Abstract
Epigenetic changes may account for the doubled risk to develop schizophrenia in individuals exposed to famine in utero. We therefore investigated DNA methylation in a unique sample of patients and healthy individuals conceived during the great famine in China. Subsequently, we examined two case-control samples without famine exposure in whole blood and brain tissue. To shed light on the causality of the relation between famine exposure and DNA methylation, we exposed human fibroblasts to nutritional deprivation. In the famine-exposed schizophrenia patients, we found significant hypermethylation of the dual specificity phosphatase 22 (DUSP22) gene promoter (Chr6:291687-293285) (N = 153, p = 0.01). In this sample, DUSP22 methylation was also significantly higher in patients independent of famine exposure (p = 0.025), suggesting that hypermethylation of DUSP22 is also more generally involved in schizophrenia risk. Similarly, DUSP22 methylation was also higher in two separate case-control samples not exposed to famine using DNA from whole blood (N = 64, p = 0.03) and postmortem brains (N = 214, p = 0.007). DUSP22 methylation showed strong genetic regulation across chromosomes by a region on chromosome 16 which was consistent with new 3D genome interaction data. The presence of a direct link between famine and DUSP22 transcription was supported by data from cultured human fibroblasts that showed increased methylation (p = 0.048) and expression (p = 0.019) in response to nutritional deprivation (N = 10). These results highlight an epigenetic locus that is genetically regulated across chromosomes and that is involved in the response to early-life exposure to famine and that is relevant for a major psychiatric disorder.
Introduction
Schizophrenia is a severe psychiatric disorder with a global life-time risk of around 1% and a typical onset in late adolescence and early adulthood. In addition to a pronounced polygenic component,1 several environmental risk factors have been identified,2 of which prenatal famine is one of the strongest: an almost two-fold increase was reported in offspring conceived during the Dutch hunger winter in 19453 and at the time of the Great Chinese famine (1959–1961).4–6
The mechanism underlying the relationship between famine exposure and schizophrenia risk remains unclear, but emerging evidence suggests that epigenetic reprogramming in response to famine exposure may play a role. Indeed, famine exposure in the first trimester of pregnancy leads to DNA methylation changes and these in turn have been found to be related to cardiovascular disorders.7–11 However, the relationship between famine-induced epigenetic changes and schizophrenia has not been studied.
We hypothesized that changes in DNA methylation play a role in the increased risk to develop schizophrenia after in utero exposure to famine. To test this hypothesis, we focused on the great famine in China between 1959 and 1961, which led to an estimated death toll of over 30 million.12 The high penetrance of famine in a large rural population during a restricted period offers an opportunity for selective sampling of schizophrenia patients and healthy controls on the basis of their exposure to famine. We also studied the role of the identified DNA methylation marks in blood and brain DNA samples of unexposed schizophrenia patients and controls. Moreover, we carried out in vitro experimental studies whereby human fibroblasts were exposed to nutritional deprivation to directly investigate methylation responses to nutritional deprivation without the potential confounds of genetic differences, medication, and other environmental factors.
Contemporary studies of DNA methylation show that much of the variability in DNA methylation is controlled by genetic variation in the same genetic region (in cis)13 as well as in genetic regions more distant from the methylation mark (in trans).14 Another point of increasing interest is the relation of the DNA methylation differences with gene expression, as a relevant functional readout of methylation differences.15 Further analyses therefore investigated the relationship of identified methylation differences with genotypes as well as their relations with expression.
Results
Four samples were included in the current paper. (1) The Chinese famine sample in which the relation between famine and schizophrenia was studied. (2) A case-control blood sample from the Netherlands in which we analyzed differences between unexposed schizophrenia patients and unaffected individuals using DNA from whole blood. (3) The case-control brain sample in which we replicated case-control differences using DNA from brain tissue and analyzed the relations with genotype and gene expression. (4) Fibroblasts cultures obtained from schizophrenia patients and healthy controls. Table 1 gives an overview of the characteristics of these four samples.
Table 1.
Sample characteristics of study samples
| Sample 1: Chinese famine | Sample 2: Case-control blood | Sample 3: Case-control brain | Sample 4: Fibroblasts | |||||
|---|---|---|---|---|---|---|---|---|
| Schizophrenia | Controls | Schizophrenia | Controls | Schizophrenia | Controls | Schizophrenia | Controls | |
| N | 74 | 79 | 15 | 49 | 91 | 123 | 5 | 5 |
| Male (%) | 46 (62%) | 31 (39%) | 9 (60%) | 4 (14%) | 55% | 67% | 3 (60%) | 2 (40%) |
| Mean age (sd) | 47.3 (0.7) | 47.9 (0.8) | 40.1 (13.8) | 35.9 (17.0) | 52.6 (5.2) | 45.9 (16.8) | 39.0 (10.3) | 36.5 (6.5) |
| Famine exposure (%) | 23 (31%) | 25 (32%) | – | – | – | – | In vitro | In vitro |
| Tissue source | Blood | Blood | Brain (DLPFC) | Fibroblast culture | ||||
| Methylation | 450K BeadChip array | 450K BeadChip array | 450K BeadChip array | EPIC BeadChip array | ||||
| Reference workflow | – | – | Jaffe et al.4 | – | ||||
SCZ schizophrenia, DLPFC dorsolateral prefrontal cortex
The genome-wide methylation analysis of the Chinese famine sample identified one single region containing the dual specificity phosphatase 22 (DUSP22) gene promoter with higher DNA methylation levels in the famine-exposed schizophrenia patients compared to all other groups (Chr6: 291687-293285, Family Wise Error Rate (FWER) = 0.01). DUSP22 methylation was also significantly higher in Chinese schizophrenia patients independent of famine exposure (B = 0.07, p = 0.025). DUSP22 hypermethylation in the same region was also significant in blood DNA samples of Dutch schizophrenia patients (N = 15, mean methylation = 0.43, sd = 0.10) compared to healthy controls (N = 49, mean methylation = 0.32, sd = 0.20, p = 0.03). DUSP22 methylation was also higher in postmortem prefrontal cortex (PFC) tissue of schizophrenia patients (N = 91; mean methylation = 0.40, sd = 0.10) compared to unaffected controls (N = 123; mean methylation = 0.37, sd = 0.13) (B = 0.35, p = 0.007). Figure 1 and Table 2 show the results from the association analysis.
Fig. 1.
Identification and replication of a differentially methylated region in DUSP22 in blood. Overview of the 3000 bp area downstream and upstream of the dual specificity phosphatase 22 (DUSP22) differentially methylated region (DMR). The top panel displays the blood DNA methylation levels per group in the Chinese famine discovery sample (first panel). The second and third panels contain the DNA methylation levels for schizophrenia patients and healthy controls in blood or brain tissue, respectively. The other panels indicate the presence of coding exons (blue blocks) and non-coding introns (gray line) of the DUSP22 gene (fourth panel), and the location of a CpG island (fifth panel) based on information extracted for genome build Hg19 from the UCSC website41 with the gviz R package 42. The DUSP22 DMR is indicated across all panels with a light-blue rectangle; chr, chromosome
Table 2.
Detailed data by exposure and schizophrenia status
| Chinese famine cohort | ||||
|---|---|---|---|---|
| Schizophrenia | Controls | |||
| Exposed | Unexposed | Exposed | Unexposed | |
| N | 23 | 51 | 25 | 54 |
| Age | 50.1 (0.6) | 46.7 (0.8) | 50.3 (0.5) | 46.8 (1.0) |
| Male (%) | 18 (69%) | 28 (54%) | 10 (40%) | 21 (39%) |
| DUSP22 Methylation (sd) | 0.46 (0.04) | 0.35 (0.17) | 0.31 (0.19) | 0.33 (0.18) |
| Case-control blood cohort | ||
|---|---|---|
| Schizophrenia | Controls | |
| N | 15 | 49 |
| Age, mean (sd) | 40.1 (13.8) | 35.9 (17.0) |
| Male (%) | 60% | 10% |
| DUSP22 Methylation (sd) | 0.43 (0.10) | 0.32 (0.20) |
| Case-control brain cohort | ||
|---|---|---|
| Schizophrenia | Controls | |
| N | 91 | 123 |
| Age, mean (sd) | 52.6 (5.2) | 45.9 (16.8) |
| Male (%) | 55% | 67% |
| DUSP22 Methylation (sd) | 0.40 (0.10) | 0.37 (0.13) |
Genetic control of DUSP22 DNA methylation
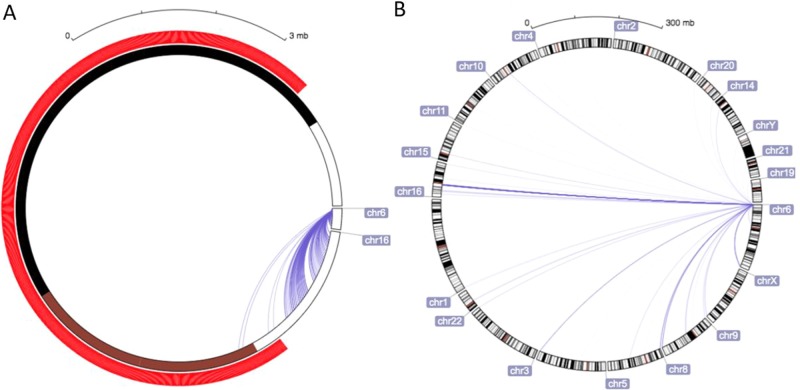
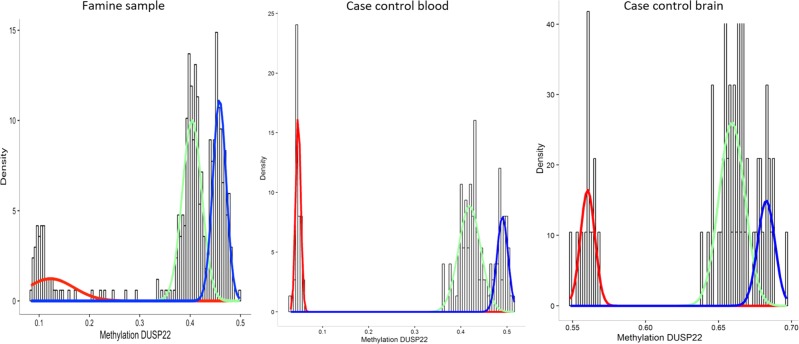
In the brain case-control sample, the association between genetic loci and methylation levels at the ten loci (CpGs) in the DUSP22 differentially methylated region (DMR) was examined for 7.5 million observed + imputed single nucleotide polymorphisms (SNPs) with minor allele frequency (MAF >5%). We identified 69 SNPs, all on chromosome 16, that were associated with all ten methylation loci at the p < 10–20 significance level (see supplementary table 1). This genetic regulation outside the DUSP22 region (in trans) is consistent with previous studies14 and three online databases of genetic variants associated with methylation methylation Quantitative Trait Locus (mQTLs) for either fetal brain tissue (http://epigenetics.essex.ac.uk/mQTL/16) or whole blood (http://genenetwork.nl/biosqtlbrowser/17and http://www.mqtldb.org/18), reporting five trans-SNPs on chromosome 16 (rs1433753, rs9674439, rs12923277, rs12927233, and rs12933929). The underlying genetic background spans a region on chromosome 16 (Chr16:34190042-46441560) as large as 12 Mb, and the previously reported five trans-SNPs are at least 15,000 base pairs away from each other. It is worth noticing that data on chromosome interactions (obtained using Hybridization Capture—Hi-C) in postmortem neurons confirm the strong interaction between the DUSP22 DMR and the mQTL region on chromosome 6. The strongest interaction is present between the 3′ end on chromosome 16 (chr16:34160000-34200000) and the region chr6:280000-320000 (see Fig. 2a, b). Visual inspection of the data of Rao et al.19 from human GM12878 B-lymphoblastoid cells also indicates that the entire region containing these trans-SNPs is in interchromosomal contact with the DUSP22 promoter region (see supplemental data 1). Subsequent mixture analysis identified three underlying Gaussian distributions in the samples of the study (see Fig. 3) that we used as an indicator of genetic determinant of the methylation levels, subsequently coined “genetic background”.
Fig. 2.
Overview of the chromosome–chromosome interactions measured with in situ Hi-C. Panel a zooms into the DUSP22 DMR, while b provides an overview of the chromosome interactions. A darker blue indicates more frequent interactions
Fig. 3.
Density plot of the average methylation at the DUSP22 differentially methylated region (DMR) in the four population samples: the Chinese famine sample, the case-control blood sample, the blood genomics sample, and the brain case-control sample. The colored lines represent the estimation of the underlying distributions in the respective samples
Influence of genotype on schizophrenia risk
The Chinese famine sample showed a significant gene–environment interaction between the genetic background and famine exposure on schizophrenia risk (B = 1.20, p = 0.042). After adjustment for genetic background, the relationship of DUSP22 methylation with famine and schizophrenia was only present in a selection of participants without the 35 participants with a genetic predisposition for low invariable methylation levels (N = 118, B = 0.028, p = 0.072). In the Dutch case-control sample, the association between DUSP22 promoter methylation and schizophrenia persisted after adjustment for genetic background (B = 0.741, p = 0.025). In the postmortem brains, the association between DUSP22 promoter methylation and schizophrenia was attenuated after adjustment for genetic background (B = 0.13, t = 1.745, p = 0.083).
Ethnicity
Ethnicity influenced the relationship between DUSP22 methylation and schizophrenia in the brain case-control sample (Schizophrenia by Race interaction: B = 0.680, p = 0.007). The genetic background of DUSP22 methylation was significantly different between African–American and Caucasian subjects (p < 0.001 in Pearson’s χ2 test). The absence of the low methylation genotype in the African–Americans contributes to the lower variation in DUSP22 DMR methylation levels (see Table 3). This is in line with the stratified analyses for ethnicity that show an association between DUSP22 methylation and schizophrenia in the Caucasians only (B = 0.692, p = 0.002), and not in the African–Americans (B = −0.086, p = 0.436).
Table 3.
Sample characteristics of the brain dataset per ethnicity
| African–American | Caucasian | All | |
|---|---|---|---|
| N | 99 | 115 | 214 |
| Age (mean (sd)) | 49.24 (16.30) | 48.28 (16.62) | 48.73 (16.44) |
| Male (%) | 58 (58.6) | 74 (64.3) | 132 (61.7) |
| Schizophrenia (%) | 38 (38.4) | 53 (46.1) | 91 (42.5) |
| DUSP22 DMR (mean (sd)) | 0.43 (0.07) | 0.34 (0.14)* | 0.38 (0.12) |
| DUSP22 expression (mean (sd)) | 2.83 (0.33) | 2.81 (0.33) | 2.82 (0.33) |
| Genetic background (%) | |||
| 1 | 3 (3.1) | 27 (23.5)* | 30 (14.0) |
| 2 | 35 (36.4) | 65 (56.5)* | 100 (46.7) |
| 3 | 61 (63.5) | 23 (20.0)* | 84 (39.3) |
*p < 0.001 in either a Pearson’s χ2 test (for genotype) or t-test (for methylation levels)
Smoking and urban background
In the Chinese famine sample, as expected, smoking was more frequent in the schizophrenia patients (B = 0.01, p = 0.046). No differences between the famine and non-famine groups were present (p = 0.9). The DUSP22 promoter methylation was also not associated with the smoking proxy (B = 0.028, p = 0.112), nor is it highlighted in previous association studies into smoking.20,21 Also inclusion of urban background in the model did not alter the results. In the case-control blood samples, five of 15 schizophrenia patients were current smokers in contrast to three of 49 smokers in the healthy controls (χ2 test, p = 0.02), DUSP22 promoter methylation was not associated with schizophrenia status (B = −0.79, p = 0.223). Smoking was dealt with in the case-control brain samples by adjusting the methylation data using the first principle components.22 Supplemental data Figs. 1,2, and 4 show the relations of smoking with the variables of interest.
DUSP22 methylation and expression
A correlation between DUSP22 methylation and expression was not present in the brain samples, nor was genetic background associated with DUSP22 expression. In contrast, the schizophrenia patients had significantly lower DUSP22 transcript levels (schizophrenia: N = 60, DUSP22 expression = 2.97, sd = 0.17; controls: N = 96, DUSP22 expression = 2.87, sd = 0.21) (B = −0.09, p = 0.005), also after adjustment for genetic background (B = −0.14, p < 0.001), suggesting transcriptional regulation of DUSP22 by other factors than DNA methylation in the adult brain of schizophrenia patients.
Fibroblast
Depriving fibroblasts from nutrition by withholding 15% fetal bovine serum (FBS) resulted in a significant increase in DUSP22 methylation (Paired Wilcoxon, p-value = 0.049) and an almost two-fold increase in DUSP22 expression after 72 h (Paired Wilcoxon, p-value = 0.019). Analysis of the methylation difference between the famine and control conditions showed no significant difference in response comparing fibroblasts from schizophrenia patients and controls. Removal of one member of the included healthy homozygous twin pair slightly reduced the significance (p = 0.054 or p = 0.063 depending on which member was removed).
Discussion
Genome-wide analysis of DNA methylation in whole blood from a sample of schizophrenia patients (N = 74) and controls (N = 79), where one-third of both groups were exposed in utero to the great famine in China (1959–1961), identified one region in the promoter of the dual specificity phosphatase 22 (DUSP22) gene (Chr6:291687-293285) with significantly higher DNA methylation levels in famine-exposed schizophrenia patients compared to unexposed patients and healthy controls. In this sample, patients also had significant hypermethylation independent of famine exposure suggesting that DUSP22 hypermethylation is primarily involved in schizophrenia. In an independent but unexposed Dutch sample, a similarly unbiased genome-wide analysis of whole-blood DNA identified the same hypermethylated DUSP22 region in schizophrenia patients (N = 15) as compared to healthy individuals (N = 49). In the postmortem tissue from the PFC, significant DUSP22 hypermethylation was also found in schizophrenia patients (N = 91) compared to unaffected individuals (N = 123). Support for a direct relationship between famine exposure and DUSP22 methylation was obtained by depriving the fibroblasts of schizophrenia patients (N = 5) and controls (N = 5) from nutrition. DUSP22 methylation and expression significantly increased in response to nutritional deprivation.
The hypermethylated region in DUSP22 encompasses a CpG island in the promoter as well as the histone marks H3K27ac and H3K4me3 that are indicative of active gene transcription in both PFC brain and blood cells.23,24 This region contains ten CpG loci and showed clustering (banding) of the methylation levels indicative of genetic regulation. In the postmortem brains, we identified 69 SNPs on chromosome 16 with highly significant associations with DUSP22 methylation, which included previously reported mQTLs in blood17 and brain.16 This interaction between the DUSP22 DMR on chromosome 6 and the SNPs on chromosome 16 (Chr16:34190042-46441560) is consistent with new data of Hi-C proximity maps from human postmortem brains as well as previously published Hi-C data on human lymphoblastoid cells.19 These data highlight the importance of SNPs that physically interact in three-dimensions (3D) with chromatin and influence target transcript levels.14 The identified mQTLs are not associated with schizophrenia in the most recent GWAS meta-analysis4 suggesting that these genetic variants are not primary risk alleles for schizophrenia.
The biological relevance of DUSP22 methylation could not be substantiated by a correlation between DUSP22 methylation and expression in the brain samples, nor was genetic background associated with DUSP22 expression. In the brain samples, the schizophrenia patients had significantly lower DUSP22 transcript levels uncorrelated with DUSP22 methylation and genetic background suggesting that in the adult brain of schizophrenia patients, transcriptional regulation of DUSP22 is independent of DNA methylation or that other factors interfere with the relationship between DUSP22 methylation and expression. There are several factors that may confound the reported relationships, and although we provided replication and have investigated several potential confounders including smoking, the relatively small sample sizes and residual confounding remain a limitation. Also urban origin was defined by the current dwelling, and it is possible that it is not an accurate reflection of the birthplace.
The results of this series of experiments suggest that altered transcriptional regulation of DUSP22 in response to famine is a schizophrenia susceptibility factor. DUSP22 is a recently identified Dual Specificity Phosphatase. Studies on the hippocampus of patients with Alzheimer’s disease have linked promoter hypermethylation of DUSP22 to changes in TAU phosphorylation,25 which in turn has been connected to nutritional deprivation.26 A further link between DUSP22 methylation and nutrition is supported by trial data showing that low DUSP22 DNA methylation at baseline predicted high weight loss in response to a dietary intervention.27
Notwithstanding these pre-existing links between DUSP22 methylation and nutrition, the presence of higher DUSP22 methylation in the blood and brain of schizophrenia patients not exposed to famine also suggest that aberrant DUSP22 methylation is more generally involved in the neurodevelopmental processes underlying the etiology of schizophrenia. The absence of any signal of DUSP22 methylation in recent large EWAS studies in schizophrenia28–32 and the absence of an association with the genetic variation associated with these methylation differences4 points out that DUSP22 is not a primary risk gene for schizophrenia. Instead the evidence from this and other studies point to a role of DUSP22 methylation in regulating responses to a variety of environmental stressors33–36 that may deviate early developmental processes of the brain.37,52 It is possible that such vulnerability in combination with particular environmental insults increase the risk of schizophrenia in a subgroup. In support, a recent study by Vitale et al.38 showed that one of the CpG in our DMR (cg11235426, Chr6: 292522) was differentially methylated in induced pluripotent stem cells (iPSCs) from schizophrenia patients as compared to controls (logFC = −2.44, FDR = 0.04) and in schizophrenia patients prenatally exposed to diethylstilbestrol (DES), DUSP22 methylation (Chr6:291687-293331) was higher compared to exposed controls (unadjusted p-value = 0.00018).39
Further, of note is the strong trans genetic regulation that stretches over 30 Mb on a different chromosome that coincides with chromosome-chromosome interactions.19 The data fit a model where genetic background determines environmental susceptibility of the DUSP22 gene and schizophrenia risk. Famine changes the epigenetic regulation of the DUSP22 promoter in those that are genetically vulnerable. This putative epigenetic mechanism of early-life environmental influences on brain development is likely to be important, not only for understanding the etiology of schizophrenia, but also because it opens new perspectives on mechanisms of gene–environment interactions.
Material and methods
Chinese famine sample
The Chinese famine started suddenly in 1959 as the result of agriculture reforms by Mao and lasted until 1961. Whereas the onset was sudden, the exact end dates vary by geographical location due to a subsequent drought that affected the northern provinces.40 In collaboration with the University in Changchun in the Northern Province of Jilin, we included schizophrenia patients and healthy controls that had been exposed to famine within the first 3 months of gestation based on a birth date between January 1960 and September 1961. All participants gave written informed consent. In order to balance the potential influence of urban background,5 recruitment was stratified for rural and city hospitals. A total of 74 schizophrenia patients and 81 healthy controls approximately matched for famine exposure were assessed. Information on medication was obtained using a structured interview. Diagnosis was made using a full psychiatric evaluation according to DSM IV criteria by licensed psychiatrists. Patients diagnosed with schizophrenia according to DSM IV included those with 295.x Schizophrenia, 295.4 Schizophreniform disorder, 295.7 Schizoaffective disorder, 297.1 Delusional disorder, but excluded 298.8 Brief psychotic disorder, 297.3 Shared psychotic disorder, 293.x Psychotic disorder due to a medical condition, 293.x Substance-induced psychotic disorders, and 298.9 Psychotic disorder not otherwise specified. The absence of a mental health disorder in the healthy controls was assessed by a Chinese translation of the Mental Health Screening Form-III (MHSF-III).41,42 If more than 10% of the patients used a specific medication type, we investigated its association with DNA methylation (using the first principal component of the methylation measures). Based on this criterion, clozapine (n = 37) and chlorpromazine (n = 26) use were examined as potential confounders in the Chinese famine discovery sample by analyzing the correlation with main determinants (famine and diagnosis) and outcome (methylation). Smoking was addressed similarly as to Hannon et al.43 whereby a smoking proxy was calculated based on DNA methylation values for CpGs previously associated with smoking.20,21 In cases where these correlations were significant and changes in the coefficient were larger than 10%, the variable was considered a potential confounder and models were adjusted by including it as covariate (see Extended Data Fig. 1). Based on their correlation with general DNA methylation levels (Extended Data Fig. 2), covariates included: age, gender, the first two DNA methylation-based ancestry principal components as well as the cell-type proportion estimates based on the Houseman algorithm.44 We separately investigated the potential role of urban versus rural background by including this as covariate in the analysis. We excluded one sample based on gender mismatch with the gender prediction from DNA methylation levels (one non-famine-exposed healthy control) and one sample based on the use of insulin (one famine-exposed healthy control).
Case-control blood samples
Whole-blood DNA samples were collected in 2016 from 15 schizophrenia patients (9 male, mean age = 40.1, sd = 13.8) and 49 healthy controls (4 male, mean age = 35.9, sd = 17.0) at the University Medical Center, Utrecht, The Netherlands. Participants were of Dutch origin with three or more Dutch grandparents. Patients were outpatients from the University Medical Center. Eligibility was assessed by their treating psychiatrist and inclusion was done by research staff (LH). Healthy controls were recruited in the general population using online and paper advertisements. All participants gave written informed consent. Diagnosis of schizophrenia (295.x) (N = 11) or schizoaffective disorder (295.7) (N = 4) according to DSM IV criteria was verified in medical records, supplied by the treating physician or established with the Structured Clinical Interview for DSM-IV (SCID).45 In the healthy controls, the absence of a DSM-IV diagnosis was assessed with the Mini International Neuropsychiatric Interview (MINI) plus46 by at least one well-trained rater. Medication use was collected using a self-report questionnaire. All schizophrenia patients were on a stable (at least 1 month) dosing schedule of psychotropic medication.
Case-control brain samples
We investigated DUSP22 DNA methylation, mRNA, and genotype in 214 adult postmortem dorsolateral PFC samples of patients with schizophrenia according to DSM-IV (N = 91, 50 male, mean age = 52.6, sd = 5.2) and unaffected controls (N = 123, 82 male, mean age = 45.9, sd = 16.8) from the Lieber Institute.22 All participants gave written informed consent. A majority was Caucasian (Patients: N = 53, 27 male, age = 50.4 ± 15.6; Controls: N = 62, 47 male, age = 46.4 ± 17.4) but a sizable proportion (46%) was of Afro-American descent (Patients: N = 38, 23 male, age = 55.5 ± 14.2; Controls: N = 61, 35 male, age = 45.3 ± 16.4). Postmortem diagnosis according to DSM-IV was obtained when two board-certified psychiatrists reached consensus after reviewing data from as many sources as possible (i.e., multiple psychiatric records, police reports, neuropathology reports, medical examiner’s information, toxicology screen, postmortem family interview).47 Medication was assessed via a chart review and/or toxicology on brain tissue.22
Fibroblast cell lines
Fibroblast cell lines were established using skin biopsies from five schizophrenia patients (2 male, mean age = 39.0, sd = 10.3) and five age-matched healthy controls (1 male, mean age = 38.4, sd = 7.0). All participants gave written informed consent. One healthy control donor (control 6) turned out to be the homozygous co-twin of another (control 8). All participants were Dutch and had three or more Dutch grandparents. Schizophrenia (295.x) was diagnosed according to DSM-IV as established using the Comprehensive Assessment of Psychiatric symptoms and History (CASH).48 The mental health status of healthy controls was checked using the MINIplus interview.46
General analysis
All data were obtained after written, informed consent from all participants and local medical ethical approval. This research was conducted in accordance with all relevant guidelines and procedures, and the work was approved by the University Medical Center medical ethical review board. Statistical analyses were carried out using R version 3.2.3.49 For DNA methylation, β values (the ratio between methylated and unmethylated probe intensities as a measure of methylation percentage) were used for graphical display and reporting because they are more intuitive, but analyses were carried out using M-values (log2 of β values), which have better statistical validity,50 but give similar results.
Genome wide analysis of DNA methylation
All details of quality control, batch effect removal and analysis are reported in the supplemental materials. In the Chinese famine and the case-control blood sample, quality control included filtering for detection of p-values, low bead count, cross hybridizing- and non-autosomal probes. Normalization was done using functional normalization as implemented in the minfi R package.51 Batch effects were limited as a result of the distribution of the samples of the array; we only removed a small remaining batch effect for position. Methylation levels were adjusted for cell-type composition estimates derived using the Houseman algorithm.44 Linear regression was used to identify associations of single CpG with famine and schizophrenia and the bumphunter algorithm52 for differential methylated regions (details in supplement).
The quality control and analysis of the brain case-control sample were described previously22 and include adjustment for neuronal proportion and adjustment for technical batches.
Banding and trans genetic regulation of DUSP22 methylation
At the DUSP22 DMR, there were different DNA methylation bands in all four population samples: the Chinese famine sample, the case-control blood sample, the blood genomics sample, and the brain case-control sample (see Fig. 1). Measurement errors due to genetic variants in cis that can interfere with hybridization13,53 were ruled out by showing that five out of 12 SNPs that are located on the DNA sequence of the DUSP22 DMR in the dbSNP142 database and that were available in both the MRS and brain case-control samples (rs860102, rs3734780, rs117766562, rs9503164, and rs148619589) had a minor allele frequency (MAF) <5% and were not associated at the 5% level with DUSP22 DMR methylation in either the blood genomics samples or the brain case-control sample. Therefore, we did not examine these cis-SNPs any further. To examine the influence of genetic background on DNA methylation levels, we investigated the association of the mean methylation of the ten CpG in the DUSP22 DMR with genotype. Moreover, we used finite mixture modeling as implemented in the mixtools R package to derive the underlying genetic background based on the mixture of three distributions in the methylation levels of the DUSP22 DMR. In all samples, each participant was allocated to one of the three “genetic” groups based on the level of methylation. To investigate the potential confounding influence of genotype, we used membership to the methylation distribution (1,2,3) as ordinal indicator in χ2 tests, logistic regression, or linear regression as appropriate (see supplement 1). For the stratified analysis by genotype levels (excluding participants with membership to low methylation genetic background), genetic background was not added to the models.
Analysis of expression in brain and blood
Details of the analysis of the relationship between DUSP22 methylation and expression of transcript levels can be found in the supplement. In short, we analyzed the association between DNA methylation levels of the DUSP22 DMR and DUSP22 expression and adjusted for the underlying genetic background.
In situ Hi-C data from human postmortem brain tissue
Flash frozen postmortem brain tissue was obtained from the Human Brain Collection Core (HBCC) of the National Institute of Mental Health (NIMH), US. The anterior cingulate cortex sample used in this study is from a 35-year-old non-psychiatric female. Nuclei isolation through extraction, purification, and fluorescence-activated nuclear sorting (FANS) was performed with minor adjustments per Kundakovic et al.54 (see supplement).
Nutritional deprivation of fibroblasts
Details of the procedures and analysis are available in the supplemental materials. In short, fibroblasts from ten donors were cultured in Minimum Essential Medium (MEM) (Gibco®) with or without 15% FBS to mimic famine. DNA and mRNA were extracted and analyzed using EPIC methylation arrays and qPCR. Non-parametric analysis of average methylation of all DUSP22 CpGs for the paired observations of all ten donors for the 72-h nutritional deprivation compared to the condition with FBS was done using a paired Wilcoxon Signed Rank Test.
Electronic supplementary material
Acknowledgements
We are grateful to all participants in our study. We thank Roel de Rijk for his help with genotyping; Ruben van het Slot and Bobby Koeleman for their help with the EPIC arrays; Marc Bohlken and Annet van Bergen for helping with fibroblast collection. Leonard Schalkwyk kindly provided advice regarding R coding issues. Beijing Gene-Square Square Biotech Ltd. facilitated the methylation measurement of the Chinese samples. Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org) hosted by SURFsara and financially supported by the Netherlands Scientific Organization (NWO 480-05-003 PI: Posthuma) along with a supplement from the Dutch Brain Foundation and the VU University Amsterdam. This project was facilitated by the EpiChemBio COST Action CM-1406. The funders had no role in the design and reporting of the study.
Author contributions
M.P.B., Z.X., and R.S.K conceived the study. M.P.B., L.C.H., G.U., A.X.M., P.R., and A.E.J. conducted the statistical analysis, Q.Y., H.X., Y.W., J.P.S., H.E.H.P., J.E.K., D.G.B., C.H.V., C.M.N., Y.H., and J.Y. collated the data, Y.H., L.D.W., P.R., and E.S. carried out the laboratory analysis, L.D.W., E.M.H., B.P.F.R., S.A., C.H.V., D.R.W., J.Y., and R.S.K. oversaw the methodology, M.P.B. and L.C.H. wrote the first draft. A.E.J. provided statistical advice. All authors contributed to and approved the final manuscript.
Data Availability
The DNA methylation datasets generated during and/or analyzed during the current study are available in the GEO repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116380
Clinical data are not available due to ethical restrictions.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies the paper on the npj Schizophrenia website (10.1038/s41537-018-0058-4).
References
- 1.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature511, 421–427 (2014). [DOI] [PMC free article] [PubMed]
- 2.Van Os J, et al. Identifying gene-environment interactions in schizophrenia: contemporary challenges for integrated, large-scale investigations. Schizophr. Bull. 2014;40:729–736. doi: 10.1093/schbul/sbu069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Susser E, et al. Schizophrenia after prenatal famine. Further evidence. Arch. Gen. Psychiatry. 1996;53:25–31. doi: 10.1001/archpsyc.1996.01830010027005. [DOI] [PubMed] [Google Scholar]
- 4.St. Clair D, He L. Rates of adult schizophrenia following of 1959–1961. JAMA. 2005;294:557–562. doi: 10.1001/jama.294.5.557. [DOI] [PubMed] [Google Scholar]
- 5.Xu MQ, et al. Prenatal malnutrition and adult schizophrenia: further evidence from the 1959–1961 Chinese famine. Schizophr. Bull. 2009;35:568–576. doi: 10.1093/schbul/sbn168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Susser E, St Clair D. Prenatal famine and adult mental illness: interpreting concordant and discordant results from the Dutch and Chinese Famines. Soc. Sci. Med. 2013;97:325–330. doi: 10.1016/j.socscimed.2013.02.049. [DOI] [PubMed] [Google Scholar]
- 7.Roseboom T, de RS, Painter R. The Dutch famine and its long-term consequences for adult health. Early Hum. Dev. 2006;82:485–491. doi: 10.1016/j.earlhumdev.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Heijmans, B. T. et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl Acad. Sci. USA105, 17046–17049 (2008). [DOI] [PMC free article] [PubMed]
- 9.Tobi EW, et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat. Commun. 2014;5:5592. doi: 10.1038/ncomms6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C, Lumey LH. Exposure to the Chinese famine of 1959-61 in early life and long-term health conditions: a systematic review and meta-analysis. Int. J. Epidemiol. 2017;46:1157–1170. doi: 10.1093/ije/dyx013. [DOI] [PubMed] [Google Scholar]
- 11.Tobi EW, et al. DNA methylation as a mediator of the association between prenatal adversity and risk factors for metabolic disease in adulthood. Sci. Adv. 2018;4:eaao4364. doi: 10.1126/sciadv.aao4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker, J. Hungry Ghosts: China’s Secret Famine (Free Press, New York, 1996).
- 13.Boks MP, et al. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS ONE. 2009;4:e6767. doi: 10.1371/journal.pone.0006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemire M, et al. Long-range epigenetic regulation is conferred by genetic variation located at thousands of independent loci. Nat. Commun. 2015;6:6326. doi: 10.1038/ncomms7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Eijk, K. R. et al. Genetic analysis of DNA methylation and gene expression levels in whole blood of healthy human subjects. BMC Genomics. 10.1186/1471-2164-13-636 (2012). [DOI] [PMC free article] [PubMed]
- 16.Hannon, E., Lunnon, K., Schalkwyk, L. & Mill, J. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics10, 1024–1032 (2015). [DOI] [PMC free article] [PubMed]
- 17.Bonder MJ, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 2017;49:131–138. doi: 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 18.Gaunt TR, et al. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016;17:61. doi: 10.1186/s13059-016-0926-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao SSP, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeilinger S, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliott HR, et al. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin. Epigenetics. 2014;6:4. doi: 10.1186/1868-7083-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaffe AE, et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat. Neurosci. 2016;19:40–47. doi: 10.1038/nn.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez-Mut JV, et al. Promoter hypermethylation of the phosphatase DUSP22 mediates PKA-dependent TAU phosphorylation and CREB activation in Alzheimer’s disease. Hippocampus. 2014;24:363–368. doi: 10.1002/hipo.22245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanagisawa M, Planel E, Ishiguro K, Fujita SC. Starvation induces tau hyperphosphorylation in mouse brain: implications for Alzheimer’s disease. FEBS Lett. 1999;461:329–333. doi: 10.1016/S0014-5793(99)01480-5. [DOI] [PubMed] [Google Scholar]
- 27.Moleres A, et al. Differential DNA methylation patterns between high and low responders to a weight loss intervention in overweight or obese adolescents: the EVASYON study. FASEB J. 2013;27:2504–2512. doi: 10.1096/fj.12-215566. [DOI] [PubMed] [Google Scholar]
- 28.Viana J, et al. Schizophrenia-associated methylomic variation: molecular signatures of disease and polygenic risk burden across multiple brain regions. Hum. Mol. Genet. 2017;26:210–225. doi: 10.1093/hmg/ddw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao Y, et al. The DNA methylome and transcriptome of different brain regions in schizophrenia and bipolar disorder. PLoS ONE. 2014;9:e95875. doi: 10.1371/journal.pone.0095875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Numata S, Ye T, Herman M, Lipska BK. DNA methylation changes in the postmortem dorsolateral prefrontal cortex of patients with schizophrenia. Front. Genet. 2014;5:280. doi: 10.3389/fgene.2014.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wockner LF, et al. Brain-specific epigenetic markers of schizophrenia. Transl. Psychiatry. 2015;5:e680. doi: 10.1038/tp.2015.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montano C, et al. Association of DNA methylation differences with schizophrenia in an epigenome-wide association study. JAMA Psychiatry. 2016;73:506. doi: 10.1001/jamapsychiatry.2016.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lountos GT, Cherry S, Tropea JE, Waugh DS. Structural analysis of human dual-specificity phosphatase 22 complexed with a phosphotyrosine-like substrate. Acta Crystallogr. F Struct. Biol. Commun. 2015;71:199–205. doi: 10.1107/S2053230X15000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouyang B, et al. Hypomethylation of dual specificity phosphatase 22 promoter correlates with duration of service in firefighters and is inducible by low-dose benzo[a]pyrene. J. Occup. Environ. Med. 2012;54:774–780. doi: 10.1097/JOM.0b013e31825296bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen AJ, et al. The dual specificity JKAP specifically activates the c-Jun N-terminal kinase pathway. J. Biol. Chem. 2002;277:36592–36601. doi: 10.1074/jbc.M200453200. [DOI] [PubMed] [Google Scholar]
- 36.Rutten, B. P. F. et al. Longitudinal analyses of the DNA methylome in deployed military servicemen identify susceptibility loci for post-traumatic stress disorder. Mol. Psychiatry. 10.1038/mp.2017.120 (2017). [DOI] [PMC free article] [PubMed]
- 37.Pidsley R, et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol. 2014;15:483. doi: 10.1186/s13059-014-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitale AM, et al. DNA methylation in schizophrenia in different patient-derived cell types. npj Schizophrenia. 2017;3:6. doi: 10.1038/s41537-016-0006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivollier F, et al. Methylomic changes in individuals with psychosis, prenatally exposed to endocrine disrupting compounds: lessons from diethylstilbestrol. PLoS ONE. 2017;12:e0174783. doi: 10.1371/journal.pone.0174783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smil V. China’s great famine: 40 years later. BMJ. 1999;319:1619–1621. doi: 10.1136/bmj.319.7225.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiz MA, Peters RH, Sanchez GM, Bates JP. Psychometric properties of the Mental Health Screening Form Iii within a metropolitan jail. Crim. Justice Behav. 2009;36:607–619. doi: 10.1177/0093854809334013. [DOI] [Google Scholar]
- 42.Carroll JFX. Development of the Mental Health Screening Form III. Int. J. Ment. Health Addict. 2008;6:72–76. doi: 10.1007/s11469-007-9102-8. [DOI] [Google Scholar]
- 43.Hannon E, et al. An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 2016;17:176. doi: 10.1186/s13059-016-1041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houseman EA, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.First, M. B., Spitzer, R. L., Gibbon, M. & Williams, J. B. W. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Patient edition with Psychotic Screen (SCID-I/P W/PSY SCREEN). (BiometricsResearch, New York State Psychiatric Insitute, New York, 2002).
- 46.Sheehan DV, et al. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J. Clin. Psychiatry. 1998;59(Suppl 2):22–33. [PubMed] [Google Scholar]
- 47.Lipska BK, et al. Critical factors in gene expression in postmortem human brain: focus on studies in schizophrenia. Biol. Psychiatry. 2006;60:650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 48.Andreasen, N. C., Flaum, M. & Arndt, S. The Comprehensive Assessment of Symptoms and History (CASH). An instrument for assessing diagnosis and psychopathology. Arch. Gen. Psychiatry49, 615–623 (1992). [DOI] [PubMed]
- 49.R Core Team (2017). R: A language and environment for statistical computing. R Foundation forStatistical Computing, Vienna, Austria. https://www.R-project.org/.
- 50.Du P, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aryee MJ, et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jaffe AE, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidemiol. 2012;41:200–209. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bell JT, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kundakovic M, et al. Practical guidelines for high-resolution epigenomic profiling of nucleosomal histones in postmortem human brain tissue. Biol. Psychiatry. 2017;81:162–170. doi: 10.1016/j.biopsych.2016.03.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA methylation datasets generated during and/or analyzed during the current study are available in the GEO repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116380
Clinical data are not available due to ethical restrictions.