Abstract
Introduction
Amyotrophic lateral sclerosis (ALS) is a debilitating neurologic disorder with poor survival rates and no clear biomarkers for disease diagnosis and prognosis.
Methods
We compared serum miRNA expression from patients with ALS to healthy controls and patients with multiple sclerosis and Alzheimer’s disease. We also correlated miRNA expression in cross-sectional and longitudinal cohorts of ALS patients with clinical parameters.
Results
We identified 7 microRNAs (miR-192-5p, miR-192-3p, miR-1, miR-133a-3p, miR-133b, miR-144-5p and miR-19a-3p) that were upregulated and 6 microRNAs (miR-320c, miR-320a, let-7d-3p, miR-425-5p, miR-320b and miR-139-5p) that were downregulated in ALS patients compared to healthy controls, Alzheimer’s disease and multiple sclerosis patients. Changes in 4 miRNAs (miR-136-3p, miR-30b-5p, miR-331-3p, and miR-496) correlated positively and change in 1 microRNA (miR-2110) correlated negatively with changes in clinical parameters in longitudinal analysis.
Discussion
Our findings identified serum microRNAs that can serve as biomarkers for ALS diagnosis and progression.
Keywords: microRNA, serum, biomarkers, ALS, longitudinal analysis, disease comparisons
Introduction
Amyotrophic lateral sclerosis (ALS) is a debilitating neurodegenerative disorder characterized by the loss of motor neurons in the central nervous system (CNS) 1. Currently, the two treatment options available, riluzole and edravone have modest effects on survival and rate of progression, respectively2,3,4. Both these treatments appear to be more effective when started early in the disease2,4. Hence, the current average diagnostic delay of approximately 12 months is potentially deleterious5. Furthermore, our lack of understanding of disease pathogenesis hinders target identification and novel ALS therapy development6. Biomarkers for use in early diagnosis, prognosis, or disease progression have the potential to hasten drug development and improve clinical trial design.
Although the majority of ALS cases occur sporadically (sALS) with incompletely understood etiology, approximately 10% are familial by pedigree (familial ALS; fALS)1. Many genes have now been correlated to ALS pathology – some of the most common and best understood include SOD1, C9orf72, TARDBP and FUS7,8. Although these genetic mutations are associated with fALS cases, they also occur in approximately 10% of sALS patients suggesting an underlying genetic component to the disease9,10.
Interestingly, two of the most commonly mutated genes in the disease, TARDBP and FUS are implicated in the biogenesis of microRNA (miRNA)11–13, thereby suggesting that dysregulation of miRNA expression might be a valuable tool to predict disease onset and progression. miRNAs are short (18–25 nucleotides) non-coding RNA that regulate gene expression by inhibiting mRNA translation or affecting mRNA degradation14. Recent studies suggest that several diseases including neurodegenerative disorders are associated with the altered expression of miRNAs15. Consistent with this, differentially expressed miRNAs have been identified in plasma, serum, spinal cord and cerebrospinal fluid (CSF) of ALS patients16. However, small patient cohorts and the absence of longitudinal evaluation have posed major limitations to the translation of these findings to clinical settings.
The utility of miRNAs for treatment and diagnosis is an active area of research. Several miRNA-based therapeutics are currently in phase I and II clinical trials for numerous disease indications including hepatitis C infections, type 2 diabetes, non-alcoholic fatty liver disease, cutaneous T cell lymphoma, and mesothelioema17–19. In addition, specific miRNAs are known to differentiate healthy versus disease states that has led to testing miRNAs as diagnostic biomarkers in the clinic for cardiovascular, neurological, pulmonary disorders and various types of cancer19,20.
We aimed to identify miRNAs differentially expressed in serum samples from participants with ALS compared to both healthy participants and those with other neurologic diseases. We also studied the association between miRNA expression with disease progression. Finally, using available longitudinal data, we investigated whether changes in miRNA expression correlated with changes in clinical measures over time.
Methods
Participants
Patients included in the study met the El Escorial World Federation of Neurology Criteria21 for clinically definite, probable, probable laboratory-supported or possible ALS. Participants were enrolled as a convenience sample from the Massachusetts General Hospital multidisciplinary ALS clinic and were assessed clinically at each visit. ALS disease duration was calculated from the day of onset of the first weakness or spasticity symptoms. The two parameters recorded at each visit were the revised ALS Functional Rating Scale (ALSFRS-R), and vital capacity (VC)22,23. Samples and clinical scores were collected longitudinally at approximately 3 to 6 month intervals. Of the 23 ALS patients enrolled, 3 were fALS and 20 were sALS). Two of the fALS patients had C9orf72 repeat expansions and 1 was positive for an SOD1 mutation. 17 patients (15 sALS and 2 fALS) were treated with riluzole. The remaining 6 patients were untreated. Table 1 includes the detailed information about ALS diagnosis, treatment regimen and the number of visits for each patient.
Table 1.
Detailed characteristics of ALS patients
| El-Escorial criteria | ALS | Age | Gender | Treatment (Riluzole) | No. of visits |
|---|---|---|---|---|---|
| Probable Laboratory Supported | sporadic | 70.8 | Female | Yes | 5 |
| Definite | sporadic | 43.1 | Male | Yes | 4 |
| Definite | sporadic | 44.2 | Male | Yes | 2 |
| Probable | sporadic | 65.3 | Female | No | 7 |
| Possible | sporadic | 57.4 | Male | Yes | 6 |
| Probable Laboratory Supported | sporadic | 75.8 | Male | Yes | 2 |
| Probable | sporadic | 71.4 | Male | Yes | 2 |
| Probable Laboratory Supported | sporadic | 53.3 | Male | Yes | 6 |
| Possible | sporadic | 54.5 | Male | No | 6 |
| Probable Laboratory Supported | sporadic | 68.2 | Male | No | 2 |
| Probable Laboratory Supported | sporadic | 57.8 | Male | Yes | 4 |
| Definite | sporadic | 57.9 | Female | Yes | 1 |
| Probable Laboratory Supported | sporadic | 49.3 | Male | Yes | 3 |
| Probable | sporadic | 57.9 | Female | Yes | 3 |
| Probable | sporadic | 40.7 | Female | Yes | 5 |
| Probable | sporadic | 53 | Male | Yes | 1 |
| Definite | familial | 54.8 | Male | Yes | 1 |
| Definite | sporadic | 50.7 | Male | Yes | 5 |
| Probable | sporadic | 48.5 | Male | No | 2 |
| Definite | familial | 50.8 | Female | No | 1 |
| Definite | sporadic | 55.4 | Male | No | 2 |
| Probable | familial | 41.6 | Female | Yes | 1 |
| Probable | sporadic | 54.4 | Male | Yes | 1 |
Healthy controls (HC) (30 participants, 9 males and 21 females, average age 43±12.2 years) were obtained from the Brigham PhenoGenetic cohort study and from healthy participants enrolled in the Comprehensive Longitudinal Investigation of Multiple sclerosis at Brigham and Women’s Hospital and Partners MS Center (CLIMB, IRB 2013P002181/BWH). These subjects were recruited from the general population of Boston, are over 18 years of age, and have no chronic inflammatory, infectious or metabolic diseases24,25. Multiple sclerosis (MS) patient samples were obtained from the CLIMB study, and included relapsing-remitting (RRMS) (29 patients, 12 male and 17 female, average age 36±8.0 years ) secondary progressive (SPMS) (26 patients, 6 male and 20 female, average age 47±8.0 years), and primary progressive patients (PPMS) (18 patients, 9 male and 9 female, average age 51±8.0 years) who met the 2010 revisions to the McDonald criteria26. Alzheimer’s disease (AD) patient samples (30 patients, 9 male and 21 female, average age 74±3.6 years) were obtained in collaboration with Dr. David Bennett, Rush Lab, Chicago. AD patients recruited for the study were cognitively non-impaired at the time of enrollment and ranged in age from 65 to 9024.
A signed informed consent was received from all of the participants. Secondary use approval for other disease samples was obtained (IRB 2013P002181/BWH).
Samples and analysis
Blood samples were collected in glass red top serum vacutainer tubes without additives (BD Biosciences). Each sample was centrifuged at 2000 RPM for 10 minutes to separate serum and then stored at −80°C until RNA extraction. Serum was frozen within 2 hours of the blood draw. Multiple freeze–thaw cycles were avoided by storing the samples in aliquots.
RNA was isolated using the miRcury kit (Exiqon, Waltham, MA) and converted to cDNA using a synthesis kit from Exiqon (Woburn, MA). Prepared cDNAs were stored at −20°C until use. Locked Nucleic Acid (LNA)-SYBR- green-based real time PCR (RT-PCR) Custom made panel based on a prior study25 containing 191 miRNAs, were used for profiling in the first set. Normalization was performed using the mean expression of miRNA with the best stability index, i.e., those that were stably expressed in our sample set. NormFinder software was used to calculate the stability index. We used four normalizing miRNAs (miR-15b-5p, miR-19a-3p, miR-126-3p and miR-425-5p). The following formula was used to calculate the normalized Cq values:
Statistical analysis
The expression of circulating miRNAs was compared between HCs, ALS, MS and AD patients.
For the ALS patients, the following three comparisons were made: a) baseline miRNA expression to baseline ALSFRS-R, baseline VC and disease duration, b) baseline miRNA expression to change in ALSFRS-R and VC measured over time during multiple patient visits, and c) changes in miRNA expression to changes in ALSFRS-R and VC measured over time.
For each miRNA, the number of subjects who had detectable expression level and the mean expression level was calculated for each group (ALS, HC, MS and AD). ALS patients were then compared to each of the three comparison groups separately using a Wilcoxon rank sum test. Familial and sporadic ALS patients were also compared to each other using a Wilcoxon rank sum test.
A Wilcoxon rank sum test was used so that participants with miRNA levels below the limit of detection (missing or undetected values) could contribute to the analysis. Undetected expression values were assigned a value 1 less than the minimum detectable miRNA level across all subjects. The summary statistics and p-values were calculated for the three group comparisons (Supplementary Tables 1–3) In addition to the individual miRNA comparisons, two logistic regression models were built with ALS vs. all other groups as the outcomes and combinations of miRNAs. The first logistic regression model included all miRNAs that were significantly upregulated in ALS patients in all three group comparisons (ALS vs. HC, ALS vs. MS, ALS vs. AD), and the second logistic regression model included all miRNAs that were significantly downregulated in ALS patients. Subsequent logistic regression models were generated by comparing these miRNAs expression specifically within the individual groups (ALS vs. HC, ALS vs. MS and ALS vs. AD). The miRNAs chosen for inclusion in the multivariable logistic regression model were significantly associated with ALS in univariate analyses as shown in Supplementary Tables 1–3. The logistic regression coefficients and area under the ROC curve were calculated.
To investigate the clinical utility of miRNAs in ALS patients, we assessed both the cross-sectional and longitudinal associations between miRNAs and clinical outcomes. To assess the cross-sectional relationship between miRNAs and clinical measures, we estimated Spearman’s correlation coefficient between the baseline measurement of each miRNA and the ALSFRS-R, VC and ALS disease duration. In addition to the cross-sectional comparison, the change in each of the clinical measures over the course of follow-up was calculated by fitting a linear regression with the ALSFRS-R or VC as the outcome and the time since the first measurement as the predictor. To assess whether baseline miRNA was predictive of change in clinical measures, we estimated Spearman’s correlation coefficient between the baseline miRNA measurement and the slope in clinical measure. To assess whether change in miRNA was associated with change in clinical measures, we calculated the slope for each miRNA using a linear regression model and then estimated Spearman’s correlation coefficient between the two slope estimates. If the miRNA and clinical measure changed in the same direction i.e. both increased or decreased, then they were positively correlated; however if one parameter decreased while the other increased they were negatively correlated. In addition to the main analyses, we also completed the same analyses adjusting for use of riluzole and the results were largely unchanged so only the unadjusted results are reported.
In all the comparisons, p-values were also adjusted for multiple comparisons using false discovery rate (FDR), and these values are provided in the supplementary material (Supplementary Tables 1–3). Statistical analysis was completed using the statistical packages R (www.r-project.org) and Stata/IC version 14 (www.stata.com).
In all analyses, a p-value less than 0.05 was considered statistically significant. KEGG pathway analysis with miRNAs was carried out using DIANA miRPath27.
Results
Comparison of miRNAs expression between ALS patients, HC and other diseases
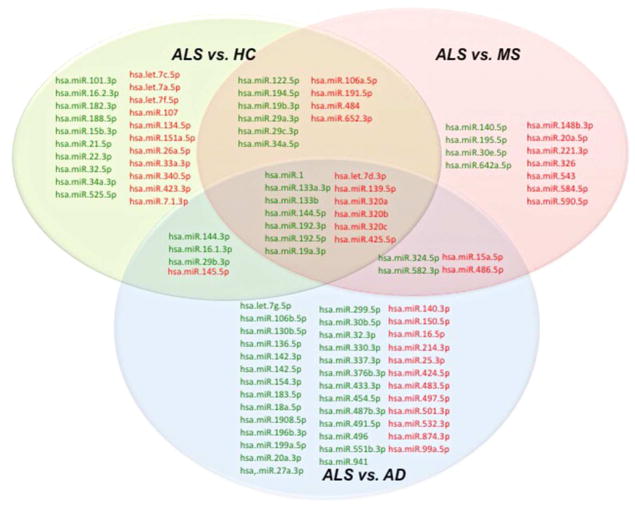
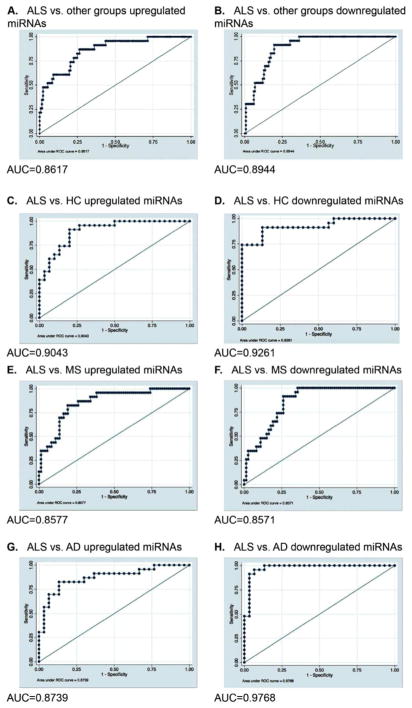
We found that 102 miRNAs were differentially expressed in at least one group comparison. Figure 1 shows the group comparison and the miRNAs that were specifically up- or downregulated in a single comparison and in comparison with one other group. We found 21 miRNAs that were only differentially expressed between ALS and HC, 11 miRNAs were only differentially expressed between patients with ALS and MS and 38 miRNAs were only differentially expressed between patients with ALS and AD. In addition to the miRNAs that were unique to a particular group comparison, 13 miRNAs were differentially expressed specifically in ALS patients compared to all other groups. Seven miRNAs were upregulated in ALS patients and 6 miRNAs were downregulated in ALS patients as compared to the three groups (Table 2). A logistic regression model was built with the 7 miRNAs that were upregulated in ALS in all comparisons. The estimated logistic regression coefficients are provided in Table 2 and the ROC curve is shown in Figure 2a. The area under the ROC curve for this model was 0.86, showing clear discrimination between ALS and other groups. The 6 miRNAs that were downregulated in ALS in all comparisons were used to build a single model with the estimated logistic regression coefficients provided in Table 2, and the ROC curve is shown in Figure 2b. The area under the ROC curve for this model was 0.89. Additionally, logistic regression models were generated for specific group comparisons using the combination of 7 miRNAs that were upregulated in ALS in all three comparisons and the 6 miRNAs that were downregulated in ALS compared to all other group comparisons (Figure 2).
Figure 1. Venn diagram showing the serum miRNAs differentially expressed in ALS compared to Healthy controls (HC), Multiple sclerosis (MS) and Alzheimer’s disease (AD).
Upregulated miRNAs are indicated in green and downregulated miRNAs are indicated in red.
Table 2.
Estimated coefficients from multivariable logistic regression model including all miRNAs upregulated and downregulated in ALS versus other diseases.
| miRNA | Coefficient | SE | p-value | Lower limit of 95% CI | Upper limit of 95% CI |
|---|---|---|---|---|---|
| Upregulated miRNAs | |||||
| has-miR-1 | 0.219 | 0.249 | 0.378 | −0.26854 | 0.70750 |
| has-miR-133a-3p | 0.183 | 0.3385 | 0.588 | −0.47924 | 0.84573 |
| has-miR-133b | 0.392 | 0.355 | 0.27 | −0.30404 | 1.08813 |
| has-miR-144-5p | 0.797 | 0.408 | 0.051 | −0.00297 | 1.59599 |
| has-miR-192-3p | 0.320 | 0.162 | 0.048 | 0.002112 | 0.63876 |
| has-miR-192-5p | 0.600 | 0.380 | 0.115 | −0.14526 | 1.34572 |
| has-miR-19a-3p | 2.092 | 0.790 | 0.008 | 0.54564 | 3.63861 |
| Downregulated miRNAs | |||||
| hsa-let-7d-3p | −0.430 | 0.510 | 0.4 | −1.42977 | 0.57114 |
| hsa-mir-139-5p | −0.490 | 0.316 | 0.121 | −1.11012 | 0.12977 |
| hsa-miR-320a | 1.652 | 1.156 | 0.153 | −0.61316 | 3.91649 |
| hsa-miR-320b | −0.271 | 1.054 | 0.797 | −2.33706 | 1.79411 |
| hsa-miR-320c | −3.718 | 1.397 | 0.008 | −6.4574 | −0.97951 |
| hsa-miR-425-5p | −3.003 | 1.353 | 0.026 | −5.65544 | −0.35143 |
SE = Standard error, CI = Confidence interval
Figure 2. ROC curves for miRNAs differentially expressed in ALS versus other groups (HC,MS and AD).
ROC curve for a combination of 7 miRNAs ( miR-1, miR-133a-3p, miR-133b, miR-144-5p, miR-192-3p, miR-195-5p and miR-19a-3p) upregulated in (A) ALS vs. all 3 groups, (C) ALS vs. HC, (E) ALS vs. MS, (G) ALS vs. AD . ROC curve for a combination of 6 miRNAs (let-7d-3p, miR-320a, miR-320b, miR-320c, miR-425-5p and miR-139-5p) downregulated in (B) ALS vs. all 3 groups, (D) ALS vs. HC, (F) ALS vs. MS, (H) ALS vs. AD ROC= Receiver Operating Characteristic, HC=healthy controls, ALS= Amyotrophic lateral sclerosis, MS= Multiple sclerosis, AD=Alzheimer’s disease, AUC = Area under the curve.
Correlation of miRNAs with clinical parameters
The estimated Spearman’s correlation coefficient comparing baseline miRNAs expression with baseline ALSFRS-R, VC and disease duration is provided in Supplementary Table 4, and a summary of the significant miRNAs is provided in Table 3. A positive correlation indicated that increased miRNA expression correlated with higher ALSFRS-R and VC values (Table 3). 4 miRNAs significantly correlated with ALSFRS-R score - 1 positively and 3 negatively (Table 3). 8 miRNAs negatively correlated with VC, and 6 positively correlated (Table 3). For disease duration, 7 miRNAs were negatively correlated (a decreased expression of these miRNAs is associated with longer disease duration), and 1 miRNA was positively correlated with disease duration (Table 3).
Table 3.
Serum miRNAs correlating with baseline clinical parameters.
| miRNA | Correlation coefficient | p-value | |
|---|---|---|---|
| ALSFRS-R | hsa-miR-32-5p | 0.468 | 0.043 |
| hsa-miR-127-3p | −0.463 | 0.046 | |
| hsa-miR-191-5p | −0.579 | 0.01 | |
| hsa-miR-770-5p | −0.482 | 0.036 | |
| VC | hsa-miR-101-3p | 0.606 | 0.006 |
| hsa-miR-15b-3p | 0.528 | 0.020 | |
| hsa-miR-19a-3p | 0.635 | 0.003 | |
| hsa-miR-361-3p | 0.518 | 0.023 | |
| hsa-miR-451a | 0.622 | 0.004 | |
| hsa-miR-92a-1-5p | 0.494 | 0.032 | |
| hsa-miR-1 | −0.510 | 0.026 | |
| hsa-miR-133a-3p | −0.729 | 0.0004 | |
| hsa-miR-133b | −0.620 | 0.005 | |
| hsa-miR-182-3p | −0.514 | 0.024 | |
| hsa-miR-221-3p | −0.605 | 0.006 | |
| hsa-miR-330-3p | −0.475 | 0.04 | |
| hsa-miR-584-5p | −0.575 | 0.010 | |
| hsa-miR-589-3p | −0.492 | 0.033 | |
| Disease duration | hsa-miR-9-3p | 0.455 | 0.044 |
| hsa-miR-142-3p | −0.558 | 0.012 | |
| hsa-miR-21-5p | −0.457 | 0.044 | |
| hsa-miR-33a-5p | −0.450 | 0.048 | |
| hsa-miR-34a-5p | −0.535 | 0.016 | |
| hsa-miR-376b-3p | −0.464 | 0.039 | |
| hsa-miR-491-5p | −0.473 | 0.035 |
ALSFRS-R = revised ALS Functional rating scale, VC = vital capacity
The estimated Spearman’s correlation coefficients comparing baseline miRNAs and longitudinal change in ALSFRS-R and VC are provided in Supplementary Table 5. A positive correlation indicated that a high baseline expression is associated with a higher slope value or a lesser decrease with time, while a negative correlation indicated a high baseline miRNA expression is associated with a lower slope value or a greater decrease with time. 4 miRNAs correlated positively, and 2 miRNAs correlated negatively with ALSFRS-R change (Table 4). 8 miRNAs correlated positively and no miRNAs correlated negatively with changes in VC over time (Table 4).
Table 4.
Serum miRNAs correlating with clinical parameters in longitudinal analysis
| Longitudinal analysis | miRNA | Correlation coefficient | p-value |
|---|---|---|---|
| Baseline miRNA / slope ALSFRS-R | hsa-miR-10a-3p | 0.702 | 0.007 |
| hsa-miR-30e-3p | 0.742 | 0.005 | |
| hsa-miR-525-5p | 0.613 | 0.026 | |
| hsa-miR-598-3p | 0.564 | 0.045 | |
| hsa-miR-199a-5p | −0.610 | 0.030 | |
| hsa-miR-27a-3p | −0.654 | 0.018 | |
| Baseline miRNA / slope VC | hsa-miR-133a-3p | 0.725 | 0.007 |
| hsa-miR-133b | 0.632 | 0.024 | |
| hsa-miR-155-5p | 0.560 | 0.05 | |
| hsa-miR-182-3p | 0.775 | 0.003 | |
| hsa-miR-1913 | 0.558 | 0.05 | |
| hsa-miR-22-5p | 0.593 | 0.036 | |
| hsa-miR-584-5p | 0.610 | 0.030 | |
| hsa-miR-99a-5p | 0.731 | 0.006 | |
| Slope miRNA / Slope ALSFRS-R | hsa-miR-148b-3p | 0.66 | 0.017 |
| hsa-miR-151a-5p | 0.643 | 0.021 | |
| hsa-miR-181c-3p | 0.747 | 0.005 | |
| hsa-miR-199a-3p | 0.593 | 0.036 | |
| hsa-miR-199a-5p | 0.703 | 0.01 | |
| hsa-miR-301b | 0.588 | 0.038 | |
| hsa-miR-324-5p | 0.725 | 0.007 | |
| hsa-miR-326 | 0.654 | 0.018 | |
| hsa-miR-376b-3p | 0.571 | 0.045 | |
| Slope miRNA / Slope VC | hsa-let-7c-5p | 0.560 | 0.05 |
| hsa-miR-107 | 0.687 | 0.012 | |
| hsa-miR-142-3p | 0.610 | 0.030 | |
| hsa-miR-154-3p | 0.577 | 0.043 | |
| hsa-miR-17-3p | 0.118 | 0.700 | |
| hsa-miR-181a-2-3p | 0.648 | 0.02 | |
| hsa-miR-423-3p | 0.593 | 0.04 | |
| hsa-miR-595 | 0.566 | 0.047 | |
| hsa-miR-652-3p | 0.604 | 0.032 | |
| hsa-miR-320b | −0.610 | 0.030 | |
| hsa-miR-483-5p | −0.577 | 0.043 | |
| hsa-miR-532-3p | −0.626 | 0.025 | |
| hsa-miR-589-3p | −0.599 | 0.034 | |
| hsa-miR-874-3p | −0.626 | 0.025 |
ALSFRS-R = revised ALS Functional rating scale, VC = vital capacity
Finally, we compared the change in miRNAs and the change in each clinical measure over time to assess whether miRNAs can be used as biomarkers of disease progression (Supplementary Table 5). The change in 4 miRNAs correlated positively with the change in both clinical measures and the change in miR-2110 correlated negatively with the change in both ALSFRS-R and VC (Table 5).
Table 5.
miRNAs correlating with changes in ALSFRS-R and VC
| miRNA | Correlation coefficient ALSFRS-R score | p-value ALSFRS-R score | Correlation coefficient VC | p-value VC |
|---|---|---|---|---|
| hsa-miR-136-3p | 0.637 | 0.022 | 0.687 | 0.012 |
| hsa-miR-30b-5p | 0.577 | 0.043 | 0.604 | 0.032 |
| hsa-miR-331-3p | 0.577 | 0.043 | 0.593 | 0.036 |
| hsa-miR-496 | 0.687 | 0.012 | 0.571 | 0.045 |
| hsa-miR-2110 | -0.571 | 0.045 | −0.687 | 0.012 |
ALSFRS-R = revised ALS Functional rating scale, VC = vital capacity
In a sub-analysis between sporadic and familial ALS patients, we found that the expression of miR-574-3p was significantly higher in sALS patients compared to fALS and miR-628-3p was significantly lower in sALS patients compared to fALS (Supplementary Table 6).
Discussion
ALS biomarkers have the potential to help elucidate the pathogenesis of ALS, identify relevant pathways for drug development, provide biological patient strata, and after much validation, even act as surrogate endpoints in clinical trials. No single biomarker can fill all these roles; in fact, each of these biomarker applications might require different characteristics. But as a group, miRNAs, which are highly varied but tightly controlled, broadly relevant and ubiquitously involved in cellular processes, could potentially serve many of these roles. Already, several studies have identified miRNAs in CSF, serum, plasma, leukocytes, spinal cord and skeletal muscle biopsies from ALS patients16. Using easily accessible serum samples, we have identified unique miRNA signatures that can distinguish participants with ALS from healthy participants and those with AD or MS. We also identified other miRNAs expression changes that correlate with disease state at baseline and over time and still others that predict rate of ALS progression as determined by changes in miRNA expression with changes in clinical measures such as ALSFRS-R score, VC and disease duration.
We observed some overlap with previously reported miRNAs (reviewed in16). For instance, miR-1, miR-133a-3p and miR-133b were previously found to be upregulated in plasma and serum samples from ALS patients28. In addition, miR-133b was also found to be downregulated in spinal cord biopsies29. Here, we report that miR-1, miR-133a-3p and miR-133b are upregulated in serum from ALS patients compared to other groups and consistent with this, their expression negatively correlates with VC. Another interesting miRNA, miR-451 has been reported to be downregulated in blood leukocytes from ALS patients30 and we found that its expression positively correlates with VC indicating that lower expression is associated with worse disease. miR-142-3p was found to be upregulated in spinal cord tissue samples from ALS patients29,31. In our analysis, miR-142-3p negatively correlated with DD and low miR-142-3p expression correlated with rapid change in VC in our longitudinal studies.
In addition to intersecting with previously identified miRNAs, we also found striking physiological relevance for other miRNAs recognized in this study. Of the 7 upregulated miRNAs, miR-1, miR-133a-3p and miR-133b are myomiRs or muscle–specific microRNAs that are critical for the development and maintenance of skeletal muscles32,33. Previous studies have indicated an enhanced expression of Myogenic Differentiation 1 (MyoD1) and myogenin (MyoG) in ALS patients suggesting an active differentiation process in these patients34. Muscle atrophy is prevalent in ALS patients and an enhancement of homeostatic regulators of myogenesis might be a mechanism to cope with the muscular atrophy. miR-133b and miR-206 are encoded by a bicistronic transcript and have been studied in the G93A SOD1 mouse model. Upregulation of these miRNAs is associated with the onset of neurological symptoms in mice as a compensatory mechanism to regenerate neuromuscular synapses35. Our observation of enhanced expression of myomiRs in serum samples from ALS patients might be related to muscle atrophy and therefore serve as markers of disease onset and prognosis.
Among the downregulated miRNAS, miR-320a is known to target Aquaporins 1 and 4(AQP1 and AQP4)36. The overexpression of AQP4 mostly in perivascular astrocytes has been observed in mouse and rat models of ALS and is associated with breakdown of the blood brain barrier (BBB)37. Impairment of the BBB though subtle, has been described in ALS patients38. Other members of the miR-320 family, miR-320b and miR-320c were also found to be downregulated in our samples. Interestingly, miR-320a, b and c are proposed diagnostic markers for schizophrenia as their expression is downregulated in the peripheral blood of schizophrenia patients39. However, the molecular mechanism by which they might be contributing to schizophrenia remains to be investigated. An analysis of the common pathways regulated by miR-320a, miR-320b, miR-320c, miR-425-5p, let-7d-3p and mir-139-5p points to their role in the Hippo signaling pathway27. Mammalian sterile 20-like kinase 1 (MST1) is a protein kinase implicated in the Hippo pathway and its homozygous deletion delays the onset of ALS in mice40. One potential hypothesis for this effect might be that MST-1 induces neuronal cell death and muscle degeneration upon phosphorylation. While MST-1 is not a direct target of these miRNAs, they target upstream regulators such as large tumor suppressor kinase (LATS1, LATS2), and Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma (YWHAG) implicated in the Hippo pathway27,41.
Previous studies have shown that miR-574-3p is implicated in TDP-43 binding and is downregulated in lymphoblast cell lines in all patients except those with SOD1 mutation12. Furthermore, both miR-628-3p and miR-574-3p are implicated in regulating DNA methyltransferases. There is growing evidence to indicate epigenetic changes in the promoter region of genes involved in processes such as calcium homeostasis, excitotoxicity and oxidative stress in patients with sALS42. The comparison of sporadic versus familial ALS was not the primary focus of the current investigation. Therefore, these studies warrant further investigation with larger patient groups to identify epigenetic signatures that might potentially distinguish groups of ALS patients.
Our longitudinal analysis suggests that miRNAs may be useful as prognostic biomarkers to provide information about expected rate of disease progression. We identified 5 miRNAs whose change in expression over time correlated with both changes in ALSFRS-R score and VC. Of these, the low expression of miR-136-3p, miR-30b-5p, miR-331-3p and miR-496 and high expression of miR-2110 correlated with more rapid disease progression. A KEGG pathway analysis of miR-136-3p, miR-30b-5p, miR-331-3p showed that these miRNAs are involved in the extracellular matrix (ECM) receptor pathway. Gene expression profile analysis in fibroblasts from patients with fALS indicated an enrichment in ECM receptor pathway and focal adhesion pathway molecules compared to healthy controls43. Accordingly, an upregulation of ECM-related genes was also observed in astrocytes from SOD1 G93A mice, and these genes were downregulated in the spinal cord from presymptomatic SOD1 G93A mice compared to WT mice. Some of the genes in this pathway that are upregulated in these disease models include Tumor necrosis factor (TNF), Endothelin1 (END1), and Angiotensin (AGT)44,45.
In a recent study comparing 12 different commercially available miRNA expression platforms for studying differential expression in serum, the LNA-based quantitative PCR platform from Exiqon, showed good reproducibility, accuracy and the highest specificity46. Therefore, we believe this is a robust system to measure the expression of miRNAs. However, due to the sample size and the exploratory nature of this study there were some limitations: 1) While serum samples from patients with MS and AD were used as controls to identify differentially expressing miRNAs in ALS, the use of serum samples from patients with other neurological disorders whose phenotypes overlap with that of ALS such as inclusion body myositis and multifocal motor neuropathy will provide better differential diagnostic information, 2) not all miRNAs were expressed in every sample. In the group comparisons, the diverse age groups of participants in the AD group compared to others might account for this loss of miRNA expression and 3) only a subset of miRNAs remained significant after correcting for multiple comparisons using the false discovery rate (Supplementary Tables 1–3).
This study correlates changes in miRNA expression with changes in clinical parameters over time, providing insight into disease onset and progression. It provides the groundwork for confirmatory work in the future with a comprehensive validation in a larger cohort of patients, ideally with genomic information, collected in a multi-center study. This will enable the confirmation of these miRNAs as markers of diagnosis, disease prognosis as well as surrogate markers to serve as primary and secondary outcomes in clinical trials.
Supplementary Material
Acknowledgments
This study was funded by NIH common funds via NCATS UH2/UH3 grant TR000890. We thank all the patients who consented to participate in this study.
Abbreviations
- ALS
Amyotrophic lateral sclerosis
- CNS
Central Nervous System
- sALS
Sporadic ALS
- fALS
Familial ALS
- miRNA
MicroRNA
- CSF
Cerebrospinal fluid
- HC
Healthy controls
- CLIMB
Comprehensive Longitudinal Investigation of Multiple sclerosis at Brigham and Women’s Hospital and Partners MS Center
- ALSFRS-R
ALS Functional Rating Scale
- VC
Vital capacity
- MyoD1
Myogenic Differentiation 1
- MyoG
Myogenin
- AQP
Aquaporins
- BBB
Blood brain barrier
- MST1
Mammalian sterile 20-like kinase 1
- LATS1, LATS2
Large tumor suppressor kinase
- YWHAG
Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma
- ECM
Extracellular matrix
- TNF
Tumor necrosis factor
- END1
Endothelin 1
- AGT
Angiotensin
Footnotes
Ethical Publication Statement: We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Disclosures of conflict of interest:R. Raheja, K. Regev, M.A Mazzola, V Beynon, A. Paul report no disclosures. B.C. Healy served on the scientific advisory board for Biogen, Worldwide Medical Biostatistics, Multiple Sclerosis, served on the editorial board for Statistical Methods in Medical Research, received research support from Merck Serono, Genzyme, Novartis, Google Life Sciences, NIH, National Multiple Sclerosis Society. F. Glenn received research support from Sao Paulo Research Foundation–Brazil. C. Diaz-Cruz received research support from Merck Serono, Google Life Sciences. T. Gholipur received research support from Merck Serono, compensation as a reviewer from Boehringer Ingelheim, their spouse received compensation as a reviewer from Boehringer Ingelheim. B.I. Glanz received research support from Merck Serono SA, NIH, NMSS. P. Kivisakk received research support from EMD Serono, Sanofi Genzyme, Verily Life Sciences. T. Chitnis served on clinical trial advisory boards for Novartis Pharmaceuticals, Genzyme-Sanofi, consulted for Biogen Idec, Novartis, Genzyme-Sanofi, Genentech Roche, received research support from EMD Serono, Novartis, Biogen, Verily, National Multiple Sclerosis Society, the Peabody Foundation, the Consortium for MS Centers, Guthy-Jackson Charitable Foundation. H.L. Weiner served on the scientific advisory board for the Guthy-Jackson Charitable Foundation, Teva Pharmaceutical Industries Ltd., Biogen Idec, Novartis, Sanofi-Aventis, consulted for Therapix, Biogen, Novartis, Serono, Teva, Sanofi, National Multiple Sclerosis Society. J.D Berry has consulted with Biogen, Denali Therapeutics and Neuraltus Pharmaceuticals, and has received research support from Voyager Therapeutics, GSK, Cytokinetics, Brainstorm Cell Therapeutics, and Novartis. R. Gandhi served on the editorial board for Journal of Neurological Disorders & Stroke, received research support from Novartis, Biogen, EMD Serono, Sanofi NIH, Nancy Davis Foundation, Harvard NeuroDiscovery Center, National Multiple Sclerosis Society.
References
- 1.Paez-Colasante X, Figueroa-Romero C, Sakowski SA, Goutman SA, Feldman EL. Amyotrophic lateral sclerosis: mechanisms and therapeutics in the epigenomic era. Nat Rev Neurol. 2015;11(5):266–279. doi: 10.1038/nrneurol.2015.57. [DOI] [PubMed] [Google Scholar]
- 2.Bensimon G, Lacomblez L, Meininger V. A controlled trial of riluzole in amyotrophic lateral sclerosis. ALS/Riluzole Study Group. N Engl J Med. 1994;330(9):585–591. doi: 10.1056/NEJM199403033300901. [DOI] [PubMed] [Google Scholar]
- 3.Writing G, Edaravone ALSSG. Safety and efficacy of edaravone in well defined patients with amyotrophic lateral sclerosis: a randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2017;16(7):505–512. doi: 10.1016/S1474-4422(17)30115-1. [DOI] [PubMed] [Google Scholar]
- 4.Petrov D, Mansfield C, Moussy A, Hermine O. ALS Clinical Trials Review: 20 Years of Failure. Are We Any Closer to Registering a New Treatment? Front Aging Neurosci. 2017;9:68. doi: 10.3389/fnagi.2017.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cellura E, Spataro R, Taiello AC, La Bella V. Factors affecting the diagnostic delay in amyotrophic lateral sclerosis. Clin Neurol Neurosurg. 2012;114(6):550–554. doi: 10.1016/j.clineuro.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 6.Mitsumoto H, Brooks BR, Silani V. Clinical trials in amyotrophic lateral sclerosis: why so many negative trials and how can trials be improved? Lancet Neurol. 2014;13(11):1127–1138. doi: 10.1016/S1474-4422(14)70129-2. [DOI] [PubMed] [Google Scholar]
- 7.Al-Chalabi A, Hardiman O. The epidemiology of ALS: a conspiracy of genes, environment and time. Nat Rev Neurol. 2013;9(11):617–628. doi: 10.1038/nrneurol.2013.203. [DOI] [PubMed] [Google Scholar]
- 8.Marangi G, Traynor BJ. Genetic causes of amyotrophic lateral sclerosis: new genetic analysis methodologies entailing new opportunities and challenges. Brain Res. 2015;1607:75–93. doi: 10.1016/j.brainres.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renton AE, Chio A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17(1):17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su XW, Broach JR, Connor JR, Gerhard GS, Simmons Z. Genetic heterogeneity of amyotrophic lateral sclerosis: implications for clinical practice and research. Muscle Nerve. 2014;49(6):786–803. doi: 10.1002/mus.24198. [DOI] [PubMed] [Google Scholar]
- 11.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci U S A. 2012;109(9):3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freischmidt A, Muller K, Ludolph AC, Weishaupt JH. Systemic dysregulation of TDP-43 binding microRNAs in amyotrophic lateral sclerosis. Acta Neuropathol Commun. 2013;1:42. doi: 10.1186/2051-5960-1-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morlando M, Dini Modigliani S, Torrelli G, Rosa A, Di Carlo V, Caffarelli E, Bozzoni I. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2012;31(24):4502–4510. doi: 10.1038/emboj.2012.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 15.Grasso M, Piscopo P, Confaloni A, Denti MA. Circulating miRNAs as biomarkers for neurodegenerative disorders. Molecules. 2014;19(5):6891–6910. doi: 10.3390/molecules19056891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cloutier F, Marrero A, O’Connell C, Morin P., Jr MicroRNAs as potential circulating biomarkers for amyotrophic lateral sclerosis. J Mol Neurosci. 2015;56(1):102–112. doi: 10.1007/s12031-014-0471-8. [DOI] [PubMed] [Google Scholar]
- 17.Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee SS. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol Ther Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Chen J, Sen S. MicroRNA as Biomarkers and Diagnostics. J Cell Physiol. 2016;231(1):25–30. doi: 10.1002/jcp.25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12–23. doi: 10.1016/j.critrevonc.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Brooks BR. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. J Neurol Sci. 1994;124(Suppl):96–107. doi: 10.1016/0022-510x(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 22.Cedarbaum JM, Stambler N, Malta E, Fuller C, Hilt D, Thurmond B, Nakanishi A. The ALSFRS-R: a revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS Study Group (Phase III) J Neurol Sci. 1999;169(1–2):13–21. doi: 10.1016/s0022-510x(99)00210-5. [DOI] [PubMed] [Google Scholar]
- 23.Czaplinski A, Yen AA, Appel SH. Forced vital capacity (FVC) as an indicator of survival and disease progression in an ALS clinic population. J Neurol Neurosurg Psychiatry. 2006;77(3):390–392. doi: 10.1136/jnnp.2005.072660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan G, White CC, Winn PA, Cimpean M, Replogle JM, Glick LR, Cuerdon NE, Ryan KJ, Johnson KA, Schneider JA, Bennett DA, Chibnik LB, Sperling RA, De Jager PL, Bradshaw EM. Trans-pQTL study identifies immune crosstalk between Parkinson and Alzheimer loci. Neurol Genet. 2016;2(4):e90. doi: 10.1212/NXG.0000000000000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regev K, Paul A, Healy B, von Glenn F, Diaz-Cruz C, Gholipour T, Mazzola MA, Raheja R, Nejad P, Glanz BI, Kivisakk P, Chitnis T, Weiner HL, Gandhi R. Comprehensive evaluation of serum microRNAs as biomarkers in multiple sclerosis. Neurol Neuroimmunol Neuroinflamm. 2016;3(5):e267. doi: 10.1212/NXI.0000000000000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polman CH, Reingold SC, Banwell B, Clanet M, Cohen JA, Filippi M, Fujihara K, Havrdova E, Hutchinson M, Kappos L, Lublin FD, Montalban X, O’Connor P, Sandberg-Wollheim M, Thompson AJ, Waubant E, Weinshenker B, Wolinsky JS. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69(2):292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3. 0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tasca E, Pegoraro V, Merico A, Angelini C. Circulating microRNAs as biomarkers of muscle differentiation and atrophy in ALS. Clin Neuropathol. 2016;35(1):22–30. doi: 10.5414/NP300889. [DOI] [PubMed] [Google Scholar]
- 29.Figueroa-Romero C, Hur J, Lunn JS, Paez-Colasante X, Bender DE, Yung R, Sakowski SA, Feldman EL. Expression of microRNAs in human post-mortem amyotrophic lateral sclerosis spinal cords provides insight into disease mechanisms. Mol Cell Neurosci. 2016;71:34–45. doi: 10.1016/j.mcn.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Wei Q, Chen X, Li C, Cao B, Ou R, Hadano S, Shang HF. Aberration of miRNAs Expression in Leukocytes from Sporadic Amyotrophic Lateral Sclerosis. Front Mol Neurosci. 2016;9:69. doi: 10.3389/fnmol.2016.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koval ED, Shaner C, Zhang P, du Maine X, Fischer K, Tay J, Chau BN, Wu GF, Miller TM. Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice. Hum Mol Genet. 2013;22(20):4127–4135. doi: 10.1093/hmg/ddt261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Townley-Tilson WH, Callis TE, Wang D. MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int J Biochem Cell Biol. 2010;42(8):1252–1255. doi: 10.1016/j.biocel.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen L, Jorgensen LH, Bech RD, Frandsen U, Schroder HD. Skeletal Muscle Remodelling as a Function of Disease Progression in Amyotrophic Lateral Sclerosis. Biomed Res Int. 2016;2016:5930621. doi: 10.1155/2016/5930621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326(5959):1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sepramaniam S, Armugam A, Lim KY, Karolina DS, Swaminathan P, Tan JR, Jeyaseelan K. MicroRNA 320a functions as a novel endogenous modulator of aquaporins 1 and 4 as well as a potential therapeutic target in cerebral ischemia. J Biol Chem. 2010;285(38):29223–29230. doi: 10.1074/jbc.M110.144576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomas-Camardiel M, Venero JL, Herrera AJ, De Pablos RM, Pintor-Toro JA, Machado A, Cano J. Blood-brain barrier disruption highly induces aquaporin-4 mRNA and protein in perivascular and parenchymal astrocytes: protective effect by estradiol treatment in ovariectomized animals. J Neurosci Res. 2005;80(2):235–246. doi: 10.1002/jnr.20443. [DOI] [PubMed] [Google Scholar]
- 38.Garbuzova-Davis S, Sanberg PR. Blood-CNS Barrier Impairment in ALS patients versus an animal model. Front Cell Neurosci. 2014;8:21. doi: 10.3389/fncel.2014.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vachev TIPN, Stoyanova VK, Ivanov HY, Minchev DS. Downregulation of miR-320 gene family memners in the peropheral blood f schizophrenia patients. Int J Curr Microbiol All Sci. 2016;5(1):221–230. [Google Scholar]
- 40.Lee JK, Shin JH, Hwang SG, Gwag BJ, McKee AC, Lee J, Kowall NW, Ryu H, Lim DS, Choi EJ. MST1 functions as a key modulator of neurodegeneration in a mouse model of ALS. Proc Natl Acad Sci U S A. 2013;110(29):12066–12071. doi: 10.1073/pnas.1300894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Granucci E. Theses dissertation. Boston University; 2016. Evaluating the role of the Hippo pathway in the onset and disease progression of the SOD1 mouse model of anyotrophic lateral sclerosis. [Google Scholar]
- 42.Morahan JM, Yu B, Trent RJ, Pamphlett R. A genome-wide analysis of brain DNA methylation identifies new candidate genes for sporadic amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2009;10(5–6):418–429. doi: 10.3109/17482960802635397. [DOI] [PubMed] [Google Scholar]
- 43.Kotni MK, Zhao M, Wei DQ. Gene expression profiles and protein-protein interaction networks in amyotrophic lateral sclerosis patients with C9orf72 mutation. Orphanet J Rare Dis. 2016;11(1):148. doi: 10.1186/s13023-016-0531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phatnani HP, Guarnieri P, Friedman BA, Carrasco MA, Muratet M, O’Keeffe S, Nwakeze C, Pauli-Behn F, Newberry KM, Meadows SK, Tapia JC, Myers RM, Maniatis T. Intricate interplay between astrocytes and motor neurons in ALS. Proc Natl Acad Sci U S A. 2013;110(8):E756–765. doi: 10.1073/pnas.1222361110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Oliveira GP, Alves CJ, Chadi G. Early gene expression changes in spinal cord from SOD1(G93A) Amyotrophic Lateral Sclerosis animal model. Front Cell Neurosci. 2013;7:216. doi: 10.3389/fncel.2013.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, Cheo D, D’Andrade P, DeMayo M, Dennis L, Derveaux S, Feng Y, Fulmer-Smentek S, Gerstmayer B, Gouffon J, Grimley C, Lader E, Lee KY, Luo S, Mouritzen P, Narayanan A, Patel S, Peiffer S, Ruberg S, Schroth G, Schuster D, Shaffer JM, Shelton EJ, Silveria S, Ulmanella U, Veeramachaneni V, Staedtler F, Peters T, Guettouche T, Wong L, Vandesompele J. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11(8):809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.