Abstract
Grain traits are important agronomic attributes with the market value as well as milling yield of bread wheat. In the present study, quantitative trait loci (QTL) regulating grain traits in wheat were identified. Data for grain area size (GAS), grain width (GWid), factor form density (FFD), grain length-width ratio (GLWR), thousand grain weight (TGW), grain perimeter length (GPL) and grain length (GL) were recorded on a recombinant inbred line derived from the cross of NW1014 × HUW468 at Meerut and Varanasi locations. A linkage map of 55 simple sequence repeat markers for 8 wheat chromosomes was used for QTL analysis by Composite interval mapping. Eighteen QTLs distributed on 8 chromosomes were identified for seven grain traits. Of these, five QTLs for GLWR were found on chromosomes 1A, 6A, 2B, and 7B, three QTLs for GPL were located on chromosomes 4A, 5A and 7B and three QTLs for GAS were mapped on 5D and 7D. Two QTLs were identified on chromosomes 4A and 5A for GL and two QTLs for GWid were identified on chromosomes 7D and 6A. Similarly, two QTLs for FFD were found on chromosomes 1A and 5D. A solitary QTL for TGW was identified on chromosome 2B. For several traits, QTLs were also co-localized on chromosomes 2B, 4A, 5A, 6A, 5D, 7B and 7D. The QTLs detected in the present study may be validated for specific crosses and then used for marker-assisted selection to improve grain quality in bread wheat.
Electronic supplementary material
The online version of this article (10.1007/s12298-018-0552-1) contains supplementary material, which is available to authorized users.
Keywords: Quantitative trait loci, Grain traits, Molecular markers, Linkage map
Introduction
All over the world, bread wheat (Triticum aestivum L.) is one of the major staple crop and it provides ~ 20% calories in human diet (http://faostat.fao.org). A primary objective of all major wheat breeding programs is to enhance grain yield in order to ensure global food security. Grain yield has the following three major components: grain number per spike, spike number per unit area and 1000-grain weight (TGW). TGW is positively correlated with grain size (Wu et al. 2016) and is an important trait due to its phenotypic stability and high heritability (Kuchel et al. 2007). Selection for higher TGW has been carried out during domestication process, and therefore it has been argued that in cereals, grain size is a component of domestication syndrome (Fuller 2007; Brown et al. 2009). Despite polygenic control of TGW, phenotypic selection for this trait is effective (Sidwell et al. 1976). However, phenotypic selection is laborious and time consuming. Therefore, the use of molecular markers for indirect marker-assisted selection (MAS) should be a convenient alternative to phenotypic selection. TGW is mainly determined by grain length (GL), grain width (GWid) and grain thickness (GT) (Campbell et al. 1999; Dholakia et al. 2003; Breseghello and Sorrells 2006a, b; Sun et al. 2009). Larger grains generally have favorable effect on TGW, seedling vigor and flour yield characteristics (Chastain et al. 1995). In the past, more than 100 QTLs for TGW, GL and GWid in wheat have been identified, which are distributed on all the 21 chromosomes of wheat (Varshney et al. 2000; Börner et al. 2002; Groos et al. 2003; Huang et al. 2003, 2004, 2006; Narasimhamoorthy et al. 2006; Breseghello and Sorrells 2006a, b, 2007; Kumar et al. 2006; Sun et al. 2009; Ramya et al. 2010; Mir et al. 2012; Williams et al. 2013; Okamoto et al. 2013; Patil et al. 2013; Tyagi et al. 2014; Simmonds et al. 2014; Zhang et al. 2015). However, many of the QTLs for TGW are not detectable across environments, explain only a small proportion of the phenotypic variation for grain weight, and also exhibit high QTL × genotype and QTL × QTL epistatic interactions (Campbell et al. 2003; Gupta et al. 2007; Prashant et al. 2012; Patil et al. 2013; Cabral et al. 2018). Together these factors, make such QTL unsuitable for use in MAS for wheat breeding. Also, QTLs for other important grain traits such as grain length-width ratio (GLWR), grain width (GWid), grain area size (GAS), grain perimeter length (GPL) and factor form density (FFD) have been rarely reported (Giura and Saulescu 1996; Dholakia et al. 2003; Sun et al. 2009; Okamoto et al. 2013; Li et al. 2015). Therefore, the present study was designed to conduct QTL analysis for seven important grain traits (TGW, GL, GLWR, GWid, GAS, GPL and FFD) using a RIL mapping population derived from the cross NW1014 × HUW468, so as to identify important QTL and associated markers for improvement of grain traits using MAS during wheat breeding.
Materials and methods
Plant material
A set of 106 RILs derived from the cross NW1014 × HUW468 was used for QTL analysis of seven grain traits in the present study. The seed for the mapping population was procured from Dr. Arun Joshi and colleagues, who developed the mapping population at the Institute of Agricultural Sciences, Banaras Hindu University, Varanasi, India.
SSR markers
For construction of genetic maps, a set of 376 SSRs (gwm, wmc, swm, barc and cfd) involving only 8 wheat chromosomes (1A, 2B, 4A, 5A, 5D, 6A, 7B and 7D) was tried during the present study, because only these 8 chromosomes are shown to be associated with QTLs for grain traits repeatedly (Dholakia et al. 2003; Groos et al. 2003; Huang et al. 2006; Kumar et al. 2006; Wang et al. 2009; Gegas et al. 2010; Ramya et al. 2010; Mir et al. 2012; Tyagi et al. 2014; Zhang et al. 2015; Li et al. 2015). Out of a total of 376 SSR markers, 55 SSR markers that were polymorphic between the parental genotypes of the RIL mapping population were used for preparation of the molecular maps. Out of the 55 polymorphic SSR markers, primer sequences of the 38 SSRs are given in Supplementary Table 1. The aliquots for the remaining 17 polymorphic gwm SSRs were kindly provided by Dr. M.S Röder, Leibniz-Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben, Correnstrasse-3, Germany and their primer sequences are proprietary and hence are not listed in Supplementary Table 1. The primer aliquots for the remaining SSRs were synthesized by Integrated DNA Technologies, (IDT), USA.
Table 1.
Descriptive statistics of the parental genotypes and RILs for different grain traits in two environments
| Trait | Environment | Parents | RILs | ||||
|---|---|---|---|---|---|---|---|
| NW1014 | HUW468 | Range | Average | SD | CV% | ||
| Grain area size | E1 | 12.43 | 14.09 | 8.30–13.80 | 10.85 | 1.22 | 11.20 |
| (GAS) (mm2) | E2 | 11.90 | 14.20 | 4.10–13.30 | 9.93 | 1.24 | 12.49 |
| Grain perimeter length | E1 | 14.97 | 16.08 | 12.30–16.30 | 14.33 | 0.81 | 5.62 |
| (GPL) (mm) | E2 | 14.50 | 16.50 | 6.80–15.30 | 13.70 | 0.98 | 7.19 |
| Grain length | E1 | 6.01 | 6.19 | 4.80–6.40 | 5.67 | 0.35 | 6.23 |
| (GL) (mm) | E2 | 5.70 | 6.00 | 2.60–6.00 | 5.36 | 0.41 | 7.57 |
| Grain width | E1 | 2.71 | 3.02 | 2.10–3.00 | 2.49 | 0.19 | 7.47 |
| (GWid) (mm) | E2 | 2.80 | 2.90 | 1.20–2.90 | 2.37 | 0.22 | 9.39 |
| Grain length–width | E1 | 2.23 | 2.06 | 1.90–3.20 | 2.30 | 0.18 | 7.77 |
| Ratio (GLWR) | E2 | 2.10 | 2.00 | 2.00–2.30 | 2.01 | 0.04 | 2.03 |
| Factor form density | E1 | 2.70 | 2.95 | 1.30–4.10 | 2.44 | 0.52 | 21.43 |
| (FFD) | E2 | 2.50 | 2.90 | 1.30–10.40 | 2.40 | 1.07 | 44.43 |
| 1000-grain weight | E1 | 44.00 | 55.33 | 16.00–51.00 | 34.03 | 5.67 | 16.67 |
| (TGW) (g) | E2 | 43.50 | 54.30 | 14.30–65.60 | 29.80 | 9.49 | 31.85 |
E1 Meerut, E2 Varanasi, SD standard deviation, CV coefficient of variance
Field experiment and recording of data (including genotyping)
RILs of the mapping population (NW1014 × HUW468) and two parental genotypes were evaluated in augmented block design experiments during the crop season 2013–2014 at two locations, namely Ch. Charan Singh University, Meerut located at 28.9664°N and 77.8367°E latitude and longitude and BHU, Varanasi located at 25.3176°N and 82.9739°E latitude and longitude. Standard agronomic practices were followed for raising the crop with application of 120 kg N/ha and the irrigation as per the requirements of the crop.
The data of grain traits were collected on 50 grains each of 106 RILs and the parental genotypes using Smart Grain software ver. 1.2 (Tanabata et al. 2012). Smart Grain uses image to determine grain shape. It automatically recognizes all grains within a digital image, detects outlines, and then calculates all the grain size parameters including grain area size (GAS), grain width (GWid), grain length-width ratio (GLWR), grain perimeter length (GPL) and grain length (GL). TGW in grams (g) was determined by weighing 1000 grains of each of the 106 RILs and the two parental genotypes. The data on grain traits were also utilized for the calculation of factor form density (FFD), , which describes the grain density differences and the deviation of the shape from a cylindrical form (Giura and Saulescu 1996).
SSRs were used for genotyping of the mapping population and the two parental genotypes. For this purpose, SSRs were amplified using PCR and the amplified products were resolved on PAGE. The PCR reaction mixture composition and profile of PCR for amplification of SSRs were the same as given by Röder et al. (1998).
Construction of linkage map
As mentioned above, only 8 wheat chromosomes were used for construction of map and QTL analyses. The linkage maps for individual chromosomes were prepared by MAPMAKER/EXP v3.0b (Lander et al. 1987). The ‘Try’ and ‘Compare’ commands were used to anchor additional markers onto the maps.
QTL analysis
QTL Cartographer v 2.5 was used for performing composite interval mapping i.e. CIM (Wang et al. 2005). Threshold LOD was calculated experiment wise on the basis of 1000 permutations (Churchill and Doerge 1994; Doerge and Churchill 1996). QTL were detected for individual environments and also jointly for both the environments. QTL detected in both the environments were considered as stable, and the QTL explaining > 20% PV were considered as major QTL.
Statistical analysis
Values of means, standard deviation, correlation coefficients and box plots showing the distribution of phenotypic data for different traits were determined using SPSS version 20.
Results and discussion
To examine the genetic architecture of the grain weight in wheat, a number of studies involving QTL analyses have been conducted in the past. However, only limited studies involving QTL mapping of other grain traits such as grain area size (GAS), grain width (GWid), grain length-width ratio (GLWR), thousand grain weight (TGW), grain perimeter length (GPL), grain length (GL) and factor form density (FFD) have been conducted (Campbell et al.1999; Ramya et al. 2010; Gegas et al. 2010; Prashant et al. 2012; Williams et al. 2013; Okamoto et al. 2013; Williams and Sorrells 2014; Tyagi et al. 2014; Rasheed et al. 2014; Zhang et al. 2015; Wu et al. 2016). In view of this, the present study involving QTL analyses for seven grain traits in wheat was undertaken. The results pertaining to the phenotypic variation in grain traits in parental genotypes and the RIL population, correlations among different traits in the RIL population and QTL analyses are presented and discussed.
Phenotypic variation in grain traits
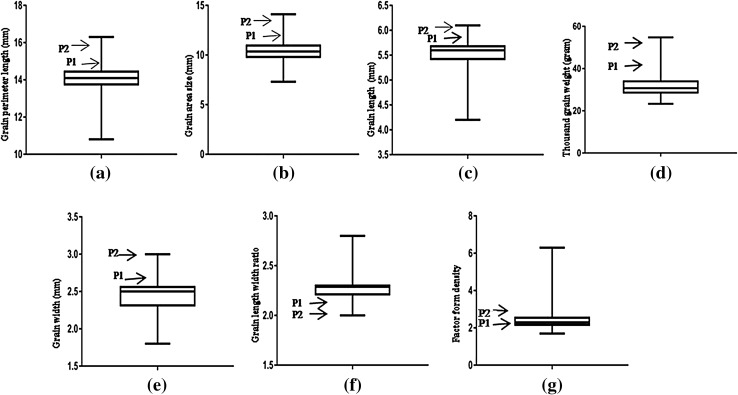
The parental genotype HUW468 of the mapping population showed consistently higher mean values for all the grain traits in both the environments except for the GLWR, which was higher in the parental genotype NW1014. The data for different traits in the RIL populations showed a wide range and continuous distributions for each of the seven traits (Fig. 1). This suggested that each of the seven traits is controlled by multiple loci. The range of distribution of data in the first or the fourth quartile of the box plots was much wider for most of the traits, suggesting skewed distribution of the data. Transgressive segregation for most of the traits was observed, where mean values of individual RILs transgressed the mean values of either one or both the parental genotypes (Table 1; Fig. 1). Relatively more variation was witnessed for the following three traits: GAS, FFD and TGW.
Fig. 1.
Box plots showing the distribution of data of the seven grain traits for 106 RILs derived from the cross NW1014 × HUW468. Arrows labelled as P1 (NW1014) and P2 (HUW468) represent the mean values of the parental genotypes; a grain perimeter length; b grain area size; c grain length; d thousand grain weight; e grain width; f grain length-width ratio; g factor form density
Correlations among traits
Details of the correlation coefficient (r) values involving seven grain traits individually at Meerut and Varanasi and also for the pooled data of the two environments are presented in Table 2. Most pairs of traits exhibited significant correlations (negative in some cases) in the individual environments as well as in the pooled data. The correlation of TGW with other traits was inconsistent over the two environments. In the pooled analyses, TGW had positive and significant association with only FFD. Significant positive correlation among GAS, GPL, GL and GWid indicate that these traits together contribute to grain size as reported earlier (Okamoto et al. 2013). However, correlation between these four traits and GLWR were not significant with TGW (except for few weak associations). This suggested that the above five traits and the TGW are each controlled by different sets of loci. This is in agreement with previous reports indicating that genetic control of these traits is largely independent (Gegas et al. 2010; Kumar et al. 2016).
Table 2.
Correlation coefficients among seven grain traits in the RIL population of bread wheat in two different environments as well as in pooled data over the environments
| GAS | GPL | GL | GWid | GLWR | FFD | |
|---|---|---|---|---|---|---|
| 1. GPL | ||||||
| E1 | 0.85** | |||||
| E2 | 0.85** | |||||
| AE | 0.84** | |||||
| 2. GL | ||||||
| E1 | 0.77** | 0.97** | ||||
| E2 | 0.78** | 0.97** | ||||
| AE | 0.76** | 0.96** | ||||
| 3. GWid | ||||||
| E1 | 0.92** | 0.68** | 0.56** | |||
| E2 | 0.93** | 0.76** | 0.65** | |||
| AE | 0.93** | 0.74** | 0.61** | |||
| 4. GLWR | ||||||
| E1 | − 0.28** | 0.20** | 0.34** | − 0.51** | ||
| E2 | − 0.06 | − 0.16 | − 0.16 | − 0.1 | ||
| AE | − 0.40** | 0.04 | 0.21 | − 0.56** | ||
| 5. FFD | ||||||
| E1 | − 0.55** | − 0.49** | − 0.48** | − 0.52** | 0.19* | |
| E2 | − 0.39** | − 0.59** | − 0.58** | − 0.39** | 0.15 | |
| AE | − 0.48** | − 0.53** | − 0.52** | − 0.49** | 0.12 | |
| 6. TGW | ||||||
| E1 | 0.04 | 0.09 | 0.06 | 0.01 | 0.16 | 0.79** |
| E2 | 0.25* | 0.12 | 0.08 | 0.26** | − 0.02 | 0.64** |
| AE | 0.09 | 0.13 | 0.1 | 0.09 | 0.02 | 0.66** |
E1 Meerut, E2 Varanasi, AE pooled data of the two environments, GAS grain area size, GPL grain perimeter length, GL grain length, GW grain width, GLWR grain length-width ratio, FFD factor form density, TGW thousand grain weight
*Significant at P = 0.05; **significant at P = 0.01
Significant negative correlation between GLWR and GWid both in the individual environments and in the pooled data indicated that wider grains have reduced GLWR, this type of association may be responsible for grain roundness (Okamoto et al. 2013). Factor form density (FFD) is negatively associated with both GL and GWid, which is in agreement with an earlier report (Dholakia et al. 2003). However, FFD and TGW are positively correlated, which is also in agreement with two earlier reports (Dholakia et al. 2003; Gegas et al. 2010).
SSR polymorphism and construction of linkage maps
As mentioned earlier, 376 SSRs were tried for genotyping, but only 120 SSRs were polymorphic between the two parental genotypes. Fifty five SSRs showed a good fit to 1:1 segregation in the RIL mapping population and the remaining 65 SSRs showed huge segregation distortion and thus the genetic maps were prepared using genotyping data of 55 SSRs only. The lengths of genetic maps for individual chromosomes ranged from 154.3 cM (chromosome 4A) to 460.9 cM (chromosome 2B) and number of mapped SSR loci ranged from three (chromosome 4A) to 10 (chromosome 2B).
The linkage maps for each of the six chromosomes 2B, 4A, 5A, 5D, 6A and 7B carried large gap between the mapped markers making the above maps relatively longer. The large gaps may either represent genomic regions that lack polymorphism among the parental genotypes of the RIL population or these gaps may also be due to exclusion of markers showing segregation distortion. The (unobservable) distorted segregation of these loci causes the observed markers to deviate from the normal Mendelian segregation ratio (1:1) of the RIL population. In all kinds of mapping populations like F2, DH and RILs, segregation distortion generally occurs. Among these mapping populations, RILs exhibit the maximum probability of distortions due to continued selfing for 5–6 generations (Singh et al. 2007). In addition several studies reported deviation from the expected Mendelian segregation ratios in wheat (Faris et al. 2000; Peng et al. 2000; Kumar et al. 2007), rice (Harushima et al. 1996; Xu 1997; Lyttle 1991), barley (Graner et al. 1991; Kleinhofs et al. 1993; Devaux et al. 1995) and maize (Wendel et al. 1987; Dufour et al. 2001; Lu et al. 2002).
QTLs for grain traits
Eighteen QTLs were detected for the seven grain traits following analyses of data of two individual environments (Meerut and Varanasi) and the data pooled over both the environment (AE) (Table 3; Fig. 2). The LOD score values for the individual QTLs ranged between 1.83 and 10.85, and the 18 QTLs were located on eight chromosomes. Fourteen of these QTLs were detected in Meerut environment and only 4 QTLs were detected in Varanasi environment and none of the QTL was detected in both the environments. This suggested significant QTL × environment interaction, as also reported in some previous studies (Campbell et al. 2003; Kumar et al. 2009).
Table 3.
QTLs for grain traits detected by composite interval mapping in a RIL population of bread wheat
| Trait/QTL | Environmenta | Flanking markersb | Positionc | LOD | Ad | R2 (%)e |
|---|---|---|---|---|---|---|
| 1. Grain length | ||||||
| QGl.ccsu-4A.1 | E1 | Xgwm397-Xgwm601 (Xgwm397) | 25.00 | 2.40 | − 0.26 | 53.00 |
| QGl.ccsu-5A.1 | E1 | Xgwm126-Xwms1171 (Xgwm126) | 33.00 | 3.36 | 0.26 | 53.84 |
| 2. Grain width | ||||||
| QGwid.ccsu-7D.1 | E1, AE | Xgwm635-Xgwm37 (Xgwm37) | 372.50, 379.50 | 2.28, 2.45 | − 0.13, 0.10 | 47.12–33.29 |
| QGwid.ccsu-6A.1 | E2 | Xwmc398-Xgwm169 (Xwmc398) | 0.00 | 2.08 | 0.06 | 7.66 |
| 3. Grain length width ratio | ||||||
| QGlwr.ccsu-1A.1 | E1 | Xgwm99-Xgwm633 (Xgwm99) | 228.70 | 2.96 | − 0.09 | 13.87 |
| QGlwr.ccsu-2B.1 | E2 | Xgwm972-Xgwm148 (Xgwm972) | 99.20 | 2.30 | − 0.08 | 17.80 |
| QGlwr.ccsu-2B.2 | E1, AE | Xgwm410-Xgwm429 (Xgwm410) | 110.10, 109.10 | 2.08, 2.22 | − 0.05, 0.04 | 7.98–7.74 |
| QGlwr.ccsu-6A.1 | E2 | Xwmc398-Xgwm169 (Xwmc398) | 18.00 | 2.45 | − 0.12 | 36.70 |
| QGlwr.ccsu-7B.1 | E1 | Xgwm146-Xgwm573 (Xgwm146) | 130.60 | 3.28 | 0.44 | 21.83 |
| 4. Grain perimeter length | ||||||
| QGpl.ccsu-4A.1 | E1 | Xgwm397-Xgwm601 (Xgwm397) | 27.00 | 2.50 | − 0.60 | 52.50 |
| QGpl.ccsu-5A.1 | E1 | Xgwm126-Xwms1171 (Xgwm126) | 43.00 | 2.53 | 0.61 | 55.86 |
| QGpl.ccsu-7B.1 | E2 | Xgwm146-Xgwm573 (Xgwm146) | 130.60 | 10.85 | 3.45 | 41.62 |
| 5. Grain area size | ||||||
| QGas.ccsu-5D.1 | E1 | Xwmc318-Xcfd3 (Xcfd3) | 335.90 | 2.29 | − 0.94 | 58.15 |
| QGas.ccsu-7D.1 | E1 | Xbarc 0092-Xgwm635 (Xgwm635) | 321.60 | 2.58 | − 0.91 | 52.91 |
| QGas.ccsu-7D.2 | E1 | Xgwm635-Xgwm37 (Xgwm635) | 368.50 | 2.57 | − 0.84 | 63.66 |
| 6. Factor form density | ||||||
| QFfd.ccsu-1A.1 | E1 | Xwmc611-Xwmc93 (Xwmc611) | 119.5 | 1.83 | 0.33 | 7.70 |
| QFfd.ccsu-5D.1 | E1 | Xwmc318-Xcfd40 (Xcfd40) | 379.9 | 1.85 | 0.31 | 7.90 |
| 7. Thousand grain weight | ||||||
| QTgw.ccsu-2B.1 | E1 | Xgwm972-Xgwm148 (Xgwm972) | 84.7 | 2.98 | − 3.14 | 30.13 |
aE1 Meerut, E2 Varanasi, AE pooled data of the two environment
bMarkers in parentheses represent closest marker of the QTL
cPosition = distance (cM) between the QTL and the first marker of the relevant chromosome
dAdditive effect of the QTL (a positive value indicates the HUW468 allele having a positive effect on the trait and negative value indicate that the NW1014 allele having a positive effect on the trait), e Percentage of phenotypic variation explained (PVE) by the QTL
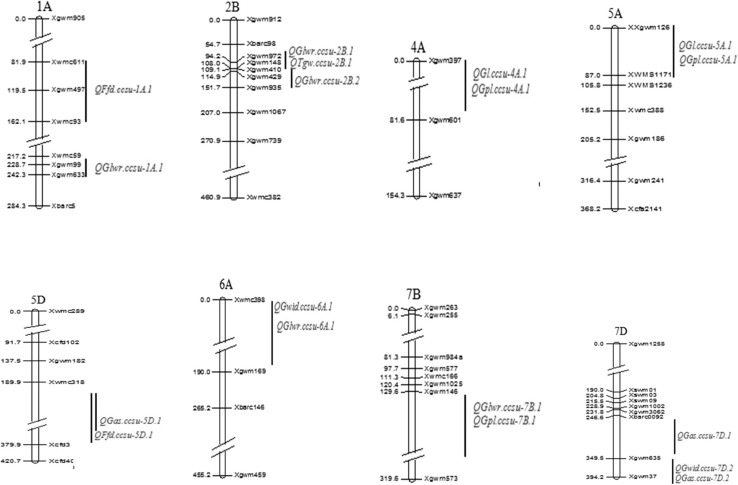
Fig. 2.
Important genomic regions harbouring QTLs for grain traits in bread wheat detected using RIL mapping population derived from the cross NW1014 × HUW468. In each case, the genomic region containing the QTL is indicated by black vertical bar followed by the name of the QTL (highlighted with red color). The marker loci are indicated on the right, and the genetic distances (cM) are shown on the left (color figure online)
The number of QTLs mapped on individual chromosomes ranged from two QTLs each on chromosomes 1A, 4A, 5A, 6A, 7B and 5D to three QTLs each on chromosomes 7D and 2B (for details see Table 3; Fig. 3). Individual QTLs explained 7.70–63.66% of PV for the concerned trait. Twelve QTLs with PV exceeding 20% were considered as major QTLs. These included two QTLs each for both GL and GLWR, three QTLs each for both GPL and GAS and one QTL for both TGW and GWid (Table 3).
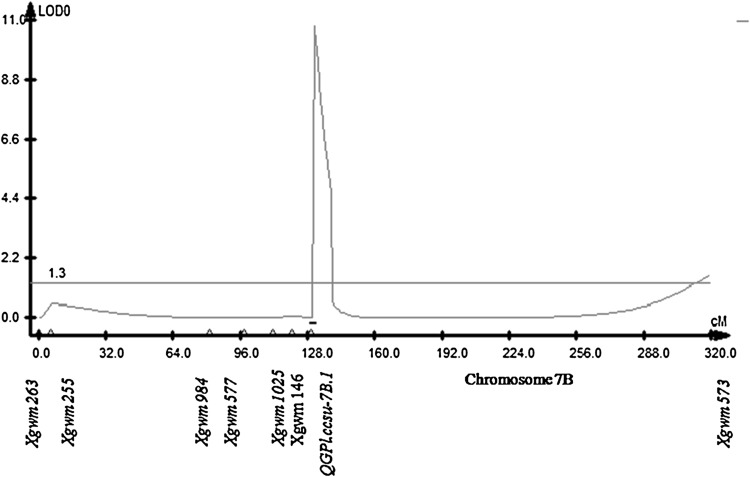
Fig. 3.
A representative QTL Cartographer plot for QTL (QGPl.ccsu-7B.1) for grain perimeter length identified on chromosome 7B of bread wheat
In pooled data as well as in Meerut environment, two QTLs, one each for both GWid and GLWR were detected. Out of these two QTLs, QTL for GWLR (QGlwr.ccsu-2B.2) is a minor QTL, since it explained only up to 7.98% PV, but the QTL for GWid (QGwid.ccsu-7D.1) located in marker interval Xgwm635-Xgwm37 is a major QTL (range of PVE = 33.29–47.12%). This particular QTL is the most important among all the QTLs identified in this study. The allele for higher GWid for this QTL was contributed by the parental genotype HUW468.
Co-localized QTLs for grain traits
In earlier studies also, the QTLs that are co-located for grain related traits were reported (Peng et al. 2003; Groos et al. 2003; Marza et al. 2006; Quarrie et al. 2005). In our study, maximum number of QTLs for different traits were co-located and grouped in particular genomic regions in the chromosomes 4A, 5A, 6A, 2B, 7B, 5D and 7D (except chromosome 1A). For instance, out of the two QTLs for GL, one QTL each was co-located with two separate QTLs for GPL. One QTL for GWid and GPL were co-located with separate QTLs for GLWR and similarly a QTL for GWid was co-located with QTL for GAS. A QTL each for TGW and GAS were co-located with a QTL for GLWR and FFD, respectively. These co-located QTLs may represent pleiotropic QTLs for more than one trait (for details see Table 4).
Table 4.
Co-localized QTLs for different grain traits in wheat
| Chromosome | Traits for co-localized QTL | Marker interval | Parental allele |
|---|---|---|---|
| 2B | GLWR and TGW | Xgwm972-Xgwm148 | −/− |
| 4A | GL and GPL | Xgwm397-Xgwm601 | −/− |
| 5A | GL and GPL | Xgwm126-Xwms1171 | +/+ |
| 6A | Gwid and GLWR | Xwmc398-Xgwm169 | +/− |
| 5D | GAS and FFD | Xwmc318-Xcfd3–Xcfd40 | −/+ |
| 7B | GLWR and GPL | Xgwm146-Xgwm573 | +/+ |
| 7D | Gwid and GAS | Xgwm635-Xgwm37 | −/− |
GLWR grain length-width ratio, TGW thousand grain weight, GL grain length, GPL grain perimeter length, GWid grain width, GAS grain area size, FFD factor form density, + sign = HUW468 allele contributed for increased trait value, − sign = NW1014 allele contributed for increased trait value
As stated above, the two QTLs, one each for GL (QGl.ccsu-4A.1) and GPL (QGPl.ccsu-4A.1) were co-localized on chromosome 4A. The closest marker Xgwm397 associated with these co-localized QTLs was also earlier reported to be the closest marker to the QTL for GL and VP (vertical perimeter) (Williams et al. 2013; Breseghello and Sorrells 2007). Each of these two co-localized QTLs were detected only in the Meerut environment, although each QTL explained > 50% of the PV for the traits. The positive alleles for the above two QTLs were contributed by the parental genotype NW1014 and therefore QTL alleles from the parental genotypes NW1014 have the potential for exploitation in MAS for improvement of GL and GPL traits in wheat. Although none of these two traits are significantly correlated with TGW. This suggested that these traits were controlled by independent loci which is in agreement with an earlier report by Gegas et al. (2010).
Similarly, two other QTLs, one each for GL (QGl.ccsu-5A.1) and GPL (QGPl.ccsu-5A.1) located on chromosome 5A were also co-located and the marker closest to these QTLs is Xgwm126. These two QTLs were also detected at Meerut environment and each QTL also explained > 50% PV for the respective traits. A comparison of the genetic map of chromosome 5A prepared during the present study with that reported by Ganal and Röder (2007) showed that the above two QTLs and the QTL for GL (QGl.ccsu-5A.1) earlier identified by Tyagi et al. (2014) and Ramya et al. (Ramya et al. 2010) were present in the adjacent regions, suggesting that the genomic regions harbouring these QTLs possess genes that control GL and GPL in wheat. Although, the marker interval containing the above two co-located QTLs is more than 80 cM. However, this large interval between the two flanking markers may be due to absence of polymorphic markers among the markers tried during the present study or the failure to map polymorphic markers in this region due to segregation distortion. Hence, there is need to screen more markers in future studies so as to find more polymorphic markers for this region in order to map additional markers to more precisely map the QTLs in this region. Similar to above co-localized QTLs for GL and GPL on chromosome 4A, the co-localized QTLs for these two traits on chromosome 5A are also important for use in MAS for improving grain traits in wheat. However, the positive alleles for these two QTLs were contributed by the parental genotype HUW468.
A QTL each for GWid (QGwid.ccsu-7D.1) and GAS (QGAs.ccsu-7D.2) were mapped in the marker interval Xgwm635-Xgwm37. Marker Xcfd69 associated with QTLs for GWid and TGW reported by Williams et al. (2013) was located at 4 cM from the marker Xgwm37 in the consensus map of Somers et al. (2004). Further, a gene for grain size in wheat (TaGS-D1), which is an ortholog of rice gene OsGS3 affecting grain length and weight in wheat is located adjacent to the co-localized QTL discussed above in the present study (Zhang et al. 2014). Thus, the genomic region encompassed by the markers Xgwm635 and Xcfd69 seems to be important for controlling grain traits. Further, in the consensus map of Somers et al. (2004), the marker Xgwm 37 associated with the above co-located QTL for GWid (QGwid.ccsu-7D.1) and GAS (QGAs.ccsu-7D.2) was placed adjacent to the marker Xwmc273 at a distance of 2 cM. The marker Xwmc273 was reported to be associated with the grain filling rate (GFR) in wheat under heat stress using bulked segregant analysis (Barakat et al. 2012). The physiological mechanisms underlying GFR have been widely explored (Egli et al. 1989), and the GFR plays an important role in determining grain size and consequently the grain yield. Therefore, the genomic region harbouring the markers Xgwm635, Xgwm37, Xwmc273 and Xcfd69 could be exploited for improvement of grain traits during wheat breeding.
Two QTLs, one each for GLWR (QGlwr.ccsu-7B.1) and GPL (QGpl.ccsu-7B.1) were mapped on chromosome 7B in the marker interval Xgwm146-Xgwm573 during the present study. Two QTLs (QPv.sdau-7B and QPet.sdau-7B) for starch traits were also reported on chromosome 7B in an earlier study by Sun et al. (2008). On the consensus map reported by Somers et al. (2004), the markers Xgwm644 and Xgwm 577 associated with the QTLs QPv.sdau-7B and QPet.sdau-7B for starch traits are at a distance of 6 cM and 13 cM from the markers Xgwm573 and Xgwm 146, respectively, flanking the above QTLs for GLWR and GPL reported during the present study. Hence the genomic region on the chromosome 7B harbouring the above QTLs is important as it controls GLWR, GPL and also starch traits, which contributes to grain processing and end-use quality (Hurkman et al. 2003; Yamamori and Quynh 2000; Tetlow et al. 2004; Yamamori 2009).
The closest marker (Xcfd3) to the QTL for GAS (QGAs.ccsu-5D.1) reported during the present study and marker Xcfd81 closest to the QTL for GAS reported by Okamoto et al. (2013) on chromosome 5D are located at an interval of 29 cM in the consensus map of Somers et al. (2004). Therefore, the QTL for GAS identified in the current study is different than the QTL for GAS identified by Okamoto et al. (2013). However, the marker Xcfd3 that is closest to the QTL QGAs.ccsu-5D.1 for GAS is also reported to be associated with a QTL, QGfd.nfcri-5D for grain filling duration (GFD) (Wang et al. 2009). Since the grain filling is a crucial and dynamic process, its duration and rate determine the grain related traits such as grain size, grain weight and consequently the GFD indirectly determines grain yield in wheat (Gebeyehou et al. 1982; Li and Pan 2005). Hence, the genomic region harbouring the above QTLs on chromosome 5D seems to be important in controlling grain traits as well as physiological processes such as GFD and are thus valuable for use in MAS.
Two QTLs are located on chromosome 6A, one each for GWid (QGwid.ccsu-6A.1) and GLWR (QGLwr.ccsu-6A.1) and their closest marker is Xwmc398. In an earlier study by Su et al. (2011), a wheat ortholog (TaGW2) of rice gene GW2 was mapped on chromosome 6A and the closest marker to this gene is cfd 80.2. This marker is co-located with the marker Xwmc398 that is closest to the above QTLs for GWid and GLWR and thus, it is possible that the above two QTLs may represent the TaGW2 for grain weight in wheat. The QTL for GLWR also explained ~ 37% PV for the trait making this an important candidate for use in MAS.
Relative to the QTLs for above discussed traits, one QTL each for FFD were mapped on chromosomes 1A and 5D, both these QTLs were minor as they explained 7.70 and 7.90% PV. One of these QTLs, namely QFfd.ccsu-5D.1 was flanked closely by the marker Xcfd40. This particular marker was also closest to the QTL (QKl.ncl-5D.1) for GL reported by Ramya et al. (2010). The marker Xwmc93 flanking the QTL (QFfd.ccsu-1A.1) detected during present study was co-located with Xwmc312 in the consensus map of Somers et al. (2004). The marker Xwmc312 was reported to be linked with QTL for TGW, GWid, GL and GLW (Li et al. 2015). Therefore, the genomic regions close to these QTLs are important in controlling several grain traits in wheat. Similarly, Xgwm 99, the closest marker to the QTL for GLWR (QGlwr.ccsu-1A.1) reported in this study was earlier shown to be associated with a QTL for TGW by Elangovan et al. (2011) and Mir et al. (2012). Hence the genomic region on the chromosome 1A is important as it controls different grain traits.
In a region adjacent to QTL for GLWR (QGlwr.ccsu-2B.2), a QTL for TGW (QTkw.ncl-2B.1) was reported by Ramya et al. (2010). A QTL for TGW (QTgw.ccsu-2B.1) was also found on chromosome 2B in a genomic region flanked by markers Xgwm 972 and Xgwm148. This particular QTL explained 30.13% PV and hence is a major QTL controlling TGW. This QTL and the QTL for TGW (QKw.ncl-2B.2) reported by Ramya et al. (2010) were present adjacent to each other in the consensus map of Somers et al. (2004). The desirable allele for higher TGW for this QTL was contributed by the parental genotype NW1014, which may be exploited in MAS for improvement of grain traits.
Two QTL for GLWR (QGlwr.ccsu-2B.1 and QGlwr.ccsu-2B.2) and a QTL for TGW (QTgw.ccsu-2B.1) were co-located in the genomic region flanked by markers Xgwm972 and Xgwm 148 in the present study. A QTL (QFLL-2B) for flag leaf length (FLL) earlier reported on chromosome 2B by Liu et al. (2018) was flanked by markers Xbarc 318 and Xwmc 344. Markers Xgwm972 and gwm148 flanking the above three QTL for grain traits and the marker Xwmc344 associated to the QTL for FLL are located in adjacent region in the consensus maps of Somers et al. (2004) and Ganal and Röder (2007). Hence, the above genomic region of 2B covering QTLs for GLWR, TGW and FLL is an important region as the flag leaf in wheat play an important role in photosynthesis and contributes 45–58% of photosynthates during the grain-filling stage (Khaliq et al. 2008). FLL is also strongly positively correlated with spike length, kernel number, and weight per spike, indicating that larger flag leaves contribute to the yield-related traits (Liu et al. 2018).
Two QTLs, one each for GLWR (QGlwr.ccsu-7B.1) and GPL (QGpl.ccsu-7B.1) were novel and reported in the present study for the first time.
Conclusions
QTL analysis for grain traits provides a basis for improvement of grain quality in wheat using marker-assisted selection. Present study provides information on genomic regions regulating grain size in wheat. Out of a number of QTLs for different traits reported during the present study, a QTL for GWid (QGwid.ccsu-7D.1) was stable and explained 33.29–47.12% PV. This QTL was co-located with a QTL for GAS (QGas.ccsu-7D.2), which explained 63.66% PV for grain area size. Therefore, markers Xgwm635 and Xgwm37 flanking these QTLs could be used for MAS for improving the grain quality in bread wheat.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors like to thank The Head, Department of Genetics and Plant Breeding, CCS University (Meerut, India) for providing facilities. PKG and HSB were each awarded the position of INSA Senior Scientist. PKG was also awarded a National Academy of Sciences India (NASI) Senior Scientist Platinum Jubilee Fellowship during the tenure of this research work. SK and VJ each were awarded a JRF/SRF in research projects sanctioned by the Department of Biotechnology, Government of India, New Delhi. Primer aliquots for 17 SSRs provided by Dr. M.S Röder, Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany, is gratefully acknowledged.
References
- Barakat MN, Al-Doss A, Elshafei AA, et al. Bulked segregant analysis to detect quantitative trait loci (QTL) related to heat tolerance at grain filling rate in wheat using simple sequence repeat (SSR) markers. Afr J Biotechnol. 2012;11:12436–12442. [Google Scholar]
- Börner A, Schumann E, Fürste A, et al. Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.) Theor Appl Genet. 2002;105:921–936. doi: 10.1007/s00122-002-0994-1. [DOI] [PubMed] [Google Scholar]
- Breseghello F, Sorrells ME. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics. 2006;172:1165–1177. doi: 10.1534/genetics.105.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breseghello F, Sorrells ME. Association analysis as a strategy for improvement of quantitative traits in plants. Crop Sci. 2006;46:1323–1330. doi: 10.2135/cropsci2005.09-0305. [DOI] [Google Scholar]
- Breseghello F, Sorrells ME. QTL analysis of kernel size and shape in two hexaploid wheat mapping populations. Field Crops Res. 2007;101:172–179. doi: 10.1016/j.fcr.2006.11.008. [DOI] [Google Scholar]
- Brown TA, Jones MK, Powell W, Allaby RG. The complex origins of domesticated crops in the Fertile Crescent. Trends Ecol Evol. 2009;24:103–109. doi: 10.1016/j.tree.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Cabral AL, Jordan MC, Larson G, et al. Relationship between QTL for grain shape, grain weight, test weight, milling yield, and plant height in the spring wheat cross RL4452/‘AC Domain’. PLoS ONE. 2018;13(1):e0190681. doi: 10.1371/journal.pone.0190681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KG, Bergman CJ, Gualberto DG, et al. Quantitative trait loci associated with kernel traits in soft × hard wheat cross. Crop Sci. 1999;39:1184–1195. doi: 10.2135/cropsci1999.0011183X003900040039x. [DOI] [Google Scholar]
- Campbell BT, Baenziger PS, Gill KS, et al. Identification of QTLs and environmental interactions associated with agronomic traits on chromosome 3A of wheat. Crop Sci. 2003;43:1493–1505. doi: 10.2135/cropsci2003.1493. [DOI] [Google Scholar]
- Chastain TG, Ward KJ, Wysocki DJ. Stand establishment responses of soft white winter wheat to seedbed residue and seed size. Crop Sci. 1995;35:213–218. doi: 10.2135/cropsci1995.0011183X003500010040x. [DOI] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux P, Kilian A, Kleinhofs A. Comparative mapping of the barley genome with male and female recombination derived, doubled haploid populations. Mol Gen Genet. 1995;249:600–608. doi: 10.1007/BF00418029. [DOI] [PubMed] [Google Scholar]
- Dholakia BB, Ammiraju JSS, Singh H, et al. Molecular marker analysis of kernel size and shape in bread wheat. Plant Breed. 2003;122:392–395. doi: 10.1046/j.1439-0523.2003.00896.x. [DOI] [Google Scholar]
- Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour P, Johnsson C, Antoine-Michard S, et al. Segregation distortion at marker loci: variation during microspore embryogenesis in maize. Theor Appl Genet. 2001;102:993–1001. doi: 10.1007/s001220100584. [DOI] [Google Scholar]
- Egli DE, Ramseur EL, Yu ZW, et al. Source-sink alterations affect the number of cells in soybean cotyledons. Crop Sci. 1989;29:732–735. doi: 10.2135/cropsci1989.0011183X002900030039x. [DOI] [Google Scholar]
- Elangovan M, Dholakia BB, Rai R, et al. Mapping QTL associated with agronomic traits in bread wheat (Triticum aestivum L.) J Wheat Res. 2011;3:14–23. [Google Scholar]
- Faris JD, Haen KM, Gill BS. Saturation mapping of a gene-rich recombination hot spot region in wheat. Genetics. 2000;154:823–835. doi: 10.1093/genetics/154.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DQ. Contrasting patterns in crop domestication and domestication rates: recent archaeobotanical insights from the old world. Ann Bot (Lond.) 2007;100:903–924. doi: 10.1093/aob/mcm048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganal MW, Röder MS. Microsatellite and SNP markers in wheat breeding. In: Varshney RK, Tuberosa R, editors. Genomics-assisted crop improvement: genomics applications in crops. Dordrecht: Springer; 2007. pp. 1–24. [Google Scholar]
- Gebeyehou G, Knott DR, Baker RJ. Rate and duration of grain filling in durum wheat cultivars. Crop Sci. 1982;22:337–340. doi: 10.2135/cropsci1982.0011183X002200020033x. [DOI] [Google Scholar]
- Gegas VC, Nazari A, Griffiths S, et al. A genetic framework for grain size and shape variation in wheat. Plant Cell. 2010;22:1046–1056. doi: 10.1105/tpc.110.074153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giura A, Saulescu NN. Chromosomal location of genes controlling grain size in a large grained selection of wheat (Triticum aestivum L.) Euphytica. 1996;89:77–80. doi: 10.1007/BF00015722. [DOI] [Google Scholar]
- Graner A, Jahoor A, Schondelmaier J, et al. Construction of an RFLP map of barley. Theor Appl Genet. 1991;83:250–256. doi: 10.1007/BF00226259. [DOI] [PubMed] [Google Scholar]
- Groos C, Robert N, Bervas E, Charmet G. Genetic analysis of grain protein content, grain yield and thousand-kernel weight in bread wheat. Theor Appl Genet. 2003;106:1032–1040. doi: 10.1007/s00122-002-1111-1. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Balyan HS, Kulwal PL, et al. QTL analysis for some quantitative traits in bread wheat. J Zhejiang Univ Sci. 2007;8:807–814. doi: 10.1631/jzus.2007.B0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima Y, Kurata N, Yano M, et al. Detection of segregation distortions in an indica-japonica rice cross using a high-resolution molecular map. Theor Appl Genet. 1996;92:145–150. doi: 10.1007/BF00223368. [DOI] [PubMed] [Google Scholar]
- Huang XQ, Coster H, Ganal MW, Röder MS. Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (Triticum aestivum L.) Theor Appl Genet. 2003;106:1379–1389. doi: 10.1007/s00122-002-1179-7. [DOI] [PubMed] [Google Scholar]
- Huang XQ, Kempf H, Ganal MW, Röder MS. Advanced backcross QTL analysis in progenies derived from a cross between a German elite winter wheat variety and a synthetic wheat (Triticum aestivum L.) Theor Appl Genet. 2004;109:933–943. doi: 10.1007/s00122-004-1708-7. [DOI] [PubMed] [Google Scholar]
- Huang XQ, Cloutier S, Lycar L, et al. Molecular detection of QTLs for agronomic and quality traits in a doubled haploid population derived from two Canadian wheats (Triticum aestivum L.) Theor Appl Genet. 2006;113:753–766. doi: 10.1007/s00122-006-0346-7. [DOI] [PubMed] [Google Scholar]
- Hurkman WJ, McCue KF, Altenbach SB, et al. Effect of temperature on expression of genes encoding enzymes for starch biosynthesis in developing wheat endosperm. Plant Sci. 2003;164:873–881. doi: 10.1016/S0168-9452(03)00076-1. [DOI] [Google Scholar]
- Khaliq I, Irshad A, Ahsan M. Awns and flag leaf contribution towards grain yield in spring wheat (Triticum aestivum L.) Cereal Res Commun. 2008;36:65–76. doi: 10.1556/CRC.36.2008.1.7. [DOI] [Google Scholar]
- Kleinhofs A, Kilian A, SaghaiMaroof MA, et al. A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor Appl Genet. 1993;86:705–717. doi: 10.1007/BF00222660. [DOI] [PubMed] [Google Scholar]
- Kuchel H, Williams KJ, Langridge P, et al. Genetic dissection of grain yield in bread wheat: I. QTL analysis. Theor Appl Genet. 2007;115:1029–1041. doi: 10.1007/s00122-007-0629-7. [DOI] [PubMed] [Google Scholar]
- Kumar N, Kulwal PL, Gaur A, et al. QTL analysis for grain weight in common wheat. Euphytica. 2006;151:135–144. doi: 10.1007/s10681-006-9133-4. [DOI] [Google Scholar]
- Kumar S, Gill BS, Faris JD. Identification and characterization of segregation distortion loci along chromosome 5B in tetraploid wheat. Mol Genet Genomics. 2007;278:187–196. doi: 10.1007/s00438-007-0248-7. [DOI] [PubMed] [Google Scholar]
- Kumar A, Kumar J, Singh R, et al. QTL analysis for grain colour and pre-harvest sprouting in bread wheat. Plant Sci. 2009;177:114–122. doi: 10.1016/j.plantsci.2009.04.004. [DOI] [Google Scholar]
- Kumar A, Mantovani EE, Seetan R, et al. Dissection of genetic factors underlying wheat kernel shape and size in an elite × non adapted cross using a high density SNP linkage map. Plant Genome. 2016;9:1–22. doi: 10.3835/plantgenome2015.09.0081. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Li XJ, Pan ZD. A study on the grain filling characteristic of different weight wheat. Rev China Agric Sci Technol. 2005;7:26–30. [Google Scholar]
- Li M, Wang Z, Liang Z, et al. Quantitative trait loci analysis for kernel-related characteristics in common wheat (Triticum aestivum L.) Crop Sci. 2015;55:1–9. doi: 10.2135/cropsci2014.03.0249. [DOI] [Google Scholar]
- Liu K, Xu H, Liu G, et al. QTL mapping of flag leaf-related traits in wheat (Triticum aestivum L.) Theor Appl Genet. 2018;131:839–849. doi: 10.1007/s00122-017-3040-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Romero-Severson J, Bernardo R. Chromosomal regions associated with segregation distortion in maize. Theor Appl Genet. 2002;105:622–628. doi: 10.1007/s00122-002-0970-9. [DOI] [PubMed] [Google Scholar]
- Lyttle TW. Segregation distorters. Annu Rev Genet. 1991;25:511–557. doi: 10.1146/annurev.ge.25.120191.002455. [DOI] [PubMed] [Google Scholar]
- Marza F, Bai GH, Carver BF, Zhou WC. Quantitative trait loci for yield and related traits in the wheat population Ning7840 × Clark. Theor Appl Genet. 2006;112:688–698. doi: 10.1007/s00122-005-0172-3. [DOI] [PubMed] [Google Scholar]
- Mir RR, Kumar N, Jaiswal V, et al. Genetic dissection of grain weight in bread wheat through quantitative trait locus interval and association mapping. Mol Breed. 2012;29:963–972. doi: 10.1007/s11032-011-9693-4. [DOI] [Google Scholar]
- Narasimhamoorthy B, Gill BS, Fritz AK, et al. Advanced backcross QTL analysis of a hard winter wheat × synthetic wheat population. Theor Appl Genet. 2006;112:787–796. doi: 10.1007/s00122-005-0159-0. [DOI] [PubMed] [Google Scholar]
- Okamoto Y, Nguyen AT, Yoshioka M, et al. Identification of quantitative trait loci controlling grain size and shape in the D genome of synthetic hexaploid wheat lines. Breed Sci. 2013;63:423–429. doi: 10.1270/jsbbs.63.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil RM, Tamhankar SA, Oak MD, et al. Mapping of QTL for agronomic traits and kernel characters in durum wheat (Triticum durum Desf.) Euphytica. 2013;190:117–129. doi: 10.1007/s10681-012-0785-y. [DOI] [Google Scholar]
- Peng J, Korol AB, Fahima T, et al. Molecular genetic maps in wild emmer wheat, Triticum dicoccoides: genome-wide coverage, massive negative interference, and putative quasi-linkage. Genome Res. 2000;10:1509–1531. doi: 10.1101/gr.150300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Ronin Y, Fahima T, et al. Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc Natl Acad Sci USA. 2003;100:2489–2494. doi: 10.1073/pnas.252763199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashant R, Kadoo N, Desale C, et al. Kernel morphometric traits in hexaploid wheat (Triticum aestivum L.) are modulated by intricate QTL × QTL and genotype × environment interactions. J Cereal Sci. 2012;56:432–439. doi: 10.1016/j.jcs.2012.05.010. [DOI] [Google Scholar]
- Quarrie SA, Steed A, Calestani C, et al. A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring × SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor Appl Genet. 2005;110:865–880. doi: 10.1007/s00122-004-1902-7. [DOI] [PubMed] [Google Scholar]
- Ramya P, Chaubal A, Kulkarni K, et al. QTL mapping of 1000-kernel weight, kernel length, and kernel width in bread wheat (Triticum aestivum L.) J Appl Genet. 2010;51:421–429. doi: 10.1007/BF03208872. [DOI] [PubMed] [Google Scholar]
- Rasheed A, Xia X, Ogbonnaya F, et al. Genome wide association for grain morphology in synthetic hexaploid wheats using digital imaging analysis. BMC Plant Biol. 2014;14:128. doi: 10.1186/1471-2229-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röder MS, Korzun V, Wendehake K, et al. A microsatellite map of wheat. Genetics. 1998;149:2007–2023. doi: 10.1093/genetics/149.4.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidwell RJ, Smith EL, McNew RW. Inheritance and interrelationships of grain yield and selected yield-related traits in a hard red winter wheat cross. Crop Sci. 1976;16:650–654. doi: 10.2135/cropsci1976.0011183X001600050013x. [DOI] [Google Scholar]
- Simmonds J, Scott P, Leverington-Waite M, et al. Identification and independent validation of a stable yield and thousand grain weight QTL on chromosome 6A of hexaploid wheat (Triticum aestivum L.) BMC Plant Biol. 2014;14:191. doi: 10.1186/s12870-014-0191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Ghai M, Garg M, et al. An integrated molecular linkage map of diploid wheat based on a Triticum boeoticum × T. monococcum RIL population. Theor Appl Genet. 2007;115:301–312. doi: 10.1007/s00122-007-0543-z. [DOI] [PubMed] [Google Scholar]
- Somers DJ, Isaac P, Edwards K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.) Theor Appl Genet. 2004;109:1105–1114. doi: 10.1007/s00122-004-1740-7. [DOI] [PubMed] [Google Scholar]
- Su Z, Hao C, Wang L, et al. Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.) Theor Appl Genet. 2011;122:211–223. doi: 10.1007/s00122-010-1437-z. [DOI] [PubMed] [Google Scholar]
- Sun H, Lu J, Fan Y, et al. Quantitative trait loci (QTLs) for quality traits related to protein and starch in wheat. Prog Nat Sci. 2008;18:825–831. doi: 10.1016/j.pnsc.2007.12.013. [DOI] [Google Scholar]
- Sun XY, Wu K, Zhao Y, et al. QTL analysis of kernel shape and weight using recombinant inbred lines in wheat. Euphytica. 2009;165:615–624. doi: 10.1007/s10681-008-9794-2. [DOI] [Google Scholar]
- Tanabata T, Shibaya T, Hori K, et al. SmartGrain: high-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol. 2012;160:1871–1880. doi: 10.1104/pp.112.205120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow IJ, Morell MK, Emes MJ. Recent developments in understanding the regulation of starch metabolism in higher plants. J Exp Bot. 2004;55:2131–2145. doi: 10.1093/jxb/erh248. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Mir RR, Balyan HS, Gupta PK. Interval mapping and meta-QTL analysis of grain traits in common wheat (Triticum aestivum L.) Euphytica. 2014;201:367–380. doi: 10.1007/s10681-014-1217-y. [DOI] [Google Scholar]
- Varshney R, Prasad M, Roy JK, Kumar N, Singh H, Dhaliwal HS, Balyan HS, Gupta PK. Identification of eight chromosomes and a microsatellite marker on 1AS associated with QTL for grain weight in bread wheat. Theor Appl Genet. 2000;100:1290–1294. doi: 10.1007/s001220051437. [DOI] [Google Scholar]
- Wang S, Basten CJ, Zeng ZB (2005) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
- Wang RX, Hai L, Zhang XY, et al. QTL mapping for grain filling rate and yield related traits in RILs of the Chinese winter population Heshangmai × Yu8679. Theor Appl Genet. 2009;118:313–325. doi: 10.1007/s00122-008-0901-5. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Edwards MD, Stuber CW. Evidence for multilocus genetic control of preferential fertilization in maize. Heredity. 1987;58:297–302. doi: 10.1038/hdy.1987.44. [DOI] [PubMed] [Google Scholar]
- Williams K, Sorrells ME. Three-dimensional seed size and shape QTL in hexaploid wheat (Triticum aestivum L.) populations. Crop Sci. 2014;54:98–110. doi: 10.2135/cropsci2012.10.0609. [DOI] [Google Scholar]
- Williams K, Munkvold J, Sorrells M. Comparison of digital image analysis using elliptic Fourier descriptors and major dimensions to phenotype seed shape in hexaploid wheat (Triticum aestivum L.) Euphytica. 2013;190:99–116. doi: 10.1007/s10681-012-0783-0. [DOI] [Google Scholar]
- Wu Y, Yongcai Fu, Zhao S, et al. Clustered primary branch1, a new allele of dwarf11, controls panicle architecture and seed size in rice. Plant Biotechnol J. 2016;14:377–386. doi: 10.1111/pbi.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. Quantitative trait loci: separating, pyramiding, and cloning. Plant Breed Rev. 1997;15:85–139. [Google Scholar]
- Yamamori M. Amylose content and starch properties generated by five variant Wx alleles for granule-bound starch synthase in common wheat (Triticum aestivum L.) Euphytica. 2009;165:607–614. doi: 10.1007/s10681-008-9793-3. [DOI] [Google Scholar]
- Yamamori M, Quynh NT. Diversity effects of Wx-A1, -B1 and -D1 protein deficiencies on apparent amylose content and starch pasting properties in common wheat. Theor Appl Genet. 2000;100:32–38. doi: 10.1007/s001220050005. [DOI] [Google Scholar]
- Zhang Y, Liu J, Xia X, He Z. TaGS-D1, an ortholog of rice OsGS3, is associated with grain weight and grain length in common wheat. Mol Breed. 2014;34:1097–1107. doi: 10.1007/s11032-014-0102-7. [DOI] [Google Scholar]
- Zhang G, Wang Y, Guo Y, et al. Characterization and mapping of QTLs on chromosome 2D for grain size and yield traits using a mutant line induced by EMS in wheat. Crop Sci. 2015;3:135–144. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.