Abstract
Cysteine dioxygenase (CDO) plays an essential role in sulfur metabolism by regulating homeostatic levels of cysteine. Human CDO contains a posttranslationally generated Cys93–Tyr157 crosslinked cofactor. Here, we investigated this Cys-Tyr crosslinking by incorporating unnatural tyrosines in place of Tyr157 via a genetic method. The catalytically active variants were obtained with a thioether bond between Cys93 and the halogen-substituted Tyr157, and we determined crystal structures of both wild-type and engineered CDO variants in the purely uncrosslinked form and with a mature cofactor. Along with mass spectrometry and 19F NMR, these data indicated that the enzyme could catalyze oxidative C–F or C–Cl bond cleavage, resulting in a substantial conformational change of both Cys93 and Tyr157 during cofactor assembly. These findings provide insights into the mechanism of Cys-Tyr cofactor biogenesis and may aid the development of bioinspired aromatic carbon–halogen bond activation.
Posttranslational modification increases the number of possible molecular variations of proteins in living cells by several orders of magnitude and hence is known as “the chemistry of proteome diversifications”1,2. While reversible protein modifications play central roles in cellular regulation, unidirectional posttranslational modifications generate novel cofactors to enhance or expand the catalytic repertoire of enzymes3–5. Irreversible posttranslational modifications have become a fundamental challenge for chemists to predict protein structures and functions. A protein-derived Cys-Tyr cofactor has recently been found in mammalian cysteine dioxygenase (CDO, EC 1.13.11.20)6,7. Such a cofactor is only known in a few proteins8–12. CDO is a nonheme iron enzyme that catalyzes the conversion of L-cysteine to cysteine sulfinic acid (CSA) (Supplementary Fig. 1). The product, CSA, is ultimately catabolized to taurine and sulfate13. The Cys-Tyr cofactor contains a thioether (C-S) bond between the side chains of a cysteine residue (Cys93, human CDO numbering) and a tyrosine residue (Tyr157)14. The presence of such a Cys-Tyr cofactor boosts the catalytic efficiency of CDO by one order of magnitude15, due to concerted redox action of the metal ion and the protein-derived cofactor.
CDO occupies a central position in biological sulfur metabolism; its enzymatic activity is crucial for maintaining proper cysteine levels for protein synthesis and for initiating cysteine catabolism. It has been established that the metabolites derived from cysteine and the ratio of cysteine to sulfate and taurine exert a wide variety of physiological effects in cells, including energy balance, fat metabolism16, autoimmune and neurological conditions6,17, oxidative stress18, O2 sensing and hypoxia19–21, and the production of the signaling molecule H2S22. Reduced activity of CDO results in elevated serum levels of neuroexcitatory cysteine, which has been associated with rheumatoid arthritis,23 breast cancer24, and several neurological disease states, such as Alzheimer’s and Parkinson’s diseases25. In mouse models, knockout of CDO results in increased activity of the alternative desulfhydration pathways, causing increased H2S production and cytotoxicity22.
When recombinant CDO is isolated, both crosslinked and uncrosslinked forms are present. Separation of the two forms can be achieved in the denatured state by SDS-PAGE; two distinct bands are observable with the faster-migrating band identified as the crosslinked form.15 The crosslinking reaction takes place during catalysis as an autocatalytic reaction due to an uncoupled oxygen activation at the nonheme iron center of the enzyme, where O2 activation is not linked to the oxidation of the substrate cysteine but rather to its own residues. It takes an unclarified number, likely hundreds to thousands, of turnovers to obtain a fully mature enzyme15. Depending on the substrate concentrations (L-cysteine and O2), pH, and temperature, the time to reach the fully crosslinked form in solution varies. Once the Cys-Tyr cofactor is generated, the mature form of CDO performs the coupled dioxygenation reaction more efficiently with an increased capability of metabolizing high levels of cysteine.
To date, understanding of the mechanism of Cys-Tyr crosslink biogenesis in CDO has stagnated due to several technical challenges. First, the uncrosslinked form of active CDO, the starting material for studying cofactor biogenesis, is difficult to isolate, though it is observable in denatured gels. Thus, little spectroscopic characterization has been done, and no structural information is currently available for the uncrosslinked form. Second, traditional site-directed mutagenesis approaches can support the catalytic importance of the cofactor, but mutation of Cys93 or Tyr157 leads to disruption of the formation of the Cys-Tyr crosslink and the inability of the protein variants to generate a mature enzyme active site, thus offering limited information15,26–28.
In the present work, we surmounted these challenges by employing a genetic method for site-specific incorporation of unnatural amino acids29 into human CDO (hCDO) at position 157 of the protein sequence. The resulting protein variants were characterized by enzymatic activity and biophysical techniques to explore the mechanism of cofactor biogenesis in CDO. An oxidative carbon-fluorine (C-F) or carbon-chlorine (C-Cl) bond cleavage was observed during cofactor formation in the di-halogen-substituted enzyme variants, indicating that the nonheme iron center generates an oxidant that is strong enough to oxidize the adjacent second coordination sphere residues for crosslinking whether a stronger or weaker bond is present or the chemical properties of the tyrosine are altered. When the engineered protein with a mono-chlorine-substituted tyrosine was exploited, the structural and mass spectrometry analyses showed that steric hindrance is the next factor governing selectivity for generating a new C-S bond.
RESULTS
Structures of ligand-free and substrate-bound human CDO
To investigate the formation and key role of the Cys93–Tyr157 crosslink, we determined the crystal structures of mature hCDO in both the ligand-free status and in complex with L-cysteine (Fig. 1a), refined to 2.20 and 1.80 Å resolution, respectively (Supplementary Table 1). Prior to this work, CDO research was primarily focused on the mouse and rat gene products (mouse and rat CDOs have identical protein sequence). Only one human CDO structure was reported at 2.7 Å resolution14, and whether this is a ligand-bound structure is ambiguous30. The ligand-free structure we determined belongs to the P65 space group with each asymmetric unit containing one CDO molecule. The electron density of the enzyme active site shows that the obtained structures are 100% crosslinked with Cys93 and Tyr157 covalently connected via a thioether bond. The overall structure is analogous to that of the mouse and rat CDO described previously7. Three water ligands were found to coordinate with the Fe ion in all the resting-state wild-type (WT) CDO structures (Supplementary Fig. 2) that we obtained at various resolutions, consistent with the published spectroscopic data from mouse/rat CDO indicating that the iron ion exhibits a pseudo-octahedral coordination geometry7,31. The first crystal structure, determined from Ni2+-reconstituted mouse CDO shows three water ligands6, while the rat CDO structure and Mössbauer spectroscopy show a tetra-coordinate iron center with only one or two water ligand(s)7,31,32. The number of the water ligands is relevant to mechanistic understanding because the first step of the catalytic cycle is substrate-ligation to the iron ion. The crystal structure of CDO in complex with L-cysteine (Fig. 1a) shows that the substrate L-cysteine displaces two water molecules with both the amino and thiol groups coordinating to the ferrous ion, and the remaining water ligand is only weakly associated to the Fe ion, at a distance of 2.6 Å, and is hydrogen-bonded with the hydroxyl group of the Cys-Tyr cofactor. The substrate L-cysteine forms a salt bridge with Arg60, and is hydrogen-bonded to Tyr58 and the Cys-Tyr cofactor in addition to its bidentate coordination to the Fe(II) ion.
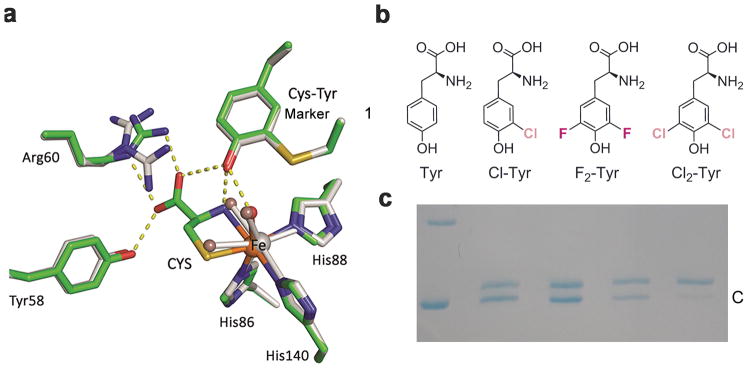
Figure 1. Crystal structures of human CDO and incorporation of unnatural amino acids into the catalytic active site tyrosine.
a, Overlay of the active site of the ligand-free (grey) and substrate (CYS)-bound complex (green) crystal structures of human CDO (PDB entries: 6BGF & 6BGM). Arg60 presents two conformations in the ligand-free structure. The omit Fo-Fc electron densities for the water ligands (WAT) and the L-cysteine are shown in Supplementary Figure 2a,b. b, Native tyrosine and halogen-substituted unnatural tyrosine analogous used in this study, i.e., L-Tyr, 3-Cl-L-Tyr (Cl-Tyr, 1), 3,5-F2-L-Tyr (F2-Tyr, 2), and 3,5-Cl2-L-Tyr (Cl2-Tyr, 3). c, SDS-PAGE shows two bands, with the slower moving band corresponding to uncrosslinked enzyme (U) and faster moving band to the mature CDO with a crosslinked cofactor (C). The lanes from left to right of the molecular weight marker are as follows: 1) wild-type CDO, 2) Cl-Tyr157 CDO, 3) F2-Tyr157 CDO, and 4) Cl2-Tyr157 CDO, respectively. The full gel image is displayed in Supplementary Figure 5b,c.
Genetic incorporation of unnatural amino acids
In order to investigate the biogenesis mechanism in CDO we genetically substituted Tyr157 with three unnatural amino acids: 3-chloro-L-tyrosine (Cl-Tyr, 1), 3,5-difluoro-L-tyrosine (F2-Tyr, 2), and 3,5-dichloro-L-tyrosine (Cl2-Tyr, 3) (Fig. 1b). The advantage of this approach is that it allows for less perturbation to the enzyme active site structure than site-directed mutagenesis while specifically and chemically altering the amino acid residue of interest. The genetic incorporation of unnatural amino acids was site-specific, and all six other tyrosine residues in hCDO were not altered. The CDO protein isolated was 100% modified at position 157 with specific tyrosine analogues. This efficient genetic incorporation of unnatural tyrosines into a specific tyrosine residue in a protein has been previously used for the study of other systems including a nonheme Fe enzyme, ribonucleotide reductase33–35.
Of the unnatural tyrosine compounds available, F2-Tyr is the most interesting for this study because fluorine is highly electronegative and small in size. Thus, we expect that F2-Tyr157 would mimic the native human enzyme with minimal structural perturbation while specifically attenuating the hydrogen-bonding potential of the tyrosyl hydroxyl group due to the strong electron-withdrawing properties of fluorine. To selectively incorporate F2-Tyr into hCDO, a mutant of Methanocaldococcus jannaschii tyrosyl amber suppressor tRNA(MjtRNATyrCUA)/F2-TyrRS (Y32R, L65Y, H70G, F108N, Q109C, D158N, L162S) pair was used to target the TAG codon, as previously reported36. We expressed the hCDO variant bearing F2-Tyr at position 157 in Escherichia coli, using pEVOL-F2-TyrRS and PV16-hCDO157TAG plasmids. Likewise, we also genetically incorporated Cl-Tyr and Cl2-Tyr into the 157 position of hCDO by a similar method using pEVOL-Cl-TyrRS and pEVOL-Cl2-TyrRS37.
The ESI mass spectra of the WT, Cl-Tyr157, F2-Tyr157, and Cl2-Tyr157 variants under denaturing solution conditions showed an ensemble of charge states ranging from [M+8H+]+8 to [M+33H+]+33. Deconvolution of these mass spectra result in experimentally determined masses for these proteins of 22,913.20, 22,947.11, 22,949.16, and 22982.07 Da for WT, Cl-Tyr157, F2-Tyr157 and Cl2-Tyr157 CDO, respectively (Supplementary Fig. 3). These measured mass values are consistent with the predicted masses of 22,913.21, 22,947.17, 22,949.19 and 22982.13 Da from protein sequence, respectively. These masses of Cl-Tyr157, F2-Tyr157, and Cl2-Tyr157 reflect a +34, 36 or 69 Da difference compared to WT CDO, as would be expected for the substitution of one or more halogen atoms for hydrogen at Tyr157.
The unnatural amino acids’ function in human CDO
The fluorine- and chlorine-bearing CDO variants were catalytically active, with the as-isolated F2-Tyr157 variant having about 10% activity, and Cl/Cl2-Tyr157 having 2% activity, of that observed for WT CDO (Supplementary Fig. 4 and Supplementary Table 2). The question motivating the next step of this study is whether or not the crosslinked Cys-Tyr cofactor could be formed in the F2-Tyr157 and Cl2-Tyr157 CDO variants. Based on bond dissociation energies, the formation of the C-S bond needed for cofactor assembly was expected to be hindered by the fluorine (but not chlorine) substitution. Because the C-F bond is often thought to be one of the strongest single bonds (second only to the C-Si bond) between formally neutral atoms,38 one may expect that F2-Tyr157 substitution would greatly hinder, or prevent, the formation of the thioether bond for cofactor assembly. Oxidative formation of the crosslink with F2-Tyr157 would require breaking a C-F bond and an S-H bond, and forming a new C-S bond. Without knowing the mechanism of the cofactor formation, it was difficult to predict a priori whether or not the crosslinked Cys-Tyr cofactor would still be formed in the halogen-substituted CDO variants.
The as-isolated WT, Cl-Tyr, F2-Tyr157, and Cl2-Tyr157 CDO variants all exhibited two bands by SDS-PAGE, indicating that the substitutions were unable to abrogate crosslink formation (Fig. 1c). Based on gel analysis, we estimated that 55%, 54%, 41%, and 16% crosslink were present in the as-isolated CDO samples, respectively. Further processing of the as-isolated halogen-substituted CDO variants with L-cysteine and O2 led to the mainly crosslinked enzyme concomitant with the weakening of the slower-migrating band (Supplementary Fig. 5). Upon closer analysis of the ESI mass spectra, we found that F2-Tyr157 CDO in the +10 charge state and below contained an additional peak corresponding to 20 Da lower mass (Supplementary Fig. 6), which fits the expected mass of the crosslinked form of F2-Tyr157 CDO. After crosslink formation, the protein is more resistant to acid and organic solvent denaturation, and thus is more compact, so it carries less protonic charge and appears at lower charge states.
Oxidative C-F bond cleavage during cofactor biogenesis
The gel analysis and the ESI data suggested that the Cys-Tyr cofactor was formed in the CDO variants with unnatural amino acids. Next, we considered if they do so by breaking the C-F or C-Cl bond or forming a differently configured Cys-Tyr, i.e., linking the thiol group to C2 or C6. In order to rigorously determine the nature and position of the crosslink in F2-Tyr157 CDO, an MS/MS analysis was conducted (Fig. 2a).
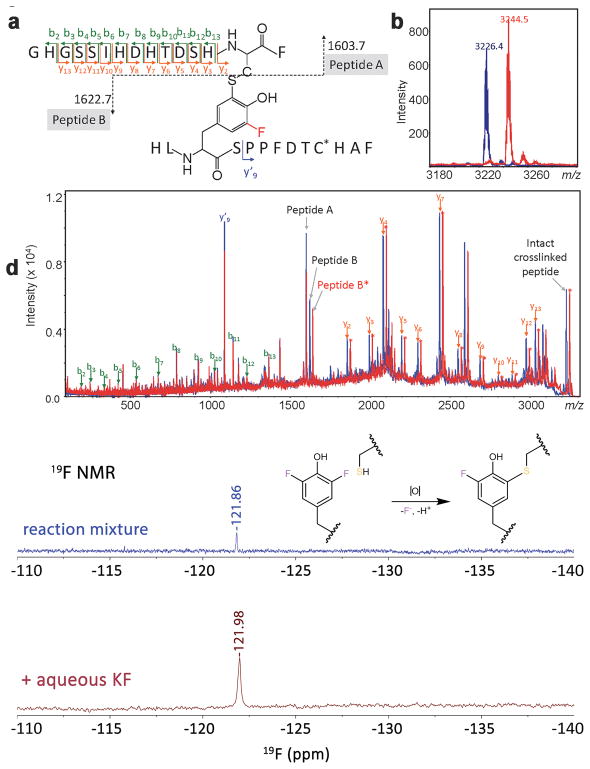
Figure 2. MS/MS spectra of crosslinked peptides of WT and F2-Tyr157 CDO proteins and 19F NMR detection of leaving fluoride.
a, Proposed crosslink of peptides Gly80–Phe94 and His155–Phe167 through Cys93 and F2-Tyr157. b, Mass spectrometry of the crosslinked peptide Gly80–Phe94/His155–Phe167 including one carboxyamidomethyl cysteine at m/z 3,226.4 for WT and m/z 3,244.5 for F2-Tyr157 CDO. c, Collision-induced dissociation (CID)–MS/MS spectrum of precursor ion m/z 3,226.4 and m/z 3,244.5. d, In situ detection of fluoride during cofactor biogenesis in F2-Tyr157 CDO. 19F NMR spectra of the fluoride ion in solution from the cross-linking reaction of F2-Tyr157 CDO after acid precipitation of the protein. Top: 19F NMR spectrum after the conversion of F2-Tyr157 CDO to the all-cross-linked form after 12,480 transients. Bottom: 19F NMR spectrum after spiking in aqueous KF to confirm that the signal is F– after 128 transients. Both spectra are referenced to internal trifluoroacetic acid. Mass spectrometry experiments were repeated one time, and 19F NMR experiments were repeated two times with similar results. The F2-Tyr157 CDO was also recorded once by 19F NMR before the autocatalytic processing reaction (a negative control) and the resulting spectrum is shown in Supplementary Figure 7.
After conversion to the all-crosslink form, the WT and F2-Tyr157 CDO proteins were digested by three enzymes: trypsin, followed by GluC, and lastly chymotrypsin. As expected, the crosslinked peptide from WT hCDO was detected at m/z 3,226.4 (Fig. 2b). In samples of F2-Tyr157 CDO, no peak was detected around m/z 3,262 corresponding to a crosslinked peptide containing two fluorine atoms on Tyr157. Instead, a peak at m/z 3,244.5 was found, which is consistent with the crosslinked peptide containing only one fluorine atom (Fig. 2b).
To confirm the location of the crosslink and displacement of a fluorine atom upon crosslinking, tandem mass spectrometry utilizing LIFT™ technology39 on a MALDI TOF/TOF mass spectrometer was conducted. An illustration of the crosslinked peptide indicating the major fragment ions detected is depicted in Figure 2a. Based on this schematic, any N-terminal fragment (b-ion, green colored) before His92 of peptide A (residues Gly80 through His92) and any C-terminal ion (y-ion, orange colored) of peptide B (residues Ser158 through Phe167) after Ser158 would have the same mass (or m/z value) between the crosslinked WT and F2-Tyr157 CDO peptides. The mass spectrometry data showed that this is indeed the case. Our data also showed an increase of +18 Da in F2-Tyr157 relative to the WT peptide for any fragment containing Tyr157 (Fig. 2c). These results unambiguously confirmed the crosslink between Cys93 and Tyr157, and that Tyr157 contains one, and only one, fluorine atom in the mature form, the other having been displaced during the formation of the crosslink. These mass spectrometry analyses further suggested that the cofactor formed in F2-Tyr157 CDO is analogous to the Cys-Tyr cofactor found in the native protein. The fluorine atom released during oxidative conversion was detected by 19F NMR (Fig. 2d). Since fluorine release should be stoichiometric with the protein, over 10,000 scans over the course of 20 h on a 500 MHz NMR spectrometer were collected. Spiking in aqueous KF confirmed that the signal for the released fluorine is F– (Fig. 2d). Such a fluoride signal is not present in the 19F NMR spectrum of F2-Tyr157 CDO (Supplementary Fig. 7).
Achieving cofactor biogenesis in crystallo
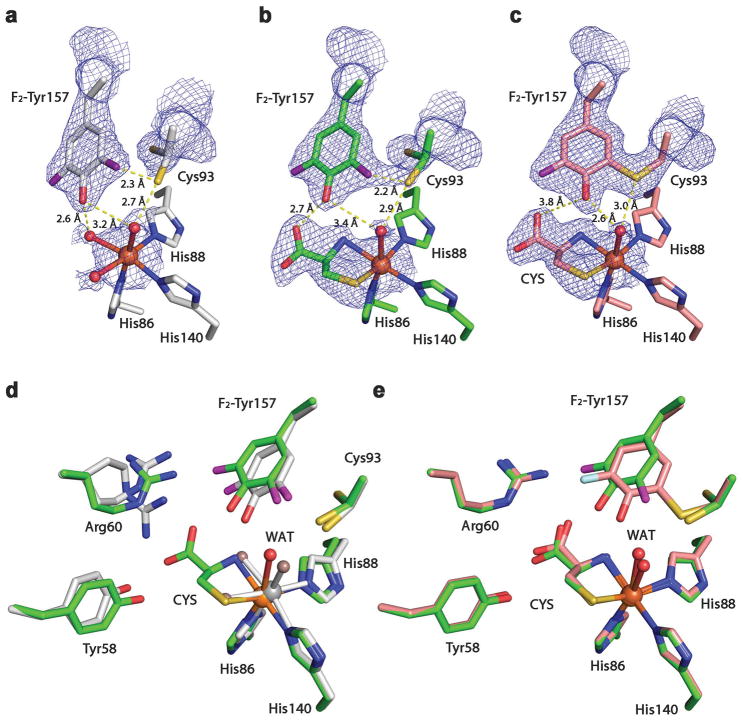
We determined the crystal structures of F2-Tyr157 hCDO and Cl2-Tyr157 hCDO (Fig. 3, Supplementary Fig. 8 and Supplementary Table 1) in the absence of and in complex with L-cysteine. When F2-Tyr157 crystals were grown anaerobically, only the uncrosslinked form crystallizes, and we thus obtained the long-sought, 100% uncrosslinked, structure of CDO, refined to 2.40 Å resolution (Fig. 3a). Cys93 exhibits two alternative conformations in the uncrosslinked CDO structure, with occupancy in a 73:27 ratio, and the sulfhydryl group in the major conformation points away from the iron ion. The catalytic Fe(II) ion is ligated by the Nε atoms of His86, His88, and His140 and three water molecules. The substrate-bound, uncrosslinked crystal structure was refined to 2.10 Å resolution (Fig. 3b). A substantial rotation of F2-Tyr157 is noted (Supplementary Fig. 9a). As shown in the wild-type structure, three water ligands are bound to the Fe ion in all the ligand-free structures of the CDO variants, and only one water remains associated with the iron ion in the substrate-bound structure (Supplementary Fig. 2). The b-factor of the water ligand changes from 15.5 in the ligand-free structure to 37.8 in the substrate-bound structure, indicating that the water ligand becomes flexible when L-cysteine binds to the metal ion.
Figure 3. Crystal structures of F2-Tyr157 CDO.
2Fo – Fc electron density was contoured at 1.2 σrms, with potential H-bond interactions (broken lines) shown. a, The active site of the 100% uncrosslinked F2-Tyr157 CDO structure in the substrate-free form. b, The active site of the 100% uncrosslinked F2-Tyr157 CDO structure in the substrate-bound form. c, The active site of the mature F2-Tyr157 CDO structure in the substrate-bound form. d, Overlaid active site of the 100% uncrosslinked F2-Tyr157 CDO structure in the substrate-free (2.40 Å resolution, grey) and substrate-bound (2.10 Å resolution, green) forms. e, Superimposition of the substrate-bound mature F2-Tyr CDO (1.95 Å resolution, pink) and 100% uncrosslinked structures (green). The L-cysteine substrate is labeled as CYS. The omit Fo-Fc electron densities for the water ligands (WAT) and the L-cysteine are shown in Supplementary Figure 2.
We also obtained a crystal structure with both crosslinked and uncrosslinked forms in the same crystal structure from the as-isolated, ligand free, F2-Tyr157 hCDO crystalized under aerobic conditions and refined it to 1.80 Å resolution. The crosslinked and uncrosslinked forms of the cofactor were present in a 53:47 ratio in this structure based on their respective occupancy (Supplementary Fig. 8a,b). The crosslinked conformation of Cys93 showed a connecting density to F2-Tyr157 indicating that the crosslinked cofactor is present. Notably, Cys93 also exhibited two alternative conformations in this structure. In the uncrosslinked conformation, the sulfhydryl group of Cys93 points away from the iron center, while in the crosslinked form, the sulfur atom of Cys93 turns toward the iron center by nearly 3 Å. The F2-Tyr157 ring also rotated about 46 degrees along the axis through Cβ and the OH-group, with a nearly 1 Å shift towards Cys93, allowing C-S bond formation (Supplementary Fig. 9b). We also found a similar phenomenon in the crystal structure from the as-isolated Cl2-Tyr157 CDO crystallized under aerobic conditions (Supplementary Figs. 8c,d and 9c).
Taking advantage of the uncrosslinked crystals, we subsequently investigated the possible uncoupled reaction in crystallo that leads to the oxidation of the second coordination sphere residues and the formation of the cofactor. After reacting the F2-Tyr157 variant crystals with L-cysteine (100 mM) under aerobic conditions for 5 h, the single crystals of F2-Tyr157 hCDO showed a fully mature, cysteine-bound active site with 100% crosslink between Cys93 and Tyr157 (Fig. 3c), and the structure was refined to 1.95 Å resolution (Supplementary Table 1). An overlay of the uncrosslinked F2-Tyr157 CDO structures with and without substrate cysteine bound at the active site is shown in Figure 3d, and an overlay of the crosslinked and uncrosslinked F2-Tyr157 CDO structures in complex with L-cysteine is shown in Figure 3e and Supplementary Figure 9d. A substantial rotation of F2-Tyr157 is noted between the uncrosslinked and mature forms of the enzyme. Cys93 shows only one conformation in the mature form40 but two conformations in each of the uncrosslinked structures. Likewise, in crystallo cofactor maturation was also observed in Cl2-Tyr157 CDO protein crystals.
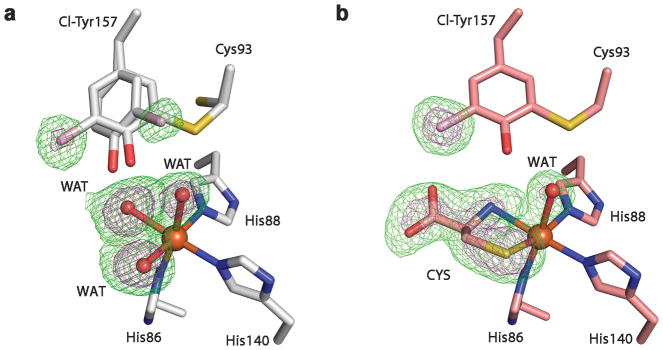
In order to discern which bond would be cleaved when both C-H and C-Cl bonds are available, we analyzed the mono-chlorine-substituted Cl-Tyr157 variant by mass spectrometry and determined its crystal structures from the purely uncrosslinked and the fully crosslinked forms, respectively (Supplementary Table 1). In the as-isolated form, the uncrosslinked and crosslinked portions are present in a similar occupancy. Figure 4a shows the overlay of the active site of the ligand-free Cl-Tyr157 CDO structures in the crosslinked and uncrosslinked forms. The most revealing structural information for mechanistic understanding is the uncrosslinked form. The crystal structure shows in the uncrosslinked form that equal populations of the C-H and C-Cl bond of the Tyr157 are positioned towards Cys93 and the iron ion, indicating that the C-H and C-Cl bonds have similar opportunity for oxidative cleavage. In the fully processed form with a mature cofactor and bound L-cysteine substrate at the nonheme Fe center, only a single conformation of the cofactor is present with 100% occupancy of the chlorine (Fig. 4b). As corroborated by the mass spectrometry analysis (Supplementary Fig. 10), the C-H bond is cleaved but not the C-Cl bond. Therefore, we conclude that the large size of chlorine makes the C-Cl bond less accessible for oxidation.
Figure 4. Crystal structures of Cl-Tyr157 CDO.
The omit Fo-Fc electron densities (green) of the WT CDO & mutants contoured at 3 σ (green) and 6 σ (purple), respectively. a, The active site of the Cl-Tyr157 CDO structure in the crosslinked and uncrosslinked forms in the absence of the substrate. b, The active site of the substrate-bound Cl-Tyr157 CDO structure in the crosslinked form. CYS represents the L-cysteine substrate bound at the nonheme iron center.
DISCUSSION
The success in obtaining a purely uncrosslinked crystal structure of CDO is an important advancement towards unfolding the cofactor biogenesis mechanism. Previously, the uncrosslinked CDO structure was obtained from the C93A mutant.28 The key difference is that the uncrosslinked structure we report is a fully capable protein for cofactor assembly whereas the mutant is a permanently disabled variant in terms of cofactor biogenesis. In both the substrate-free and substrate-bound mutant structures, a buffer-derived chloride ion is ligated to the iron ion in the mutant structures, but the chloride ligation is not seen in our structures. Our purely uncrosslinked structure revealed important details for mechanistic understanding that were not possible to detect in the mutant structures. Cys93 shows two conformations before the crosslink reaction, and the sulfur atom of Cys93 undergoes a nearly 3 Å rotation toward the iron center during autocatalytic cofactor biogenesis. The findings presented in this work may be useful in considering the mechanism of the Cys-Tyr biogenesis.
The ability of CDO to catalyze aromatic C-F bond cleavage is not a predictable result considering that the energy of an aliphatic C-F bond (485 kJ mol−1) is substantially higher than that of the corresponding aliphatic C-H bond (411 kJ mol−1).41 Since the bond energy of an aliphatic C-Cl bond (i.e., 327 kJ mol−1) is lower than that of a C-H bond, the cleavage of the C-Cl bond in Cl2-Tyr157 CDO is no surprise. However, it is an interesting observation that the C-H bond is preferentially cleaved in mono-ClTyr157 CDO even though both C-H and C-Cl are equally populated towards Cys93 and the iron center.
It should be noted that the C-F bond in this case may not be as strong as in an aliphatic molecule. However, the aromatic C-F bond is expected to be stronger than that of the corresponding C-H bond. When a hydroxyl group is attached to the C4 ring carbon of a tyrosine, it activates the C3 and C5 positions. We propose that the C-F bond may be cleaved during a putative radical step where Tyr157 is oxidized, either directly by the iron-bound oxidant or a Cys93 radical (Supplementary Fig. 11). Once a tyrosyl radical is formed, it is well-known that the radical is delocalized to specific positions,4 including the ring C3 and C5. Thus, the C-H or C-F bond activation in Tyr157 would be easier than generally thought for a C-H or C-F bond in hydrocarbons. This is especially true after a tyrosyl radical is formed during cofactor biogenesis.
The crosslink formation is apparently less efficient for the Cl2-Tyr157 derivative than the F2-Tyr157 variant and the wild-type protein. Because a C-Cl bond is less durable than a C-F bond, the less efficient crosslinking of Cl2-Tyr157 CDO suggests that there may be an additional factor governing bond cleavage. We hypothesize that the size of chlorine atom adversely affects access of the C-Cl bond. The crosslink formation in mono-substituted Cl-Tyr157 CDO is as efficient as in the native enzyme, and in the uncrosslinked structure, the C-H and C-Cl of Cl-Tyr157 are equally populated towards Cys93 and the Fe ion. However, it is the stronger C-H bond that is cleaved and the weaker C-Cl bond that is retained in the mature Cl-Tyr157 CDO variant. These results suggest that the size of chlorine makes it more difficult for Cl-Tyr157 to position closer to a putative Cys93 radical or an iron-bound oxidant relative to the fluorinated tyrosine derivative. Because the small size of fluorine, there is no tRNA synthetase available for producing mono-fluorine substituted tyrosine derivative. The heterogeneous substitution of chlorine and fluorine at the ring 3 and 5 positions of tyrosine (3-chloro-5-fluoro tyrosine) is also inaccessible by the genetic incorporation approach. Based on the current results, we envision that the C-F bond would be more likely cleaved in a hypothetical 3-chloro-5-fluoro-Tyr157 CDO variant.
The data presented in this work collectively indicate an O2-dependent carbon–halogen bond cleavage by a protein-bound iron center under mild, physiologically relevant conditions. Even though the C-F bond is one of the strongest covalent bonds in organic chemistry and stronger than corresponding C-H bonds, CDO is able to cleave a C-F bond for the purpose of cofactor biogenesis. The cleavage of an aromatic C-F bond by nitrogen-ligated nonheme iron model complexes has been described in two recent papers42,43. However, to our knowledge, the observation described here for CDO is the first example of an oxidative C-F bond cleavage by a protein-bound iron center. The activation and functionalization of C-F bonds is an important area of inorganic, organic, and medicinal chemistry. In biological systems, the only C-F bond cleavage previously known was the oxygen-independent reactions catalyzed by fluoroacetate dehalogenase and BzCoA reductase44,45. This unexpected finding during the study of CDO cofactor biogenesis reveals new insights into the power of the nonheme iron ion in a dioxygenase and sheds light onto the interplay between the catalytic metal ion and the cofactor of CDO. It further increases our expectation of the oxidizing power of nonheme Fe enzymes. The oxidative C-F bond cleavage described in this study broadens the range of fluorine chemistry and further extends potential industrial applications of natural or engineered Fe-dependent proteins.
ONLINE METHODS
General methods
Primers were synthetized by Integrated DNA Technologies. Reagents were purchased from Sigma-Aldrich, New England Biolabs (NEB), and Thermo, Inc. DNA manipulations in Escherichia coli were carried out according to standard procedures. Ampicillin (100 μg/mL), and chloramphenicol (30 μg/mL) were used as antibiotics for selection of recombinant strains.
Synthesis of 3,5-difluorotyrosine
To synthesize F2-Tyr, we transformed 2,6-difluorophenol to F2-Tyr by using Citrobacter Freundii (ATCC8090) tyrosine phenol lyase (TPL) following established methods46. One liter solution containing 30 mM ammonium acetate, 60 mM sodium pyruvate, 5 mM β-mercaptoethanol, 40 μM PLP, 10 mg of TPL enzyme, pH 8.0, was stirred in the dark for 3 days at room temperature. The aqueous phase was then concentrated, and was purified by a C-18 column from 10% to 90% methanol, 0.1% trifluoroacetic acid (v/v) in water. To test for the presence of the unnatural amino acid, a ninhydrin solution consisting of 0.2% (w/v) ninhydrin, 95% (v/v) N-butanol, 0.5% (v/v) acetic acid and 4.3% (v/v) water was prepared. The enzymatic reaction mixture was first mixed with the ninhydrin solution at a 1:1 ratio, and then spotted onto a TLC plate. A purple/pink spot indicates the presence of the unnatural amino acid. The F2-Tyr prepared was used in the next experiment for genetic incorporation into hCDO.
Cloning, expression, purification and protein analysis
Expression vector pVP16-hCDO was bought from the DNASU Plasmid Repository. Expression and purification of His-MBP-tagged enzymes hCDO was performed as follows: the culture was grown at 37 °C in Luria Bertani (LB) media in a baffled flask at 200 rpm with the appropriate antibiotic to an optical density of 0.8 AU at 600 nm. Isopropyl-β-thiogalactoside (IPTG) was added to a final concentration of 500 μM and the temperature was lowered to 28 °C. After 12 h of further incubation, the cells were harvested and resuspended in lysis buffer (300 mM NaCl, 50 mM Tris-HCl, 1 mM ferrous ammonium sulfate, pH 8.0) and then disrupted by a Microfluidizer LM20 cell disruptor, and the supernatant was recovered after centrifugation (13,000g for 30 min). His-MBP-tagged protein was separated using nickel-nitrilotriacetic acid resin. After being washed with washing buffer (300 mM NaCl, 50 mM Tris-HCl, 20 mM imidazole, pH 8.0), protein was eluted with elution buffer (300 mM NaCl, 50 mM Tris-HCl, 250 mM imidazole, pH 8.0). Purified protein-containing fractions were confirmed by SDS-PAGE and then dialyzed into the storage buffer (50 mM Tris-HCl, 100 mM NaCl, 10% glycerol, pH 8.0). The dialyzed protein was concentrated and stored at −80 °C for further use. Protein concentration was determined by UV-vis absorbance at 280 nm (ε280 nm = 25,440 cm−1M−1). For the expression of F2-Tyr157, Cl-Tyr157, and Cl2-Tyr157 hCDO proteins, pEVOL-F2-TyrRS (or pEVOL-Cl-TyrRS and pEVOL-Cl2-TyrRS) was co-transformed with pVP16-hCDO157TAG into BL21 (DE3), respectively. A single colony was grown over night at 37 °C in 4 mL of LB medium. The transformed cells were induced with 0.5 mM IPTG and 0.02% L-arabinose at OD600 nm of 0.8 in the presence of 0.5 mM F2-Tyr, Cl-Tyr157, or Cl2-Tyr. After growing for 12 h at 30 °C, the F2-Tyr157, Cl-Tyr157, and Cl2-Tyr157 hCDO proteins were purified using the protocol described for WT hCDO. The MBP tag was removed by cleavage with TEV protease during the purification of the MBP-fused hCDO. The liberated native CDO and mutants were further purified by gel-filtration chromatography in 20 mM Tris-HCl, 50 mM NaCl (pH 8.0) and were ultrafiltrated to the required concentration for subsequent catalytic assays, and crystallization. We used the GelAnalysis (http://www.gelanalyzer.com) to estimate the ratio of the cross-linked form.
Catalytic assay & maturation assay
L-Cysteine was dissolved in hCDO storage buffer. Freshly prepared hCDO and L-cysteine solutions were incubated in a 37 °C water bath for 5 minutes. Appropriate amounts of these two solutions were well mixed to initiate the reaction in a 37 °C water bath. The activity assay protocol for hCDO was derived from that previously described for CDO15,47,48. A typical assay was conducted in a total volume of 300 μL as follow: 0.2 – 0.5 μM as-purified hCDO, 50 mM MES buffer pH 6.1, 0.3 mM ammonium iron sulfate, 62.5 μM bathocuproine disulfonate and varying concentrations of L-cysteine (0 – 25 mM). The reaction was initiated by addition of cysteine with shaking to ensure proper oxygenation using a shaker (220 rpm). The hCDO-catalyzed reaction was quenched by the addition of 2 μL HFBA and centrifugation for removal of precipitated proteins. All aliquots were immediately ultrafiltered through Microcon (Millipore, Bedford, MA) 10,000 Da molecular weight cut-off ultrafiltration tubes. The flow-through was kept in an ice water bath for further high-performance liquid chromatography analysis. To detect the dioxygenase product CSA, reactions aliquots were applied to a reverse-phase column (Thermo InertSustain C-18 column, 100 x 4.6 mm, 5 μM) with a mobile phase of 20 mM sodium acetate, 0.6% methanol, 0.3% heptofluorobutyric acid, pH 2.0. Curves of initial reaction velocity versus cysteine starting concentration were fitted with the Michaelis-Menten equation in OriginPro (OriginLab, Northampton, MA) to determine kcat and Km values. The maturation assay was performed in 100 mM Tris-HCl (pH 8.0) buffer with 25 mM L-cysteine and 75 μM enzyme at 37 °C. The SDS-PAGE was replicated at least three times by the independent experiments to ensure reproducibility.
Crystallization
A 2-μL aliquot of the enzyme solution (10 mg/ml) was mixed with 2- μL of reservoir solution containing 0.1 M MES (pH 6.5), 2 M ammonium sulfate, 2% PEG 400 and crystallized by the hanging drop, vapor-diffusion method at 22 °C. The crystals grew as rods in 3 days. After soaking in a cryoprotectant containing reservoir solution plus 20% glycerol for 0.5 min, the crystal was flash-frozen and stored by liquid nitrogen for data collection using synchrotron radiation. The substrate-bound structures were obtained by soaking 100 mM L-cysteine to the CDO crystals. The anaerobic crystallization for obtaining pure uncrosslinked CDO was conducted in an O2-free COY anaerobic chamber.
Data collection, structure determination and refinement
Crystallographic data were acquired at 100 K temperature at Stanford Synchrotron Radiation Lightsource beamline BL9-2 (PDB code, wavelength (Å); 6BGM, 0.97946; 6BGF, 1.0000; 6BPU, 0.97946; 6BPV, 0.97622; 6BPW, 0.97946; 6BPX, 1.0000; 6CDH, 1.0000; 6CDN, 1.0000) and at the Advanced Photon Sources (Argonne National Laboratory, Argonne, IL) beamline 19BM (PDB code, wavelength (Å); 6BPT, 0.97919; and 6BPS, 0.97919). All X-ray diffraction intensity data were integrated, scaled, and merged using HKL200049. Molecular replacement was performed with Phenix50 using the crystal structure of CDO as a starting model (Protein Data Bank entry 2ICl)14. The final model was manually adjusted and refined with Coot51 and Phenix. Ramachandran statistics were analyzed using MolProbity (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php). All the phi and psi angles were located in the preferred and allowed regions without any outliers. We generated all the molecular model figures using PyMOL (W.L. deLano, The PyMOL Molecular Graphics System version 1.8.6.0. Schrödinger LLC, http://www.pymol.org/; 2002).
Mass Spectrometry
Solutions of the intact protein were exhaustively desalted by filtration with 10 mM ammonium acetate through centrifugal filters with 10 kDa membrane cut-off. For ESI-MS analysis, samples were diluted to approximately 2 μM in a solution containing 50% methanol and 0.1% acetic acid. Mass spectra were collected on a maXis plus quadrupole-time of flight mass spectrometer equipped with an electrospray ionization source (Bruker Daltonics) and operated in the positive ionization mode. Samples were introduced via syringe pump at a constant flow rate of 3 μL/min. Source parameters are summarized as follows: capillary voltage, 3,500 V with a set end plate offset of 500 V; nebulizer gas pressure, 0.4 bar; dry gas flow rate, 4.0 L/min; source temperature, 200 °C. Mass spectra were averages of one minute of scans collected at a rate of 1 scan per second in the range 50 ≤ m/z ≤ 3000. Compass Data Analysis software version 4.3 (Bruker Daltonics) was used to process all mass spectra.
To generate cross-linked peptide species within a mass range suitable for efficient fragmentation and large coverage of the peptide sequence, a multiple enzyme digestion approach was used as previously described by Kleffmann and coworkers52 with minor modification. In short, the WT and F2-Tyr157 CDO proteins were digested in solution first with trypsin, followed by GluC protease and then chymotrypsin. Proteomics-grade enzymes were used in 1:25 enzyme:substrate ratio and incubated at 37 °C with each enzyme overnight. For MALDI-TOF/TOF mass spectrometry, a supersaturated matrix solution was prepared by dissolving α-cyano-4-hydroxycinnamic acid (Bruker Daltonics) in a mixture of 50% acetonitrile, 0.1% TFA in water. Reconstituted digests were spotted onto a stainless-steel target using the sandwich method where 1 μL of matrix solution was spotted and dried, followed by 1 μL of sample solution, and another 1 μL of matrix solution. All MALDI-TOF/TOF mass spectra were collected on an ultrafleXtreme MALDI-TOF/TOF mass spectrometer equipped with a smartbeam II laser (Bruker Daltonics) in the positive ionization and reflector modes. The instrument was calibrated using the peptide calibration standard mixture (from Bruker Daltonics containing angiotensin II, angiotensin I, substance P, ACTH clip 1–17, ACTH clip 18–39, and somatostatin 28); and acquisition optimized in the mass range from 500 to 4500 m/z. Approximately 3000–8000 shots were acquired per MS and LIFT-MS/MS spectrum using 1000 Hz acquisition speed. FlexAnalysis 3.3 software (Bruker Daltonics) was used for data processing.
Nuclear magnetic resonance spectroscopy
19F NMR spectra were recorded on an Agilent (Billerica, MA) DD2 500 MHz spectrometer equipped with a 5-mm inverse detect probe at 300 K. Spectra were recorded in 90/10 buffer/D2O and referenced to internal trifluoroacetic acid (−76.5 ppm). One-dimension 19F spectra (s2pul) were recorded with 5s relaxation delay, 64 k data points, and multiplied with an exponential function for a line-broadening of 5 Hz before Fourier transformation. All NMR data were processed using MestReNova NMR v11.0.3 software. The condition for conversion of F2-Tyr157 CDO to the all cross-linked form was 50 mM pH 6.5 MES buffer, 25 mM L-cysteine, and 300 μM protein.
Statistics and reproducibility
All experiments, including the assays and SDS-PAGE, were obtained from replicated independent experiments to ensure reproducibility. For the representative results the numbers of independent experiments are labelled in the figure legends or associated text to the corresponding tables.
Data Availability
Structure factors and coordinates for the crystal structures solved in this work have been deposited in the Protein Data Bank under accession codes 6BGM, 6BGF, 6BPU, 6BPV, 6BPW, 6BPX, 6CDH, 6CDN, 6BPT, and 6BPS.
Supplementary Material
Acknowledgments
The work is supported in part by the National Institutes of Health Grants GM107529, GM108988, and MH107985, the National Science Foundation grant CHE-1623856, and the Lutcher Brown Distinguished Chair Endowment fund (to A.L.). J.W. acknowledges the support of the National Science Foundation of China grants (91527302, 31370016, and U1532150). The mass spectrometry facility is sponsored by National Institutes of Health Grant G12MD007591. The NMR spectrometer is a shared instrument sponsored by the National Science Foundation under the award number #1625963. X-ray synchrotron data were collected at the beamlines of the Advanced Photon Source Section 19, Structural Biology Center user program GUP-48198, Argonne National Laboratory and at the beamline BL9-2 of the Stanford Synchrotron Radiation Lightsource (SSRL) under the user program #5B14, SLAC National Accelerator Laboratory. The beamline staff scientists are acknowledged for the assistance of the remote data collections. The Advanced Photon Source is a U.S. Department of Energy, Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. SSRL is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515 and by the National Institutes of Health (P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of National Institutes of Health or National Science Foundation.
Footnotes
Author contributions:
Genetic incorporation of unnatural amino acids was performed by J.L. (cloning, protein expression and purification, enzyme assays). J.W. and F.L. provided TyrRS. W.G. and J.L. conducted the mass spectrometry analyses. J.L. obtained all protein crystals, collected X-ray diffraction data, and interpreted and refined the structural data together with I.S. The mechanistic models were proposed and refined by J.L., I.D., Y.W, and A.L. Y.W. participated in the unnatural amino acids production and isolation by an enzymatic method. D.J.W. performed the 19F NMR analysis. A.L. conceived the research and wrote the manuscript together with J.L. All authors contributed to data analysis and to the writing and editing of the manuscript.
Competing Financial Interests Statement:
None
References
- 1.Walsh CT. Posttranslational Modification of Proteins: Expanding Nature’s Inventory. Roberts & Co; 2006. [Google Scholar]
- 2.Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Protein posttranslational modifications: The chemistry of proteome diversifications. Angew Chem Int Ed. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 3.Klinman JP, Bonnot F. Intrigues and intricacies of the biosynthetic pathways for the enzymatic quinocofactors: PQQ, TTQ, CTQ, TPQ, and LTQ. Chem Rev. 2014;114:4343–4365. doi: 10.1021/cr400475g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stubbe J, van Der Donk WA. Protein radicals in enzyme catalysis. Chem Rev. 1998;98:705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 5.Krebs C, Bollinger JM, Jr, Booker SJ. Cyanobacterial alkane biosynthesis further expands the catalytic repertoire of the ferritin-like ‘di-iron-carboxylate’ proteins. Curr Opin Chem Biol. 2011;15:291–303. doi: 10.1016/j.cbpa.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCoy JG, et al. Structure and mechanism of mouse cysteine dioxygenase. Proc Natl Acad Sci U S A. 2006;103:3084–3089. doi: 10.1073/pnas.0509262103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simmons CR, et al. Crystal structure of mammalian cysteine dioxygenase. A novel mononuclear iron center for cysteine thiol oxidation. J Biol Chem. 2006;281:18723–18733. doi: 10.1074/jbc.M601555200. [DOI] [PubMed] [Google Scholar]
- 8.Whittaker JW. Free radical catalysis by galactose oxidase. Chem Rev. 2003;103:2347–2363. doi: 10.1021/cr020425z. [DOI] [PubMed] [Google Scholar]
- 9.Cowley RE, et al. Structure of the reduced copper active site in preprocessed galactose oxidase: Ligand tuning for one-electron O2 activation in cofactor biogenesis. J Am Chem Soc. 2016;138:13219–13229. doi: 10.1021/jacs.6b05792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnell R, Sandalova T, Hellman U, Lindqvist Y, Schneider G. Siroheme- and [Fe4-S4]-dependent NirA from Mycobacterium tuberculosis is a sulfite reductase with a covalent Cys-Tyr bond in the active site. J Biol Chem. 2005;280:27319–27328. doi: 10.1074/jbc.M502560200. [DOI] [PubMed] [Google Scholar]
- 11.Polyakov KM, et al. High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens . J Mol Biol. 2009;389:846–862. doi: 10.1016/j.jmb.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 12.Hromada SE, et al. Protein oxidation involved in Cys-Tyr post-translational modification. J Inorg Biochem. 2017;176:168–174. doi: 10.1016/j.jinorgbio.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 13.Stipanuk MH, Ueki I, Dominy JE, Jr, Simmons CR, Hirschberger LL. Cysteine dioxygenase: a robust system for regulation of cellular cysteine levels. Amino Acids. 2009;37:55–63. doi: 10.1007/s00726-008-0202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye S, et al. An insight into the mechanism of human cysteine dioxygenase - Key roles of the thioether-bonded tyrosine-cysteine cofactor. J Biol Chem. 2007;282:3391–3402. doi: 10.1074/jbc.M609337200. [DOI] [PubMed] [Google Scholar]
- 15.Dominy JE, Jr, et al. Synthesis of amino acid cofactor in cysteine dioxygenase is regulated by substrate and represents a novel post-translational regulation of activity. J Biol Chem. 2008;283:12188–12201. doi: 10.1074/jbc.M800044200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niewiadomski J, et al. Effects of a block in cysteine catabolism on energy balance and fat metabolism in mice. Ann N Y Acad Sci. 2016;1363:99–115. doi: 10.1111/nyas.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon C, Bradley H, Waring RH, Emery P. Abnormal sulphur oxidation in systemic lupus erythematosus. Lancet. 1992;339:25–26. doi: 10.1016/0140-6736(92)90144-r. [DOI] [PubMed] [Google Scholar]
- 18.Kwon DY, et al. Impaired sulfur-amino acid metabolism and oxidative stress in nonalcoholic fatty liver are alleviated by betaine supplementation in rats. J Nutr. 2009;139:63–68. doi: 10.3945/jn.108.094771. [DOI] [PubMed] [Google Scholar]
- 19.Olson KR, et al. Thiosulfate: a readily accessible source of hydrogen sulfide in oxygen sensing. Am J Physiol. 2013;305:R592–R603. doi: 10.1152/ajpregu.00421.2012. [DOI] [PubMed] [Google Scholar]
- 20.Weits DA, et al. Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat Commun. 2014;5:4425/4421–4435/4410. doi: 10.1038/ncomms4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White MD, et al. Plant cysteine oxidases are dioxygenases that directly enable arginyl transferase-catalysed arginylation of N-end rule targets. Nat Commun. 2017;8:14690. doi: 10.1038/ncomms14690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueki I, et al. Knockout of the murine cysteine dioxygenase gene results in severe impairment in ability to synthesize taurine and an increased catabolism of cysteine to hydrogen sulfide. Am J Physiol Endocrinol Metab. 2011;301:E668–684. doi: 10.1152/ajpendo.00151.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradley H, et al. Sulfate metabolism is abnormal in patients with rheumatoid arthritis. Confirmation by in vivo biochemical findings. J Rheumatol. 1994;21:1192–1196. [PubMed] [Google Scholar]
- 24.Jeschke J, et al. Frequent inactivation of cysteine dioxygenase type 1 contributes to survival of breast cancer cells and resistance to anthracyclines. Clin Cancer Res. 2013;19:3201–3211. doi: 10.1158/1078-0432.CCR-12-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heafield MT, et al. Plasma cysteine and sulphate levels in patients with motor neurone, Parkinson’s and Alzheimer’s disease. Neurosci Lett. 1990;110:216–220. doi: 10.1016/0304-3940(90)90814-p. [DOI] [PubMed] [Google Scholar]
- 26.Li W, Blaesi EJ, Pecore MD, Crowell JK, Pierce BS. Second-sphere interactions between the C93-Y157 cross-link and the substrate-bound Fe site influence the O2 coupling efficiency in mouse cysteine dioxygenase. Biochemistry. 2013;52:9104–9119. doi: 10.1021/bi4010232. [DOI] [PubMed] [Google Scholar]
- 27.Davies CG, Fellner M, Tchesnokov EP, Wilbanks SM, Jameson GN. The Cys-Tyr cross-link of cysteine dioxygenase changes the optimal pH of the reaction without a structural change. Biochemistry. 2014;53:7961–7968. doi: 10.1021/bi501277a. [DOI] [PubMed] [Google Scholar]
- 28.Driggers CM, et al. Structure-based insights into the role of the Cys-Tyr crosslink and inhibitor recognition by mammalian cysteine dioxygenase. J Mol Biol. 2016;28:3999–4012. doi: 10.1016/j.jmb.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 30.Driggers CM, et al. Cysteine dioxygenase structures from pH 4 to 9: Consistent Cys-persulfenate formation at intermediate pH and a Cys-bound enzyme at higher pH. J Mol Biol. 2013;425:3121–3136. doi: 10.1016/j.jmb.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Souness RJ, et al. Mechanistic implications of persulfenate and persulfide binding in the active site of cysteine dioxygenase. Biochemistry. 2013;52:7606–7617. doi: 10.1021/bi400661a. [DOI] [PubMed] [Google Scholar]
- 32.Tchesnokov EP, et al. An iron-oxygen intermediate formed during the catalytic cycle of cysteine dioxygenase. Chem Commun. 2016;52:8814–8817. doi: 10.1039/c6cc03904a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oyala PH, et al. Biophysical characterization of fluorotyrosine probes site-specifically incorporated into enzymes: E. coli ribonucleotide reductase as an example. J Am Chem Soc. 2016;138:7951–7964. doi: 10.1021/jacs.6b03605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ravichandran KR, et al. Formal reduction potentials of difluorotyrosine and trifluorotyrosine protein residues: Defining the thermodynamics of multistep radical transfer. J Am Chem Soc. 2017;139:2994–3004. doi: 10.1021/jacs.6b11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minnihan EC, Young DD, Schultz PG, Stubbe J. Incorporation of fluorotyrosines into ribonucleotide reductase using an evolved, polyspecific aminoacyl-tRNA synthetase. J Am Chem Soc. 2011;133:15942–15945. doi: 10.1021/ja207719f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li F, et al. A genetically encoded 19F NMR probe for tyrosine phosphorylation. Angew Chem Int Ed Engl. 2013;52:3958–3962. doi: 10.1002/anie.201300463. [DOI] [PubMed] [Google Scholar]
- 37.Liu X, et al. Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers. J Am Chem Soc. 2014;136:13094–13097. doi: 10.1021/ja505219r. [DOI] [PubMed] [Google Scholar]
- 38.O’Hagan D. Understanding organofluorine chemistry. An introduction to the C-F bond. Chem Soc Rev. 2008;37:308–319. doi: 10.1039/b711844a. [DOI] [PubMed] [Google Scholar]
- 39.Suckau D, et al. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal Bioanal Chem. 2003;376:952–965. doi: 10.1007/s00216-003-2057-0. [DOI] [PubMed] [Google Scholar]
- 40.Fellner M, Aloi S, Tchesnokov EP, Wilbanks SM, Jameson GN. Substrate and pH-dependent kinetic profile of 3-mercaptopropionate dioxygenase from Pseudomonas aeruginosa. Biochemistry. 2016;55:1362–1371. doi: 10.1021/acs.biochem.5b01203. [DOI] [PubMed] [Google Scholar]
- 41.Cottrell TL. The Strengths of Chemical Bonds. 2. Butterworths Scientific; 1958. p. 317. [Google Scholar]
- 42.Sahu S, et al. Aromatic C-F hydroxylation by nonheme iron(IV)-oxo complexes: Structural, spectroscopic, and mechanistic Investigations. J Am Chem Soc. 2016;138:12791–12802. doi: 10.1021/jacs.6b03346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sahu S, et al. Direct observation of a nonheme iron(IV)-oxo complex that mediates aromatic C-F hydroxylation. J Am Chem Soc. 2014;136:13542–13545. doi: 10.1021/ja507346t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan PWY, Yakunin AF, Edwards EA, Pai EF. Mapping the reaction coordinates of enzymatic defluorination. J Am Chem Soc. 2011;133:7461–7468. doi: 10.1021/ja200277d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiedt O, et al. ATP-dependent C-F bond cleavage allows the complete degradation of 4-fluoroaromatics without oxygen. MBio. 2016;7:e00990-00916. doi: 10.1128/mBio.00990-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seyedsayamdost MR, Yee CS, Stubbe J. Site-specific incorporation of fluorotyrosines into the R2 subunit of E-coli ribonucleotide reductase by expressed protein ligation. Nat Protoc. 2007;2:1225–1235. doi: 10.1038/nprot.2007.159. [DOI] [PubMed] [Google Scholar]
- 47.Arjune S, Schwarz G, Belaidi AA. Involvement of the Cys-Tyr cofactor on iron binding in the active site of human cysteine dioxygenase. Amino Acids. 2015;47:55–63. doi: 10.1007/s00726-014-1843-7. [DOI] [PubMed] [Google Scholar]
- 48.Stipanuk MH, Dominy JE, Jr, Ueki I, Hirschberger LL. Measurement of cysteine dioxygenase activity and protein abundance. Curr Protoc Toxicol. 2008;38:6.15.11–16.15.25. doi: 10.1002/0471140856.tx0615s38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst D-Biol Crystal. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst D-Biol Crystal. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Kleffmann T, Jongkees SAK, Fairweather G, Wilbanks SM, Jameson GNL. Mass-spectrometric characterization of two posttranslational modifications of cysteine dioxygenase. J Biol Inorg Chem. 2009;14:913–921. doi: 10.1007/s00775-009-0504-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure factors and coordinates for the crystal structures solved in this work have been deposited in the Protein Data Bank under accession codes 6BGM, 6BGF, 6BPU, 6BPV, 6BPW, 6BPX, 6CDH, 6CDN, 6BPT, and 6BPS.