Abstract
The presented procedure allows rapid isolation of extracellular vesicles (EVs) from plasma using size-exclusion chromatography (SEC). Additionally, an approach for reducing the lipid and salt content of the EV isolate in preparation for mass spectrometry (MS)-based proteomic analysis is presented. An example setup for proteomic profiling of the processed samples by nanoflow liquid chromatography coupled to tandem mass spectrometry (nLC-MS/MS) is also presented. Approximately 1000 protein groups in blood plasma-derived EVs can be identified and quantitated following this procedure and using the described instrumentation.
Keywords: Extracellular vesicles, EVs, Microparticles, Exosomes, Proteomics, Size-exclusion chromatography, Mass spectrometry, Nano-LC-MS, Sample preparation, Proteomic profiling
1. Introduction
Extracellular vesicles (EVs) are now the accepted nomenclature for phospholipid bilayer enclosed particles (exosomes, microparticles, etc.) secreted by most, likely all, cells [1, 2]. EVs are found in all physiological fluids and are implicated in disease progression, immune response, inflammation, intercellular communication and other biological phenomena. EVs have diagnostic potential as “liquid biopsies,” because disease-associated EVs can be detected in physiological fluids without invasive surgery [3–5]. Conventional EV isolation techniques by ultracentrifugation and density gradient centrifugation [6] are low-throughput and impractical in a clinical setting. Alternatively, size-exclusion chromatography (SEC) can be used to isolate EVs in minutes without specialized equipment [7]. SEC uses porous low binding beads to allow large particles (i.e., EVs), which cannot enter the pores, to readily pass through the bed of the stationary phase, while smaller sample constituents (e.g., proteins) are slowed by the tortuous path through the pores. SEC is a low-resolution chromatographic technique, and SEC-based EV isolations may also contain other particles of similar size (30–800 nm) and low levels of plasma protein complexes and aggregates [3]. While higher purity isolation can be obtained using more time- and resource-consuming procedures, the presented approach is capable of rapid enrichment of EVs from blood plasma and may be applied to other highly complex physiological fluids, and samples of other types (e.g., cell culture media).
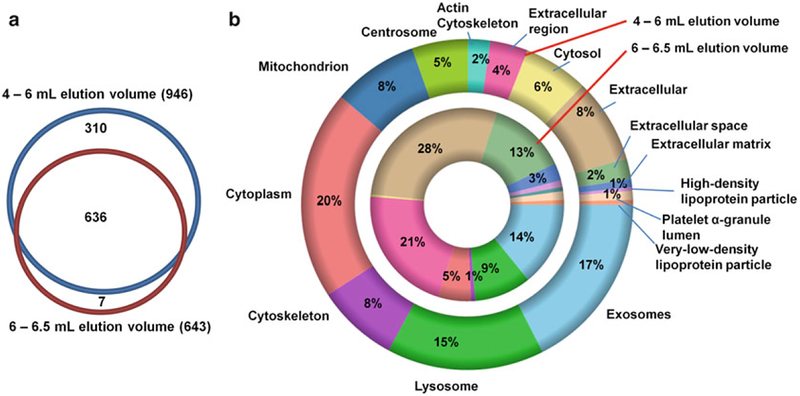
The described protocol greatly accelerates EV isolation and allows a reasonable depth of proteomic profiling. The SEC columns for gravity-driven EV isolation are easy to pack, and demonstrate reproducible performance. Figure 1 compares the gene ontology (GO) terms between the proteins identified in the early eluting 4–6 mL and 6–6.5 mL fractions obtained using a 10-mL SEC column. The increase in the representation of GO terms associated with plasma proteins in the later eluting fraction indicates that the content of plasma proteins is increased, and EVs and plasma proteins are indeed separated by size. The described Bligh–Dyer liquid–liquid extraction [8] separates the lipid and protein components of the collected fraction while reducing the salt content. This improves the reproducibility of nLC-MS/MS-based proteomic profiling and extends the life span of the LC columns. The resulting lipid-rich fractions can be used for lipidomic profiling of EV isolates.
Fig. 1.
Baseline proteome and Gene Ontology (GO) term enrichment comparison between the 4–6 mL elution volumes and 6–6.5 mL elution volumes. Frozen, cell-free, plasma was processed using the described workflow. Nearly 1000 protein groups were identified (<1% FDR) and quantified by MaxQuant. The overlap in protein identifications is presented in the Venn diagram (panel a). The GO cellular localization terms correspond to enriched proteins (2× intensity compared to the other fraction; panel b). The additional proteins identified in the 4–6 mL elution volume contribute to increased intracellular, and Exosome GO localization terms (panel b). The results are consistent with reduced concentration of plasma proteins in the earlier eluting fraction which improves detection of EV proteins and deeper proteomic profiling
2. Materials
Use HPLC-grade solvents (water, isopropanol) and sterile Dulbecco’s phosphate buffered saline (DPBS), pH 7.4 (Sigma-Aldrich, St. Louis MO) for SEC column preparation and separation of plasma. Use LC-MS grade water, acetonitrile, and formic acid for LC-MS analysis (Sigma-Aldrich). Filter the solvents and buffers by passing them through a 0.22 μm filter before the SEC preparation to mitigate possible contamination with the microparticulate material. Prepare the single use SEC column within hours of planned isolation by packing an empty 10 mL centrifuge column (e.g., Pierce pt. # 89898 10 mL columns with a 13 mL reservoir) with a slurry of cross-linked Sepharose CL-2B particles (Sigma-Aldrich pt.# CL2B300) shipped in 50% isopropanol.
Rinse the 5–10 mL disposable syringe with a Luer-lock adapter (e.g., BD pt.#.309604) with filtered DPBS. Condition the 30 mm diameter 0.8 μm pore cellulose acetate membrane filters with the Luer adapter (e.g., Cameo syringe filters Sigma-Aldrich pt. #741,675 or #741864) with sterile DPBS before use.
Blood drawing system:
Use BD Vacutainer® UltraTouch™ Push Button Blood Collection Set consisting of a 21 gauge 0.75″ needle, 7″ tubing (pt#367393) and 10 mL EDTA-coated Vacutainer® tube (pt# 367856) (BD Life Sciences, Franklin Lakes, NJ) for drawing blood.
Sample preparation reagents:
Use HPLC grade chloroform, methanol, water, 2,2,2-triflouroethanol, and highest purity iodoacetamide (IAA), Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), formic acid and other reagents from Sigma-Aldrich, unless specified otherwise. Use MS-grade Trypsin/Lys-C mix from Promega (Madison, WI pt.#V5073) or similar. Use low retention Eppendorf tubes from VWR (Radnor, PA) or other sources.
3. Methods
All steps should be performed at room temperature unless otherwise indicated. It is advised to rinse the Eppendorf tubes with LCMS grade methanol and dry in a clean environment before use. NOTE: Use freshly prepared 50 mM TCEP and 100 mM IAA stock solutions. Store the IAA solution in the dark as light degrades the reagent.
3.1. Preparation of Single-Use SEC Columns
1. Open the top cap and the bottom stopper and rinse the empty 10 mL column with 10 mL of 50% aqueous isopropanol.
2. Pour 12 mL of the CL-2B slurry and allow the slurry to settle down and condense as the beads are retained by the filter (30 μm pore size) and the solvent flows through.
3. If the bed volume is below the 10 mL mark on the side of the column, add more slurry until the bed volume is at 10 mL. If the bed volume exceeds 10 mL, the excess slurry can be scooped out using a clean spatula.
4. Fill the 13 mL reservoir with 50% isopropanol and allow all of the liquid to flow through. Repeat once more with 50% isopropanol and twice more with water.
5. Condition the column with DPBS allowing a total of 50 mL to flow through. Seal the top and bottom of the column so that it does not dry out; it is advised to prepare the column within a few hours of isolation. NOTE: If the stationary phase becomes dry, then cracks may form and lead to channeling effects. Drying can also change the porosity of the stationary phase, which can reduce the separation performance of the columns resulting in increased plasma protein contamination of the EV isolate.
3.2. SEC Isolation of EVs
1. Collect blood by percutaneous cubital venipuncture drawn into an EDTA-containing Vacutainer (BD Biosciences) and discard the first bolus to reduce artifacts from piercing the skin.
2. Collect 8 mL of plasma in the 10 mL EDTA-coated Vacutainer.
3. Centrifuge at 2000 × for 5 min. During centrifugation or before starting the procedure, rinse with DPBS the 5 mL syringe and disk membrane filter to be used.
4. Discard the top 100–200 μL of plasma.
5. Aspirate 2 mL of plasma in a Luer-lock 5–10 mL syringe (e.g., BD pt.# 309,604). NOTE: Avoid any contact of the sample with the syringe plunger.
6. Attach the DPBS rinsed 0.8 μm cellulose acetate disk filter (Sterlitech, Kent, WA) to the syringe.
7. Open the column stopper and slowly inject 1 mL of plasma through the filter onto a SEC column so that the bed of the stationary phase is not disturbed.
8. Allow the injected volume to enter the stationary phase; the yellow hue of the plasma can easily be observed through the container wall. Close the stopper and immediately proceed to step 9 to minimize longitudinal sample diffusion.
9. Add 10 mL of DPBS into the reservoir, open the stopper, and start collecting the eluate. Use 2 mL Eppendorf tubes with a labeled volume scale (or similar) to measure the eluate volume.
10. The EV population generally elutes by gravity flow in the 2–7 mL elution volume, and the peak is between 4 and 6 mL while the protein fraction starts to elute at around 6 mL. Collect the fraction required for analysis. Use the fraction of 4–6 mL that contains a high level of EVs with a reduced level of plasma proteins for LC-MS proteomic profiling. Close the stopper after collecting the EV-rich fraction(s). The collected EV isolate can be now used for qualitative and quantitative imaging techniques (microscopy, nano-flow cytometry, etc.).
11. The column, syringe, filter, and blood collection system can now be discarded following appropriate biohazard protocols. For quantitative comparison it is advised to estimate vesicle concentration in each sample using nanopore, brownian motion (e.g. NanoSight, Malvern), or dynamic light scattering, and normalize loading based on these measurements. Plasma composition is heterogeneous and normalization based on net protein concentration may not represent EV levels due to plasma protein contamination.
12. The samples can now be either frozen and stored at −80 °C, or processed immediately.
3.3. Lipid Depletion Using Bligh–Dyer Liquid–Liquid Extraction Followed by Tryptic Digestion of the EV Isolate
1. If frozen, thaw the samples at room temperature.
2. Concentrate the samples in the SpeedVac to a volume of approximately 200 μL.
3. Add 400 μL of methanol and 200 μL of chloroform. Vortex for 1 min and then sonicate in the ultrasonication water bath for 5 min at room temperature.
4. Add 200 μL of chloroform and vortex for 1 min.
5. Add 200 μL of water and vortex for 1 min.
6. Centrifuge at 14,000 × for 10 min at room temperature.
7. Transfer the sample tube into a −20 °C freezer and incubate for 4 h (the cold temperature and centrifugation are used to focus the precipitated proteins in the interface between the two phases).
8. Centrifuge at 14,000 × for 10 min at room temperature.
9. If a complementary metabolomics or lipidomics analysis is desired, the chloroform (lower) phase should be transferred into a clean Eppendorf container; otherwise, it can be discarded.
10. Carefully discard the methanol/water (upper) phase.
11. Lyophilize the remaining protein pellet to dryness in a SpeedVac.
12. Add 50 μL of 30 mM ammonium bicarbonate pH 8.0 in 50% aqueous 2,2,2-triflouroethanol to solubilize the precipitated proteins, sonicate for 5 min in the water ultrasonication bath at room temperature. NOTE: Protein concentration of the sample can be assayed after this step using a sample aliquot.
13. Add 6 μL of the 50 mM TCEP in 125 mM ammonium bicarbonate, pH 8 (to the TCEP concentration of approximately 5 mM in the sample) and incubate at 37 °C for 30 min. Check the pH using a submicroliter aliquot and a pH strip and make sure that the pH of the sample is approximately 7–8 after adding TCEP.
14. Add 7 μL of freshly prepared 100 mM IAA in 125 mM ammonium bicarbonate, pH 8 (to the IAA concentration of approximately 10 mM in the sample) and incubate in the shaker for 45 min in the dark.
15. Add 457 μL of 25 mM ammonium bicarbonate pH 7.5 and check the pH.
16. Confirm that the pH is approximately 7–8 and add 2 μg of Trypsin/Lys-C Mix, MS-grade (Promega), corresponding to the 1:20–1:50 enzyme to protein ratio.
17. Incubate in the shaker at 40 °C for 16 h or overnight.
18. Add formic acid to the concentration of 0.1% to quench digestion. NOTE: Peptide concentration of the sample can be assayed after this step using a sample aliquot.
19. Concentrate to the volume of approximately 50 μL using a SpeedVac.
20. Inject 1 μL of the resulting digest on to the nano-LC-MS column. Such injection volume will correspond to the amount of EV isolate derived from 20 μL of plasma. The sample amount to subject to molecular profiling can be adjusted appropriately according to the goals of the study.
3.4. nLC-MS/MS Analysis
Due to the presence of residual salts and lipids, it is advised to desalt online with a trapping column before LC-MS analysis. An example LC-MS protocol is presented below:
Column:
a 25 cm long bed packed with 3 μm Magic C18 AQ, pore size 200 Å stationary phase in a fused silica capillary of the 75 μm internal diameter and 365 μm outer diameter with a porous Kasil frit polymerized in-house [9] or acquired from commercial sources (e.g., New Objective pt. # IF360–75–50-N-5). To generate stable nanoelectrospray, use electrospray voltage of 1.9–2.4 kV applied through a distal coated fused capillary tip 20 μm ID pulled to 10 μm opening (New Objective pt.# FS360–20–10-D) connected to the end of the column with a 250 μm Teflon sleeve (Thermo Fisher Scientific, pt.# 160486).
Mass spectrometry:
Use Top 15 data-dependent acquisition (DDA) on a QExactive with the following settings. MS1: automated gain control (AGC) 1e6, maximum ion accumulation time (IT): 30 ms, resolving power at m/z 200 (R): 70 k. MS2: AGC 1e5, maximum IT: 50 ms, R: 17.5 k. Dynamic Exclusion 60 s. Isolation window: 2 m/z and optimized HCD energy for the instrument used (e.g., 28). Use the signal for dodecamethylcyclohexasiloxane ions (445.12 m/z) as an internal calibrant in lock-mass mode.
Liquid chromatography:
Use the mobile phase consisting of Solvent A (0.1% formic acid in water) and Solvent B (0.1% formic acid in acetonitrile). Inject 1 μL of the sample directly onto the column at a 250 nL/min flow rate and 2% B.
Gradient (250 nL/min):
desalting for 15 min at 2%B; linear ramp to 42% B over 90 min; linear ramp to 95% B over 15 min, 10 min hold at 95%B; drop to 2%B in 1 min, hold at 2%B for 14 min. NOTE: Longer linear gradients can be used to improve the depth of proteomic profiling.
Data analysis:
Use MaxQuant (v.1.5.3.28 or later) or similar database search software. Use the most current SwissProt/UniProt human protein sequence database appended with sequences of common contaminants (e.g., “the common Repository of Adventitious Proteins, cRAP”, http://www.thegpm.org/crap/). Apply the following settings for the first search: 20 ppm allowed MS1 and MS2 mass tolerance; enzyme specificity: tryptic; carbamidomethylation of cysteine as a static modification. Main search settings: 4.5 ppm precursor mass and 8 ppm fragment mass errors; carbamidomethylation of cysteine set as a static modification, oxidation of methionine set as a variable modification; digestion specificity: semi-tryptic with two allowed missed cleavages. Filter the results to the peptide-level FDR of 1% or less.
Acknowledgments
This work was supported by the ASMS 2015 Research Award (ARI), NIH 1R01GM120272–01 (ARI), and the Dana-Farber Cancer Institute/Northeastern University Joint Seed Funding Program in Cancer Drug Development (ARI). The authors thank Dr. Ionita Ghiran (Beth Israel Deaconess Medical Center) for fruitful discussions and his assistance with blood collection.
References
- 1.Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW, Thery C (2014) Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 3:26913. doi: 10.3402/jev.v3.26913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gould SJ, Raposo G (2013) As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles 2. doi: 10.3402/jev.v2i0.20389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kreimer S, Belov AM, Ghiran I, Murthy SK, Frank DA, Ivanov AR (2015) Mass spectrometry-based molecular characterization of extracellular vesicles: lipidomics and proteomics. J Proteome Res 14(6):2367–2384 [DOI] [PubMed] [Google Scholar]
- 4.Colombo M, Raposo G, Thery C (2014) Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30:255–289. doi: 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- 5.Thery C (2015) Cancer: diagnosis by extracellular vesicles. Nature 523(7559):161–162. doi: 10.1038/nature14626 [DOI] [PubMed] [Google Scholar]
- 6.Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, Simpson RJ (2012) Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 56 (2):293–304. doi: 10.1016/j.ymeth.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 7.Boing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A, Nieuwland R (2014) Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles 3. doi: 10.3402/jev.v3.23430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37(8):911–917 [DOI] [PubMed] [Google Scholar]
- 9.Cortes HJ, Pfeiffer CD, Richter BE, Stevens TS (1987) Porous ceramic bed supports for fused silica packed capillary columns used in liquid chromatography. J High Resol Chromatogr 10 (8):446–448 [Google Scholar]