Abstract
Ewing sarcoma (ES) is the second most common bone malignancy affecting children and young adults with poor prognosis due to high metastasis incidence. Our group previously described that EphA2, a tyrosine kinase receptor, promotes angiogenesis in Ewing sarcoma (ES) cells via ligand-dependent signaling. Now we wanted to explore EphA2 ligand-independent activity, controlled upon phosphorylation at S897 (p-EphA2S897), as it has been linked to metastasis in several malignancies. By reverse genetic engineering we explored the phenotypic changes after EphA2 removal or reintroduction. Gene expression microarray was used to identify key players in EphA2 signaling. Mice were employed to reproduce metastatic processes from orthotopically implanted engineered cells. We established a correlation between ES cells aggressiveness and p-EphA2S897. Moreover, stable overexpression of EphA2 in low EphA2 expression ES cells enhanced proliferation and migration, but not a non-phosphorylable mutant (S987A). Consistently, silencing of EphA2 reduced tumorigenicity, migration and invasion in vitro, and lung metastasis incidence in experimental and spontaneous metastasis assays in vivo. A gene expression microarray revealed the implication of EphA2 in cell signaling, cellular movement and survival. ADAM19 knockdown by siRNA technology strongly reproduced the negative effects on cell migration observed after EphA2 silencing. Altogether, our results suggest that p-EphA2S897 correlates with aggressiveness in ES, so blocking its function may be a promising treatment.
Keywords: EphA2, Ewing sarcoma, metastasis, ADAM19
Introduction
Generally, Ewing sarcoma (ES) tumors, the second most frequent bone tumors among children and adolescents, are very aggressive and, although chemosensitive, tend to recur and metastasize. The survival of ES patients presenting with metastasis at diagnosis is poor (20%) 1. The lack of knowledge regarding the molecular mechanisms that regulate the metastatic process is the main reason for the lack of efficient therapeutics for these patients.
Ephrin (Eph) receptors are the most extensive subfamily of receptor tyrosine-kinases (RTKs) involved in several processes, including angiogenesis, tissue-border formation, cell migration, and cell plasticity 2. These receptors are well-established mediators in cell-cell interactions and motility and are expressed in human cancers, including melanoma, prostate, breast, colon, lung, and esophageal carcinomas 3. Among these receptors, EphA2 plays important roles in oncogenesis, metastasis, and treatment resistance 4. In ES, we demonstrated the involvement of EphA2 in tumor progression by promoting angiogenesis in a ligand-dependent manner 5. Moreover, EphA2 has been previously implicated in vascular mimicry, an important process that contributes to ES malignancy and worse prognosis 6. However, EphA2 also exhibit tumor progression properties by acting in a ligand-independent fashion, promoting metastasis in several types of tumors 7–9.
Caveolin-1 (CAV1), a structural protein responsible for the formation of caveolae, plays a crucial role in the development of metastasis in melanomas, prostate, breast, and colon cancer 10. Our group also demonstrated the implication of CAV1 in the development of ES metastasis 11,12. Additionally, we also showed that CAV1 interacts with EphA2 in ES cells to promote angiogenesis in a kinase-dependent manner 5, underscoring the importance of this receptor in the complex signaling pathways that control metastasis through, perhaps, kinase-independent activities.
Here, we show that EphA2 is indeed phosphorylated at S897 in most ES cell lines. CAV1 knockdown results in a significant loss of S897 phosphorylation, demonstrating a clear relationship between both proteins. Furthermore, using site-directed mutagenesis and gain- and loss-of-function experiments, we demonstrated that EphA2 is essential for the aggressive properties of ES in a kinase-independent manner. Therefore, blocking EphA2 expression or its function with drugs or genetic tools may be of therapeutic use for the treatment of ES.
Material and Methods
Cell culture and transfection
Ewing Sarcoma cell lines: A673, STA-ET-1, TC252, and CADO-ES (gifts from Dr. Heinrich Kovar), EW7 (gift from Dr. Olivier Delattre), RH1 (gift from Dr. Peter Houghton), A4573 (gift from Dr. Santiago Ramón y Cajal), SK-N-MC, TC71, RD-ES, and SK-ES-1 (bought from Leibniz Institute DSMZ). Cell lines were cultured in RPMI 1640-GlutaMAX (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Life Technologies) and 1% penicillin-streptomycin (Life Technologies). All cell lines were incubated at 37°C in a humidified atmosphere of 5% CO2 in air and checked regularly for mycoplasma infection. Exponentially growing cells were used for all experiments.
Cells were stably transfected using Lipofectamine 2000 (Life Technologies) following the manufacturer’s protocol. RH1 transfected cells stably expressing pCMV6-EphA2 Myc-DKK tagged vector (Origene #RC205725) were selected with 0.6 mg/mL of neomycin (Life Technologies). A673 and TC252 cells stably transfected with pRS-shEphA2 vector (Origene #TR320327) were selected with 0.5 μg/mL and 0.2 μg/mL of puromycin (Sigma-Aldrich), respectively, for 14 days. Antibiotic-resistant pools and individual clones were isolated for further analysis and maintained in the presence of antibiotics.
For transient gene silencing, cells were transfected using Dharmafect (GE Healthcare) using 100 nM customized siRNA for ADAM19 (5′-GCUCCUUCCUACACAGAAA-3′).
Clinical material
For testing the expression of EphA2 and p-EphA2S897 in Tissue Micro Arrays (TMAs), tumor samples from a recent published study were used 13. Written informed consent was obtained from each patient at the Hospital Universitario Virgen del Rocio, Seville, Spain. Staining of EphA2 and p-EphA2S897 in the TMA was scored by a blinded trained pathologist on a positive-negative scale. EphA2 staining at endothelia was considered as an internal positive control. Immunohistochemical (IHC) techniques were done as previously described 11. Expression of EphA2 and p-EphA2S897 in TMA’s was analyzed using the same antibodies used for western blotting.
Cell treatments
For MEK and Akt inhibitors treatments, cells were seeded and incubated with the corresponding drug 24 h later. MEK inhibitors: 20 μM U0126 (LC Laboratories) for 24–48 h and 50 nM Trametinib (LC Laboratories) for 24 h. Akt inhibitors: BKM-120 (Selleckchem) and MK22-06 (Selleckchem) for 24 h.
Site-directed mutagenesis
Mutagenesis was performed using QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies) following the manufacturer’s protocol. pCMV6-EphA2 Myc-DKK tagged vector was used as a template. In order to prevent residue phosphorylation, EphA2 serine (S) 897 residue was changed into alanine (A), using the following forward (5′-GTGTCTATCCGGCTCCCCGCCACGAGCGGCTCGGAGG-3′) and reverse (5′-CCTCCGAGCCGCTCGTGGCGGGGAGCCGGATAGACAC-3′) primers. After mutagenesis, the full cDNA was sequenced to verify the S897 to A897 mutation and to confirm the absence of other possible unspecific mutations in the coding sequence. The mutant vector was named S897A.
Clonogenic assay
For clonogenic assays, 500 cells were seeded in the wells of a 6-well plate. When colonies reached saturation, approximately 14 days after seeding, cells were fixed with cold methanol for 10 min, washed with Dulbecco’s Phosphate Buffered Saline (PBS, Biowest), stained with crystal violet (Sigma-Aldrich) for 20 min, and washed with water. The total colony number was manually counted using ImageJ. In some cases, colonies were discolored with a 10% glacial acetic acid solution and crystal violet was quantified by spectrometry.
Proliferation assay
For proliferation assays, 5000 cells were seeded in the wells of a 96-well plate. At 24, 48, 72, and 96 h after seeding, the culture medium was removed and 100 μL of a 1:10 dilution of Water Soluble Tetrazolium (WST-1, Roche) in the medium was added to each well. After 60 and 120 min, cell viability was quantified by spectrometry and extrapolated as a measure of cell proliferation.
Transwell migration assay
Cells were harvested as usual. After an additional wash with RPMI, 1.5 × 105 cells in 150 μL serum-free medium were added to the top chamber of 8-μm pore polycarbonate transwells (Transwell Permeable Supports-Corning). Meanwhile, in the bottom chamber, 500 μL of complete medium (10% FBS) were added. For the migration assays in the presence of 50 nM Trametinib, cells were pre-treated with the drug the 24 h prior seeding and Trametinib was added to both chambers. For the migration assays in the presence of siRNA, cells were transfected 6 h prior tripsinization and seeding. After 24 h for A673, 48 h for TC252, 72 h for RH1, and 48 h for the cellular panel comparison, cells on the upper chamber were removed with a cotton swab. Migrating cells still attached on the membrane’s underside were fixed for 30 min using 70% ethanol and stained with crystal violet. Transwell membranes were collected and 5 pictures of each transwell were acquired by optical microscopy (100×). Generally, membranes were discolored with a 10% glacial acetic acid solution and crystal violet was quantified by spectrometry. In some instances we opted for a direct manual counting of the number of migrating cells in the membrane using ImageJ. Results are presented as the percentage of a designated control condition.
Matrigel invasion assay
The procedure was the same to the migration assay, but transwells were previously coated with 50 μL of cold Matrigel (BD Biosciences) diluted 1:20 in RPMI and placed in a 37°C incubator for 6 h. After Matrigel polymerization, cells were seeded, stained, and counted as in the migration assay. The reaction was stopped after 48 h for A673 and 72 h for TC252 and RH1. For the assays in the presence of 50 nM Trametinib, cells were pre-treated with the drug the 24 h prior seeding and Trametinib was added to both chambers. For the assays in the presence of siRNA, cells were transfected 6 h prior tripsinization and seeding.
Real-Time Cell Analysis of invasive behavior
Cell invasion was also analyzed using RTCA (Real-Time Cell Analysis) xCELLigence system (ACEA Biosciences). Cells (4 × 104) in 100 μL RPMI medium (FBS-free) were seeded onto CIM (cell invasion and migration) plates (ACEA Biosciences) previously coated with 30 μL of cold Matrigel (BD Biosciences) diluted 1:20 in RPMI. One hundred microliters of RPMI containing a 10% FBS were added to the bottom chamber.
CIM plates consist of Boyden chambers coupled to microelectronic sensors in the bottom part of the membrane that measure electrical impedance. Results are reported as cell index, a dimensionless parameter correlated with cell migration. Continuous measures were taken every 15 min, allowing monitoring of the cell invasion status of different cell lines simultaneously over time.
Western blot
Cells were lysed with RIPA Buffer (ThermoFisher Scientific) containing protease inhibitors (Complete, Mini; Protease Inhibitor Cocktail Tablets, Roche) and phosphatase inhibitors (PhosStop, Phosphatase Inhibitor Cocktail Tablets, Roche) for 30 min on ice. Lysates were sonicated, centrifuged at 13000 rpm at 4°C for 30 min, and supernatants recovered. Samples (50 μg) were resolved by 8, 10, or 12% SDS-PAGE and transferred onto nitrocellulose membranes (0.2 μm, Bio-Rad). Membrane blocking was performed with 5% skimmed milk in PBS containing 0.1% Tween20 (Sigma-Aldrich) at room temperature for 1 h. Next, membranes were incubated overnight at 4°C with the appropriate primary antibody (EphA2 1:1000 #6997, Phospho-EphA2 Ser897 1:1000 #6347, ERK1/2 1:2000 #4695, Phospho-ERK1/2 Thr202/Tyr204 1:1000 #4376, Akt 1:2000 #9272, Phospho-Akt Ser473 1:1000 #4060 from Cell Signaling; CAV1 1:10000 #610059 from BD Transduction Lab; and ADAM19 1:500 #ab104800 from Abcam;). Blots were then incubated at room temperature for 1 h with a horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit and goat anti-mouse, Life Technologies) and the peroxidase activity was detected by enhanced chemiluminescence (Pierce) following the manufacturer’s instructions. Immunodetection of α-tubulin (#ab28439) or β-actin (#ab49900) from Abcam was used as a loading control.
RNA extraction and reverse transcription-PCR (RT-PCR)
Total RNA (2 μg), extracted by using the NucleoSpin RNA or the NucleoSpin miRNA (for Microarray purpose) from Macherey-Nagel, was used for cDNA synthesis with SuperScript II Reverse Transcriptase (Life Technologies).
Gene expression analysis-Microarray
Gene expression microarray was performed at Servei d’Anàlisi de Microarrays (Institut Hospital del Mar d’Investigacions Mèdiques, Barcelona, Spain). Amplification, labeling, and hybridization were performed according to the GeneChip WT PLUS Reagent kit protocol, and then the samples were hybridized to GeneChip Human Gene 2.0 ST Array (Affymetrix) in a GeneChip Hybridization Oven 640. Washing and scanning were performed using the Expression Wash, Stain, and Scan Kit and the GeneChip System of Affymetrix (GeneChip Fluidics Station 450 and GeneChip Scanner 3000 7G). After quality control of raw data, they were background corrected, quantile-normalized, and summarized to a gene-level using the robust multi-chip average (RMA) 14 obtaining a total of 48,144 transcript clusters, excluding controls, which roughly correspond to genes or other mRNAs as miRNAs or lincRNAs. Linear Models for Microarray (limma) 15, a moderated t-statistics model, was used for detecting differentially expressed genes between the conditions. Correction for multiple comparisons was performed using false discovery rate (FDR) 16. Genes with a p-value less than 0.05 with an absolute fold change above 1.5 were selected as significant. Functional analyses were performed with Ingenuity Pathway Analysis v 9.0 (Ingenuity® Systems, www.qiagenbioinformatics.com) and Gene Set Enrichment Analysis 17,18 with collections C2, C4, and C7. All analyses were performed in R (v 3.1.1, http://www.R-project.org/) with packages aroma.affymetrix 19, Biobase 20, and limma 21.
Quantitative Real Time PCR
Quantitative reverse transcription-PCR (qRT-PCR) was performed under universal cycling conditions on LightCycler 480 II instrument (Roche) using TaqMan PCR Mastermix and TaqMan probes from Life Technologies (ACTB 4333762F, ADAM19 Hs00224960_m1, CCL2 Hs00234140_m1, LUM Hs00929860_m1, PCDH8 Hs04187285_g1, PI3KCG Hs00277090_m1, and PTPN21 Hs00234784_m1). Cycle threshold (CT) values were normalized to that of β-actin. Relative expression level of the target gene among the different samples was calculated using the ΔΔCT method 22.
Subcutaneous tumor induction
In vivo tumors were induced with subcutaneous injections of 5 × 106 A673 wild type or EphA2 silenced cells, resuspended in 100 μL of RPMI. Injections were performed in the hind legs of 6-week-old female athymic Balb/cnu/nu nude mice (Harlan). In each mouse, control cells were injected in the left flank and EphA2 silenced cells in the right one. Tumor volumes were measured three times per week and calculated according to the following equation: volume = 1/2 × (length × width2). When tumors reached a volume of 1 cm3, mice were euthanized and tumors were recovered for further analysis. Tumors were cut in half and one half was frozen, while the other half was fixed in paraformaldehyde at 4% and embedded in paraffin. Animals were cared for according to the Institutional Guidelines for the Care and Use of Laboratory Animals. Ethics approval was provided by local appointed ethics committee from IDIBELL, Barcelona, Spain. n = 8 for each condition.
Experimental metastasis assay
A673 wild type or EphA2 silenced cells (2 × 106), resuspended in 100 μL of PBS were injected intravenously in the tail vein of 6-week-old female athymic BALB/cnu/nu nude mice (Harlan). The presence of morbidity symptoms was checked every 48 h. Sixty days after injection, mice were euthanized and lungs were recovered to examine the presence of metastasis. Lungs were fixed in 4% paraformaldehyde and embedded in paraffin. Lung sections were stained with hematoxylin & eosin and metastases were counted under an optical microscope. Animal care procedures were followed as described above. n = 10 for each condition.
Orthotopic xenograft metastasis assay
This assay was performed as previously described 12. Briefly, 2 × 106 cells resuspended in 100 μL of PBS were injected into the gastrocnemius muscles of 6-week-old female athymic nude mice (BALB/cnu/nu) from Harlan. Once primary tumor-bearing limbs reached a volume of 800 mm3, the gastrocnemius muscles were surgically resected. At day 60 after injection, mice were euthanized and lungs were fixed in 4% paraformaldehyde and embedded in paraffin. Lung sections were stained with hematoxylin & eosin and metastases were counted under an optical microscope. n = 7 for each condition.
Statistical analysis
Unless otherwise stated, data were analyzed for statistical significance using Student’s t test. Fisher’s exact test was used for evaluating differences in lung metastasis incidence in mice. Experiments were performed thrice; p ≤ 0.05 was regarded as significant.
Results
Phosphorylation of EphA2 at serine 897 correlates with the migratory capacity of ES cells
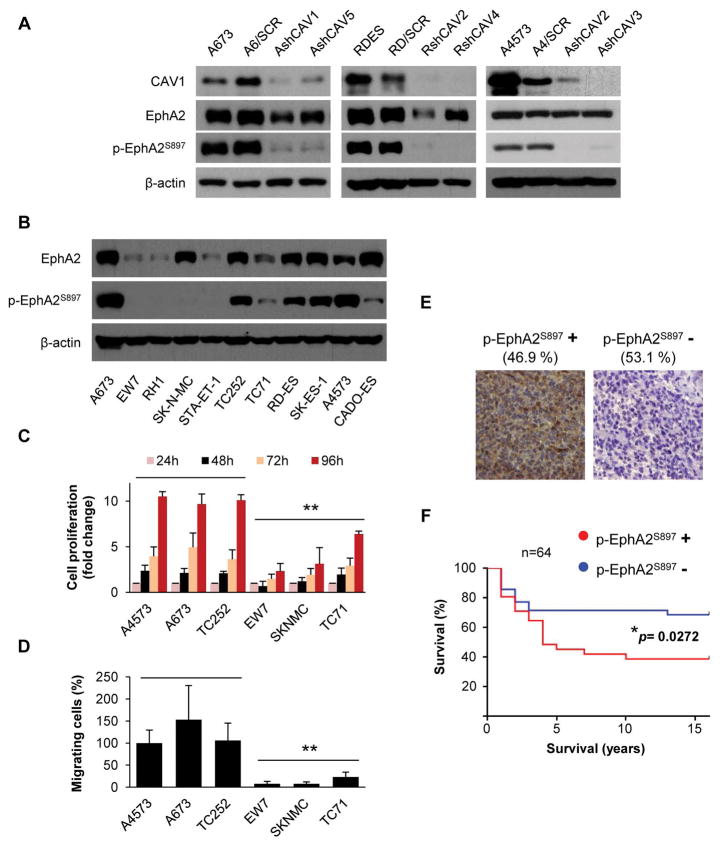
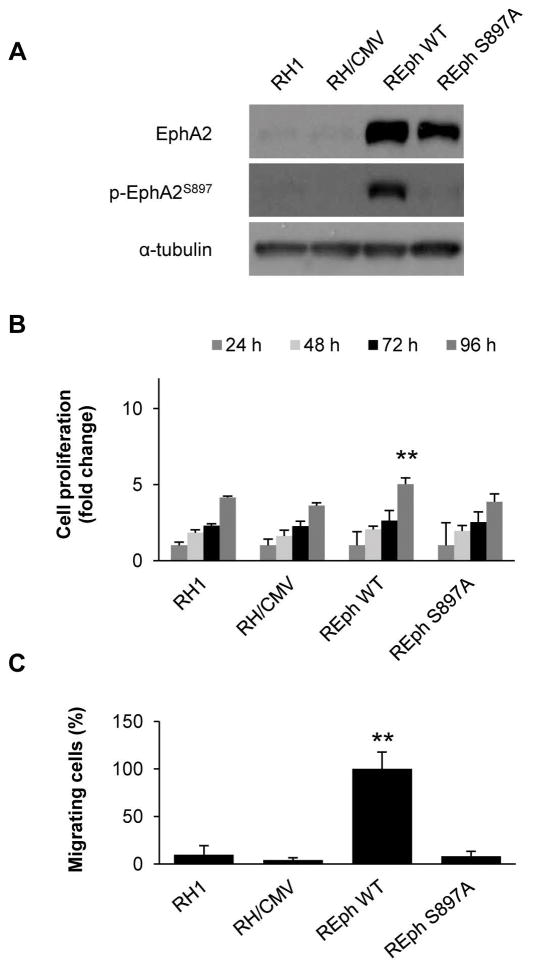
Phosphorylation of EphA2 on S897 (p-EphA2S897) has been linked to a higher migratory capacity of cancer cells 23. This property is independent of ligand binding to the receptor 7–9. Our group previously showed that CAV1 promotes metastasis in ES 11 and demonstrated its relationship with EphA2, promoting angiogenesis 5. Indeed, CAV1 silencing resulted in de-phosphorylation of EphA2 in three previously published models (Fig. 1A). Moreover, EphA2 phosphorylation levels at S897 were high in a panel of ES cell lines (Fig. 1B). We compared the proliferation and migration abilities between three ES cell lines with highly phosphorylated EphA2 and three ES cell lines with lower levels of phosphorylation, showing that cells with high levels of p-EphA2S897 proliferated and migrated significantly more than those with lower phosphorylation levels (Fig.1C–D), suggesting that p-EphA2S897 confers advantage towards a more aggressive phenotype in ES. We further used IHC on TMAs to analyze the expression and phosphorylation status of EphA2 in a panel of 86 ES patients (Fig. S1A and Fig. 1E). Most of the evaluable ES samples (73) expressed EphA2 (90.4%). However, only 46.9% of the 64 evaluable samples were positive for p-EphA2S897. Interestingly, Kaplan-Meier method compared by Long-rank (Mantel-Cox test) analysis showed only lower overall survival in patients that were positive for p-EphA2S897, p=0.0272 (Figure S1B and Fig. 1F). Moreover, 14 out of the 64 evaluated patients presented metastasis. Of those patients, 71.4% died because of disease and 60% of them (10) were positive for p-EphA2S897. In contrast, 75% of the patients with metastasis and still alive (3 out of 4) were negative for p-EphA2S897. To further demonstrate the relationship between p-EphA2S897 and a more aggressive phenotype, we stably transfected RH1 cells with a wild type (wt) EphA2 construct (REph WT) and a non-phosphorylatable S897A EphA2 mutant construct (REph S897A) (Figs. S2 and 2A). Indeed, only cells transfected with the EphA2 wt construct gained significant proliferation and migration abilities (Fig. 2B–C).
Figure 1. EphA2 receptor phosphorylation on S897 correlates with aggressiveness.
(A) Representative western blot showing EphA2 phosphorylation in Caveolin-1 (CAV1) silenced clones of A673, RDES, and A4573 cells. SCR = control scrambled sequence. (B) Representative western blot showing total EphA2 expression and its phosphorylation at S897 residue in a panel of ES cell lines. (C) WST1 tetrazolium-based proliferation assay comparing cell lines with high levels of p-EphA2S897 (A4573, A673, and TC252) versus those with low p-EphA2S897 (EW7, TC71, and SK-N-MC). (D) Migration assay in Boyden chambers in the selected panel of high or low p-EphA2S897 expression cell lines. A4573 cell line was set as reference. (E) Sample micrographs from the Ewing sarcoma tissue microarray showing differential expression pattern of p-EphA2S897. Magnification: 40×. Data are presented as means ± SD. Statistical significance was achieved by the Student’s t test from at least three different experiments: *p ≤ 0.05 **p ≤ 0.01. (F) Kaplan-Meier curve comparing differential survival of Ewing sarcoma patients in function of p-EphA2S897 expression. Long-rank (Mantel-Cox test) analysis was used to generate p value.
Figure 2. Only reintroduction of a phosphorylable form of EphA2 is able to promote aggressiveness in RH1 cells.
(A) Western blot showing EphA2 expression and its phosphorylation at S897 in RH1 reintroduction model. CMV = empty vector. (B) WST1 tetrazolium-based proliferation assay for the RH1 EphA2 reintroduction model. (C) Migration assay in Boyden chambers for the RH1 EphA2 reintroduction model. REphWT was set as reference. Data are presented as means ± SD. Statistical significance was achieved by the Student’s t test from at least three different experiments: *p ≤ 0.05 **p ≤ 0.01.
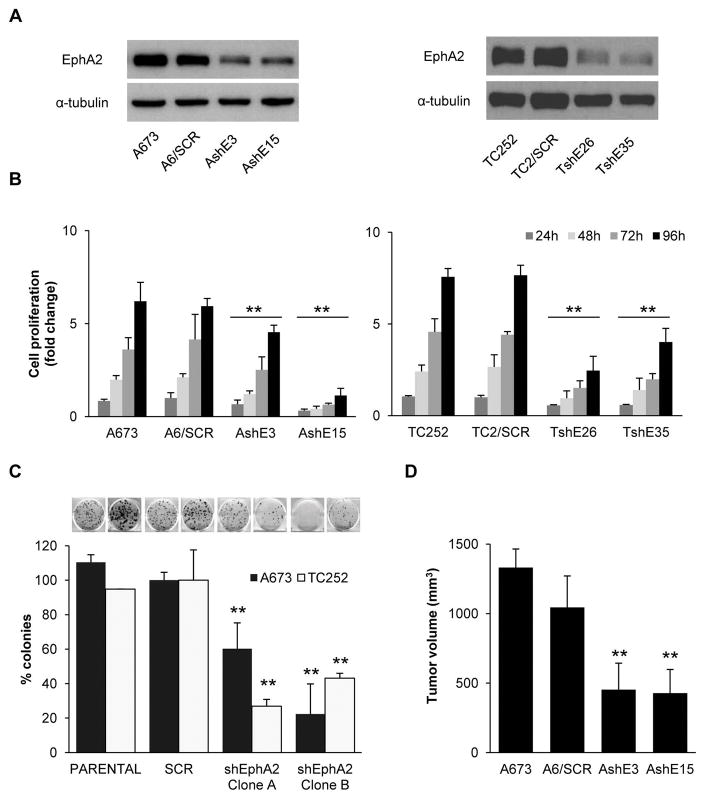
EphA2 silencing results in decreased cell viability, clonogenic capacity, and tumor growth
To confirm the involvement of EphA2 in the progression of ES, we established two silenced models of EphA2 using A673 and TC252 cells by transfecting them with a shRNA against EphA2 or a shRNA control (Fig. 3A). EphA2 knockdown in both cell lines significantly reduced both cell proliferation, as measured by the WST1 tetrazolium-based assay, and clonogenic capacity (Fig. 3B–C). Furthermore, EphA2 silencing resulted in reduced tumor growth in nude mice (Fig. 3D). EphA2 knockdown in the resulting tumors was demonstrated by western blot (Fig. S3).
Figure 3. EphA2 knockdown reduces ES cell viability.
(A) Representative western blot showing EphA2 expression in silencing models generated from A673 and TC252 cell lines. (B) Proliferation assay using the WST1 tetrazolium-based assay. (C) Quantification of clonogenic assay. Representative images are shown above the bars. “Clone A” and “B” denote clones AshE3 and TshE26 and AshE15 and TshE35, respectively. (D) In vivo tumor growth in immunodepressed mice after subcutaneous injection of cells. Tumor size at the time of euthanasia is shown; n=8. SCR = control scrambled sequence. AshE#/TshE# = EphA2 silenced selected clones. Data are presented as means ± SD. Statistical significance was achieved by the Student’s t test from at least three different experiments: *p ≤ 0.05 **p ≤ 0.01.
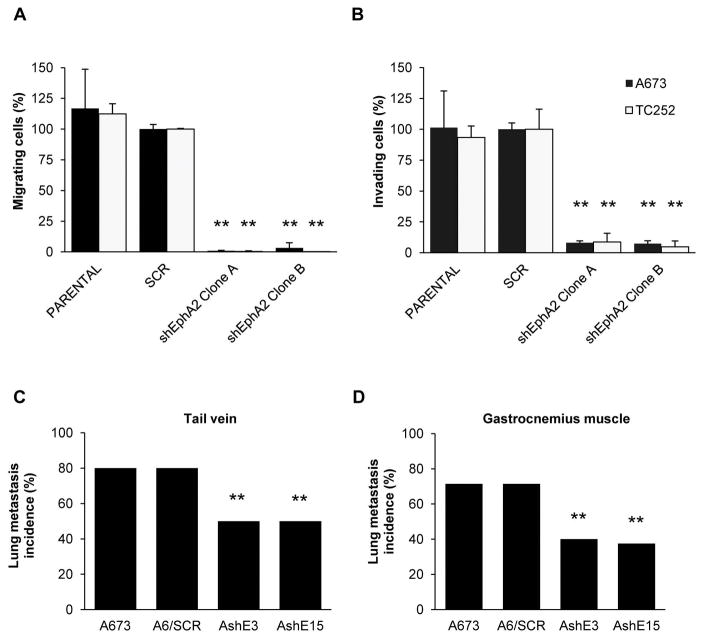
EphA2 silencing results in decreased cell migration, invasion and metastasis in vivo
As observed with CAV1 silencing 5, EphA2 knockdown resulted in a significant reduction of the migratory and invasive capacity of ES cells in vitro (Fig. 4A–B and S4A). Moreover, we performed an experimental metastasis assay, injecting tumor cells through the tail vein of nude mice and observed a reduction in the incidence of lung metastasis when EphA2 silenced cells were injected (Fig. 4C). To further study this process, we used a previously described orthotopic model 12, in which tumor cells were injected in the gastrocnemius muscle. Consistently, a significant decrease in the incidence of lung metastasis (Fig. 4D and S4B) and in the number of metastasis per mice was observed (Fig. S4C). These results strongly suggest that aggressive progression of ES cells highly depends on EphA2 signaling.
Figure 4. EphA2 knockdown reduces ES cells motility.
(A) Migration assay in Boyden chambers using EphA2 silenced models. “Clone A” and “B” denote clones AshE3 and TshE26 and AshE15 and TshE35, respectively. SCR models were set as reference. (B) Invasion assay in Matrigel-coated Boyden chambers using EphA2 silenced models. “Clone A” and “B” denote clones AshE3 and TshE26 and AshE15 and TshE35, respectively. SCR models were set as reference. (C) Lung metastasis incidence in immunodepressed mice after tail-vein injection of control or EphA2 silenced A673 cells; n = 10. (D) Lung metastasis incidence in immunodepressed mice after injection of tumor cells in the gastrocnemius; n = 7. SCR = control scrambled sequence. Data are presented as means ± SD. Statistical significance was achieved by the Student’s t test from at least three different experiments. Fisher’s exact test was used for evaluating differences in lung metastasis incidence in mice: *p ≤ 0.05 **p ≤ 0.01.
EphA2 ligand-independent signaling induces a reciprocal regulatory feedback loop involving the ERK signaling pathway
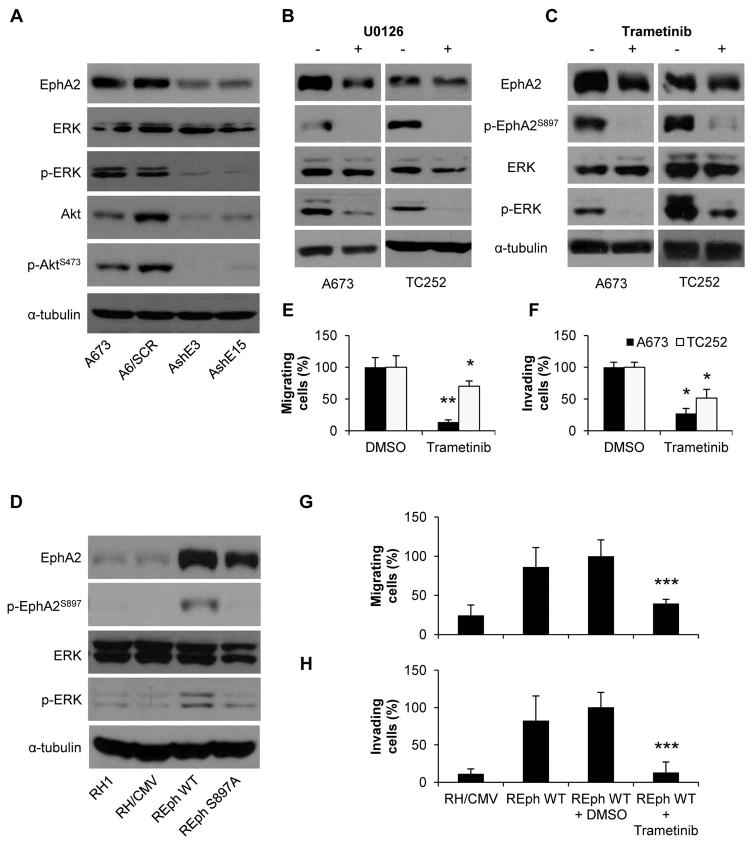
The EphA2 receptor, through its ligand-independent signaling, induces a reciprocal regulatory feedback loop involving the Akt kinase. This loop leads to a tumor progressive phenotype due to activation of the RAS/ERK and PI3K/Akt signaling pathways 8. To test whether this was the case in our model, we screened for changes in either Akt or ERK kinase pathway after EphA2 knockdown. Results showed that Akt expression was altered and so its phosphorylation. However, in the case of ERK, only the phosphorylation was affected by the loss of EphA2 (Fig. 5A). To verify the possibility of a reciprocal regulatory feedback loop involving ERK and/or Akt kinases, we treated A673 and TC252 cells with two MEK inhibitors (known for decreasing ERK phosphorylation, U0126 and Trametinib) and two Akt inhibitors (MK-2206 and BKM120). Inhibition of the MAPK pathway by both inhibitors greatly reduced the phosphorylation of EphA2 at S897 (Fig. 5B–C). In contrast, only in TC252 some effect was observed in cells treated with BKM120 (Fig. S5A–B). In addition, only REph WT cells showed an increase in ERK phosphorylation (Fig. 5D) that correlated with their augmented aggressive behavior depicted in Figure 1. No such clear increase was observed in AKT phosphorylation (Fig. S5C). Moreover, Trametinib treatment abrogates both motility and invasive capacity either in EphA2 naturally expressing cell lines (Figs. 5E–F) or in the EphA2 knock-in RH1 model (Fig. 5G–H). Taken together, these results strongly suggest the presence of a relevant reciprocal regulatory loop between EphA2 and ERK, with non-ligated EphA2 functioning as a downstream substrate and effector of ERK kinase.
Figure 5. EphA2 has a reciprocal regulatory phosphorylation feed-back loop with ERK, independent of the presence of the ligand.
(A) Representative western blot showing ERK, p-ERK, Akt, and p-AktS473 levels in EphA2 silenced clones of the A673 cell line. SCR = control scrambled sequence. AshE#/TshE# = EphA2 silenced selected clones. (B–C) Representative western blots showing EphA2 and p-EphA2S897 levels after U0126 (B) and Trametinib (C) treatments in A673 and TC252 cell lines. Phosphorylation of ERK (B and C) is shown as a control of treatment efficiency. (D) Representative western blot showing EphA2, p-EphA2897, ERK, and p-ERK in the RH1 reintroduction model. CMV = empty vector. (E) Migration assay in Boyden chambers for A673 and TC252 cell lines after 50 nM Trametinib treatment. DMSO (vehicle) conditions were set as reference. (F) Invasion assay in Matrigel-coated Boyden chambers for A673 and TC252 cell lines after 50 nM Trametinib treatment. DMSO (vehicle) conditions were set as reference. (G) Migration assay in Boyden chambers in the RH1 EphA2 reintroduction model. REphWT plus DMSO (vehicle) condition was set as reference. (H) Invasion assay in Matrigel-coated Boyden chambers in the RH1 EphA2 reintroduction model. REphWT plus DMSO (vehicle) condition was set as reference. Data are presented as means ± SD. Statistical significance was achieved by the Student’s t test from at least three different experiments. Fisher’s exact test was used for evaluating differences in lung metastasis incidence in mice: *p ≤ 0.05 **p ≤ 0.01 ***p ≤ 0.001.
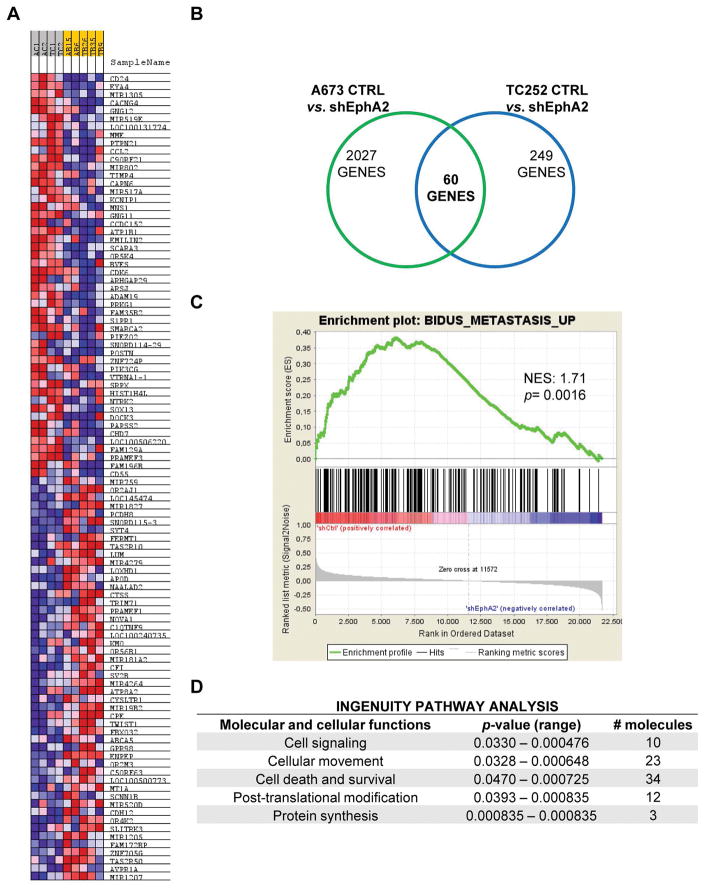
Microarray analysis confirms an implication of EphA2 in cell signaling, cellular movement, and survival
To gain insight into the molecular mechanism of EphA2-mediated tumorigenesis and metastasis, we identified EphA2 regulated genes by microarray profiling comparing A673 and TC252 control cells with EphA2 silenced cells (Fig. 6A). Sixty genes that commonly overlapped between both cell lines were differently expressed in EphA2 silenced cells (Fig 6B and Supporting Table 1). To test whether EphA2 silencing in silico reproduces the phenotypic effects on motility and invasion, we used gene set enrichment analysis (GSEA). Genes that positively correlated with metastasis were associated with EphA2 expression (Fig. 6C). Of note, a strong correlation with EGFR signaling was observed (Fig. S6), suggesting a possible crosstalk between the EphA2 and EGFR pathways. By using another network/pathway enrichment analysis, the Ingenuity Pathway Analysis (IPA), we observed that genes downregulated after EphA2 inhibition were mainly enriched in cell signaling, cellular movement, and survival categories (Fig. 6D).
Figure 6. EphA2 participates in several regulatory pathways linked to metastasis.
(A) Heatmap of up- (red) and down- (blue) regulated genes in control cells vs. EphA2 silenced cells. (B) Venn Diagram showing the number of common genes differentially expressed after EphA2 knockdown in A673 and TC252 cell lines. (C) Gene Set Enrichment Analysis (GSEA) showing a correlation between genes differentially expressed after EphA2 silencing and those involved in metastasis. NES = Normalized Enrichment Score. (D) Ingenuity pathway analysis showing cellular processes significantly altered by EphA2 knockdown.
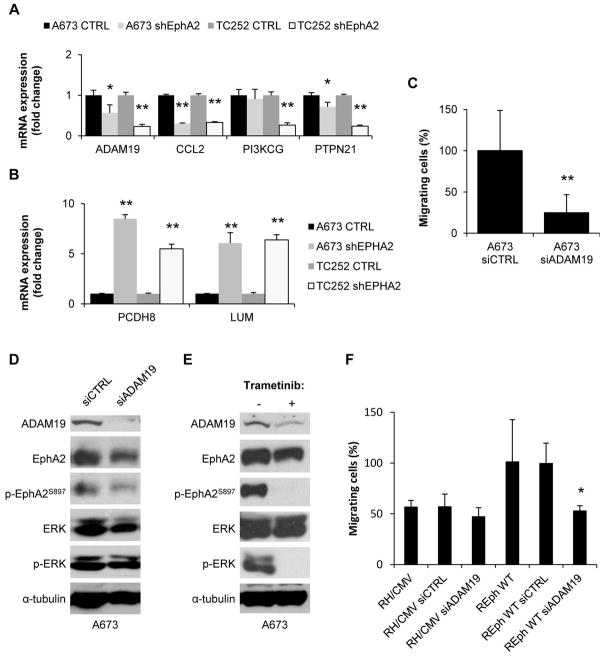
We validated a subset of these genes with quantitative reverse-transcriptase polymerase chain reaction (qRT–PCR) following EphA2 knockdown (Fig. 7A–B). This gene set included 4 downregulated genes (CCL2, ADAM19, PIK3CG, and PTPN21) and 2 upregulated genes (PCDH8 and LUM). We focused our attention on the metalloproteinase disintegrin, ADAM19, which is associated with invasiveness in several types of cancer and has not been studied in ES. ADAM19 knockdown by siRNA technology strongly reproduced the negative effects on cell migration observed after EphA2 silencing in A673 cells (Fig. 7C and Fig. S7A) but it had no significant effects on ERK and EphA2 phosphorylation (Fig. 7D) However, inhibition of the MAPK pathway by Trametinib resulted in ADAM19 downregulation in A673 cells (Fig. 7E and Fig. S7B). Furthermore, ADAM19 knockdown also reversed the aggressive phenotype induced by wild type EphA2 in the RH1 model (Fig. 7F and Fig. S7C). Again, this phenotype occurs without changes on ERK and EphA2 phosphorylation. Taking together, these results suggest that ADAM19 is critical for Ewing sarcoma aggressiveness and that EphA2 promotes its expression through MAPK signaling.
Figure 7. ADAM19 participates in the migratory behavior induced by EphA2 in ES cells.
(A–B) Validation of up- (A) and down- (B) regulated targets after EphA2 silencing by RT-qPCR in A673 and TC252 models. CTRL represents the mean from both the parental and the control vector transfected pool. shEphA2 represents the mean from the two representative clones employed through the previous experiments. (C) Migration assay in Boyden chambers after ADAM19 silencing in A673 cell line. siCTRL (= non-targeting siRNA) was set as reference. (D) Representative western blot showing ADAM19 expression and phosphorylation of EphA2 and ERK after siRNA silencing in A673 cell line. siCTRL = non-targeting siRNA. (E) Representative western blot showing ADAM19 expression and EphA2 phosphorylation after treatment with 50 nM Trametinib in the A673 cell line. ERK and p-ERK are shown as treatment efficiency controls. (F) Migration assay in Boyden chambers after ADAM19 silencing in RH1 EphA2 reintroduction model. REphWT plus siCTRL (= non-targeting siRNA) was set as reference. Data are presented as means ± SD. Statistical significance was achieved by the Student’s t test from at least three different experiments: *p ≤ 0.05 **p ≤ 0.01.
Discussion
The present study demonstrates that EphA2-expressing cells are essential for tumor propagation in ES, highlighting the importance of ligand-independent kinase activity of EphA2 in this process. Interestingly, our findings also indicate that EphA2 is reciprocally regulated by a feedback loop involving the ERK signaling pathway. The involvement of ERK in this feedback loop is novel and relevant to sarcomas. Transcriptome profiling revealed that EphA2 signaling activates a specific subset of genes that promote cell migration. EphA2 downregulation leads to a significant delay in ES onset and progression. Altogether, our findings suggest a rationale for inhibiting EphA2 expression and/or activity to block ES growth and migration. Our data represent a proof of principle for targeting EphA2-expressing cells for anti-metastatic therapy.
RTKs are known as key regulators of cancer cell proliferation, migration, invasion, and metastatic spread. Ligand-binding to the extracellular domain triggers canonical activation of the intracellular tyrosine-kinase domain. In contrast, it has become evident that RTKs are also regulated by non-canonical tyrosine-kinase independent mechanisms via phosphorylation of their serine/threonine residues 24. Here, we report, for the first time, the ligand-independent function of the EphA2 receptor during ES tumor progression. Indeed, ES cells with a more aggressive nature showed higher levels of EphA2 phosphorylation on S897 under the same culture conditions. Moreover, exogenous introduction of a wild type EphA2 construct, but not a S897A mutant, into low expressing cells increased their malignant phenotype. This phosphorylation site has been directly associated with aggressive progression in other cancer types 7,8,25 and is phosphorylated by activation of either the MAPK or Akt signaling pathway 8. In our models, EphA2 phosphorylation on S897 depends mostly on the MEK/ERK pathway as only MEK inhibitors were able to greatly block phosphorylation on this site. To further demonstrate the role of EphA2 in ES tumor progression, we established a low expressing model in two ES cell lines (A673 and TC252). Although widely affecting the behavior of ES cells, EphA2 silencing affected more prominently migration and invasion in vitro and tumor growth and metastasis spread in vivo. Consistently, the most relevant effect associated with the tumor progression properties of EphA2 in cancer cells is higher metastatic capacity 26. Furthermore, this trait has been linked to the ligand-independent activity associated with p-EphA2S897 7. Interestingly, EphA2 silencing in ES cells resulted in the dephosphorylation of ERK1/2, suggesting the existence of a reciprocal regulatory feed-back loop involving the ERK signaling pathway. We recently showed that MAPK signaling is important for the migration of ES cells in a model linking CAV1 expression and phosphorylation of ERK1/2 12. Here, p-EphA2S897 was significantly reduced in CAV1-silenced models underscoring the relationship between these proteins in the progression of ES.
To gain insights into the molecular mechanisms related to the EphA2 properties during ES progression, we performed expression arrays comparing EphA2 high expressing cells with EphA2 silenced cells. GSEA analysis showed a strong correlation between EphA2 and EGFR signaling pathways, consistent with several studies that indicated a relationship between these two pathways in which EphA2 promotes survival of cancer cells resistant to EGFR inhibition 27,28. There is no data about EGFR signaling during the progression of ES. Nevertheless, ErbB4, a member of the epidermal growth factor receptor subfamily, confers metastatic capacity to ES cells 29,30. Therefore, one could speculate that a crosstalk exists between EphA2 and ErbB4 during the progression of ES. GSEA and IPA analyses also confirmed a correlation between EphA2 and molecular traits related to metastasis progression. One of the top processes detected in the IPA analysis was axonal guidance. The axonal guidance pathway plays an important role in neuronal extension and location during embryo development 31. Recently, accumulating evidence indicates that the axonal guidance pathway is also involved in tumor development and progression by regulating tumor cell migration, cell death, and angiogenesis in various cancers 32.
Of a common set of 60 genes, we selected 6 of them for validation purposes. Four genes (CCL2, ADAM19, PIK3CG, and PTPN21), associated with tumor progression, were downregulated in EphA2 silenced cells, and 2 genes (PCDH8 and LUM), considered as tumor suppressors, were upregulated in EphA2 silenced cells. PCDH8 is a member of the cadherin family and is known to function as a candidate tumor suppressor, which is inactivated in many cancers 33,34. LUM is a member of the small leucine-rich proteoglycan (SLRP) family and significantly decreases cell migration, invasion, and anchorage-independent growth in vitro and metastasis in vivo 35. CCL2 is an inflammatory chemokine closely connected with tumor associated macrophage (TAM) infiltration and cancer progression. CCL2 is overexpressed in various cancer types such as lung, breast, and prostate cancer 36. CCL2 secreted by tumor cells can recruit monocytes and TAMs. These tumor-infiltrating inflammatory cells form a tumor-protective microenvironment, thus enhancing tumor growth and metastasis. PIK3CG and PTPN21 are cell signaling-associated proteins with roles in tumor progression 37,38. We focused on ADAM19, a member of the disintegrin and metalloproteinase family of proteins (ADAMs), which are involved in various biological functions such as fertilization, embryonic development, cell adhesion, cell migration, cell signaling, proteolytic shedding and proteolysis 39. Loss of function experiments demonstrated that ADAM19 knockdown reproduced the effects of EphA2 silencing on cell migration but not on cellular signaling. Our results suggest that ADAM19 participates in the metastatic progression of ES and is a downstream effector of the EphA2 ligand-independent signaling. In fact, deregulation of many ADAM proteins has been observed in the regulation of growth factor activities and integrin functions, leading to the promotion of cell growth and invasion in human tumors 40.
Our data suggest that EphA2 expression is necessary for ES metastatic spread. Interestingly, we demonstrated that EphA2 function is mediated via its ligand-independent kinase activity and that EphA2 is mostly reciprocally regulated by a feedback loop involving the ERK signaling pathway. Although, in this study, we focused on demonstrating that ADAM19 is a downstream effector of the EphA2 ligand-independent signaling in the metastatic progression of ES, we cannot disregard the involvement of other genes identified in the EphA2-dependent GSEA dataset. In fact, our data suggest a crosstalk between EphA2 and EGFR signaling pathways. In addition, CCL2 might be critical for promoting an ideal niche for metastatic progression. These results highlight the need for additional studies to ascertain the contribution of CCL2 in the metastatic process using humanized models and suggest the potential role of the immune system in metastatic progression. These findings suggest targeting of EphA2 and EphA2-mediated events as novel targets for ES therapy, opening an opportunity to test EphA2 small molecule agonists 41 alone or in combination with either antiangiogenic agents or targeted therapy against EGFR-related components overexpressed in the most aggressive forms of ES.
Supplementary Material
Novelty and Impact statement.
Ewing sarcoma (ES) tumors, the second most frequent bone tumors among children and adolescents, are very aggressive and, although chemosensitive, tend to recur and metastasize. Lack of knowledge regarding the molecular mechanisms that regulate the metastatic process is the main reason for the lack of efficient therapeutics for these patients. Here, by using gain and loss of function experiments and in vitro and in vivo assays, we established a correlation between ES aggressiveness and EphA2 phosphorylation.
Acknowledgments
The authors thank Giselle Blanco for her assistance editing the manuscript.
Funding: OMT: Fundación Alba Pérez lucha contra el cáncer infantil. Instituto de Salud Carlos III (CES12/021; PI11/00038; PI15/00035; AC14/00026) and EU’s Fondo Europeo de Desarrollo Regional (FEDER) “Una manera de hacer Europa/A way to achieve Europe”. SG-M & SR-V: Fundación Alba Pérez lucha contra el cáncer infantil. OA-R & DH-M: Fundación Científica de la AECC. PHG: National Institutes of Health (R01CA138503). Mary Kay Foundation (9033-12 & 001-09) and The Roy J Carver Charitable Trust (RJCCT 01-224).
Footnotes
Author contributions: OMT conceived and supervised the study. SG-M designed and performed the main experiments and coordinated the data analysis. SG-M and RL-A conceived and developed the orthotopical metastasis model. RL-A, OA-R, DH-M, JH-M, LL-T, PA-P, and SR-V contributed to the design and conduct of experiments. LH-P and EdA provided the TMA and contributed to the analysis. JM and EdA provided expertise and feedback. SG-M, SR-V, PHG, and OMT wrote the manuscript. All authors approved the final manuscript.
Competing interests: Authors declare no competing interests.
Availability of data and material: Additional data are presented as Supplementary Figures and Spreadsheets. The datasets supporting the conclusions of this article are available in the GEO repository; accessible as GSE106480.
References
- 1.Williams RF, Fernandez-Pineda I, Gosain A. Pediatric Sarcomas. Surg Clin North Am. 2016;96:1107–25. doi: 10.1016/j.suc.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Kania A, Klein R. Mechanisms of ephrin–Eph signalling in development, physiology and disease. Nat Rev Mol Cell Biol. 2016;17:240–56. doi: 10.1038/nrm.2015.16. [DOI] [PubMed] [Google Scholar]
- 3.Shiuan E, Chen J. Eph Receptor Tyrosine Kinases in Tumor Immunity. Cancer Res. 2016;76:6452–7. doi: 10.1158/0008-5472.CAN-16-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heinzlmeir S, Kudlinzki D, Sreeramulu S, Klaeger S, Gande SL, Linhard V, Wilhelm M, Qiao H, Helm D, Ruprecht B, Saxena K, Médard G, et al. Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs. ACS Chem Biol. 2016;11:3400–11. doi: 10.1021/acschembio.6b00709. [DOI] [PubMed] [Google Scholar]
- 5.Sáinz-Jaspeado M, Huertas-Martinez J, Lagares-Tena L, Martin Liberal J, Mateo-Lozano S, de Alava E, de Torres C, Mora J, Del Muro XG, Tirado OM. EphA2-induced angiogenesis in ewing sarcoma cells works through bFGF production and is dependent on caveolin-1. PLoS One. 2013;8:e71449. doi: 10.1371/journal.pone.0071449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Schaft DWJ, Hillen F, Pauwels P, Kirschmann DA, Castermans K, oude Egbrink MGA, Tran MGB, Sciot R, Hauben E, Hogendoorn PCW, Delattre O, Maxwell PH, et al. Tumor Cell Plasticity in Ewing Sarcoma, an Alternative Circulatory System Stimulated by Hypoxia. Cancer Res. 2005;65:11520–8. doi: 10.1158/0008-5472.CAN-05-2468. [DOI] [PubMed] [Google Scholar]
- 7.Paraiso KHT, Das Thakur M, Fang B, Koomen JM, Fedorenko IV, John JK, Tsao H, Flaherty KT, Sondak VK, Messina JL, Pasquale EB, Villagra A, et al. Ligand-independent EPHA2 signaling drives the adoption of a targeted therapy-mediated metastatic melanoma phenotype. Cancer Discov. 2015;5:264–73. doi: 10.1158/2159-8290.CD-14-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miao H, Li D-Q, Mukherjee A, Guo H, Petty A, Cutter J, Basilion JP, Sedor J, Wu J, Danielpour D, Sloan AE, Cohen ML, et al. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell. 2009;16:9–20. doi: 10.1016/j.ccr.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taddei ML, Parri M, Angelucci A, Onnis B, Bianchini F, Giannoni E, Raugei G, Calorini L, Rucci N, Teti A, Bologna M, Chiarugi P. Kinase-Dependent and -Independent Roles of EphA2 in the Regulation of Prostate Cancer Invasion and Metastasis. Am J Pathol. 2009;174:1492–503. doi: 10.2353/ajpath.2009.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Núñez-Wehinger S, Ortiz RJ, Díaz N, Díaz J, Lobos-González L, Quest AFG. Caveolin-1 in cell migration and metastasis. Curr Mol Med. 2014;14:255–74. doi: 10.2174/1566524014666140128112827. [DOI] [PubMed] [Google Scholar]
- 11.Sáinz-Jaspeado M, Lagares-Tena L, Lasheras J, Navid F, Rodriguez-Galindo C, Mateo-Lozano S, Notario V, Sanjuan X, Garcia Del Muro X, Fabra A, Tirado OM. Caveolin-1 modulates the ability of Ewing’s sarcoma to metastasize. Mol Cancer Res. 2010;8:1489–500. doi: 10.1158/1541-7786.MCR-10-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagares-Tena L, García-Monclús S, López-Alemany R, Almacellas-Rabaiget O, Huertas-Martínez J, Sáinz-Jaspeado M, Mateo-Lozano S, Rodríguez-Galindo C, Rello-Varona S, Herrero-Martín D, Tirado OM. Caveolin-1 promotes Ewing sarcoma metastasis regulating MMP-9 expression through MAPK/ERK pathway. Oncotarget. 2016;7:56889–903. doi: 10.18632/oncotarget.10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huertas-Martínez J, Court F, Rello-Varona S, Herrero-Martín D, Almacellas-Rabaiget O, Sáinz-Jaspeado M, Garcia-Monclús S, Lagares-Tena L, Buj R, Hontecillas-Prieto L, Sastre A, Azorin D, et al. DNA methylation profiling identifies PTRF/Cavin-1 as a novel tumor suppressor in Ewing sarcoma when co-expressed with caveolin-1. Cancer Lett. 2017;386:196–207. doi: 10.1016/j.canlet.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 14.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 19.Bengtsson H, Simpson K, Bullard J, Hansen K. aroma. affymetrix: A generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory. Berkeley: 2008. pp. 1–9. [Google Scholar]
- 20.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–21. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Yamada N, Tanaka T, Hori T, Yokoyama S, Hayakawa Y, Yano S, Fukuoka J, Koizumi K, Saiki I, Sakurai H. Crucial roles of RSK in cell motility by catalysing serine phosphorylation of EphA2. Nat Commun. 2015;6:7679. doi: 10.1038/ncomms8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen M-K, Hung M-C. Proteolytic cleavage, trafficking, and functions of nuclear receptor tyrosine kinases. FEBS J. 2015;282:3693–721. doi: 10.1111/febs.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tawadros T, Brown MD, Hart CA, Clarke NW. Ligand-independent activation of EphA2 by arachidonic acid induces metastasis-like behaviour in prostate cancer cells. Br J Cancer. 2012;107:1737–44. doi: 10.1038/bjc.2012.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunne PD, Dasgupta S, Blayney JK, McArt DG, Redmond KL, Weir J-A, Bradley CA, Sasazuki T, Shirasawa S, Wang T, Srivastava S, Ong CW, et al. EphA2 Expression Is a Key Driver of Migration and Invasion and a Poor Prognostic Marker in Colorectal Cancer. Clin Cancer Res. 2016;22:230–42. doi: 10.1158/1078-0432.CCR-15-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amato KR, Wang S, Tan L, Hastings AK, Song W, Lovly CM, Meador CB, Ye F, Lu P, Balko JM, Colvin DC, Cates JM, et al. EPHA2 Blockade Overcomes Acquired Resistance to EGFR Kinase Inhibitors in Lung Cancer. Cancer Res. 2016;76:305–18. doi: 10.1158/0008-5472.CAN-15-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch H, Busto MEDC, Kramer K, Médard G, Kuster B. Chemical Proteomics Uncovers EPHA2 as a Mechanism of Acquired Resistance to Small Molecule EGFR Kinase Inhibition. J Proteome Res. 2015;14:2617–25. doi: 10.1021/acs.jproteome.5b00161. [DOI] [PubMed] [Google Scholar]
- 29.Kang H-G, Jenabi JM, Zhang J, Keshelava N, Shimada H, May WA, Ng T, Reynolds CP, Triche TJ, Sorensen PHB. E-cadherin cell-cell adhesion in ewing tumor cells mediates suppression of anoikis through activation of the ErbB4 tyrosine kinase. Cancer Res. 2007;67:3094–105. doi: 10.1158/0008-5472.CAN-06-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendoza-Naranjo A, El-Naggar A, Wai DH, Mistry P, Lazic N, Ayala FRR, da Cunha IW, Rodriguez-Viciana P, Cheng H, Tavares Guerreiro Fregnani JH, Reynolds P, Arceci RJ, et al. ERBB4 confers metastatic capacity in Ewing sarcoma. EMBO Mol Med. 2013;5:1019–34. doi: 10.1002/emmm.201202343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nugent AA, Kolpak AL, Engle EC. Human disorders of axon guidance. Curr Opin Neurobiol. 2012;22:837–43. doi: 10.1016/j.conb.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehlen P, Delloye-Bourgeois C, Chédotal A. Novel roles for Slits and netrins: axon guidance cues as anticancer targets? Nat Rev Cancer. 2011;11:188–97. doi: 10.1038/nrc3005. [DOI] [PubMed] [Google Scholar]
- 33.Morris MR, Ricketts CJ, Gentle D, McRonald F, Carli N, Khalili H, Brown M, Kishida T, Yao M, Banks RE, Clarke N, Latif F, et al. Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene. 2011;30:1390–401. doi: 10.1038/onc.2010.525. [DOI] [PubMed] [Google Scholar]
- 34.Yu JS, Koujak S, Nagase S, Li C-M, Su T, Wang X, Keniry M, Memeo L, Rojtman A, Mansukhani M, Hibshoosh H, Tycko B, et al. PCDH8, the human homolog of PAPC, is a candidate tumor suppressor of breast cancer. Oncogene. 2008;27:4657–65. doi: 10.1038/onc.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikitovic D, Katonis P, Tsatsakis A, Karamanos NK, Tzanakakis GN. Lumican, a small leucine-rich proteoglycan. IUBMB Life. 2008;60:818–23. doi: 10.1002/iub.131. [DOI] [PubMed] [Google Scholar]
- 36.Borsig L, Wolf MJ, Roblek M, Lorentzen A, Heikenwalder M. Inflammatory chemokines and metastasis--tracing the accessory. Oncogene. 2014;33:3217–24. doi: 10.1038/onc.2013.272. [DOI] [PubMed] [Google Scholar]
- 37.Subramaniam PS, Whye DW, Efimenko E, Chen J, Tosello V, De Keersmaecker K, Kashishian A, Thompson MA, Castillo M, Cordon-Cardo C, Davé UP, Ferrando A, et al. Targeting nonclassical oncogenes for therapy in T-ALL. Cancer Cell. 2012;21:459–72. doi: 10.1016/j.ccr.2012.02.029. [DOI] [PubMed] [Google Scholar]
- 38.Warabi M, Nemoto T, Ohashi K, Kitagawa M, Hirokawa K. Expression of protein tyrosine phosphatases and its significance in esophageal cancer. Exp Mol Pathol. 2000;68:187–95. doi: 10.1006/exmp.2000.2303. [DOI] [PubMed] [Google Scholar]
- 39.Giebeler N, Zigrino P. A Disintegrin and Metalloprotease (ADAM): Historical Overview of Their Functions. Toxins (Basel) 2016;8:122. doi: 10.3390/toxins8040122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duffy MJ, McKiernan E, O’Donovan N, McGowan PM. Role of ADAMs in cancer formation and progression. Clin Cancer Res. 2009;15:1140–4. doi: 10.1158/1078-0432.CCR-08-1585. [DOI] [PubMed] [Google Scholar]
- 41.Petty A, Idippily N, Bobba V, Geldenhuys WJ, Zhong B, Su B, Wang B. Design and synthesis of small molecule agonists of EphA2 receptor. Eur J Med Chem. 2018;143:1261–76. doi: 10.1016/j.ejmech.2017.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.