Abstract
Fom3, the antepenultimate enzyme in the fosfomycin biosynthetic pathway in Streptomyces spp., is a class B cobalamin-dependent radical SAM methyltransferase that catalyzes methylation of (5′-cytidylyl)-2-hydroxyethylphosphonate (2-HEP-CMP) to form (5′-cytidylyl)-2-hydroxypropylphosphonate (2-HPP-CMP). Previously, the reaction of Fom3 with 2-HEP-CMP produced 2-HPP-CMP with mixed stereochemistry at C2. Mechanistic characterization has been challenging because of the insoluble expression and poor cobalamin (B12) incorporation in Escherichia coli. Recently, soluble E. coli expression and incorporation of cobalamin into Fom3 were achieved by overexpression of the BtuCEDFB cobalamin uptake system. Herein, we use this new method to obtain Fom3 from Streptomyces wedmorensis. We show that the initiator 5′-deoxyadenosyl radical stereospecifically abstracts the pro-R hydrogen atom from the C2 position of 2-HEP-CMP and use the downstream enzymes FomD and Fom4 to demonstrate that our preparation of Fom3 produces only (2S)-2-HPP-CMP. Additionally, we show that the added methyl group originates from SAM under multiple-turnover conditions, but the first turnover uses a methyl donor already present on the enzyme; furthermore, cobalamin isolated from Fom3 reaction mixtures contains methyl groups derived from SAM. These results are consistent with a model in which Fom3 catalyzes methyl transfer from SAM to cobalamin and the resulting methylcobalamin (MeCbl) is the ultimate methyl source for the reaction.
Fosfomycin is a broad-spectrum phosphonate antibiotic that disrupts peptidoglycan biosynthesis by irreversibly inactivating UDP-N-acetylglucosamine enolpyruvyltransferase.1,2 Under the name Monurol, fosfomycin is used for uncomplicated urinary tract infections and cystitis.3,4 The compound is produced by strains of Streptomyces and Pseudomonas, but the two genera use different biosynthetic pathways that share only the first and last steps.5–11 Both pathways involve conversion of phosphoenolpyruvate (PEP) to phosphonopyruvate by the PEP mutase Fom1 (Scheme 1).12,13 In Streptomyces, subsequent decarboxylation by Fom2 produces phosphonoacetaldehyde (PnAA).14 FomC then catalyzes the NADPH-dependent reduction of PnAA to 2-hydroxyethylphosphonate,7,15 which undergoes cytidylylation by a cytidylyltransferase domain (CyT) on Fom1 to form (5′-cytidylyl)-2-hydroxyethylphosphonate (2-HEP-CMP).10 Methylation of 2-HEP-CMP by the cobalamin (B12)-dependent class B radical SAM methyltransferase16 Fom3 yields (5′-cytidylyl)-2-hydroxypropylphosphonate (2-HPP-CMP), which is proposed to undergo hydrolysis catalyzed by FomD to form CMP and 2-hydroxypropylphosphonate (2-HPP).11 In the last step, the epoxidase Fom4 converts exclusively the S enantiomer of 2-HPP to fosfomycin.6,17,18 The stereospecificity of this reaction has been demonstrated both by feeding stereospecifically 2H-labeled 2-HPP to fosfomycin-producing Streptomyces fradiae and by in vitro enzyme assays,6,18–20 which have also shown that Fom4 converts (R)-2-HPP to 2-oxopropylphosphonate (2-OPP).18,20
Scheme 1.
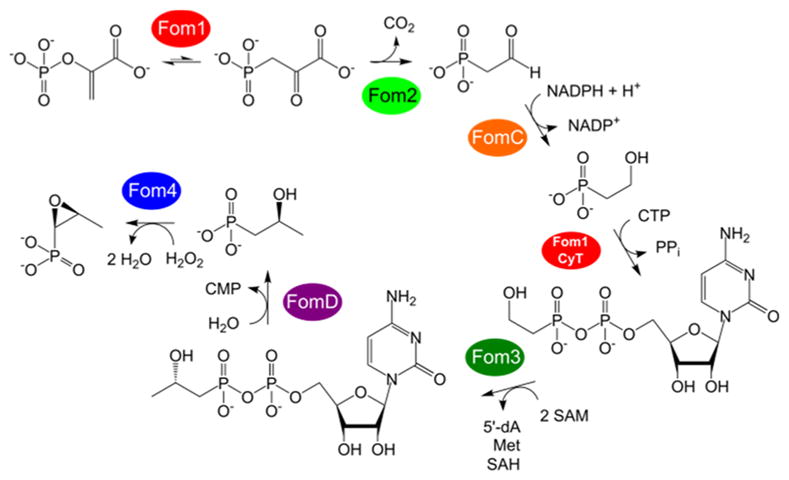
Biosynthesis of Fosfomycin in Streptomyces spp.
Previously, Fom3 was believed to catalyze the methylation of 2-HEP, a hypothesis consistent with isotope labeling and genetic studies.7,8 Seminal work by Kuzuyama et al. recently identified the correct substrate by the observation that fosfomycin production in a fom1 knockout strain could not be rescued by supplementation with the ostensibly downstream metabolite 2-HEP.10 Subsequently, a cytidylyltransferase domain of Fom1 that had no existing assigned function was found to catalyze cytidylylation of 2-HEP to form 2-HEP-CMP.10 Fom3 converted 2-HEP-CMP to 2-HPP-CMP in vitro, but curiously, the transformation was not stereospecific and produced comparable amounts of (2R)- and (2S)-2-HPP-CMP. This finding was unexpected given that (R)-2-HPP cannot be converted to fosfomycin by Fom4.
To examine the mechanism of the Fom3 reaction, we used the cytidylyltransferase domain of Fom1 (UniProtKB P96074) to enzymatically synthesize 2-HEP-CMP (Figure S1) and standards of (2R)- and (2S)-2-HPP-CMP.11 Coexpression of N-terminally His6-tagged Fom3 from Streptomyces wedmorensis (UniProtKB Q56184) with the Escherichia coli suf iron–sulfur cluster assembly system yielded Fom3 that lacked cobalamin but exhibited methylation activity in reactions supplemented with methylcobalamin (MeCbl) or hydroxocobalamin (HOCbl) (Figure S2). We hypothesized that incorporation of B12 during overexpression might improve both the yield and the activity and attempted to promote incorporation of cobalamin into Fom3 by coexpression with the E. coli B12 uptake system, composed of the outer membrane permease BtuB, solute binding protein BtuF, and ATP-dependent inner membrane transporter BtuCD, along with the functionally uncharacterized protein BtuE.21–23 Our expression construct placed the btu genes under an arabinose promoter and incorporated an artificial ribosome binding site (RBS) in front of btuF, btuB, and btuCED, which was cloned as an intact genomic fragment (Figure S3a). Coexpression of this construct with N-terminally MBP-tagged Fom3 did not improve the protein yield or activity.
Rather than troubleshoot these findings, we adopted the method recently reported by Booker and co-workers using a similar strategy that was successful with Fom3.24 These authors constructed a different plasmid expressing BtuCEDFB (Figure S3b) and overexpressed SUMO-tagged Fom3 in E. coli grown in M9-ethanolamine medium. This procedure resulted in Fom3 containing 0.86 equiv of B12 (82% MeCbl and 18% HOCbl). We mutated the btuCEDFB region of our plasmid to match the published design, which used a different artificial RBS sequence, included an additional RBS between btuC and btuE, removed the intergenic regions, and generated a 1 bp overlap between the stop codon of one gene and the start codon of the next for btuDFB (Figure S3c). Expression and purification using this construct yielded SUMO-tagged Fom3 containing 0.7 equiv of B12 [30% MeCbl and 70% HOCbl (Figure S4)]. The activity of this protein was much greater than that of our previous preparation, and addition of exogenous MeCbl or HOCbl was not necessary for catalysis (Figure S5). SUMO-Fom3 prepared by this method was used for all subsequent experiments.
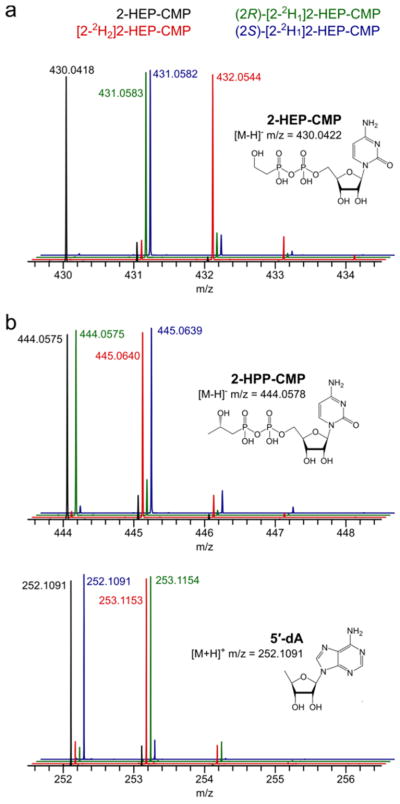
The previously observed formation of both diastereomers of 2-HPP-CMP could be caused either by abstraction of a hydrogen atom from both prochiral positions on C2 of 2-HEP-CMP or by addition of a methyl group to both faces of the resulting radical. To distinguish between these two possibilities, we enzymatically cytidylylated 2-HEP isotopologues deuterated at the pro-R position, the pro-S position, and both positions on C2.25 The products of SUMO-Fom3 reactions with these substrates were analyzed by liquid chromatography–mass spectrometry (LC–MS). Reactions with (2S)-[2-2H1]-2-HEP-CMP showed label retention in the 2-HPP-CMP product, (2R)-[2-2H1]-2-HEP-CMP caused transfer of the deuterium label to 5′-deoxyadenosine (5′-dA), and [2-2H2]-2-HEP-CMP retained one label and transferred one to 5′-dA (Figure 1). These results complement previous studies by Hammerschmidt et al. showing that feeding (S)-[2-2H1]-2-HEP to S. fradiae results in the incorporation of deuterium into fosfomycin while feeding (R)-[2-2H1]-2-HEP does not19 and are consistent with stereospecific abstraction of the pro-R hydrogen atom at C2 of 2-HEP-CMP by a 5′-deoxyadenosyl radical.
Figure 1.
(a) Liquid chromatography–mass spectrometry (LC–MS) analysis of 2-HEP-CMP (black), [2-2H2]-2-HEP-CMP (red), (2R)-[2-2H1]-2-HEP-CMP (green), and (2S)-[2-2H1]-2-HEP-CMP (blue). (b) LC–MS analysis of 2-HPP-CMP (left) and 5′-dA (right) produced from SUMO-Fom3 reactions with 2-HEP-CMP isotopologues.
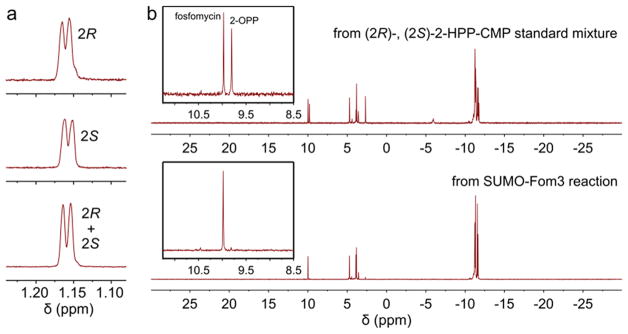
After ruling out nonstereospecificity in H atom abstraction to explain the formation of two product diastereomers, we attempted to confirm production of (2R)- and (2S)-2-HPP-CMP by SUMO-Fom3 using 1H and 31P nuclear magnetic resonance (NMR) spectroscopy. Unfortunately, the C3 proton signals of a mixture of (2R)- and (2S)-2-HPP-CMP standards could not be resolved (Figure 2a) and neither could the 31P signals (Figure S6). We therefore developed another method for distinguishing the two diastereomers using the differential activity of Fom4 with respect to (R)- and (S)-2-HPP. To convert 2-HPP-CMP to 2-HPP, we expressed N-terminally His6-tagged FomD from S. wedmorensis (UniProtKB O83033), the previously suggested candidate for catalysis of the hydrolysis step.10,11 FomD indeed hydrolyzed both the putative native substrate (2S)-2-HPP-CMP and the 2R diastereomer (Figure S7). This substrate tolerance enabled stereochemical analysis of 2-HPP-CMP by the two-step enzymatic conversion of (2R)- and (2S)-2-HPP-CMP to 2-OPP and fosfomycin, respectively (Scheme 2).
Figure 2.
Stereochemistry determination of the Fom3 product. (a) Methyl signals in 1H NMR spectra (D2O, 600 MHz) of standards of (2R)-2-HPP-CMP, (2S)-2-HPP-CMP, and a mixture of (2R)- and (2S)-2-HPP-CMP. (b) 31P NMR spectra (D2O, 600 MHz) of the products after reaction of FomD and Fom4 with a mixture of (2R)- and (2S)-2-HPP-CMP standards (top) and with 2-HPP-CMP produced by Fom3 (bottom), demonstrating that only fosfomycin is formed from the Fom3 product and not 2-OPP.
Scheme 2.
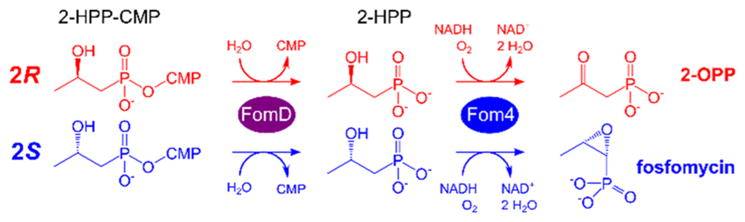
Differentiation of (2R)- and (2S)-2-HPP-CMP
Thus, the SUMO-Fom3 reaction product and an equimolar mixture of (2R)- and (2S)-2-HPP-CMP standards were each treated sequentially with FomD and Fom4, and the resulting products were characterized by 31P NMR spectroscopy (Figure 2b and Figure S8). Fosfomycin was the dominant phosphonate species formed from the SUMO-Fom3 reaction product, indicating the generation of (2S)-2-HPP-CMP and not (2R)-2-HPP-CMP under our assay conditions.
To determine the ultimate source of the methyl group, SUMO-Fom3 was reacted with excess 2-HEP-CMP in the presence of [methyl-2H3]SAM (CD3-SAM) and MeCbl and conversely with unlabeled SAM and [methyl-2H3]MeCbl (CD3Cbl). Minimal methyl transfer from MeCbl or CD3Cbl was observed in the product, even with concentrations of MeCbl or CD3Cbl that were 10-fold higher than that of SAM (Figure 3a), implicating SAM as the methyl donor for steady state catalysis. However, under single-turnover conditions with CD3-SAM and no added MeCbl, a significant amount of unlabeled product was formed, indicating the presence of transferable methyl groups on SUMO-Fom3 (Figure 3b). Furthermore, reaction mixtures containing excess CD3-SAM and HOCbl produced CD3Cbl (Figure S9). Together, these results suggest methyl transfer from SAM to 2-HEP-CMP via enzyme-bound cobalamin.
Figure 3.
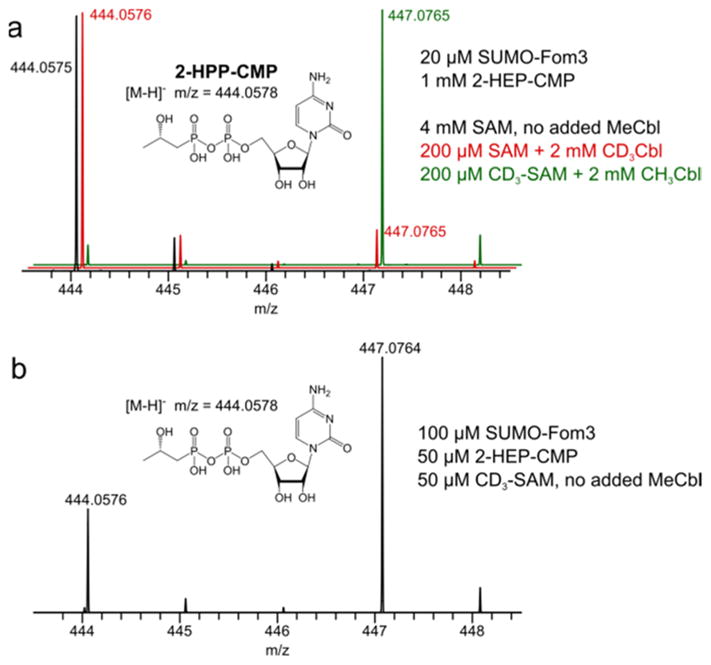
LC–MS analysis of 2-HPP-CMP from SUMO-Fom3 reactions with CD3-SAM and/or CD3Cbl under (a) multiple-turnover conditions (20 μM SUMO-Fom3 and 1 mM 2-HEP-CMP) and (b) single-turnover conditions (100 μM SUMO-Fom3 and 50 μM 2-HEP-CMP). Formation of the unlabeled product in single-turnover reactions with CD3-SAM and no added MeCbl demonstrates that as-isolated SUMO-Fom3 contains transferable methyl groups.
In summary, we show that abstraction of a hydrogen atom from 2-HEP-CMP by SUMO-Fom3 is stereospecific and that the hydrogen is transferred to 5′-deoxyadenosine. The transfer of methyl to the resulting proposed substrate radical is also stereospecific, causing inversion of configuration at C2 and forming (2S)-2-HPP-CMP. Added methyl- or hydroxocobalamin is not necessary for Fom3 activity, and during steady state turnover, the methyl group is provided by SAM, which was confirmed by using deuterium-labeled SAM. This result is consistent with observations for other class B radical SAM methyltransferases.26–29 However, single-turnover studies demonstrate that the methyl group is likely transferred from SAM to cobalamin and then to the substrate. This observation is consistent with the observed net retention of stereochemistry in the methyl group of fosfomycin when methionine with a chiral methyl group was fed to the producing organism.30 Collectively, these observations are consistent with the proposed mechanism depicted in Scheme 3.7
Scheme 3.
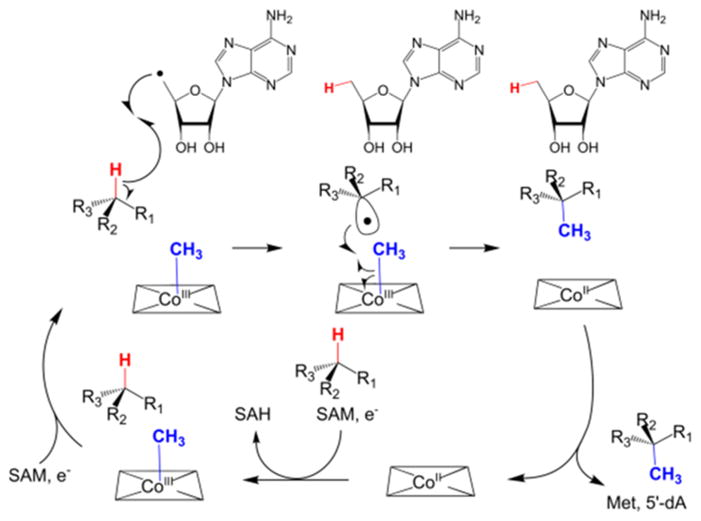
Proposed Mechanism for Fom3
Supplementary Material
Acknowledgments
The authors thank Profs. C. Cameron, V. Bandarian, and J. E. Cronan for E. coli strains and plasmids. The authors thank Dr. Z. Li of the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign for LC–MS analysis on a Q Exactive instrument.
Funding
This study was supported by the National Institutes of Health (Grant P01 GM077596 to W.A.v.d.D.) and the National Science Foundation Graduate Research Fellowship Program (Grant DGE-1144245 to M.I.M.). A 600 MHz NMR spectrometer was funded by National Institutes of Health Grant S10-RR028833.
ABBREVIATIONS
- 2-HEP
2-hydroxyethylphosphonate
- 2-HEP-CMP
(5′-cytidylyl)-2-hydroxyethylphosphonate
- 2-HPP
2-hydroxypropyl-phosphonate
- 2-HPP-CMP
(5′-cytidylyl)-2-hydroxypropyl-phosphonate
- 2-OPP
2-oxopropylphosphonate
- 5′-dA
5′-deoxyadenosine
- CyT
cytidylyltransferase
- HOCbl
hydroxocobalamin
- LC–MS
liquid chromatography–mass spectrometry
- MBP
maltose binding protein
- MeCbl
methylcobalamin
- PEP
phosphoenolpyruvate
- PnAA
phosphonoacetaldehyde
- RBS
ribosome binding site
- SAM
S-adenosyl-L-methionine
- SUMO
small ubiquitin-like modifier
Footnotes
ORCID
Wilfred A. van der Donk: 0000-0002-5467-7071
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.8b00616.
Materials, methods, Figures S1–S9, and Table S1 (PDF)
References
- 1.Kahan FM, Kahan JS, Cassidy PJ, Kropp H. The mechanism of action of fosfomycin (phosphonomycin) Ann N Y Acad Sci. 1974;235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 2.Marquardt JL, Brown ED, Lane WS, Haley TM, Ichikawa Y, Wong CH, Walsh CT. Kinetics, stoichiometry, and identification of the reactive thiolate in the inactivation of UDP-GlcNAc enolpyruvoyl transferase by the antibiotic fosfomycin. Biochemistry. 1994;33:10646–10651. doi: 10.1021/bi00201a011. [DOI] [PubMed] [Google Scholar]
- 3.Stein GE. Single-dose treatment of acute cystitis with fosfomycin tromethamine. Ann Pharmacother. 1998;32:215–219. doi: 10.1345/aph.17227. [DOI] [PubMed] [Google Scholar]
- 4.Bailey RR. Management of lower urinary tract infections. Drugs. 1993;45(Suppl 3):139–144. doi: 10.2165/00003495-199300453-00023. [DOI] [PubMed] [Google Scholar]
- 5.Seto H, Kuzuyama T. Bioactive Natural Products with Carbon-Phosphorus Bonds and Their Biosynthesis. Nat Prod Rep. 1999;16:589–596. doi: 10.1039/a809398i. [DOI] [PubMed] [Google Scholar]
- 6.Liu P, Murakami K, Seki T, He X, Yeung SM, Kuzuyama T, Seto H, Liu HW. Protein purification and function assignment of the epoxidase catalyzing the formation of fosfomycin. J Am Chem Soc. 2001;123:4619–4620. doi: 10.1021/ja004153y. [DOI] [PubMed] [Google Scholar]
- 7.Woodyer RD, Li G, Zhao H, van der Donk WA. New insight into the mechanism of methyl transfer during the biosynthesis of fosfomycin. Chem Commun. 2007:359–361. doi: 10.1039/b614678c. [DOI] [PubMed] [Google Scholar]
- 8.Woodyer RD, Shao Z, Thomas PM, Kelleher NL, Blodgett JA, Metcalf WW, van der Donk WA, Zhao H. Heterologous production of fosfomycin and identification of the minimal biosynthetic gene cluster. Chem Biol. 2006;13:1171–1182. doi: 10.1016/j.chembiol.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Kim SY, Ju KS, Metcalf WW, Evans BS, Kuzuyama T, van der Donk WA. Different biosynthetic pathways to fosfomycin in Pseudomonas syringae and Streptomyces species. Antimicrob Agents Chemother. 2012;56:4175–4183. doi: 10.1128/AAC.06478-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho SH, Kim SY, Tomita T, Shiraishi T, Park JS, Sato S, Kudo F, Eguchi T, Funa N, Nishiyama M, Kuzuyama T. Fosfomycin biosynthesis via transient cytidylylation of 2-hydroxyethylphosphonate by the bifunctional Fom1 enzyme. ACS Chem Biol. 2017;12:2209–2215. doi: 10.1021/acschembio.7b00419. [DOI] [PubMed] [Google Scholar]
- 11.Sato S, Kudo F, Kim SY, Kuzuyama T, Eguchi T. Methylcobalamin-dependent radical SAM C-methyltransferase Fom3 recognizes cytidylyl-2-hydroxyethylphosphonate and catalyzes the nonstereoselective C-methylation in fosfomycin biosynthesis. Biochemistry. 2017;56:3519–3522. doi: 10.1021/acs.biochem.7b00472. [DOI] [PubMed] [Google Scholar]
- 12.Hidaka T, Iwakura H, Imai S, Seto H. Studies on the biosynthesis of fosfomycin. 3 Detection of phosphoenol-pyruvate phosphomutase activity in a fosfomycin high-producing strain of Streptomyces wedmorensis and characterization of its blocked mutant NP-7. J Antibiot. 1992;45:1008–1010. doi: 10.7164/antibiotics.45.1008. [DOI] [PubMed] [Google Scholar]
- 13.Kuzuyama T, Hidaka T, Imai S, Seto H. Studies on the biosynthesis of fosfomycin. V Cloning of genes for fosfomycin biosynthesis. J Antibiot. 1993;46:1478–1480. doi: 10.7164/antibiotics.46.1478. [DOI] [PubMed] [Google Scholar]
- 14.Nakashita H, Kozuka K, Hidaka T, Hara O, Seto H. Identification and expression of the gene encoding phosphonopyruvate decarboxylase of Streptomyces hygroscopicus. Biochim Biophys Acta, Gene Struct Expression. 2000;1490:159–162. doi: 10.1016/s0167-4781(99)00249-3. [DOI] [PubMed] [Google Scholar]
- 15.Shao Z, Blodgett JA, Circello BT, Eliot AC, Woodyer R, Li G, van der Donk WA, Metcalf WW, Zhao H. Biosynthesis of 2-hydroxyethylphosphonate, an unexpected intermediate common to multiple phosphonate biosynthetic pathways. J Biol Chem. 2008;283:23161–23168. doi: 10.1074/jbc.M801788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou S, Alkhalaf LM, de Los Santos EL, Challis GL. Mechanistic insights into class B radical-S-adenosylmethionine methylases: ubiquitous tailoring enzymes in natural product biosynthesis. Curr Opin Chem Biol. 2016;35:73–79. doi: 10.1016/j.cbpa.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 17.Hammerschmidt F. Biosynthesis of Natural Products with a P-C Bond. Part 8: On the Origin of the Oxirane Oxygen Atom of Fosfomycin in Streptomyces fradiae. J Chem Soc, Perkin Trans. 1991;1:1993–1996. [Google Scholar]
- 18.Zhao ZB, Liu PH, Murakami K, Kuzuyama T, Seto H, Liu HW. Mechanistic studies of HPP epoxidase: Configuration of the substrate governs its enzymatic fate. Angew Chem, Int Ed. 2002;41:4529–4532. doi: 10.1002/1521-3773(20021202)41:23<4529::AID-ANIE4529>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 19.Hammerschmidt F, Kählig H. Biosynthesis of natural products with a P-C bond 7 Synthesis of [1,1-2H2]-, [2,2-2H2]-, (R)- and (S)-[1-2H1](2-hydroxyethyl)phosphonic Acid and (R,S)-[1-2H1](1,2-dihydroxyethyl)phosphonic acid and incorporation studies into fosfomycin in Streptomyces fradiae. J Org Chem. 1991;56:2364–2370. [Google Scholar]
- 20.Olivares P, Ulrich EC, Chekan JR, van der Donk WA, Nair SK. Characterization of two late-stage enzymes involved in fosfomycin biosynthesis in pseudomonads. ACS Chem Biol. 2017;12:456–463. doi: 10.1021/acschembio.6b00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cadieux N, Bradbeer C, Reeger-Schneider E, Koster W, Mohanty AK, Wiener MC, Kadner RJ. Identification of the Periplasmic Cobalamin-Binding Protein BtuF of Escherichia coli. J Bacteriol. 2002;184:706–717. doi: 10.1128/JB.184.3.706-717.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Veaux LC, Clevenson DS, Bradbeer C, Kadner RJ. Identification of the BtuCED Polypeptides and Evidence for Their Role in Vitamin B12 Transport in Escherichia coli. J Bacteriol. 1986;167:920–927. doi: 10.1128/jb.167.3.920-927.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bassford PJ, Jr, Kadner RJ. Genetic Analysis of Components Involved in Vitamin B12 Uptake in Escherichia coli. J Bacteriol. 1977;132:796–805. doi: 10.1128/jb.132.3.796-805.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanz ND, Blaszczyk AJ, McCarthy EL, Wang B, Wang RX, Jones BS, Booker SJ. Enhanced solubilization of class B radical S-adenosylmethionine methylases by improved cobalamin uptake in Escherichia coli. Biochemistry. 2018;57:1475–1490. doi: 10.1021/acs.biochem.7b01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitteck JT, Malova P, Peck SC, Cicchillo RM, Hammerschmidt F, van der Donk WA. On the stereochemistry of 2-hydroxyethylphosphonate dioxygenase. J Am Chem Soc. 2011;133:4236–4239. doi: 10.1021/ja1113326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marous DR, Lloyd EP, Buller AR, Moshos KA, Grove TL, Blaszczyk AJ, Booker SJ, Townsend CA. Consecutive radical S-adenosylmethionine methylations form the ethyl side chain in thienamycin biosynthesis. Proc Natl Acad Sci U S A. 2015;112:10354–10358. doi: 10.1073/pnas.1508615112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HJ, McCarty RM, Ogasawara Y, Liu YN, Mansoorabadi SO, LeVieux J, Liu HW. GenK-catalyzed C-6′ methylation in the biosynthesis of gentamicin: isolation and characterization of a cobalamin-dependent radical SAM enzyme. J Am Chem Soc. 2013;135:8093–8096. doi: 10.1021/ja312641f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parent A, Guillot A, Benjdia A, Chartier G, Leprince J, Berteau O. The B12-radical SAM enzyme PoyC catalyzes valine Cβ-methylation during polytheonamide biosynthesis. J Am Chem Soc. 2016;138:15515–15518. doi: 10.1021/jacs.6b06697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Schnell B, Baumann S, Müller R, Begley TP. Biosynthesis of Branched Alkoxy Groups: Iterative Methyl Group Alkylation by a Cobalamin-Dependent Radical SAM Enzyme. J Am Chem Soc. 2017;139:1742–1745. doi: 10.1021/jacs.6b10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schweifer A, Hammerschmidt F. Stereochemical course of methyl transfer by cobalamin-dependent radical SAM methyltransferase in fosfomycin biosynthesis. Biochemistry. 2018;57:2069–2073. doi: 10.1021/acs.biochem.8b00264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.