ABSTRACT
Chaperone-mediated autophagy (CMA) is a major pathway of lysosomal proteolysis essential for the control of intermediary metabolism. So far, the absence of any identifiable LAMP2A – a necessary and limiting protein for CMA – outside of the tetrapod clade, led to the paradigm that this cellular function was (presumably) restricted to mammals and birds. However, after we identified expressed sequences displaying high sequence homology with the mammalian LAMP2A in several fish species, our findings challenge that view and suggest that CMA likely appeared much earlier during evolution than initially thought. Hence, our results do not only shed an entirely new light on the evolution of CMA, but also bring new perspectives on the possible use of complementary genetic models, such as zebrafish or medaka for studying CMA function from a comparative angle/view.
KEYWORDS: Chaperone-mediated autophagy, evolution, fish, LAMP2A, RNA-seq
Being one of the main pathways involved in lysosomal proteolysis, chaperone-mediated autophagy (CMA) has been described as a selective mechanism for the degradation of specific soluble proteins within lysosomes [1]. Besides a well described role in protein quality control (resulting from its ability to selectively target damaged or non-functional proteins for degradation), the diversity of the sub-proteome degraded by CMA also associates this function with the regulation of transcriptional programs [2], cell death and cell survival mechanisms [3–5], DNA repair and cell cycle progression [6], as well as a variety of intracellular processes related to the control of cellular energetics [7–10]. Over the last few years, CMA has thus emerged as a major core component in the control of cellular homeostasis [11].
In detail, cytosolic proteins bearing a KFERQ-like motif are first recognized by the chaperone HSPA8/HSC70 [12]. The substrate/chaperone complex then docks at the lysosomal membrane through specific binding to the cytosolic tail of LAMP2A (lysosome-associated membrane protein 2A). Multimerization of LAMP2A will then result in the formation of a translocation complex, and promote translocation of substrate proteins [13]. Following unfolding and internalization, substrate proteins are then rapidly degraded by lysosomal proteases. Next, LAMP2A disassembles from the translocation complex, allowing a new cycle of substrate binding and translocation [14].
LAMP2A originates from the alternative splicing of the LAMP2 gene, giving rise to 3 different splice variants. These splice variants all share a common lumenal domain but display different cytosolic and transmembrane regions. CMA activity is tightly correlated with (i) the level of LAMP2A (and not those of the 2 other splice variants) at the lysosomal membrane [15] and (ii) the efficiency of assembly/disassembly of LAMP2A in this compartment [14]. As such, LAMP2A is considered to be the necessary and limiting component for CMA activity [15].
In this context, because LAMP2A has, so far, been characterized only in birds and mammals, and not in other clades, functional CMA is thought to be restricted to tetrapods [16]. Interestingly, in Drosophila, selective endosomal microautophagy (eMI), a recently identified form of microautophagy which shares together with CMA the dependence on KFERQ-like motifs and HSPA8/HSC70 for substrate targeting, has been suggested to constitute an alternative to CMA [16,17]. Although it is tempting to speculate that in non-tetrapod species eMI might be an ancestral form of selective autophagy for the degradation of substrates that in tetrapods are shared between eMI and CMA, our data reveal that the picture is probably much more complex.
Indeed, homology-based searches on in house-developed RNA-seq databases (PhyloFish), providing consistent and exhaustive gene expression data from 23 different ray-finned fish species [18], resulted in the identification of several contigs displaying high sequence homology with mammalian LAMP2A. In detail, the inferred amino acid sequences of the fish Lamp2A contigs not only displayed high homology when compared to the transmembrane and cytoplasmic domains of mammalian LAMP2A, but also high conservation of key motifs shown to be essential for proper function of the protein (Figure 1(A)). Hence, fish Lamp2A sequences display the typical GY dipeptide conserved in all forms of LAMP2 and required for targeting to lysosomes [19]. Fish Lamp2A present also the hydrophobic phenylalanine (F) residue (not conserved in the other forms of Lamp2) important for lysosomal targeting [20]. Additionally, 3 of the 4 positively charged amino acids necessary for binding substrate proteins [15] are also present in fish sequences, as are the 2 glycine residues (G) involved in the multimeric pattern of Lamp2A [13], at least in some fish species (Figure 1(A)).
Figure 1.
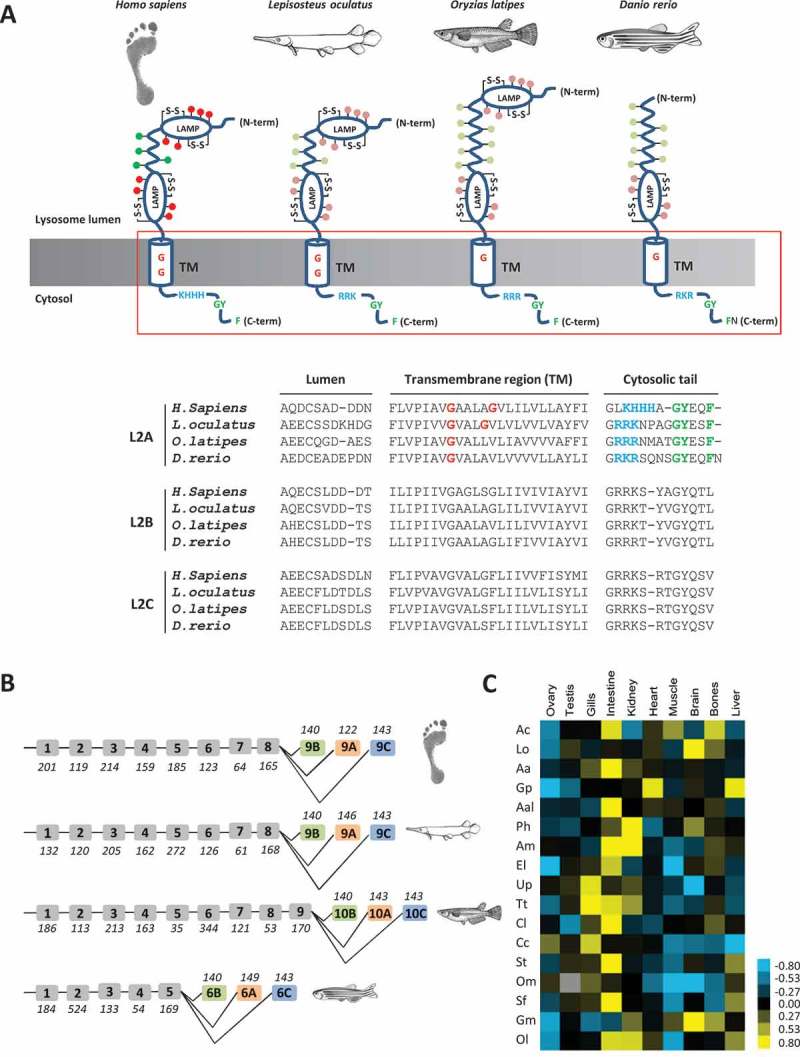
Protein Structure, gene organization and mRNA expression of Lamp2A in fish (A) Schematic drawing of the structure of vertebrate LAMP2A. The human LAMP2A is used as reference. It shows a lumenal region comprising 2 N-glycosylated LAMP domains of approximately 160 residues (each with 2 disulfide bonds, S-S) separated by a proline-rich, O-glycosylated ‘hinge’ region of approximately 30 amino acid residues [21]. O- and N-linked glycosylation are indicated in green and red, respectively. The transmembrane (TM) domain, harboring 2 glycine residues (red G) involved in the multimeric pattern of LAMP2A, is followed by a short, C-terminal cytosolic tail that is comprised of 4 positively-charged amino acids (KHHH in blue) required for the binding of substrate proteins as well as motifs for lysosomal targeting (in green). The schematic drawing of fish Lamp2A has been done on the basis of sequence complementarity with the human LAMP2A. Potential O- and N-linked glycosylation are indicated in light green and pink, respectively. Sequence alignment of the boxed region of the 3 LAMP2/Lamp2 variants is shown below. The positively-charged amino acids required for the binding of substrate proteins are colored in blue. The GY dipeptide as well as the hydrophobic F required for targeting of LAMP2A to lysosomes are in green. The glycine residues (G) involved in the multimeric pattern of LAMP2A are in red. (B) The genomic structure of LAMP2/lamp2 is conserved in vertebrates and contains the 3 alternative exons (B, A and C) encoding the transmembrane domain and cytoplasmic tail specific for each isoform. The size of exons (in base pairs) is shown in italics below or above each exon. (C) Data from RNAseq show that lamp2a is expressed in different tissues of a large number of ray-finned fish. Relative expression of lamp2a was expressed in number of reads per kilobase per million reads per species, after normalization of data by the total number of sequences obtained for each tissue and species. The obtained values were then log transformed and centered to the median (set at 0.00). Ac, Amia calva (bowfin); Lo, Lepisosteus oculatus (spotted gar); Aa, Anguilla anguilla (European eel); Gp, Gnathonemus petersi (elephantnose fish); Aal, Alosa alosa (allis shad); Ph, Pangasianodon hypophthalmus (striped Catfish); Am, Astyanax mexicanus (cave Mexican tetra); El, Esox lucius (northern pike); Up, Umbra pygmae (eastern mudminnow); Tt, Thymallus thymallus (grayling); Cl, Coregonus lavaretus (European whitefish); Cc, Coregonus clupeaformis (American whitefish); St, Salmo trutta (brown trout); Om, Oncorhynchus mykiss (rainbow trout); Sf, Salvelinus fontinalis (brook trout); Gm, Gadus morhua (Atlantic cod); Ol, Oryzias latipes (medaka).
Furthermore, sequence analysis of the complete genomes of several fish species also revealed high conservation of the genomic organization of the LAMP2 gene across vertebrates with notably the presence of the 3 alternative exons (B, A and C) encoding the transmembrane domain and cytoplasmic tail specific of each isoform (LAMP2B, LAMP2A and LAMP2C, respectively) (Figure 1(B)). However, the number and size of exons encoding the lumenal region are only moderately conserved among species (Figure 1(B)), leading to lower homology of the corresponding region (from 15 to 20% between analyzed species) compared to the cytoplasmic tail (from 21 to 85%) (Table S1), thus accounting for different structure (Figure 1(A)) and possibly functional variations of Lamp2A among species.
Finally, fine expression analysis of the newly characterized lamp2a splice variants in different fish species shows that they are expressed in different tissues of a large variety of ray-finned fish species, including the medaka (Oryzias latipes), a model species widely used in biomedical research (Figure 1(C)). Although no lamp2a transcripts could be identified in zebrafish (Danio rerio), RT-PCR analysis using specific primers targeting a conserved region of the lamp2a exon from fish nevertheless revealed significant zebrafish lamp2a expression in different tissues, including intestine, kidney and liver (data not shown).
Overall, our data show for the first time the existence as well as the expression of lamp2a transcripts in different tissues of a large variety of ray-finned fish species, and therefore imply that CMA function might have appeared much earlier during evolution than initially thought. A number of issues remain now to be addressed about the functionality of the Lamp2A sequences found in fish. This includes (i) whether or not in fish lamp2a mRNA can be successfully translated to protein, (ii) whether or not these Lamp2A proteins properly localize to lysosomes, and (iii) whether or not the polymerized fish Lamp2A regulates the relocation of the substrates. Further understanding of the structure-function relationship between fish Lamp2A displaying differences in key motifs will also help to complete this picture. The presence of Lamp2A outside of the tetrapod clades opens up new perspectives in autophagy research. Comparative approaches across phylogenetic distant species (fish versus tetrapods for instance) will certainly provide new insights on selective autophagy by exploring the extent to which CMA, but also the crosstalk between CMA and other components of the cellular proteostasis networks (in particular eMI), diverged during vertebrate evolution.
Funding Statement
This study was supported by the INRA ‘Animal Physiology and Livestock Systems’ Division, and the French National Research Agency (ANR-17-CE20-0033 ‘Fish-and-Chap’).
Acknowledgments
We thank AM Cuervo (Albert Einstein College of Medicine, NY) for helpful advice.
Competing financial interests
The authors declare no competing financial interests.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Kaushik S, Cuervo AM.. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22(8):407–417. PMID: 22748206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Valdor R, Mocholi E, Botbol Y, et al. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat Immunol. 2014;15(11):1046–1054. PMID: 25263126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ferreira JV, Fofo H, Bejarano E, et al. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy. 2013;9(9):1349–1366. PMID: 23880665. [DOI] [PubMed] [Google Scholar]
- [4].Xie W, Zhang L, Jiao H, et al. Chaperone-mediated autophagy prevents apoptosis by degrading BBC3/PUMA. Autophagy. 2015;11(9):1623–1635. PMID: 26212789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yang Q, She H, Gearing M, et al. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323(5910):124–127. PMID: 19119233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Park C, Suh Y, Cuervo AM. Regulated degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage. Nat Commun. 2015;6:6823 PMID: 25880015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17(6):759–770. PMID: 25961502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lv L, Li D, Zhao D, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42(6):719–730. PMID: 21700219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schneider JL, Suh Y, Cuervo AM. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014;20(3):417–432. PMID: 25043815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Xia HG, Najafov A, Geng J, et al. Degradation of HK2 by chaperone-mediated autophagy promotes metabolic catastrophe and cell death. J Cell Biol. 2015;210(5):705–716. PMID: 26323688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tasset I, Cuervo AM. Role of chaperone-mediated autophagy in metabolism. FEBS J. 2016;283(13):2403–2413. PMID: 26854402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chiang HL, Terlecky SR, Plant CP, et al. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246(4928):382–385. PMID: 2799391. [DOI] [PubMed] [Google Scholar]
- [13].Bandyopadhyay U, Kaushik S, Varticovski L, et al. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28(18):5747–5763. PMID: 18644871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bandyopadhyay U, Sridhar S, Kaushik S, et al. Identification of regulators of chaperone-mediated autophagy. Mol Cell. 2010;39(4):535–547. PMID: 20797626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000;113(Pt 24):4441–4450. PMID: 11082038. [DOI] [PubMed] [Google Scholar]
- [16].Tekirdag KA, Cuervo AM. Chaperone-mediated autophagy and endosomal microautophagy: joint by a chaperone. J Biol Chem. 2017. PMID: 29247007 DOI: 10.1074/jbc.R117.818237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mukherjee A, Patel B, Koga H, et al. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy. 2016;12(11):1984–1999. PMID: 27487474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pasquier J, Cabau C, Nguyen T, et al. Gene evolution and gene expression after whole genome duplication in fish: the PhyloFish database. BMC Genomics. 2016;17:368 PMID: 27189481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Williams MA, Fukuda M. Accumulation of membrane glycoproteins in lysosomes requires a tyrosine residue at a particular position in the cytoplasmic tail. J Cell Biol. 1990;111(3):955–966. PMID: 2391371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Guarnieri FG, Arterburn LM, Penno MB, et al. The motif Tyr-X-X-hydrophobic residue mediates lysosomal membrane targeting of lysosome-associated membrane protein 1. J Biol Chem. 1993;268(3):1941–1946. PMID: 8420968. [PubMed] [Google Scholar]
- [21].Wilke S, Krausze J, Bussow K. Crystal structure of the conserved domain of the DC lysosomal associated membrane protein: implications for the lysosomal glycocalyx. BMC Biol. 2012;10:62 PMID: 22809326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


