Abstract
Breast development occurs through well-defined stages representing ‘windows of susceptibility’ to adverse environmental exposures that potentially modify breast cancer risk. Systematic characterization of morphology and transcriptome during normal breast development lays the foundation of our understanding of cancer etiology. We examined mammary glands in female Sprague Dawley rats across six developmental stages – pre-pubertal, peri-pubertal, pubertal, lactation, adult parous and adult nulliparous. We investigated histology by Hematoxylin and Eosin and Mallory’s Trichrome stain, proliferative and apoptotic rate by immunohistochemistry and whole-transcriptome by microarrays. We identified differentially expressed genes between adjacent developmental stages by linear models, underlying pathways by gene ontology analysis and gene networks and hubs active across developmental stages by coexpression network analysis. Mammary gland development was associated with large-scale changes in the transcriptome; particularly from pre-pubertal to peri-pubertal period and the lactation period were characterized by distinct patterns of gene expression with unique biological functions such as immune processes during pre-pubertal development and cholesterol biosynthesis during lactation. These changes were reflective of the shift in mammary gland histology, from a rudimentary organ during early stages to a secretory organ during lactation followed by regression with age. Hub genes within mammary gene networks included metabolic genes such as Pparg during the pre-pubertal stage and tight junction-related genes claudins and occludins in lactating mammary glands. Transcriptome profile paired with histology enhanced our understanding of mammary development, which is fundamental in understanding the etiologic mechanism of breast cancer, especially pertaining to windows of susceptibility to environmental exposures that may alter breast cancer risk.
Keywords: Mammary gland development, Sprague Dawley rat, Breast cancer, Mammary gland histology, Mammary gland transcriptome, Coexpression network
BACKGROUND
Breast cancer is a developmental disease that is estimated to afflict one out of every eight women in the United States [1]. Animal models, especially rodent models, have provided useful insights regarding cellular and molecular aspects of breast cancer, guided better therapeutic strategies and have been instrumental in demonstrating ‘windows of susceptibility’ to environmental exposures leading to enhanced breast cancer susceptibility [2]. These windows of susceptibility represent key stages of mammary gland development during which large scale structural, functional and molecular alterations take place [3]. Development-driven architectural changes in the mammary gland have been thought to influence susceptibility to breast cancer [4]. Normal breast development is fundamentally linked to breast cancer because the same hormones and signaling molecules that drive breast development are dysregulated in breast cancer [5–7]. Investigating molecular mechanisms that underlie structural and functional changes during natural mammary gland development in rodent models is crucial to understanding how timing of adverse environmental exposures alters breast cancer risk, especially in light of using animals as a human-equivalent model.
Two main tissue compartments make up the mammary gland: the epithelium and the stroma. The epithelium constitutes luminal epithelial cells that form a ductal network and milk-secreting alveoli, and basal epithelial cells that form the outer layer of the gland. The stroma consists mainly of adipocytes that make up the fat pad in which the extensive system of ducts and alveoli is embedded, as well as endothelial cells, fibroblasts and immune cells [3]. Breast development involves intricate crosstalk between the epithelium and stroma, and is regulated by hormones and growth factors. Breast tissue is one of the few tissues that undergo major developmental changes after birth. These stages, embryonic, pubertal, pregnancy, lactation and involution, are characterized by profound alterations in mammary gland structure and function, proliferative and apoptotic process, accompanied by genomic and epigenomic changes [3].
Existing studies have investigated gene expression profiles guiding specific stages of mouse mammary gland development including embryonic [8], puberty [9] and the pregnancy-lactation-involution period [10,11]. However, in rats, a commonly used animal model for breast cancer research, there is a lack of studies that investigated large-scale mammary development from early post-natal period to puberty and into adulthood systematically combining structural as well as molecular changes and gene network analysis. Herein, we profiled both the histology and whole-transcriptomes of developing mammary glands in female Sprague Dawley rats, across six comprehensive stages of development: pre-pubertal, peri-pubertal, pubertal, lactation, adult parous and adult nulliparous. We combined histological analyses with differential gene expression and coexpression network analysis with the goal of providing useful insights that not only aid our understanding of breast development but also to highlight histologic and gene expression patterns that characterize crucial developmental time periods that may constitute windows of susceptibility on the pathway to breast cancer.
METHODS
Animals
Animal studies were carried out at Cesare Maltoni Cancer Research Centre/Ramazzini Institute (CMCRC/RI) (Bentivoglio, Italy) in accordance with the rules of Italian law for Animal Welfare (at the time Decreto Legislativo 116, 1992), following the principles of Good Laboratory Practices and Standard Operating Procedures of the CMCRC/RI facility, which include authorization by the ethical committee. The investigation focuses only on female Sprague Dawley (SD) rats which belong to the colony that has been used for over 40 years in the laboratory of the CMCRC/RI. Animals were housed in makrolon cages (41x25x15 cm) at 2 or 3 per cage, with a stainless steel wire top and a shallow layer of white wood shavings as bedding (Giuseppe Bordignon, S.a.s. Treviso, Italy). All animals were kept in a single room at 23 ± 3°C and at 40–60% relative humidity. Lighting was artificial and the light/dark cycles were tended to be 12 hours each. All animals were given the same standard “Corticella” pellet diet (Piccioni Laboratory, Milan, Italy). Feed and tap water were available ad libitum and were both periodically analyzed to exclude biological and chemical contamination (phytoestrogens, mycotoxins, pesticides, arsenic, lead, mercury, selenium).
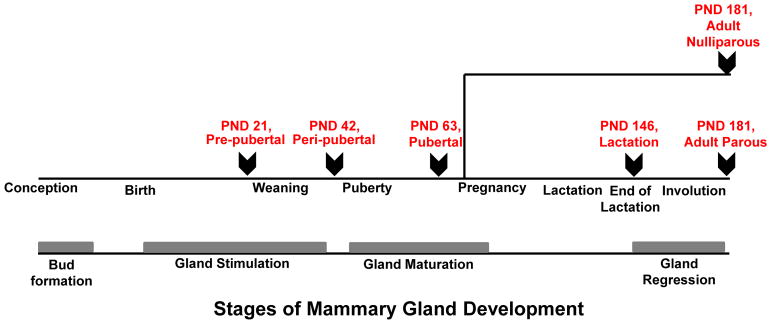
Animals examined for histology and microarrays in this study are part of the “control group” for a study on effects of environmental chemicals on developing mammary glands [12]. These control rats were gavaged orally with olive oil, the vehicle for chemical treatment in the parent study. Olive oil (lot #111275, Montalbano Agricola Alimentare Toscana, Italy) was stored in the dark at room temperature (20°C) in glass containers and administered by gastric intubation using glass syringes. Animals were sacrificed at six developmental time-points reflective of distinct stages of mammary gland development – pre-pubertal (n=10), peri-pubertal (n=5), pubertal (n=5), lactation (n=3), adult parous (n=5) and adult nulliparous (n=5) (Fig. 1). Pre-pubertal animals were sacrificed at postnatal day (PND) 21, peri-pubertal group at PND 42 and pubertal group at PND 63. Animals in the lactation group were sacrificed at the end of lactation at lactating day 28 (PND 146). Adult parous and age-matched nulliparous animals were sacrificed at PND 181, corresponding to 35 days after the end of lactation in parous animals. Animals were housed with their dams until weaning at PND 28. Pre-pubertal, peri-pubertal and pubertal animals received olive oil for 20 days consecutively. Animals in the lactation group and adult parous and nulliparous animals received olive oil daily until weaning, then thrice a week until sacrifice. Animals received 1 mL olive oil through dams until weaning, 0.5 mL from PND 28 – PND 63, and 1 mL after PND 63 until sacrifice. Animals were euthanized via carbon dioxide inhalation and necropsy was immediately performed for collection of selected tissues. All animals belonging to the same developmental group were sacrificed on the same day, regardless of the stage of their estrous cycle.
Figure 1. Study design.
Female Sprague Dawley rats were sacrificed at six distinctive stages of mammary gland development for both histology and transcriptome analyses – pre-pubertal (n=10), peri-pubertal (n=5), pubertal (n=5), lactation (n=3), adult parous (n=5) and adult nulliparous (n=5). PND: Postnatal Day; Arrows indicate point of sacrifice.
Further 20 untreated SD female rats from the CMCRC/RI breeding facility were sacrificed at other developmental stages of interest for mammary glands and were included as part of histology and/or immunohistochemistry experiments, but not microarrays. These stages were: weaning, corresponding to PND 28 (n=5); 8 days after the end of lactation (n=5) referred as involution day 8 (INVO 8); and at reproductive senescence both in parous (n=5) and nulliparous (n=5) rats (aging parous and aging nulliparous, PND 540). The same procedures for necropsy and tissue collection were applied for these animals. Totally 53 female SD rats were included in the study (Table 1).
Table 1.
Animal experimental plan and approaches used to investigate physiological mammary gland development across life-stages. PND: Postnatal Day; INVO: Involution; H&E: Hematoxylin and eosin, MT: Mallory’s Trichrome, TGFβ3: Transforming growth factor β 3
| Animals | Morphologic evaluation | Transcriptome | |||||
|---|---|---|---|---|---|---|---|
| No of animals. | Developmental stage | Age at necropsy (days) | H&E | MT | Ki-67 | TGFβ3 | |
| 10 | Pre-pubertal: PND 21 | 21 | X | X | _ | _ | X |
| 5 | Weaning: PND 28 | 28 | X | X | X | _ | _ |
| 5 | Peri-pubertal: PND 42 | 42 | X | X | X | _ | X |
| 5 | Pubertal: PND 63 | 63 | X | X | X | _ | X |
| 3 | Lactation: Lactation day 28/PND 146 | 146 | X | X | X | X | X |
| 5 | Involution: Involution day 8 (INVO 8) | 154 | X | X | X | X | _ |
| 5 | Adult Parous: PND 181 | 181 | X | X | X | X | X |
| 5 | Adult Nulliparous: PND 181 | 181 | X | X | X | X | X |
| 5 | Aging parous: PND 540 | >540 | X | X | _ | _ | _ |
| 5 | Aging nulliparous: PND 540 | >540 | X | X | _ | _ | _ |
TOTAL n: 53
Histology
Mammary gland histology was performed following the standard operating procedures of the CMCRC/RI laboratory. Second axillary (right and left) mammary glands were dissected without the inclusion of lymph nodes, cut in longitudinal sections [13], placed on cardboard to fix them flat and fixed in alcohol 70% for at least 48h for histopathological examination. The tissues were embedded in paraffin blocks and sections of 3–6 μm were cut for each specimen. Oven-dried sections were deparaffinized with xylene and dehydrated through a graded series of ethanol (100%, 95% and 80% ethanol) and distilled water. One section of each tissue was routinely stained with Hematoxylin and Eosin (H&E). Another section was stained with Mallory’s Trichrome (MT) [20]. Weigert's iron hematoxylin stains the nuclei in black, Orange G stains erythrocytes and muscle fibers in red and phosphotungstic and phosphomolybdic acid with aniline blue stains collagen in blue (Bio Optica, Italy). Sections were microscopically examined by two pathologists in a blinded fashion. A qualitative and semi-quantitative assessment included examinations of a representative section of the gland. Morphology assessment included overall morphology and nuclear, cytoplasmic and membrane details in tissues.
Immunohistochemistry
Immunohistochemistry (IHC) of tissue sections from alcohol fixed mammary glands was performed to measure Ki-67, a marker for proliferating cells, and transforming growth factor β3 (TGFβ3) as an apoptotic marker. Air dried sections were deparaffinized with xylene and dehydrated through a graded series of ethanol (100%, 95% and 80% ethanol) and distilled water. Before performing IHC staining, alcohol fixed paraffin-embedded samples were post-fixed in 10% neutral buffered formalin (NBF) for 30 minutes at 4°C and then transferred in 70% alcohol for 18–24 hours. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide for 15 minutes.
Slides were placed into a Tissue Tek® container filled with a preheated 1X solution of Rodent Decloaker (pH 6) (Biocare Medical, Pacheco, CA) and decloacked for 6 minutes at 300 watts into the microwaves. Non-specific binding sites were blocked with the appropriate serum for 20 minutes in a humidified chamber. Sections were incubated with Ki-67 (Novus Biological, Littleton, CO) or TGFβ3 (Novus Biological, Littleton, CO), at the dilution of 1:10 or 1:20, respectively. Positive and negative controls (omission of the primary antibody and IgG-matched serum) were included for each immunohistochemical run. Afterwards, slices were incubated with ImmPRESSTM (Peroxidase) Polymer Anti-Rabbit IgG (vector Laboratories, Burlingame,CA) for 30 minutes at room temperature. The entire antibody-enzyme complex was then made visible by the reaction with diaminobenzidine until adequate color development was seen. Finally, sections were rinsed in distilled water, counterstained with hematoxylin, dehydrated, and cleared in xylene. Cover slips and mountant were applied for optical microscopy analysis. Two pathologists did the evaluation of the slides independently. Criteria for sufficient staining were antibody binding specificity, tissue morphology and overall staining quality. Ki-67 and TGFβ3 were classified as positive or negative without any score. Digital photos were acquired with a Nikon Coolpix 995 (Nital SpA, Turin, Italy) mounted on a light microscope (MZ6, Leica Microsystems, Milan, Italy).
Transcriptome Profiling
The fifth left and right caudal mammary glands of olive-oil treated animals were collected without the inclusion of lymph nodes, flash-frozen in liquid nitrogen and total RNA was extracted using Maxwell 16 LEV simplyRNA Blood kit (Promega, WI) or by Direct-zol RNA MiniPrep kit (Zymo Research, CA). RNA concentration was determined using Nanodrop (Thermo Scientific, MA) and RNA quality was assessed using a 2100 Bioanalyzer (Agilent Technologies, CA). Samples with RNA Integrity Number 7 were used for microarrays. Transcriptomes were profiled by GeneChip Rat Gene 2.0 ST arrays (Affymetrix, CA) at the Yale Center for Genome Analysis (Yale School of Medicine, CT). Biotinylated cRNA were prepared according to the standard Affymetrix protocol from 250 ng total RNA (Manual Target Preparation for GeneChip® Whole Transcript (WT) Expression Arrays, Affymetrix 2015). Following fragmentation, 5.2 ug of cRNA were hybridized for 16 hr at 45¼C on GeneChip Rat Gene 2.0 ST arrays. GeneChips were washed and stained in the Affymetrix Fluidics Station 450. Arrays were scanned using the GeneChip Scanner 3000. Quality control of .CEL files and preprocessing by robust multiarray average (RMA) method were done using expression console software (Affymetrix, CA). Batch effects due to different RNA extraction methods were removed using ComBat package [14] in RStudio (R version 3.0.2). We applied a signal intensity filter to retain only those probesets with high and stable expression (signal value > 30th percentile in at least 1 experimental group). A variance-based filter was used to retain the top 50% of probesets with high interquartile range resulting in a final dataset containing 7,831 genes. Principal components analysis (PCA) was carried out using the function prcomp () in RStudio. Differential gene expression analysis between developmental groups was performed using linear models for microarray data (limma) package [15] in RStudio. A false discovery rate (FDR) of 5% using Benjamini-Hochberg correction and a fold change of ≥2 fold was used. Functional enrichment analysis of differentially expressed genes was carried out using gene ontology (GO) [16] via DAVID Bioinformatics Resources 6.7 [17]. Fisher’s exact test was used to assess significance of enrichment at FDR of 5%. Microarray data have been deposited in NCBI’s Gene Expression Omnibus (GEO) [18] and are accessible through GEO Series accession number GSE87613 - (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=mlefgyuappcplgz&acc=GSE87613)
Weighted Gene Coexpression Network Analysis (WGCNA)
WGCNA [19] was used to identify modules of genes significantly associated with stages of mammary gland development – spanning 7,831 genes. The coexpression network was constructed as follows. Briefly, pairwise Pearson correlations were calculated for each gene pair and transformed into a signed adjacency matrix using a soft threshold power of 12 to maximize gene-gene correlations while maintaining a large number of gene-gene connections in the network. These connections were converted into a topological overlap matrix (TOM), which is used to quantitatively measure the coexpression of genes. For example, a pair of genes (network nodes) that share many ‘neighbors’ will have high topological overlap. Hierarchical clustering was performed using 1-TOM as the distance measure and modules were identified using WGCNA’s dynamic tree-cutting algorithm. Modules whose expression profiles were very similar were merged together. Module eigengenes, defined as the first principal component of a given module, were used to correlate modules with mammary gland developmental progression. Significant modules were defined as being highly correlated with mammary gland developmental progression (Pearson correlation > 0.5 or < −0.5 and p-value < 0.05). GO analysis was performed on gene sets contained within these modules. ‘Hub’ genes within significant modules were defined as having an absolute module membership (MM) > 0.9, with p-value < 0.05. Additionally, hub genes also had to have a gene significance (GS) value correlated with the direction of the eigengene of the corresponding module. The GS is a measure of the association of a given gene’s expression with development. Genes with high MM and high GS are therefore genes that are highly connected to other genes in the module and are also significantly correlated with mammary development.
Quantitative reverse transcriptase PCR (q-RT-PCR)
One hub gene per module that contributed to enrichment of relevant GO terms was chosen for validation by q-RT-PCR. cDNA was synthesized from total RNA using AffinityScriptMultiple Temperature Reverse Transcriptase kit (Agilent Technologies, CA). Gene-specific primers were designed using Primer-BLAST and synthesized by MWG Operon (Eurofins Genomics, KY) (Table 2). Forty cycles of PCR amplification were performed as follows: denature at 95¼C for 15 sec, anneal at 55¼C for 20 sec, extend at 72¼C for 30 sec. Assays were performed in triplicate using SYBR Green (Life Technologies, CA) on ABI PRISM 7900HT sequence detection system (Applied Biosystems, CA). Data were normalized using expression values of ribosomal protein S11 (Rps11) and alpha-tubulin (Tuba1a) as reference genes. Statistical differences between adjacent developmental stages were determined using t-test.
Table 2.
Gene and primer information for qPCR validation
| Gene Name | Primer sequence (Forward, Reverse) |
|---|---|
| Col1a1 | TGGTACATCAGCCCAAACC, GAACCTTCGCTTCCATACTCG |
| Pparg | CGAAGAACCATCCGATTGAAGC, AAGGCACTTCTGAAACCGACA |
| Tgfb3 | AGGTTTTCCGTTTCAATGTGTCC, ATTCTCTGCTCTGTGCGCTTG |
| Xbp1 | AGCTGGGCATCTCAAACCTG, TTCGTTGGCAAAAGTGTCCTC |
| Ar | GCCCATGCCAGAGAAGTAGT, TTCAGGAAAGTCCACGCTCA |
| Tuba1a | GGAAGTGGATGCGAGGGTA, AGCGCCCAACCTACACTAAC |
| Rps11 | CGAGGGCACCTACATAGACA, GAGATAGTCCCCGGCGGATGA |
RESULTS
Histology of mammary glands at different developmental stages
Animals were sacrificed at the following developmental time-points reflective of distinct stages of mammary gland development pre-pubertal (PND 21), weaning (PND 28), peri-pubertal (PND 42), pubertal (PND 63), lactation (PND 146, corresponding to lactation day 28), involution (INVO 8, corresponding to 8 days after the end of lactation or PND 154), adult parous (PND 181, corresponding to 35 days after the end of lactation) and adult nulliparous (PND 181), aging parous and aging nulliparous (PND 540). Mammary gland sections were stained with Hematoxylin and Eosin (H&E) to visualize the overall morphology and with Mallory’s Trichrome to assess collagen distribution and to determine the relative proportion of each tissue type (glandular, adipose, connective and secretory) at each developmental stage (Fig. 2–3). To better understand the mechanisms of mammary gland remodeling, we investigated the cell proliferation and apoptosis in the developmental windows more susceptible to cellular turnover by immunohistochemistry for Ki-67 and TGFβ3 antibodies. Ki-67 is a nuclear protein essential for cellular proliferation, and is associated with ribosomal RNA transcription. It can be detected within the cell nucleus during interphase. Ki-67 is present throughout the active phases of the cell cycle, but is absent in resting cells, and thus is considered to be an index of cell proliferation [20]. TGFβ3 is a local mammary-derived signaling factor synthesized in response to milk stasis that induces apoptotic cell death during the first phase of involution post weaning [21]. IHC of tissue sections from alcohol fixed mammary glands was employed to measure Ki-67 at PND 28, 42, 63, 146, INVO 8, PND 181 and TGFβ3 at PND 146, INVO 8 and PND 181.
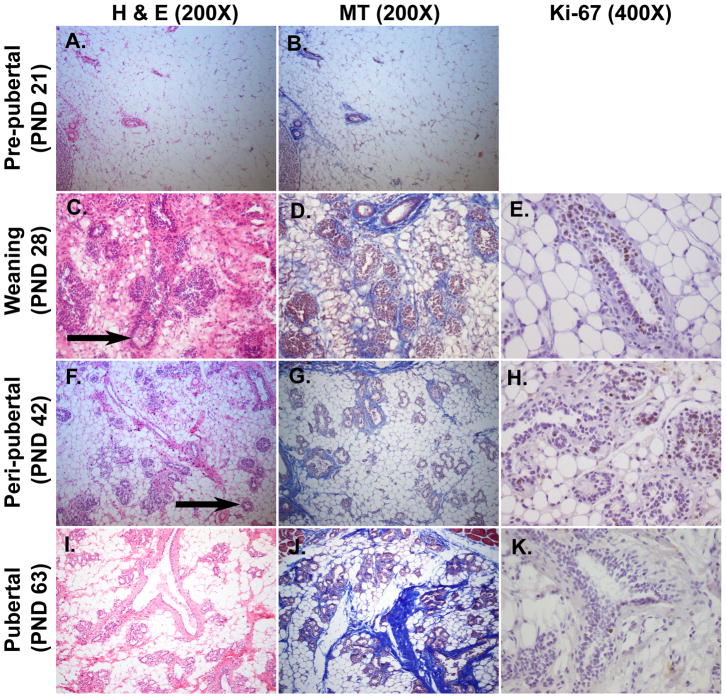
Figure 2. Histological features of mammary glands during early stage of development (PND 21-PND 63).
Panels A-C-F-I were stained with Hematoxylin and Eosin (H&E, 200X) and panels B-D-G-J with Mallory’s Trichrome (MT, 200X). Panels E-H-K were immunostained with Ki-67 antibody (400X). The second axillary mammary glands were dissected from female Sprague Dawley rats. A–B: The stromal compartment is predominant and filled with adipocytes. C-D: Ductal growth is visible as evidenced by formation of TEBs (arrow). E: TEBs are the sites of ductal elongation and branching and represent the sites of highest proliferation in the gland, as revealed by the positivity to immunohistochemistry for the marker of proliferation Ki-67. F–G: TEBs are characterized by a single layer of cap cells and multilayered pre-luminal body cells (arrow). H: Duct elongation is characterized as a proliferative process as revealed by immunohistochemistry for Ki-67 antibody. I-J: Glands are arranged into lobules of compound branched alveolar glands, separated by dense interlobular connective tissue and fat. K: Ki-67 is poorly expressed at pubertal stage, indicating that the exponential development is already occurred.
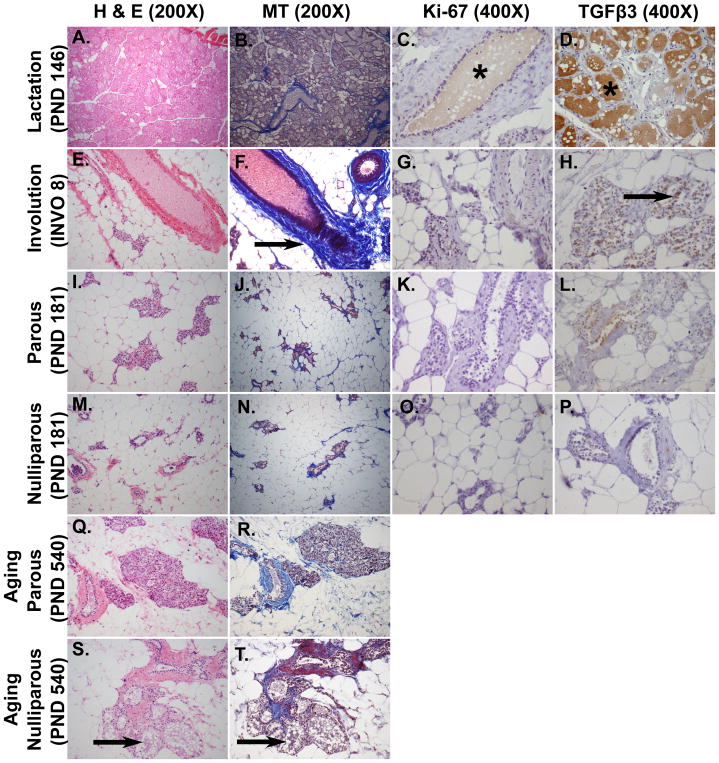
Figure 3. Histological features of mammary glands during and after lactation (PND 146, INVO 8 and PND 181), in nulliparous age-matched rats (PND 181) and aging parous and aging nulliparous rats (PND 540).
Panels A-E-I-M-Q-S were stained with Hematoxylin and Eosin (H&E, 200X) and panels B-F-J-N-R-T with Mallory’s Trichrome (MT, 200X). Panels C-G-K-O were immunostained with Ki-67 antibody (400X) and panels D-H-L-P with TGFβ3 antibody (400X). The second axillary mammary glands were dissected from female Sprague Dawley rats. A–B: Secretory lobulo-alveolar structures fill the mammary fat pad. C: Antibody against Ki-67 demonstrates a poor proliferation rate during late lactation (brown milk staining, indicated with an asterisk, might be an artefact). D: TGFβ3 staining is negative although it seemed olive oil treatment induced a brown artefact in the lumen of secreting glands (asterisk). E–F: Dense stroma (arrow) is visible around the ducts and the stroma appears increased around the clusters of collapsed alveoli. G: The proliferative index using Ki-67 is almost negative. H: There is an increased apoptotic rate during the involution process as revealed by a positive TGFβ3 immunostaining (arrow). I–J: Most of the parenchyma is replaced by adipose tissue. Few remaining collapsed lobulo-alveoli are lined by non-secretory epithelial cells. K: The proliferative rate is negative. L: TGFβ3 is expressed at lower levels in the mammary glands of adult parous animals 35 days after the end of lactation (PND 181) compared to eight days after the end of lactation (INVO 8), indicating that the majority of cell death has already occurred. M–N: Majority of the gland is made of adipocytes, resembling I–J, but the number of lobulo-alveoli is lower. O: The proliferative rate is negative. P: TGFβ3 staining is not observed. Q-R-S-T: An inappropriate but physiological secretory activity (galactoceles, arrowed) is observed in aging nulliparous rats compared to aging parous rats.
Distinct histological features were observed during early developmental stages. In the pre-pubertal group (PND 21, Fig. 2A–B) the parenchyma was rudimentary and consisted of a small ductal tree. Each branch was composed of a single layer of epithelial cells surrounding a central lumen. The stromal compartment was predominant and the gland was filled with adipocytes. At PND 28, corresponding to weaning (Fig. 2C–D), the mammary glands revealed a rapid growth and the primary epithelial structure in the glands at the start of the ductal growth was the terminal end bud (TEB). TEBs consist of multiple layers of epithelium with an outer layer of undifferentiated pluripotent stem cells called cap cells, which sit on the basal lamina. These structures are the sites of ductal elongation and branching and represent the sites of highest proliferation in the gland [3], as revealed by positive Ki-67 staining (Fig. 2E). During peri-puberty (PND 42, Fig. 2F–G), which is considered the growth phase, the proliferative edge of the TEB was not surrounded by connective tissue and therefore proliferation and cell migration resulted in invasion into the fat pad and elongation of the duct. The basement membrane and the stroma was formed by fibroblasts surrounding the ducts. Epithelial cells exhibited a positive Ki-67 staining in mammary TEBs (Fig. 2H). The pubertal mammary gland (PND 63, Fig. 2I–J) was characterized by increased ductal branching and arrangement into lobulo-alveolar structures that were separated by connective tissue and fat. Ki-67 was poorly expressed at pubertal stage, indicating that exponential development had already occurred (Fig. 2K).
Histological features of adult mammary glands also exhibited distinctive patterns depending on stage of development. At PND 146, corresponding to lactation day 28 in dams (Fig. 3A–B), extensive growth and alveolar maturation occurred to form milk-producing glands. As the alveoli expanded to completely fill the gland, there was a simultaneous reduction in the amount of connective tissue as revealed by MT staining, and fat contained within adipocytes was metabolized. The rate of cell renewal appeared negative with low to absent Ki-67 expression (Fig. 3C) and TGFβ3 was also not expressed (Fig. 3D). After weaning, the gland goes through a process of death and remodeling termed involution (INVO). The lobular alveolar structures that produce milk are still present but the glands rapidly regress to a virgin-like state through apoptosis. At 8 days from the end of lactation (INVO 8, Fig. 3E–F), there was residual milk in the alveoli but the gland appeared histologically different from the lactating gland. Dense stroma was visible around the ducts and around the clusters of collapsed alveoli. There was a reduced proliferative index as revealed by Ki-67 (Fig. 3G), while conversely, an increased apoptotic rate was observed as revealed by positive TGFβ3 immunostaining (Fig. 3H). The mammary gland of adult parous animals (PND 181, Fig. 3I–J) representing 35 days after the end of lactation, was completely remodeled, resembling the pre-pregnant mature gland. The few remaining alveoli were lined by cuboidal non-secreting epithelial cells. The abundance of interstitial and connective tissue was increased around the collapsed alveoli and the parenchyma was replaced by adipose tissue. The mammary glands did not contain TEBs therefore the proliferative rate appeared negative (Fig. 3K). TGFβ3 was expressed at low levels indicating that the majority of cell death had already occurred (Fig. 3L). Adult nulliparous mammary glands (PND 181, Fig. 3M–N) had a general appearance similar to age-matched parous glands, but with a lower number of lobules and more adipose tissue compared to adult parous glands. Mitotic and apoptotic cells were not observed as demonstrated by the negative expression of Ki-67 (Fig. 3O) and TGFβ3, respectively. (Fig. 3P). During reproductive senescence (PND 540), aging parous rats appeared histologically normal (Fig. 3Q–R). However, an age-related inappropriate secretory activity was observed in aging nulliparous rats (PND 540, Fig. 3S–T), where ducts and alveoli became dilated and cystic and filled with proteinaceous secretory fluid (galactoceles).
Transcriptome changes between adjacent stages of developing mammary glands
For whole-transcriptome analyses, we focused on the following developmental stages: pre-pubertal (PND 21), peri-pubertal (PND 42), pubertal (PND 63), lactation (PND 146), adult parous and age-matched adult nulliparous (PND 181).
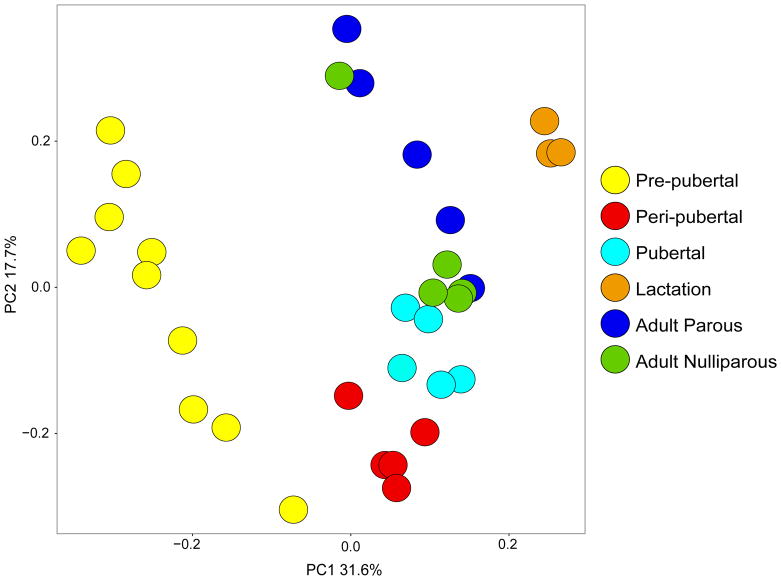
Accompanied with significant changes in mammary gland histology, we also observed dynamic changes in mammary transcriptome from pre-pubertal stage to adulthood in both parous and nulliparous rats. To visualize the whole-transcriptome changes of the developing mammary gland, we performed principal components analysis (PCA) (Fig. 4). The first principal component (PC) separated rats into three clusters– pre-pubertal rats fell into the first cluster, rats in the peri-pubertal, pubertal, adult parous and age-matched nulliparous groups formed a second cluster and rats in the lactation group formed a third cluster. Along the second PC axis, there was a progression from early to later development starting from peri-pubertal to pubertal to adult parous and age-matched nulliparous groups.
Figure 4. Principal components analysis (PCA) of mammary tissues from different developmental stages spanning 7,831 genes.
Clustering of animals by developmental stage is evident.
We performed differential gene expression analysis to examine transcriptome differences between two adjacent stages of development followed by gene ontology (GO) enrichment (Table 3). From pre-pubertal (PND 21) to peri-pubertal (PND 42) stage, 697 genes were differentially expressed with 322 genes up-regulated and 375 down-regulated. Among the top up-regulated genes (FDR < 0.05, fold change > 4) were caseins Csn3, Csn2 and Csn1s2a and mammary gland receptors such as progesterone receptor (Pgr), prolactin receptor (Prlr) and erb-b2 receptor tyrosine kinase 4 (Erbb4); up-regulated genes were enriched for ‘gland development’ by GO analysis. Down-regulated genes on the other hand were enriched for immune signaling pathways with top genes (FDR < 0.05, fold change > 4) including phosphatases (Ptprc and Ptpn22), tyrosine kinases (Lck), adaptor proteins (Skap1 and Trat1), Cd molecules (Cd3e, Cd19, Cd28, Cd69) and interleukins (Il7r). From peri-pubertal (PND 42) to pubertal stage (PND 63), the number of altered genes was much less with only 38 differentially expressed genes (26 up; 12 down), and these genes were not enriched for any functional categories. There were 1042 genes that were altered from puberty (PND 63) to lactation (PND 146); 362 genes were up-regulated and were enriched in ‘cholesterol biosynthetic process’, where top up-regulated genes (FDR < 0.05, fold change > 4) included cholesterol biosynthesis genes 24-dehydrocholesterol reductase (Dhcr24) and isopentenyldiphosphate delta isomerase 1 (Idi1) as well as lactation-related genes such as caseins (Csn1s2a and Csn1s2b) and whey acidic protein (Wap). There were 680 genes that were down-regulated from puberty to lactation and were enriched for ‘extracellular matrix organization’ (ECM). Several collagen genes were among the most down-regulated (FDR < 0.05, fold change > 4), for example Col1a1, Col1a2, Col3a1, Col5a1, Col5a2. There were 1022 genes altered from glands at lactation (PND 146) to adult parous glands (PND 181). Up-regulated genes (n=623) were enriched for ‘regulation of cell proliferation’; top up-regulated genes (FDR < 0.05, fold change > 4) were related to immune function (Cd molecules, T-cell-specific transcription factor 1 (Tcf7) and tumor necrosis factor receptor superfamily, member 14 (Tnfrsf14)) as well as mammary gland development (amphiregulin (Areg) and Pgr). Interestingly, genes down-regulated (n=399) from lactation to adult parous glands had a large overlap (~70%) with genes that were up-regulated from puberty to lactation with similar enrichment for ‘cholesterol biosynthetic process’. We also examined the mammary transcriptome of adult nulliparous rats and observed only 5 differentially expressed genes between the two groups (Reg3b, Pcdh17, Spt1, Igh-6, RGD1563231).
Table 3.
Differential gene expression analysis between adjacent developmental stages. Number of significantly differentially expressed genes (DEGs) (FDR < 0.05, fold change ≥2) and gene ontology (GO) enrichment analysis (FDR < 0.05) between adjacent developmental stages by limma. Genes in bold are changed by more than 4 fold.
| Number of DEGs | Top GO term | Genes from dataset involved in the GO term | |
|---|---|---|---|
|
| |||
| Pre-pubertal to Peri- pubertal | Total: 697 | ||
| Up: 322 | GO:0048732; gland development | Egfr, Fgfr2, Elf3, Rxfp1, Tbx3, Erbb4, Erbb3, Elf5, Foxa1, Tgfb3, Tp63, Cdh1, Mdk, Muc4, Pgr, Vdr, Prlr, Serpinb5, Areg, Tph1, Bmp7 | |
| Down: 375 | GO:0002684; positive regulation of immune system process | Itgal, Il27Ra, Mmp9, Fcer2, Fcnb, Cd247, Klrk1, Nfkbia, Ptpn22, Il7r, Skap1, Vcam1, Sh2d1a, Klhl6, Cd69, Zap70, Il2rg, Inpp5d, Cd5, Spn, Cd28, Ptprc, Il2ra, Gimap5, Il7, Cd3e, Foxp3, Trat1, Cd38, Prkcq, Coro1a, Cd37, Cd19, Cd80, Lax1, Cd40lg, Lck, Cd79b, Cd226, Sash3 | |
|
| |||
| Peri-pubertal to Pubertal | Total: 38 | ||
| Up: 26 | NA | NA | |
| Down: 12 | NA | NA | |
|
| |||
| Pubertal to Lactation | Total: 1042 | ||
| Up: 362 | GO:0006695; cholesterol biosynthetic process | G6pd, Hmgcs2, Fdps, Prkaa2, Idi1, Hsd17b7, Nsdhl, Dhcr24 | |
| Down: 680 | GO:0030198; extracellular matrix organization | Mia, Col3a1, Eln, Col2a1, Postn, Vit, Serpinh1, Tnfrsf11b, Smoc1, Lox, Fn1, Dpt, Reck, Ccdc80, Olfml2a, Nid1, Hspg2, Col5a2, Col5a1, Lama2, Fbln1, Serpinb5, Col1a2, Pdgfra, Col1a1, Lamc1, Adamts2 | |
|
| |||
| Lactation to Adult Parous | Total: 1022 | ||
| Up: 623 | GO:0042127; regulation of cell proliferation | Fgf7, Tacr1, Prrx1, Fgf10, Zeb1, Gli3, Pgr, Gpc3, Ptges, Apoe, Pdgfc, Fgf1, Spn, Hyal1, Ar, Tnfrsf14, Prkcq, Ccnd1, Cd80, Ccnd2, Cd40lg, Pla2g2a, Pmp22, Wnt5a, Fxyd2, Rbp4, Erbb4, Egln3, St8sia1, Kitlg, Il34, Aldh3a1, Vcam1, Stat4, Rac2, Bcl11b, Bcl2, Zap70, Figf, Cd28, Ptprc, Tcf7, Il2ra, Tbx3, Il7, Ctla4, Tnfrsf13c, Igf1, Rogdi, Foxp3, Shox2, Btla, Fcgr2b, Dbp, Areg, Eaf2, Mab21l1, Igfbp3 | |
| Down: 399 | GO:0006695; cholesterol biosynthetic process | G6pd, Hmgcs2, Fdps, Idi1, Hsd17b7, Nsdhl, Fdft1, Dhcr24 | |
|
| |||
| Adult Nulliparous vs. Adult Parous | Total: 5 | ||
| Up: 2 | NA | NA | |
| Down: 3 | NA | NA | |
Gene networks of developing mammary glands
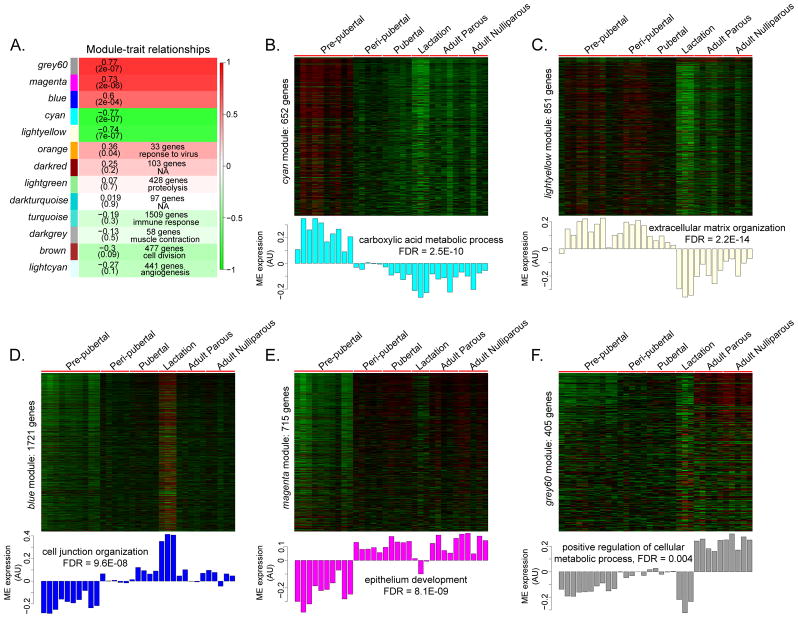
To systematically examine the entire transcriptome in mammary glands, we used WGCNA to group highly correlated genes into co-expression ‘modules’. We identified 13 gene modules using our dataset of 7,831 genes; 341 genes did not load into any specific module (Fig. 5A). Size of modules ranged from 33 genes in the orange module to 1,721 genes in the blue module. Five of the 13 modules showed a significant developmental stage-specific gene expression pattern – cyan, lightyellow, blue, magenta and grey60, i.e., module eigengenes of these modules were significantly correlated with mammary gland developmental progression (|Pearson correlation| > 0.5, p < 0.05) (Fig. 5B–F). Altogether these five modules constituted 4,344 genes, which may represent the total gene set which can be used to characterize mammary glands transitioning from pre-pubertal period into adulthood.
Figure 5. WGCNA of developing mammary glands.
A. Heatmap showing association between modules and developmental groups. Each cell contains the Pearson correlation of the module eigengene (ME) (the first principal component of genes in a module) with developmental stage and p-values are indicated in brackets. For non-significant modules, number of genes within the module and where applicable, top GO term is indicated in the corresponding cell. Red corresponds to modules that contain genes whose expression increases during mammary gland development and green corresponds to modules containing genes with decreasing expression during development; color intensity is proportional to the strength of the correlation. B–F. Heatmaps and corresponding ME profile of significant modules. Genes are in rows, samples in columns. Top GO enrichment result for each module and associated FDR are indicated. AU: Arbitrary units.
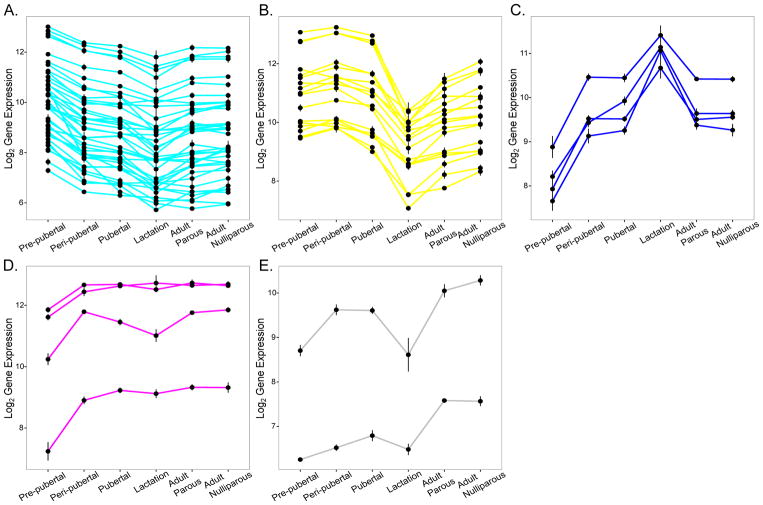
The cyan (Fig. 5B) and lightyellow (Fig. 5C) modules comprised genes showing a downward trend in expression pattern through development. The cyan module constituted 652 genes whose expression was markedly elevated during pre-pubertal stage only, and subsequently remained down-regulated through the rest of mammary gland development. We identified 143 hub genes within the cyan module, of which 39 were associated with “metabolic function” by GO analysis; these included genes such as peroxisome proliferator-activated receptor gamma (Pparg), CCAAT/enhancer binding protein (C/EBP), alpha (Cebpa) and aldehyde dehydrogenase 6 family, member A1 (Aldh6a1) among others (module membership (MM) > 0.95, p < 3.5E-06) (Fig. 6A). The lightyellow module consisted of 851 genes with high expression from pre-pubertal to pubertal stages, followed by marked down-regulation during lactation and sustained low expression in adult parous and adult nulliparous rats. There were 75 hub genes, of which 13 belonged to the collagen family (Col1a1, Col1a2, Col3a1, Col4a1, Col4a2, Col5a1, Col5a2, Col5a3, Col6a1, Col6a2, Col6a3, Col14a1, Col15a1; MM > 0.91, p < 0.005) and several were involved in extracellular matrix (ECM) function by GO annotation, such as laminins Lamc1 and Lama2 and fibrillin 1 (Fbn1) (MM > 0.93, p < 0.0005) (Fig. 6B).
Figure 6. Selected hub gene profiles from each module.
Plots showing gene expression profiles of hub genes from each module, selected on the basis of involvement in GO enrichment terms associated with each module. A. 39 cyan module hub genes with GO annotation ‘metabolic function’. B. 17 lightyellow module hub genes belonging to collagen family or with GO annotation ‘extracellular matrix’. C. 4 blue module hub genes with GO annotation ‘junction’ – occludin and claudins. D. 4 magenta module hub genes – with GO annotation related to mammary gland (Gata3, Xbp1, Igfbp6, Tfap2c). E. 2 grey60 module hub genes Esrrb and Ar.
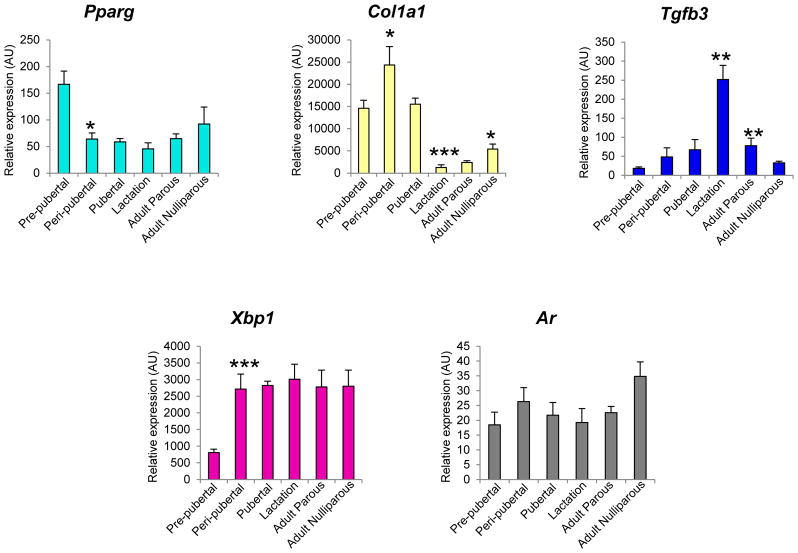
Blue (Fig. 5D), magenta (Fig. 5E) and grey60 (Fig. 5F) modules represented genes with an upward trend of expression across mammary gland development. The blue module had 1,721 genes that exhibited low expression from the pre-pubertal period into puberty but were uniquely up-regulated during lactation, reverting to low expression in adulthood in both parous and nulliparous rats. 179 hub genes were found, including tight junction genes such as transforming growth factor beta 3 (Tgfb3; MM = 0.91, p = 4.28E-07) occludin (Ocln; MM = 0.98, p = 9.98E-05) and claudins (Cldn3, Cldn4, Cldn7; MM > 0.9, p < 0.0005) (Fig. 6C). Magenta module represented a group of 715 genes that exhibited low expression pattern during the pre-pubertal stage followed by sustained high expression during subsequent developmental stages (except lactation stage). There were 90 hub genes present in this module, of which many had functions in mammary gland development by GO annotation (for example GATA binding protein 3 (Gata3), transcription factor AP-2 gamma (Tfap2c), insulin-like growth factor binding protein 5 (Igfbp5) and X-box binding protein 1 (Xbp1) [22]; MM > 0.91, p < 5E-05) (Fig. 6D). Finally the grey60 module was made of 405 genes whose expression was consistently low from pre-pubertal to lactation stages, with marked up-regulation in adult parous and adult nulliparous groups only. We identified 8 hub genes belonging to this module. Although we did not identify hub genes with known functional roles in metabolic processes (top module GO enrichment), we found genes such as estrogen-related receptor beta (Esrrb) [23] and androgen receptor (Ar) [24] to be interesting hub gene candidates, since they have functional roles in the mammary gland (MM > 0.9, p < 6.8E-06) (Fig. 6E). The expression of module hub genes was validated by qPCR (Fig. 7).
Figure 7. Module hub genes validated by qPCR.
One hub gene per module was selected for validation by qPCR. cyan module: Ppar-gamma, lightyellow module: Col1a1, blue module: Tgfb3, magenta module: Xbp1, grey60 module: Ar. Bars represent mean ± standard error. Statistical differences between adjacent developmental stages were determined using t-test; *indicates p<0.05, **p<0.01 and ***p<0.0005. AU: Arbitrary units.
DISCUSSION
Understanding breast development is central to our understanding of breast cancer since the same hormones and signaling pathways that govern breast development become dysregulated during breast cancer development [5,7] and stages of breast development represent windows of susceptibility to environmental exposures that can alter breast cancer susceptibility [2]. Since examining normal breast tissue at various stages of development in humans is limited by availability of tissues, animal models, especially rodent models provide a useful system to study this process. Both human and rat mammary development follow the same sequence of events, although the timing of these events is condensed in rats due to their shorter lifespan. In both species mammary epithelia undergo a short burst of growth just before birth and subsequently grow at an isometric rate until just before puberty [25]. Rats have been widely used to study breast cancer since the premalignant stages of mammary cancer in rats closely recapitulate the human disease [26]. In particular, the Sprague Dawley rat model has been shown to be physiologically relevant and genetically well-defined for studying human breast cancer given similar age-equivalent distribution of mammary tumors as in the human population; mammary carcinomas from this animal model also share many morphological and molecular features with human breast cancer, including estrogen-dependence, chromosomal instability and cell cycle dysregulation [27]. One caveat of our study is that some of the animals were gavaged with olive oil, since this was part of a larger study looking at the effects of environmental chemical exposures on mammary gland development which used olive oil as a vehicle control. Nevertheless, our findings provide important insights about mammary gland development trajectories given that olive oil is a common orally administered vehicle in toxicology studies, particularly for lipophilic compounds.
To our knowledge, this is the first systematic survey combining mammary histology, transcriptome and gene network profiles during six stages of mammary gland development – pre-pubertal, peri-pubertal, pubertal, lactation and adulthood (parous and nulliparous) stages – in SD rats. We observed significant histological changes accompanied by dynamic transcriptome changes during mammary gland development. Specifically, the period from pre-pubertal to peri-pubertal development, and the period from puberty to lactation to post-lactation adulthood are highly dynamic and characterized by distinct patterns of gene expression with unique biological functions. These changes were reflective of the shift in mammary gland structure and function as confirmed by histology results, from a quiescent organ during the pre-pubertal period to a ductal, branched organ during puberty, to a lobuloalveolar, secretory organ during pregnancy and lactation and finally a remodeled gland similar to age-matched nulliparous counterparts following involution and regression. Mammary tissues were not micro-dissected for transcriptome analyses; particularly for the young animals during early developmental stages, i.e. the pre-pubertal and peri-pubertal stages, mammary glands were very small making dissection challenging. Given that mixed cell types including surrounding stroma may have been included in our analysis, changes in transcriptome profiles may in part also be reflective of changes in proportion of different cell types.
From late embryonic period to right before puberty, mammary gland development undergoes isometric growth, keeping up with overall development, until the influence of hormones and growth factors at the beginning of puberty drives the next stage of development [3]. This period of mammary development represents a window of heightened sensitivity to environmental insults that have the potential to modify cancer risk [28]. During the pre-pubertal period, the stromal compartment was dominant, with a preponderance of adipocytes. This was supported by the presence of cyan module hub genes such as Pparg and Cebpa, both of which are key transcription factors in adipogenesis [29]. Differential gene expression analysis revealed that immune-related signaling genes were up-regulated during this period, which could also be reflective of the high stromal tissue content during this period. The magenta module consisting of genes uniquely under-expressed during pre-pubertal period was enriched for functions relevant to mammary gland development. Little is known about the molecular factors involved between embryonic and pubertal mammary development. Our results reflect that the mammary gland at pre-puberty is actively participating in basic metabolic functions to promote growth, while not yet having achieved more specialized functions.
During pubertal mammary gland development, TEBs promote ductal elongation and branching [30]. We observed extensive epithelial proliferation as revealed by IHC with Ki67 cell proliferation marker. TEBs display enhanced sensitivity to carcinogens [4], and the pubertal period has been shown to be a window of heightened sensitivity to environmental exposures that may alter cancer risk [2,12]. Both histology and gene expression analysis revealed that peri-pubertal and pubertal mammary glands in SD rats were similar, but differed from the preceding and the subsequent stages. Higher glandular tissue with TEB formation and lower adipose tissue was evident during peri-puberty and puberty compared to pre-pubertal period. Correspondingly, WGCNA revealed that genes related to mammary gland development such as Gata3 and Xbp1 (magenta module) and ECM organization such as collagens and laminins (lightyellow module) were active during the pubertal period; these genes are highly expressed in pubertal mammary epithelium and are critical for maintaining TEB structure and function [22,31,32]. These findings demonstrate that we were able to identify key regulatory molecules at a specific stage of mammary gland developmental using WGCNA.
During pregnancy massive epithelial proliferation drives ductal branching to produce a network of lobulo-alveoli – clusters of secretory epithelial cells – that are capable of milk synthesis and secretion [33]. We previously showed that lactating mammary glands were more sensitive to chemical exposures compared to glands at later stages of development [34]. Histology showed the presence of lobulo-alveolar structures and the abundance of glandular and secretory tissue components at lactation (PND 146). Mammary growth during lactation may occur during early lactation (lactation day 1–14), where milk secretion increases to peak due to the continued differentiation of mammary secretory cells and increased secretory activity per cell [35]. During established lactation, lack of cell division and proliferation as indicated by the absence of Ki-67 expression in our data suggests that the tissue is already working at its maximum capacity [35]. Secreted milk contains significant amounts of cholesterol [36]. Correspondingly we found activation of genes enriched for cholesterol biosynthetic process. Conversely there was reduced expression of genes with roles in ECM, including several collagen genes. Expansion of the epithelial compartment with concomitant reduction of the stromal compartment was shown to cause a massive decline in stromal genes including collagens during pregnancy and lactation [10]. The blue module represented a group of over a 1000 genes that were active during lactation that included hub genes Ocln and claudins, the two major structural protein families that constitute tight junctions [37], which are crucial for maintaining structural integrity of mammary epithelial cells during lactation [38]. Interestingly, Tgfβ3 was also a blue module hub gene found to be active during lactation, while protein expression of TGFβ3 determined by IHC was negative. TGFβ3 has been shown to be subject to extensive translational regulation such that RNA levels may not correspond directly to protein levels [39].
Pregnancy is thought to be required for the breast to achieve a fully differentiated state [40] and structural and genomic changes arising from completion of pregnancy and lactation may be protective against breast cancer [41]. Parous mammary tissues have been shown to exhibit distinct gene expression patterns compared to nulliparous counterparts in both humans [42] and rats [43,44]. Surprisingly in our study, adult parous and age-matched nulliparous mammary glands at PND 181 did not show dramatic histologic or transcriptome differences. The adult parous group in our study may be reflective of the fully regressed mammary gland since this group was sacrificed 35 days after the end of lactation. The initial phase of involution occurs within a few days of end of lactation [45]. During post-lactational involution, apoptosis of the epithelial structures remodels the mammary gland back to a simple ductal architecture, although the gland remains more differentiated compared to its virgin counterpart [3]. Our result showed that at day 8 of involution (INVO 8) the alveoli were mostly collapsed and the stroma increased around the collapsed structures. There is a dramatic induction of TGFβ expression at the beginning of involution and relatively high levels are maintained for up to 9 days post-weaning [46,47]. Our results revealed that TGFβ3 protein expression was high during the first stage of involution (INVO 8), but expressed at much lower levels by late involution at PND 181, consistent with earlier studies [21]. Cholesterol biosynthesis was down-regulated from lactation to adult parous group at PND 181, indicating the cessation of milk secretion and the shift in mammary structure to more adipose and lesser glandular and secretory components. WGCNA revealed that gene expression changes occurred concurrently in parous and nulliparous tissues at PND 181, with there being no distinctive patterns between the two groups. The magenta and grey60 modules, which represented genes that were active in both parous and nulliparous tissues, and were related to epithelium development and metabolic process, respectively, could reflect the simple ductal architecture and quiescent state of the regressed mammary gland. The lightyellow module had a group of genes commonly under-expressed in both parous and nulliparous rats, with functions enriched for ECM organization, such as collagens and laminins, which have been shown to be markedly down-regulated in fully regressed as well as nulliparous mammary glands [48]. Taken together our results indicate that the structure and molecular make-up of fully regressed mammary glands are similar to their age-matched nulliparous counterparts at PND 181, explaining the lack of histologic and transcriptomic differences between these two groups in our study.
During reproductive senescence (PND 540) an inappropriate secretory activity was observed in virgin SD rats. During the normal aging process, ovulation does not occur, and the levels of estradiol remain persistently elevated. Evaluation of vaginal cytology in this condition revealed a persistent estrous state. The elevated estradiol levels stimulate prolactin release, producing mammary duct ectasia, and ultimately, galactoceles. In humans, such changes are considered dysplastic. Therefore, when evaluating mammary glands in rodent bioassays, it is important to interpret histologic features with caution and understanding these physiological mechanisms of mammary gland development may help clarify treatment-related changes [13].
Supplementary Material
Acknowledgments
This study was supported by grants from NIEHS/NCI (1U01ES019459 and R01 CA172460), and Susan G. Komen Italia onlus.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Howlader N, Noone AM, Krapcho M, Miller D, Bishop K, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA, editors. SEER Cancer Statistics Review, 1975–2014. National Cancer Institute; Bethesda, MD: Apr, 2017. https://seer.cancer.gov/csr/1975_2014/, based on November 2016 SEER data submission, posted to the SEER web site. [Google Scholar]
- 2.Rudel RA, Fenton SE, Ackerman JM, Euling SY, Makris SL. Environmental Exposures and Mammary Gland Development: State of the Science, Public Health Implications, and Research Recommendations. Environ Health Perspect. 2011;119:1053–61. doi: 10.1289/ehp.1002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macias H, Hinck L. Mammary gland development. Wiley Interdiscip Rev Dev Biol. 2012;1:533–57. doi: 10.1002/wdev.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russo J, Lynch H, Russo IH. Mammary gland architecture as a determining factor in the susceptibility of the human breast to cancer. Breast J. 2001;7:278–91. doi: 10.1046/j.1524-4741.2001.21033.x. [DOI] [PubMed] [Google Scholar]
- 5.Lanigan F, O’Connor D, Martin F, Gallagher WM. Molecular links between mammary gland development and breast cancer. Cell Mol Life Sci CMLS. 2007;64:3159–84. doi: 10.1007/s00018-007-7386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malhotra GK, Zhao X, Band H, Band V. Shared signaling pathways in normal and breast cancer stem cells. J Carcinog. 2011;10:38. doi: 10.4103/1477-3163.91413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hynes NE, Watson CJ. Mammary gland growth factors: roles in normal development and in cancer. Cold Spring Harb Perspect Biol. 2010;2:a003186. doi: 10.1101/cshperspect.a003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowin P, Wysolmerski J. Molecular mechanisms guiding embryonic mammary gland development. Cold Spring Harb Perspect Biol. 2010;2:a003251. doi: 10.1101/cshperspect.a003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNally S, Martin F. Molecular regulators of pubertal mammary gland development. Ann Med. 2011;43:212–34. doi: 10.3109/07853890.2011.554425. [DOI] [PubMed] [Google Scholar]
- 10.Rudolph MC, McManaman JL, Hunter L, Phang T, Neville MC. Functional development of the mammary gland: use of expression profiling and trajectory clustering to reveal changes in gene expression during pregnancy, lactation, and involution. J Mammary Gland Biol Neoplasia. 2003;8:287–307. doi: 10.1023/b:jomg.0000010030.73983.57. [DOI] [PubMed] [Google Scholar]
- 11.Clarkson RWE, Wayland MT, Lee J, Freeman T, Watson CJ. Gene expression profiling of mammary gland development reveals putative roles for death receptors and immune mediators in post-lactational regression. Breast Cancer Res BCR. 2004;6:R92–109. doi: 10.1186/bcr754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gopalakrishnan K, Teitelbaum SL, Lambertini L, Wetmur J, Manservisi F, Falcioni L, et al. Changes in mammary histology and transcriptome profiles by low-dose exposure to environmental phenols at critical windows of development. Environ Res. 2017;152:233–43. doi: 10.1016/j.envres.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis B, Fenton S. Chapter 61 - Mammary Gland. In: Haschek WM, Rousseaux CG, Wallig MA, editors. Haschek Rousseauxs Handb Toxicol Pathol Third Ed [Internet] Boston: Academic Press; 2013. pp. 2665–94. cited 2018 Mar 19. Available from: https://www.sciencedirect.com/science/article/pii/B9780124157590000613. [Google Scholar]
- 14.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinforma Oxf Engl. 2012;28:882–3. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 16.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 20.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–22. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen AV, Pollard JW. Transforming growth factor beta3 induces cell death during the first stage of mammary gland involution. Dev Camb Engl. 2000;127:3107–18. doi: 10.1242/dev.127.14.3107. [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa D, Calvo V, Avivar-Valderas A, Lade A, Chou H-I, Lee YA, et al. Epithelial Xbp1 is required for cellular proliferation and differentiation during mammary gland development. Mol Cell Biol. 2015;35:1543–56. doi: 10.1128/MCB.00136-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Twigger A-J, Hepworth AR, Lai CT, Chetwynd E, Stuebe AM, Blancafort P, et al. Gene expression in breastmilk cells is associated with maternal and infant characteristics. Sci Rep. 2015;5:12933. doi: 10.1038/srep12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hickey TE, Robinson JLL, Carroll JS, Tilley WD. Minireview: The androgen receptor in breast tissues: growth inhibitor, tumor suppressor, oncogene? Mol Endocrinol Baltim Md. 2012;26:1252–67. doi: 10.1210/me.2012-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neville MC, Daniel CW. The Mammary Gland: Development, Regulation, and Function. Springer; 1987. [Google Scholar]
- 26.Russo J, Gusterson BA, Rogers AE, Russo IH, Wellings SR, van Zwieten MJ. Comparative study of human and rat mammary tumorigenesis. Lab Investig J Tech Methods Pathol. 1990;62:244–78. [PubMed] [Google Scholar]
- 27.Maltoni C, Minardi F, Pinto C, Belpoggi F, Bua L. Results of three life-span experimental carcinogenicity and anticarcinogenicity studies on tamoxifen in rats. Ann N Y Acad Sci. 1997;837:469–512. doi: 10.1111/j.1749-6632.1997.tb56895.x. [DOI] [PubMed] [Google Scholar]
- 28.Fenton SE, Reed C, Newbold RR. Perinatal environmental exposures affect mammary development, function, and cancer risk in adulthood. Annu Rev Pharmacol Toxicol. 2012;52:455–79. doi: 10.1146/annurev-pharmtox-010611-134659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosen ED, Hsu C-H, Wang X, Sakai S, Freeman MW, Gonzalez FJ, et al. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 2002;16:22–6. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson CJ, Khaled WT. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Dev Camb Engl. 2008;135:995–1003. doi: 10.1242/dev.005439. [DOI] [PubMed] [Google Scholar]
- 31.Klinowska TC, Soriano JV, Edwards GM, Oliver JM, Valentijn AJ, Montesano R, et al. Laminin and beta1 integrins are crucial for normal mammary gland development in the mouse. Dev Biol. 1999;215:13–32. doi: 10.1006/dbio.1999.9435. [DOI] [PubMed] [Google Scholar]
- 32.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–55. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oakes SR, Hilton HN, Ormandy CJ. The alveolar switch: coordinating the proliferative cues and cell fate decisions that drive the formation of lobuloalveoli from ductal epithelium. Breast Cancer Res BCR. 2006;8:207. doi: 10.1186/bcr1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manservisi F, Gopalakrishnan K, Tibaldi E, Hysi A, Iezzi M, Lambertini L, et al. Effect of maternal exposure to endocrine disrupting chemicals on reproduction and mammary gland development in female Sprague-Dawley rats. Reprod Toxicol Elmsford N. 2015;54:110–9. doi: 10.1016/j.reprotox.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capuco AV, Wood DL, Baldwin R, Mcleod K, Paape MJ. Mammary cell number, proliferation, and apoptosis during a bovine lactation: relation to milk production and effect of bST. J Dairy Sci. 2001;84:2177–87. doi: 10.3168/jds.S0022-0302(01)74664-4. [DOI] [PubMed] [Google Scholar]
- 36.Clarenburg R, Chaikoff IL. Origin of milk cholesterol in the rat: dietary versus endogenous sources. J Lipid Res. 1966;7:27–37. [PubMed] [Google Scholar]
- 37.Chiba H, Osanai M, Murata M, Kojima T, Sawada N. Transmembrane proteins of tight junctions. Biochim Biophys Acta. 2008;1778:588–600. doi: 10.1016/j.bbamem.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 38.Stelwagen K, Singh K. The role of tight junctions in mammary gland function. J Mammary Gland Biol Neoplasia. 2014;19:131–8. doi: 10.1007/s10911-013-9309-1. [DOI] [PubMed] [Google Scholar]
- 39.Flanders KC, Wakefield LM. Transforming growth factor-βs and mammary gland involution; functional roles and implications for cancer progression. J Mammary Gland Biol Neoplasia. 2009;14:131–44. doi: 10.1007/s10911-009-9122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russo J, Russo IH. Development of the human breast. Maturitas. 2004;49:2–15. doi: 10.1016/j.maturitas.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 41.Russo IH, Russo J. Pregnancy-induced changes in breast cancer risk. J Mammary Gland Biol Neoplasia. 2011;16:221–33. doi: 10.1007/s10911-011-9228-y. [DOI] [PubMed] [Google Scholar]
- 42.Peri S, de Cicco RL, Santucci-Pereira J, Slifker M, Ross EA, Russo IH, et al. Defining the genomic signature of the parous breast. BMC Med Genomics. 2012;5:46. doi: 10.1186/1755-8794-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uehara N, Unami A, Kiyozuka Y, Shikata N, Oishi Y, Tsubura A. Parous mammary glands exhibit distinct alterations in gene expression and proliferation responsiveness to carcinogenic stimuli in Lewis rats. Oncol Rep. 2006;15:903–11. [PubMed] [Google Scholar]
- 44.Blakely CM, Stoddard AJ, Belka GK, Dugan KD, Notarfrancesco KL, Moody SE, et al. Hormone-Induced Protection against Mammary Tumorigenesis Is Conserved in Multiple Rat Strains and Identifies a Core Gene Expression Signature Induced by Pregnancy. Cancer Res. 2006;66:6421–31. doi: 10.1158/0008-5472.CAN-05-4235. [DOI] [PubMed] [Google Scholar]
- 45.Watson CJ. Involution: apoptosis and tissue remodelling that convert the mammary gland from milk factory to a quiescent organ. Breast Cancer Res BCR. 2006;8:203. doi: 10.1186/bcr1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faure E, Heisterkamp N, Groffen J, Kaartinen V. Differential expression of TGF-beta isoforms during postlactational mammary gland involution. Cell Tissue Res. 2000;300:89–95. doi: 10.1007/s004410000183. [DOI] [PubMed] [Google Scholar]
- 47.D’Cruz CM, Moody SE, Master SR, Hartman JL, Keiper EA, Imielinski MB, et al. Persistent parity-induced changes in growth factors, TGF-beta3, and differentiation in the rodent mammary gland. Mol Endocrinol Baltim Md. 2002;16:2034–51. doi: 10.1210/me.2002-0073. [DOI] [PubMed] [Google Scholar]
- 48.Schedin P, Mitrenga T, McDaniel S, Kaeck M. Mammary ECM composition and function are altered by reproductive state. Mol Carcinog. 2004;41:207–20. doi: 10.1002/mc.20058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.