Abstract
To develop a new strategy that controls vascular pathogen infections in economic crops, we examined a possible enhancer of the vascular activity of XYLOGEN PROTEIN 1 promoter (Px). This protein is specifically expressed in the vascular tissues of Arabidopsis thaliana and plays an important role in xylem development. Although Px is predicted as vascular-specific, its activity is hard to detect and highly susceptible to plant and environmental conditions. The cauliflower mosaic virus 35S promoter (35S) is highly active in directing transgene expression. To test if 35S could enhance Px activity, while vascular specificity of the promoter is retained, we examined the expression of the uidA reporter gene, which encodes β-glucuronidase (GUS), under the control of a chimeric promoter (35S-Px) or Px by generating 35S-Px-GUS and Px-GUS constructs, which were transformed into tobacco seedlings. Both 35S-Px and Px regulated gene expression in vascular tissues. However, GUS expression driven by 35S-Px was not detected in 30- and 60-day-old plants. Quantitative real-time PCR analysis showed that GUS gene expression regulated by 35S-Px was 6.2–14.9-fold higher in vascular tissues than in leaves. Histochemical GUS staining demonstrated that 35S-Px was strongly active in the xylem and phloem. Thus, fusion of 35S and Px might considerably enhance the strength of Px and increase its vascular specificity. In addition to confirming that 35S enhances the activity of a low-level tissue-specific promoter, these findings provide information for further improving the activity of such promoters, which might be useful for engineering new types of resistant genes against vascular infections.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1379-8) contains supplementary material, which is available to authorized users.
Keywords: Plant vascular pathogens, XYLOGEN PROTEIN 1, CaMV 35S promoter, Chimeric promoters, Promoter engineering
Introduction
Vascular pathogens can infect a wide range of economic crops, may destroy vascular tissues, and spread quickly throughout the whole plant via xylem vessels, causing large-scale wilting symptoms and plant death (Mansfield et al. 2012). The control of vascular bacterial and fungal diseases mainly depends on breeding resistant varieties, which is a lengthy and continuous process as resistance may soon be lost due to the emergence of new pathovars. The discovery of resistance genes, such as antimicrobial T4 lysozyme (During 1993; Rivero et al. 2012), cecropins (Florack et al. 1995), avirulence (avr) Xa21(Wang et al. 1996), avrA (Lorang et al. 1994), and quenching quorum-sensing aiiA (Dong et al. 2002), provides an alternative method for controlling microbial vascular infections by genetic engineering. A new type of resistance genes, such as aiiA (Dong et al. 2002), is expected to play a role against pathogen infections, although these genes not necessarily present higher expression under vascular infection conditions. Identification of low-level promoters that can regulate the transgenic economic expression of resistance genes in plant vascular or xylem tissues is critical for using these new types of resistance genes against microbial vascular infections.
The promoter of XYLOGEN PROTEIN 1 (Px) is predicted to be xylem-specific, as the Arabidopsis thaliana XYLOGEN PROTEIN 1 (AtXYP1) plays an important role in xylem development (Fukuda 2004) and its homologous protein in Zinnia elegans (ZeXYP1) is located in xylem cells (Motose et al. 2004). However, it remains unknown whether Px can direct gene expression in plant vascular tissues. Bioinformatics revealed that Px contains three cis-elements involved in salicylic acid (SA) response, namely, two TCA elements (Tyagi et al. 2005) and one ocs-element, responsible for SA binding (Zhang and Singh 1994; Gómez-Ros et al. 2012). Salicylic acid is a phenolic compound that plays an important role in plant growth as well as in defense response against pathogens (Boatwright and Pajerowska-Mukhtar 2013; Campos et al. 2014). Thus, Px might be involved in vascular/xylem-specific and SA-inducible expression of resistance genes against vascular-invading microbes and be modified by genetic engineering.
However, there are few reports on SA response by vascular-specific promoters, even for those extensively investigated such as the phenylalanine ammonia-lyase (PAL) gene promoters from loblolly pine and poplar (Gray-Mitsumune et al. 1999; Osakabe and Chiang 2009), the celery coenzyme A ligase (4CL) gene promoter (Hauffe et al. 1993), and the glycine-rich protein glucuronidase (GRP1.8) gene promoter (Keller et al. 1989; Keller and Heierli 1994). Moreover, few reports have considered using vascular promoters in vascular disease protection.
A study using a Px-β-glucuronidase (GUS) gene-fusion construct indicated that Px presented a low total expression in plant organs, and suggested that its expression is more concentrated in the xylem (Kobayashi et al. 2011) compared with that of promoters with stronger GUS visible activity (Bevan et al. 1989; Hauffe et al. 1991; Osakabe and Chiang 2009; Xu et al. 2018). Although few studies have considered the improvement of tissue-specific plant promoters such as Px to avoid susceptibility associated with circumscribed tissues and lower expression, improved synthetic promoters are important tools to fine-tune gene expression in plant biotechnology to meet the challenges of modern agriculture (Liu et al. 2013).
The constitutive cauliflower mosaic virus (CaMV) 35S promoter (here termed 35S) is well-known for its high activity in directing transgene expression in all tissues (Odell et al. 1985), although resulting in some side effects (Robert et al. 2013). The 35S truncated at − 89 has been used to test specificity upstream of other promoter elements (Keller and Heierli 1994; Liu et al. 2003), and using 35S with two upstream enhancers (at − 460) increased its activity by three fold. Combining 35S with the mannopine synthase (MAS) promoter (region from + 65 to − 301) can increase its activity about sixfold in relation to that observed with enhancers (Comai et al. 1990). These results suggest that the interaction of two different promoters can increase their activity. However, there are few reports on whether using the 35S full-length promoter upstream from lower level tissue-specific promoters can enhance gene expression and maintain the tissue specificity of the promoter.
In the present study, we tested if fusing Px and 35S promoters could retain the tissue specificity of Px and enhance transgene expression.
Methods
Promoter construction and cloning
For identifying the effective promoter region of AtXYP1 (i.e., At5g64080), we analyzed the non-coding region upstream the gene using PLANTCARE (Lescot et al. 2002; Rombauts et al. 1999) and JASPAR (Sandelin et al. 2004).
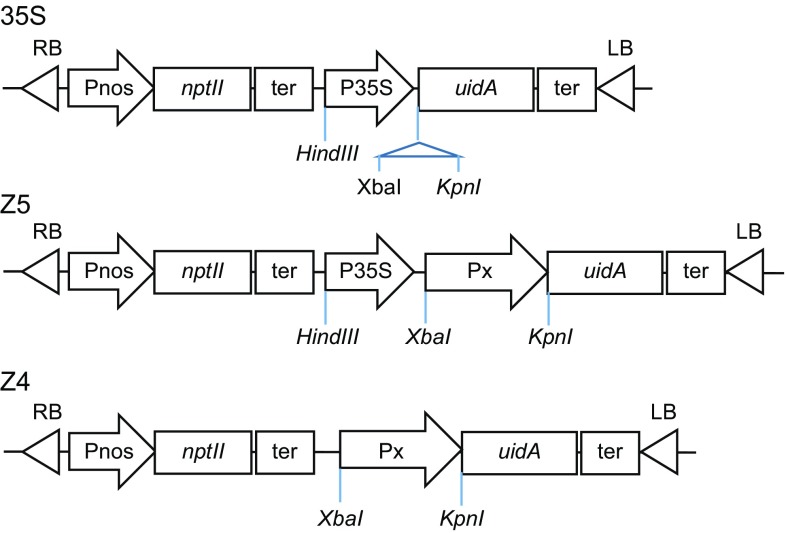
Total genomic DNA was extracted from three A. thaliana samples using the Tiangen plant DNAeasy kit (TransGen Biotech, Beijing, China). A fragment of about 2.4 kb of the Px promoter of AtXYP1 (At5g64080) was then PCR amplified using primers Px-3, 5′-GCTCTAGATTCAAGCTTAGACCAAGCCG-3′, and Px-5, 5′-CATGGTACCGCTCGAAAACGAAGGATAAACA-3′ (underlined sequences are XbaI and KpnI restriction enzyme sites, respectively) and the extracted genomic DNA samples. Promoter Px was then cloned into the plant expression vector pBI121-35S-GUS (35S) (Fig. 1) to generate the expression construct pBI121-35S-Px-GUS (designated as Z5) (Fig. 1). The fragment corresponding to promoter 35S was removed from construct Z5 using the restriction enzymes HindIII and XbaI, resulting in the single promoter construct pBI121-Px-GUS (designated Z4) (Fig. 1).
Fig. 1.
Structure of expression vectors used in this study. (35S) Construct 35S in which the GUS reporter gene uidA was under the control of 35S promoter. (Z5) Construct Z5 in which uidA was under the control of chimeric promoter 35S-Px. (Z4) Construct Z4 in which uidA was driven by Px promoter. Symbol: Px, the promoter of AtPxy1; uidA, the gene encoding β-glucuronidase (GUS); P35S, cauliflower mosaic virus 35S promoter; Pnos, the promoter of nopaline synthase; nptII, neomycin phosphotransferase gene; ter, nopaline synthase transcription terminator
The generated constructs were verified by PCR analysis using primers Px-3 and Px-5, and the Px PCR fragments were sequenced (Thermo Fisher Scientific, Shanghai, China).
Transgenic plant generation and selection
Constructs Z4, Z5, and 35S were transformed into the Agrobacterium tumefaciens strain GV3101 by the freeze and thaw method. Briefly, competent bacteria were freeze-dried in liquid nitrogen, thawed in a water bath at 37 °C (5 min), and then frozen in an ice bath (5 min). Leaf discs of Nicotianatabacum variant K326 were transformed as previously described (Horsch 1985). Transformed tobacco callus tissues and seedlings were selected based on kanamycin resistance, which was encoded by the nptII gene included in the expression cassette (Fig. 1), using a Murashige and Skoog medium containing 100 mg/L kanamycin and 400 mg/L cefotaxime. Wild-type (WT) tobacco leaf discs were used as negative controls in the selection of transgenic lines. Transgenic explants were grown at 25 °C under a 16/8 h light/dark cycle until 8 cm height and then potted and maintained in a greenhouse at 25 °C. For each construct, 13–15 independent tobacco transgenic lines were obtained and analyzed.
For verification of transgenic plants, total genomic DNA was isolated from the leaves of kanamycin-resistant tobacco lines using the Tiangen plant DNAeasy kit (TransGen Biotech, Beijing, China). The presence of Px and GUS gene uidA sequences in the transgenic plants was verified by PCR analysis with the uidA primer pairs GUS17-F (5′-TGGATCGCGAAAACTGTGGA-3′) and GUS17-R(5′-TCATTGTTTGCCTCCCTGCT-3′) and the Px primer pairs Px-5 and Px-3.
Gene expression analysis
Vascular tissues containing primary veins were isolated from the area between the first and second leaf of young stems of various transgenic lines, and total RNA was extracted using the EASY spin plus plant RNA Kit (Aidlab, Beijing, China) following the manufacturer’s instructions. Residual genomic DNA removal and cDNA synthesis were performed using the TRAN Easy Script kit (TransGen Biotech, Beijing, China) following the manufacturer’s instructions. Reverse transcription (RT)-PCR amplification of uidA was performed using the TransTaq High Fidelity (HiFi) PCR Supermix (TransGen Biotech, Beijing, China ) and the forward and reverse primers GUS90-F (5′-TGGCAGTGAAGGGCGAAC-3′) and GUS90-R (5′-CGGTCGCGAGTGAAGATCC-3′).A tubulin gene (GenBank accession number: LOC107797065) fragment of about 250 bp was amplified as a loading control using primers TU-F (5′-GACGACCAAAGCCAGTAAAG-3′) and TU-R (5′-CCGGGTGAAGGTTATCTCTA-3′).
Quantitative real-time PCR (qPCR) was performed using the Luna Universal qPCR Master Mix (New England Biolabs, Ipswich, MA, USA) and 0.5 µL of synthesized cDNA as template following the manufacturer’s protocol. Several reference genes, namely, 18S ribosomal RNA, β-actin2, elongation factor1α, ubiquitin, and tubulin, were compared in pre-experiments. Tubulin was selected as the refence gene for the qPCR because this gene is stably expressed in vascular tissues and leaves, with a similar amplification efficiency to that of uidA. The forward GUS77-F (5′-GACGACCAAAGCCAGTAAAG-3′) and the reverse GUS77-R(5′-CCGGGTGAAGGTTATCTCTA-3′) primers, specific for the uidA coding sequence, were used in the qPCR analysis. The amplified fragment was 176 bp in length. The GUS calibration curve showed a correlation coefficient (r2) of 0.995 and a − 3.302 slope, and amplification efficiency was 100.84%. The tubulin calibration curve displayed an r2 of 0.993 and a − 3.429 slope, and the amplification efficiency was 95.77%. Relative fold changes were calculated using the comparative CT method (Schmittgen and Livak 2008; Bustin et al. 2009).
Transcription expression on vascular and leaf tissues was analyzed by SPSS 14 (IBM, Armonk, NY, USA) based on paired-samples t test and one-way analysis of variance (ANOVA) followed by Duncan’s comparisons. Significance was established at P < 0.05.
Protein extraction and immunoblot analysis
Immunoblotting was performed as previously described (Janes 2015) with minor modifications. Briefly, the vascular tissues of young stems between the first and fourth leaves and their primary veins were cut and frozen in liquid nitrogen. Proteins were extracted using the RIPA lysis buffer with the Protease Inhibitor Cocktail (both Cell Signaling Technology Inc., Beijing, China) following the manufacturer’s instructions. Protein concentration was measured with the Pierce Bradford Kit (Thermo Fisher Scientific). Each protein sample (about 30 µg) was loaded and separated by electrophoresis on a 5–10% gradient sodium dodecyl sulfate polyacrylamide gel (SDS–PAGE) (Bio-Rad, Hercules, CA, USA). After completion of electrophoresis, proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad), and then incubated with the primary antibody anti-β-glucuronidase (1:1000 dilution; Sigma–Aldrich, St. Louis, MI, USA) or with anti-β-actin-HRP anti serum (1:10,000 dilution; Shanghai Kangchen Technology Co. Ltd., Shanghai, China; loading control). The PVDF membrane was then incubated with the anti-rabbit secondary antibody (1:10,000 dilution; Sigma Aldrich), followed by detection using the Super Signal West Pico Chemiluminescent substrate (Pierce, Thermo Fisher Scientific).
Quantitative GUS activity assay
For quantifying GUS activity, soluble proteins were extracted from vascular tissues (containing leaf vein and stem between the first and second node) using a buffer (pH 7.5) containing 50 Mm NaH2PO4 (pH 7.0), 10 mM EDTA, 0.1% Triton X-100, 0.1% sodium lauroyl sarcosine, and 10 mM 3-mercaptoethanol (Jefferson et al. 1987). The extracts were centrifuged at 4 °C and 10,000×g for 30 min and the resulting supernatants were filtered and condensed using an Amicon Ultra-15 30K (Merck Millipore, Burlington, MA,USA) filter to remove small molecules, which might produce auto-fluorescence. Protein concentrations were determined using the Bradford Kit (Pierce, Thermo Fisher Scientific). A fluorescent assay was performed for quantifying GUS activity from the extracted vascular protein samples using 1 mM 4-methylumbelliferyl glucuronide (Sigma Aldrich) as substrate. The enzymatic reaction was performed at 37 °C for 4 h and stopped with 900 µL ice-cold 0.2 M Na2CO3. Fluorescence was measured on a SYNERGY microplate reader (BioTek, Winooski, VT, USA) at 365 nm (excitation wavelength) and 455 nm (emission wavelength). Experiments were performed in triplicate (biological replicates) and enzyme activity was based on a standard curve (Y = 227.79X + 660.04, r2 = 0.99) of fluorescence intensity and 4-methylumbelliferone (4-MU) concentration. Data are mean enzyme activity ± standard deviation. Statistical analyses (t tests) were conducted using SPSS 14.
Histochemical GUS staining
Vein pieces (0.5 cm) were fixed in 90% ethanol for 30 min, washed 3–5 times with 100 mM phosphate buffer (pH 8.0), and then incubated at 45 °C for 8 h in a buffer containing 2 mg/mL 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (X-gluc) (Jefferson et al. 1987), 1 mM K3[Fe(CN)6], 1 mM K4[Fe(CN)]6, 20 mM EDTA, and 20% methyl alcohol. Dehydrated materials were then embedded in paraffin and cut into 10-µm slices with a rotary microtome. Slices were dewaxed, counter-stained with 0.1% safranin for 10 s, and further dehydrated and mounted in Permount TM Mounting Medium (Sanson Biotech Company, Shanghai, China). Plant tissues were then observed and photographed under a light microscope equipped with a digital camera (ZEISS, Oberkochen, Germany) at 50× and 100× magnifications. Stained xylem vessel elements were counted at 100× magnification. Data are averages of three sections from three Z4 lines.
Results and discussion
Promoter analysis and generation of transgenic tobacco lines
Because Px probably drives xylem-specific expression of xylogen protein, as evidenced by the function of protein AtXYP1 and immunoblot analysis results obtained for its homologue ZeXYP1 (Motose et al. 2004), we cloned Px and analyzed its expression before examining the enhanced vascular expression provided by the chimeric promoter 35S-Px.
The 2.2 kb fragment upstream of Px contains several core promoter elements (Fig. S1), including the putative CAAT box “GGCCAT” (− 1305 to − 1300 bp) and the TATA box “TATATAA” (− 1020 to − 1014 bp), and two element “TCACCAACC” (− 1140 to − 1132 bp and − 1601 to − 1608 bp) responsible for the vascular-specific activity in PAL and 4CL promoters (Hauffe et al. 1993; Osakabe and Chiang 2009). In addition, this region contained the elements involved in SA response, such as TCA elements (− 106 to − 99 bp, − 1127 to − 1120 bp) (Tyagi et al. 2005) and ocs-elements (− 246 to − 239 bp, − 1765 to − 1758 bp) (Zhang and Singh 1994; Gómez-Ros et al. 2012). Because the coding region of AtXYP1 may also contain regulatory elements, 177 bp of this region were included in the analysis of Px (Fig. S1).
The successful introduction of target sequences in constructs Z5 and Z4 was verified by PCR using primers specific for uidA and Px, which showed that all Z4 lines (14), 12 of the 13 Z5 lines, and all the 35S lines integrated the exogenous GUS sequence into their genomes (Fig. S2a–c). The Px sequence was also detected in the 14 Z4 lines and in 12 of the 13 Z5 lines (Fig. S2a, b).
Chimeric 35S-Px and Px promoters drive uidA expression in vascular tissues
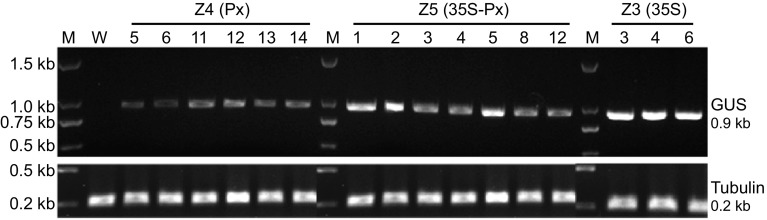
To test if uidA was expressed in the vascular tissues of transgenic lines, we extracted total RNA from their vascular tissues and used it for RT-PCR analysis. Transcripts of uidA were detected in lines Z4-5, -6, -11, -12, -13, and -14, in lines Z5-1, -2, -3, -4, -5, -8, -10, and -12, and in lines 35S-3, -4, and -6 (Fig. 2). Based on the intensity of the bands of RT-PCR products, uidA expression levels were highest in 35S transgenic lines followed by Z5 (35S-Px) and Z4 (Px) lines (Fig. 2).
Fig. 2.
Transcription analysis of β-glucuronidase (GUS) reporter gene uidA expression in the vascular tissues of transgenic tobacco plants: In lines, Z4 uidAis regulated by the promoter of Arabidopsis thaliana XYLOGEN PROTEIN 1; in lines, 35S uidA is regulated by the cauliflower mosaic virus 35S promoter; in lines, Z5 uidA is regulated by the chimeric promoter 35S-Px. M, DNA markers; W, wild type (negative control). The endogenous tubulin gene was amplified under the same conditions as a loading control
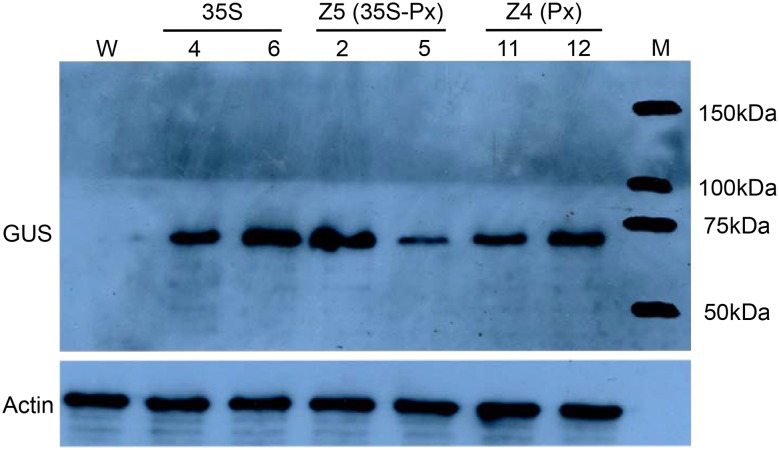
Next, we examined the expression of the protein encoded by uidA, i.e., GUS, in vascular tissues using the transgenic lines 35S-4 and -6, Z5-2 and -5, Z4-11 and -12, as these showed relatively higher levels of uidA transcription according to the RT-PCR analysis. The GUS protein was detected in the six selected transgenic lines, while WT lines showed no signal (Fig. 3). The detected bands were of the same molecular mass (68 kDa) as that of the bacterial GUS protein (Jefferson et al. 1987). GUS expression was highest in 35S transgenic lines, followed by that in Z5-2, Z4-11, Z4-12, and Z5-5, in this order, although no statistical difference was detected. In general, these results agreed with the quantitative transcriptional assay, except for the transgenic line Z5-5, whose GUS enzyme activity level was lower than expected based on the transcriptional assay (Fig. 2).
Fig. 3.
Immunoblot analysis of β-glucuronidase (GUS) expression in the vascular tissues of transgenic tobacco plants: In lines 35S, uidA is regulated by the cauliflower mosaic virus 35S promoter; in lines Z4, uidA is regulated by the promoter of Arabidopsis thaliana XYLOGEN PROTEIN 1; in lines Z5, uidA is regulated by the chimeric promoter 35S-Px.M, protein markers; W, wild type tobacco (negative control). The endogenous actin protein was detected using an actin-specific antibody and used as a loading control
According to the results of RT-PCR and immunoblotting, uidA was expressed in the vascular tissues of the tested Z5 and Z4 transgenic lines, suggesting that both 35S-Px and Px drive gene expression in vascular tissues.
The chimeric 35S-Px promoter expands Px vascular specificity
Promoter activity and specificity in transgenic tobacco were assessed based on the activity of GUS. Young transgenic tobacco plants (30 and 60-old) were stained using the GUS substrate X-gluc; WT plants were used as negative controls. Clear GUS activity was detected in all the 35S-transformed lines, mostly in leaves, but it was hardly visible in all vegetative organs of transgenic lines with high GUS transcription expression (i.e., Z5-1, -2, and -5; Z4-11, -12, and -14; Fig. 4).
Fig. 4.
Histochemical activity of β-glucuronidase (GUS) in transgenic plants: a–d 60-day-old transgenic tobacco plants belonging to lines Z4, Z5, and 35S, and wild-type (WT), respectively; e–k 30-day-old transgenic tobacco plants belonging to lines Z4, Z5, and 35S, and WT, respectively. In lines Z4, uidA is regulated by the promoter of Arabidopsis thaliana XYLOGEN PROTEIN 1; in lines 35S, uidA is regulated by the cauliflower mosaic virus 35S promoter; in lines Z5, uidA is regulated by the chimeric promoter 35S-Px. All the transgenic lines listed in Fig. 3 were stained and their representatives are shown here. Plants were stained in a buffer containing GUS substrate X-gluc (2 mg/mL) at 37 °C for 8 h
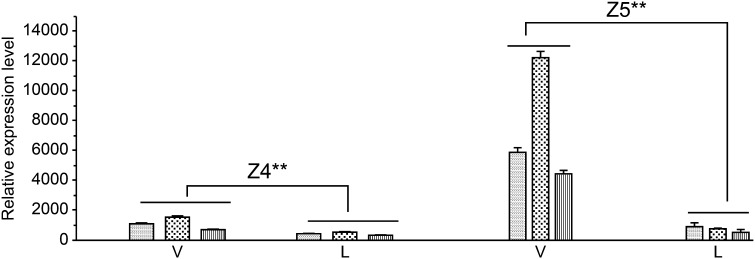
Because uidA expression levels were relatively low in Z5 and Z4 lines and it was difficult to compare promoter activity among plant organs based on GUS staining, we performed a qPCR to analyze the expression pattern of uidA under the control of promoters 35S-Px and Px based on the total RNA extracted from the leaves (the primary vein was removed) of Z4 and Z5 transgenic lines. As shown in Fig. 5, the relative expression of uidA in Z5 and Z4 seedlings was 6.2–14.9-fold (P = 0.001) and 2.2–2.8-fold (P = 0.001) higher in vascular tissues than in leaf tissues, respectively. Because the leaf tissues used in this assay contained secondary and minor veins, which were difficult to separate and remove, the basal expression level of uidA in leaf tissues might, at least partially, be influenced by these minor veins. The above results suggest that uidA expression driven by the chimeric promoter 35S-Px and by Px is more concentrated in internal vascular tissues (e.g., xylem) than in other tissues.
Fig. 5.
Quantitative PCR analysis of β-glucuronidase (GUS) transcriptional expression in vascular and leaf tissues: In lines Z4, uidA is regulated by the promoter of Arabidopsis thaliana XYLOGEN PROTEIN 1; In lines Z5, uidA is regulated by the chimeric promoter 35S-Px. WT, wild-type tobacco; V, vascular tissues of 50- to 60-day-old plants; L, leaves with main vein removed. Experiments were performed in triplicate (biological replicates) and data represent mean relative expression ± standard deviation. Transcription expression between vascular and leaf tissues (paired-samples t-test): For Z4 and Z5lines, *** P = 0.001
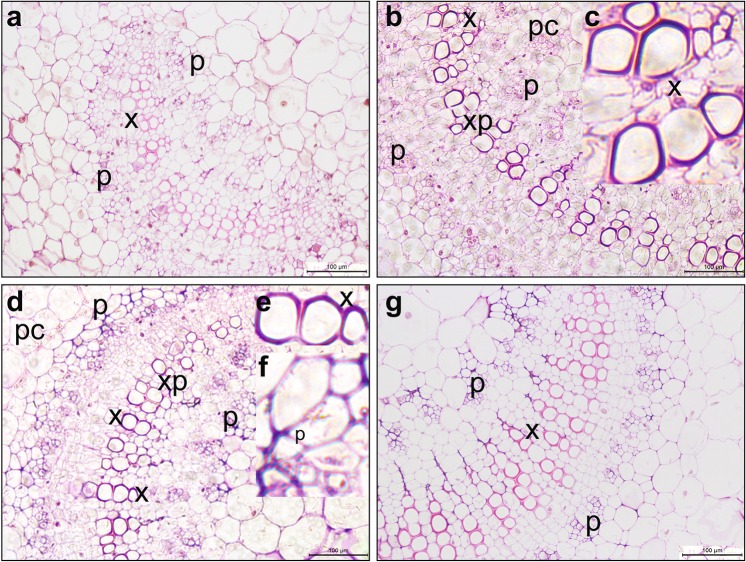
To examine the specific vascular location of the expressed GUS enzyme driven by 35S-Px and Px, X-gluc–stained cross-sections of the vascular tissues of the second young leaves from transgenic Z5-1, -2, and -5 and Z4-11, -12, and-14 seedlings were observed under the microscope. Although GUS activity was observed in the vascular tissues of all transgenic lines tested (Fig. 6), Px showed xylem-specific expression. While no obvious blue color was observed in WT tissues (Fig. 6a), blue staining was observed in some xylem vessel elements (Fig. 6b, c). Weaker GUS staining was observed in phloem (p), parenchymal cells (pc), and cortex of Z4 leaves, also indicating that Px is xylem-specific. Enhanced GUS expression driven by 35S-Px was clearly observed in xylem vessel elements and phloem, but it was weaker in parenchyma cells and cortex (Fig. 6d, e, f). Overall, our results showed that, in Z5 lines, the 35S-Px promoter displayed stronger activity in phloem cells than the single Px promoter, while maintaining vascular specificity.
Fig. 6.
Histochemical staining of β-glucuronidase (GUS) and microscopic observations. a non-transgenic tobacco; b transgenic line Z4-12, uidA is regulated by the promoter of Arabidopsis thaliana XYLOGEN PROTEIN 1; c enlargement of the xylem in (b) showing GUS enzyme activity in xylem elements; d transgenic line Z5-2, uidA is regulated by the chimeric promoter 35S-Px; e enlargement of the xylem in (d) showing GUS enzyme activity in xylem elements; f enlargement of the external phloem in (d) showing GUS enzyme activity in phloem cells; g transgenic line 35S-6, uidA is regulated by the cauliflower mosaic virus 35S promoter. Tobacco vein sections were stained with X-gluc. Scale bar = 100. x xylem; p phloem; M meristem; xp xylem ray parenchymal cell; pc parenchymal cell
In contrast, GUS staining was extremely variable in 35S lines, as it was observed in all (Fig. 6g) or in part of the vascular tissues. Thus, the GUS expression driven by 35S was random even among lines.
Chimeric 35S-Px enhances vascular expression level
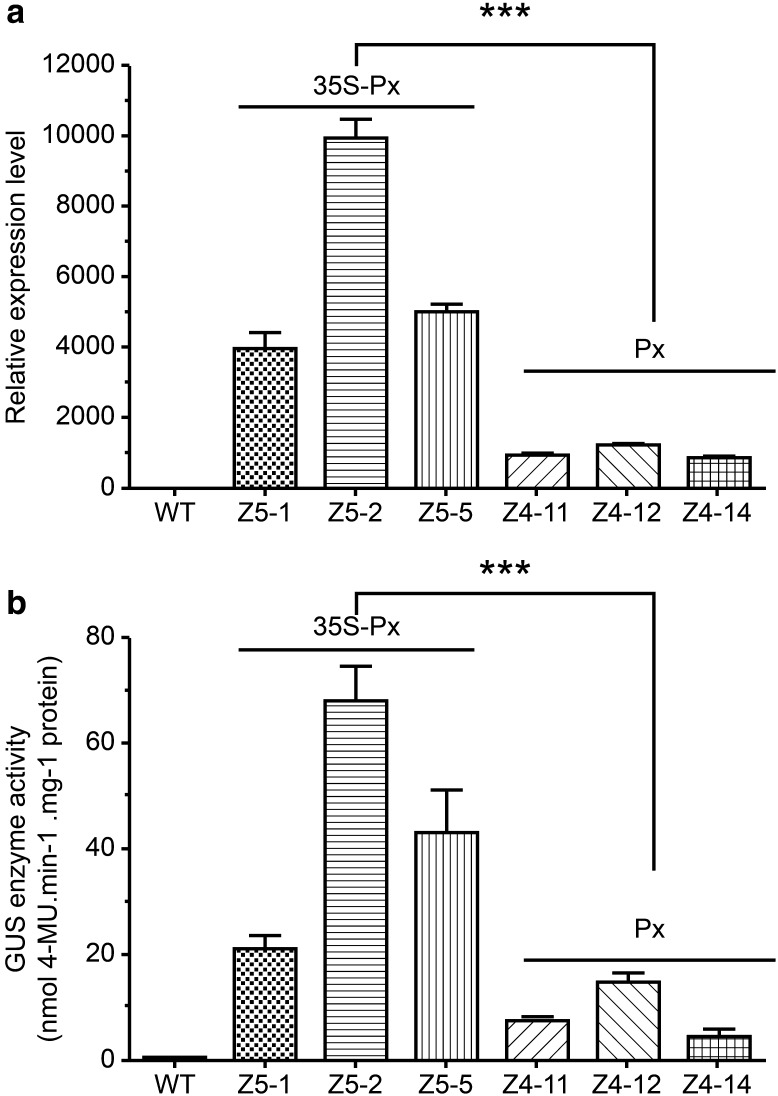
We tested the relative enhancement provided by 35S-Px in relation to Px using RT-PCR. Results showed that, while the transcriptional expression levels of the reporter gene uidA in the vascular tissues of the three Z4 lines were similar, variations in uidA transcription were obvious among the three Z5 transgenic lines: it was significantly higher in Z5-2 (P = 0.000) than in Z5-1 and Z5-5 lines, and did not differ between these two lines (P = 0.11) (Fig. 7a); this trend is similar to that found in the immunoblot analysis (Fig. 3). Notably, the uidA expression levels in the vascular tissues of the three Z5 transgenic lines (35S-Px) were significantly higher (3.97–10.05-fold) than in the Z4 transgenic lines (Px) (P = 0.000) (Fig. 5a).
Fig. 7.
Strength of promoters Px and 35S-Px. a Quantitative PCR analysis of β-glucuronidase (GUS) transcriptional expression. Total RNA was extracted from the apical vascular tissues of 50–60-day-old plants. Relative transcription was calculated using the comparative CT method (Schmittgen and Livak 2008; Bustin et al. 2009). ***P = 0.000 (Duncan’s test). b Quantitative enzymatic assay of GUS activity in transgenic tobacco lines. The apical vascular tissues of 50–60-day-old plants were also used for soluble protein extraction and measurement of GUS activity. The reaction was carried out at 37 °C for 4 h, and 0.43 mg soluble protein was obtained from each sample. The experiment was repeated at least three times (biological replicates). Bars and whiskers represent mean GUS activity ± standard deviation (SD). ***P = 0.000 (Duncan’s test)
To verify the above findings, we conducted a quantitative enzymatic assay to determine GUS activity in the transgenic lines using 4-MU as the reaction substrate (Jefferson et al. 1987). Generally, the results agreed with that of the quantitative transcriptional assay, except for Z5-1, as its GUS enzyme activity level was lower than expected based on the transcriptional assay. Enzyme activities in the other two Z5 lines were about 3.06–9.92-fold higher (P = 0.000) than in Z4 transgenic lines (Fig. 7b).
Conclusions
The present study identified and characterized the chimeric 35S-Px promoter and the Px promoter of the gene encoding AtXYP1 based on bioinformatics and transgenic tobacco plants generated using 35S-Px-uidA and Px-uidA reporter gene constructs. Results showed that 35S-Px enhanced the expression of uidA in vascular tissues compared to that displayed under the control by Px. Histochemical staining of GUS activity of vein sections from Z5 (35S-Px) transgenic plants evidenced that GUS was mainly active in the xylem and phloem of vascular tissues. This expression pattern agrees with the function reported for the AtXYP1 protein, which is responsible for direct xylogen accumulation in xylem-differentiated elements (Fukuda 2004; Motose et al. 2004). The low expression level of the reporter gene driven by Px is similar to the expression pattern of XYP1 in A. thaliana (Kobayashi et al. 2011), thereby suggesting that the Px identified in the present study is an intact and functional promoter maintaining its original characteristics and strength. Our results also showed that the tandem 35S-Px promoter displayed enhanced phloem specificity compared to the xylem expression of the Px promoter. This agrees with the 35S domain (− 89 to − 460) containing strong vascular-specific elements (Benfey et al. 1989, 1990). The strength of 35S-Px is substantially higher than that of Px. Thus, placing the full-length 35S promoter upstream the xylem-specific promoter Px enhances its strength and expands its vascular specificity.
In summary, the fusion of the intact 35S promoter to the Px promoter can expand the vascular specificity of Px and substantially improve its strength. According to previous and present results, the enhancement provided by the tandem promoter is closely related to promoters’ activity, length, and interaction (Benfey et al. 1990; Comai et al. 1990; Liu et al. 2003).The strength of Px might be further improved by removing certain repressor elements or cis-elements as previously reported (Hauffe et al. 1993; Keller and Heierli 1994). Alternatively, addition of the 35S might enhance the strength of Px in vascular tissues (Benfey et al. 1990), although further verification is necessary, given that the expression of the chimeric 35S-Px promoter was still significantly lower than that of the 35S promoter and easily affected by plant vigor and environmental factors.
Details on SA regulation by Px remain unclear, although our preliminary data indicated that Px-GUS expression was SA-inducible. However, more specifically designed deletion assays need to be performed before reaching a solid conclusion. The potential of Px as a plant resistance gene expression tool should be verified in a challenge assay of transgenic lines using vascular pathogens such as Ralstonia solanacearum, Xanthomona campestris pv. campestris, or Erwinia carotovora ssp. carotovora. Overall, we identified and validated a useful and vascular-specific chimeric plant promoter (35S-Px) that might be used in further transgenic engineering procedures for optimizing crop resistance.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1: Figure S1 DNA sequence of the AtXYP1 promoter (Px). The promoter sequence contains part of AtXYP1 coding region and its 5′region. Symbol: +1, the first codon of AtXYP1 coding region. Important cis-elements were identified and labeled: (1) CAAT box, core promoter element; (2) TATA box, core promoter element; (3) AC elements, AC-rich region responsible for vascular-specific activity; (4) TCA element, element involved in salicylic acid response; (5) Ocs-element, element involved in auxin and salicylic acid response. (TIF 4874 KB)
Supplementary material 2: Figure S2 PCR analysis of the reporter sequences with T1 generation of transgenic tobacco. a, Detection of GUS gene (upper panel, 1.7 kb) and Px sequence (lower panel, 2.4 kb) by PCR amplification in Z4 (Px-GUS) transgenic lines (lanes 1–14). b, Detection of GUS gene (upper panel) and Px sequence (lower panel) by PCR amplification in Z5 (35S-Px-GUS) transgenic lines (lanes 1–13). c, Detection of GUS gene by PCR amplification in 35S (35S-GUS) transgenic lines (lanes 1–15). Symbols: M, DNA size markers; W, wild type tobacco as a negative control; P, Z5 plasmid as a positive control. (EPS 4748 KB)
Acknowledgments
The study was financed by the National Basic Research Program of China 973 (Grant No. 2015CB150600).
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Contributor Information
Yu-mei Chen, Email: 508376557@qq.com.
Yi-hu Dong, Email: dongyh@imcb.a-star.edu.sg.
Zhi-bin Liang, Email: 798693704@qq.com.
Lian-hui Zhang, Phone: +86-13570466460, Email: Lianhui01@scau.edu.cn.
Yi-zhen Deng, Phone: +86-18814113691, Email: 18cyz05@sina.com.
References
- Benfey PN, Ren L, Chua NH. The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J. 1989;8(8):2195–2202. doi: 10.1002/j.1460-2075.1989.tb08342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Ren L, Chua NH. Combinatorial and synergistic properties of CaMV 35S enhancer subdomains. EMBO J. 1990;9(6):1685–1696. doi: 10.1002/j.1460-2075.1990.tb08292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M, Shuffle bottom D, Edwards K, et al. Tissue- and cell- specific activity of a phenylalanine ammonia-lyase promoter in transgenic plants. EMBO J. 1989;8(7):1899–1906. doi: 10.1002/j.1460-2075.1989.tb03592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatwright JL, Pajerowska-Mukhtar K. Salicylic acid: an old hormone up to new tricks. Mol Plant Pathol. 2013;14(6):623–634. doi: 10.1111/mpp.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Campos L, Granell P, Tárraga S, et al. Salicylic acid and gentisic acid induce RNA silencing-related genes and plant resistance to RNA pathogens. Plant Physiol Biochem. 2014;77:35–43. doi: 10.1016/j.plaphy.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Comai L, Moran P, Maslyar D. Novel and useful properties of a chimeric plant promoter combining CaMV 35S and MAS elements. Plant Mol Biol. 1990;15(3):373–381. doi: 10.1007/BF00019155. [DOI] [PubMed] [Google Scholar]
- Dong Y, Wang L, Xu J. Quenching quorum-sensing dependent bacterial infection by an N-acyl homoserine lactonase. Nature. 2002;411:813–816. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- During K. Can lysozymes mediate antibacterial resistance in plants? Plant Mol Biol. 1993;23(1):209–214. doi: 10.1007/BF00021432. [DOI] [PubMed] [Google Scholar]
- Florack D, Allefs S, Bollen R, et al. Expression of giant silk moth cecropin B genes in tobacco. Transgenic Res. 1995;4(2):132–141. doi: 10.1007/BF01969415. [DOI] [PubMed] [Google Scholar]
- Fukuda H. Plant cell biology: Signals that control plant vascular cell differentiation. Nature Rev Mol Cell Biol. 2004;5(5):379–391. doi: 10.1038/nrm1364. [DOI] [PubMed] [Google Scholar]
- Gómez-Ros LV, Gabaldón C, López Núñez-Flores MJ, et al. The promoter region of the Zinnia elegans basic peroxidase isoenzyme gene contains cis-elements responsive to nitric oxide and hydrogen peroxide. Planta. 2012;236(2):327–342. doi: 10.1007/s00425-012-1604-3. [DOI] [PubMed] [Google Scholar]
- Gray-Mitsumune M, Molitor EK, Cukovic D, et al. Developmentally regulated patterns of expression directed by poplar PAL promoters in transgenic tobacco and poplar. Plant Mol Biol. 1999;39(4):657–669. doi: 10.1023/A:1006148715050. [DOI] [PubMed] [Google Scholar]
- Hauffe KD, Paszkowski U, Schulze-Lefert P, et al. A parsley 4CL-1 promoter fragment specifies complex expression patterns in transgenic tobacco. Plant Cell. 1991;3(5):435–443. doi: 10.1105/tpc.3.5.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauffe K, Lee SP, Subramaniam R. Combinatorial interactions between positive and negative cis-acting elements control spatial patterns of 4CL-1 expression in transgenic tobacco. Plant J. 1993;4(2):235–253. doi: 10.1046/j.1365-313X.1993.04020235.x. [DOI] [PubMed] [Google Scholar]
- Horsch R. A simple and general method for transferring genes into plants. Science. 1985;227(4691):1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Janes KA. An analysis of critical factors for quantitative immunoblotting. Sci Signal. 2015;8(371):rs2. doi: 10.1126/scisignal.2005966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller B, Heierli D. Vascular expression of the grp1.8 promoter is controlled by three specific regulatory elements and one unspecific activating sequence. Plant MolBiol. 1994;26:747–756. doi: 10.1007/BF00013759. [DOI] [PubMed] [Google Scholar]
- Keller B, Schmid J, Lamb CJ. Vascular expression of a bean cell wall glycine-rich protein–β-glucuronidase gene fusion in transgenic tobacco. EMBO J. 1989;8(5):1309–1314. doi: 10.1002/j.1460-2075.1989.tb03510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Motose H, Iwamoto K, et al. Expression and genome-wide analysis of the xylogen-type gene family. Plant Cell Physiol. 2011;52(6):1095–1106. doi: 10.1093/pcp/pcr060. [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, et al. Plant CARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZZ, Wang JL, Huang X, et al. The promoter of a rice glycine-rich protein gene, Osgrp-2, confers vascular-specific expression in transgenic plants. Planta. 2003;216(5):824–833. doi: 10.1007/s00425-002-0934-y. [DOI] [PubMed] [Google Scholar]
- Liu W, Yuan JS, Stewart CJ. Advanced genetic tools for plant biotechnology. Nat Rev Genet. 2013;14(11):781–793. doi: 10.1038/nrg3583. [DOI] [PubMed] [Google Scholar]
- Lorang JM, Shen H, Kobayashi D, et al. avrA and avrE in Pseudomonas syringaepv. tomato PT23 play a role in virulence on tomato plants. Mol Plant-Microbe Interact. 1994;7(4):508–515. doi: 10.1094/MPMI-7-0508. [DOI] [Google Scholar]
- Mansfield J, Genin S, Magori S, et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 2012;13(6):614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motose H, Sugiyama M, Fukuda H. A proteoglycan mediates inductive interaction during plant vascular development. Nature. 2004;429(6994):873–878. doi: 10.1038/nature02613. [DOI] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua NH. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature. 1985;313(6005):810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Chiang K. Isolation of 4-coumarate co-A ligase gene promoter from Loblolly pine (Pinus taeda) and characterization of tissue-specific activity in transgenic tobacco. Plant PhysiolBiochem. 2009;11(47):1031–1036. doi: 10.1016/j.plaphy.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Rivero M, Furman N, Mencacci N. Stacking of antimicrobial genes in potato transgenic plants confers increased resistance to bacterial and fungal pathogens. J Biotechnol. 2012;157(2):334–343. doi: 10.1016/j.jbiotec.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Robert CA, Erb M, Hitpold I, et al. Genetically engineered maize plants reveal distinct costs and benefits of constitutive volatile emissions in the field. Plant Biotechnol J. 2013;11(5):628–639. doi: 10.1111/pbi.12053. [DOI] [PubMed] [Google Scholar]
- Rombauts S, Déhais P, Van Montagu M, Rouzé P. Plant CARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999;27(1):295–296. doi: 10.1093/nar/27.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32(Database issue):91–94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Tyagi W, Rajagopal D, Singla-Pareek SL, et al. Cloning and regulation of a stress-regulated Pennisetum glaucum vacuolar ATPase c gene and characterization of its promoter that is expressed in shoot hairs and floral organs. Plant Cell Physiol. 2005;46(8):1411–1422. doi: 10.1093/pcp/pci154. [DOI] [PubMed] [Google Scholar]
- Wang GL, Song WY, Ruan DL, et al. The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv. oryzae isolates in transgenic plants. Mol Plant Microbe Interact. 1996;9(9):850–855. doi: 10.1094/MPMI-9-0850. [DOI] [PubMed] [Google Scholar]
- Xu W, Liu W, Ye R, et al. A profilin gene promoter from switchgrass (Panicum virgatum L.) directs strong and specific transgene expression to vascular bundles in rice. Plant Cell Rep. 2018;37(4):587–597. doi: 10.1007/s00299-018-2253-1. [DOI] [PubMed] [Google Scholar]
- Zhang B, Singh KB. ocs element promoter sequences are activated by auxin and salicylic acid in Arabidopsis. Proc Natl Acad Sci USA. 1994;91(7):2507–2511. doi: 10.1073/pnas.91.7.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1: Figure S1 DNA sequence of the AtXYP1 promoter (Px). The promoter sequence contains part of AtXYP1 coding region and its 5′region. Symbol: +1, the first codon of AtXYP1 coding region. Important cis-elements were identified and labeled: (1) CAAT box, core promoter element; (2) TATA box, core promoter element; (3) AC elements, AC-rich region responsible for vascular-specific activity; (4) TCA element, element involved in salicylic acid response; (5) Ocs-element, element involved in auxin and salicylic acid response. (TIF 4874 KB)
Supplementary material 2: Figure S2 PCR analysis of the reporter sequences with T1 generation of transgenic tobacco. a, Detection of GUS gene (upper panel, 1.7 kb) and Px sequence (lower panel, 2.4 kb) by PCR amplification in Z4 (Px-GUS) transgenic lines (lanes 1–14). b, Detection of GUS gene (upper panel) and Px sequence (lower panel) by PCR amplification in Z5 (35S-Px-GUS) transgenic lines (lanes 1–13). c, Detection of GUS gene by PCR amplification in 35S (35S-GUS) transgenic lines (lanes 1–15). Symbols: M, DNA size markers; W, wild type tobacco as a negative control; P, Z5 plasmid as a positive control. (EPS 4748 KB)