Abstract
Fusion and fission of cellular membranes involve dramatic, protein-mediated changes in membrane curvature. Many of the experimental methods useful for investigating curvature sensing or generation require specialized equipment. We have developed a system based on supported lipid bilayers (SLBs) in which lipid tubules are simple to produce and several types of membrane remodeling events can be readily imaged using widely available instrumentation (e.g., tubule fission and/or membrane budding). Briefly, high ionic strength during lipid bilayer deposition results in incorporation of excess lipids in the SLB. After sequentially washing with water and physiological ionic strength buffer solutions, lipid tubules form spontaneously. We find that tubule formation results from solution-dependent spreading of the SLB; washing from water into physiological ionic strength buffer solution leads to expansion of the bilayer and formation of tubules. Conversely, washing from physiological buffer into water results in contraction of the membrane and loss of tubules. We demonstrate the utility of these supported tubulated bilayers, termed “STuBs,” with an investigation of Sar1B, a small Ras family G-protein known to influence membrane curvature. The addition of Sar1B to STuBs results in dramatic changes in tubule topology and eventual tubule fission. Overall, STuBs are a simple experimental system, useful for monitoring protein-mediated effects on membrane topology in real time, under physiologically relevant conditions.
Keywords: Supported bilayer, vesicle budding, membrane fission, exocytosis, endocytosis
GRAPHICAL ABSTRACT
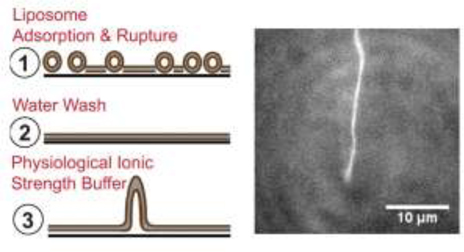
Introduction
The execution of a variety of fundamental biological processes including vesicle budding, fission, and fusion, depends on extensive changes to the morphology of lipid membranes (Chernomordik et al., 2006, McMahon and Boucrot, 2015). Such dynamic remodeling likely requires the participation of proteins, which by scaffolding (Hinshaw and Schmid, 1995), active insertion (Peter et al., 2004, Frost et al., 2008), or modulating tension (Sheetz et al., 2006, Sheetz, 2001) exert their influence on membrane structure.
Because of the immense interest in these topics, a number of techniques have recently been developed to monitor curvature sensing or generation – but generally not both simultaneously – in controlled experimental systems. For example, a fluorescence-based microscopy assay allows quantification of curvature dependence of protein binding using differentially sized liposomes tethered to a glass surface (Hatzakis et al., 2009). Supported lipid bilayers (SLBs) formed on substrates patterned with nanoparticles of defined size have also been used to quantify curvature sensing (Alnaas et al., 2017). Generation of highly curved lipid tubules from pipette-immobilized giant unilamellar vesicles (GUVs) has been reported for a number of proteins, a technique which allows careful control of membrane tension but requires specialized instrumentation (Shi and Baumgart, 2015). Microbead-supported membranes with excess lipid reservoir (SUPER templates) can be used to detect curvature generation from flexible synthetic lipid bilayers based on formation of budded vesicles or tubules (Pucadyil and Schmid, 2010, Pucadyil and Schmid, 2008). With skill, the latter technique can be adapted to detect binding to pre-formed tubules. Another recently developed technique for quantitative, high-throughput analysis of membrane fission is supported membrane tubules (SMrT templates), which consist of arrays of long lipid tubules prepared using a flow cell and oriented parallel to a glass surface (Dar et al., 2017, Dar et al., 2015). Our goal was to develop a simple, broadly accessible assay system, based on SLBs and complementary to the techniques described above, for visualizing protein-mediated curvature sensing and generation. In this study, we describe such a system characterized by the presence of stable lipid tubules on a planar glass surface – a system we name supported tubulated bilayers (“STuBs”).
In the context of intracellular trafficking, membrane remodeling results in the sequestration of membrane-enclosed spaces for the transport of proteins and lipids. Transport from the endoplasmic reticulum (ER) to the Golgi is mediated by coat protein complex II (COPII)-coated vesicles (Barlowe et al., 1994). Key components in their formation include the inner and outer coat complex proteins Sec23/24 and Sec13/31 and the small GTPase Sar1B (Barlowe and Schekman, 1993, Saito et al., 2017, Futai et al., 2004, Hanna et al., 2016). Recent studies in cell-based and cell-free systems suggest that Sar1B has a direct role in both sensing and driving membrane curvature (Hanna et al., 2016), can form tubules from GUVs (Hariri et al., 2014, Bacia et al., 2011), and localizes to vesicles with COPII (Kurokawa et al., 2016) – activities which have been characterized using a variety of experimental platforms. In this study, we demonstrate how the STuBs platform can be used to detect several membrane-dependent activities of Sar1B in a single experimental setup.
In the first part of the study, we describe the STuBs system and identify the key factors that lead to tubule formation. The formation of tubules is sensitive to the ionic conditions used during SLB deposition and subsequent washing. In particular, deposition of an SLB under high ionic strength followed by sequential washes with water and a buffer containing physiological salt concentrations promotes bilayer spreading and as a result, tubule formation. Inclusion of 0.5 mM Mg2+ in the physiological buffer, required for the study of GTPases, slightly decreases the extent of bilayer spreading, but nevertheless produces ample tubules for studying membrane remodeling by Mg2+-dependent proteins such as Sar1B.
We then use STuBs to capture a variety of functions of recombinant human Sar1B protein. We show that addition of Sar1B to STuBs results in the fission of pre-formed tubules and induction of stable Sar1B-dependent lipid structures. These Sar1B-induced structures are detected using a combination of epifluorescence and total internal reflection fluorescence (TIRF) microscopy. Dual-color imaging of fluorescent Sar1B and fluorescent lipid tracers provides evidence of protein localization to tubules and protein-driven fission in the presence of GTP. Overall, STuBs allow observation and quantification of protein effects on single tubules or many tubules in parallel, allowing imaging of tubule fission events and insight into processes of membrane remodeling.
2. Materials and Methods
2.1 Materials
1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycerol-3- phospho-L-serine (sodium salt) (DOPS), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (LRB- DOPE), 1,2-dioleoyl-sn-glycero-3-[phosphoinositol-4,5-bisphosphate] (DOPIP2), 1- palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) in chloroform were from Avanti Polar Lipids (Alabaster, AL). 1,1’-dioctadecyl-3,3,3’,3’-tetramethylindodicarbocyanine perchlorate (DiD) was from Life Technologies (Eugene, OR). 2-mercaptoethanol was from Calbiochem (EMD Millipore, Billerica, MA). GTP, GDP, and GMP-PNP were from Sigma Aldrich (St. Louis, MO). Alexa Fluor 488 (AF488) TFP ester was from ThermoFisher Scientific (Waltham, MA). All other chemicals were ACS grade or higher.
2.2 Protein Expression and Purification
A GST-Sar1B fusion protein (Hanna et al., 2016) was expressed from a pGEX6P-1 plasmid in BL21 DE3 E. coli by induction with 50 µM IPTG for 4 hr at 22 ºC. The cells were isolated by centrifugation, resuspended in lysis buffer (phosphate-buffered saline with 1 mM EDTA, 10 µM GDP, and protease inhibitors), and lysed with chicken egg white lysozyme for 45 min. The lysate was clarified by centrifugation at 30,000 x g and loaded by gravity onto a glutathione Sepharose 4B resin column. Sar1B was released from the column by an overnight 4 ºC digestion with PreScission protease (GE Healthcare). The Sar1B sample was concentrated in a 10,000 MWCO Amicon ultrafiltration device and resolved over a Superdex 200 10/300 GL column in Sar1B storage buffer (50 mM HEPES, 100 mM NaCl, 1 mM dithiothreitol, 10 µM GDP, pH 7.6) using an Akta FPLC (GE Healthcare). The monodisperse protein was collected and re-concentrated, prior to being flash frozen and stored at –80ºC (Figure S1). For experiments, the protein was diluted > 100-fold from this storage buffer into the indicated experimental solution conditions. Sar1BGTP indicates Sar1B in in the presence of 5 µM GTP in Buffer A + 0.5 mM MgCl2, Sar1BGDP indicates Sar1B in in the presence of 5 µM GDP in Buffer A + 0.5 mM MgCl2, and Sar1BGMP-PNP indicates Sar1B in in the presence of 5 µM GMP-PNP in Buffer A + 0.5 mM MgCl2. A portion of the purified Sar1B was labeled with AF488 TFP ester following the manufacturer’s protocol (ThermoFisher Scientific).
2.3 Liposome Preparation
Chloroform suspensions of lipids were mixed in the indicated molar ratios along with either DiD or LRB-DOPE as a fluorescent tracer, evaporated under a stream of N2 and kept under a vacuum for a minimum time of 1 h, and then resuspended in Buffer A (140 mM KCl, 15 mM NaCl, 20 mM HEPES, pH 7.4) to a total lipid concentration of 3.0 mM. The resuspended multilamellar vesicles were then converted to liposomes via either sonication (Sonics VibraCell) or extrusion (Avanti Mini Extruder) using a polycarbonate filter of 100 nm pore size. The use of sonicated versus extruded liposomes had no noticeable effect on tubule formation.
2.4 Preparation of SLBs and Supported Tubulated Bilayers
Glass coverslips (#1.5 thickness; Fisher or VWR) were etched through immersion of each cover glass in a 3:1 mixture of H2SO4:H2O2 (30% solution) for 1 h. The coverslips were thoroughly rinsed and stored in deionized water for ≤ 24 h prior to use. Before depositing each SLB, the coverslips were dried with N2 and fitted with a 9-mm diameter silicone-gasketed perfusion chamber with a height of 0.6 mm (CoverWell, Grace BioLabs).
SLBs and STuBs were then formed via the vesicle fusion method. Liposome suspensions were mixed with bilayer deposition buffer and pipetted into the perfusion chamber mounted on the coverslip. Several bilayer deposition buffers were tested. Our standard condition for formation of STuBs included a final concentration of 1 M NaCl in Buffer A. For some samples, fluorescently-labeled and unlabeled liposomes were prepared separately and mixed together during bilayer deposition to the desired molar ratio of fluorophore. After incubation at room temperature for 30–40 min, each SLB sample was washed extensively with deionized water, taking care not to introduce air bubbles. For samples not imaged under perfusion, buffer was introduced into the samples by 4 sequential additions of 1 chamber volume of the desired buffer solution through the holes on the top of the perfusion chamber, mixing extensively upon each addition.
2.5 Fluorescence Microscopy
Total internal reflection fluorescence (TIRF) or traditional widefield fluorescence microscopy was utilized to obtain high resolution, real-time images of the SLBs and STuBs using three separate instruments: an Axio Observer TIRF microscope with 100x, 1.46 NA oil immersion objective (Zeiss) with illumination from either a 20-mW 552-nm laser or a 100-mW 640-nm laser (Intelligent Imaging Innovations) and detection with an Evolve 512×512 EMCCD camera (Photometrics); an Olympus CellTIRF microscope with a 100x, 1.49 NA oil immersion objective (Olympus) with illumination from either a 50-mW 561-nm laser or a 60-mW 488-nm laser (Olympus) and detection with an iXon Ultra EMCCD camera (Andor); and an Olympus IX81 inverted fluorescence microscope with a 60x, 1.49 NA oil immersion TIRF objective and illumination from a 561 nm Sapphire laser (Coherent) and detection with an iXon 3 EMCCD camera (Andor)(Rao et al., 2017). Image acquisition was controlled with SlideBook 6 software (Intelligent Imaging Innovations) for the Axio Observer system, CellSens software (Olympus) for the CellTIRF system, and MetaMorph for the Olympus IX81 system. An exposure time of 50 ms was used for single color imaging and 100 ms for dual-color imaging.
Bilayer expansion experiments were conducted using the Olympus CellTIRF system equipped with a perfusion system (ALA Scientific Instruments). STuBs labeled with 0.1% LRB-DOPE were prepared and washed into the desired starting buffer following a water wash. Image sequences were then acquired while perfusing the sample into deionized water. The final image sequence was acquired while perfusing into Buffer A with or without 500 µM MgCl2. Photobleach correction and fluorescence intensity analysis were performed using custom software written in MATLAB (Mathworks).
Tubule counting experiments were conducted using the Olympus CellTIRF system. Samples were initially imaged in Buffer A with 500 µM MgCl2 and 5 µM of either GDP, GTP, or GMP-PNP. Following image acquisition of the initial state in the absence of protein, addition of Sar1B was performed as indicated through the holes in the perfusion chambers and image sequences were captured. Five regions of interest (ROIs) were imaged per sample using ≥ 3 samples as indicated in the text. In order to visualize tubules that project perpendicular to the planar support, images were acquired with illumination vertical or at a subcritical angle with the focus up and away from the surface of the SLB. Initial number of starting tubules in each field sampled prior to protein addition was 96 ± 11 (SEM).
Polarized TIRF (pTIRF) experiments were conducted using the Olympus IX81 system as previously described (Passmore et al., 2014). The p and s polarizations of the 561 nm Sapphire laser were split into separate paths, each controlled by a separate Lambda SC shutter (Sutter Instruments). STuBs including 0.2% DiD as a fluorescent tracer were prepared in Buffer A with 500 µM MgCl2 and 5 µM GTP. Images were acquired in alternating s- and p-polarization while pipetting Sar1B into the perfusion chamber. Membrane deformations were identified and analysis was performed using custom software written in IDL (ITT, Boulder, CO).
Two color TIRF experiments were conducted using the Olympus CellTIRF system(Rao et al., 2017). The 561-nm laser was at a vertical illumination to image full length rhodamine labeled tubules and the 488-nm laser was in TIR to selectively image Alexa Fluor 488 labeled Sar1B bound to the membrane, significantly increasing the signal to noise ratio. Emission was separated on an OptoSplit II system from Cairn Research. Optical filters in the OptoSplit were obtained from Semrock (FF01–514/30–25, FF02–617/73, and dichroic FF580-FDi01).
2.6 Image Analysis
TIRF images were analyzed using ImageJ (NIH, Bethesda, MD). Tubule quantification was achieved through counting each tubular structure and averaging across the five ROIs. For each of the conditions, the average number of tubules prior to protein addition was normalized to a value of 1. Two color imaging experiments were first rolling ball background subtracted and binned in the time axis across 5 neighboring frames prior to analysis. Student t-tests were performed using Prism 6 software (GraphPad, La Jolla, CA) comparing each data point to the protein-free condition. Computer simulations (Anantharam et al., 2010) were used to predict the structure of indentations observed via pTIRF imaging.
2.7 Tubule width estimation
The width of DOPC tubules was estimated from geometrical considerations, based on the observation of some tubules maturing into large, flattened structures with diameters of several micrometers (Figure S2). First, fluorescence intensities of the large, flattened structures were quantified by measuring linescans across > 50 such structures (Figure S2D-F). Most had intensities ~4x that of the surrounding SLB (Figure S2F), suggesting that the structures consist of three stacked bilayers on top of the SLB as schematically illustrated in Figure S2G. Second, for six separate maturation events that were captured on video, the total length of each tubule was measured as described in Figure S3 and compared to the surface area of the corresponding post-maturation structure. Assuming that the tubule is a cylinder, that the post-maturation structure is three stacked bilayers, and that both contain the same total amount of lipid, then the tubule diameter d is given by
| (1) |
where l is the measured tubule length and a is the measured post-maturation surface area (Figure S3).
3. Results
3.1 Observation of tubulated bilayers
Supported tubulated bilayers form readily using a straightforward modification of commonly used SLB preparation techniques. For this study, tubulated and non-tubulated SLBs were prepared by vesicle fusion and visualized using fluorescence microscopy with fluorescent lipid tracers. Briefly, glass coverslips were etched with piranha solution [3:1 H2SO4: H2O2 (30% solution)] to remove organic adsorbents and render the surface hydrophilic. SLB formation was induced by the addition of unilamellar liposomes in buffer to a silicon-gasketed perfusion chamber. To promote tubulation, the liposomes were incubated in the perfusion chamber in an environment of high ionic strength (0.5 – 1 M NaCl), which has been shown to drive accelerated adsorption of liposomes to the glass support (Pucadyil and Schmid, 2010). Following incubation, SLBs were rinsed extensively with water to remove unincorporated liposomes and then imaged in a buffered solution at physiological ionic strength. Of note, tubulation was observed consistently in two authors’ laboratories (Anantharam and Knight) using the same general protocols but different stock solutions, materials, and reagents.
Two lipid compositions were tested: DOPC and 3:1 DOPC/DOPS, each containing 0.1% LRB-DOPE as a fluorescent tracer. After water rinsing, DOPC SLBs displayed uniform fluorescence with occasional bright puncta indicative of adhered liposomes, but no tubules (Figure 1A). Addition of a HEPES buffer at physiological ionic strength (Buffer A; see Methods) initiated the growth of flexible tubules, which could be observed growing and lying on the membrane surface (Figure 1B). Over several minutes, some of the tubules matured into larger, flattened vesicle-like structures on the SLB surface (Figure S2). Quantitation of LRB-DOPE tracer lipid intensity profiles across these structures suggests that most consisted of three stacked bilayers on top of the SLB (Figure S2). Notably, the SLB under such structures was intact, as revealed by single-molecule diffusion of a second distinct tracer lipid (DiD) included in the SLB at 100 parts per billion (ppb) concentration (Movie S2). Based on comparing the length of tubules before maturation to the area of flattened vesicle-like structures after maturation events, the tubule diameter was estimated to be 100 ± 20 nm (Figure S3). Thus, the diameter of DOPC tubules is consistent with bilayer rather than micellar structure.
Figure 1: Spontaneous formation of lipid tubules from SLBs.
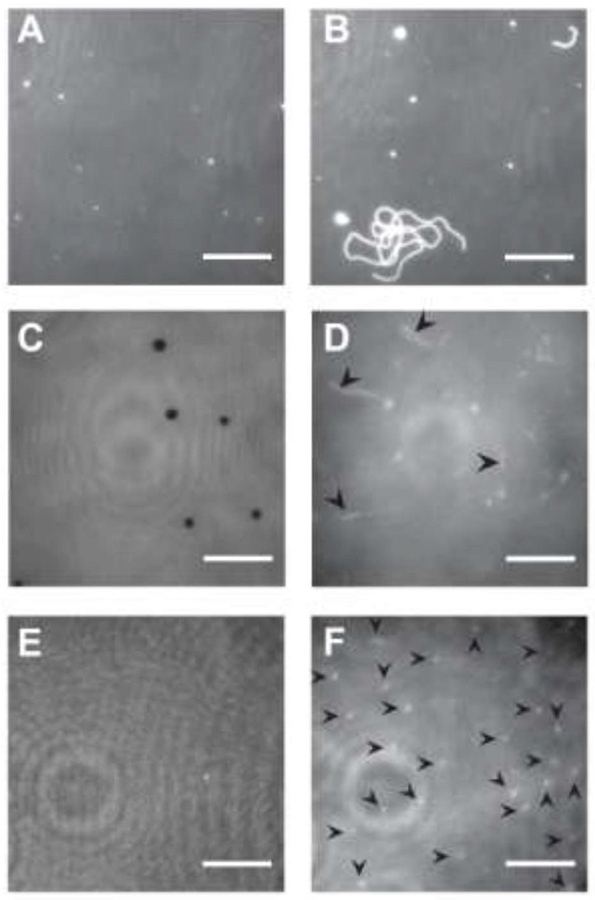
(A) Bilayer of DOPC, 0.1% LRB-DOPE in water. (B) Same sample as A, after adding Buffer A. (C) Bilayer of 3:1 DOPC/DOPS, 0.1% LRB-DOPE in water. Some regions of the surface are depleted of lipid (dark spots). (D) Same sample as C, after adding Buffer A. Arrows highlight longer tubules. Focus is away from surface in order to highlight tubules that extend vertically away from the supported bilayer. (E) A second bilayer of 3:1 DOPC/DOPS, 0.1% LRB-DOPE in water. (F) Same sample as E, containing shorter, more abundant tubules. Each arrow indicates a tubule. Image intensities have been scaled to visualize tubules, including adjusting the contrast in panels D and F. There is no indication that LRB-DOPE preferentially partitions into tubules relative to the SLB. Movie S1 corresponding to panel D is available in the Supporting Information. Scale bars 10 μm.
Tubulation was also observed from SLBs of other lipid compositions upon addition of Buffer A. After washing with water, 3:1 DOPC/DOPS bilayers displayed planar topology sometimes containing µm-sized regions of depleted fluorescence (Figure 1C), consistent with a previous report of holes in anionic SLBs at low ionic strength (Kalb et al., 1992). These holes disappeared and tubules appeared after the bilayer was exposed to Buffer A. Tubules from 3:1 DOPC/DOPS typically appeared flexible,extended vertically away from to the SLB, and persisted stably for ≥ 1 h without maturing into larger structures (Figure 1D and F). Tubules were also observed from SLBs composed of POPC, 3:1 POPC/POPS, and 3:1 DOPC/DOPS with 1% DOPIP2. Tubulation was not dependent on the presence of LRB-DOPE, as the phenomenon was also observed when 0.2% DiD was used as the fluorescent tracer. Due to the high temporal stability of tubules generated from 3:1 DOPC/DOPS, all subsequent experiments used this lipid composition.
3.2 Tubule formation depends strongly on ionic strength during SLB deposition
Because an increase in liposome adhesion rate has been reported with increasing ionic strength (Kalb et al., 1992), we tested the impact of NaCl concentration on tubule formation. For this experiment, SLBs were deposited in Buffer A with varying amounts of added NaCl, washed extensively with water, and then imaged in Buffer A. SLBs deposited under low to moderate NaCl concentration (< 0.5 M) seldom formed tubules. Tubules were more abundant (i.e., more tubules per x-y area) and longer when using higher NaCl concentrations during bilayer deposition (Figure 2) We did not observe tubules on SLBs imaged in water; rather, tubules appear upon exchanging the SLB from water into an imaging buffer containing physiological salt concentrations. Thus, tubule formation is highly sensitive to ionic strength both during and after SLB deposition.
Figure 2: Effect of NaCl concentration on tubulation.
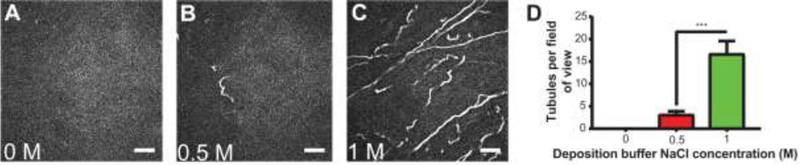
Bilayers (3:1 DOPC/DOPS with 0.1% LRB-DOPE) were deposited using Buffer A plus NaCl at concentrations of (A) 0 M, (B) 0.50 M, or (C) 1 M, then washed thoroughly with water prior to imaging in Buffer A. Representative images are shown from triplicate samples, after background subtraction. Focus is adjusted up from the surface in order to capture tubules extending into solution. Scale bars, 10 μm. (D) When using 0 M NaCl in liposome deposition buffer tubules do not form. Increasing NaCl concentration from 0.5 M to 1 M significantly increases the number of tubules observed per field of view from 3.0 ± 0.9 to 16.6 ± 3.2. Error bars are ± SEM. ***: P < 0.001 using student’s t-test.
3.3 Tubulation is driven by expansion of the supported lipid bilayer
One way that salts could drive tubulation is by decreasing the equilibrium packing density of lipids in the x-y plane of the substrate, which would cause the bilayer to expand laterally. To test for this possibility, we observed the ability of SLBs to spread into voids induced by scratching an SLB immersed in water with tweezers. Using TIRF microscopy, a time-lapse image sequence was acquired while perfusing Buffer A into the sample chamber (Movie S3). Within approximately 30 seconds, the supported bilayer completely filled the scratched region (Figure 3A-C). Tubule formation was observed a few seconds later in the same region of the SLB, suggesting that the forces promoting SLB expansion are also responsible for driving tubulation.
Figure 3: Tubulation is driven by expansion of the SLB.
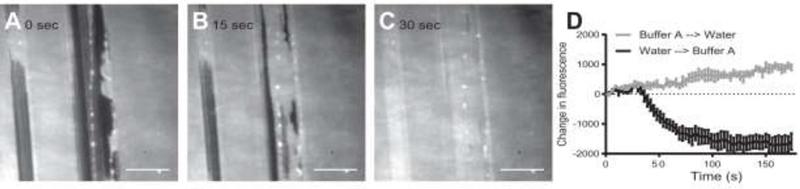
(A-C) Images of a scratched SLB at various time points during exchange of the bathing solution by perfusion from water into Buffer A. Further expansion of the bilayer beyond that which was necessary to fill the scratched region results in tubule formation. Scale bars, 10 µm. (D) Changes in average fluorescence intensity per unit area upon exchange of the bathing solution from Buffer A into water (gray) or vice versa (black). SLB retraction is indicated by an increase in fluorescence (2.7 ± 0.3 percent increase exchanging from Buffer A into water). Expansion is indicated by decreased fluorescence (4.9 ± 0.9 percent decrease exchanging from water into Buffer A). All SLBs were 3:1 DOPC/DOPS with 0.1% LRB-DOPE. Error bars are ± SEM.
To quantify the extent of expansion, unscratched SLBs were deposited in high salt, washed into deionized water and then into Buffer A, and imaged with TIRF microscopy during perfusion with water. The fluorescence per unit area was observed to increase over time for several minutes, corresponding to a retraction of the bilayer (Figure 3D, gray). Subsequent washing of the same samples with Buffer A resulted in a decrease in fluorescence per unit area (Figure 3D, black), corresponding to an expansion of the bilayer. The effect is qualitatively independent of the total amount of lipid in the bilayer, as the same pattern was observed using SLBs deposited in physiological ionic strength buffer (Figure S4). The effect is also independent of the presence of Mg2+ in the perfusion buffer, as the same pattern was observed using SLBs deposited in high ionic strength buffer and perfused with Buffer A + 0.5 mM MgCl2 (Figure S5). The magnitude of the fluorescence change in samples with low ionic strength deposition buffer was smaller than for the SLBs deposited under high salt conditions, consistent with our earlier observation that tubulation is less extensive from SLBs deposited in lower salt conditions (Figure 2).
3.4 STuBs as a platform to monitor Sar1B-mediated lipid remodeling
Sar1B is a well-characterized vesicle trafficking protein that is known to bind and remodel lipid membranes in a GTP-dependent manner (Hanna et al., 2016, Bacia et al., 2011, Long et al., 2010, Kuge et al., 1994). Sar1B dimers bind lipid membranes via insertion of an amphipathic helix into the lipid bilayer, driving membrane deformations in a manner dependent upon the nucleotide state of the protein (Futai et al., 2004, Hariri et al., 2014). GTP-bound Sar1B has been shown to form a scaffold of dimers that dissociate upon GTP hydrolysis (Futai et al., 2004, Hariri et al., 2014). Addition of GTP-bound Sar1B to supported bilayers or GUVs is known to drive curvature-dependent membrane remodeling and vesiculation (Hanna et al., 2016, Bacia et al., 2011, Long et al., 2010). To visualize Sar1B-driven membrane remodeling using STuBs, wild-type human Sar1B bound to GTP (Sar1BGTP) was added to tubulated bilayers of 3:1 DOPC/DOPS labeled with 0.1% LRB-DOPE, and the lipid tubules were imaged using fluorescence microscopy. Several tubule morphologies were observed upon Sar1BGTP addition (Figure 4 and 5). We observed tubules becoming rigid upon addition of increased Sar1BGTP (Figure 4A-B), tubules that terminated in bulbous vesicle-like structures reminiscent of previously-described “pseudo-vesiculated tubules” (Hariri et al., 2014) (Figure 4C), and tubules with a beads-on-a-string morphology (Figure 4D and 5C). The pseudo-vesiculated tubules were the most commonly observed of the non-flexible tubule morphologies (Figure 4F). In some cases, tubules exhibited ombinations of these morphologies; the most complex structure observed was a bifurcated, rigid tubule with a beads-on-a-string morphology (Figure 4D). Many of the vesicle-like “beads” exhibited fluidity along their respective tubules (Movie S4). The overall likelihood of observing flexible tubules declined dramatically with increasing concentrations of Sar1BGTP (Figure 4E).
Figure 4: Sar1B membrane remodeling.
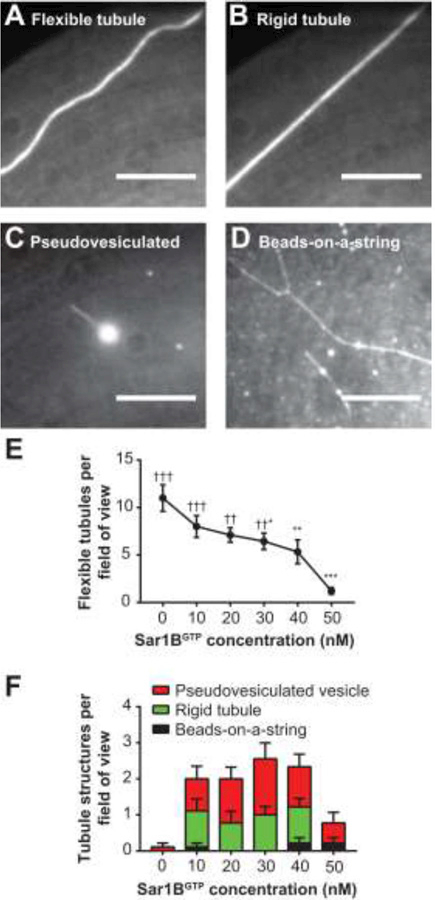
The addition of Sar1BGTP to STuBs alters the morphology of lipid tubules. (A) A flexible tubule. (B) Same tubule as in (A) following addition of Sar1BGTP, exhibiting a rigid morphology. (C) A pseudo-vesiculated tubule. (D) A rigid, bifurcated tubule and a tubule exhibiting a beads-on-a-string morphology. (E) As Sar1BGTP concentration increases, the number of flexible tubules per field of view significantly decreases. (F) The prevalence of Sar1BGTP-induced structures changes as a function of concentration. Statistics by 1-way ANOVA. * indicates comparison to 0-nM Sar condition, † indicates comparison to 50-nM Sar1B condition. Adjusted P Values; *, †: P < 0.05, **, †† P: < 0.01, ***, †††: P < 0.001. All are in Buffer A + 0.5 mM MgCl2 + 5 μM GTP. Scale bars 10 µm. Error bars ± SEM.
Figure 5: Fluorescently labeled Sar1B binds tubules.
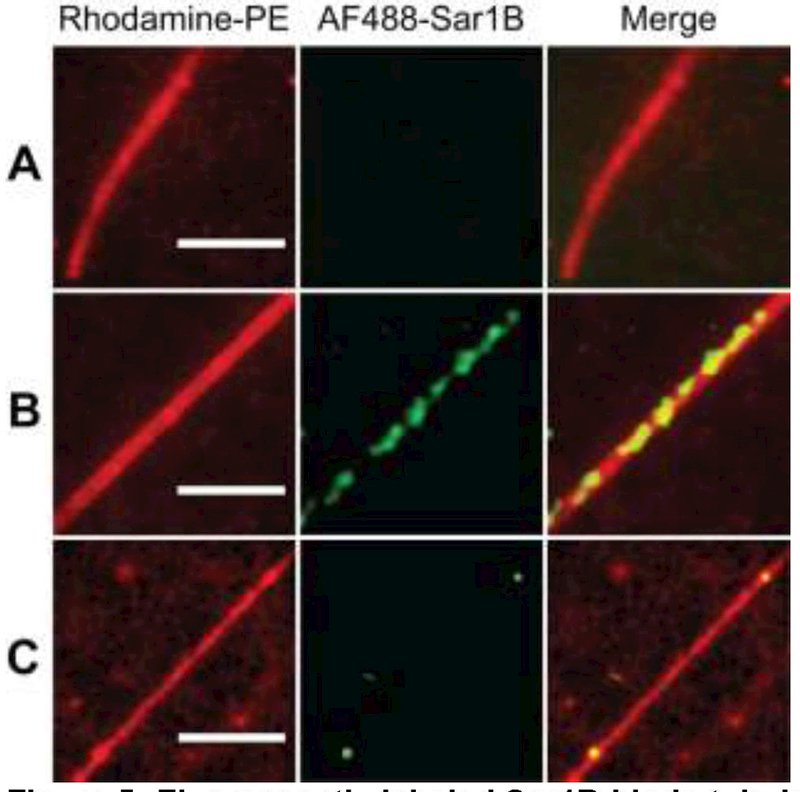
(A) A flexible tubule with no bound Sar1B. (B) A rigid tubule with Sar1B decorating its length. (C) A tubule exhibiting a beads-on-a-string morphology with Sar1B bound at the “beads”. Conditions: 1 nM AF488-Sar1B, 10 nM unlabeled Sar1BGTP in Buffer A + 0.5 mM MgCl2 + 5 µM GTP. Scale bars 5 µm.
To visualize sites of action of Sar1BGTP, purified protein labeled on surface-exposed lysine residues (using the Alexa Fluor 488 TFP ester) was added to 3:1 DOPC/DOPS STuBs containing 0.1% LRB-DOPE. Fluorescent protein was observed to colocalize with many tubules, which is consistent with our previous report of Sar1B binding to curved membranes (Figure 5) (Hanna et al., 2016). As before, tubules without associated AF488-Sar1BGTP fluorescence were flexible (Figure 5A). Tubules with associated AF488-Sar1BGTP fluorescence adopted one of the structures described in Figure 4 (Figures 5B, C).
3.5 Imaging fission events mediated by Sar1BGTP
Sar1B-catalyzed fission of tubules was also detected using STuBs. Figure 6A-D shows an event in which AF488-Sar1BGTP is observed initially clustering on the side of a tubule (Figure 6B), then migrating to the base and inducing a conversion of the tubule into a vesicle (Figure 6C, D). Quantification of AF488-Sar1BGTP fluorescence from this event shows that Sar1B accumulated on the structure over time (Figure 6E). Averaging over several such events aligned to the moment of fission (time “0”) suggests that Sar1B triggers fission at a relatively low threshold concentration and then continues to accumulate post-fission (Figure 6F).
Figure 6: Two color imaging shows real time tubule fission at site of Sar1B binding.
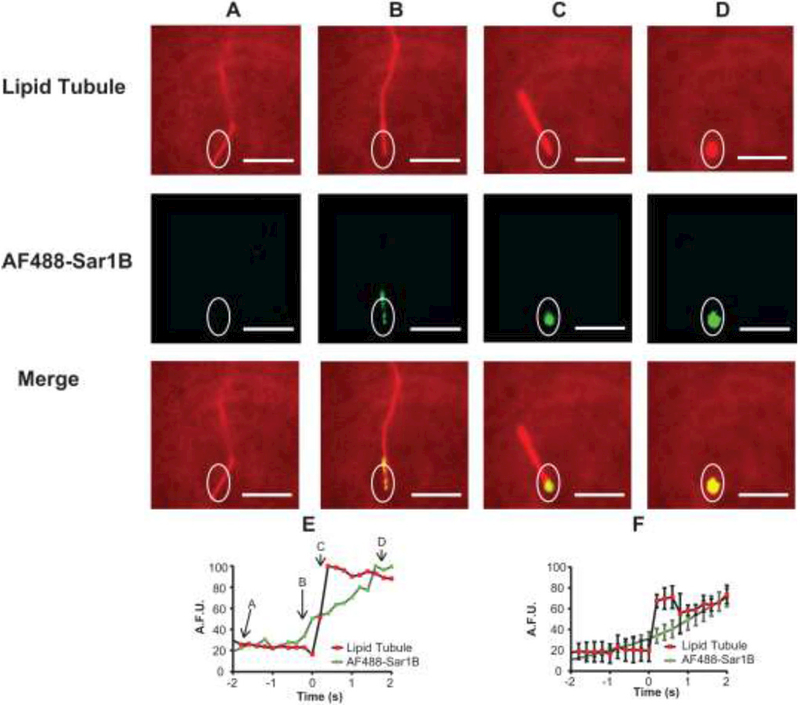
Images are shown from a single image sequence of a tubule undergoing fission. Results show a tubule prior to significant AF488-Sar1BGTP binding (A) and after binding AF488-Sar1BGTP (B-C). After the AF488-Sar1BGTP localized to the tubule base (C), the tubule disassembled into a vesicle-like structure (D), presumably due to fission. The lipid bilayer was labeled with 0.1% LRB-DOPE and excited by 561-nm laser in an epifluorescence geometry. The AF488-Sar1BGTP was excited by a 488-nm laser in TIR. (E) Fluorescence intensity vs. time graph of tubule fission event shown in (A-D). (F) Average fluorescence intensity vs. time of 5 tubule fission events. Error bars ± SEM. Scale bars 10 μm.
The physiological function of Sar1B involves the budding of vesicles from the endoplasmic reticulum (Kuge et al., 1994). Thus, we sought to determine whether Sar1B could form membrane buds by bending planar regions of STuBs. To monitor such phenomena, we used a form of TIRF microscopy (polarized TIRFM, or pTIRFM) which is curvature-sensitive (Anantharam et al., 2010). STuBs labeled with 0.2% DiD were exposed to 10 nM Sar1BGTP while imaging continuously. DiD fluorescence was excited by sequential exposure to orthogonal p- and s-polarized beams. From these images, pixel-to-pixel P/S and P+2S images were calculated (Figure 7A). The P/S ratio image reports on local membrane deviations from parallelism with the glass coverslip (Anantharam et al., 2010). The linear combination P+2S approximately reports total DiD emission, which is proportional to the amount of DiD in the evanescent field at the imaged location convolved with the evanescent field intensity. Membrane deformations (i.e., tubules and buds) involve both perpendicularly oriented lipids and a greater amount of total lipid in a narrow vertical column within the brightest part of the evanescent field. Therefore, budding events manifest as simultaneous increases in both P/S (Figure 7B, 7D) and P+2S (Figure 7C, 7E).
Figure 7: Sar1B bends membranes in a GTP-dependent manner.
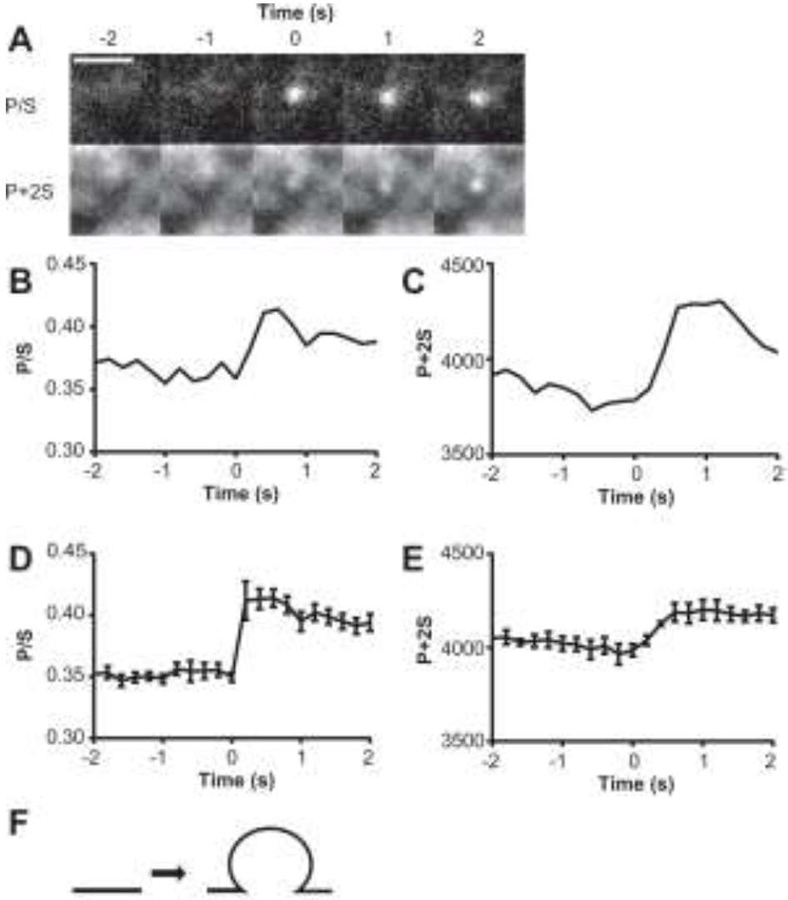
(A) Increases in DiD-labeled membrane curvature (P/S) and concentration (P + 2S) observed after 10 nM Sar1BGTP is added to STuBs. (B-C) Graphs corresponding to the event shown in panel A. Averaged P/S (D) and P+2S (E) changes for several membrane budding events observed in the presence of Sar1BGTP (n=5). Error bars ± SEM. No similar events were observed in otherwise identical samples with Sar1BGDP or lacking protein (n ≥ 3). (G) Predicted structure of the detected membrane indentations based on computer simulations (Anantharam et al., 2010). Scale bars 2 µm.
An example of a Sar1BGTP-mediated bending event detected with pTIRFM is shown in Figure 7. Here, both the P/S and P+2S at a single diffraction-limited spot increase dramatically within 60 seconds of Sar1BGTP addition. The bud formed in under 360 ms (two frames) and persisted for the ~15-s remainder of the image sequence. A structure that is consistent with computer simulations (Anantharam et al., 2010) and reflecting changes in DiD emission is shown in Figure 7F. The cartoon represents a persistent deformation (long-lived increase in P/S) with more membrane in the region of interest (increase in P+2S) than prior to Sar1BGTP addition. These observations support the idea that Sar1B drives membrane topological changes (fission and membrane bending), and illustrate the utility of STuBs for imaging real-time, protein-mediated changes in lipid morphology.
3.6 Quantitation of changes in tubule density
Addition of Sar1BGTP induces both tubule fission and formation of stable Sar1B-dependent lipid structures (Figures 4, 6, and 7). In order to assess which of these processes dominates, we quantified changes in the density of tubules upon Sar1B addition. Five minutes after the addition of Sar1B (in the presence of different guanosine nucleotides) to STuBs, five randomly chosen 82 μm x 82 μm regions of interest were imaged. The density of tubules was determined by counting tubules at each region, and data for each sample were normalized with respect to tubule density prior to protein addition (Figure 8).
Figure 8: Sar1B drives tubule fission in a GTP-dependent manner.
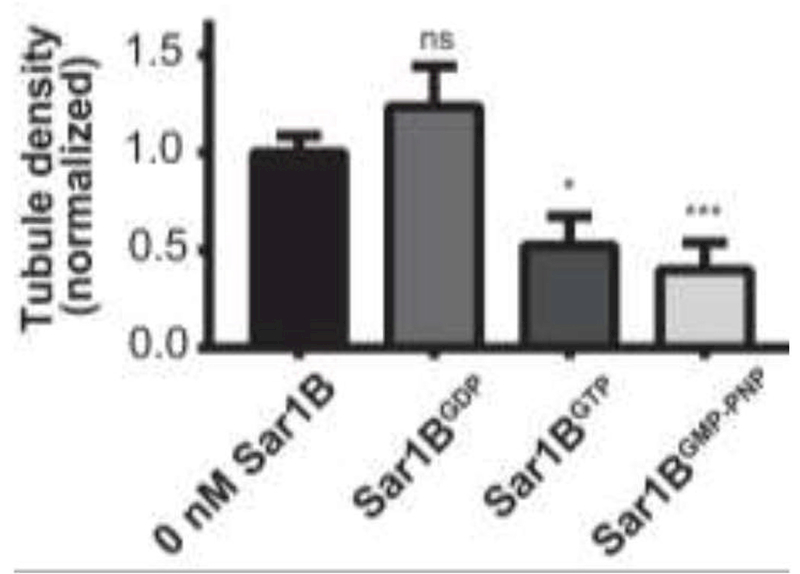
Effect of wild-type Sar1B on tubule density in the presence of 5 µM GDP, 5 µM GTP, and 5 µM GMP-PNP. In these experiments, 50 nM Sar1B was titrated onto an SLB containing approximately 100 pre-formed tubules in each field of view sampled. *:P < 0.05, **: P < 0.01, ***:P < 0.001, ****:P < 0.0001 relative to the absence of protein. Error bars ± SEM. To test for significance, data were compared to 0 nM condition using Student’s t-test. All samples used 3:1 DOPC/DOPS with 0.1% LRB-DOPE as the fluorescent tracer.
Despite the observed ability of Sar1BGTP to generate curvature from a planar bilayer (Figure 7), the bulk density measurements showed fission to be the dominant process; addition of Sar1BGTP in the nanomolar range resulted in a statistically significant net loss of tubules (Figure 8). Notably, no significant difference in tubule density was observed with Sar1BGDP over the same concentration range (Figure 8). Sar1BGDP is known to bind membranes, but with lower affinity than the GTP-bound state. As a result, Sar1BGDP has been used as control for membrane fission induced by protein crowding (Snead et al., 2017). Recently, there have been conflicting reports regarding the role of GTP hydrolysis in Sar1B-driven membrane fission (Hanna et al., 2016, Hariri et al., 2014, Adolf et al., 2013). Therefore, the experiment was also conducted in the presence of GMP-PNP, a non-hydrolyzable GTP analog. The addition of Sar1BGMP-PNP resulted in a significant decrease in the density of pre-formed tubules, similar to the effect of Sar1BGTP (Figure 8). This result is consistent with prior reports of GTP hydrolysis-independent Sar1B-driven vesicle scission on thin membrane tubules in vitro (Hariri et al., 2014) and formation and release of COPII transport carriers in semi-intact cell systems (Adolf et al., 2013).
4.Discussion
4.1 A simple new method for detecting protein-membrane remodeling
Here we report the development and first application of STuBs to study protein-mediated effects on membrane curvature and fission. STuBs are readily and reproducibly obtained from commonly used lipid compositions at physiological pH and ionic strength. Tubules are stable on the minutes to hours timescale and can be imaged to measure effects of proteins on membrane tubule morphology, budding, and fission both qualitatively and quantitatively. One key feature of STuBs is its simplicity: tubule formation requires no specialized equipment and is compatible with detection via wide-field fluorescence microscopy and polarized TIRF microscopy as demonstrated here. Using two-color measurements with labeled protein and lipid, we observe real-time membrane remodeling and fission of individual tubules driven by Sar1B. By quantifying changes in tubule density upon addition of protein, we provide evidence that the fission activity of Sar1B is correlated with GTP binding rather than hydrolysis. These results with Sar1B demonstrate that STuBs are a useful system for investigating protein effects on membrane tubules.
4.2 Model for STuBs formation
The mechanism by which STuBs form in this study likely parallels that of a complementary model system reported previously, SUPER templates (Pucadyil and Schmid, 2010, Pucadyil and Schmid, 2008). Like SUPER templates, STuBs form following deposition of an SLB in high salt, followed by extensive washing in deionized water and subsequent addition of physiological buffer (Figure 9) (Pucadyil and Schmid, 2010). The presence of high salt during bilayer deposition leads to incorporation of lipid beyond that which can be accommodated by a planar membrane within the available substrate area. Essentially, the rate of liposome adsorption to the glass surface exceeds the rate of rupture and spreading to form a bilayer (Pucadyil and Schmid, 2010, Kalb et al., 1992). Notable differences between STuBs and SUPER templates include the substrate geometry and preparation method: SUPER templates form around commercially prepared silica microbeads, while STuBs form within the confined area of a perfusion chamber mounted directly on a highly hydrophilic piranha-etched glass coverslip. For a supported bilayer with a confined substrate surface area, any excess lipid incorporated during deposition must ultimately result in the formation of structure in the z-dimension. We do not observe tubules immediately after washing in water (Figure 1A/C/E); on the contrary, holes are sometimes observed (Figure 1C). This observation is consistent with previous reports that excess liposomes can be removed by washing in this manner (Kalb et al., 1992). However, the SLB must contain excess lipid after the water wash step, because tubules subsequently form following addition of physiological buffer (Figure 1B/D/F). In principle, the excess lipid could be accommodated in water through a decreased area per lipid (increased molecular packing density) (Helm et al., 1986) and/or by an increased effective substrate area (e.g., conforming to the nanometer-scale undulations of the piranha-etched glass substrate) (Seu et al., 2007). The first model is consistent with studies on anionic phospholipid monolayers, in which the area per lipid at the gas/fluid transition has been shown to increase by ~10% between 10 μM and 1 M NaCl (Helm et al., 1986). The second model is consistent with prior reports that salts and buffers induce swelling of stacked lipid bilayers in the z-dimension (Pabst et al., 2007, Koerner et al., 2011, Petrache et al., 2006). Both models assume the presence of microscopic defects in the SLB that allow rapid transbilayer equilibration of small solutes; such defects have been shown to characterize SLBs in contrast to freestanding vesicles (Kendall et al., 2010, Pucadyil and Schmid, 2010, Lee et al., 1999).
Figure 9: Model for STuBs formation and effects.
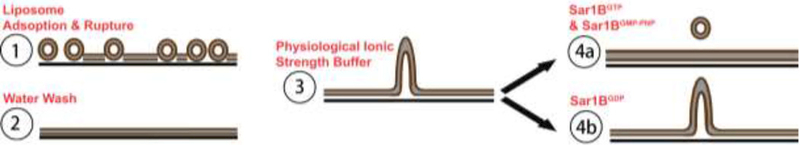
(1) SLB deposition occurs via liposome fusion. In the presence of high salt, liposomes adsorb to the glass surface faster than they rupture and spread, resulting in a bilayer containing excess lipid. (2) Washing with water removes unincorporated liposomes; although the bilayer still contains excess lipid, it appears flat for reasons that could involve decreased area per lipid and/or stronger lipid-glass interaction in water (see text). (3) Following addition of physiological ionic strength buffer, the excess lipid is no longer accommodated in the planar bilayer and tubules form. (4a-b) Effects of Sar1B addition with various nucleotides.
Addition of physiological buffer overcomes the accommodation of excess lipid, leading to spreading of the SLB into any voids, such as the holes visible in Figure 1C or the scratch introduced in Figure 3A. After voids are filled, the continued expansion results in formation of tubular protrusions in the z-dimension (“3” in Figure 9). Tubule formation is likely favored over blebbing or budding due to the support geometry and in order to minimize the loss of favorable contacts, as described previously (Lipowsky, 2013). STuBs morphologies range from relatively dense fields of short tubules (e.g., Figure 1F) to sparse, longer tubules; the reasons for this variability are not yet clear but presumably arise from rates of stochastic tubule budding versus extension.
We observed tubule formation from every lipid composition tested, including zwitterionic DOPC and POPC as well as mixtures containing 25% DOPS or POPS. These lipids possess little intrinsic curvature and our SLBs are symmetric; thus, the tubule formation observed here is not likely driven by membrane asymmetry or intrinsic curvature effects as in some other systems (Sendecki et al., 2017, Kreutzberger et al., 2017). Rather, the presence of excess lipid in a confined substrate area appears to be the major driving force. The tubules formed from DOPS-containing bilayers tended to orient more vertically than pure DOPC tubules, likely due to electrostatic repulsion between the tubules and the planar SLB, and were therefore chosen as the system for our investigation of Sar1B. We note that the concentration of PS in the intracellular ER membranes Sar1B normally encounters is likely lower than in the lipid compositions used here (Bollen and Higgins, 1980). Nevertheless, Sar1B is effective at remodeling these higher PS containing membranes (Hariri et al., 2014).
4.3 Connection to previous SLB studies
Spontaneous tubulation has been reported since the earliest studies with SLBs (Tamm and McConnell, 1985); however, there have been surprisingly few efforts to use these tubules as platforms for studying protein-lipid interactions. Anecdotally, researchers working on SLBs have often worked to eliminate tubules in order to focus studies on uniform, planar lipid bilayers. In particular, tubules and other 3-dimensional structures have been reported previously in studies that used high salt concentrations for SLB deposition (Kalb et al., 1992, Cambrea and Hovis, 2007). As noted above, high-salt deposition is also used for incorporating an excess lipid reservoir in SUPER templates (Pucadyil and Schmid, 2010). However, a survey of the literature suggests that a large majority of research groups use physiological or lower salt concentrations when forming SLBs, conditions which do not generally produce tubules (Figure 2) (Anderson et al., 2009, Cremer and Boxer, 1999, Kiessling and Tamm, 2003). SLBs containing a high percentage of the curvature-inducing lipid phosphatidylethanolamine (PE) appear to be an exception to this rule, as it was recently reported that 50% or higher POPE content leads to spontaneous formation of tubules and other 3-dimensional structures from SLBs deposited under near-physiological ionic strength (Sendecki et al., 2017). It is not clear whether the reported PE-rich tubules arise from incorporation of excess lipid into the SLB analogous to STuBs, or from other mechanisms. PE is a significant cellular component with interesting effects on membrane fusion (Kreutzberger et al., 2017). Nevertheless, our results with low intrinsic curvature lipids show that curvature strain is not necessary for SLB tubulation when high salt conditions are used for the bilayer deposition.
4.4 Complementarity to existing methods for measuring protein effects on membrane curvature
STuBs comprise a simple experimental platform for studying protein effects on membrane morphology, and complement several other systems that have been developed in recent years for studying the ability of proteins to sense membrane curvature (Hatzakis et al., 2009, Wang et al., 2012), induce tubulation from GUVs (Shi and Baumgart, 2015, Stachowiak et al., 2012), remove lipid from SUPER templates (Neumann et al., 2013), or induce scission of pre-formed tubules (Dar et al., 2017). Compared to existing methods for generating pre-formed tubules, STuBs are simple to prepare and require no specialized equipment other than a fluorescence microscope for imaging. Conceivably, STuBs could be used as a straightforward initial approach for identifying and characterizing proteins that remodel tubulated membranes, as a precursor to detailed kinetic and/or thermodynamic studies using SMrT templates, pipette-aspirated GUVs, or other techniques. STuBs are also advantageous for situations in which it is desirable to directly observe the points of contact where tubules emerge from the underlying planar lipid bilayer. Because STuBs allow for observation of membrane budding as well as proteins interacting with existing tubules within the same sample, this system could in principle be used to monitor curvature sensing and curvature generation simultaneously. STuBs are easy to produce, and several samples can be prepared and scanned for the desired tubule density and length distributions prior to adding a protein of interest.
4.5 Effect of Sar1B on STuBs
Here we demonstrate the utility of STuBs by investigating Sar1B-driven membrane remodeling. Measurements include imaging colocalization of tubules and fluorophore-labeled Sar1B (Figures 5 and 6), capturing individual tubule fission events and membrane budding (Figures 6 and 7), and counting tubules to determine net effects of the protein on tubule density (Figure 8). The use of fluorescent lipid tracers allows for observation of membrane remodeling events with video frame rate resolution using any form of widefield fluorescence microscopy. Addition of fluorophore-tagged protein allows for direct imaging of colocalization, but is not necessary for imaging morphology changes (Figure 4) or quantifying effects of protein addition on tubule formation and fission (Figure 8). Overall, STuBs enable both simultaneous observation and quantification of protein effects on many tubules, and imaging of individual tubulation and fission events.
The remodeling we observe using Sar1BGTP on STuBs is consistent with previous reports, with the advantage that we directly observe fission of tubules from the underlying SLB in real-time (Hanna et al., 2016, Simunovic et al., 2017). Most prior studies on the membrane remodeling properties of Sar1B have been conducted using model systems such as GUVs and liposomes, with remodeling detected either using bulk spectroscopic methods or electron microscopy (Futai et al., 2004, Hariri et al., 2014). Sar1B has been previously observed to remodel SLBs on mica, using atomic force microscopy (AFM) at relatively low time resolution (Hanna et al., 2016). Here we observe Sar1B-mediated fission events in real time using 2-color imaging (Figure 6) and quantify the extent of fission upon titration of Sar1B with various ligands (Figure 8). The ability of both Sar1BGTP and Sar1BGMP-PNP to remove tubules supports the hypothesis that interactions between Sar1B proteins in their active conformations and the membranes with which they are associated are sufficient to drive membrane remodeling, ultimately leading to fission (Hariri et al., 2014, Adolf et al., 2013). This effect was observed previously using AFM, appearing as a transient remodeling of SLBs that was arrested within a few seconds after addition of GMP-PNP (Hanna et al., 2016). Here, the higher temporal resolution afforded by STuBs provides additional insight into this brief event. In contrast, the GDP-bound form of Sar1B does expose its amphipathic helix, but is incapable of stable membrane penetration and is not significantly associated with tubule fission (Figure 8) (Hanna et al., 2016). This insight demonstrates the potential of STuBs as a simple, versatile platform for detecting changes in membrane curvature in real-time, using widely available fluorescence microscopy techniques.
Supplementary Material
HIGHLIGHTS.
Depositing liposomes under high ionic strength incorporates excess lipids into supported lipid bilayers
Excess lipid is accommodated in the z-direction by lipid tubules, forming Supported Tubulated Bilayers (STuBs)
Pre-formed lipid tubules can be used to measure protein-mediated membrane remodeling in real time easily and quantitatively
The GTPase Sar1B when bound to GTP significantly remodels membranes whereas Sar1B bound to GDP does not
ACKNOWLEDGMENT
The authors thank Dr. Ron Holz and Dr. Dan Axelrod for critical feedback of this manuscript, Dr. Lee Cambrea for advice on SLB formation, Dr. Bob Fuller and members of the University of Michigan trafficking club for helpful comments and suggestions during early stages of this project, and Dr. Horia Petrache, Dr. Dan Schwartz, Joseph Vasquez, and Alexandra H. Ranski for helpful discussions.
Funding Sources
This work was supported by National Institutes of Health (GM111997 to A. Anantharam, GM102866 to J.D.K., GM088242 to G.G.T. and GM110567 to A. Audhya), American Heart Association (SDG14420049 to A. Anantharam), The Michigan Undergraduate Research Opportunity Program (to A. Anantharam and N.A.S.), and the Richard Barber Summer Research Program (to A. Anantharam and P.J.D.).
Abbreviations:
- TIRF
Total Internal Reflection Fluorescence
- pTIRF
Polarized Total Internal Reflection Fluorescence
- SLB
Supported Lipid Bilayer
- STuBs
Supported Tubulated Bilayers
- COPII
Coat Protein complex II
- SMrT
Supported Membrane Tubules
- SUPER
Supported Membranes with Extra Reservoir
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPS
1,2-dioleoyl-sn-glycerol-3-phospho-L-serine (sodium salt)
- LRB-DOPE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl)
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPS
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine
- DiD
1,1’-dioctadecyl-3,3,3’,3’-tetramethylindodicarbocyanine perchlorate
- AF488
Alexa Fluor 488
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information. Five supplemental figures and four movies are available in the Supporting Information.
The following files are available free of charge.
Supplemental Figures and Legends (PDF)
Supplemental Movies S1-S4 (each as a separate .AVI file)
Author Contributions
N.A.S., P.J.D., J.D.K., and A. Anantharam planned and designed experiments. M.G.H. and A. Audhya provided reagents. N.A.S., P.J.D., G.G.T., and J.D.K. performed experiments. N.A.S., P.J.D., J.D.K., and A. Anantharam analyzed data and wrote the manuscript with contributions from all authors. All authors have given approval to the final version of the manuscript.
Competing Interests:
The authors have no competing interests to declare, other than the grant support noted above.
REFERENCES
- ADOLF F, HERRMANN A, HELLWIG A, BECK R, BRUGGER B & WIELAND FT 2013. Scission of COPI and COPII vesicles is independent of GTP hydrolysis. Traffic, 14, 922–32. [DOI] [PubMed] [Google Scholar]
- ALNAAS AA, MOON CL, ALTON M, REED SM & KNOWLES MK 2017. Conformational Changes in C-Reactive Protein Affect Binding to Curved Membranes in a Lipid Bilayer Model of the Apoptotic Cell Surface. The Journal of Physical Chemistry B, 121, 2631–2639. [DOI] [PubMed] [Google Scholar]
- ANANTHARAM A, ONOA B, EDWARDS RH, HOLZ RW & AXELROD D 2010. Localized topological changes of the plasma membrane upon exocytosis visualized by polarized TIRFM. J Cell Biol, 188, 415–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANDERSON TH, MIN Y, WEIRICH KL, ZENG H, FYGENSON D & ISRAELACHVILI JN 2009. Formation of supported bilayers on silica substrates. Langmuir, 25, 6997–7005. [DOI] [PubMed] [Google Scholar]
- BACIA K, FUTAI E, PRINZ S, MEISTER A, DAUM S, GLATTE D, BRIGGS JA & SCHEKMAN R 2011. Multibudded tubules formed by COPII on artificial liposomes. Sci Rep, 1, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARLOWE C, ORCI L, YEUNG T, HOSOBUCHI M, HAMAMOTO S, SALAMA N, REXACH MF, RAVAZZOLA M, AMHERDT M & SCHEKMAN R 1994. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell, 77, 895–907. [DOI] [PubMed] [Google Scholar]
- BARLOWE C & SCHEKMAN R 1993. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature, 365, 347–9. [DOI] [PubMed] [Google Scholar]
- BOLLEN IC & HIGGINS JA 1980. Phospholipid asymmetry in rough- and smooth-endoplasmic-reticulum membranes of untreated and phenobarbital-treated rat liver. Biochem J, 189, 475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAMBREA LR & HOVIS JS 2007. Formation of three-dimensional structures in supported lipid bilayers. Biophys J, 92, 3587–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHERNOMORDIK LV, ZIMMERBERG J & KOZLOV MM 2006. Membranes of the world unite! J Cell Biol, 175, 201–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CREMER PS & BOXER SG 1999. Formation and spreading of lipid bilayers on planar glass supports. Journal of Physical Chemistry B, 103, 2554–2559. [Google Scholar]
- DAR S, KAMERKAR SC & PUCADYIL TJ 2015. A high-throughput platform for real-time analysis of membrane fission reactions reveals dynamin function. Nat Cell Biol, 17, 1588–96. [DOI] [PubMed] [Google Scholar]
- DAR S, KAMERKAR SC & PUCADYIL TJ 2017. Use of the supported membrane tube assay system for real-time analysis of membrane fission reactions. Nat Protoc, 12, 390– 400. [DOI] [PubMed] [Google Scholar]
- FROST A, PERERA R, ROUX A, SPASOV K, DESTAING O, EGELMAN EH, DE CAMILLI P & UNGER VM 2008. Structural basis of membrane invagination by F-BAR domains. Cell, 132, 807–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FUTAI E, HAMAMOTO S, ORCI L & SCHEKMAN R 2004. GTP/GDP exchange by Sec12p enables COPII vesicle bud formation on synthetic liposomes. EMBO J, 23, 4146– 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANNA MGT, MELA I, WANG L, HENDERSON RM, CHAPMAN ER, EDWARDSON JM & AUDHYA A 2016. Sar1 GTPase Activity Is Regulated by Membrane Curvature. J Biol Chem, 291, 1014–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARIRI H, BHATTACHARYA N, JOHNSON K, NOBLE AJ & STAGG SM 2014. Insights into the mechanisms of membrane curvature and vesicle scission by the small GTPase Sar1 in the early secretory pathway. J Mol Biol, 426, 3811–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HATZAKIS NS, BHATIA VK, LARSEN J, MADSEN KL, BOLINGER PY, KUNDING AH, CASTILLO J, GETHER U, HEDEGARD P & STAMOU D 2009. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat Chem Biol, 5, 835–41. [DOI] [PubMed] [Google Scholar]
- HELM CA, LAXHUBER L, LOSCHE M & MOHWALD H 1986. Electrostatic Interactions in Phospholipid-Membranes .1. Influence of Mono-Valent Ions. Colloid and Polymer Science, 264, 46–55. [Google Scholar]
- HINSHAW JE & SCHMID SL 1995. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature, 374, 190–2. [DOI] [PubMed] [Google Scholar]
- KALB E, FREY S & TAMM LK 1992. Formation of supported planar bilayers by fusion of vesicles to supported phospholipid monolayers. Biochim Biophys Acta, 1103, 307–16. [DOI] [PubMed] [Google Scholar]
- KENDALL EL, MILLS E, LIU JW, JIANG XM, BRINKER CJ & PARIKH AN 2010. Salt-induced lipid transfer between colloidal supported lipid bilayers. Soft Matter, 6, 2628–2632. [Google Scholar]
- KIESSLING V & TAMM LK 2003. Measuring distances in supported bilayers by fluorescence interference-contrast microscopy: polymer supports and SNARE proteins. Biophys J, 84, 408–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOERNER MM, PALACIO LA, WRIGHT JW, SCHWEITZER KS, RAY BD & PETRACHE HI 2011. Electrodynamics of lipid membrane interactions in the presence of zwitterionic buffers. Biophys J, 101, 362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KREUTZBERGER AJB, KIESSLING V, LIANG B, YANG ST, CASTLE JD & TAMM LK 2017. Asymmetric Phosphatidylethanolamine Distribution Controls Fusion Pore Lifetime and Probability. Biophys J, 113, 1912–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUGE O, DASCHER C, ORCI L, ROWE T, AMHERDT M, PLUTNER H, RAVAZZOLA M, TANIGAWA G, ROTHMAN JE & BALCH WE 1994. Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J Cell Biol, 125, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUROKAWA K, SUDA Y & NAKANO A 2016. Sar1 localizes at the rims of COPII-coated membranes in vivo. J Cell Sci, 129, 3231–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE JB, PETROV PG & DOBEREINER H 1999. Curvature of zwitterionic membranes in transverse pH gradients. Langmuir, 15, 8543–8546. [Google Scholar]
- LIPOWSKY R 2013. Spontaneous tubulation of membranes and vesicles reveals membrane tension generated by spontaneous curvature. Faraday Discuss, 161, 305–31; discussion 419–59. [DOI] [PubMed] [Google Scholar]
- LONG KR, YAMAMOTO Y, BAKER AL, WATKINS SC, COYNE CB, CONWAY JF & ARIDOR M 2010. Sar1 assembly regulates membrane constriction and ER export. J Cell Biol, 190, 115–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCMAHON HT & BOUCROT E 2015. Membrane curvature at a glance. J Cell Sci, 128, 1065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEUMANN S, PUCADYIL TJ & SCHMID SL 2013. Analyzing membrane remodeling and fission using supported bilayers with excess membrane reservoir. Nature Protocols, 8, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PABST G, HODZIC A, STRANCAR J, DANNER S, RAPPOLT M & LAGGNER P 2007. Rigidification of neutral lipid bilayers in the presence of salts. Biophys J, 93, 2688– 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PASSMORE DR, RAO TC, PELEMAN AR & ANANTHARAM A 2014. Imaging plasma membrane deformations with pTIRFM. J Vis Exp, e51334. [DOI] [PMC free article] [PubMed]
- PETER BJ, KENT HM, MILLS IG, VALLIS Y, BUTLER PJ, EVANS PR & MCMAHON HT 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science, 303, 495–9. [DOI] [PubMed] [Google Scholar]
- PETRACHE HI, TRISTRAM-NAGLE S, HARRIES D, KUCERKA N, NAGLE JF & PARSEGIAN VA 2006. Swelling of phospholipids by monovalent salt. J Lipid Res, 47, 302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PUCADYIL TJ & SCHMID SL 2008. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell, 135, 1263–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PUCADYIL TJ & SCHMID SL 2010. Supported bilayers with excess membrane reservoir: a template for reconstituting membrane budding and fission. Biophys J, 99, 517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAO TC, SANTANA RODRIGUEZ Z, BRADBERRY MM, RANSKI AH, DAHL PJ, SCHMIDTKE MW, JENKINS PM, AXELROD D, CHAPMAN ER, GIOVANNUCCI DR & ANANTHARAM A 2017. Synaptotagmin isoforms confer distinct activation kinetics and dynamics to chromaffin cell granules. J Gen Physiol, 149, 763–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAITO K, MAEDA M & KATADA T 2017. Regulation of the Sar1 GTPase Cycle Is Necessary for Large Cargo Secretion from the Endoplasmic Reticulum. Front Cell Dev Biol, 5, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SENDECKI AM, POYTON MF, BAXTER AJ, YANG T & CREMER PS 2017. Supported Lipid Bilayers with Phosphatidylethanolamine as the Major Component. Langmuir, 33, 13423–13429. [DOI] [PubMed] [Google Scholar]
- SEU KJ, PANDEY AP, HAQUE F, PROCTOR EA, RIBBE AE & HOVIS JS 2007. Effect of surface treatment on diffusion and domain formation in supported lipid bilayers. Biophys J, 92, 2445–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHEETZ MP 2001. Cell control by membrane-cytoskeleton adhesion. Nat Rev Mol Cell Biol, 2, 392–6. [DOI] [PubMed] [Google Scholar]
- SHEETZ MP, SABLE JE & DOBEREINER HG 2006. Continuous membrane cytoskeleton adhesion requires continuous accommodation to lipid and cytoskeleton dynamics. Annu Rev Biophys Biomol Struct, 35, 417–34. [DOI] [PubMed] [Google Scholar]
- SHI Z & BAUMGART T 2015. Membrane tension and peripheral protein density mediate membrane shape transitions. Nat Commun, 6, 5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMUNOVIC M, MANNEVILLE JB, RENARD HF, EVERGREN E, RAGHUNATHAN K, BHATIA D, KENWORTHY AK, VOTH GA, PROST J, MCMAHON HT, JOHANNES L, BASSEREAU P & CALLAN-JONES A 2017. Friction Mediates Scission of Tubular Membranes Scaffolded by BAR Proteins. Cell, 170, 172–184 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SNEAD WT, HAYDEN CC, GADOK AK, ZHAO C, LAFER EM, RANGAMANI P & STACHOWIAK JC 2017. Membrane fission by protein crowding. Proc Natl Acad Sci U S A, 114, E3258–E3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STACHOWIAK JC, SCHMID EM, RYAN CJ, ANN HS, SASAKI DY, SHERMAN MB, GEISSLER PL, FLETCHER DA & HAYDEN CC 2012. Membrane bending by protein-protein crowding. Nat Cell Biol, 14, 944–9. [DOI] [PubMed] [Google Scholar]
- TAMM LK & MCCONNELL HM 1985. Supported phospholipid bilayers. Biophys J, 47, 105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG MS, MESSERSMITH RE & REED SM 2012. Membrane curvature recognition by C-reactive protein using lipoprotein mimics. Soft Matter, 8, 7909–7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


