Abstract
Replication, validation and extension of experiments are crucial for scientific progress. Computational experiments are inherently scriptable and should be easy to reproduce. However, it remains difficult and time consuming to reproduce computational results because analyses are designed and run in a specific computing environment, which may be difficult or impossible to match from written instructions. We report a workflow named continuous analysis that can build reproducibility into computational analyses. Continuous analysis combines Docker, a container technology akin to virtual machines, with continuous integration, a software development technique, to automatically re-run a computational analysis whenever updates or improvements are made to source code or data. This allows results to be accurately reproduced without needing to contact the study authors. Continuous analysis allows reviewers, editors or readers to verify reproducibility without manually downloading and re-running any code and can provide an audit trail for analyses of data that cannot be shared.
Leading scientific journals have highlighted a need for improved reproducibility in order to increase confidence in results and reduce the number of retractions1–5. In a recent survey, 90% of researchers acknowledged that there ‘is a reproducibility crisis’6. Computational reproducibility is the ability to exactly reproduce results given the same data, as opposed to replication, which requires a new independent experiment. Computational protocols used for research should be readily reproducible because all of the steps are scripted into a machine-readable format. However, results can often only be reproduced with help from the original authors, and reproducing results requires a substantial time investment. Garijo et al.7 estimated that it would take 280 hours for a non-expert to reproduce the computational construction of a drug-target network for Mycobacterium tuberculosis8. Written descriptions of computational approaches can be difficult to understand and may lack sufficient details, including data preprocessing, model parameter selection and software versions, which are crucial for reproducibility. Indeed, Ioannidis et al.9 indicated that the outputs of 56% of microarray gene expression experiments could not be reproduced and another 33% could only be reproduced with discrepancies. Additionally, Hothorn and Leisch found that more than 80% of manuscripts did not report software versions10.
It has been proposed that open science could aid reproducibility3,11. In open science the data and source code are shared. Intermediate results and project planning are sometimes also shared12. Sharing data and source code are necessary, but not sufficient, to make research reproducible. Even when code and data are shared, it remains difficult to reproduce results due to variability in computing environments, operating systems, and the versions of software used during the original analysis. It is common to use one or more software libraries during a project. Using these libraries creates a dependency to a particular version of the library; research code often only works with old versions of these libraries13. Developers of newer versions may have renamed functions, resulting in broken code, or changed the way a function works to yield a slightly different result without returning an error. For example, Python 2 would perform integer division by default, so 5/2 would return 2. Python 3 performs floating-point division by default, so the same 5/2 command now returns 2.5. In addition, old or broken dependencies can mean that it is not possible for readers or reviewers to recreate the computational environment used by the authors of a study. In this case it becomes impossible to validate or extend results.
We first illustrate, using a practical example, the problem of reproducibility of computational studies. Then we describe the development and validation of a method named continuous analysis that can address this problem.
RESULTS
One example illustrating how data-sharing does not automatically make science reproducible can be found in routine analyses of differential gene expression. Differential expression analyses are performed first by quantifying RNA levels in two or more conditions and then identifying the transcripts with expression levels that are altered by the experiment. When a DNA microarray is used to measure transcript expression levels, positions on the array correspond to oligonucleotides of certain sequences, termed probes. A certain set of probes is used to estimate the expression level of each gene or transcript. As our understanding of the genome changes, the optimal mapping of probes to genes or transcripts can change as well.
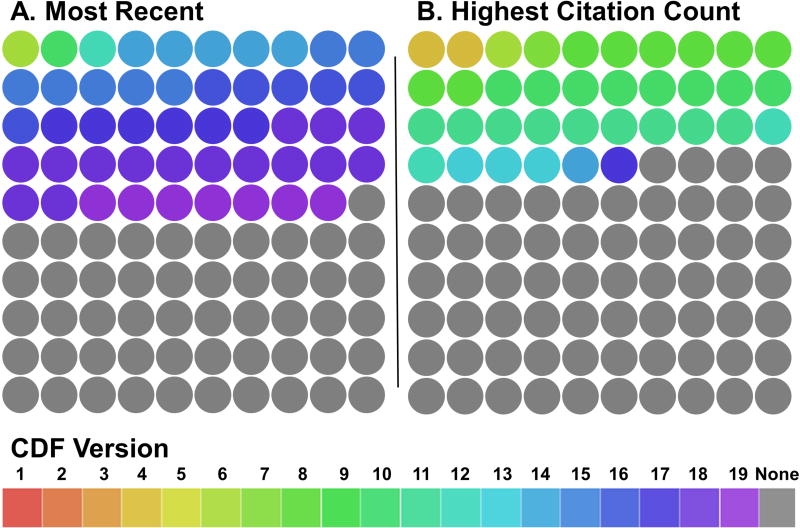
Dai et al.14 publish and maintain a popular source of probe set description files that are routinely updated (BrainArray Custom CDF). Analyses that fail to report the probe set version, or that were performed with probe set definitions that are now missing, can never be reproduced. We set out to ascertain the extent of this problem through a literature search. We analyzed the one hundred most recently published papers citing Dai et al. that were accessible at our institution (Supplementary Data 1). We identified these manuscripts using Web of Science on November 14, 2016. We recorded the number of papers that cited a version of Custom CDF, including which version was cited. These articles adhered to expectations of citing methods appropriately because they cited the source of their probe set definitions. Of these 100 papers, 49 (49%) specified which version was used (Figure 1A). These manuscripts reported the use of versions 6, 10, 12, 14, 15, 16, 17, 18, and 19 of the BrainArray Custom CDF. As of November 14, 2016 versions 6 and 12 were no longer available for download on the BrainArray web site. To determine the extent to which a lack of version reporting of the probe set affects high impact papers, we analyzed at the one hundred most cited papers that cite Dai et al. (Supplementary Data 2). We determined the one hundred most cited papers using Web of Science on November 14, 2016. Of these 100 papers, 36 (36%) specified which version of the Custom CDF was used (Figure 1B). These manuscripts used versions 4, 6, 7, 8, 10, 11, 12, 13, 14 and 17. Versions 4, 6, 7, 11 and 12 were not available for download as of November 14, 2016.
Figure 1. Reporting of custom CDF file descriptors in published papers.
Works citing custom chip description files (Custom CDF) frequently do not cite the version. Each manuscript is represented by a circle in which color indicates the version used by each paper. A.) 51 of the 100 most recent papers citing Dai et al.14 do not list a version (4 additional papers were excluded from analysis because they cited Dai et al.14 but do not use the Custom CDF). B.) 64 of the 100 most cited papers which cite Dai et al.14 do not list a version. (15 additional papers were excluded from analysis because they cited Dai et al.14 but do not use Custom CDF).
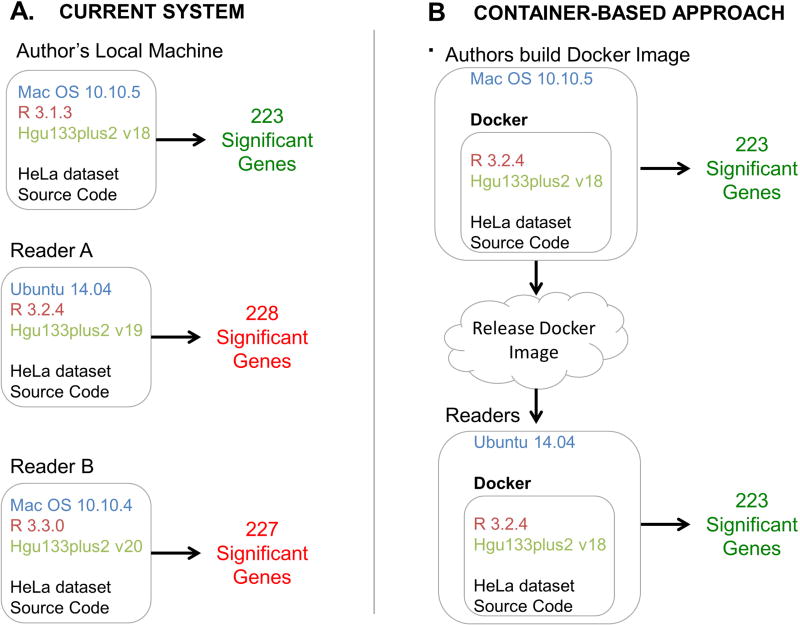
In order to evaluate how different BrainArray Custom CDF versions affect the outcomes of standard analyses, we downloaded a recently published gene expression dataset (GEO accession number GSE47664). The reported experiment with this dataset measured gene expression in normal HeLa cells and HeLa cells with TIA1 and TIAR knocked down15. We ran the same source code using the same dataset altering only the version of the BrainArray Custom CDF library (versions 18, 19 and 20). Each version identified a different number of significantly altered genes (Figure 2A). There were 15 genes identified as significant in version 19 that were not identified in version 18, and 10 genes identified as significant in version 18 that were not identified version 19. There were 18 genes identified as significant in version 20 that were not found in version 18 and 14 genes identified as significant in version 18 that were not identified in version 20. These results indicate that study outcomes are not reproducible without an accurate version number.
Figure 2. Research computing versus container-based approaches.
A.) The status quo requires a reader or reviewer to find and install specific versions of dependencies. These dependencies can become difficult to find and may become incompatible with newer versions of other software packages. Different versions of packages identify different numbers of significantly differentially expressed genes from the same source code and data. B.) Containers define a computing environment that captures dependencies. In containerbased systems, the results are the same regardless of the host system.
Using Docker containers improves reproducibility
To improve reproducibility, researchers can maintain dependencies using the free open-source software tool Docker13,16. Docker can be used to create an “image” that allows users to download and run a container, which is a minimalist virtual machine with a predefined computing environment. Docker images can be several gigabytes in size, but once downloaded can be started in a matter of seconds and have minimal overhead13. This technology has been widely adopted and is now supported by many popular cloud providers including Amazon, Google and Microsoft.
Docker wraps software into a container that includes everything the software needs to run (operating system, system tools, installed software libraries etc.). This allows the software to run the exact same way in any environment. Boettiger introduced Docker containers as a path to reproducible research by eliminating dependency management, remediating issues caused by imprecise documentation, limiting the effects of code rot (dependencies to specific software library versions) and eliminating barriers to software reuse13. In addition, Docker images can be tagged to coincide with software releases and paper revisions. This means that even as software is updated, the exact computing environment of a specific older version can be available through the tag of the container’s revision history.
In order to assess whether using Docker containers could improve reproducibility of the same experiment we also carried out an analysis of differential gene expression using Docker containers on mismatched machines17. This process allows versions to be matched, and produces the same number and set of differentially expressed genes (Figure 2B).
Docker is a useful starting point for reproducible workflows. While it helps to match the computing environment, users must manually rebuild the environment, rerun the analysis pipeline, and update the container after each relevant change. In addition, it does not produce logs of exactly what was run. It also does not automatically track results alongside the specific versions of the code and data that generated them. In summary, Docker can provide manual reproducibility when it is used appropriately.
Continuous Analysis
Our goal when developing continuous analysis was to produce an automatic and verifiable end-to-end run for computational analyses with minimal start-up costs. The status quo process requires researchers to perform an analysis and then diligently describe each step and communicate exact versions of software library dependencies used, which can be hundreds or thousands of packages for modern operating systems. To sidestep requiring readers and reviewers to download and install multiple software packages and datasets, continuous analysis preserves the exact computing environment used for the original analysis. A Docker container is built at the time of original analysis and thus includes the exact versions used by the original authors without the risk of packages later becoming inaccessible. The continuous aspect refers to an analysis being rerun, the results saved in version control, and the container being automatically updated after any relevant changes to the software script or data.
Continuous analysis is an extension of continuous integration18. Continuous integration is widely used in software development. In this workflow, whenever developers update code in a source control repository, an automated build process is triggered. This automated build process first runs any existing test scripts in an attempt to catch bugs introduced into software. If there are no tests or the software passes the tests, the software is automatically sent to remote servers so that users worldwide can access it.
Our continuous analysis workflows (Figure 3) use continuous integration services to run computational analyses, update figures, and publish changes to online repositories whenever changes are made to the source code used in an experiment. We provide continuous analysis workflows for popular continuous integration systems that can be used with multiple types of computing environments including local computing and cloud computing.
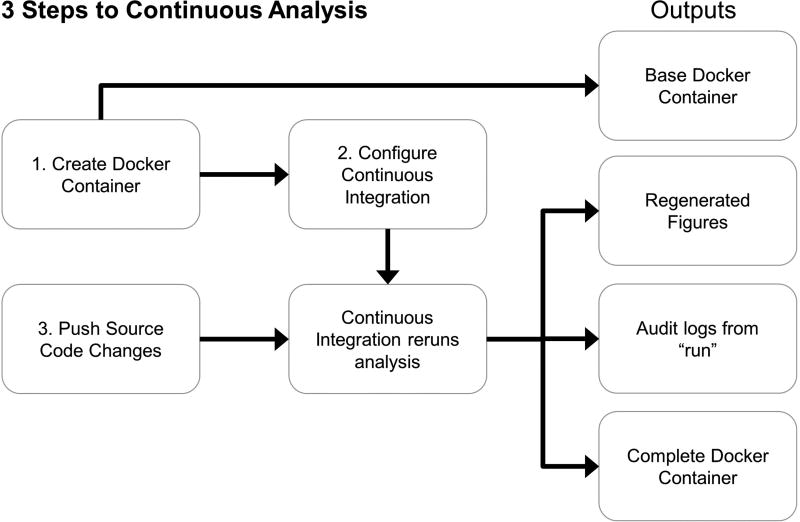
Figure 3. Setting up continuous analysis.
Continuous analysis can be set up in three primary steps (numbered 1, 2, and 3). (1) The researcher creates a Docker container with the required software. (2) The researcher configures a continuous integration service to use this Docker image. (3) The researcher pushes code that includes a script capable of running the analyses from start to finish. The continuous integration provider runs the latest version of code in the specified Docker environment without manual intervention. This generates a Docker container with intermediate results that allows anyone to rerun analysis in the same environment, produces updated figures, and stores logs describing everything that occurred. Example configurations are available in the online methods our online repository (https://github.com/greenelab/continuous_analysis). Because code is run in an independent, reproducible computing environment and produces detailed logs of what was executed, this practice reduces or eliminates the need for reviewers to re-run code to verify reproducibility.
In the continuous analysis workflows that we developed, a continuous integration service is used to monitor the source code repository. Whenever a change is made to a user-specified branch of the repository, the service re-runs the scientific analysis. Workflows are defined in files written in YAML that specify the configuration parameters and commands that should be run. The YAML language is widely used by continuous integration services to specify a human-readable set of instructions. The continuous analysis YAML files that we have developed for multiple services to enable users to employ local computing, cloud-based computing, or commercial service providers.
Each workflow begins by specifying a base Docker image to replicate the researcher’s computing environment. The YAML files that we developed provide a place for researchers to choose a base Docker image. Using Docker allows other researchers to re-run code in a computing environment that matches what the original authors used, even if they do not duplicate the original authors’ continuous integration configuration. Next, the continuous analysis workflow YAML files specify one or more shell commands required to perform the analysis. Researchers can replace the commands in our examples with their own analytical code. Executing these steps generates the relevant figures from the analysis. Our YAML implementation of continuous analysis then updates the remote source code repository by adding figures and results generated during the run. Finally, a Docker container with the final computing environment is automatically updated. This continuous analysis process allows changes to be automatically tracked as a project proceeds and pairs each result with the source code, data, and Docker container used to generate it.
Using continuous integration in this fashion automatically generates a log of exactly what code was run that is synchronized to the code, data, and computing environment (Supplemental Figure 1). Version control systems allow for images to be easily compared, which provides continuous analysis users with the ability to observe results before and after changes (Figure 4). Interactive development tools, such as Jupyter19,20, RMarkdown21,22 and Sweave23 can be incorporated to present the code and analysis in a logical graphical manner. For example, we recently used Jupyter with continuous analysis in our own publication24 and corresponding repository25. Reviewers can follow what was done in an audit fashion without having to install and run software while having confidence that analyses are reproducible.
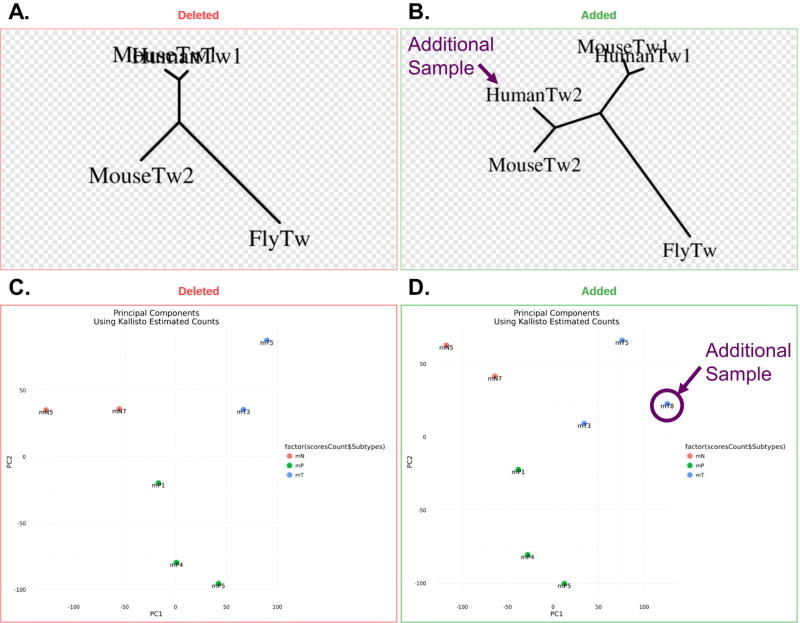
Figure 4. Reproducible workflows with continuous analysis.
Resulting figures from the run are committed back to Github where changes between runs can be viewed. A, B.) The effect of adding an additional gene (HumanTw2) to a phylogenetic tree-building is shown. C, D.) The effect of adding an additional sample (mt8) to an RNA-seq differential expression experiment PCA plot is shown.
When an author is ready to publish their work, they should archive their repository, which contains the automatically generated results alongside the analytical source code and scripted commands for data retrieval. With our continuous analysis workflows, the authors can use the `docker save` command to export the latest static container, which should also be archived. There are an increasing number of services that allow digital artifacts to be archived and distributed, including Figshare or Zenodo. Journals may also allow authors to upload these files as supplementary elements. If the archiving service used by the authors provides a digital object identifier, future users can easily cite the computing environment and source code. For example, our continuous analysis environment26 and results27 are available in this fashion with results and our source code is provided as Supplementary Source Code 1.
This system imposes minimal cost in terms of time and money on the researcher. Continuous analysis is set up once per project, and will then run automatically for the life of the project. We provide example YAML workflows for commonly used services. Researchers can replace the steps in our example analyses with their own commands to enable automatic reproducibility for their own projects.
Setting up continuous analysis
To set up continuous analysis, a researcher needs to do three things. First they must create a Dockerfile, which specifies the software required for their analysis. Second, they need to connect a continuous integration service to their version control system and add a continuous analysis command script to run their analysis. Finally, they need to save their changes to the version control system. Many researchers already perform the first and third tasks in the course of standard procedures for computational research.
The continuous integration system (Figure 3) will automatically rerun the specified analysis with each change, precisely matching the source code and results. It can also be set to listen and run only when changes are marked in a specific way, e.g. by committing to a specific ‘staging’ branch. For the first project, continuous analysis can be set up in less than a day. For subsequent projects, the continuous analysis protocol can be amended in less than an hour.
We have set up continuous analysis using the free and open source Drone software on a PC and connected it to the GitHub version control service (detailed instructions are available in Online Methods). This method is free to users. Our GitHub repository and Supplementary Source Code include continuous analysis YAML scripts for local, cloud-based, and full-service paid configurations27. However, it is important to note that while full service providers can be set up in minutes, they may impose computational resource limits or monthly fees. Private installations require configuration but can scale to a local cluster or cloud service to match the computational complexity of all types of research. With free, open-source continuous integration software28, computing resources are the only associated costs.
We suggest a development workflow where continuous analysis runs only on a selected branch (Supplemental Figure 2). Our example setup configures a “staging” branch for this purpose. Researchers can push to this branch whenever they would like to generate results files and figures. If the updates to this branch succeed, the changes – along with the results of analyses – are then automatically carried over to the master or production branch and released.
Reproducible workflows
Following initial setup, continuous analysis can be adopted into existing workflows that use source control systems. We used continuous analysis in our work using neural networks to stratify patients based on their electronic health records25. In addition, we provide two example analyses using continuous analysis: a phylogenetic tree building and RNA-seq differential expression analysis.
The phylogenetic tree-building example (detailed in Online Methods) aligned 4 mRNA sequences (MouseTw1, HumanTw1, MouseTw2 and FlyTw) using MAFFT29 and built a phylogenetic tree with these alignments using PHYLIP30 (Figure 4A). After adding an additional sample (HumanTw2), continuous analysis rebuilt the tree (Figure 4B).
The RNA-seq example (detailed in Online Methods) demonstrated differential expression analysis between three different organoid models of pancreatic cancer in mice based on Boi et al.31 (GEO accession number GSE63348) while reusing source code from Balli32. This analysis used kallisto33, limma34,35, and sleuth36 to quantify the transcript counts, performed principal components analysis and ran a differential expression analysis. The analysis was initially performed with 7 samples (Figure 4C). An 8th sample was added to show how continuous analysis tracked results (Figure 4D). This example also demonstrated the ability of continuous analysis to scale to the analysis of large datasets – this GEO accession includes 150GB of data (approximately 480 million reads).
Discussion
Continuous analysis provides a verifiable end-to-end run of scientific software in a fully specified environment, thereby enabling true reproduction of computational analyses. Because continuous analysis runs automatically, it can be set up at the start of any project to provide an audit trail that allows reviewers, editors and readers to assess reproducibility without a large time commitment. If readers or reviewers need to re-run the code on their own (e.g. to change a parameter and evaluate the impact on results), they can easily do so with a Docker container containing the final computing environment and results that has been automatically kept up to date. Version control systems enable automatic notification of code updates and new runs to those who “star” or “watch” a repository on services such as Github, Gitlab, and Bitbucket. Wide adoption of continuous analysis could conceivably be linked with the peer review and publication process allowing interested parties to be notified of updates.
Continuous analysis can also be applied to closed data that cannot be released e.g. patient data. Without continuous analysis, reproducing or replicating computational analyses based on closed data is dependent on the original authors completely describing each step, which often becomes relegated to supplementary information. Readers and reviewers must then diligently follow complex written instructions without any confirmation they are on the right track. The pairing of automatically updated containers, source code, and results with the audit log provides readers with confidence that results would be replicable if the data were available. This allows independent researchers to attempt to replicate findings in their own non-public datasets without worrying that a failure to replicate could be caused by source code or environment differences.
Continuous analysis currently has limitations. It may be impractical to use continuous analysis at every commit for generic preprocessing steps involving very large data or analyses requiring particularly high computational costs. In particular, steps that take days to run or incur substantial costs in computational resources (e.g. genotype imputation) may be too expensive for existing providers37. We demonstrate continuous analysis on a user-defined “staging” branch. This enables researchers to control costs by choosing when to trigger analyses that are automatically reproducible. We strongly recommend the use of continuous analysis whenever a single machine can be used. For workflows that require cluster computing, employing continuous analysis is technically feasible but requires significant systems administration expertise because the cluster must be provisioned automatically. For work involving cluster computing, researchers may elect to employ continuous analysis for steps that do not require a cluster. In this case, researchers should carefully report which steps in their workflows the process covers. It is conceivable that continuous analysis systems could be specifically designed for scientific workflows to facilitate reproducible cluster-based analyses with the same ease as single-machine analyses.
For small datasets and less intensive computational workflows it is easiest to use a full service continuous integration service. These services have the shortest setup times – often only requiring a user to enable the service and add a single file to their source code. With private data or for analyses that include large datasets or require significant computing, cloud-based or locally hosted continuous integration server can be employed.
Reproducibility could have wide-reaching benefits for the advancement of science. For authors, reproducible work is credible. Stodden et al.38 highlight the importance of capturing and sharing data, software, and the computational environments. Continuous analysis addresses reproducibility in this narrow sense by automatically capturing the computational reagents needed to generate the same results from the same inputs. It does not solve reproducibility in the broader sense: how robust results are to parameter settings, starting conditions and partitions in the data. By automating narrow-sense reproducibility, continuous analysis lays the groundwork needed to address questions related to the reproducibility and robustness of findings in the broad sense.
ONLINE METHODS
Assessment of reporting of Custom CDF versions
We performed a literature analysis of the 104 most recently published articles citing Dai et al.14 that were accessible at our institution using the Web of Science on November 14, 2016. We aimed to capture 100 articles that included expression analysis and used this resource. We excluded four articles that did not perform expression analysis using the Custom CDF and thus would not be expected to cite a version.
We repeated this process for the 115 highest cited articles that cited Dai et al.14 and were accessible at our institution using the Web of Science on November 14, 2016. We again aimed to capture 100 articles that included expression analysis. We excluded fifteen articles that did not quantify expression with the Custom CDF and would not be expected to cite a version.
For the 100 articles in each set that included expression analysis, we searched for the citation or mention of Custom CDF and Dai et al. and examined the methods section and any supplementary materials to determine which, if any, Custom CDF version was specified.
HeLa cell differential expression analysis using BrainArray Custom CDF
We compared the results of a differential expression using versions 18, 19 and 20 of the BrainArray Custom CDF files14. We performed the differential expression analysis between wild type HeLa cells and HeLa cells with a double knockdown of T-cell intracellular antigen 1 (TIA1) and TIA1 related/like (TIAR/TIAL1) proteins (GEO accession number GSE47664). The experiment included 3 biological replicates of the wild type HeLA cells, and 3 biological replicates of the HeLA cells with double knockdown.
Statistical analysis
Differential expression analysis was performed using the Bioconductor multtest R package39 to perform a two-sided t-test. The Bonferroni adjustment was used for multiple testing correction. The complete P values for Custom CDF versions 18, 19 and 20 are included (Supplemental Data 3, 4, 5 respectively). For this analysis, an alpha threshold of 10−5 was used. Continuous analysis was used for this example and is available17 (https://github.com/greenelab/continuous_analysis_brainarray).
Setting up Continuous Analysis
Installing and Configuring Drone
We used a private local continuous integration server with Drone 0.4 for the examples in this work. A detailed walkthrough is available27. The first step to set up a local continuous integration server is to install Docker on the local machine. We pulled the drone image via the Docker command:
sudo docker pull drone/drone:0.4
We created a new application within Github (https://github.com/settings/developers). The homepage URL was the local machine’s IP address and the callback URL was the local machine’s IP address followed by “/authorize”.
Homepage URL: http://IP-HERE/
Callback URL: http://IP-HERE/authorize/
After creating the application we noted of the Client ID and Client Secret generated by Github. Next, we created the drone configuration file on our local machine at “/etc/drone/dronerc”. First we created a directory:
sudo mkdir /etc/drone
Then, we created a new file named “dronerc” in the newly directory with the client information in the following format. We filled in the Client ID and Secret with the information obtained from GitHub:
REMOTE_DRIVER=github
REMOTE_CONFIG=https://github.com?client_id=....&client_secret=....
The drone instance was ready to run:
sudo docker run \
--volume /var/lib/drone:/var/lib/drone \
--volume /var/run/docker.sock:/var/run/docker.sock \
--env-file /etc/drone/dronerc \
--restart=always \
--publish=80:8000 \
--detach=true \
--name=drone \
drone/drone:0.4
The continuous integration server was accessed at ip-address/login. We clicked the login button and chose the repository we wished to activate for continuous analysis. This automatically created a webhook in GitHub to notify Drone when new changes were made to the repository.
Creating a Docker image to replicate the research environment
We used a Dockerfile to define a Docker image with the software required by the analysis (examples available40–42). This file was named “Dockerfile.” The first line of the file specified a base image to pull from. We used Ubuntu 14.04, so we set the first line to:
FROM Ubuntu:14.04
We specified the maintainer with contact information on the next line as:
MAINTAINER “Brett Beaulieu-Jones” brettbe@med.upenn.edu
Next, commands to install the appropriate packages were added to the file:
RUN apt-get install –y git python-numpy wget gcc python-dev python-setuptools python-dev build-essential
After constructing the Dockerfile, we built the Docker image using the Docker build command.
sudo docker build –t username/image_name
After building the image, we pushed it to Docker Hub in order to share the exact computing environment with others. We used the following commands replacing the username and image_name with the appropriate values.
sudo docker login
sudo docker push username/image_name
In addition to pushing the tagged docker images to Docker Hub, we saved static versions of the docker images before and after analysis. We uploaded these to Zenodo to receive a digital object identifier26.
docker save brettbj/continuous_analysis_base > continuous_analysis_base
docker save brettbj/continuous_analysis > continuous_analysis
Running a continuous analysis
After configuration, the analysis was run using a file entitled “.drone.yml”. When this file was added to a repository, Drone performed all of the commands within it (Supplemental Figure 3). Whenever changes are made to the source code, Drone repeats these steps. Example “.drone.yml” files are available40–42.
Within continuous analysis, all commands were executed within a build section of the .drone.yml file. First, the base Docker image created previously was pulled.
- build:
- image: username/image_name
Then shell scripting was used to re-run the appropriate analysis.
- commands:
-
-mkdir –p output
-
-…
-
-
To run continuous analysis only on the staging branch, a final section was added.
- branches:
-
-staging
-
-
Phylogenetic tree-building example
This example aligned 5mRNA sequences and uses these alignments to build simple phylogenies. We used 5 mRNA sequences accessible from the NCBI nucleotide database: Twist – Fly (NM_079092, splice form A), Twist1 - Human (NM_000474), Twist1 - Mouse (NM_011658), Twist2 - Human (NM_057179), Twist2 - Mouse (NM_007855).
This analysis was performed twice. The first analysis did not include: Twist2 – Human (NM_05179). The second analysis included all five sequences (Supplemental Figure 4).
The sequences were aligned using MAFFT29 and then converted to PHYLIP interleaved format using EMBOSS Seqret43. A maximum parsimony tree was generated for the sequences using PHYLIP30 DNAPARS and a representation of this tree was drawn with PHYLIP drawtree. PHYLIP Seqboot was used to assess robustness of the generated tree and PHYLIP consense to determine the consensus tree form the bootstrapped trees. The complete continuous analysis runs42 and Docker images before and after analysis are available26.
RNA-seq differential analysis of mouse models for pancreatic cancer
This workflow was based on Balli32 and uses mouse organoid cultures generated by Boj et al.31. Boj et al. generated organoids from three different tissues: normal pancreas (mN), early stage lesions (mP) and pancreatic adenocarcinoma (mT). The authors then performed RNA-seq on these organoids (GEO accession number GSE63348; SRA accession number SRP049959).
We performed differential expression analysis initially on 7 samples: 2 normal, 3 mP and 2 mT (150gb FASTQ format, 480 million reads. An eighth sample (mT) was added in the second run to demonstrate the differences under continuous analysis. Four preprocessing steps were performed prior to beginning continuous analysis.
These steps were performed to reduce the amount of time continuous analysis runs for and to limit the necessary bandwidth. Pre-processing steps should only be performed with time and/or resource heavy tasks that follow a standard easy to follow workflow. First, the samples were downloaded from the Sequence Read Archive (SRA accession number SRP049959). The SRA36 toolkit was used to split the .sra files in to FASTQ files. The mouse reference genome (mm10) assembly was downloaded. Finally, these files were stored in a folder accessible to local FTP so they would not need to be re-downloaded with each run.
Within the continuous analysis process, kallisto36 was used to generate an index file from the reference mouse genome and then to quantify abundances of transcripts for each RNA-seq sample. Next, lowly expressed genes were filtered out (<1) and a principal components plot was generated (2-components) (Supplemental Figure 5). Finally, a limma34 linear model was fit for differential gene expression analysis and the results were plotted in the form of a volcano plot (Supplemental Figure 6).
The complete continuous analysis runs40 and Docker images before and after analysis were uploaded and made available26.
Data Availability
The HeLA cells (wild type and double knockout) are available through GEO (accession number GSE47664). The genes used in the Phylogenetic tree-building example are available via the NCBI nucleotide database (IDs: NM_079092, NM_000474, NM_011658, NM_057179, NM_007855). The RNA-seq data used for the larger differential expression analysis are available in the Sequence Read Archive (Accession number SRP049959).
Code Availability
The Docker images for all three experiments are available on figshare (10.6084/m9.figshare.3545156.v1). The Continuous Analysis examples and instructions accompanying this manuscript are available on Zenodo (http://doi.org/10.5281/zenodo.178613). Instuctions and examples will be periodically updated on Github and contributions are welcomed (https://github.com/greenelab/continuous_analysis).
Supplementary Material
Acknowledgments
This work was supported by the Gordon and Betty Moore Foundation under a Data Driven Discovery Investigator Award to CSG (GBMF 4552). A Commonwealth Universal Research Enhancement (CURE) Program grant from the Pennsylvania Department of Health supported BKB. We would like to thank D. Balli for providing the RNA-seq analysis design, K. Siewert for providing the phylogenetic analysis design, and A. Whan for contributing a Travis-CI implementation. We also thank M. Paul, Y. Park, G. Way, A. Campbell, J. Taroni, and L. Zhou for serving as usability testers during the implementation of continuous analysis.
Footnotes
Competing Financial Interests
The authors have no competing financial interests to declare.
References
- 1.Rebooting review. Nat Biotech. 2015;33(4):319. doi: 10.1038/nbt.3202. [DOI] [PubMed] [Google Scholar]
- 2.Software with impact. Nat Meth. 2014;11(3):211. doi: 10.1038/nmeth.2880. [DOI] [PubMed] [Google Scholar]
- 3.Peng RD. Reproducible Research in Computational Science. Science (80-) 2011;334(6060):1226–1227. doi: 10.1126/science.1213847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNutt M. Reproducibility. Science (80-) 2014;343(6168):229. doi: 10.1126/science.1250475. http://science.sciencemag.org/content/343/6168/229.abstract. [DOI] [PubMed] [Google Scholar]
- 5.Illuminating the black box. Nature. 2006;442(7098):1. doi: 10.1038/442001a. [DOI] [PubMed] [Google Scholar]
- 6.Baker M. 1,500 scientists lift the lid on reproducibility. Nature. 2016;533(7604):452–454. doi: 10.1038/533452a. [DOI] [PubMed] [Google Scholar]
- 7.Garijo D, Kinnings S, Xie LL, et al. Quantifying reproducibility in computational biology: The case of the tuberculosis drugome. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0080278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinnings SL, Xie LL, Fung KH, Jackson RM, Xie LL, Bourne PE. The Mycobacterium tuberculosis drugome and its polypharmacological implications. PLoS Comput Biol. 2010;6(11) doi: 10.1371/journal.pcbi.1000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ioannidis JPA, Allison DB, Ball CA, et al. Repeatability of published microarray gene expression analyses. Nat Genet. 2009;41(2):149–155. doi: 10.1038/ng.295. [DOI] [PubMed] [Google Scholar]
- 10.Hothorn T, Leisch F. Case studies in reproducibility. Brief Bioinform. 2011;12(3):288–300. doi: 10.1093/bib/bbq084. [DOI] [PubMed] [Google Scholar]
- 11.Groves T, Godlee F. Open science and reproducible research. BMJ. 2012;344 doi: 10.1136/bmj.e4383. [DOI] [PubMed] [Google Scholar]
- 12. [Accessed January 1, 2016];ThinkLab. https://thinklab.com/
- 13.Boettiger C. An introduction to Docker for reproducible research, with examples from the R environment. ACM SIGOPS Oper Syst Rev Spec Issue Repeatability Shar Exp Artifacts. 2015;49(1):71–79. doi: 10.1145/2723872.2723882. [DOI] [Google Scholar]
- 14.Dai M, Wang P, Boyd AD, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nunez M, Sanchez-Jimenez C, Alcalde J, Izquierdo JM. Long-term reduction of T-cell intracellular antigens reveals a transcriptome associated with extracellular matrix and cell adhesion components. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0113141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Docker. Docker. https://www.docker.com.
- 17.Beaulieu-Jones B, Greene C. Continuous Analysis BrainArray: Submission Release Continuous Analysis BrainArray: Submission Release. 2016 Aug; doi: 10.5281/zenodo.59892. [DOI] [Google Scholar]
- 18.Duvall P, Matyas S, Glover A. Continuous Integration: Improving Software Quality and Reducing Risk. 2007 http://portal.acm.org/citation.cfm?id=1406212.
- 19.Pérez F, Granger BE. {IP}ython: a System for Interactive Scientific Computing. Comput Sci Eng. 2007;9(3):21–29. doi: 10.1109/MCSE.2007.53. [DOI] [Google Scholar]
- 20. [Accessed January 8, 2016];Jupyter. http://jupyter.org/. Published 2016.
- 21.RStudio. RStudio: Integrated development environment for R (Version 0.97.311) J Wildl Manage. 2011;75(8):1753–1766. doi: 10.1002/jwmg.232. [DOI] [Google Scholar]
- 22.Baumer B, Cetinkaya-Rundel M, Bray A, Loi L, Horton NJ. R Markdown: Integrating A Reproducible Analysis Tool into Introductory Statistics. Technol Innov Stat Educ. 2014;8(1):20. doi: 10.5811/westjem.2011.5.6700. [DOI] [Google Scholar]
- 23.Leisch Friedrich. Sweave: Dynamic generation of statistical reports using literate data analysis. Compstat 2002 - Proc Comput Stat. 2002;(69):575–580. doi:10.1.1.20.2737. [Google Scholar]
- 24.Beaulieu-Jones BK, Greene CS. Semi-supervised learning of the electronic health record for phenotype stratification. J Biomed Inform. 2016 Dec;64:168–178. doi: 10.1016/j.jbi.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Beaulieu-Jones BK. Denoising Autoencoders for Phenotype Stratification (DAPS): Preprint Release. Zenodo. 2016 Jan; doi: 10.5281/zenodo.46165. [DOI] [Google Scholar]
- 26.Beaulieu-Jones BK, Greene CS. Continuous Analysis Example Docker Images. 2016 doi: 10.6084/m9.figshare.3545156.v1. [DOI] [Google Scholar]
- 27.Beaulieu-Jones BK, Whan A, Greene CS. greenelab/continuous_analysis: Continuous Analysis v1.0 [Data set] Zenodo. doi: 10.5281/zenodo.178613. . Published 2016. [DOI]
- 28.Drone.io. https://drone.io/
- 29.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plotree D, Plotgram D. PHYLIP-phylogeny inference package (version 3.2) [Accessed August 2, 2016];cladistics. 1989 http://onlinelibrary.wiley.com/doi/10.1111/j.1096-0031.1989.tb00562.x/abstract.
- 31.Boj SF, Hwang C-I, Baker LA, et al. Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell. 2015;160(1):324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balli D. [Accessed August 1, 2016];Using Kallisto for expression analysis of published RNA-seq data. https://benchtobioinformatics.wordpress.com/2015/07/10/using-kallisto-for-gene-expression-analysis-of-published-rnaseq-data/. Published 2015.
- 33.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNAseq quantification. Nat Biotechnol. 2016;34(5):525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 34.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 36.Pimentel HJ, Bray N, Puente S, Melsted P, Pachter L. Differential analysis of RNA-Seq incorporating quantification uncertainty. bioRxiv. 2016 doi: 10.1101/058164. [DOI] [PubMed] [Google Scholar]
- 37.Souilmi Y, Lancaster AK, Jung J-Y, et al. Scalable and cost-effective NGS genotyping in the cloud. BMC Med Genomics. 2015;8(1):64. doi: 10.1186/s12920-015-0134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stodden V, McNutt M, Bailey DH, et al. Enhancing reproducibility for computational methods. Science. 2016;354(6317) doi: 10.1126/science.aah6168. [DOI] [PubMed] [Google Scholar]
- 39.Pollard KS, Dudoit S, van der Laan MJ. Multiple Testing Procedures: the multtest Package and Applications to Genomics. Springer New York: 2005. pp. 249–271. [DOI] [Google Scholar]
- 40.Beaulieu-Jones BK, Greene CS. Continuous Analysis RNA-seq Example. GitHub repository. https://github.com/greenelab/continuous_analysis_rnaseq. Published 2016.
- 41.Beaulieu-Jones BK, Greene CS. Continuous Analysis BrainArray Example. GitHub repository. https://github.com/greenelab/continuous_analysis_brainarray. Published 2016.
- 42.Beaulieu-Jones BK, Greene CS. Continuous Analysis Phylogenetic Tree Building Example. GitHub repository. https://github.com/greenelab/continuous_analysis_phylo. Published 2016.
- 43.Rice P, Longden I, Bleasby A, et al. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The HeLA cells (wild type and double knockout) are available through GEO (accession number GSE47664). The genes used in the Phylogenetic tree-building example are available via the NCBI nucleotide database (IDs: NM_079092, NM_000474, NM_011658, NM_057179, NM_007855). The RNA-seq data used for the larger differential expression analysis are available in the Sequence Read Archive (Accession number SRP049959).