module in the AMBER 12 suit of programs.39
Molecular dynamics simulation
Molecular Dynamics simulation (MDS) was performed using the AMBER 12 software and FF14SB force field.39,40 The complex models of WT / H130R T2-TrpRS bound with EC1-2 domain of chicken VE-cadherin were independently immersed into the center of a truncated octahedron box of TIP3P water molecules with a margin distance of 12.0 Å, potassium counterions were added by using the AMBER XLEAP module to keep system in electric neutrality.39 Each binding complex was firstly energy minimized by the steepest descent method for 2000 steps with WT / H130R T2-TrpRS and EC1-2 restricted by a harmonic constraint of 100 kcal·mol−1Å−2. A further conjugate gradient minimization of 2000 steps was performed with no constraint. Then the system was gradually heated from 0 K to 300 K under the NVT ensemble over a period of 200 ps, during which the Langevin thermostat with a coupling coefficient of 1.0 ps and a weak constraint of 10 kcal·mol−1Å−2 on proteins was applied. Each model was subsequently subjected to an equilibrium simulation for 200 ps by removing all constraints. Finally, a 20 ns production MDS for each model was conducted under NPT ensemble. Periodic boundary conditions were applied. System temperature was kept 300 K using the Berendsen thermostat with a time constant of 1 ps. Isotropic constant pressure was maintained by Berendsen pressure coupling algorithm with a time constant of 1 ps. Hydrogen involved covalent bonds were constrained by the SHAKE algorithm.41 The long-range electrostatic interactions were treated by the Particle Mesh Ewald (PME) method.42 The cutoffs for long-range electrostatic and Van der Waals interactions were both set to 12.0 Å. The time step was set to 2 fs in all simulations, and the coordinates were saved every 1 ps to record the MD trajectories.
Pull-down assays
Lysates of E. coli cells that transformed with genes expressing 6 × His tagged full-length or T2-TrpRS (WT and H130R) were mixed with lysates from melanoma C8161 cells expressing VE-cadherin. The mixtures were incubated at 4°C overnight, followed by incubation with Ni-NTA resin for 1 hour to capture histidine-tagged TrpRS proteins and VE-cadherin complex. Input samples were taken out prior to the addition of Ni-NTA resin. The resin was washed 3 times with wash buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM DTT, 20 mM imidazole). VE-cadherin pull downed by TrpRS was eluted with SDS-PAGE reducing sample buffer and subjected to SDS-PAGE followed by western blot with rabbit anti VE-cadherin (Cell Signaling Technology) and HRP-conjugated 6xHis (Proteintech) antibodies to detect VE-cadherin and TrpRS proteins, respectively.
Surface plasmon resonance
The EC1-2 domain of VE-cadherin was expressed in a Bac-to-Bac baculovirus system using Sf9 insect cells. The EC1-2 domain was purified and immobilized on a CM5 chip as bait ligand though amine coupling. The WT or H130R T2-TrpRS expressed and purified from E.coli was injected as prey analyte in HBS-P buffer containing 20 mM Hepes pH7.5, 150 mM NaCl, 0.02% surfactant P-20. The response unit is plotted over injection time to probe the association and dissociation of T2-TrpRS with immobilized EC1-2 domain. Interestingly, the binding only can be detected when 0.1 mM ZnCl2 was present in the HBS-P buffer. After optimizing the association and dissociation parameters, the binding sensorgram, kinetic analysis was performed by injecting the WT or H130R T2-TrpRS at series concentrations. The binding constant was calculated from the sensorgram of WT T2-TrpRS ranges from 12.5 nM to 200 nM and the H130R T2-TrpRS from 0.78 nM to 25 nM in a 2 fold dilution.
Supplementary Material
Acknowledgment
This work was supported by grants from US National Institutes of Health (R01 GM088278 and R01 NS085092), the National Natural Science Foundation of China (31400630 and 31570738), and Zhejiang Provincial Natural Science Foundation of China (LY14C050002), fellowships from the National Foundation for Cancer Research (US), and funding from aTyr Pharma through an agreement with The Scripps Research Institute.
References
- 1.Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Ann Rev Biochem. 1987;56:125–58. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- 2.Guo M, Schimmel P, Yang XL. Functional expansion of human tRNA synthetases achieved by structural inventions. FEBS Lett. 2010;584(2):434–42. doi: 10.1016/j.febslet.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol. 2010;11(9):668–74. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang XL, Guo M, Kapoor M, Ewalt KL, Otero FJ, Skene RJ, McRee DE, Schimmel P. Functional and crystal structure analysis of active site adaptations of a potent anti-angiogenic human tRNA synthetase. Structure. 2007;15(7):793–805. doi: 10.1016/j.str.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otani A, Slike BM, Dorrell MI, Hood J, Kinder K, Ewalt KL, Cheresh D, Schimmel P, Friedlander M. A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc Natl Acad Sci U S A. 2002;99(1):178–83. doi: 10.1073/pnas.012601899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wakasugi K, Slike BM, Hood J, Otani A, Ewalt KL, Friedlander M, Cheresh DA, Schimmel P. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc Natl Acad Sci U S A. 2002;99(1):173–7. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzima E, Reader JS, Irani-Tehrani M, Ewalt KL, Schwartz MA, Schimmel P. VE-cadherin links tRNA synthetase cytokine to anti-angiogenic function. J Biol Chem. 2005;280(4):2405–8. doi: 10.1074/jbc.C400431200. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Q, Kapoor M, Guo M, Belani R, Xu X, Kiosses WB, Hanan M, Park C, Armour E, Do MH, et al.. Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat Struct Mol Biol. 2010;17(1):57–61. doi: 10.1038/nsmb.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng R, Chen YC, Zeng Z, Liu XX, Liu R, Qiang O, Li X. Inhibition of mini-TyrRS-induced angiogenesis response in endothelial cells by VE-cadherin-dependent mini-TrpRS. Heart Vessels. 2012;27(2):193–201. doi: 10.1007/s00380-011-0137-1. [DOI] [PubMed] [Google Scholar]
- 10.Kise Y, Lee SW, Park SG, Fukai S, Sengoku T, Ishii R, Yokoyama S, Kim S, Nureki O. A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase. Nat Struct Mol Biol. 2004;11(2):149–56. doi: 10.1038/nsmb722. [DOI] [PubMed] [Google Scholar]
- 11.Riordan JF. Biochemistry of zinc. Med Clin North Am. 1976;60(4):661–74. doi: 10.1016/S0025-7125(16)31851-X. [DOI] [PubMed] [Google Scholar]
- 12.Coleman JE. Zinc proteins: enzymes, storage proteins, transcription factors, and replication proteins. Annu Rev Biochem. 1992;61:897–946. doi: 10.1146/annurev.bi.61.070192.004341. [DOI] [PubMed] [Google Scholar]
- 13.Saghiri MA, Asatourian A, Orangi J, Sorenson CM, Sheibani N. Functional role of inorganic trace elements in angiogenesis-Part II: Cr, Si, Zn, Cu, and S. Crit Rev Oncol Hematol. 2015;96(1):143–55. doi: 10.1016/j.critrevonc.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 14.Kaji T, Fujiwara Y, Yamamoto C, Sakamoto M, Kozuka H. Stimulation by zinc of cultured vascular endothelial cell proliferation: possible involvement of endogenous basic fibroblast growth factor. Life Sci. 1994;55(23):1781–7. doi: 10.1016/0024-3205(94)90088-4. [DOI] [PubMed] [Google Scholar]
- 15.Uzzo RG, Crispen PL, Golovine K, Makhov P, Horwitz EM, Kolenko VM. Diverse effects of zinc on NF-kappaB and AP-1 transcription factors: implications for prostate cancer progression. Carcinogenesis. 2006;27(10):1980–90. doi: 10.1093/carcin/bgl034. [DOI] [PubMed] [Google Scholar]
- 16.Boehm T, O'Reilly M S, Keough K, Shiloach J, Shapiro R, Folkman J. Zinc-binding of endostatin is essential for its antiangiogenic activity. Biochem Biophys Res Commun. 1998;252(1):190–4. doi: 10.1006/bbrc.1998.9617. [DOI] [PubMed] [Google Scholar]
- 17.Nurbekov MK, Favorova OO, Dmitrenko SG, Bolotina IA, Kiselev LL. [Role of zinc ions in the functioning of bovine tryptophanyl-tRNA-synthetase]. Mol Biol (Mosk). 1981;15(5):1000–10. [PubMed] [Google Scholar]
- 18.Kisselev LL, Favorova OO, Nurbekov MK, Dmitriyenko SG, Engelhardt WA. Bovine tryptophanyl-tRNA synthetase. A zinc metalloenzyme. Eur J Biochem. 1981;120(3):511–7. doi: 10.1111/j.1432-1033.1981.tb05729.x. [DOI] [PubMed] [Google Scholar]
- 19.Wakasugi K. Human tryptophanyl-tRNA synthetase binds with heme to enhance its aminoacylation activity. Biochemistry. 2007;46(40):11291–8. doi: 10.1021/bi7012068. [DOI] [PubMed] [Google Scholar]
- 20.Doublie S, Bricogne G, Gilmore C, Carter CW Jr.. Tryptophanyl-tRNA synthetase crystal structure reveals an unexpected homology to tyrosyl-tRNA synthetase. Structure. 1995;3(1):17–31. doi: 10.1016/S0969-2126(01)00132-0. [DOI] [PubMed] [Google Scholar]
- 21.Ilyin VA, Temple B, Hu M, Li G, Yin Y, Vachette P, Carter CW Jr.. 2.9 A crystal structure of ligand-free tryptophanyl-tRNA synthetase: domain movements fragment the adenine nucleotide binding site. Protein Sci: a publication of the Protein Society. 2000;9(2):218–31. doi: 10.1110/ps.9.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Y, Liu Y, Shen N, Xu X, Xu F, Jia J, Jin Y, Arnold E, Ding J. Crystal structure of human tryptophanyl-tRNA synthetase catalytic fragment: insights into substrate recognition, tRNA binding, and angiogenesis activity. J Biol Chem. 2004;279(9):8378–88. doi: 10.1074/jbc.M311284200M311284200. [DOI] [PubMed] [Google Scholar]
- 23.Retailleau P, Weinreb V, Hu M, Carter CW Jr. Crystal structure of tryptophanyl-tRNA synthetase complexed with adenosine-5′ tetraphosphate: evidence for distributed use of catalytic binding energy in amino acid activation by class I aminoacyl-tRNA synthetases. J Mol Biol. 2007;369(1):108–28. doi: 10.1016/j.jmb.2007.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakasugi K. Species-specific differences in the regulation of the aminoacylation activity of mammalian tryptophanyl-tRNA synthetases. FEBS Lett. 2010;584(1):229–32. doi:S0014-5793(09)00990-9. [DOI] [PubMed] [Google Scholar]
- 25.Shen N, Zhou M, Yang B, Yu Y, Dong X, Ding J. Catalytic mechanism of the tryptophan activation reaction revealed by crystal structures of human tryptophanyl-tRNA synthetase in different enzymatic states. Nucleic Acids Res. 2008;36(4):1288–99. doi:gkm1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen N, Guo L, Yang B, Jin Y, Ding J. Structure of human tryptophanyl-tRNA synthetase in complex with tRNATrp reveals the molecular basis of tRNA recognition and specificity. Nucleic Acids Res. 2006;34(11):3246–58. doi:34/11/3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brasch J, Harrison OJ, Ahlsen G, Carnally SM, Henderson RM, Honig B, Shapiro L. Structure and binding mechanism of vascular endothelial cadherin: a divergent classical cadherin. J Mol Biol. 2011;408(1):57–73. doi: 10.1016/j.jmb.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahn YH, Park S, Choi JJ, Park BK, Rhee KH, Kang E, Ahn S, Lee CH, Lee JS, Inn KS, et al.. Secreted tryptophanyl-tRNA synthetase as a primary defence system against infection. Nat Microbiol. 2016;2:16191. doi: 10.1038/nmicrobiol.2016.191. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276(Macromolecular Crystallography, part A,):307–26. [DOI] [PubMed] [Google Scholar]
- 30.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33(2):491–7. doi:0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–74. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–3. doi: 10.1107/S0907444994003112S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1): 2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–55. doi: 10.1107/S0907444996012255S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PloS one. 2011;6(9):e24657. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen R, Weng Z. Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins. 2002;47(3):281–94. doi: 10.1002/prot.10092. [DOI] [PubMed] [Google Scholar]
- 37.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52(1):80–7. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 38.Chen R, Tong W, Mintseris J, Li L, Weng Z. ZDOCK predictions for the CAPRI challenge. Proteins. 2003;52(1):68–73. doi: 10.1002/prot.10388. [DOI] [PubMed] [Google Scholar]
- 39.Case DA, Cheatham TE, 3rd Darden T, Gohlke H, Luo R, Merz KM Jr., Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26(16):1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J Chem Theory Comput. 2015;11(8):3696–713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n -alkanes. Journal of Computational Physics. 1977;23(3):327–41. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 42.Essmann U, Perera L, Berkowitz ML, Darden TA, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phy. 1995;103(19):8577–93. doi: 10.1063/1.470117. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


