Abstract
Developing precise and efficient gene editing approaches using CRISPR in primary human T cell subsets would provide an effective tool in decoding their functions. Towards this goal, we utilized lentiviral CRISPR/Cas9 systems to transduce primary human T cells to stably express the Cas9 gene and guide RNAs (gRNA) that targeted either coding or non-coding regions of genes of interest. We showed that multiple genes (CD4, CD45, CD95) could be simultaneously and stably deleted in naïve, memory, effector or regulatory (Treg) T cell susbets at very high efficiency. In addition, nuclease-deficient Cas9 (dCas9), associated with a transcriptional activator or repressor, can downregulate or increase expression of genes in T cells. For example, expression of Glycoprotein A repetitions predominant (GARP), a gene which is normally and exclusively expressed on activated Tregs, could be induced on non-Treg effector T cells by dCas9 fused to transcriptional activators. Further analysis determined that this approach could be utilized in mapping promoter sequences involved in gene transcription. Through this CRISPR/Cas9 mediated genetic editing we also demonstrated the feasibility of human T cell functional analysis in several examples: (1) CD95 deletion inhibited T cell apoptosis upon reactivation; (2) deletion of ORAI1, a Ca2+ release-activated channel, abolished Ca2+ influx and cytokine secretion, mimicking natural genetic mutations in immune-deficient patients; (3) transcriptional activation of CD25 or CD127 expression enhanced cytokine signaling by IL-2 or IL-7, respectively. Together, the application of CRISPR toolbox to human T cell subsets has important implications for decoding the mechanisms of their functional ouputs.
Introduction
T cell-based immunotherapy which utilizes the effector and regulatory functions of T cells has shown promise in the treatment of cancer, chronic viral infection (e.g., HIV) and various immune disorders (1–3). Due to the complexity in different T cell subsets and their specific roles in cell-to-cell interaction and signaling, understanding the molecular mechanisms governing the function of different human T cell subsets during immune responses is crucial (4–10). Therefore, application of methods for direct manipulation of genes are powerful tools to define T cell subset functions, support the development of assays for screening, validate T cell targeting treatments, or improve immunotherapy (11,12).
Various approaches have already been developed to genetically modify human T cells (1–6). RNA interference (RNAi) has been the predominant tool used to repress gene expression in human T cells (4, 6, 7). Recently, new tools have emerged for genome-level gene-editing, especially CRISPR/Cas9 (clustered regulatory interspaced short palindromic repeats) (8–11), which enables a relatively simple target domain construction and site-specific genome manipulation in primary cells (27, 28).
CRISPR/Cas9 consists of a poly-spacer precursor RNA that complementarily recognizes the protospacer sequence on target regions and the Cas9 nuclease protein. This chimeric nuclease, guided by a single guide RNA (sgRNA), induces site-specific double-strand DNA breaks (DSBs). The DNA repair mechanism of DSBs is followed by either non-homologous end joining (NHEJ) or homologous directed repair (HDR), which introduce random or specific mutation via nucleotides insertion, replacement, or deletion (9, 12). Further modification of the CRISPR/Cas9 system by mutating the nuclease function of Cas9 (dCas9) and fusing dCas9 to transcriptional activators or repressors (such as Vp16–p65 or KRAB domains, respectively) allows the activation or repression of gene transcription through targeting promoter or non-coding regions (13, 14). In comparison to the conventional methods of ectopic gene expression, the CRISPR/Cas9 approach enables more physiologically relevant control of gene expression through endogenous regulatory regions (13). The CRISPR/Cas9 system is also a powerful tool to identify the essential promoter regions and determine the function of non-coding elements, such as enhancers and non-coding RNAs (ncRNAs), which are involved in gene regulation (15, 16).
Here we have taken advantage of several CRISPR/Cas9 system approaches, using a lentiviral expression system, and demonstrate the feasibility of performing highly efficient and robust genetic modification in primary human T cell subsets for interrogation of their biological functions.
Materials and Methods
Lentiviral plasmid construction and viruses
LentiCRISPR v2 (Addgene plasmid #52961) (36), Lenti sgRNA(MS2)_zeo (Addgene plasmid #61427), dCas9-VP64_GFP (Addgene plasmid #61422) and Lenti MS2-P65-HSF1_Hygro (Addgene plasmid #61426) vectors were gifts from Feng Zhang (33). pHR-SFFV-KRAB-dCas9-P2A-mCherry (Addgene plasmid #60954) was a gift from Jonathan Weissman (32). TLCV2 (Addgene plasmid # 87360) was a gift from Adam Karpf. DNA sequences of all gRNAs used for gene knockout and promotor activation/repression are listed as 5′ to 3′ sequences in Supplemental Table I and Supplemental Table II respectively. The sequence of gRNAs used for gene knockout were designed using the CRISPR tool (http://crispr.mit.edu/) and the sequence of gRNAs used for targeting promoters of endogenous genes were designed using the Cas9 Activator Tool (http://sam.genome-engineering.org/database/). All sequences were selected to precede 5′-NGG protospacer-adjacent motif (PAM) sequence (27).
Cloning of gRNAs into LentiCRISPR v2 and Lenti sgRNA(MS2)_zeo was modified from Sanjana et al. (24). The lentiviral CRISPR plasmids were digested with BsmBI (Thermo Fisher Scientific) for 30 min at 37°C. The digested plasmids were gel purified using Agarose Gel and DNA Gel Extraction kit (Monarch), according to the manufacturer’s recommendations. The forward and reverse oligonucleotides that encode the gRNAs (Eurofins Genomics) were annealed and phosphorylated in the mix of T4 Ligation buffer and T4 PNK at 37°C for 30 min followed by heat inactivation at 95°C for 5 min, then ramp down to 25°C at 5°C/min. Diluted annealed oligos were ligated to digested plasmids with ligase in Ligase Buffer at room temperature for 10 min. The cloned constructs were then transformed into NEB® Stable Competent E. coli (High Efficiency) (New England Biolabs) according to the manufacturer’s protocol. The colonies were cultured overnight for plasmid DNA isolation using QIAprep Spin Miniprep Kit and Qiacube (Qiagen). Diagnostic digest was performed for comfirming the positive clones: the purified colonies with LentiCRISPR v2 and TLCV2 backbones were digested with both EcoR1 and BamHI restriction enzymes; Lenti sgRNA(MS2)_zeo vectors were digested with BciVI restriction enzyme. The colonies with positive insertion were confirmed by analyzing the resulting fragments by gel electrophoresis.
Lentivirus production and titer measurement
The lentiviruses pseudotyped with VSVG envelope were generated as previously described (37). Briefly, the confirmed plasmid DNA was co-transfected with VSVG, pLP1 and pLP2 plasmids into HEK293T cells at 80–90% confluency using Lipofectamine TM 3000 (Invitrogen) according to the manufacturer’s protocol. The transfection medium was replaced with RPMI with 10% FCS 6 hours-post transfection. Viral supernatants were collected 24-hours post-transfection and filtered through a 0.45 μm syringe filter (Millipore) to remove cellular debris. Lenti-X TM Concentrator (Takara Bio USA, Inc.) was used according to the manufacturer’s protocol to concentrate the virus 20x and the resulting lentiviral stocks were aliquoted and stored at −80 °C. To measure viral titers, virus preps were serially diluted on Jurkat cells (500 cells per well), and three days post infection, 1 μg/ml puromycin was added. After an additional 4 days of selection, when almost all control cells were, live cells were counted using flow cytometry and the number of cells transduced with 1 ml of virus supernatant was calculated as infectious units/per ml. Based on these titer values, primary T cells were transduced with a multiplicity of infection (MOI) of 5–10 as previously described (37).
T cell purification
Peripheral blood mononuclear cells from healthy individuals (New York Blood Center, New York, NY) were prepared using Ficoll-paque plus (GE Healthcare). CD4+ T cells and CD8+ T cells were isolated using Dynal CD4 Positive and CD8 Positive Isolation Kit (Invitrogen) respectively, and were >99% pure. Purified CD4+ cells were sorted in some experiments by flow cytometry (FACSAria; BD Biosciences) based on CD45RO, CCR7, CD25 and chemokine receptors expression into: 1) Naïve T cells (CD45RO− CCR7+CD25−), 2) Memory T cells (CD45RO+CD25−), 3) Naïve Tregs (CD45RO− CD25+) and 4), Th1 cells (CD45RO+CCR6−CCR4−CXCR3+), 5) Th2 cells (CD45RO+CCR6−CCR4+CXCR3−) and 6) Th17 cells (CD45RO+CCR6+). Sorted subsets were >98% pure and were kept at 37°C and 5% CO2 in complete RPMI 1640 medium (RPMI supplemented with 10% Fetal Bovine Serum (FBS, Atlanta Biologicals, Lawrenceville, GA), 8% GlutaMax (Life Technologies), 8% sodium pyruvate, 8% MEM Vitamins, 8% MEM Nonessential Amino acid and 1% penicillin/Streptomycin (all from Corning Cellgro).
T cell activation and transduction with CRISPR lentiviruses
Total CD4+ T cells, CD8+ T cells and CD4+ T cell subsets were stimulated using anti-CD3/anti-CD28 dynabeads (Invitrogen) and cultured in complete RPMI 1640 medium (Thermo Fisher Scientific) supplemented with IL-2 (10ng/ml). For gene deletion experiments, activated cells were transduced with LentiCRISPR v2 and selected with 0.4 μg/ml puromycin. For inducible gene deletion experiment, Jurkat and CD4+ T cells transduced with TLCV2 vector encoding Cas9 under inducible Tetracycline promoter and gRNA under constitutive U6 promoter. The transduced Jurkat and CD4+ T cells were then either treated with 1 μg/ml or 10 μg/ml doxycycline, respectively, one day post-transduction or left untreated. For gene promoter repression experiment, Jurkat and primary CD4+ T cells were first transduced with lentivectors pHR-SFFV-KRAB-dCas9-P2A-mCherry and Lenti-sgRNA(MS2)_zeo and then selected with 250 μg/ml zeocin 3 days post-transduction. For gene promoter activation experiments, Jurkat or primary T cells were first transduced with dCas9-VP64_GFP and Lenti_MS2-P65-HSF1_Hygro lentiviral vectors and then selected with 250 μg/ml hygromycin and expanded for 2 weeks. Then the infected and selected T cells expressing dCas9-VP64_GFP and P65 were reactivated with anti-CD3/anti-CD28 beads and transduced with lentiviruses encoding promoter-targeting gRNAs in Lenti-sgRNA_(MS2) vector and selected with 250 μg/ml zeocin 3 days post-transduction. All antibiotic selections were performed for 3 to 5 days or until more than 95% of non-transduced cells were dead. MOI of 5 and 1 were used to transduce primary T and Jurkat cells, respectively.
FACS analysis
Cell surface FACS staining was performed in FACS buffer (PBS + 2% FBS) by incubating cells with fluorochrome-conjugated antibodies for 30 min at 4°C. Antibodies used in surface staining are: CD4, CD45, CD95, CXCR3, CCR4, CCR6, CD8, GARP, LAP, IL-7R (CD127) and IL-2R (CD25) (all from Biolegend). Surface expression of human ORAI1 was determined by anti-human ORAI1 antibody (clone 29A2) (38), which recognizes the second extracellular loop of hORAI1. Mouse IgG1 antibody was used as isotype control. For detection, goat anti-mouse IgG conjugated to Alexa Fluor 647 secondary antibody (Life technologies) was used. For intracellular cytokine staining, cells were stimulated for 4 hours at 37°C with phorbol12-myristate 13-acetate (PMA; 40 ng/ml) and Ionomycin (Iono; 500 ng/ml) (both from Sigma) in the presence of GolgiStop (BD Biosciences). Cells were then stained with fixable viability dye (eBiosciences) and surface markers CD4, CD45 and CCR6 antibodies, then fixed and permeabilized using eBiosciences Fixation/ permeabilization buffers according to the manufacturer’s instructions, before staining for cytokines IFNγ, TNF, GM-CSF, IL-2, IL-4, IL-13, IL-17A and IL-22 (Biolegend). For Treg transcription factor intracellular staining, cells were stained with fixable viability dye and surface markers CD4 and CD45 antibodies, then fixed and permeabilized using eBiosciences Fixation/ permeabilization buffers before staining for Foxp3 and Helios antibodies (Biolegend). To detect TGFβ LAP protein binding to CRISPR-mediated GARP, T cells were incubated with TGFβ for 30 min at 37°C and washed with media. The cells were then stained with GARP and LAP antibodies to determine their expression on the cell surface as previously described (40). Flow cytometry analyses were performed using LSR Fortessa X-20 flow cytometer (BD Biosciences) and SP6800 Spectral Cell Analyzer (Sony Biotechnology).
Apoptosis detection
Jurkat cells and the human T cell subsets were transduced with lentiviruses targeting CD95 gene. Jurkat cells were activated by crosslinking CD95 using anti-CD95 monoclonal antibody (10 μg/ml, Serotec) coated plates. Primary CD4+ and CD8+ T cells were re-activated using anti-CD3/anti-CD28 coated beads (Invitrogen). Cells were activated for 24 hours, then analyzed for phosphatidylserine exposure by an Annexin-V-FITC (BD Bioscience) together with anti-CD95 staining according to the manufacturer’s instructions.
Cytokine stimulation and phospho-staining
Cells were stimulated with different concentrations of IL-2, IL-7 (Biolegend), or left unstimulated. One hour after cytokine stimulation, cells were washed with PBS (Phosphate Buffer Saline) and stained with CD127 and CD25 surface antibodies, and fixable viability dye (eBiosciences), then fixed using BD Cytofix (BD Biosciences) for 15 min at 37°C. Cells were then washed twice with FACS buffer and permeabilized using BD Phosflow™ Perm Buffer III (BD Bioscience) for 30 min at 4°C. Following permeabilization, cells were stained with Stat5 (pY694) antibody (BD Bioscience) for 30 min at room temperature.
Intracellular Ca2+ measurements
ORAI1-deleted or control (wild-type) Jurkat and CD4+ T cells were labeled with 2 μM Fura2-AM (Life Technologies) for 30 min in RPMI medium as described earlier (39). The cells were attached for 10 min to 96-well imaging plates (Fisher) coated with 0.01% poly-L-lysine (w/v) (Sigma) and washed twice with Ca2+-free Ringer (155 mM NaCl, 4.5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM D-glucose, and 5 mM Na-HEPES) solution. Changes in intracellular Ca2+ concentration were analyzed using a Flexstation3 plate reader (Molecular Devices) at 340 and 380 nm wavelengths. The T cells were stimulated with 1 μM thapsigargin (TG, EMD Millipore) in Ca2+-free Ringer solution, and Store-Operated Ca2+ Entry (SOCE) was analyzed after re-addition of 1 mM Ca2+ (final) Ringer solution at 400th sec.
Data Analysis
FACS data was analyzed using FlowJo (Tree Star, Ashland, OR). Statistical analyses were performed using GraphPad Prism 6.0 software (Graphpad Inc., La Jolla, CA). Erro bars represent SEM. Results were compared using two-tailed t tests. Bonferroni corrections were applied for multiple comparisons. For all experiments, significance was defined as **p < 0.01 and ***p <0.001.
Results
Deletion of multiple cell surface genes in primary human T cells via CRISPR/Cas9
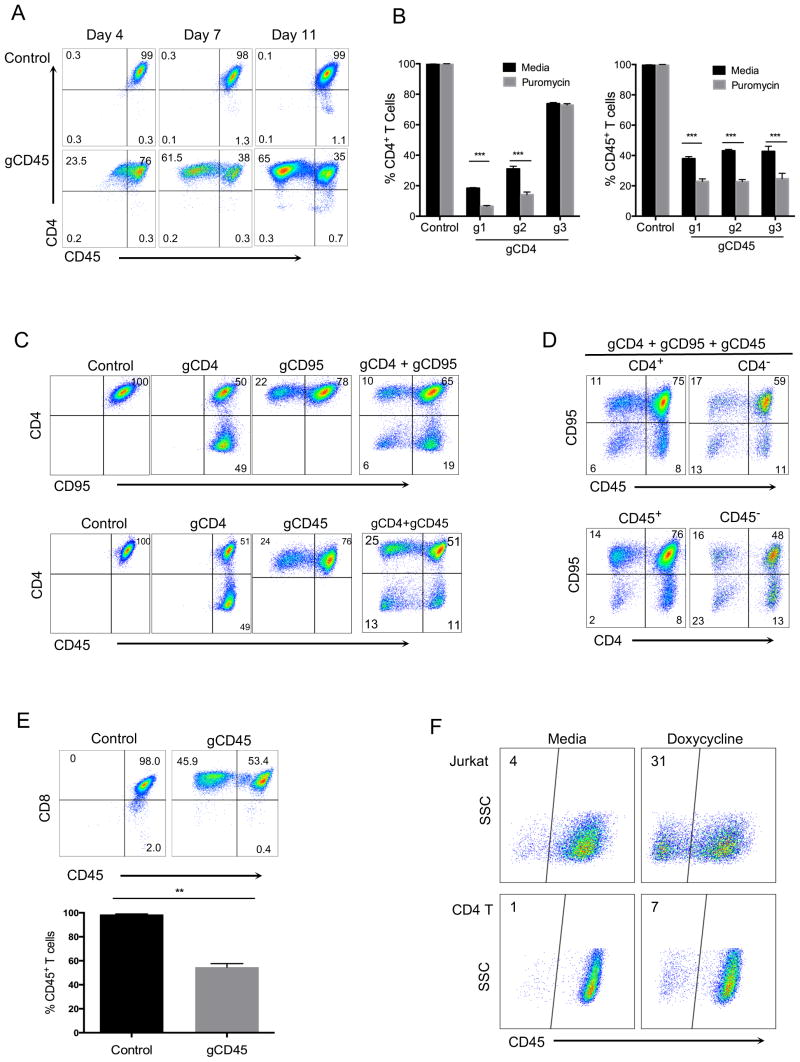
To develop an efficient and stable approach to knock out genes of interest in primary human T cells, we used a lentiviral vector (LentiCRISPR v2) which expresses both Cas9 and subcloned gene-specific gRNAs (17). We first designed gRNAs targeting three different T cell genes: CD4, CD45 and CD95. All of these proteins are highly expressed on the T cell surface under normal conditions. Purified primary CD4+ T cells were first activated using anti-CD3/anti-CD28 beads and then transduced with viruses generated by LentiCRISPR v2 vector encoding target-specific gRNAs. T cells were expanded in IL-2 and then analyzed for expression of CD4 and CD45 at different time points in culture. At 7 days post-transduction, CD45 proteins were reduced in more than half the CD4+ T cells in cells expressing Cas9 and gene specific gRNAs. This reduction was maintained stably at day 11 (Fig. 1A). Because the lentivector we used also encodes a puromycin resistance gene, we added puromycin at day 3 post-infection to a portion of the transduced cells and compared the gene knockout after 3 days to non-puromycin-selected cells. As expected, there was further statistically significant enrichment of CD4 or CD45 knockout cells with puromycin selection (Fig. 1B).
FIGURE 1. CRISPR-mediated deletion of single or multiple genes in primary human T cells.
(A) CD4 and CD45 expression in purified human CD4+ T cells. CD4+ T cells were activated with anti-CD3/anti-CD28 beads and the next day transduced with entiviruses targeting CD45 gene (gCD45-g2), as described in methods. Cells were then expanded in IL-2 and stained at indicated time points with CD4 and CD45 antibodies. (B) Comparison of knockout efficiency using three different gRNAs targeting CD4 or CD45 gene loci in human CD4+ T cells with or without puromycin selection. Puromycin (0.4 μg/ml) was added at day 3 post-transduction to a portion of activated T cells infected with the lentiviruses. Cells were then stained for CD4 or CD45 expresssion at day 7 post-transduction (day 4 post-selection). Data represent three independent experiments with cells isolated from different donors. Error bars represent SEM. ***p<0.001. (C) Double gene deletions within the same T cell. CD4+ T cells were activated and transduced with either single (gCD4, gCD95 or gCD45 alone), double (gCD4 + gCD95 or gCD4 + gCD45) lentiviruses specifically targeting CD4, CD95, or CD45 gene. (D) Analysis of triple gene deletions. T cells were simultaneously transduced with gRNAs targeting three genes (gCD4 + gCD95 + gCD45). Expression of CD45 and CD95 was determined after gating on CD4+ or CD4− T cells (top panel) and expression of CD4 and CD95 is shown after gating on CD45+ or CD45− T cells (bottom panel). Expression of surface markers was analyzed 7 days post-transduction without selection. (E) CD45 expression in human CD8+ T cells transduced with CRISPR/Cas9 targeting of CD45 gene. Purified CD8+ T cells were activated with anti-CD3/anti-CD28 beads and transduced next day with lentiCRISPR v2, as in CD4+ T cells in figure 1. Cells were cultured for another 8 days post-transduction in IL-2, without selection and stained with CD8 and CD45 antibodies. Data represent three independent experiments with cells isolated from different donors. Error bars represent SEM. **p<0.01. (F) Inducible CRISPR deletion of CD45 gene in primary human T cells and Jurkat cell line. Jurkat and CD4+ T cells transduced with a lentivector encoding Cas9 under inducible Tetracycline promoter and gRNA targeting CD45 gene (gCD45-g1). The transduced Jurkat and CD4+ T cells were either treated with 1 μg/ml or 10 μg/ml doxycycline, respectively, one day post-transduction or left untreated. After 7 day expansion in culture, without selection, cells were stained with anti-CD45 antibody to determine deletion. Data represent two independent experiments.
We next asked if we could delete two or potentially three genes simultanesouly in the same T cell. For this experiment, we transduced T cells with lentiviruses targeting CD4, CD45, and/or CD95 in a similar experiment as described above. After 7-days post transduction, combinations of gRNAs targeting either CD4 and CD45, or CD4 and CD95, showed proportionate double knockout (Fig. 1C). When viruses targeting all three genes were added together, we could also derive triple knockout cells, which were deleted for CD4, CD45 and CD95 in the same T cell (Fig. 1D).
We next applied similar CRISPR/Cas9-mediated gene deletion in primary human CD8 T cells. We chose one of the gRNAs targeting CD45 that showed highest deletion efficiency in previous CD4 T cell experiment (Figure. 1B). Purified human CD8+ T cells were similarly activated and transduced with the lentiviruses encoding Cas9 and gCD45. After 7-days post transduction, similar CRISPR-mediated gene deletion was also achieved in activated CD8+ T cells (Fig. 1E).
In order to demonstrate that the activity of CRISPR system can be regulated in human primary T cells, we engineered a version of LentiCRISPR v2 encoding Cas9 under inducible Tetracycline promoter and gRNA targeting CD45 gene (gCD45-g1). The expression of Cas9 in this vector is dependent on presence of the antibiotic doxycycline in the media. To test whether we can induce gene deletion only by after addition of doxycycline, Jurkat cells or activated CD4+ T cells were transduced with lentivirus encoding the inducible Cas9. One day after transduction, Jurkat and CD4+ T cells were either treated with 1 μg/ml or 10 μg/ml doxycycline, respectively, or left untreated as control. Cells were than expanded for 7 days in antibiotic free cell culture media and without any selection/ CD45 deletion was induced in doxycycline treated samples but very few CD45 deleted cells were present in the media treated controls (Fig. 1F).
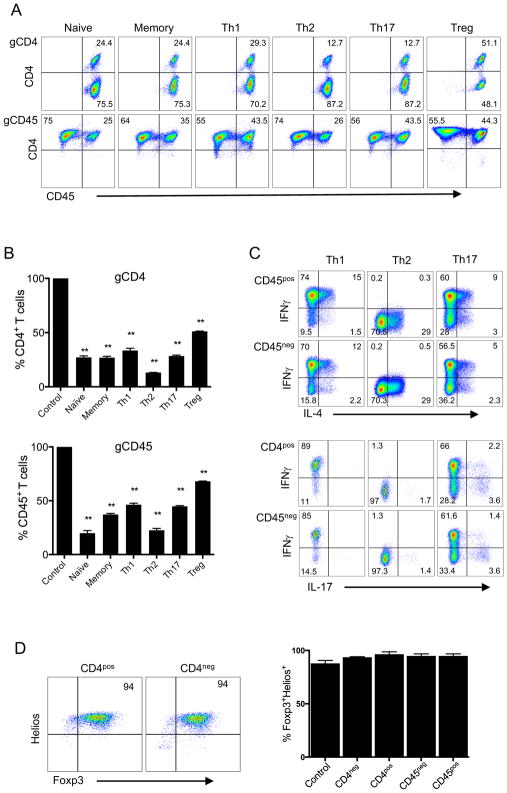
We then asked whether we could successfully employ CRISPR-mediated gene deletion in different T cell subsets. Accordingly, we first sorted CD4+ T cells into naïve (CD45RO−CCR7+CD25−), memory (CD45RO+CD25−), naïve regulatory (nTreg) (CD45RO−CD25+), and effector T cell (Th1, Th2, Th17) subsets based on chemokine receptors, namely CXCR3+ (Th1), CCR4+ (Th2) (18–20), and CCR6+ (Th17) (21). We then activated these subsets using anti-CD3/anti-CD28 beads and transduced the cells with CD4 or CD45 targeting gRNA-expressing lentivectors. Remarkably, CD4 or CD45 genes were deleted in all T cell subsets stably at high frequency, after in vitro expansion in culture (Fig. 2A and 2B). To ensure that the functional phenotype of T cells after CRISPR-mediated gene knockout was maintained, effector T cells were restimulated and then stained for the expression of secreted cytokines characteristic of each respective subtype (IFNγ for Th1, IL-4 for Th2, IL-17 for Th17). Cells that were deleted for CD45 or CD4 showed comparable cytokine synthesis patterns to controls (Fig. 2C and Supplemental Fig. 1A) and maintained the original chemokine receptor profiles associated with their effector functions (Supplemental Fig. 1B). Based on expression of Foxp3 and Helios, as previously described (22), Tregs derived from naïve precursors also maintained their phenotype after CRISPR-mediated CD4 deletion (Fig. 2D). Together, these results demonstrate the feasibility of highly efficient and stable CRISPR-mediated deletion in different primary human T cell subsets and multiple gene knockouts simultaneously in the same T cell.
FIGURE 2. CRISPR-mediated gene deletion in primary human T cell subsets.
(A) CD4 or CD45 expression upon CRISPR/Cas9-targeted deletion of these genes in different T cell subsets. Purified CD4+ T cells were sorted via flow cytometry using the following markers: CD45RO−CCR7+CD25− (Naïve), CD45RO+CD25− (Memory), CD45RO+CCR6−CCR4−CXCR3+ (Th1), CD45RO+CCR6−CXCR3−CCR4+ (Th2), CD45RO+CCR6+ (Th17) and CD45RO−CD25+ (nTreg). CD4+ T cells were activated with anti-CD3/anti-CD28 beads and transduced with lentiviruses targeting the CD4 (gCD4-g1 top panel) or CD45 (gCD45-g1, bottom panel) genes, as in Figure 1. Transduced cells were expanded for two weeks with IL-2, without selection, and stained with CD4 and CD45 antibodies. (B) Proportion of CD4 or CD45 postive cells within T cell subsets after CRISPR/Cas9 targeted deletion. CD4 or CD45 expression was determined in each T cell subset as in 2A, after two weeks post targeting either CD4 (gCD4-g2, top panel) or CD45 (gCD45-g1, bottom panel) respectively. Expresson of CD4 or CD45 was 100% in all T cell subsets transduced with control lentiviruses (Control). Data represent three independent experiments.Error bars represent SEM. **p<0.01. (C) Cytokine expression in T cell subsets after lentiviral transduction. Th1, Th2, and Th17 subsets expanded in vitro as in A, were restimulated with PMA and Ionomycin for 4 hours in the presence of GolgiStop. The expression of IFNγ, IL-4 and IL-17 was assessed by intracellular cytokine staining. FACS plots show IL-4, IFNγ and IL-17 expression in CD45+ and CD45− populations. (D) Foxp3 and Helios expression in Tregs after lentiviral transduction. Naïve Tregs (nTregs) were activated with anti-CD3/anti-CD28 beads and transduced with gCD4 or gCD45 lentiviruses the next day. nTregs were expanded in IL-2 for 2 weeks without selection. Expanded nTregs were stained for intracellular Foxp3 and Helios expression and analyzed after gating on CD4+ or CD4− population. The bar graph shows Foxp3+Helios+ cells after gating either on CD4+ and CD4− or CD45+ and CD45− in cells transduced with lentiviruses encoding gCD4-g1 or gCD45-g1 respectively. Control in bar graph is T cells transduced with empty vector lentivirus. Data are representative of two independent experiments.
Functional consequences of CRISPR-mediated gene deletion in primary T cells
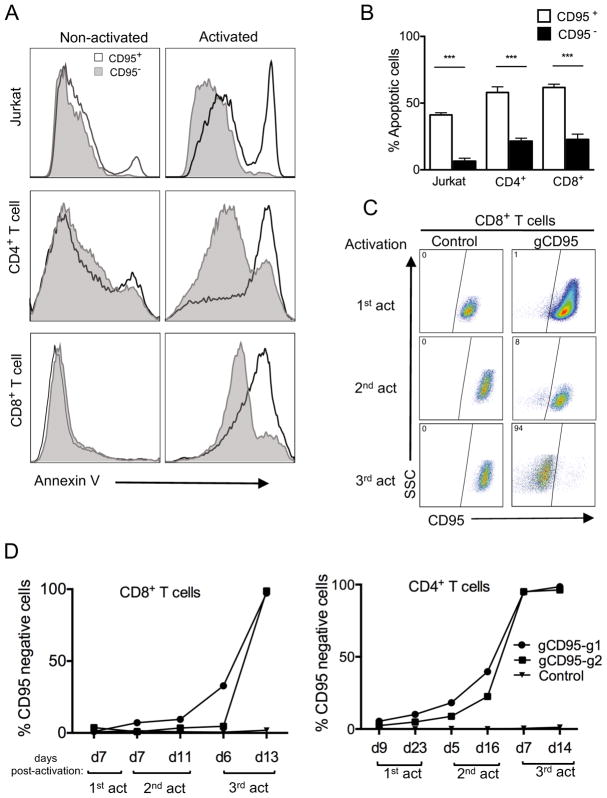
We next determined the functional outcomes in primary human T cells knocked out for several genes with known functions. First, we assessed cell death upon deletion of CD95, which induces apoptosis through signaling by its ligand, CD95L, during T cell receptor stimulation (23). In similar approaches as described above, we targeted and deleted CD95 by lentiviral transduction in both Jurkat and primary T cells. We then used a crosslinked anti-CD95 antibody on Jurkat cells, to signal through CD95. Upon 24 hours of activation apoptosis on CD95 knockout and positive cells were analyzed with apoptotic marker (Annexin V). Primary T cells with CD95 knockouts were activated by anti-CD3/anti-CD28 beads to induce apoptosis. In both Jurkat and primarty T cells, there was significantly less apoptosis in CD95 negative vs CD95 positie cells (Fig. 3A and 3B). Since CD95-deleted cells would have a survival advantage after repeated stimulations, we predicted that this would selectively enrich CRISPR-deleted subset over time. To test this, we transduced T cells with a very low MOI to start with few CD95-deleted T cells. We then monitored CD45 negative population in both CD4+ or CD8+ T cells upon repeated restimulation with anti-CD3/anti-CD28 beads and expansion for two weeks in IL-2. As predicted, CD95 negative CD4+ or CD8+ T cells increased after each round of stimulation compared to the control and by the third restimulation only surviving T cells were those deleted for CD95 expression (Fig. 3C and 3D).
FIGURE 3. Reduced apoptosis in CD95-deleted Jurkat cells and primary human T cell subsets.
(A) Analysis of CD95-induced cell death in CD95-deleted cells. Histogram overlays of Annexin V staining of CD95+ and CD95− cells in activated versus non-activated Jurkat cells and primary CD4+ and CD8+ T cells. Jurkat or primary T cells were activated and transduced with lentivirus targeting the CD95 gene (gCD95) and primary T cells were expanded in IL-2, without selection, then reactivated with anti-CD95 crosslinking antibody or anti-CD3/anti-CD28, respectively, for 24 hours. Reactivated cells were analyzed for apoptosis by Annexin V staining. (B) T cell apoptosis after CD95 gene knockout. Cells were gated on either CD95− or CD95+ 24 hours after reactivation as described in A.Data represent three independent experiments. Error bars represent SEM. ***p<0.001. (C) Preferential survival and expansion of CD95 positive population in CD8+ T cells. CD8+ T cells were transduced and reactivated as in A, through three subsequent restimulations and compared to empty vector control transduced cells. (D) Increased frequency of CD95 negative cells in CD8+ and CD4+ T cell after restimulations. Purified CD4+ or CD8+ T cell subsets were transduced with lentiviruses targeting the CD95 gene using two different gRNA clones (gCD95-g1 or gCD95-g2) and stained for CD95 expression at different time points after three re-stimulations using anti-CD3/anti-CD28 beads. Data is representative of two independent experiments.
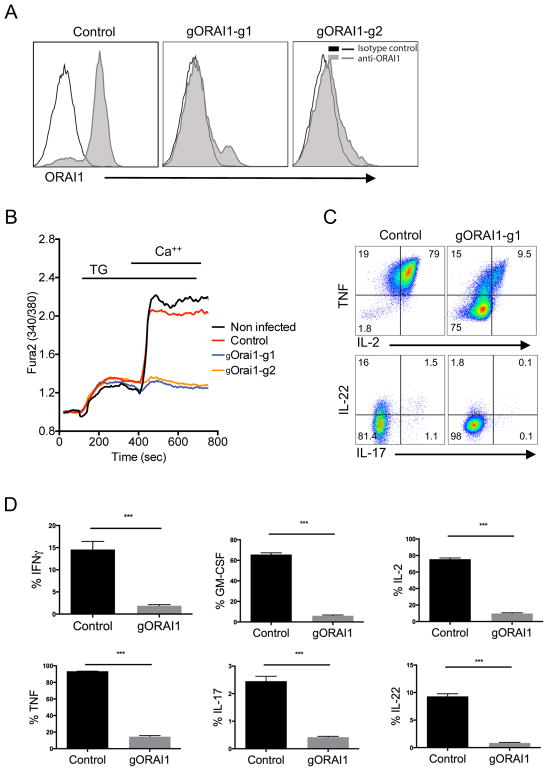
Next, we targeted and deleted the gene for the Ca2+ selective ion channel ORAI1 (24), to recapitulate the effect of deletion mutations observed on T cells in vivo. Inherited null mutations within ORAI1 in humans and targeted deletion of ORAI1 in mice severely impairs Ca2+ influx and causes severe immunodeficiency (25–27). We have also previously observed that mutation of ORAI1 or deletion of ORAI1 in human and mouse T cells, respectively, results in a strongly impaired cytokine secretory response upon activation (28, 29). The CRISPR-mediated deletion by gRNAs targeting ORAI1 were first confirmed by reduced ORAI1 protein expression in a Jurkat cell line using an antibody (Fig. 4A). Although ORAI1 expression in primary T cells was not detected with this antibody on primary T cells, extracellular Ca2+ influx after passive depletion of intracellular Ca2+ stores using the sarcoplasmic or endoplasmic reticulum Ca-ATPase family (SERCA) inhibitor thapsigargin (TG) was completely abolished in cells targeted with ORAI1 gRNAs (Fig. 4B). Remarkably, ORAI1-depleted primary CD4+ T cells displayed greatly reduced expression of pro-inflammatory cytokines, including TNF, IL-2, IL-17 and IL-22, upon activation by PMA and Ionomycin compared to control T cells (Fig. 4C and 4D). Thus, these CRISPR-modified primary human T cells recapitulate the functional phenotype of T cells from patients with a null mutation in the ORAI1 gene that causes CRAC channelopathy disease (29, 30), posing as a potential in vitro model.
FIGURE 4. Suppression of Store-Operated Ca2+ Entry (SOCE) and cytokine production in ORAI1-deleted human T cells.
(A) ORAI1 expression in Jurkat cells after CRISPR-mediated gene deletion. Jurkat cells were transduced with lentiviruses targeting the ORAI1 gene using two different gRNA clones (gORAI1-g1 or gORAI1-g2) and selected with puromycin for one week. ORAI1 protein expression was determined by anti-ORAI1 antibody staining. (B) Measurement of Ca2+ influx in ORAI1-deleted CD4+ T cells. CD4+ T cells were activated with anti-CD3/anti-CD28 beads and the next day transduced with gORAI1 lentiviruses. Puromycin was then added after 4 days post-activation and cells were selected and expanded for two weeks in culture. T cells were then loaded with Fura-2 and stimulated in 0 mM Ca2+ Ringer solution with 1 μM thapsigargin (TG) to induce passive store depletion. Ca2+ influx was measured after re-addition of 1 mM Ca2+ to the extracellular buffer at 420 sec. T cells transduced with empty lentivectors (Control) was comparable to non-infected cells. (C) Cytokine expression in ORAI1-deleted CD4+ T cells. Expression of TNF and IL-2 cytokines (top) by total CD4+ T cells, and IL-22 and IL-17 cytokines (bottom) by CD4+ CCR6+ T cells that were transduced with either empty-vector control or gORAI1-g1, expanded for 2 weeks after puromycin selection, and restimulated with PMA and Ionomycin for 4 hours in the presence of GolgiStop. Data is representative of three independent experiments. (D) Frequency of T cells expressing IFNγ, TNF, GM-CSF, IL-2, IL-17, and IL-22 analyzed from PMA and Ionomycin activation and intracellular staining as described in C. Data represent three independent experiments. Error bars represent SEM. ***p<0.001.
Modulating gene expression in primary human T cells via CRISPR/dCas9-mediated promoter targeting
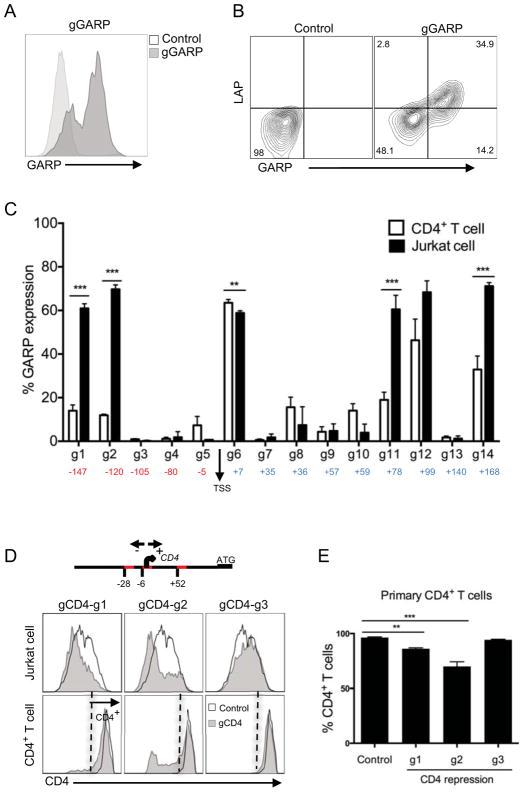
Recently it has been possible to create a nuclease deficient Cas9 protein (termed dCas9), which allows for the targeting of specific genome sequences without nucleotide changes (13, 31–33). To determine whether we can turn on or enhance gene expression through promoter-targeting using this dCas9 system in primary T cells, we used a triple lentivector system composed of dCas9 fused to VP16 tetramer activation domain (VP64), NF-κB trans-activating subunit p65 encoding lentivector, and sgRNA scaffold that includes an aptamer tetraloop for binding to the p65 NF-κB subunit to enable recruitment of additional transcription factors to the targeted promoters (14). We demonstrate the efficiency of this triple-lentivirus system by targeting Glycoprotein A Repetitions Predominant (GARP), which is exclusively expressed on activated regulatory T cells (Tregs) and is absent on other memory CD4+ T cell subsets (34). As such, we designed 14 different gRNAs targeting −200 to +200 GARP- transcriptional start site (TSS) promoter elements. We first expressed these in Jurkat cells, which are inheritently negative for GARP expression (34), and stained with an anti-GARP antibody to determine its induction. An example of one of the gRNAs that induced GARP cell surface expression is shown (Fig. 5A). We then expressed these gRNAs targeting the GARP promoter in primary T cells and selected with hygromycin and zeocin to enrich for cells expressing all three vectors as described in the methods. In these selected primary T cells, we observe robust GARP expression (Fig. 5B). Further, we have recently shown that the GARP molecule binds inactive TGFβ in Tregs, which can be detected by the antibody directed against the LAP portion of latent TGFβ (35). Indeed, similar to the activated Tregs, we found that promoter-induced GARP on primary T cells also bound exogenously added TGFβ (Fig. 5B). Upon analysis of GARP induction using all of the gRNAs targeting different regions of the GARP promoter, we found that while most GARP-TSS gRNAs (labeled g1–g14) showed induction of GARP above background levels in both Jurkat cells and primary T cells, there were major differences in gene-induction capacities not only between specific gRNAs but also between the two cell types (Fig. 5C), possibly suggesting epigenetic differences.
FIGURE 5. Efficient and functional GARP expression using CRISPR-mediated promoter activation.
(A) GARP expression in Jurkat cells after CRISPR-mediated gene activation. Jurkat were first transduced with lentiviruses expressing dCas9 and p65 proteins, selected with hygromycin. Next the dCas9 and p65 expressing cells were transduced with lentiviruses expressing gRNAs targeting promoters regions of GARP (gGARP-g12) and selected with zeocin. GARP expression in Jurkat cells was analyzed after 11 days post transduction, compared to empty vector (control), after gating on GFP+ cells (marker for dCas9 expression). (B) LAP binding to GARP molecules on primary CD4+ T cells transduced with GARP-gRNA (g12) or empty vector (control). Primary CD4+ T cells were activated with anti-CD3/anti-CD28 beads, transduced with lentiviruses expressing dCas9 and p65 proteins and selected with hygromycin. After 2 weeks expansion, dCas9 and p65 expressing cells were then reactivated with anti-CD3/anti-CD28 beads and transduced with lentiviruses bearing the gRNAs targeting different promoter regions and selected with zeocin. Analysis was performed on T cells expanded for two weeks in culture post transduction and after gating on GFP+ T cells (as marker for dCas9+ population). (C) Percentage of GARP expression level in both Jurkat (as in Figure 5A) and primary CD4+ T cells(as in Figure 5B), transduced with 14 different gRNA clones targeting the GARP promoter in the range of −200 to 200 base pairs downstream and upstream of transcription start site (TSS). Background level of GARP expression from control was subtracted. Data represent three independent experiments. Error bars represent SEM. **p<0.01, ***p<0.001. (D) CRISPR-mediated repression of CD4 expression on Jurkat and CD4+ T cells. Three guide RNAs targeting specific sites in the promoter element were determined (g1 at −28; g2 at −6; g3 at +52; TSS, transcriptional start site, defined as 0). ‘ATG’ is the start codon. CD4+ T cells were activated with anti-CD3/anti-CD28 beads and the next day transduced with respective lentiviruses expressing dCas9-KRAB (mCherry) and gRNAs targeting different sites of CD4 promotor regions. Jurkat cells were also tranduced with same viruses as in primary T cells. Data was analyzed after 7 days post transduction. Histogram overlay of CD4 expression, gated on mCherry positive cells in both Jurkat (top) and primary CD4+ T (bottom) cells. Data is representative of two independent experiments. (E) Precentage of CD4 protein depletion of primary CD4 T cells (as in Figure 5E) after repression of CD4 gene transcription by KRAB. Data represent three independent experiments. Error bars represent SEM. **p<0.01, ***p<0.001.
In a reverse approach to activating promoters, we asked if we could repress gene expression using dCas9-fused to a KRAB repression domain (13). For this, we targeted 100 base pairs upstream or downstream of the CD4 gene TSS. These three different gRNAs were cloned into a lentiviral vector that includes a zeocin resistance gene, and were transduced into primary T cells in conjunction with a lentiviral vector expressing dCas9-KRAB fusion protein with a mCherry marker (13) and selected with zeocin. One of the target gRNAs (g2) showed clearly reduced levels of cell surface CD4 expression in Jurkat and primary T cells after gating on mCherry+ cells (Fig. 5D and 5E). Together, these findings demonstrate the highly efficient induction or dampenning targeted promotor through the use of CRISPR promoter activation and repression of specific gene expression in primary human T cells.
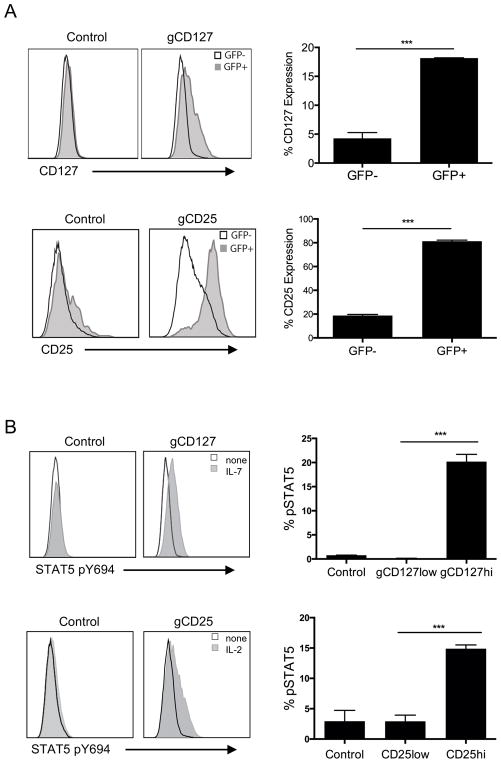
Lastly, we used this CRISPR-mediated promoter transactivation approach to test functional consequences in primary T cells by targeting promoters of cytokines with common gamma (γc) chain receptors, namely IL-7 receptor (CD127 or IL-7R) and IL-2 receptor alpha (CD25 or IL-2Ra). We first determined that both CD127 and CD25 could be upregulated through CRISPR-mediated promoter transactivation (Fig. 6A). We then asked if these receptor modulations have any functional consequence in response to respective cytokine stimulation with IL-7 or IL-2. We found that the downstream signaling through both receptors, as determined by phosphorylation of STAT5 (36), was increased upon stimulation with either IL-7 or IL-2 only in promoter-induced T cells expressing higher levels of cytokine receptors (Fig 6B and 6C). This approach highlights the feasibility of increasing endogenous gene transcription through a CRISPR/dCas9-mediated system functional interrogation in primary T cells.
FIGURE 6. Functional CD127 and CD25 gene expression on primary CD4+ T cells using CRISPR-mediated promoter activation.
(A) Expression of CD127 (top left) and CD25 (bottom left) in primary CD4+ T cells transduced with gRNA targeting CD127 or CD25 promoter. Primary CD4+ T cells were activated with anti-CD3/anti-CD28 beads, transduced with lentiviruses expressing dCas9 and p65 proteins and selected with hygromycin. After 2 weeks expansion, dCas9 and p65 expressing cells were then reactivated with anti-CD3/anti-CD28 beads and transduced with lentiviruses targeting CD127 (gCD127) or CD25 (gCD25) promoters, selected with zeocin and expanded for 2 weeks. The histograms show expressions after gating on GFP+ (gray histogram) and GFP− cells (empty histogram), GFP being the marker for dCas9 expression. Percentgate of CD127 (top right) or CD25 (bottom right) expression in primary CD4+ T cells transduced with CD25− or CD127− promoter targeting gRNAs, between GFP+ or GFP− population (as in Figure 6A). Data represent three independent experiments. Error bars represent SEM. ***p<0.001. (B) Increased phosphorylated (pY694) Stat5 expression in promoter-induced cytokine receptor populations in primary CD4+ T cells transduced with gRNA targeting CD127 or CD25 promoter. Histogram overlays of phosphorylated (pY694) Stat5 expression in CD127hi population in CD127-expression induced CD4+ T cells stimulated with 3ng/ml IL-7 (gray histogram) compared to non-stimulated (empty histogram) and CD25hi population in CD25-expression induced CD4+ T cells stimulated with 1ng/ml IL-2 (gray histogram) compared to non-stimulated (empty histogram). Bottom panel represents pSTAT5 induction in T cells transduced with empty vector (control) gated on total cells, gCD127 (gated on CD127low and CD127hi, top) or gCD25 (gated on CD25low and CD25hi, bottom). Data represent three independent experiments. Error bars represent SEM. ***p<0.001.
Discussion
CRISPR/Cas9 genome editing technology has the potential for decoding T cell functionality and modeling a variety of immunological diseases. Here we report several CRISPR-mediated gene editing methods, using stable lentiviral transduction, to demonstrate the feasibility of functional interrogation of primary human T cell subsets. Lentiviral delivery of Cas9 with gene-specific gRNAs was highly efficient in various human T cell subsets (CD4+, CD8+ T cells), differentiation states (naive, memory) and functional subsets (Treg, Th1, Th2, Th17). It was also possible to knockout up to three different genes (CD4, CD45, CD95) in the same T cell and maintain stably for prolonged periods in culture, even after several T cell restimulations. Further, using a nuclease dead version of Cas9 (dCas9) fused to a transcriptional activator domain, we successfully induced transcription of a gene restricted only to Tregs (GARP) on effector T cells, and amplified cytokine signaling through increased IL-7R and IL-2R expression. Collectively, our studies demonstrate the feasibility of CRISPR/Cas9-mediated genetic manipulations in primary human T cells, suitable for applications in a broad range of functional assays as well as engineering T cells for specific immunotherapeutic approaches.
Several recent reports have used transient transfection methods for the transfer of Cas9 and gRNAs either as a plasmid or ribonucleoprotein complex into primary T cells (5, 37). Plasmid transfection approaches have low efficiency and result in relatively high T cell death that confounds the activation of T cells (data not shown). Transient Cas9-gRNA transfection as a ribonucleoprotein (Cas9 RNP)-gRNA complex has the advantage of lower toxicity and higher efficiency in primary T cells, possibly due to a high abundance of Cas9 protein with gRNA and more optimal nuclear translocation as complexes (5), and may have the advantage of safer gene knockout or mutations for therapeutic applications (38). However, Cas9 RNPs rapidly degrade within 24 hours and thus have a limited temporal window. Our approach of using lentiviral transduction of Cas9-gRNA vectors, stably expressed in primary human T cells, overcomes this problem and results in highly efficient gene targeting for even multiple genes in the same T cells. The stable expression of Cas9 is especially necessary in approaches that target promoters or non-coding regulatory regions, or in inducible conditions.
Null mutations in many genes are associated with inherited genetic disorders, such as calcium release-activated channels (CRAC) channelopathy (39). Patients with null mutations in ORAI1 or STIM1 genes lack Ca2+ influx in T cells in response to T cell receptor stimulation (26, 30). Ca2+ influx through ORAI1 channels is crucial for T cell function, and patients with null mutations suffer from severe, life threatening immunodeficiencies. In this study, we could recapitulate the phenotype of null mutations in ORAI1 by specifically targeting and deleting ORAI1 in the Jurkat cell line and primary human CD4+ T cells. Using the CRISPR/Cas9 approach in primary human T cells will facilitate the analysis of the genetic and molecular mechanisms underlying rare inherited diseases (e.g CRAC channelopathy) for which patient samples are difficult to obtain, thus limiting studies using human cell culture. The CRISPR/Cas9 approach can bypass this bottleneck and provide cells to study the effects of mutations in genes underlying known immunodeficiency diseases such as Omenn syndrome (mutations in RAG1/RAG2 genes) (40) or Wiskott-Aldrich syndrome (mutations in WASP gene) (41). This editing method will enable us in the future to explore the roles of other genes disrupted in human genetic diseases, in impacting T cell functions.
In addition to gene-deletion mutations, it is also possible to perturb T cell functions without permanent genome-editing via promoter-targeted dCas9 molecules fused either to activator or repressor transcription factor domains. Unlike Cas9-gRNA-mediated knockout approach, for transcriptional activation or repression of a targeted gene, stable dCas9-gRNA expression would be necessary for continuously binding to promoter elements to either recruit or block transcription factor binding to promoter elements in primary human T cells. This transcriptional turn-on/off system enables tunable perturbation of gene expression for functional interrogations. As an example, we targeted GARP to achieve transcriptional activation, since we have previously shown that expression of GARP is highly specific to regulatory T cells (34), and is completely absent in other T cell subsets. GARP is the receptor for the latency-associated peptide (LAP), which binds as LAP-TGFβ complex to produce a suppressive signal to target T cells expressing TGFβ receptors (35). Ectopic expression of GARP on conventional T cells induces Foxp3 expression possibly because it can hold surface TGFβ, which can induce Foxp3 (42). While using promoter-targeted activator CRISPR/dCas9-P65 system, most of the gRNAs specific to GARP in +/− 200 base pairs of the TSS could induce GARP in effector T cells and Jurkat cells, albeit major differences between these cells with some of the gRNAs noted previously. This discrepancy may be a result of the activity of enhancer sites that bind through transcription factors on the promoter elements. This wide coverage of promoter elements to determine GARP expression in T cells revealed further complexity in transcriptional activation due to the free binding capability of dCas9/gRNA complexes, which might be from modifications to the promoter or enhancer sites in the these cells. It was also surprising that, although recognition sites of several gRNAs were close to each other, it showed very different transcriptional activation capability, perhaps partly due to nucleosome positioning effects (43). Alternatively, dCas9 binding position on the sites of promoter elements can either directly block RNA polymerase activity or transcription factors binding to enhancer sites (44). It is also possible that transcription factor binding motifs can be in proximity with the dCas9/gRNA complex recognition sites, blocking binding of dCas9 to the these sequences (45). In future studies, screening of other promoter elements using overlapping gRNAs could be an effective way to map the binding of transcription factors and enhancer elements on promoters. Specifically, this CRISPR-dCas9 mediated reprogramming of T cells to induce GARP may also have potential therapeutic uses in converting effector T cells into Tregs within transplantation or autoimmune disease settings.
Feasible and robust usage of lentiviral CRISPR/Cas9 toolbox in primary human T cells also has clinical potential, such as in improving cancer immunotherapy. As we have shown, deletion of CD95 can render effector T cells more resistant to cell death and could be developed to enhance anti-tumor T cell engineering. However, this approach would involve further development of safety-kill switches to avoid uncontrolled T cell growth in vivo. Our approach to increase expression of CD25 and CD127 through CRISPR-mediated transcriptional activation enhances signaling via IL-2 or IL-7, respectively, which could be used to enhance survival of T cell memory or engineered anti-tumor T cells in combination with the deletion of checkpoint inhibitors (e.g. PD1). Genetic engineering using a combination of factors to create a more effective T cell response in cancer patients may significantly improve the efficiency of immunotherapies in the future (46).
In summary, our findings show the feasibility of CRISPR-mediated stable manipulation of T cells for extensive and rapid functional interrogation studies. A wide range of adaptations and applications of CRISPR-mediated genome engineering in human T cells also holds great promise for cell-based therapies for cancer, microbial infections, immune deficiencies, autoimmune and chronic inflammatory diseases.
Supplementary Material
Acknowledgments
The research in this study was supported by National Institute of Health (NIH) grants R01AI121920 and U54NS105539 to DU.
References
- 1.Chi S, Weiss A, Wang H. A CRISPR-Based Toolbox for Studying T Cell Signal Transduction. Biomed Res Int. 2016;2016:5052369. doi: 10.1155/2016/5052369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavalieri S, Cazzaniga S, Geuna M, Magnani Z, Bordignon C, Naldini L, Bonini C. Human T lymphocytes transduced by lentiviral vectors in the absence of TCR activation maintain an intact immune competence. Blood. 2003;102:497–505. doi: 10.1182/blood-2003-01-0297. [DOI] [PubMed] [Google Scholar]
- 3.Didigu CA, Wilen CB, Wang J, Duong J, Secreto AJ, Danet-Desnoyers GA, Riley JL, Gregory PD, June CH, Holmes MC, Doms RW. Simultaneous zinc-finger nuclease editing of the HIV coreceptors ccr5 and cxcr4 protects CD4+ T cells from HIV-1 infection. Blood. 2014;123:61–69. doi: 10.1182/blood-2013-08-521229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Liu S, Shen Q, Zha X, Zheng H, Yang L, Chen S, Wu X, Li B, Li Y. Differential gene expression profiles of PPP2R5C-siRNA-treated malignant T cells. DNA Cell Biol. 2013;32:573–581. doi: 10.1089/dna.2013.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE, Haliburton GE, Ye CJ, Bluestone JA, Doudna JA, Marson A. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci U S A. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nat Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- 7.Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, Agami R, Ge W, Cavet G, Linsley PS, Beijersbergen RL, Bernards R. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 8.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koike-Yusa H, Li Y, Tan EP, del Velasco-Herrera CM, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 11.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, Engreitz JM. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 2016;354:769–773. doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCaffrey J, Sibert J, Zhang B, Zhang Y, Hu W, Riethman H, Xiao M. CRISPR-CAS9 D10A nickase target-specific fluorescent labeling of double strand DNA for whole genome mapping and structural variation analysis. Nucleic Acids Res. 2016;44:e11. doi: 10.1093/nar/gkv878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sallusto F, Lenig D, Mackay CR, Lanzavecchia A. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J Exp Med. 1998;187:875–883. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin S, Rottman JB, Myers P, Kassam N, Weinblatt M, Loetscher M, Koch AE, Moser B, Mackay CR. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J Clin Invest. 1998;101:746–754. doi: 10.1172/JCI1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Ambrosio D, Iellem A, Bonecchi R, Mazzeo D, Sozzani S, Mantovani A, Sinigaglia F. Selective up-regulation of chemokine receptors CCR4 and CCR8 upon activation of polarized human type 2 Th cells. J Immunol. 1998;161:5111–5115. [PubMed] [Google Scholar]
- 21.Singh SP, Zhang HH, Foley JF, Hedrick MN, Farber JM. Human T cells that are able to produce IL-17 express the chemokine receptor CCR6. J Immunol. 2008;180:214–221. doi: 10.4049/jimmunol.180.1.214. [DOI] [PubMed] [Google Scholar]
- 22.Khaitan A, Kravietz A, Mwamzuka M, Marshed F, Ilmet T, Said S, Ahmed A, Borkowsky W, Unutmaz D. FOXP3+Helios+ Regulatory T Cells, Immune Activation, and Advancing Disease in HIV-Infected Children. J Acquir Immune Defic Syndr. 2016;72:474–484. doi: 10.1097/QAI.0000000000001000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–192. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prakriya M, Feske S, Gwack Y, Srikanth S, Rao A, Hogan PG. Orai1 is an essential pore subunit of the CRAC channel. Nature. 2006;443:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- 25.McCarl CA, Picard C, Khalil S, Kawasaki T, Rother J, Papolos A, Kutok J, Hivroz C, Ledeist F, Plogmann K, Ehl S, Notheis G, Albert MH, Belohradsky BH, Kirschner J, Rao A, Fischer A, Feske S. ORAI1 deficiency and lack of store-operated Ca2+ entry cause immunodeficiency, myopathy, and ectodermal dysplasia. J Allergy Clin Immunol. 2009;124:1311–1318e1317. doi: 10.1016/j.jaci.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feske S. Immunodeficiency due to defects in store-operated calcium entry. Ann N Y Acad Sci. 2011;1238:74–90. doi: 10.1111/j.1749-6632.2011.06240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaeth M, Yang J, Yamashita M, Zee I, Eckstein M, Knosp C, Kaufmann U, Karoly Jani P, Lacruz RS, Flockerzi V, Kacskovics I, Prakriya M, Feske S. ORAI2 modulates store-operated calcium entry and T cell-mediated immunity. Nat Commun. 2017;8:14714. doi: 10.1038/ncomms14714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maul-Pavicic A, Chiang SC, Rensing-Ehl A, Jessen B, Fauriat C, Wood SM, Sjoqvist S, Hufnagel M, Schulze I, Bass T, Schamel WW, Fuchs S, Pircher H, McCarl CA, Mikoshiba K, Schwarz K, Feske S, Bryceson YT, Ehl S. ORAI1-mediated calcium influx is required for human cytotoxic lymphocyte degranulation and target cell lysis. Proc Natl Acad Sci U S A. 2011;108:3324–3329. doi: 10.1073/pnas.1013285108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gwack Y, Srikanth S, Feske S, Cruz-Guilloty F, Oh-hora M, Neems DS, Hogan PG, Rao A. Biochemical and functional characterization of Orai proteins. J Biol Chem. 2007;282:16232–16243. doi: 10.1074/jbc.M609630200. [DOI] [PubMed] [Google Scholar]
- 30.Feske S. CRAC channelopathies. Pflugers Arch. 2010;460:417–435. doi: 10.1007/s00424-009-0777-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R, Kozhaya L, Mercer F, Khaitan A, Fujii H, Unutmaz D. Expression of GARP selectively identifies activated human FOXP3+ regulatory T cells. Proc Natl Acad Sci U S A. 2009;106:13439–13444. doi: 10.1073/pnas.0901965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran DQ, Andersson J, Wang R, Ramsey H, Unutmaz D, Shevach EM. GARP (LRRC32) is essential for the surface expression of latent TGF-beta on platelets and activated FOXP3+ regulatory T cells. Proc Natl Acad Sci U S A. 2009;106:13445–13450. doi: 10.1073/pnas.0901944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owen DL, Farrar MA. STAT5 and CD4 + T Cell Immunity. F1000Res. 2017;6:32. doi: 10.12688/f1000research.9838.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou P, Chen S, Wang S, Yu X, Chen Y, Jiang M, Zhuang K, Ho W, Hou W, Huang J, Guo D. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci Rep. 2015;5:15577. doi: 10.1038/srep15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lacruz RS, Feske S. Diseases caused by mutations in ORAI1 and STIM1. Ann N Y Acad Sci. 2015;1356:45–79. doi: 10.1111/nyas.12938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang YQ, Cui YX, Feng J. Clinical phenotype and gene diagnostic analysis of Omenn syndrome. Zhonghua Er Ke Za Zhi. 2013;51:64–68. [PubMed] [Google Scholar]
- 41.Derry JM, Ochs HD, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;78:635–644. doi: 10.1016/0092-8674(94)90528-2. [DOI] [PubMed] [Google Scholar]
- 42.Wang R, Wan Q, Kozhaya L, Fujii H, Unutmaz D. Identification of a regulatory T cell specific cell surface molecule that mediates suppressive signals and induces Foxp3 expression. PLoS One. 2008;3:e2705. doi: 10.1371/journal.pone.0002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Radman-Livaja M, Rando OJ. Nucleosome positioning: how is it established, and why does it matter? Dev Biol. 2010;339:258–266. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitfield TW, Wang J, Collins PJ, Partridge EC, Aldred SF, Trinklein ND, Myers RM, Weng Z. Functional analysis of transcription factor binding sites in human promoters. Genome Biol. 2012;13:R50. doi: 10.1186/gb-2012-13-9-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Golubovskaya V, Wu L. Different Subsets of T Cells, Memory, Effector Functions, and CAR-T Immunotherapy. Cancers (Basel) 2016:8. doi: 10.3390/cancers8030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.