Abstract
Genome-scale metabolic network models can be used for various analyses including the prediction of metabolic responses to changes in the environment. Legumes are well known for their rhizobial symbiosis that introduces nitrogen into the global nutrient cycle. Here, we describe a fully compartmentalised, mass and charge-balanced, genome-scale model of the clover Medicago truncatula, which has been adopted as a model organism for legumes. We employed flux balance analysis to demonstrate that the network is capable of producing biomass components in experimentally observed proportions, during day and night. By connecting the plant model to a model of its rhizobial symbiont, Sinorhizobium meliloti, we were able to investigate the effects of the symbiosis on metabolic fluxes and plant growth and could demonstrate how oxygen availability influences metabolic exchanges between plant and symbiont, thus elucidating potential benefits of inter organism amino acid cycling. We thus provide a modelling framework, in which the interlinked metabolism of plants and nodules can be studied from a theoretical perspective.
Introduction
Nitrogen belongs to the elements which are absolutely crucial for life, because it is contained in all amino acids and many other essential biomolecules. Whereas nitrogen is the most abundant element in the earth’s atmosphere, most of it is present in the inert form of nitrogen gas (N2) constituting approximately 80% of the total atmosphere. Therefore, despite its high abundance, nitrogen is often a limiting factor for growth in plants. To overcome this in agriculture, large amounts of nitrogen are applied to the soil in the form of artificial fertilisers to promote plant growth. Before the industrial revolution, farmers applied a crop rotation scheme to their fields and left the field barren or planted it with beans or peas every few years to let the soil ‘recover’. We now know that this strategy re-introduces bio-available nitrogen into the soil. This is due to ability of the planted crops (beans or peas) or plants quickly occupying barren fields, such as clovers, to form a nitrogen fixing symbiosis with rhizobia. Rhizobia carry a gene for the highly oxygen sensitive nitrogenase enzyme, which catalyses the reduction of atmospheric nitrogen to ammonium, and the plants provide the symbiont with an environment protecting it from damaging oxygen. In this way atmospheric nitrogen is introduced into the global organic nutrient cycle. Simultaneously the nitrogenase requires large amounts of energy and reductants to catalyse nitrogen fixation. This has led to the development of a tightly regulated oxygen supply for oxidative phosphorylation by the plant to the symbiont1,2. This symbiosis has seen the most attention and is best understood in plants of the Fabacaea (or Leguminosae) family, to which beans, peas and clovers belong3–7. In recent years, Medicago truncatula has become a model plant for the legume-rhizobia symbiosis8. While there is a plethora of rhizobial strains, only a limited number interact which each host9,10. Sinorhizobium meliloti is a well-studied symbiont of M. truncatula11 and therefore an optimal organism to model this symbiosis. Low nitrogen availability will trigger the recruitment of rhizobia to the plant roots and initiate the nodulation, in which rhizobia invade the plant root and nodules are formed. In these nodules, the rhizobia are taken up by plant cells, surrounded by a membrane and differentiate into bacteroids. Upon completing differentiation they begin fixing nitrogen, which is made available to the plant primarily in the reduced form of ammonium. In return, the plant provides organic acids as nutrients to the rhizobia4,12. In addition there is evidence that amino acid cycling is essential for nitrogen fixation at least in some rhizobial strains13,14. Obviously, the mutual dependence of the metabolism of plant and rhizobium is crucial for this symbiotic interaction. During the establishment of the nodules, the metabolic fluxes in both plant and rhizobium change drastically15.
To explore, understand and analyse the changes in the metabolic fluxes during nodulation, computer simulations have become increasingly established in modern biology research. For this, detailed high quality metabolic network reconstructions have to be established. Whereas there exist now over a hundred16,17 genome-scale metabolic network reconstructions for a wide range of organisms, and a number of steps in the development of these models can now efficiently be automated18,19, the reconstruction of genome-scale metabolic networks still involves numerous manual curation steps and therefore is very time-consuming20. However, even before a reconstruction is finished and established simulation techniques, such as flux balance analysis (FBA) can be employed, the building process itself provides considerable insight into the metabolic capabilities of the investigated organism and allows to refine its genome annotation. Once a highly curated metabolic network reconstruction is established, it presents a theoretical framework allowing to query the system and understand its functional properties. The numerous possible theoretical investigations21 are for example useful to help understanding the structure and regulation of metabolic networks22,23, identify essential genes24, predict putative drug targets25 or support engineering of novel pathways26 producing desired compounds of technological or economic interest. In the plant sciences, genome-scale models with different degrees of accuracy and completeness exist for the model unicellular green alga Chlamydomonas reinhardtii27, the model species Arabidopsis thaliana22,28, rice Oryza sativa29 and others30,31. While initially plant models were unicellular representations of metabolism, not distinguishing between different tissues, recent efforts tend to refine these models and generate multi-tissue representations32. While the network structure and catalysed reactions are now fairly well established and can be extracted using the genome annotation, most enzyme kinetics are still unknown. This makes the use of ODE based kinetic models impossible on genome-scale networks, as thousands of parameters would have to be estimated. FBA is a modelling technique that addresses this issue by imposing a steady state assumption on the fluxes, indicating that there is no change of internal metabolite concentrations in the network33. FBA further imposes an objective that the model should optimise (e.g. minimal effort to produce a compound or maximisation of biomass34), which yields a unique optimal value. Thus the network can be investigated for the effects of a changing environment without the need for extensive parameter fitting and optimisation. Unfortunately, the flux distribution itself (in contrast to the optimal value) is not necessarily unique in FBA35. This can however be addressed by using minimisation of the overall quadratic flux, which represents a solution with efficient enzyme usage, for a given optimal FBA value, at which point the flux distribution becomes unique.
In this paper, we focus on the presentation of a highly curated genome-scale metabolic reconstruction of M. truncatula and provide an analysis of general metabolic properties of its biochemical reaction network. For our detailed and curated reconstruction, which contains 1636 reactions in 7 compartments, we have developed an extended approach to assign metabolic reactions to sub-cellular compartments. Our approach integrates not only sequence and proteomics derived localisation information36,37 but also integrates known importers and exporters, thus taking advantage of an additional source of information. This information is integrated using the concept of network extension introduced in Christian et al.38. All reactions in our reconstruction were manually curated to achieve complete mass- and charge-balance as suggested in Thiele and Palsson39. This important property of a network allows for a system wide prediction of fluxes of protons and other charged particles over cellular membranes. To our knowledge, we present the first plant metabolic model, for which charge balancing is ensured on genome scale, while this was previously only achieved for small subsystems22 or core metabolism40,41. Charge balancing over membranes is a key prerequisite to realistically describe electron transfer chains required for ATP biosynthesis. Compartmentalisation allows for a more precise distinction between the function and necessity of isozymes which are present in multiple compartments. Omitting this information would allow the model to use reactions for which substrates are not available, because they are not transported between compartments. This would make the prediction of potentially lethal mutations less reliable. We further refined the model by integrating tissue specific information to better represent the distribution of metabolism to root and shoot of the plant.
We discuss insights gained from the reconstruction process itself and their consequences on the annotation of genes with previously unknown functions and the refinement of annotations of previously insufficiently annotated genes. We experimentally obtained tissue specific biomass data of M. truncatula, thus providing for the first time a system wide in-depth biomass composition for this plant. To investigate the interaction between plant and microbe, this highly curated plant model was connected to a model of Sinorhizobium meliloti, derived from the MetaCyc database19, and we investigated the effect of nitrogen fixation and symbiosis on the plant model. The presented model aims at simulating the nitrogen fixing state of the symbiosis and does not reflect the process of forming the symbiosis. We further addressed the question which fluxes might be most restrictive for nitrogen fixation and investigated the effect of small alterations of oxygen supply on the symbiotic nitrogen fixation capacity. The model presented in this paper provides an important resource for researchers in plant sciences in general and for studying plant-rhizobia interactions in particular. Our simulations help to interpret and understand responses of plant metabolism, which include rearrangements of metabolic fluxes and changes in energy and reductant requirements, as a result of changes in nitrogen availability.
Results
Experimental biomass Analysis leads to suggested gene annotation update
During the biomass analysis of M. truncatula (details can be found in Supplementary Table 5) we found pinitol in the plant. When trying to determine the biosynthetic pathway for this substance we found that the enzyme encoded by gene MTR_4g038440 (Entrez Gene ID: 11446905), currently annotated as a caffeic acid 3-O-methyltransferase, is very similar (87% sequence identity) to the inositol methyltransferase from Glycine max (Entrez Gene ID: 100812768). Since we could not find other candidates for this biosynthetic step, we expect that this enzyme might indeed be mis-annotated and would suggest a revision of its annotation.
A multi-tissue model of Medicago truncatula
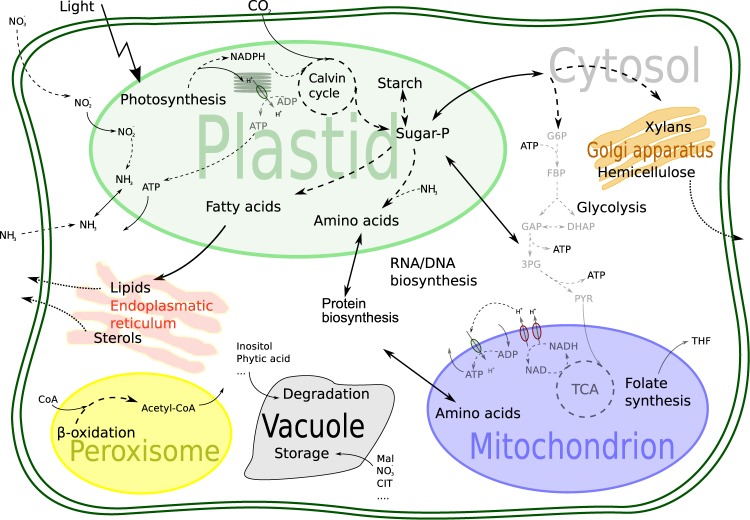
We created a genome scale model of Medicago truncatula encompassing 3403 genes coding for 2909 reactions in 8 compartments using 2780 metabolites. An overview of the model is provided in Fig. 1 and a more detailed description of this network can be found in the Supplementary Text 7.
Figure 1.
Schematic overview of the metabolic processes and compartments included in the genome-scale model of M. truncatula.
This genome-scale model encompasses our knowledge of biochemical reactions catalysed by enzymes, which are encoded in the genome along with additional necessary reactions to fulfil known functions. However, it is unable to describe processes in a particular cell type or part of the organism, as no such separation is yet included. We therefore constructed a multi-tissue model of Medicago truncatula using data from Benedito et al.42 employing the FASTCORE43 algorithm (see Materials and Methods for details).
The shoot tissue of this model was represented by 969 reactions associated with a total of 1824 genes acting on 860 metabolites and containing 126 internal transporters. The root tissue in turn consists of 958 reactions associated with a total of 1804 genes acting on 846 metabolites and including 122 transporters. These submodels were connected by 32 inter-tissue transporters and a combined biomass reaction, with the inter-tissue transporters derived from the literature32,44,45. The large reduction can partly be explained by many reactions in secondary metabolism which, in the current model formulation, are unable to carry flux, because specific exchange reactions have not been added, and because concentrations of those secondary metabolites are too small to be detected during biomass analysis. In total, the consistent part of the Medicago truncatula network contained only 1303 reactions, which are able to carry non-zero flux with the current configuration of exchange reactions, while all remaining reactions can be activated if additional exporters are included. The two-tissue model was able to sustain growth during day (using light as energy source and photosynthesis for carbon) and night (with starch as energy and carbon source). In addition, both ammonium and nitrate can be used as nitrogen sources to support growth under day and night conditions.
Computational analysis
Differences between day and night
We investigated the model under day and night conditions to determine differences in network usage under these conditions. In silico knockout experiments predict that during night conditions, 236 reactions are essential (including 26 transporters). Under phototrophic growth, 243 reactions are essential (25 transporters). In total, 232 reactions are essential under both conditions. Starch degrading reactions are essential only under night conditions, while those involved in the Calvin cycle and photosynthesis are essential only in light conditions. Interestingly, the additional transporter predicted as essential under dark conditions is the maltose exporter in the chloroplast envelope (mex). However, mex mutants are viable, albeit with a reduced growth and starch excess phenotype46. The reason that the model predicts this transporter as essential is that, in the strict mathematical sense, no flux distribution exists which fulfils the stationarity condition for maltose. Therefore, the prediction can be understood in terms of the mathematical model formulation, and the predicted phenotype (maltose accumulation) is one of the key characteristics of the mex mutant.
Nitrogen metabolism in the two tissue model
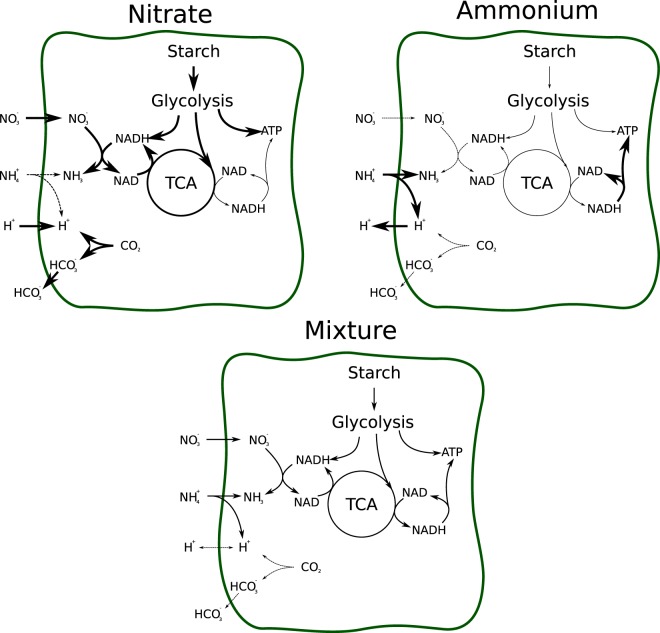
Since M. truncatula is a model organism for symbiotic nitrogen fixation, the effects of different forms of available nitrogen are of high relevance. The main sources for nitrogen are ammonium and nitrate. However, supplying both of these nitrogen sources and directly applying either flux minimisation or carbon usage minimisation would lead to the trivial result that the model selects ammonium as the source, because it is ‘cheaper’ in terms of energy and reductant (see Supplementary Text 7 and Supplemetal Dataset 1). To simulate a transition, we therefore restricted the amount of nitrogen available to the plant and scanned over a range of different ammonium vs nitrate compositions. To calculate the fluxes, we fixed the growth rate to 0.1 g/gDW per day and minimised the sum of the squares of all fluxes in the system. Figure 2 depicts different conditions of pure ammonium, mixed and pure nitrate provision.
Figure 2.
Fluxes changing with alternating nitrogen sources. Nitrate nutrition requires large amounts of reductants produced by the TCA and leads to an export of negative charges (via HCO3−). Ammonium nutrition is energetically cheaper, and allows the use of reductants for ATP synthesis, while simultaneously leads to a export of protons from the plant.
As expected the amount of required starch drops with increasing ammonium availability. At the same time, the TCA activity is reduced, as less reductant is required for nitrate reduction. Simultaneously the mitochondrial ATPase activity increases, using parts of the freed reductant. The model also indicates soil acidification during ammonium nutrition due to a high export of protons. Under mixed nutrition, this export is balanced since charge and protons are balanced when a equimolar amount of nitrate and ammonium are imported. Under pure nitrate nutrition, this exchange is actually reversed, and protons are taken up while simultaneously HCO3− is released. While a large fraction of these exported charges will in nature be balanced by the uptake of either positive or negative ions47, this computational results still nicely illustrates and at least partially explains the acidifying properties of ammonium nutrition in contrast to nitrate nutrition48.
Symbiotic nitrogen fixation
We first used the combined model (see the schematic in Fig. 3) to test under which conditions a symbiotic association with S. meliloti is beneficial to the host M. truncatula.
Figure 3.
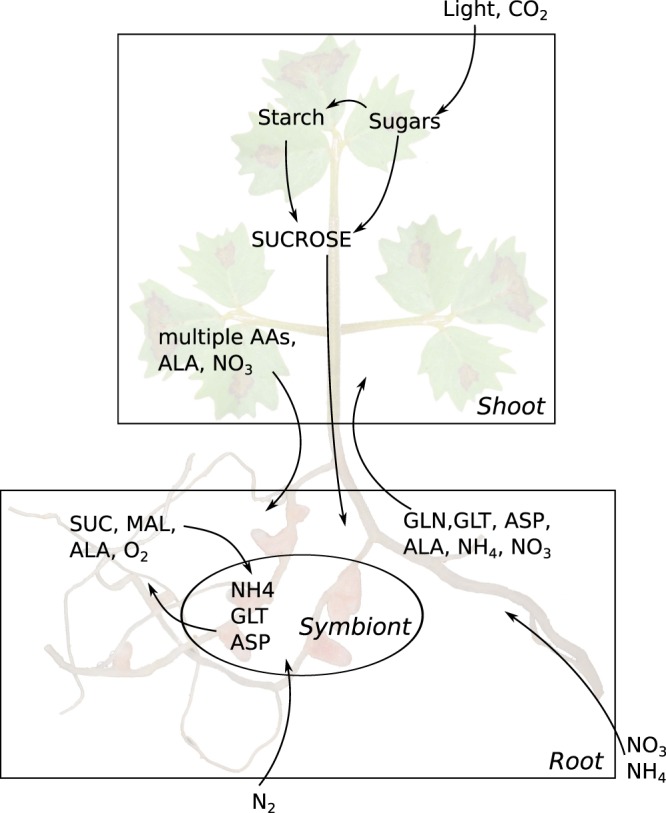
Layout of the two-tissue model combined with its symbiotic partner. The most important exchanges between the tissues and between plant and symbiont are shown. Original figure (here in background) by Ninjatacoshell (https://commons.wikimedia.org/wiki/File:Medicago_truncatula_A20_root_nodules.JPG), “Medicago truncatula A20 root nodules”. Removed the black background and used as part of the background of the combined work by Thomas Pfau, https://creativecommons.org/licenses/by-sa/3.0/legalcode.
For this, we compared the maximal predicted growth rate of the models with and without nitrogen fixing symbiont, for different external ammonium concentrations (see Fig. 4).
Figure 4.
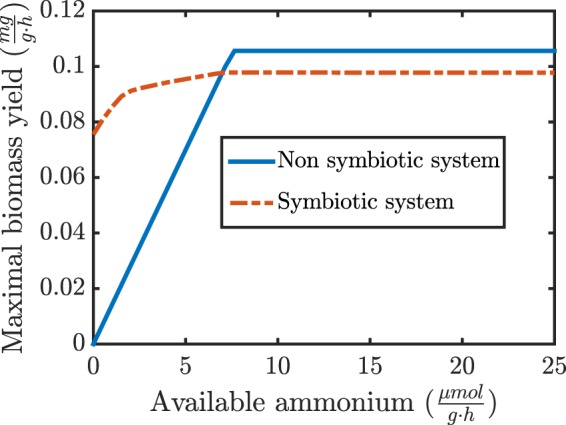
Comparison of the maximal growth of the symbiotic and non-symbiotic system. While the symbiotic system can grow without available ammonium by fixing nitrogen, this advantage necessitates a maintenance of the rhizobial symbiont, which reduces the energy available to the plant, leading to a lower maximal growth when sufficient ammonium is available.
Clearly, the growth rate is slightly lower for the symbiotic system if sufficient nitrogen is available. This can be explained by the additional energy requirement to produce organic carbon to support the symbiont. However, under low nitrogen availability (in the form of nitrate or ammonium) the ability to fix nitrogen by the rhizobium allows for plant growth even without any external nitrogen source present. Thus, this combined model can explain and to some extent quantify under which conditions a symbiotic relationship is advantageous.
There are multiple propositions in the literature concerning the transport of nitrogen from the rhizobium to the plant13,49,50. An initial investigation showed that the amount of oxygen available to the bacteroids is the main limiting factor for the amount of nitrogen that can be fixed. This is expected, because the nitrogenase requires large amounts of energy, which is most efficiently provided by oxidative phosphorylation. However, while a higher oxygen concentration provides more energy to the bacteroid, the nitrogenase complex becomes irreversibly inactivated if oxygen concentrations are too high51. Therefore the plant actively regulates the oxygen supply using leghemoglobin, ensuring sufficient oxygen for fixation, but minimising detrimental effects to the nitrogenase activity2. As such, it is interesting to see what effects slightly changing oxygen amounts would have on the symbiosis. While an increased potential biomass production is the most obvious effect of a better oxygen supply it is also interesting to investigate how the exchange fluxes between bacteroid and plant are affected if the amount of fixed nitrogen remains constant, but oxygen concentrations varies. For this, we fixed the growth rate of the plant to 80% of the maximal growth rate (determined above), and systematically varied the oxygen availability for the bacteroid and calculated the fluxes based on the assumption that enzymes are used in a maximally efficient way (see Methods).
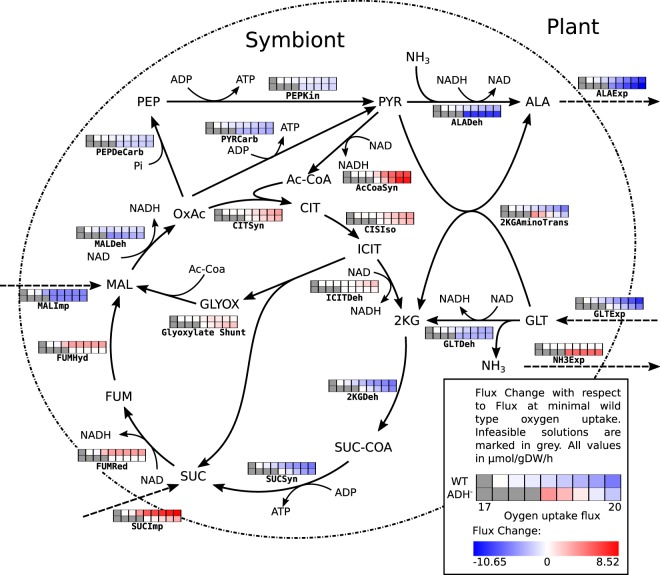
The model predicts extensive amino acid cycling between the plant and the symbiont (see Fig. 5) in agreement with earlier findings13. Alanine is predicted to be the only nitrogen-containing export product. The amino group of glutamate is transferred by transaminases to pyruvate, yielding the export product alanine and ketoglutarate. The latter is fed into the TCA cycle, where one ATP is generated by succinyl-CoA synthase. Newly fixed nitrogen is exported as alanine, which is synthesised de novo from pyruvate by alanine dehydrogenase. Pyruvate in turn is produced by reverse action of pyruvate carboxylase. The standard Gibbs free energy of reaction of pyruvate carboxylase is ΔG0 = −1.37 kcal/mol (according to MetaCyc), but due to the comparatively large amounts of dicarboxylic acids and the high demand for ATP it appears plausible that the equilibrium can easily be shifted to pyruvate. The imported carbon source required for pyruvate synthesis depends strongly on the available oxygen. For low oxygen supply, the capacity of the respiratory chain is limited and import of malate minimises the amount of produced NADH. With increasing oxygen availability, ATP production by oxidative phosphorylation is increasingly efficient. This explains the observed switch from malate to succinate uptake, because introduction of succinate into the TCA cycle provides one additional reductant as compared to malate. Therefore succinate is only used when sufficient oxygen is available (or when no malate is provided).
Figure 5.
Changes of fluxes for changing oxygen uptake by the symbiont. Grey values indicate no feasible solution (i.e. insufficient oxygen for sufficient nitrogen fixation). The upper row corresponds to wild type solutions while the lower row represents alanine dehydrogenase knockout. NH3 export is only active in the mutated strain, while all nitrogen is exported as alanine in the wild type simulation (marked in orange, as the export does not change with oxygen in both conditions). With increasing oxygen availability, succinate is preferentially used as carbon source as it provides additional reductant compared to Malate. Amino acid shuttling is reduced with increasing oxygen in both conditions, due to the increased potential of complete respiration of carbon. Under oxygen limiting conditions, pyruvate is generated instead of citrate, which can either be converted to alanine (wild type), consuming reductant, or can be used as acceptor for the amino group from glutamate.
In our simulation alanine was the only nitrogen export product from the rhizobia. However, there have been experiments in which alanine dehydrogenase was knocked out, showing that de novo alanine synthesis is not required for symbiotic growth52,53. Our model allows a rapid reproduction of this experiment in silico. Our simulations (Fig. 5 and Supplementary Text 7) show, even under these conditions, a substantial export of alanine, which is directly cycled against glutamate. However, in the knock-out simulation, nitrogen is exported from the symbiont in the form of ammonia (increased flux compared to WT in Fig. 5), which is subsequently assimilated in the plant. In the knock-out conditions, the minimum amount of required oxygen increases (additional grey boxes for low Oxygen supply), which corresponds to a slightly decreased growth, a phenomenon observed by Allaway et al.52 but not by Kumar et al.53. Interestingly, the knock-out simulation does not show any uptake of dicarboxylic acids at minimal oxygen concentrations but relies solely on the use of glutamate as carbon source. This can be explained by the fact that using glutamate as carbon source allows, as described above, the use of succinyl-CoA synthase to regenerate ATP, while simultaneously producing reductants. Thus, without the ability of a de novo alanine production, an optimal ratio of ATP to reductant is provided by pure glutamate uptake. With additional oxygen, the use of succinate as reductant donor and oxidative phosphorylation as ATP source becomes more efficient.
Discussion
We have developed a genome scale compartmentalised model for the clover Medicago truncatula, a model plant for the legume–rhizobia symbiosis. We have carefully verified that our model is thermodynamically feasible, that all relevant intermediates can be replenished, and all reactions are balanced with respect to both mass and charge, which allows the interpretation of results regarding the effect of different types of nitrogen nutrition on the environmental pH. The reconstruction process was useful in its own right, as it resulted in the putative suggestion of a novel gene annotation.
However, new annotations are not the primary objective for building detailed genome-scale metabolic models. More importantly, a genome-scale model provides a theoretical framework in which experimental observations can be interpreted and understood. Computational analyses allow to query the model, assess its capabilities, and enable novel interpretations of experimental observations in a theoretical context. An interesting observation is derived by our calculation of the theoretically maximal conversion rate of starch-derived carbon into biomass at night. Considering only energy and redox requirements for the formation of biomass, a remarkably high percentage of starch carbon (92% for growth on ammonium, 79% for growth on nitrate) can be converted into biomass. These numbers show the theoretical maximal conversion rate of starch carbon into biomass carbon which will likely not reflect the real situation. However, these deliberations do allow an estimation of the non growth associated maintenance costs based on measurements of starch usage and respiration like those by Pyl et al.54. Experiments performed in A. thaliana, in which respiration and starch degradation rates were carefully measured throughout the night54, showed that at least 45% of the starch is respired. This value is still more than twice as high as the predicted minimum of 21% for the growth on nitrate. Moreover, Pyl et al.54 showed that the ratio of respired starch-derived carbon is highly dependent on the night temperature. The lowest value of 45% is observed for low (12 °C) temperatures, while for nightly temperatures of 24 °C (which was the same as the applied temperature during the day), the ratio of respired carbon increased to 75%. Estimating from our calculations that around 20% of carbon contained in starch need to be respired to build biomass, this allows to conclude that between 25% and 55% of starch-derived carbon is respired during the night for maintenance, depending on temperature and probably other external factors. This is in good agreement with previous findings of maintenance requirements of around 40% by Williams et al.55. This comparison is similar to the approach by Cheung et al.56 to estimate the non growth-related maintenance energy. However, while they based their findings on ATP fluxes in a cell culture, we base our estimates on starch consumption in planta. The derived energy is required for processes which are not directly related to growth and not included in our model, such as degradation and repolymerisation of proteins and mRNA, as well as transport processes across intracellular membranes or phosphorylation in regulatory pathways. But why the ratio is so strongly dependent on the ambient temperature remains unclear. To further understand the detailed requirements for maintenance energy, it will be necessary to formulate mathematical models of maintenance requirements based on experimental measurements of protein and mRNA turnover and intracellular transport57–59. The results of such models would allow to define further constraints on genome-scale metabolic models as the one presented here, and lead to a more profound understanding of the regulation of metabolic fluxes responding to environmental changes.
Using only a model of the host of the symbiosis it is already possible to make some investigations into the symbiotic properties. The dominating factor in the symbiosis is the transfer of nitrogen between host and symbiont. We therefore first investigated the response of metabolic fluxes in the host to changing nitrogen sources. If both nitrate and ammonium are abundant, the model predicts an exclusive uptake of ammonium, because integration into amino acids requires considerably less reductants and energy when compared to nitrate. This result is in agreement with the experimental observation that nitrate uptake is inhibited when ammonium is available60. Thus, in an evolutionary context, our model suggests that the reason for this inhibition is an increased energetic efficiency. Simultaneously proton export is observed which is in accordance with soil acidification when ammonium fertilisers are used48. If ammonium becomes limiting, the modelled fluxes change gradually to an increased uptake of nitrate, which is accompanied by a higher energy and reductant demand that is met by increased respiration during the night. The balancing of charges, using both ammonium and nitrate, might contribute to the observation that the presence of ammonium in many plants61 inhibits nitrate uptake but does not turn it off completely, while from an energetic consideration, nitrate should not be used at all. At the same time, we have to be aware of the limitations of FBA when interpreting computational results simulating changes in environmental pH. In particular, ionic barriers commonly employed by plants cannot be modelled in FBA.
By combining the plant network with a rhizobial symbiont, we were able to illustrate the evolutionary advantages in undergoing a symbiosis and to simultaneously show that it can come at a cost when formed in a rich environment. We could demonstrate that the observations of amino acid cycling and alanine as nitrogen carrier fit into a paradigm of efficiency. Further, we were able to suggest that the reason for the use of alanine as export product lies in the ability to remove some surplus reductant from the bacteroid system, but that it is not strictly necessary for symbiotic growth. However, it is interesting that even when alanine dehydrogenase is knocked out, we observed the export of alanine (now exchanged with glutamate), which supports the hypothesis that alanine is a major export product in planta, even without being the nitrogen carrier.
Rhizobia represent only one component of a highly complex and dynamic microbial community associated with plant roots62. In natural environments this community may contain beneficial microbes, delivering nutrients extracted from the soil to the plant in exchange for reduced carbon, and parasites, living on dead or live plant material. In principle, our approach to combine several network models is also applicable to more complex interaction networks63. However, the bottleneck is often our limited knowledge on the exchanged metabolites between the different players. Moreover, by increasing the complexity of the network, also the model complexity and concomitantly the possible model solutions increase dramatically. We therefore view our approach, focusing on a well-investigated model system, in which only two organisms interact, as a first step towards studying more complex interaction networks.
Materials and Methods
For a more extensive description of the Materials and Methods, we refer the reader to the Supplemental Text 7, only a short description will be provided here.
Model Reconstruction
The plant model is based on version 3.5v5 of the Medicago truncatula genome annotation64 and was reconstructed using PathwayTools65 along with the MedicCyc database66. For compartmentalisation, data from multiple databases (TAIR for homologies67, SUBA for localisations68) and recent studies69,70 in M. truncatula where used for initial localisation. Further localisations were assigned using an approach by Christian et al. to make all compartments consistent38. All seeds and targets used can be found in Supplemental Dataset 2. Transporters were added according to information from the literature71–74 and where they were necessary to allow compounds which could be produced in the uncompartmentalised network to be still producible in the compartmented network. Reaction directionalities were adjusted to avoid thermodynamically infeasible cycles.
Generation of a two tissue shoot-root Network
The two tissue model for root and shoot was created using the FASTCORE algorithm43 and gene expression data from Benedito et al.42. This was done by duplicating the model and connecting the tissues similar to an earlier approach32. For all connecting transporters see Supplemental Dataset 3. Finally, models able to grow in day and night conditions using either ammonium or nitrate as substrate were generated from the combined model and all reactions present in one of the networks were added to the final model. Data for ATP Maintenance and growth rate were extracted from De Vries and Penning75 and Lötscher et al.76, respectively.
Building a symbiotic system
A network for Sinorhizobium meliloti 1021 was extracted from MetaCyc. The model was curated to achieve mass and charge balance of all reactions and to allow it to produce most biomass components from an E.coli biomass definition77 in order to ensure a generally viable model. The model was connected to the plant root submodel using exchange reactions present in the literature (for Reviews see e.g12,15,78. and a list of used transporters can be found in Supplemental Dataset 4). Links between rhizobium fluxes and plant fluxes were generated based on existing literature information79–82.
Network analysis
Our simulations are based on the underlying assumption of a metabolic quasi steady state used in Flux Balance Analysis83, where internal concentrations do not change over time:
| 1 |
With S being the stoichiometric matrix with the position (i, j) representing the coefficient of the i-th metabolite in the j-th reaction, and v being the vector of fluxes (i.e. reaction activities). Further we in general assume that the organisms have evolved to perform whichever task they are required with a maximal efficiency. This is represented in our simulations by minimisation of the flux through the network, which corresponds to a minimal necessity for enzyme synthesis. We commonly minimise the quadratic flux values84. In contrast to minimising the simple sum of fluxes, this yields a unique solution34,85:
| 2 |
Additional constraints based on literature (e.g. maintenance ATP, maximal photon availability etc.) are introduced. To test multiple conditions, we solve this system for multiple fixed values of target reactions and compare the fluxes for these ranges. Examples of these scans can be found in the Supplemental Text 7.
Availability of scripts and reconstruction
Matlab scripts that reproduce the scans in Supplemental Text 7 underlying Figs 2 and 5 along with the CPLEX interface for ScrumPy are available online at https://github.com/sysbiolux/MedicagoScripts. The reconstruction is also available on the github site and in Supplementary Dataset 5.
Biomass composition determination
Biomass composition was determined according to adapted protocols by Williams et al.86 and Masakapalli et al.87. Plants were grown following the hydroponic culture protocol from the Medicago truncatula Handbook88. More details can be found in Supplemental Text 7. Data for biomass distribution from89 and90 was used. Cell wall composition was extracted from91,92, and93. The lipid composition was based on94. DNA GC Content was derived from95.
Electronic supplementary material
Acknowledgements
T.P. was supported by the College of Life Sciences and Medicine of the University of Aberdeen and the University of Luxembourg. Biomass determination experiments were funded through the Scottish Universities Life Science Alliance and the College of Life Sciences and Medicine, University of Aberdeen. O.E. and N.C. were supported by the Scottish Funding Council through the Scottish Universities Life Science Alliance SULSA. M.P. and O.E. are supported by the Marie-Curie Initial Training Network AccliPhot, funded by the European Union under the scheme FP7-PEOPLE-2012-ITN (Grant Agreement Number 316427). O.E. is supported by the Cluster of Excellence on Plant Sciences CEPLAS, funded by the Deutsche Forschungsgemeinschaft (EXC 1028). L.S. declared no conflict of interest. S.M. was supported by a postdoctoral fellowship by the European Union FP7 project 222716 SmartCell - Rational design of plant systems for sustainable generation of value-added industrial products.
Author Contributions
O.E., M.P. and T.P. conceived the study. N.C. and T.P. worked on the network compartmentalisation. T.P. and S.M. performed the biomass experiments. T.P. and O.E. performed the computational analysis and network reconstruction. T.P., O.E., N.C. and L.S., wrote the manuscript. All authors edited the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-30884-x.
References
- 1.Appleby CA. Leghemoglobin and rhizobium respiration. Annu. Rev. Plant Physiol. 1984;35:443–478. doi: 10.1146/annurev.pp.35.060184.002303. [DOI] [Google Scholar]
- 2.Ott T, et al. Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. biology. 2005;15:531–535. doi: 10.1016/j.cub.2005.01.042. [DOI] [PubMed] [Google Scholar]
- 3.Brewin NJ. Development of the legume root nodule. Annu Rev Cell Biol. 1991;7:191–226. doi: 10.1146/annurev.cb.07.110191.001203. [DOI] [PubMed] [Google Scholar]
- 4.Ferguson BJ, et al. Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 2010;52:61–76. doi: 10.1111/j.1744-7909.2010.00899.x. [DOI] [PubMed] [Google Scholar]
- 5.Hellriegel, H. et al. Untersuchungen über die Stickstoffnahrung- der Gramineen und Leguminosen (Berlin, Buchdruckerei der “Post” Kayssler, 1888).
- 6.Hirsch, A. M., Lum, M. R. & Downie, J. A. What makes the rhizobia-legume symbiosis so special? Plant Physiol. 127, 1484–1492, http://www.plantphysiol.org (2001). [PMC free article] [PubMed]
- 7.Drevon, J.-J. et al. The legume–rhizobia symbiosis. In Grain Legumes, 267–290 (Springer, 2015).
- 8.Cook DR. Medicago truncatula–a model in the making! Curr Opin Plant Biol. 1999;2:301–304. doi: 10.1016/S1369-5266(99)80053-3. [DOI] [PubMed] [Google Scholar]
- 9.Simsek S, Ojanen-Reuhs T, Stephens SB, Reuhs BL. Strain-ecotype specificity in sinorhizobium meliloti-medicago truncatula symbiosis is correlated to succinoglycan oligosaccharide structure. J. Bacteriol. 2007;189:7733–7740. doi: 10.1128/JB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D, Yang S, Tang F, Zhu H. Symbiosis specificity in the legume–rhizobial mutualism. Cell. microbiology. 2012;14:334–342. doi: 10.1111/j.1462-5822.2011.01736.x. [DOI] [PubMed] [Google Scholar]
- 11.Marx H, et al. A proteomic atlas of the legume medicago truncatula and its nitrogen-fixing endosymbiont sinorhizobium meliloti. Nat. biotechnology. 2016;34:1198. doi: 10.1038/nbt.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Udvardi M, Poole PS. Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant. Biol. 2013;64:781–805. doi: 10.1146/annurev-arplant-050312-120235. [DOI] [PubMed] [Google Scholar]
- 13.Lodwig EM, et al. Amino-acid cycling drives nitrogen fixation in the legume-rhizobium symbiosis. Nat. 2003;422:722–726. doi: 10.1038/nature01527. [DOI] [PubMed] [Google Scholar]
- 14.Prell J, et al. Role of symbiotic auxotrophy in the rhizobium-legume symbioses. Plos One. 2010;5:e13933. doi: 10.1371/journal.pone.0013933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prell J, Poole P. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 2006;14:161–168. doi: 10.1016/j.tim.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Baart G, Martens D. Genome-scale metabolic models: reconstruction and analysis. Methods Mol Biol. 2012;799:107–126. doi: 10.1007/978-1-61779-346-2_7. [DOI] [PubMed] [Google Scholar]
- 17.Monk J, Nogales J, Palsson BO. Optimizing genome-scale network reconstructions. Nat Biotechnol. 2014;32:447–452. doi: 10.1038/nbt.2870. [DOI] [PubMed] [Google Scholar]
- 18.Dias O, Rocha M, Ferreira EC, Rocha I. Reconstructing genome-scale metabolic models with merlin. Nucleic Acids Res. 2015;43:3899–3910. doi: 10.1093/nar/gkv294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caspi R, et al. The metacyc database of metabolic pathways and enzymes and the biocyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44:D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fell DA, Poolman MG, Gevorgyan A. Building and analysing genome-scale metabolic models. Biochem. Soc. Trans. 2010;38:1197–1201. doi: 10.1042/BST0381197. [DOI] [PubMed] [Google Scholar]
- 21.Rezola A, et al. Advances in network-based metabolic pathway analysis and gene expression data integration. Brief Bioinform. 2015;16:265–279. doi: 10.1093/bib/bbu009. [DOI] [PubMed] [Google Scholar]
- 22.Poolman MG, Miguet L, Sweetlove LJ, Fell DA. A genome-scale metabolic model of Arabidopsis and some of its properties. Plant Physiol. 2009;151:1570–1581. doi: 10.1104/pp.109.141267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikerel E, et al. Understanding regulation of metabolism through feasibility analysis. Plos One. 2012;7:e39396. doi: 10.1371/journal.pone.0039396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joyce AR, Palsson BO. Predicting gene essentiality using genome-scale in silico models. Methods Mol Biol. 2008;416:433–457. doi: 10.1007/978-1-59745-321-9_30. [DOI] [PubMed] [Google Scholar]
- 25. Perumal, D., Samal, A., Sakharkar, K. R. & Sakharkar, M. K. Targeting multiple targets in pseudomonas aeruginosa pao1 using flux balance analysis of a reconstructed genome-scale metabolic network. J. Drug Target. 19, 1–13, PMID: 20233082, 10.3109/10611861003649753 (2011). [DOI] [PubMed]
- 26.Basler G, Grimbs S, Nikoloski Z. Optimizing metabolic pathways by screening for feasible synthetic reactions. Biosystems. 2012;109:186–191. doi: 10.1016/j.biosystems.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Chang RL, et al. Metabolic network reconstruction of chlamydomonas offers insight into light-driven algal metabolism. Mol Syst Biol. 2011;7:518. doi: 10.1038/msb.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mintz-Oron, S. et al. Reconstruction of arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc. Natl. Acad. Sci. 109, 339–344, http://www.pnas.org/content/109/1/339.abstract (2012). [DOI] [PMC free article] [PubMed]
- 29.Poolman, M. G., Kundu, S., Shaw, R. & Fell, D. Responses to light intensity in a genome-scale model of rice metabolism. Plant Physiol, 10.1104/pp.113.216762 (2013). [DOI] [PMC free article] [PubMed]
- 30.de Oliveira Dal’Molin CG, Quek L-E, Palfreyman RW, Brumbley SM, Nielsen LK. Aragem, a genome-scale reconstruction of the primary metabolic network in arabidopsis. Plant Physiol. 2010;152:579–589. doi: 10.1104/pp.109.148817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saha R, Suthers PF, Maranas CD. Zea maysirs1563: a comprehensive genome-scale metabolic reconstruction of maize metabolism. Plos One. 2011;6:e21784. doi: 10.1371/journal.pone.0021784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gomes De Oliveira Dal’molin, C., Quek, L.-E., Saa, P. A. & Nielsen, L. K. A multi-tissue genome-scale metabolic modelling framework for the analysis of whole plant systems. Front. Plant. Sci. 6, 10.3389/fpls.2015.00004 (2015). [DOI] [PMC free article] [PubMed]
- 33.Palsson, B. Ø. Systems Biology - Properties of Reconstructed Networks (Cambridge University Press, 2006).
- 34.Schuetz R, Kuepfer L, Sauer U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol Syst Biol. 2007;3:119. doi: 10.1038/msb4100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mahadevan, R. & Schilling, C. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng5, 264–276, http://www.sciencedirect.com/science/article/B6WN3-4B20H5T-2/2/75ef6ffa03cf1bc9b2c9c9c81c0ebd93 (2003). [DOI] [PubMed]
- 36. Mintz-Oron, S., Aharoni, A., Ruppin, E. & Shlomi, T. Network-based prediction of metabolic enzymes’ subcellular localization. Bioinforma. 25, i247–i252, http://bioinformatics.oxfordjournals.org/content/25/12/i247.abstract (2009). [DOI] [PMC free article] [PubMed]
- 37.Agren R, et al. The raven toolbox and its use for generating a genome-scale metabolic model for penicillium chrysogenum. Plos computational biology. 2013;9:e1002980. doi: 10.1371/journal.pcbi.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christian N, May P, Kempa S, Handorf T, Ebenhöh O. An integrative approach towards completing genome-scale metabolic networks. Mol Biosyst. 2009;5:1889–1903. doi: 10.1039/b915913b. [DOI] [PubMed] [Google Scholar]
- 39.Thiele I, Palsson BØ. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnold A, Nikoloski Z. Bottom-up metabolic reconstruction of arabidopsis and its application to determining the metabolic costs of enzyme production. Plant physiology. 2014;165:1380–1391. doi: 10.1104/pp.114.235358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shameer S, Baghalian K, Cheung CM, Ratcliffe RG, Sweetlove LJ. Computational analysis of the productivity potential of cam. Nat. plants. 2018;4:165. doi: 10.1038/s41477-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 42.Benedito VA, et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008;55:504–513. doi: 10.1111/j.1365-313X.2008.03519.x. [DOI] [PubMed] [Google Scholar]
- 43.Vlassis N, Pacheco MP, Sauter T. Fast reconstruction of compact context-specific metabolic network models. Plos Comput Biol. 2014;10:e1003424. doi: 10.1371/journal.pcbi.1003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pate J. Uptake, assimilation and transport of nitrogen compounds by plants. Soil Biol. and Biochem. 1973;5:109–119. doi: 10.1016/0038-0717(73)90097-7. [DOI] [Google Scholar]
- 45.Taiz, L. & Zeiger, E. Plant Physiology, 4th edn (Sinauer Associated, Inc, 2006).
- 46.Niittylä T, et al. A previously unknown maltose transporter essential for starch degradation in leaves. Sci. 2004;303:87–89. doi: 10.1126/science.1091811. [DOI] [PubMed] [Google Scholar]
- 47.Marschner, H. Mechanisms of adaptation of plants to acid soils. In Wright, R., Baligar, V. & Murrmann, R. (eds) Developments in Plant and Soil Sciences, vol. 45, 683–702, 10.1007/978-94-011-3438-5_78 (Springer Netherlands, 1991).
- 48.Pessarakli, M. (ed.). Handbook of Plant and Crop Stress - Impact of Soil pH on Nutrient Uptake by Crop Plants, chap. 3, 51–60, 2nd edn (Dekker, 2007).
- 49.Waters JK, et al. Alanine, not ammonia, is excreted from n2-fixing soybean nodule bacteroids. Proc Natl Acad Sci USA. 1998;95:12038–12042. doi: 10.1073/pnas.95.20.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poole P, Allaway D. Carbon and nitrogen metabolism in Rhizobium. Adv Microb Physiol. 2000;43:117–163. doi: 10.1016/S0065-2911(00)43004-3. [DOI] [PubMed] [Google Scholar]
- 51.Dingler C, Oelze J. Reversible and irreversible inactivation of cellular nitrogenase upon oxygen stress in azotobacter vinelandii growing in oxygen controlled continuous culture. Arch. Microbiol. 1985;141:80–84. doi: 10.1007/BF00446744. [DOI] [PubMed] [Google Scholar]
- 52.Allaway D, et al. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol Microbiol. 2000;36:508–515. doi: 10.1046/j.1365-2958.2000.01884.x. [DOI] [PubMed] [Google Scholar]
- 53.Kumar S, Bourdès A, Poole P. De novo alanine synthesis by bacteroids of Mesorhizobium loti is not required for nitrogen transfer in the determinate nodules of Lotus corniculatus. J Bacteriol. 2005;187:5493–5495. doi: 10.1128/JB.187.15.5493-5495.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pyl E-T, et al. Metabolism and growth in arabidopsis depend on the daytime temperature but are temperature-compensated against cool nights. Plant Cell. 2012;24:2443–2469. doi: 10.1105/tpc.112.097188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams TCR, et al. A genome-scale metabolic model accurately predicts fluxes in central carbon metabolism under stress conditions. Plant Physiol. 2010;154:311–323. doi: 10.1104/pp.110.158535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cheung, C. Y. M. et al. A method for accounting for maintenance costs in flux balance analysis improves the prediction of plant cell metabolic phenotypes under stress conditions. Plant J, 10.1111/tpj.12252 (2013). [DOI] [PubMed]
- 57.Piques M, et al. Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in arabidopsis. Mol Syst Biol. 2009;5:314. doi: 10.1038/msb.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stitt M. Systems-integration of plant metabolism: means, motive and opportunity. Curr Opin Plant Biol. 2013;16:381–388. doi: 10.1016/j.pbi.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 59.Sweetlove LJ, Williams TC, Cheung C, Ratcliffe RG. Modelling metabolic co2 evolution–a fresh perspective on respiration. Plant, cell & environment. 2013;36:1631–1640. doi: 10.1111/pce.12105. [DOI] [PubMed] [Google Scholar]
- 60.Ohmori M, Ohmori K, Strotmann H. Inhibition of nitrate uptake by ammonia in a blue-green alga, anabaena cylindrica. Arch. Microbiol. 1977;114:225–229. doi: 10.1007/BF00446866. [DOI] [Google Scholar]
- 61. de la Haba, P., Agüera, E. & Maldonado, J. Differential effects of ammonium and tungsten on nitrate and nitrite uptake and reduction by sunflower plants. Plant Sci. 70, 21–26, http://www.sciencedirect.com/science/article/pii/016894529090027L (1990). [DOI] [PubMed]
- 62.Bai Y, et al. Functional overlap of the arabidopsis leaf and root microbiota. Nat. 2015;528:364–369. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
- 63.Succurro A, Moejes FW, Ebenhöh O. A diverse community to study communities: Integration of experiments and mathematical models to study microbial consortia. J. Bacteriol. 2017;199:e00865–16. doi: 10.1128/JB.00865-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young ND, et al. The medicago genome provides insight into the evolution of rhizobial symbioses. Nat. 2011;480:520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Karp, P. D. et al. Pathway tools version 13.0: integrated software for pathway/genome informatics and systems biology. Briefings Bioinforma. 11, 40–79, http://bib.oxfordjournals.org/content/11/1/40.abstract, 10.1093/bib/bbp043 (2010). [DOI] [PMC free article] [PubMed]
- 66.Urbanczyk-Wochniak, E. & Sumner, L. W. Mediccyc: a biochemical pathway database for medicago truncatula. Bioinforma. 23, 1418–1423, http://bioinformatics.oxfordjournals.org/content/23/11/1418.abstract (2007). [DOI] [PubMed]
- 67. Lamesch, P. et al. The arabidopsis information resource (tair): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210, http://nar.oxfordjournals.org/content/40/D1/D1202.abstract, 10.1093/nar/gkr1090 (2012). [DOI] [PMC free article] [PubMed]
- 68.Heazlewood, J. L., Verboom, R. E., Tonti-Filippini, J., Small, I. & Millar, A. H. Suba: the arabidopsis subcellular database. Nucleic Acids Res. 35, D213–D218, http://nar.oxfordjournals.org/content/35/suppl_1/D213.abstract, 10.1093/nar/gkl863 (2007). [DOI] [PMC free article] [PubMed]
- 69.Daher Z, et al. Proteomic analysis of medicago truncatula root plastids. Proteomics. 2010;10:2123–2137. doi: 10.1002/pmic.200900345. [DOI] [PubMed] [Google Scholar]
- 70.Dubinin, J., Braun, H.-P., Schmitz, U. & Colditz, F. The mitochondrial proteome of the model legume medicago truncatula. Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics1814, 1658–1668, http://www.sciencedirect.com/science/article/pii/S1570963911002342 (2011). [DOI] [PubMed]
- 71.Babujee, L. et al. The proteome map of spinach leaf peroxisomes indicates partial compartmentalization of phylloquinone (vitamin k1) biosynthesis in plant peroxisomes. J. Exp. Bot. 61, 1441–1453, http://jxb.oxfordjournals.org/content/61/5/1441.abstract (2010). [DOI] [PubMed]
- 72. Linka, N. & Weber, A. P. Intracellular metabolite transporters in plants. Mol. Plant3, 21–53, http://mplant.oxfordjournals.org/content/3/1/21.abstract, 10.1093/mp/ssp108 (2010). [DOI] [PubMed]
- 73.Helliwell CA, et al. A plastid envelope location of arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J. 2001;28:201–208. doi: 10.1046/j.1365-313X.2001.01150.x. [DOI] [PubMed] [Google Scholar]
- 74. León, J. & Sánchez-Serrano, J. J. Molecular biology of jasmonic acid biosynthesis in plants. Plant Physiol. Biochem. 37, 373–380, http://www.sciencedirect.com/science/article/pii/S0981942899800436 (1999).
- 75. De Vries, F. W. T. P. The cost of maintenance processes in plant cells. Annals Bot. 39, 77–92, http://aob.oxfordjournals.org/content/39/1/77.abstract (1975).
- 76.Lötscher M, Klumpp K, Schnyder H. Growth and maintenance respiration for individual plants in hierarchically structured canopies of medicago sativa and helianthus annuus: the contribution of current and old assimilates. New Phytol. 2004;164:305–316. doi: 10.1111/j.1469-8137.2004.01170.x. [DOI] [PubMed] [Google Scholar]
- 77.Orth JD, et al. A comprehensive genome-scale reconstruction of escherichia coli metabolism–2011. Mol Syst Biol. 2011;7:535. doi: 10.1038/msb.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Udvardi MK, Day DA. Metabolite transport across symbiotic membranes of legume nodules. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:493–523. doi: 10.1146/annurev.arplant.48.1.493. [DOI] [PubMed] [Google Scholar]
- 79.Vance CP, Heichel GH, Barnes DK, Bryan JW, Johnson LE. Nitrogen fixation, nodule development, and vegetative regrowth of alfalfa (Medicago sativa l.) following harvest. Plant Physiol. 1979;64:1–8. doi: 10.1104/pp.64.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aydi S, Drevon J-J, Abdelly C. Effect of salinity on root-nodule conductance to the oxygen diffusion in the Medicago truncatula-Sinorhizobium meliloti symbiosis. Plant Physiol Biochem. 2004;42:833–840. doi: 10.1016/j.plaphy.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 81.Sutton WD, Jepsen NM, Shaw BD. Changes in the number, viability, and amino-acid-incorporating activity of Rhizobium bacteroids during lupin nodule development. Plant Physiol. 1977;59:741–744. doi: 10.1104/pp.59.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bergersen, F. Physiological and biochemical aspects of nitrogen fixation by bacteroids in soybean nodule cells. Soil Biol Biochem29, 875–880, http://www.sciencedirect.com/science/article/pii/S0038071796002209 (1997).
- 83.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blank LM, Kuepfer L, Sauer U. Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biol. 2005;6:R49. doi: 10.1186/gb-2005-6-6-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grafahrend-Belau E, Schreiber F, Koschützki D, Junker BH. Flux balance analysis of barley seeds: a computational approach to study systemic properties of central metabolism. Plant Physiol. 2009;149:585–598. doi: 10.1104/pp.108.129635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Williams TCR, et al. Metabolic network fluxes in heterotrophic arabidopsis cells: stability of the flux distribution under different oxygenation conditions. Plant Physiol. 2008;148:704–718. doi: 10.1104/pp.108.125195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Masakapalli SK, Kruger NJ, Ratcliffe RG. The metabolic flux phenotype of heterotrophic arabidopsis cells reveals a complex response to changes in nitrogen supply. Plant J. 2013;74:569–582. doi: 10.1111/tpj.12142. [DOI] [PubMed] [Google Scholar]
- 88.Sumner, L. W., Journet, E. P. & Ulrike, M. (eds). The Medicago truncatula handbook, chap. Growing M. truncatula: choice of substrates and growth conditions, 1–26 (The Medicago Consortium, 2006).
- 89.Mettupalli, D. R. Impact of Arbuscular Mycorrhiza symbiosis on photosynthesis in Medicago truncatula. Ph.D. thesis, Göteborgs Universitet (2011).
- 90.Yousfi N, Slama I, Ghnaya T, Savouré A, Abdelly C. Effects of water deficit stress on growth, water relations and osmolyte accumulation in Medicago truncatula and M. laciniata populations. C R Biol. 2010;333:205–213. doi: 10.1016/j.crvi.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 91.Jung, H. & Engels, F. Alfalfa stem tissues: cell wall deposition, composition, and degradability. Crop science. 42 (2), 524–534, http://hdl.handle.net/10113/11745 (2002).
- 92.Nakashima J, Chen F, Jackson L, Shadle G, Dixon RA. Multi-site genetic modification of monolignol biosynthesis in alfalfa (medicago sativa): effects on lignin composition in specific cell types. New Phytol. 2008;179:738–750. doi: 10.1111/j.1469-8137.2008.02502.x. [DOI] [PubMed] [Google Scholar]
- 93.Johnson, J. M.-F., Barbour, N. W. & Weyers, S. L. Chemical composition of crop biomass impacts its decomposition. Soil Sci. Soc. Am. J. 71, 155–162, https://www.soils.org/publications/sssaj/abstracts/71/1/155, 10.2136/sssaj2005.0419 (2007).
- 94.Bakoglu A, Bagci E, Kocak A, Yuce E. Fatty acid composition of some medicago l. (fabaceae) species from turkey. Asian J. Chem. 2010;22:651–656. [Google Scholar]
- 95.Blondon F, Marie D, Brown S, Kondorosi A. Genome size and base composition in Medicago sativa and M. truncatula species. Genome. 1994;37:264–270. doi: 10.1139/g94-037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.