Abstract
Purpose
Secondary findings from genomic sequencing are becoming more common. We compared how healthcare providers with and without specialized genetics training anticipated responding to different types of secondary findings.
Methods
Providers with genomic sequencing experience reviewed five secondary findings reports and reported attitudes and potential clinical follow-up. Analyses compared genetic specialists and physicians without specialized genetics training, and examined how responses varied by secondary finding.
Results
Genetic specialists scored higher than other providers on 4-point scales assessing understandings of reports (3.89 vs 3.42, p=0.0002), and lower on scales assessing reporting obligations (2.60 vs 3.51, p<0.0001) and burdens of responding (1.73 vs 2.70, p<0.0001). Nearly all attitudes differed between findings, although genetic specialists were more likely to assert that laboratories had no obligations when findings had less-established actionability (p<0.0001 in interaction tests). The importance of reviewing personal and family histories, documenting findings, learning more about the variant and recommending familial discussions also varied according to finding (all p<0.0001).
Conclusion
Genetic specialists felt better prepared to respond to secondary findings than providers without specialized genetics training, but perceived fewer obligations for laboratories to report them, and the two groups anticipated similar clinical responses. Findings may inform development of targeted education and support.
INTRODUCTION
The use of genomic sequencing in patient care is increasing in a variety of settings, raising questions about whether and how to report secondary findings.1–6 The American College of Medical Genetics and Genomics (ACMG) currently recommends that laboratories offer screening of at least 59 genes for pathogenic variants whenever genome-scale sequencing is ordered, regardless of indication.7,8 Some laboratories will offer to screen hundreds of additional genes for monogenic disease risks, and provide other types of secondary results such as carrier status for autosomal recessive conditions. Advocates assert that screening for genomic secondary findings is a low-cost opportunity to identify life-threatening risk factors that might never be identified otherwise.7,9 Opponents raise concerns that healthcare providers, especially those without specialized genomics training, may be unprepared to respond to unsolicited findings,10 or may be unsure about their obligations to respond.11–15 These concerns are especially relevant regarding variants where evidence is lacking about the implications for asymptomatic patients.16
Data to inform these issues from the provider perspective are lacking. To help fill this gap, in an exploratory study, we presented five secondary finding reports to healthcare providers with experience disclosing genomic sequencing results, and asked them how they anticipated responding. The goal of this study was to provide data from early adopters of genomic sequencing about how their attitudes towards secondary findings and the follow-up services they might order in response vary according to the type of findings identified and providers’ training.
METHODS
Participants and Study Design
In 2011, the National Institutes of Health (NIH) convened the Clinical Sequencing Exploratory Research (CSER) Consortium to investigate the application of genome-scale sequencing in different clinical settings.17 The CSER Consortium provided a unique opportunity to collect data from a group of providers with genomic sequencing experience. Coordinators and investigators from the 19 consortium studies helped to recruit a convenience sample of providers who were associated with their studies and had experience disclosing genomic sequencing results. Invitation emails provided an overview of the study and presented a link to a web-based survey hosted by Qualtrics software (Qualtrics, Provo, UT), with reminders sent approximately two weeks apart. The first page of the survey described the study, survey procedures, and potential risks and benefits, and provided information about how participants could ask the investigative team questions or express concerns. Consent was implied by survey completion. The study protocol was developed by a multidisciplinary team with experience in clinical genetics, molecular genetics, psychology and social science, and was reviewed by the Institutional Review Board of Partners HealthCare (protocol 2016P002564).
Development of the Survey Instrument
The survey instrument, including five hypothetical reports and the survey questions, was informed by a review of laboratory reports at participating CSER Consortium sites and related literature,18–22 and adapted from questionnaires administered to primary care physicians about supporting infrastructure needs and responses to genomic findings, including secondary findings.22,23 The survey was pilot tested to ensure ease-of-use and clarity of questions prior to administration on a convenience sample of 10 research assistants and providers, including two genetic counselors and two non-genetic specialists. All individuals who pilot tested the survey were familiar with genomic testing and unaffiliated with the study.
Surveys first defined important terms (e.g., “genomic sequencing,” “variant,” and “secondary findings”), and asked participants to report demographic and practice information. Participants then reviewed five secondary finding reports. Each report was followed by a series of questions to which participants reported resources they might consult, their attitudes about reporting, the importance of other clinical information, and the importance of specific follow-up actions. Closed-ended survey items were used to minimize participant burdens, and topics such as the role of clinical evidence in decision making were omitted given the length of the survey (average completion time: 50 minutes) and the absence of incentives for completion.
Presenting Scenarios and Reports
We developed presenting scenarios and genomic sequencing reports to represent prototypical examples of five types of secondary findings as summarized in Table 1. Secondary finding types included a pathogenic variant in RYR1, a gene included in ACMG recommendations for screening; a pathogenic variant in PALB2, a gene that was omitted from the ACMG list, but for which the ClinGen Actionability Working Group had developed an evidence-based summary of guidelines or recommendations;24 a pathogenic variant in CHEK2, a gene for which a ClinGen evidence-based summary was not yet available; a variant of uncertain significance (VUS) in BRCA1, a gene associated with a breast and ovarian cancer in a patient with a strong associated family history of disease (maternal or paternal lineage was unspecified); and carrier status in three genes, HFE, MUTYH, and SPG7, associated with autosomal recessive disorders. We selected RYR1, PALB2, and CHEK2 based on the existence or absence of ACMG recommendations and/or ClinGen evidence-based summaries to reflect expert opinion about the degree to which variants were considered actionable (ACMG list > ClinGen evidence-based summary > no ClinGen evidence-based summary). The design of reports was based on clinical reports used in CSER Consortium studies and modified per consensus of the study team. Although the study team chose many variants to report based on the existence or absence of ClinGen evidence-based summaries, reports deliberately omitted mention of these resources to mimic current clinical practice. An example of a report is presented in Appendix 1.
Table 1.
Overview of secondary findings presented to survey participants
| Report Number and Study Name | Gene | Condition as Reported | On ACMG List? | ClinGen Actionability Summary? | Classification | Inheritance |
|---|---|---|---|---|---|---|
| 1: “ACMG list” | RYR1 | Malignant hyperthermia susceptibility type 1 | Yes | Yes | Pathogenic | AD |
| 2: “Has ClinGen summary” |
PALB2 | PALB2-related breast cancer risk | No | Yes | Pathogenic | AD |
| 3: “No ClinGen summary” | CHEK2 | CHEK2-related cancer | No | No | Pathogenic | AD |
| 4: “VUS with family history” | BRCA1 | BRCA1-related breast cancer risk | Yes | Yes | VUS* | AD |
| 5: “Carrier status” | HFE | Hereditary Hemochromatosis |
No | Yes† | Pathogenic | AR |
| MUTYH |
MUTYH-associated polyposis |
Yes‡ | Yes† | Pathogenic | AR | |
| SPG7 | Spastic paraplegia type 7 | No | No | Pathogenic | AR | |
The introduction to this report also stated that the patient’s mother and aunt had both been diagnosed with ovarian cancer.
ClinGen Actionability Working Group evidence-based summaries were developed for biallelic HFE and MUTYH variants, rather than single heterozygous variants as presented in the study report.
ACMG recommends return of biallelic findings, rather than heterozygous findings, as presented in the study report.
We introduced each of the secondary finding reports with a presenting scenario tailored according to participants’ specialties, with pediatricians receiving reports about an 18 year old female patient and all other participants receiving reports about a 30 year old female patient. Presenting scenarios read, “During an encounter with you, a [30/18]-year old female patient with a serious health condition tells you that she received genomic sequencing. A physician confirmed that no variants were identified related to her condition, and then gave her a copy of the report. Only after the appointment did she notice that a secondary finding was identified in [gene of interest]. She shares the report below with you.” The conclusion to the presenting scenario for the BRCA1 VUS added information stating, “Her mother and an aunt had both been diagnosed with ovarian cancer.”
Outcome Measures
Attitudes.
Participants rated how strongly they agreed with seven statements, such as “The information in this report would be important to this patient’s health and healthcare” and “Genomic sequencing laboratories should be obligated to report findings like these to the ordering physician, regardless of why sequencing was ordered.” Response options on four-point scales ranged from “strongly disagree” to “strongly agree,” and a fifth “Don’t know” option was provided. Survey items were adapted from a prior survey of primary care physicians.23
Clinical follow-up.
Participants rated the importance of a universal set of nine possible responses, such as “Document this finding in the patient’s medical record” and “Recommend genetic testing of family members.” Cancer-related reports (PALB2, CHEK2, and BRCA1) had three additional actions about discussing screening, chemoprevention, and prophylactic surgery. The section related to malignant hyperthermia (RYR1) also asked about recommending that the patient carry identification of the susceptibility, and the section about carrier findings also asked about discussing reproductive plans, referral to a reproductive specialist, and testing of the patient’s partner. To facilitate comparisons by finding type and respondent training, most actions were presented because we felt that they were relevant across variants and conditions. Response options on four-point scales ranged from “not at all important” to “extremely important,” and a fifth “Don’t know” option was provided. Participants were also asked to indicate whether they, another provider, or both had obligations to initiate each action. Participants were allowed to write in additional actions they would consider initiating.
Information to interpret reports.
A series of six items asked participants to rate the importance of different clinical information for “interpreting the report,” such as the patient’s family health history and previous laboratory results. Response options on four-point scales ranged from “not at all important” to “extremely important,” and a fifth “Don’t know” option was provided. Participants could also write in additional information they would want. Items were adapted from prior work on secondary findings disclosure about Lynch syndrome.23 The same six items were displayed following each report, regardless of whether they were relevant the presenting scenario or report, to minimize the potential for survey items to cause providers to consider clinical information they might have ignored otherwise.
Resources consulted.
Surveys presented a list of nine common genetics reference sources, per the judgment of the investigative team, and asked participants to check the ones they might consult to help “understand and respond to the report.” Participants were also allowed to write in additional sources they would consult.
Data Analysis and Code Availability
Participants’ data were analyzed if they completed at least one survey item about a secondary finding report. A priori enrollment goals were not established given the exploratory nature of this study. Primary analyses examined whether attitudes and the importance of clinical follow-up differed between genetic specialists and non-genetic specialists or by the type of finding (i.e., pathogenic variant in an ACMG-list gene, pathogenic variant in a gene with a ClinGen summary of guidelines and recommendations, pathogenic variant in a gene without a ClinGen summary of guidelines and recommendations, VUS when family history suggested a genetic syndrome, or carrier status for autosomal recessive conditions). Statistical models used generalized linear models fit with generalized estimating equations, and used an exchangeable working correlation structure to account for the repeated measures within participant. Analyses of attitudes treated outcomes as continuous variables.25 Analyses of the importance of clinical follow-up dichotomized responses to compare “very important” responses against “not at all important,” “somewhat unimportant” and “somewhat important” responses because distributions were highly skewed, and used a logit link and binomial distribution. In addition to a term for genetic specialists and non-genetic specialists, models included terms for the different reports (i.e., “ACMG list,” “Has ClinGen summary,” “No ClinGen summary,” “VUS with family history,” and “carrier status). We examined whether differences between genetic specialists and non-genetic specialists depended on the specific report by including an interaction term and comparing models. Statistical significance was set at p=0.0008, per a Bonferroni correction, to account for 58 primary analyses at a Type 1 error rate of 0.05. All statistical tests were two-sided. The study protocol, statistical code, and data set are available upon request from the corresponding author.
We also performed exploratory analyses that examined whether attitudes differed between sites or between pediatricians and other types of providers by adding site or status as a pediatrician as additional terms to statistical models. We also performed exploratory analyses to examine whether the importance of clinical follow-up differed by site or pediatrician status by summing number of actions that participants rated as “very important” and running linear regression models that included site or pediatrician status, as relevant; and an additional term for genetic specialist/non-genetic specialist. Given observed differences between genetic specialists and non-genetic specialists on the item asking about understandings of reports (p=0.0002), we created a summary understanding-of-report score equal to the mean of finding-specific understanding-of-report scores, and examined whether the new variable confounded findings. Finally, we conducted subanalyses restricted to genetic specialists that compared responses of medical geneticists and genetic counselors.
Secondary analyses examined the likelihood that participants perceived personal responsibilities to initiate clinical follow-up, and focused on the proportion of actions that participants rated as “extremely important.” Secondary analyses also examined the resources that participants anticipated consulting. Secondary analyses are presented descriptively.
Available-case analyses were conducted using R version 3.4.2. We report on analyses that omitted “don’t know” responses and missing data. Additional analyses were conducted by classifying “don’t know” as missing data, assuming survey data were missing at random, and imputing missing data with fully conditional specification using R package mice 2.25,26 running 20 iterations to create each of 100 imputed datasets. Findings are highlighted in this report only when analyses of both available-case and imputed data achieved statistical significance.
RESULTS
Participant Characteristics
Characteristics of the 54 of 108 invited providers (51%) who were eligible for analyses are summarized in Table 2. Thirty seven participants (69%) were genetic specialists and 20 (43%) were investigators on CSER Consortium studies. Just over half of participants (n=29, 54%) rated their training in genetics as “more than sufficient,” and the vast majority had prior experience ordering or discussing genetic test results (93%) and sequencing results (94%).
Table 2:
Participant characteristics
| Characteristic | N (%) | Characteristic | N (%) | |
|---|---|---|---|---|
| Age | CSER investigator | 20 (43%) | ||
| 20–29 | 4 (7%) | |||
| 30–39 | 18 (33%) | CSER Study Affiliation* | ||
| 40–49 | 10 (19%) | BASIC3 | 7 (13%) | |
| 50–59 | 14 (26%) | ClinSeq | 6 (11%) | |
| 60–69 | 7 (13%) | HudsonAlpha | 4 (7%) | |
| 70+ | 1 (2%) | MedSeq | 7 (13%) | |
| MI-ONCOSEQ | 6 (11%) | |||
| Gender | NCGENES | 8 (15%) | ||
| Male | 25 (46%) | NEXT Medicine | 4 (7%) | |
| Female | 29 (54%) | NextGen | 3 (6%) | |
| PediSeq | 6 (11%) | |||
| Race | Other | 4 (7%) | ||
| White non-hispanic | 43 (80%) | |||
| Asian | 7 (13%) | Rating of genetics training | ||
| Other | 4 (7%) | Not at all sufficient | 2 (4%) | |
| Somewhat insufficient | 5 (9%) | |||
| Neutral | 3 (6%) | |||
| Specialty* | Somewhat sufficient | 15 (28%) | ||
| Genetic counseling | 21 (39%) | More than sufficient | 29 (54%) | |
| Medical genetics | 17 (31%) | |||
| Pediatrics | 14 (26%) | Tests ordered/disclosed within 12 months | ||
| Oncology | 11 (20%) | Genetic test | 50 (93%) | |
| Internal medicine | 6 (11%) | Genomic sequencing | 51 (94%) | |
| Cardiology | 4 (7%) | |||
| Other† | 9 (17%) | Practice setting* | ||
| Hospital | 41 (76%) | |||
| Clinic | 16 (30%) | |||
| Other | 12 (22%) | |||
Participants could endorse more than one response option.
Open-ended responses included neurology/neurogenetics (4), hematology (2), clinical genetics (1), biochemical genetics (1), and “pediatric, preconception, prenatal and cancer” (1).
Attitudes
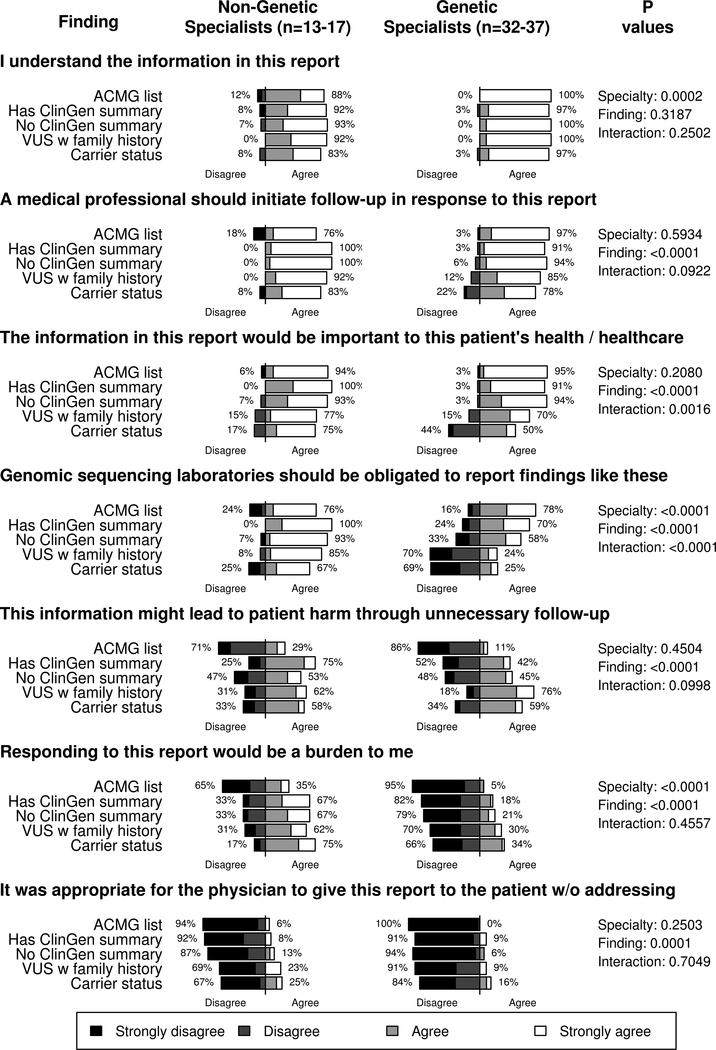
Respondents’ attitudes about reports are summarized in Figure 1. The large majority of both genetic specialists and non-genetic specialists (99% and 90% across finding types, respectively) reported that they understood the information in the report, that a medical professional should initiate follow-up (89% and 90% across finding types, respectively), and that it was not appropriate for a physician to give the report to the patient without addressing the secondary finding (92% and 83% across finding types, respectively). Across reports, mean agreement scores were higher among genetic specialists than non-genetic specialists on items about understanding the report information (3.89 vs 3.42, respectively, p<0.001), but lower on items about laboratories’ obligations to report these findings (2.60 vs 3.51, respectively, p<0.001) and whether responding to the findings would be a burden (1.73 vs 2.70, respectively, p<0.001). We also observed differences in all attitudes by the finding that was reported (all p<0.001), with the exception of whether participants believed they understood the report (p=0.319). Notably, we saw an interaction effect regarding obligations to report, where differences by finding type were observed among genetic specialists, but not non-genetic specialists (p<0.0001 on test for interaction).
Figure 1.
Attitudes about reporting, by specialty and finding. Findings include a pathogenic RYR1 variant associated with risk for malignant hyperthermia (“ACMG list”), a pathogenic PALB2 variant associated with risk for breast cancer risk (“Has ClinGen summary”), a pathogenic CHEK2 variant associated with risk for cancer (“No ClinGen summary”), a BRCA1 variant of uncertain significance in a patient with a family history of ovarian cancer (“VUS w family history”), and carrier status for three autosomal recessive conditions. P values summarize tests for non-equivalence comparing genetic specialists and non-genetic specialists (“specialty”) and comparing findings (“finding”), and include a test for interaction between specialty and finding (“interaction”).
Secondary analyses by site showed differences by site on items that collected data about whether a professional should initiate follow-up, whether the information was important to the patient’s health or healthcare, and whether information might lead to patient harm (all p<0.0004). Participants from the ClinSeq Study and MedSeq Project had the highest scores, indicating stronger agreement, on the statements about initiating follow-up and the importance of the information. In contrast, participants from the BASIC3 and MI-ONCOSEQ studies had higher agreements scores on the statement about potential harms. No differences were observed on any attitudes in models that compared pediatricians and non-pediatricians (all p≥0.14) or in subanalyses comparing genetic counselors and medical geneticists (all p≥0.02). We found no evidence that site, pediatrician status, or understandings of reports confounded findings from analyses that compared genetic specialists and non-genetic specialists.
Clinical Follow-Up and Obligations to Initiate
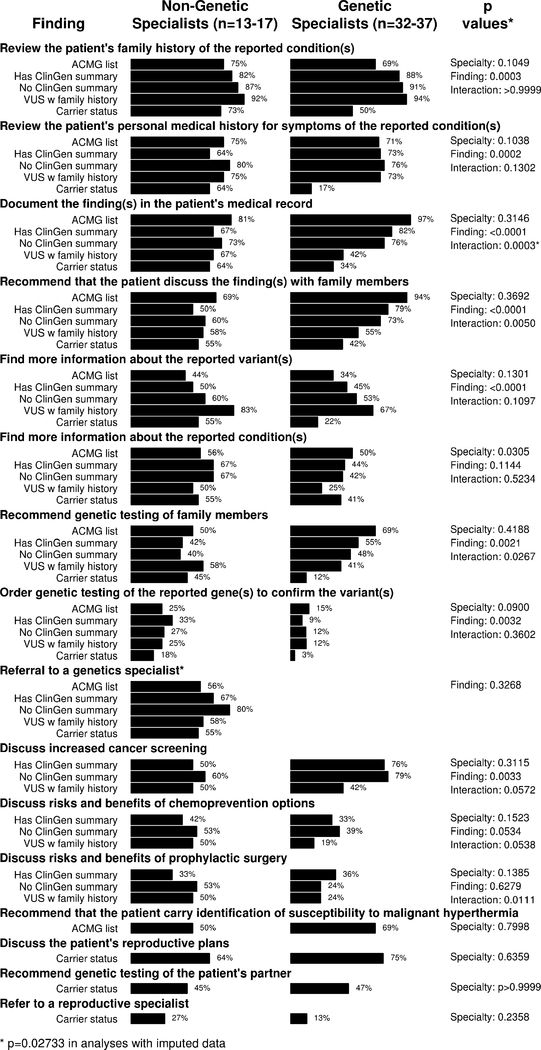
Respondents’ ratings about the importance of clinical follow-up are summarized in Figure 2. Across findings and combining genetic specialists and non-genetic specialists, participants were most likely to consider reviewing patients’ family histories and personal medical histories (79% and 66%, respectively) and least likely to rate confirmatory genetic testing and genetic testing of family members (15% and 46%, respectively) as extremely important. No comparisons between genetic specialists and non-genetic specialists achieved statistical significance. However, whether or not participants rated reviewing family histories of disease, reviewing personal medical histories, documenting findings in medical records, getting more information about the variant, and recommending that the patient discuss findings with family members as extremely important differed according to the finding (all p<0.001). Analyses that considered all response options (Appendix 2) additionally showed that the importance assigned to recommending genetic testing for family members varied by finding type.
Figure 2.
Percentage of participants rating clinical actions as “very important”, by specialty and finding. Findings include a pathogenic RYR1 variant associated with risk for malignant hyperthermia (“ACMG list”), a pathogenic PALB2 variant associated with risk for breast cancer risk (“Has ClinGen summary”), a pathogenic CHEK2 variant associated with risk for cancer (“No ClinGen summary”), a BRCA1 variant of uncertain significance in a patient with a family history of ovarian cancer (“VUS w family history”), and carrier status for three autosomal recessive conditions. P values summarize tests for non-equivalence comparing genetic specialists and non-genetic specialists (“specialty”) and comparing findings (“finding”), and include a test for interaction between specialty and finding (“interaction”).
* Genetic specialists were not presented this item.
Secondary analyses that compared whether there were differences in the total number of actions that providers rated “very important” showed no differences by site (p=0.0359) or between pediatricians and other provider types (p=0.2254). Similarly, no differences were observed between genetic counselors and medical geneticists in subanalyses of genetic specialists (p=0.4458). However, additional analyses showed that genetic specialists perceived stronger obligations to initiate follow-up than non-genetic specialists. Genetic specialists perceived personal obligations to initiate follow-up in 718 of 802 instances (90%) where they rated an action as very important. In contrast, non-genetic specialists perceived personal obligations to initiate follow-up in 226 of 403 instances (56%) where they rated an action as very important. Non-genetic specialists were also likely to perceive personal obligations to initiate referrals to genetics specialists when they deemed it important (30 of 42 instances; 71%).
Information to Interpret Reports
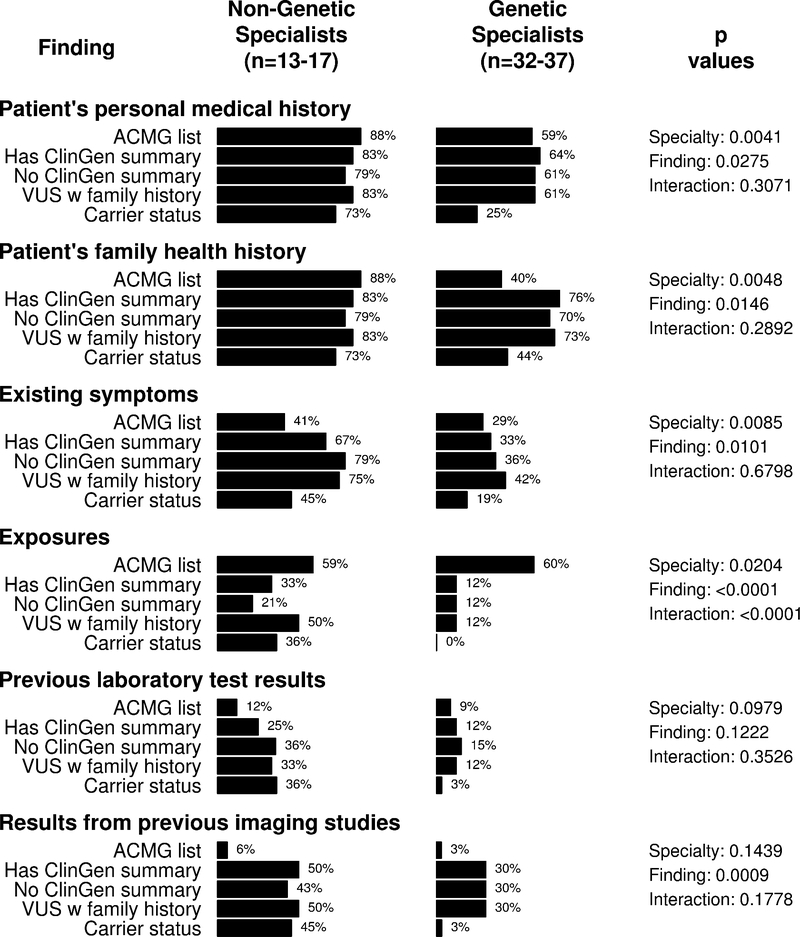
The importance that providers assigned to other clinical information for interpreting the reports is summarized in Figure 3. Across finding types and specialties, participants were most likely to consider patients’ personal and family health histories as extremely important (66% and 62%, respectively). Differences between genetic specialists and non-genetic specialists did not achieve significance on any items, although specialty did affect the association between finding type and importance of exposures (p<0.0001 in test for interaction). Sixty percent of genetic specialists and 59% non-genetic specialists rated exposures as very important to consider when considering the RYR1 variant associated with malignant hyperthermia, but genetic specialists tended to consider exposures as less important for all other reported variants. Independent effects of finding type were also observed for exposures (p≤0.0001). Analyses that considered all response options (Appendix 3) did show non-genetic specialists assigned greater importance than genetic specialists to personal histories of disease (p=0.0002), with open-ended responses suggesting that some genetic specialists were familiar with the specific variants that were presented and did not feel additional clinical information would be necessary to help them interpret the reports. Those analyses additionally showed that, while genetic specialists and non-genetic specialists assigned similar importance to imaging results when cancer-related variants (PALB2, CHEK2, BRCA1) and malignant hyperthemia (RYR1) variants were reported, genetic specialists assigned less importance to imaging results than non-genetic specialists when carrier findings were reported (p<0.0001 in interaction tests).
Figure 3.
Additional clinical information rated “very important” for interpreting the report, by specialty and finding. Findings include a pathogenic RYR1 variant associated with risk for malignant hyperthermia (“ACMG list”), a pathogenic PALB2 variant associated with risk for breast cancer risk (“Has ClinGen summary”), a pathogenic CHEK2 variant associated with risk for cancer (“No ClinGen summary”), a BRCA1 variant of uncertain significance in a patient with a family history of ovarian cancer (“VUS with family history”), and carrier status for three autosomal recessive conditions. P values summarize tests for non-equivalence comparing genetic specialists and non-genetic specialists (“specialty”) and comparing findings (“finding”), and include a test for interaction between specialty and finding (“interaction”).
Resources Consulted
Respondents generally anticipated the need to consult resources to help them understand and respond to the reports (Appendix 4). Non-genetic specialists favored discussions with colleagues/specialists (endorsed for 76% of reports) and citations in the report (endorsed for 71% of reports). Genetic specialists also favored citations in the report (endorsed for 82% of reports), but additionally anticipated using GeneReviews heavily (endorsed for 82% of reports). Open-ended responses included ClinVar, ExAC, the National Comprehensive Cancer Network, and PubMed, suggesting that some participants might conduct an independent evaluation of the reported findings.
DISCUSSION
Our study provides novel insight about how genetic specialists and non-genetic specialists with genomic sequencing experience anticipate they would respond to secondary finding reports. We found that genetic specialists were more likely to report understanding reports and less likely to perceive responding to them as a burden than non-genetic specialists. Genetic specialists were also more likely than non-genetic specialists to assert that laboratories had no obligations to report certain types of findings, such as VUSs or carrier status. Many of these findings were expected given the differences in training and job demands of genetic specialists compared to non-genetic specialists. However, these findings also highlight the challenges of integrating sequencing into medical care more broadly. The non-genetic specialists who completed this survey are likely to have been more familiar with genomics than non-genetic specialists at large and more motivated to see genomics incorporated into clinical care. It is possible that the differences we observed between groups would have been even greater if a population-based sample of non-genetic specialists had been enrolled. Our findings add urgency to calls to ensure physicians are prepared to appropriately respond to genomic sequencing reports through education and by enhancing the infrastructure supporting genomic medicine.27–30 With genomic sequencing becoming more common in primary care and other settings, current approaches to make resources such as decision support sheets available through genetics-centric repositories may not be sufficient. The genomics field will need to take steps to ensure the availability and accessibility of understandable information and resources to non-genetic specialists.
Our study also provides insight about how secondary findings with varying levels of supporting evidence are likely to be received by genetic specialists and non-genetic specialists. In the case of variants for which the evidence base about actionability was more developed, respondents were more likely to report that findings were important or that follow-up should be initiated, and less likely to report that follow-up might harm patients, that responding to the reports would be a burden, and that it was inappropriate for a physician to have provided the secondary finding report without addressing its findings. In addition, for findings with a stronger evidence base about actionability, respondents assigned more importance to documenting findings and recommending family discussions and less importance to reviewing personal and family history information and to learning about the variant. Whether these trends reflect sensitivity of respondents to the assumed actionability of the gene-disease combinations, the quantity of existing guidance, or both is unclear.
Findings in our study about carrier status merit special consideration. Even though scenarios deliberately presented women of reproductive age, participants were least enthusiastic about the value of learning about carrier status, and least likely to assign importance to clinical follow-up in response. The majority of participants reported discussing patients’ reproductive plans as very important, and it may be that participants’ responses are highly contingent to that information. Whatever the reason, providers may need to be prepared for carrier status findings, despite their reluctance, given that substantial numbers patients are likely to request this information if offered.31–33
Of additional note, genetic specialists were more likely than non-genetic specialists to perceive personal obligations to initiate follow-up even though many of them (i.e., genetic counselors) often lack ordering privileges. The majority of our items that collected data about clinical follow-up addressed actions to gain more information, such as learning more about the diseases and reviewing family history. It is possible that non-genetic specialists in our study felt that other specialists (though not necessarily specialists in genetics) would be better suited to collect and interpret such information when the findings were not within the participants’ specialty. In the large majority of cases for which non-genetic specialists rated a referral to a genetic specialist as important, they reported that they would initiate that referral. Unfortunately, we did not collect data about referrals to other types of specialties.
Limitations to our study include enrollment of a small convenience sample of providers experienced with genomics. Findings provide little insight about how physicians with no prior exposure to genomic sequencing would respond. The reports we presented are only one of many formats that are currently being used, and variants were a single exemplar for each category of secondary finding. It is possible that responses are sensitive to the specific diseases being presented rather than presence/absence of evidence summaries or the actionability of the information. A limited number of genes were presented, and biases and confounding may have been introduced by including genes that may be familiar to all providers (e.g., BRCA1 and HFE), genes that are likely to be more familiar among particular specialists (e.g., cancer-specific variants and oncologists), and genes that are likely to only be familiar to genetic specialists. Outcome measures were not validated instruments and represent hypothetical responses, and differences between groups that would be considered clinically significant were not specified.
Nevertheless, we provide important early insight about the responses of providers to genomic secondary findings that can help sequencing programs and policymakers anticipate providers’ responses and improve approaches to education and support. Such improvements will be critical as genome-scale sequencing moves beyond limited applications in subspecialty settings and becomes a more common part of medical care.
Supplementary Material
ACKNOWLEDGMENTS
The research described in this report was funded by grants U01HG006485, U01HG006487, U01HG006492, U01HG006500, U01HG006507, U01HG006546, U01HG007307, UM1HG007301, UM1HG006508, UM1HG007292, U01HG007307, ZIAHG200387, R01HG006600, and K01HG009173. The authors thank Carrie Blout, Wendy Chung, Jessica Everett, Jyoti Athanikar, Tia Kauffman, Katie Lewis, Sharon Plon, Kevin Bowling, and Myra Roche for their help recruiting participants.
REFERENCES
- 1.Quinlan-Jones E, Kilby MD, Greenfield S, et al. Prenatal whole exome sequencing: the views of clinicians, scientists, genetic counsellors and patient representatives. Prenat Diagn. 2016;36(10):935–941. [DOI] [PubMed] [Google Scholar]
- 2.Krier JB, Green RC. Management of incidental findings in clinical genomic sequencing. Curr Protoc Hum Genet. 2015;87:9.23.21–16. [DOI] [PubMed] [Google Scholar]
- 3.Vears DF, Senecal K, Borry P. Reporting practices for unsolicited and secondary findings from next generation sequencing technologies: perspectives of laboratory personnel. Hum Mutat. 2017;38(8):905–911. [DOI] [PubMed] [Google Scholar]
- 4.Friedman JM, Cornel MC, Goldenberg AJ, Lister KJ, Senecal K, Vears DF. Genomic newborn screening: public health policy considerations and recommendations. BMC Med Genomics. 2017;10(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang SK, Spector-Bagdady K, Caplan AL, Braithwaite RS. Exome and genome sequencing and parallels in radiology: searching for patient-centered management of incidental and secondary findings. J Am Coll Radiol. 2016;13(12 Pt A):1467–1472. [DOI] [PubMed] [Google Scholar]
- 6.Mackley MP, Fletcher B, Parker M, Watkins H, Ormondroyd E. Stakeholder views on secondary findings in whole-genome and whole-exome sequencing: a systematic review of quantitative and qualitative studies. Genet Med. 2017;19(3):283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15(7):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(2):249–255. [DOI] [PubMed] [Google Scholar]
- 9.Green RC, Lupski JR, Biesecker LG. Reporting genomic sequencing results to ordering clinicians: incidental, but not exceptional. JAMA. 2013;310(4):365–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGuire AL, Diaz CM, Wang T, Hilsenbeck SG. Social networkers’ attitudes toward direct-to-consumer personal genome testing. Am J Bioeth. 2009;9(6):3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen KD, Vassy JL, Jamal L, et al. Are physicians prepared for whole genome sequencing? a qualitative analysis. Clin Genet. 2016;89(2):228–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGuire AL, Burke W. An unwelcome side effect of direct-to-consumer personal genome testing: raiding the medical commons. JAMA. 2008;300(22):2669–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohane IS, Masys DR, Altman RB. The incidentalome: a threat to genomic medicine. JAMA. 2006;296(2):212–215. [DOI] [PubMed] [Google Scholar]
- 14.Brownlee S Overtreated: why too much medicine is making us sicker and poorer. New York, NY: Bloomsbury; 2007. [Google Scholar]
- 15.Kutz G Nutrigenetic Testing: Tests Purchased from Four Web Sites Mislead Consumers Testimony Before the Special Committee on Aging, U.S. Senate. Washington, DC: United States Government Accountability Office;2006. [Google Scholar]
- 16.Christensen KD, Kalia SS, Green RC. Incidental findings from genetic testing In: Raby BA, ed. UpToDate. Waltham, MA: UpToDate; 2014. [Google Scholar]
- 17.Green RC, Goddard KAB, Jarvik GP, et al. Clinical Sequencing Exploratory Research Consortium: accelerating evidence-based practice of genomic medicine. Am J Hum Genet. 2016;98(6):1051–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hindorff LA, Burke W, Laberge A, et al. Motivating factors for physician ordering of factor V Leiden genetic tests. Arch Intern Med. 2009;169(1):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheuner MT, Edelen MO, Hilborne LH, Lubin IM. Effective communication of molecular genetic test results to primary care providers. Genet Med. 2013;15(6):444–449. [DOI] [PubMed] [Google Scholar]
- 20.Haga SB, Carrig MM, O’Daniel JM, et al. Genomic risk profiling: attitudes and use in personal and clinical care of primary care physicians who offer risk profiling. J Gen Intern Med. 2011;26(8):834–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant RW, Hivert M, Pandiscio JC, Florez JC, Nathan DM, Meigs JB. The clinical application of genetic testing in type 2 diabetes: a patient and physician survey. Diabetologia. 2009;52(11):2299–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pennington JW, Karavite DJ, Krause EM, Miller J, Bernhardt BA, Grundmeier RW. Genomic decision support needs in pediatric primary care. J Am Med Inform Assoc. 2017;24(4):851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen KD, Scheuner MT, Garber JE, Rehm HL, Green RC. Responses of primary care physicians to unsolicited secondary findings about Lynch syndrome. 2015 American Society of Human Genetics Annual Meeting; Oct 22, 2015; Baltimore. [Google Scholar]
- 24.Hunter JE, Irving SA, Biesecker LG, et al. A standardized, evidence-based protocol to assess clinical actionability of genetic disorders associated with genomic variation. Genet Med. 2016;18(12):1258–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris RJ. A Primer of Multivariate Statistics. London: Psychology Press; 2001. [Google Scholar]
- 26.van Buuren S, Groothuis-Oudshoorn K. MICE: multivariate imputation by chained equations in R. J Stat Softw. 2011;45(3):1–67. [Google Scholar]
- 27.Korf BR, Berry AB, Limson M, et al. Framework for development of physician competencies in genomic medicine: report of the Competencies Working Group of the Inter-Society Coordinating Committee for Physician Education in Genomics. Genet Med. 2014;16(11):804–809. [DOI] [PubMed] [Google Scholar]
- 28.McCarthy JJ, McLeod HL, Ginsburg GS. Genomic medicine: a decade of successes, challenges, and opportunities. Sci Transl Med. 2013;5(189):189sr184–189sr184. [DOI] [PubMed] [Google Scholar]
- 29.Shirts BH, Salama JS, Aronson SJ, et al. CSER and eMERGE: current and potential state of the display of genetic information in the electronic health record. J Am Med Inform Assoc. 2015;22(6):1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vassy JL, Korf BR, Green RC. How to know when physicians are ready for genomic medicine. Sci Transl Med. 2015;7(287):287fs219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shahmirzadi L, Chao EC, Palmaer E, Parra MC, Tang S, Gonzalez KDF. Patient decisions for disclosure of secondary findings among the first 200 individuals undergoing clinical diagnostic exome sequencing. Genet Med. 2014;16(5):395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baptista NM, Christensen KD, Carere DA, Broadley SA, Roberts JS, Green RC. Adopting genetics: motivations and outcomes of personal genomic testing in adult adoptees. Genet Med. 2016;18(9):924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright MF, Lewis KL, Fisher TC, et al. Preferences for results delivery from exome sequencing/genome sequencing. Genet Med. 2014;16(6):442–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.