Abstract
The genetic, molecular and neuronal mechanism underlying circadian activity rhythms is well characterized in the brain of Drosophila. The small ventrolateral neurons (s-LNVs) and pigment dispersing factor (PDF) expressed by them are especially important for regulating circadian locomotion. Here we describe a novel gene, Dstac, which is similar to the stac genes found in vertebrates that encode adaptor proteins, which bind and regulate L-type voltage-gated Ca2+ channels. We show that Dstac is coexpressed with PDF by the s-LNVs and regulates circadian activity. Furthermore the L-type Ca2+ channel, Dmca1D, appears to be expressed by the s-LNVs. Since vertebrate Stac3 regulates an L-type Ca2+ channel we hypothesize that Dstac regulates Dmca1D in s-LNVs and circadian activity.
Keywords: Stac, pigment dispersing factor, circadian rhythm, L-type calcium channel
Introduction
The Drosophila brain contains a network of neurons that express clock genes and regulate circadian locomotion (Nitabach & Taghert, 2008). A subset of neurons that expresses the PDF neuropeptide is critical for rhythmicity of locomotor behavior. These are the large and small ventrolateral neurons (l-LNVs and s-LNVs) (Helfrich-Forster, 1997; Renn et al., 1999). Genetic ablation of PDF+ neurons, genetic silencing of PDF+ neurons and null mutations in pdf all disrupt circadian rhythms in locomotion (Renn et al., 1999; Nitabach et al., 2002; Sheeba et al., 2010). Specific knockdown of PDF in s-LNV neurons disrupts circadian locomotion; demonstrating the critical role of PDF in the s-LNV neurons for circadian rhythms (Shafer et al., 2009). Furthermore PDF, sNPF, which is another neuropeptide expressed by s-LNv neurons, and light act to set the phases of circadian neurons that are sequentially active via inhibition of Ca2+ activity in these neurons (Liang et al., 2017). This includes negative feedback onto the s-LNV neurons. Thus proper activity of PDF+ neurons is key to proper circadian rhythm.
Vertebrate genomes contain a small family of stac (SH3 and cysteine-rich domain containing protein) genes with stac3 expressed selectively by skeletal muscles (Suzuki et al., 1996; Horstick et al., 2013; Nelson et al., 2013; Reinholt et al., 2013). Stac3 is a regulator of electrical/contraction (EC) coupling in skeletal muscles and is required for release of Ca2+ from the sarcoplasmic reticulum of skeletal muscles and normal muscle contraction in both zebrafish and mice (Horstick et al., 2013; Nelson et al., 2013). Stac3 regulates EC coupling by controlling the stability, organization and voltage-dependency of the dihydropyridine receptor, a L-type Ca2+ channel (CaCh) (Linsley et al., 2017), which is the voltage sensor for EC coupling in skeletal muscles. Stac3 binds to CaV1.1, the α subunit of the skeletal muscle dihydropyridine receptor (Campiglio & Flucher, 2017; Wong et al., 2017), and a missense mutation in STAC3 is causal for the congenital Native American myopathy (Horstick et al., 2013). The other stac genes are expressed by a subset of neurons in the vertebrate nervous system (Suzuki et al., 1996; Lein et al., 2007; Legha et al., 2010) but their function is unknown. The finding that Stac3 regulates L-type CaChs, however, suggests that the other Stac proteins may also regulate CaChs in neurons. In fact Stac1 can form a molecular complex with CaV3.2 to regulate the surface expression of this T-type CaChs in mammalian cell lines (Rzhepetskyy et al., 2016), and the tandem SH3 domains of Stac2 each bind Cav1.1 and Cav1.2 (Wong et al., 2017). Thus it is possible that neuronal Stac proteins may regulate CaChs for normal nervous system function.
To better understand how the s-LNVs regulate circadian locomotion in the Drosophila brain we identified a novel gene Dstac that is similar to the vertebrate stac genes. Dstac is expressed by a subset of neurons in the Drosophila brain including the PDF+ s- and l-LNVs and regulates circadian locomotion in Drosophila. Furthermore, the L-type CaCh, Dmca1D, appears to be expressed by these same neurons. These findings define a functional role for a stac gene expressed by neurons and suggest the hypothesis that Dstac regulates the L-type CaCh, Dmca1D, in the s-LNV neurons and that this is important for the regulation of circadian rhythm by PDF.
Materials and Methods
Fly strains
Fly strains used in this paper were: w1118, yw, Pdf(M)-GAL4 (Renn et al., 1999), UAS-DCR2 (BL#24651), UAS-Dstac-RNAi (VDRC 103824 used in the behavioral analysis and VDRC 105848 used in western blot), UAS-Luciferase-RNAi (BL#31603), Mef2-GAL4 (BL#27390), Dmca1D-GAL4 (VDRC 202490), Dstac-gfp trap (BL#40742), and UAS-mCD8-tdtomato (a gift from Dr. Bing Ye, University of Michigan).
Anti-Dstac production
A Dstac cDNA that contained sequences for the CRD and SH3 domains were cloned and expressed with a His-SUMO fusion protein in BL21 (DE3) cells, and purified using Ni-NTA agarose (Invitrogen). The His-SUMO tag was cleaved by SUMO proteases to obtain the untagged Dstac proteins. The purified untagged Dstac proteins were further purified by NuPAGE 4–12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using a Bis-Tris Gel (Invitrogen) followed by excision of the appropriate Coomassie-stained band. Rabbits were immunized by a commercial vendor (ProSci) with gel slices of purified Dstac proteins. Dstac antibodies in the antiserum were purified by using a NHS-activated agarose column that was conjugated with Dstac fusion proteins. The specifity of the anti-Dstac was confirmed by Western blot analysis.
Western blot analysis
Muscles from larvae with Dstac knockdowned specifically in body wall muscles (UAS-Dstac-RNAi/+;UAS-DCR2/Mef2-GAL4, n=37) and control larvae (UAS-DCR2/+;UAS-Luciferase-RNAi/Mef2-GAL4, n=37) were dissected and frozen immediately on dry ice. Muscles were homogenized in RIPA buffer (ThermoFisher) with protease inhibitors (ThermoFisher) and centrifuged to exclude debris. The muscle lysates were loaded (seven wells/genotype) and separated by SDS-PAGE. The antibodies used for immunoblotting were rabbit anti-Dstac (1:500), mouse anti-actin (1:10,000, MP Biomedicals), anti-rabbit IgG HRP (1:2000, Santa Cruz Biotechnology), and anti-mouse IgG HRP (1:10,000, Santa Cruz Biotechnology). The protein bands were detected with Chemidoc MP imaging system (Bio-Rad). The intensities of protein bands were quantified by gel analyzer of imageJ. The sizes of the protein bands were estimated from the Western blots using a regression analysis.
Immunofluorescence labeling
Third instar larval brains of Dstac-gfp trap were dissected in ice-cold 1x phosphate buffered saline (PBS) and fixed immediately in 4% paraformaldehyde in 1x PBS for 30 minutes at room temperature (RT). We rinsed the tissues with 1xPBS with 0.1% Triton X-100 (1x PBSTX-100) 5 times, 5 minutes/time, and permeabilized with 0.25% TX-100 in 1XPBS for 15 min at RT. After 5 washes (5 minutes/wash) with 1xPBSTX-100, we blocked the brains with 1% normal goat serum (NGS), 0.2% bovine serum albumin (BSA), and 0.1% TX-100 in 1x PBS for an hour at RT. The brains were incubated with chicken anti-GFP (1:5000, Aves Laboratories) in the blocking solution at 4 ˚C for 2 days. The brains were washed with 1xPBSTX-100 for 5 times, 5 minutes/time, and then incubated with anti-chicken Alexa Fluor 488 (1:2000 Jackson ImmunoResearch). Following 5 washes (5 minutes/wash) with 1xPBSTX-100, we mounted the brains with Fluoro Gel with DABCO (Electron Microscopy Sciences).
Adult Dstac-gfp trap brains were dissected in ice-cold 1xPBS and fixed immediately in 4% paraformaldehyde in 1x PBS for 30 minutes at RT. The brains were rinsed with 1xPBSTX-100 for 5 times, 15 minutes/time. The brains were blocked with 10% NGS in 1xPBS with 2% TX-100 for 2 h at RT. The brains were incubated with mouse anti-PDF (Blau, 2005)(1:500, PDF C7 (deposited by Blau, Justin (DSHB Hybridoma Product PDF C7)) in 1xPBS with 1% NGS and 0.25% TX-100 (dilution buffer) overnight at RT. The brains were washed with 1xPBS with 3% NaCl and 1% TX-100 (wash buffer) for 3 times, 15 minutes/time. We incubated the brains with anti-mouse Alexa Fluor 647 (1:1000, Invitrogen) in dilution buffer overnight at RT. The brains were then washed with wash buffer and incubated with chicken anti-GFP (1:5000, Aves Laboratories) and anti-chicken Alexa Fluor 488 (1:2000 Jackson ImmunoResearch) following the same procedures described above. The brains were mounted with DABCO FluoroGel.
w1118 adult brains were dissected and immunolabeled following the same procedures described above except that the brains were fixed overnight with Bouin’s fixative and treated with 0.05% collagenase (Worthington Biochemical) for 10 min. The antibodies used were mouse anti-PDF (1:500, Developmental Studies Hybridoma Bank at the University of Iowa), anti-mouse Alexa Fluor 647 (1:500; Invitrogen), rabbit anti-Dstac (1:50, this paper), and anti-rabbit 488 (1:1000; Invitrogen).
The antibodies used in the labeling of UAS-mCD8-tdtomato/+;Dmca1D-GAL4/+ adult brains were mouse anti-PDF (1:500, Developmental Studies Hybridoma Bank at the University of Iowa), anti-mouse Alexa Fluor 488 (1:1000, Invitrogen), rabbit anti-RFP (1:1000, Rockland), and anti-rabbit 647 (1:1000. Invitrogen).
The Dstac-gfp trap larval/adult brains and larval muscles were imaged using an upright Leica SP5 confocal microscope with a 20x or 63x objective. The others were imaged using a Leica SP8 confocal microscope with a 63x or 100x objective and digital zoom.
in situ hybridization
A 0.28-kilobase pair fragment of the Dstac cDNA was cloned into a TA vector. The sequence of the fragment was: cgcaatggagcctccagcacagcctccgagccgctgcgtcccaatctggatggcagccaccatctgcaggagtacacctacaagaagataacggcctgcgacgtctgctcccagattctgagagggcacacacgccagggattacgctgccgcatctgcaagctgaacgcccatggagattgcgcccccaatctgccgcgctgccagccaaagcagaagctgctccggcgacagaagagcacatcggagctggagaatcgtgttgatatcgaggaagaaa.
The Dstac RNA probe was synthesized using T7 and SP6 RNA polymerases (Promega) and digoxigenin-labeled dNTPs (Roche Diagnostics). In situ hybridization was performed on the 3rd instar larval muscles of Dstac-gfp trap. The 3rd instar larval muscles were dissected in 1xPBS and fixed with 4% PFA immediately after dissection. Samples were washed by 1xPBS with 0.1% tween20 (PBT) and dehydrated with 25%, 50%, 75% and 100% methanol and rehydrated with methanol. Samples were washed with 1xPBT and incubated with 5ug/mL Proteinase K in 1xPBT for 30 minutes at RT. The samples were washed with 1xPBT and fixed again with 4% PFA and 0.25% Glutaraldehyde in 1xPBT. The samples were washed by 1xPBT, one wash with the hybridization buffer (5xSSC containing 50% formamide, 0.1% tween-20, 0.29 mg/mL tRNA (Roche Diagnostics), 0.05 mg/mL Heparin, and 9.2 mM citric acid), and incubated with hybridization buffer (HB) for 2 h at 65 ˚C. The samples were then incubated with 1.2 ng/uL Dstac RNA probes overnight at 65 ˚C. Following hybridization, the samples were washed 4 times with mixture of HB and 2xSSC with 0.1% tween-20 (SSCT), twice with 2xSSCT, and twice with 0.2xSSCT at 65 ˚C. Samples were washed with mixture of 0.2xSSCT and 1xPBT for 6 times at room temperature. Samples were blocked with 5% sheep serum and 0.2% BSA in 1xPBT for 2 h at RT. Samples were incubated with alkaline phosphatase-conjugated anti-digoxigenin Fab fragment (1:5000, Roche Diagnostics) at 4˚C overnight. Samples were washed 15 times with 1x PBT and 3 times with NTMT (0.1M NaCl, 0.1 M Tris-HCl pH 9.5, 50 mM MgCl2, 1% tween-20, and 0.24 mg/mL Levamisole). The Dstac RNA probe hybridization was visualized by incubating the samples with Fast-Red (Roche Diagnostics). The development was stopped by 2 washes of NTMT and 10 washes of 1xPBT. The samples were fixed again with 4% PFA and 0.25% Glutaraldehyde in 1xPBT. Dstac-gfp trap was labeled following the immunofluorescence labeling protocol with rabbit anti-GFP (1:1000, Torrey Pines Biolabs) and anti-rabbit Alexa Fluor 488 (1:1000, Invitrogen). Samples were washed with 50% and 70% of glycerol in 1xPBT and mounted. Samples were imaged using a Leica SP5 confocal microscope with a 20x objective.
Analysis of activity rhythms
All flies for the activity rhythm analysis were reared at 25 ˚C under a 12hr:12hr light dark cycle (LD). Adult males aged between 1 and 5 days were placed individually in glass tubes and loaded onto the Trikinetics DAM2 monitors (Waltham, MA) for locomotor activity recordings. Flies were entrained for 6 days at 25 ˚C under LD and then subjected to constant darkness (DD) for 14 days. The locomotor activity data were processed by the Drosophila Activity Monitoring system (Trikinetics, Waltham, MA) into 30 min bins for activity analysis under LD and 1 min bin under DD. The activity rhythm analysis under LD was analyzed by a counting macro (Pfeffenberger et al., 2010). The normalized morning startle response was determined as the ratio of the activity of the first bin in light/activity of the last bin in dark (Stoleru et al., 2004). Thus a ratio of 1 would signify the loss of the morning startle response. The morning and evening anticipation were calculated by the “anticipation phase score” method (ratio of the activity three hours prior to lights on/off to the activity six hours prior to lights on/off; Harrisingh et al., 2007). Clocklab (Actimetrics) was used for the circadian rhythm analysis. Dead flies were determined by loss of activity according to the actograms and were excluded from the analysis. The rhythmicity and free-runnig periods of individual flies were determined by Chi-square analysis with a confidence level of 0.01 (Sokolove & Bushell, 1978). “Power” is the peak activity in the Chi-square periodogram and “significance” is the minimum value in the periodogram where a fly is considered rhythmic. The rhythmic power was calculated by subtracting the “significance” values from the “power” values that are generated by the Chi-square periodogram analysis. A fly with a rhythmic power equal to or less than zero was considered arrhythmic. The free-running periods were analyzed with the range between 14 hours and 36 hours with 0.5-hour intervals. The analysis of free-running periods only included flies displaying significant periodicities under constant darkness and temperature.
Statistical Analysis
The rhythmic ratio (number of rhythmic flies/number of total flies x 100%) of the experimental group (Pdf-GAL4/y;UAS-dstac-RNAi/+;UAS-DCR2/+) and the other control groups was compared by Fischer’s Exact test. The free-running periods and morning and evening anticipation were analyzed by one-way ANOVA (Kruskal-Wallis test) among all the groups and Dunn’s multiple comparison tests were used to compare the control groups particularly with the experimental group. The normalized morning startle response was compared with the hypothetical value of 1 (defined as loss of morning startle response) by Wilcoxon Signed Rank Test. The protein band intensities between Dstac knockdown muscles and control muscles were compared by the Mann-Whitney U test. D’Agostino-Pearson omnibus normality test was used to decide the parametric tests or nonparametric tests to be used.
Results
Dstac is similar to the vertebrate stac genes
We identified a single Drosophila gene (CG43729) that was potentially related to the human STAC1 (SH3 and cysteine rich domain) gene with an Ensembl search (http://www.ensembl.org/Homo_sapiens/Gene/Compara_Tree?db=core;g=ENSG00000144681;r=3:36380344–36548007; Aken et al., 2017). An EnsemblMetazoa search further found potential stac genes in numerous invertebrate lineages (http://metazoa.ensembl.org/Drosophila_melanogaster/Gene/Compara_Tree/pan_compara?db=core;g=FBgn0263980;r=2R:15401096–15471884;t=FBtr0330009;collapse=none). CG43729 appears to be alternatively spliced (Fig. 1F) and encodes for a protein containing a SH3 domain and cysteine rich domain (CRD) that are the defining features unique to vertebrate stac genes (Suzuki et al., 1996). Furthermore as determined by EMBOSS (Rice et al., 2000), the amino acid sequences of the SH3 and CRD of CG43729 are well conserved with those of zebrafish Stac1 (67% and 59% similarity, respectively), human Stac1 (71% and 65%, respectively), zebrafish Stac3 (67% and 57% similarity, respectively) and human Stac3 (71% and 61%) (Figure1A, B) suggesting that CG43729 encodes for a Stac protein and thus named this gene Dstac. The putative Dstac protein also appears to contain a BAR domain normally associated with membrane curvature (reviewed in Salzer et al., 2017) in the N terminal region of Dstac that is not found in vertebrate Stac proteins but is found in invertebrate Stac proteins.
Figure 1. Dstac is similar to vertebrate Stac1 and Stac3 and expressed by a subset of neurons and by body wall muscles.
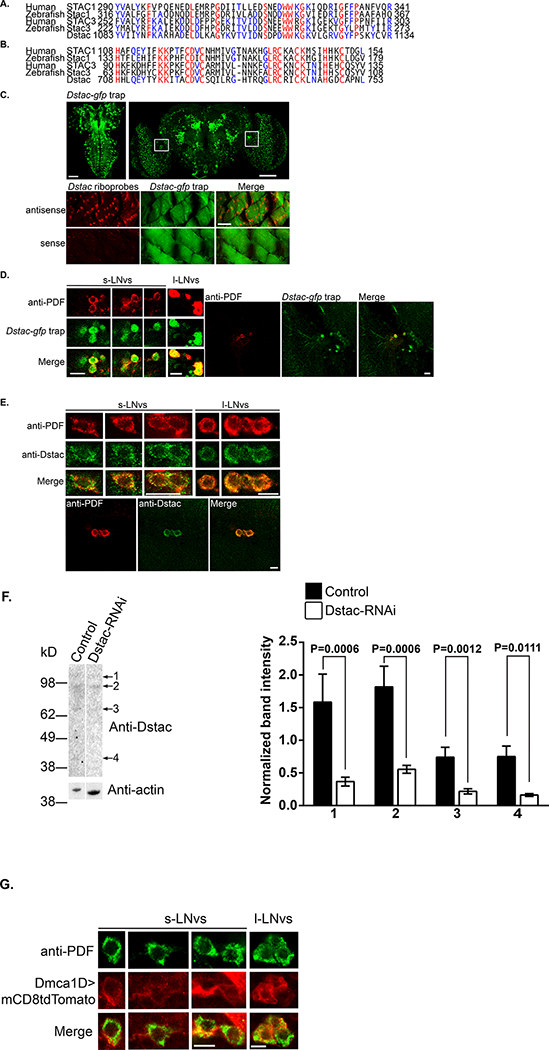
Alignment of the amino acid sequences of the SH3 domains (A) and CRD domains (B) between zebrafish Stac1, human Stac1, zebrafish Stac3, human Stac3 and Dstac. Amino acids that are identical among all three genes are highlighted in red; amino acids that are identical between Dstac and Stac1 or Stac3 are highlighted in blue. (C) The expression pattern of the Dstac-gfp trap (above) in the 3rd instar larval CNS (left) and in the adult brain (right). Expression of Dstac by body wall muscles determined by labeling by a riboprobe for Dstac (below). The larval CNS expression is from a stack of focal planes of the dorsal side of the 3rd instar larval CNS and adult brain expression from a stack of throughout the entire brain. The muscle images are a single focal plane. Boxes in the adult brain panel denote the region where the s- and l-LNV neurons are located. Scale bars, 75 μm. (D) Dstac-gfp trap adult brains labeled with anti-PDF showed colocalized expression of GFP and PDF in the s-LNV and l-LNV neurons. High magnification views of s- and l-LNVs (left panels). Lower magnification views of LNVs showing that only some Dstac-gfp neurons express PDF (right panels). Images are single focal planes. Scale bars, 10 μm. (E) Anti-PDF and anti-Dstac labeling of w1118 adult brain showed coexpression of PDF and Dstac in the s-LNVs and l-LNVs (top 3 sets of panels). Lower magnification images of l-LNVs showing that anti-Dstac labels PDF+ neurons but not other neurons in the vicinity (bottom set of panels). Images are single focal planes. Scale bars, 10 μm. (F) Western blots showed anti-Dstac labeled bands (1–4) in control larval muscles (UAS-DCR2/+;UAS-Luciferase-RNAi/Mef2-GAL4) and the intensities of all bands were significantly lower in Dstac-RNAi larval muscles (UAS-Dstac-RNAi/+;UAS-DCR2/Mef2-GAL4). The protein bands of control and Dstac-RNAi larval muscles were quantified and analyzed by Mann-Whitney U test. (G) UAS-mCD8-tdTomato/+;Dmca1D-GAL4/+ adult brain labeled with anti-PDF showed colocalized expression of tdTomato and PDF in the s-LNVs and l-LNVs. Images are single focal planes. Scale bars, 5 μm.
Dstac appears to be expressed by PDF+ neurons in the brain
Examination of a MiMIC transposon mediated transgenic line (Venken et al., 2011) in which gfp is inserted into the Dstac locus (Dstac-gfp trap) showed that Dstac appears to be expressed by both body wall muscles and a subset of neurons in the CNS of 3rd instar larvae and adult brain (Figure 1C). Interestingly in the adult brain the l-LNvs and s-LNVs that are key regulators of circadian locomotor rhythms express Dstac. In Dstac-gfp flies anti-PDF labeled l-LNvs and s-LNVs also expressed GFP (Figure 1D). Furthermore, these neurons were colabeled with anti-PDF and anti-Dstac (Figure 1E). Western blots from larval muscles probed with anti-Dstac labeled four bands (40, 69, 94 and 108 kD) suggesting that there are at least 4 isoforms of Dstac in muscles (Figure 1F). The protein levels of all four isoforms were significantly reduced when Dstac was knocked down in muscles (Figure 1F), indicating that anti-Dstac specifically detected Dstac proteins and that Dstac RNAi is effective.
The L-type Ca2+ channel, Dmca1D, appears to be expressed by PDF+ neurons in the brain
Since Stac3 regulates a L-type CaCh in vertebrate skeletal muscles (Linsley et al., 2017), we wondered whether Dstac might regulate a L-type CaCh in the PDF+ s-LNVs and l-LNVs. To see if this was possible we examined whether a L-type CaCh is coexpressed with PDF in the s-LNVs and l-LNVs. The Drosophila genome appears to contain a single L-type CaCh, Dmca1D (Zheng et al., 1995; Eberl et al., 1998). Anti-PDF labeling of Dmca1D:GAL4;UAS:mCD8tdTomato flies showed that the s-LNVs and l-LNVs coexpress mCD8tdTomato and PDF (Fig 1G). Thus it is possible that Dstac and Dmca1D are coexpressed by PDF+ s-LNVs and l-LNVs although the specificity of the Dmca1D:GAL4;UAS:mCD8tdTomato flies is as yet uncharacterized.
Knockdown of Dstac in PDF+ neurons disrupts circadian locomotion
Since Dstac is expressed by the PDF+ s-LNVs and l-LNVs, we tested whether knocking down Dstac specifically in PDF+ neurons might affect circadian locomotion (Table 1). Pdf:GAL4;UAS:DstacRNAi flies but not control flies displayed altered circadian locomotor rhythms (Figure 2A); a significant increase in arrhythmic flies in constant dark (DD) conditions and a small but significant increase in the free-running period of flies displaying significant periodicities under DD. Furthermore, under light/dark (LD) conditions, the startle response to light onset was eliminated, but there was no change in the morning nor evening anticipation (Figure 2B, D). There were no obvious morphological defects in Pdf:GAL4;UAS:DstacRNAi flies (Figure 2C) and other than the loss of the morning startle response their activity under LD was comparable to wt control flies suggesting that in these flies the altered circadian locomotor rhythm was not due to any obvious deleterious effect on locomotion in general.
Table 1.
Knocking down Dstac in PDF neurons increases arrhythmicity and free-running periods in DD and decreases morning startle response in LD.
| DD 5–13 | LD 3–6 | |||||
|---|---|---|---|---|---|---|
| Genotype | Rhythmicity | Free-running periods | Morning startle response | |||
| n rhythmic (%) |
p (Fischer’s Exact test) |
Hours Mean (±SEM) |
p (Dunn’s Multiple Comparison test) |
Normalized Mean (±SEM) |
p (Wilcoxon Signed Rank test) |
|
| yw,PDF-GAL4/y;+;+ | 32 (100) | 0.0001 | 24.11 (±0.04) |
< 0.0001 | 6.168 (±2.37) |
<0.0001 |
| yw/y;+;UAS-DCR2/+ | 29 (93) | 0.0106 | 24.16 (±0.08) |
0.0001 | 6.605 (±1.24) |
<0.0001 |
|
yw/y;UAS-Dstac- RNAi/+;+ |
29 (93) | 0.0106 | 24.24 (±0.16) |
0.0139 | 14.590 (±6.18) |
<0.0001 |
|
yw/y;UAS-Dstac- RNAi/+;UAS-DCR2/+ |
31 (96) | 0.0012 | 24.42 (±0.37) |
< 0.0001 | 7.601 (±2.85) |
<0.0001 |
|
yw,PDF-GAL4/y;+;UAS- DCR2/+ |
27 (90) | 0.0311 | 24.06 (±0.33) |
0.0368 | 1.879 (±0.11) |
<0.0001 |
|
yw,PDF-GAL4/y;UAS- Dstac-RNAi/+;+ |
28 (93) | 0.0106 | 24.80 (±0.05) |
> 0.9999 | 2.115 (±0.34) |
0.0002 |
|
yw,PDF-GAL4/y;UAS- Dstac-RNAi/+;UAS- DCR2/+ |
20 (64) | ------ | 24.80 (±0.11)# |
------ | 1.156 (±0.10) |
0.1870 |
n = number of flies tested
denotes significantly different from all other genotypes except for yw,PDF-GAL4/y;UAS-Dstac-RNAi/+;+
Figure 2. Knocking down Dstac in PDF neurons lead to arrhythmic circadian rhythms in DD and elimination of the morning startle response in LD.
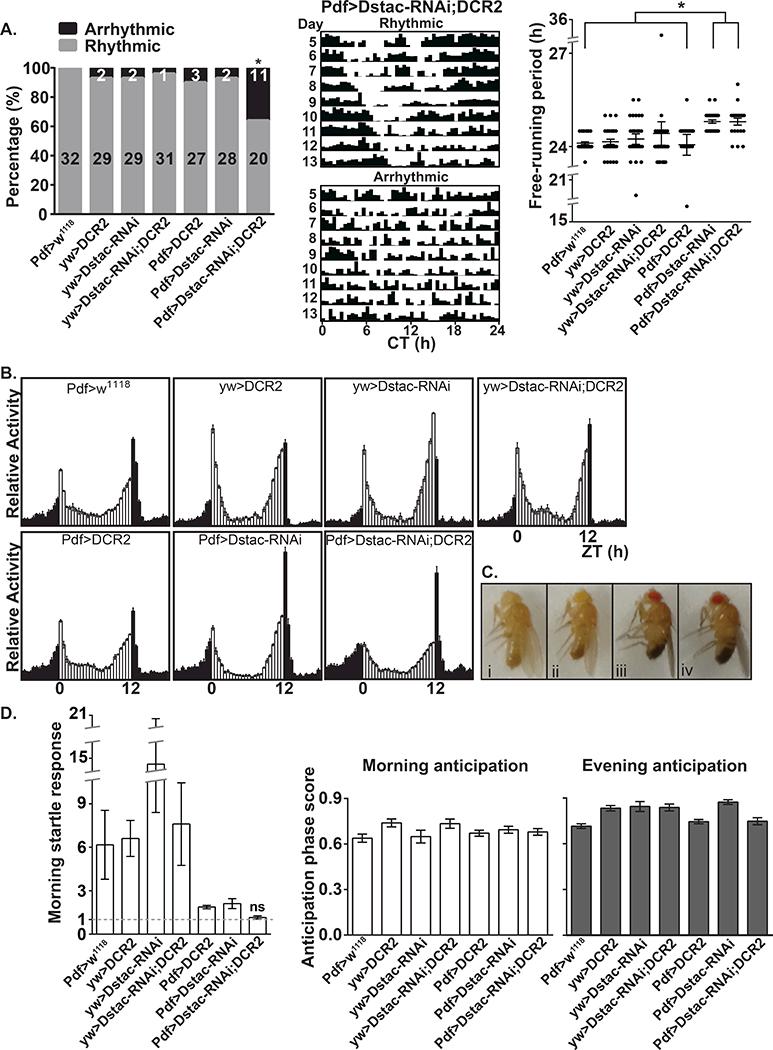
(A) Left, the percentage of rhythmic and arrhythmic flies from day 5 to 13 under DD. * denotes that the Dstac knockdown flies (Pdf-GAL4/y;UAS-Dstac-RNAi/+;UAS-DCR2/+) were significantly less rhythmic than all the other control groups (see Table 1). Numbers denote the number of flies that were rhythmic and arrhythmic. Center, actograms from a rhythmic and arrhythmic Dstac RNAi fly under DD. Right, the free-running periods of flies displaying significant periodicities under DD and constant temperature from day 5 to13 in constant darkness. * denotes that both Dstac knockdown flies with or without UAS-DCR2 displayed a slight but significant increase in the free-running periods when compared with the other control groups. Each dot represents an individual fly. (B) The activity histogram of each genotype from day 3 to 6 under LD showed no apparent change in morning and evening anticipation but a decrease morning startle response in Pdf-GAL4/y;UAS-Dstac-RNAi/+;UAS-DCR2/+ flies. (C) The morphology of Pdf:GAL4;UAS:DstacRNAi flies (2–3 days) appeared normal. (i) yw,PDF-GAL4/y;+;+. (ii) yw,PDF-GAL4/y;+;UAS-DCR2/+. (iii) yw,PDF-GAL4/y;UAS-Dstac-RNAi/+;+. (iv) yw,PDF-GAL4/y;UAS-Dstac-RNAi/+;UAS-DCR2/+. (D) Left, the value of the normalized morning startle response (see Methods) of Dstac knock down flies was not significantly (ns) different from the value of 1 by Wilcoxon Signed Rank test. The values of the normalized morning startle responses of all the other control groups were significantly larger than 1. The morning anticipation (middle) and evening anticipation (right) from day 3 to 6 under LD was not significantly different between the Dstac knock down flies and controls (Dunn’s multiple comparison test).
Discussion
The Stac proteins are defined by the unique combination of a SH3 and CRD (Suzuki et al., 1996). The stac genes are found widely both in vertebrate and invertebrate lineages. In most vertebrate genomes there is a small family of stac genes with stac3 selectively expressed by skeletal muscles while the other stacs are expressed within the nervous system. In Drosophila there appears to be a single stac gene. The conclusion that Dstac is a stac gene is based upon the presence of highly conserved SH3 and CRD of the predicted Dstac protein along with the expression of Dstac by both muscles and neurons. The latter is concordant with the expression of vertebrate stac3 by skeletal muscles and the other vertebrate stacs by neurons. One difference between invertebrate stac genes and vertebrate stac genes is the presence of a BAR domain. BAR domains are associated with membrane curvature (reviewed in Salzer et al., 2017) but the significance of the BAR domain in invertebrate stacs including Dstac is unknown. Interestingly, in vertebrates the Bin1 protein from which the BAR domain draws its name contains both BAR and a SH3 domain and like Stac3, associates with Cav1.1 in skeletal muscle where its BAR domain is associated with the formation of T-tubules in muscle (Lee et al., 2002). Mutations in the BAR domain of bin1 destabilize T-tubules and are associated with centronuclear myopathy in humans (Claeys et al., 2010). Thus the BAR domain may be critical to the function of Dstac in invertebrate muscle.
The expression of Dstac by PDF+ neurons and the disruption of circadian locomotion when Dstac is knocked down in these neurons suggest that Dstac is necessary for normal circadian locomotor rhythms in Drosophila. The disruption of such rhythms in Drosophila is the first functional phenotype described for a stac gene in neurons in any organism.
How does Dstac regulate the function of PDF+ neurons and circadian locomotion? The fact that Stac3 regulates L-type CaCh in zebrafish skeletal muscles (Linsley et al., 2017) suggests that Dstac might regulate Dmca1D, the Drosophila L-type CaCh, in PDF+ neurons. Indeed PDF+ neurons appear to express Dmca1D. However, whether Dstac regulates Dmca1D, which in turn regulates circadian locomotion is at present unknown. Testing this hypothesis will require direct examination of a circadian phenotype when Dmca1D is knocked down in the s-LNVs and analysis of Dmca1D L-type Ca2+ currents in s-LNVs in flies with Dstac knocked down selectively in these neurons.
If the hypothesis is correct, then an influx of Ca2+ into PDF+ neurons via the Dmca1D channels is critical for proper output presumably involving PDF by these neurons to the circadian network in the brain. l-LNV neurons fire Na+ channel dependent action potentials tonically or in bursts (Sheeba et al., 2008; Cao & Nitabach, 2008). 2–3 Hz membrane potential oscillations underlie the tonic pattern and slower oscillations of the bursting pattern. Both fast and slower oscillations are dependent on Na+ and Ca2+ influx. Additionally the membrane potential exhibits circadian changes with increasing hyperpolarization as the day progresses and increasing depolarization as the night progresses (Sheeba et al., 2008; Cao & Nitabach, 2008). One possibility is that Dstac and Dmca1D might be important for one or both of these changes in excitability in PDF+ neurons. Alternatively Dstac and Dmca1D might regulate the synaptic and/or paracrine response of PDF+ neurons to their inputs or the synaptic and/or paracrine output of the PDF+ neurons. For example, Dstac regulation of Dmca1D might be involved in the potential suppression of basal Ca2+ and termination of the circadian Ca2+ increase by PDF signaling in s-LNV neurons (Liang et al., 2017). However there was no effect on morning anticipation when Dstac was knocked down in the s-LNV neurons as one might expect from a disruption of signaling by these neurons so how Dstac might affect circadian rhythm may be complex. Further analysis will hopefully flesh out how Dstac and Dmca1D may regulate the function of the PDF+ neurons and thus circadian rhythms.
Acknowledgement
We thank Bethany Folk-Middlebrook, Miranda Lum and William Yau for maintaining Drosophila stocks and help with the genetics and characterization of Dstac and Cathy Collins for advise and use of facilities for the genetics.
Funding
Research was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS; RO1 AR063056) to JYK and by the National Science Foundation (IOS 1354046 to OTS. I-UH was supported in part by the Barbour Fellowship (Rackham Graduate School, University of Michigan), JWL by a Rackham Merit Fellowship (University of Michigan) and NIGMS (T32 GM007315), JV by a MCDB Undergraduate Summer Fellowship (University of Michigan) and BF-M by MPREP (NIH 5 R25 GM086262). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Declaration of interest
The authors declare no conflicts of interest.
ORCID
kuwada@umich.edu
References
- Aken BL, Achuthan P Akanni W, et al. (2017) Ensembl 2017. Nucleic Acids Research. 45:D635–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J (2005) The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J. Neurosci 25:5430–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campligio M, Flucher BE (2017) STAC3 stably interacts through its C1 domain with CaV1.1 in skeletal muscle triads. Scientific Rep. doi: 10.1038/srep41003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Nitabach MN (2008) Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci. 28:6493–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys KG, Maisonobe T, Bohm T, et al. (2010) Phenotype of a patient with recessive centronuclear myopathy and a novel BIN1 mutation. Neurol. 74:519–21. [DOI] [PubMed] [Google Scholar]
- Eberl DF, Ren D, Feng G, Lorenz LJ, Van Vactor D, Hall LM (1998) Genetic and developmental characterization of Dmca1D, a calcium channel alpha1 subunit gene in Drosophila melanogaster. Genetics. 148:1159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrisingh MC, Wu Y, Lnenicka GA, Nitabach MN (2007) Intracellular Ca2+ regulates free-running circadian clock oscillation in vivo. J. Neurosci 27:12489–12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C (1997) Development of pigment-dispersing hormone-immunoreactive neurons in the nervous system of Drosophila melanogaster. J Comp Neurol. 380:335–54. [DOI] [PubMed] [Google Scholar]
- Horstick EJ, Linsley JW, Dowling JJ et al. (2013) Stac3 is a component of the excitation-contraction coupling machinery and mutated in Native American myopathy. Nat Commun 4:1952. doi: 10.1038/ncomms2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Marcucci M, Daniell L, et al. (2202) Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science 16:1193–6. [DOI] [PubMed] [Google Scholar]
- Legha W, Gaillard S, Gascon E, Malapert P, et al. (2010) stac1 and stac2 genes define discrete and distinct subsets of dorsal root ganglia neurons. Gene Expr Patterns. 10:368–75. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature. 445:168–76. [DOI] [PubMed] [Google Scholar]
- Liang X, Holy TE, Taghert PH (2017) A series of suppressive signals within the Drosophila circadian neural circuit generates sequential daily outputs. Neuron. 94:1173–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsley JW, Hsu IU, Groom L, et al. (2017) Congenital myopathy results from misregulation of a muscle Ca2+ channel by mutant Stac3. Proc Natl Acad Sci USA. 114:228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Wu F, Liu Y, et al. (2013) Skeletal muscle-specific T-tubule protein STAC3 mediates voltage-induced Ca2+ release and contractility. Proc Natl Acad Sci USA. 110:11881–11886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH (2008) Organization of the Drosophila circadian control circuit. Curr Biol. 18:R84–93. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Blau J, Holmes TC (2002) Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 109:485–95. [DOI] [PubMed] [Google Scholar]
- Pfeiffenberger C, Lear BC, Keegan KP, Allada R (2010) Processing circadian data collected from the Drosophila Activity Monitoring (DAM) System. Cold Spring Harb Protoc. 2010;2010(11):pdb prot5519. [DOI] [PubMed] [Google Scholar]
- Reinholt BM, Ge X, Cong C, Gerrard DE, Jiang H (2013) Stac3 is a novel regulator of skeletal muscle development in mice. PLoS ONE. 8:e62760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SCP, Park JH, Rosbash M, Hall JC, Taghert PH (1999) A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 99:791–802. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: The European Molecular Biology Open Software Suite. Trends in Genetics. 16:276–7. [DOI] [PubMed] [Google Scholar]
- Rzhepetskyy Y, Lazniewska J, Proft J et al. (2016) A CaV3.2/Stac1 molecular complex controls T-type channel expression at the plasma membrane. Channels. 10:346–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer U, Kostan J, Djinovic-Carugo K (2017) Deciphering the BAR code of membrane modulators. Cell Mol Life Sci. 74:2413–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Taghert PH (2009) RNA-interference knockdown of Drosophila Pigment Dispersing Factor in neuronal subsets: the anatomical basis of a neuropeptide’s circadian functions. PLoS ONE. 4:12, e8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Gu H, Sharma VK et al. (2008) Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol. 99:976–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Holmes TC (2010) Persistence of morning anticipation behavior and high amplitude morning startle response following functional loss of small Ventral Lateral Neurons in Drosophila. PLoS ONE. 5:7, e11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolove PG, Bushell WN. (1978). The chi square periodogram: its utility for analysis of circadian rhythms. J Theor Biol. 72:131–160. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M (2001) Coupled oscillators control morning and evening locomotor behavior of Drosophila. Nature. 431:862–868. [DOI] [PubMed] [Google Scholar]
- Suzuki JK, Taga C, Yaoi T et al. (1996) Stac, a novel neuron-specific protein with cystein-rich and SH3 domains. Biochem Biophys Res Commun. 229:902–9. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Schulze KL, Haelterman NA, et al. (2011) MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nature Methods. 8:737–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong King Yuen SM, Campiglio M., Tung C, et al. (2017) Structural insights into binding of STAC proteins to voltage-gated calcium channels. Proc Natl Acad Sci USA. 114:E9520–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Feng G, Ren D et al. (1995) Cloning and characterization of a calcium channel α1 subunit from Drosophila melanogaster with similarity to the rat brain type D isoform. J Neurosci. 15:1132–43. [DOI] [PMC free article] [PubMed] [Google Scholar]


