Abstract
Members of the miR-125 family are strongly expressed in several tissues, particularly brain, but may be dysregulated in cancer including adult and pediatric glioma. In this study, miR-125 members were downregulated in pilocytic astrocytoma (PA) as a group compared to non-neoplastic brain in the Agilent platform. In the Nanostring platform, miR-125 members were downregulated primarily in pleomorphic xanthoastrocytomas and gangliogliomas. Using CISH for miR-125b, highest levels of expression were present in grade II tumors (11/33, 33% grade II tumors with 3+ expression compared to 3/70, 4% grade I tumors) (p < 0.001). When focusing on the two histologic subgroups with the largest number of samples, PA and diffuse astrocytoma (DA), the highest expression levels were present in DA, in comparison with the PA group (p = 0.01). Overexpression of miR-125b in pediatric low grade glioma (PLGG) derived cell lines (Res186, Res259, and BT66) resulted in decreased growth and invasion, as well as apoptosis. Additionally, miR-125b overexpression in BT66 resulted in senescence. These findings suggest that miR-125 is frequently underexpressed in PLGG, and overexpression results in a decrease in cell growth and induction of apoptosis, findings that deserve further investigation given its potential as a novel therapeutic strategy for PLGG.
Introduction
Pediatric low-grade glioma (PLGG) represents an heterogeneous group of primary CNS neoplasms characterized by slow growth. Glioneuronal tumors are somewhat related given that a low-grade glioma component is frequent and they are also characterized by similar demographics. The prognosis for these tumors is generally good, but complete surgical resection may not be possible in specific locations, therefore they have a potential for morbidity related to mass effect.
High resolution platforms have uncovered recurrent alterations in these tumors, i.e. BRAF-KIAA1549 fusions as the most frequent recurrent alteration in pilocytic astrocytoma (PA)1–5, which represents the most frequent subtype of PLGG. BRAF-KIAA1549 fusions lead to mitogen-activated protein kinase (MAPK)2. Interestingly, comprehensive sequencing studies reported genetic alterations in MAPK components in essentially all (100%) of PA6. PLGG also have heterogeneous histologies, including neoplasms with infiltrating astrocytic or oligodendroglial components, which may have a variety of alterations in alternative drivers (FGFR1, MYB, MYBL1)7,8.
The role of non-coding RNAs, particularly microRNAs, is an active field of study in normal physiology and a variety of disorders, including cancer. MicroRNA upregulation may target tumor suppressors, while downregulation of microRNAs may result in increased levels of oncoproteins. In the field of glioma, alterations in the expression of numerous microRNAs have been described, and they may play a role in every known process associated with glioma biology (reviewed in9). PLGG in particular are an attractive subject for microRNA profiling, given their relative lack of genomic instability in contrast to high grade tumors10. Most microRNA profiling studies in gliomas have been performed in adults, particularly high grade and diffuse subsets such as glioblastoma, where they appear to mediate a variety of important tumor properties (reviewed in9). Conversely, studies of microRNA expression in circumscribed and low grade pediatric gliomas are relatively scant. When focusing on pediatric glioblastoma, and comparing expression levels with non-neoplastic brains, studies have documented relative under-expression of miR-1224, miR-204, miR-874, miR-1296, miR-889, miR-495, miR-34c, miR-10b, miR-125a, miR-10a, while miR-617, miR-200a, miR769-3, miR-584, and miR-527 are overexpressed11,12.
When focusing on PLGG, miR-21 and miR-34 overexpression have been consistently documented in PA11–14. Several of the predicted targets of these microRNAs involve components responsible for regulation of the MAPK signaling pathway, which is universally activated, predominantly through a BRAF-KIAA1549 fusion gene product6. Our prior studies and other investigators have reported additionally relatively underexpressed microRNAs in PA compared to normal brain, including miR-124 and miR-12911,12,14,15. Functional validation of these microRNAs in PLGG has limitations, primarily given the relative lack of in vitro and in vivo models of this disease. Our group has recently documented underexpression of miR-487b in PA, and in vitro experiments demonstrated downregulation of putative targets/stem cell markers PROM1 and nestin and decreased colony formation in soft agar when miR-487b is overexpressed13. Comprehensive studies of microRNA regulation in glioneuronal tumors are even more scant, with rare reports of alterations in inflammation-associated microRNAs in ganglioglioma16,17.
miR-125b is the human homologue of one of the first microRNAs discovered, i.e. lin-4 in C. elegans18,19, and is evolutionarily conserved. The miR-125 family encompasses three homologues hsa-miR-125a, hsa-miR-125b-1 and hsa-miR-125-2, and have various roles in physiology and disease (reviewed in20). The list of putative targets is wide and contains transcripts involved in proliferation, metastasis, differentiation and immune regulation. In solid tumors, a tumor suppressor role for miR-125 in cancers of lung, ovary, breast, bladder, bone and skin has been documented. Wang et al. reported that miR-125a is regulated by EGFR signaling and leads to decreased invasion and migration in lung cancer cells21. In breast cancer, miR-125 is downregulated and affects ERBB2 and ERBB3 signaling and invasion properties22, while ectopic regulation of miR-125b regulates proliferation in hepatocellular carcinoma23. Additionally, miR-125b may play a role in apoptosis. For example, Morelli E. et al. demonstrated that miR-125b overexpression in multiple myeloma cells leads to apoptosis, likely mediated through IRF4 downregulation24.
It must be noted that miR-125b has been also found to act as an oncogene in certain contexts, including cancers of endometrium, breast and prostate20. When focusing in glioma, there have been reports of miR-125b acting as an oncogene in glioblastoma, through targeting of TP5325, and connexin4326. Additional studies have reported that miR-125b regulates the proliferation of glioma stem cells through E2F2 regulation27. The role of miR-125b in low grade glioma has not been thoroughly studied to our knowledge. Of interest, Nyholm et al. demonstrated that miR-125b leads to cellular senescence in melanoma, specifically G0/G1 cell cycle arrest, morphologic changes, beta galactosidase staining and increased levels of p53, p21 and p27.28. In the same study, there was not a significant increase in apoptosis. PLGG shares with melanoma a high prevalence of activating mutations in BRAF. Furthermore, overexpression of active BRAF leads to oncogene induced senescence in PLGG29,30, which may be one of the reasons that this disease is difficult to model in the laboratory. To our knowledge, the role of miR-125b in PLGG, and specifically senescence has not been studied. In this study, we searched for putative microRNAs associated with senescence in PLGG, and uncovered miR-125b as downregulated in low grade gliomas and glioneuronal tumors and associated with tumor suppressive functions. We therefore evaluated the expression of miR-125b in PLGG and similar BRAF activated tumors in adults and studied the functional effects of miR-125b overexpression in PLGG cells lines.
Results
miR-125b is downregulated in low grade glioma and glioneuronal tumors in microRNA expression datasets
To identify biologically relevant microRNAs for PLGG we performed a literature search of altered microRNAs in the context of senescence31,32 and dysregulated in tumors associated with BRAF alterations28,33. We searched our previously published13,14 and unpublished microRNA data, and miR-125b was consistently downregulated in low grade glioma/glioneuronal tumors when compared to normal brain.
First, we looked at a dataset of 43 PA and 5 non-neoplastic brain samples profiled with the Human miRNA Microarray V3 Agilent platform using frozen surgical tissues14 which tested for miR-125a-3p, miR-125a-5p, miR-125b, miR-125b-1 *and miR-125b-2*. In this experiment, only miR-125a-3p and miR-125b-1* were relatively underexpressed in tumors 3 fold and 6 fold respectively compared to non-neoplastic brain. The differences were most pronounced in cerebral cortex, and less so in pediatric/fetal cerebellum (Fig. 1A). In a second dataset obtained from formalin-fixed paraffin-embedded tissues, a variety of low grade glial and glioneuronal tumor pathologic subtypes (n = 41) and 4 brain samples were profiled with the Nanostring platform13, which includes miR-125b-5p, miR-125a-5p, miR-125a-3p, In this experiment, hsa-miR-125a-5p and miR-125b-5p were underexpressed 2.5 and 2 fold respectively in pleomorphic xanthoastrocytomas (n = 7), and miR-125a-5p was underexpresed 2.8 and 2.4 fold in gangliogliomas (n = 6) and rosette forming glioneuronal tumors (n = 2) respectively compared to brain (Fig. 1B). There were no significant expression differences in the other tumor types or miR-125 family members (not shown).
Figure 1.
miR-125 is downregulated in pilocytic astrocytoma (PA) and other low grade gliomas/glioneuromal tumor compared with non-neoplastic brain. MicroRNA expression data obtained through Agilent arrays demonstrated lower miR-125 expression levels in PA (n = 43) compared to non-neoplastic brain, particularly cerebral cortex (n = 3) (cbll = cerebellum n = 2). Each data point (heatmap square) represents an individual sample. (A) microRNA expression using a nanostring platform demonstrated miR-125 underexpression particularly in pleomorphic xanthoastrocytoma (PXA) (n = 7), ganglioglioma (GG) (n = 6) and rosette forming glioneuronal tumor (RFGT) (n = 2). Each point represent an individual sample. Only these 3 subgroups that demonstrated significant differences between tumor and normal brain are illustrated. Normalized counts were obtained using nSolver software (NanoString) on Raw data filtered for minimum expression threshold, and log2 transformed before analysis as previously reported13 (B).
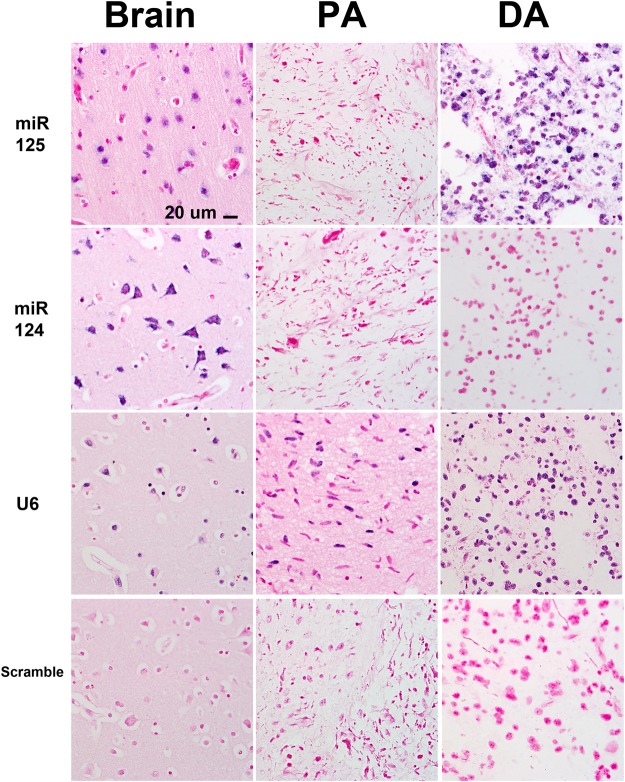
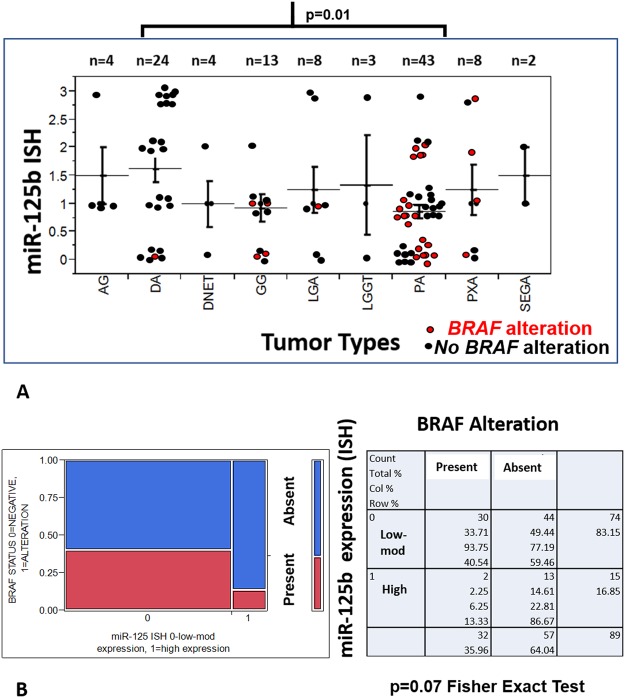
Next, we studied miR-125b expression in 125 low grade tumors of 9 different histologic subtypes using CISH, with statistical comparisons limited to the largest groups. Although absent and low reactivity were identified in both grade I and grade II tumors, highest levels of expression were present in grade II tumors (11/33, 33% grade II tumors with 3+ expression compared to 3/70, 4% grade I tumors) (p < 0.001) (Figs 2 and 3A). miR-125b expression was also preserved in non-neoplastic cells (neurons) within grade I and grade II tumors. There was a non-significant trend for high miR-125b expression in tumors lacking BRAF alterations (high expression in 13/57, 23% tumors lacking BRAF alterations vs 2/32, 6% tumors with BRAF duplication or BRAF p.V600E) (p = 0.07, 2-tail Fisher exact test) (Fig. 3B). When focusing on the two histologic subgroups with the largest number of samples (PA, DA), the highest expression levels were present in DA (n = 24), in comparison with the PA (n = 43) group (p = 0.01) (Wilcoxon Rank Sum Test).
Figure 2.
Differential miR-125b expression in low grade glioma subsets. MiR-125b expression detected by in situ hybridization was lower in pilocytic astrocytoma (PA) compared to diffuse astrocytomas (DA) and non-neoplastic brain (“brain”). Conversely, miR-124, a ubiquitous microRNA expressed in neurons was underexpressed in all gliomas tested. U6 was expressed in all tissues, while the scramble (negative) control was negative. Figures are representative of each group tested and all are taken with a 40X objective. The scale bar on the upper left figure applies to all figures in the panel.
Figure 3.
miR-125b expression is downregulated in pilocytic astrocytoma, and other circumscribed gliomas compared to diffuse astrocytoma. In situ hybridization studies using probes targetting miR-125b demonstrated variable levels in low grade gliomas and glioneuronal tumors, but levels were highest in diffuse astrocytoma (DA) and lowest in pilocytic astrocytoma (PA) (p = 0.01) (Wilcoxon Rank Sum Test). Strength of ISH hybridization was scored using a 4 tiered scale semiquantitative scale, ranging from 3 (strong, similar to brain), 2 (moderate), 1 (weak), 0 (negative) (y-axis). Each dot represents an individual tumor, and BRAF mutated tumors are in red color (AG = angiocentric glioma, DNET = dysembryoplastic neuroepithelial tumor, GG = ganglioglioma, LGA = low grade astrocytoma, LGGT = low grade glioneuronal tumor, PXA = pleomorphic xanthoastrocytoma, SEGA = subependymal giant cell astrocytoma) (A). There was a non-significant trend for high miR-125b expression in tumors lacking BRAF alterations (BRAF duplications or BRAF p.V600E) as demonstrated by the two tailed Fisher exact test and contingency table (B).
miR-125b expression is frequent in non-neoplastic brain and lower in neural stem cell compartments
We next assessed miR-125b expression levels in non-neoplastic human tissues. First we looked at a published comprehensive dataset of microRNA expression in 114 non-neoplastic and cancer tissues and cells lines (“the human microRNome”) obtained through small RNA-seq reads34. Increased expression levels, as represented in reads per million (RPM), were overrepresented in central nervous system (CNS) derived tissues and cells, particularly prefrontal cortex (22,005 RPM) and retinal pigment epithelium (RPE) (67150 RPM). Increased expression was also noted in skin (58488 RPM), cervix (30022 RPM), valve interstitial cell (23430 RPM) and osteoblasts (21736 RPM) (Supplementary Table 3).
To better define the range of miR-125b expression in mature brain and development we studied various brain regions from 6 patients with non-neoplastic processes using CISH which was uniformly expressed in all samples. (Supplementary Fig. 1 shows representative images of expression in mature non-neoplastic brain. In mature brain, strong miR-125b expression (3+) was evident in neurons of the neocortex (Supplementary Fig. 1A), spinal cord, dentate nucleus, cerebellum (Purkinje cells, dentate nucleus, internal granular layer), reactive astrocytes, arachnoidal cells and ependymal cells. miR-125b was weak to absent in white matter (Supplementary Fig. 1B).
In both fetal samples examined, miR-125b was expressed in the cortical plate, cerebellar internal granular cell layer, and hippocampus (dentate gyrus and cortex). Expression was absent in the external granular cell layer and ventricular zone (representative images of expression in both samples illustrated in Fig. 4). These findings suggest that miR-125b expression is a feature of mature neuronal and astrocytic cells, but is decreased in fetal neural stem cell compartments.
Figure 4.
miR-125 expression varies in different fetal brain cell compartments. miR-125b expression studied in a 20 week-old fetal brain. miR-125b expression was preserved in maturing CNS cellular elements, including the cortical plate and the internal granular cell layer of the cerebellum (CBLL) (asterisks), but lost in stem cell niches, such as the ventricular zone (VZ) and the external granular cell layer (arrows). miR-125b expression was also preserved in neurons of the dentate nucleus and developing white matter. Scale bars for low power and high power figures on top apply to all correspondng low and high power figures below. Low power views are taking using a 10X objective while high power views are taken with a 20X objective.
miR-125b overexpression halts cell growth in pediatric low grade (PLGG)-derived cell lines
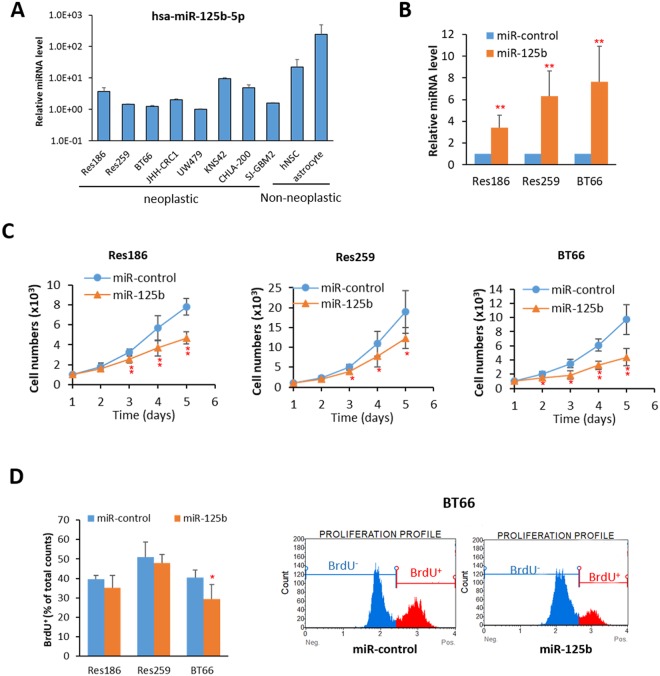
Next we studied various glioma cell lines for miR-125b-5p expression using qRT-PCR. The expression of miR-125b-5p was variable across the glioma cell lines but uniformly lower than non-neoplastic cell cultures, including mature astrocytes and human neural stem cells (hNSC) (Fig. 5A). In contrast, expression of miR-125b-1-3p and miR-125b-2-3p was similar to non-neoplastic cells, with the exception of decreases in miR-125b-1-3p in UW479 and miR-125b-2-3p in BT66 and UW479 (Supplementary Fig. 2). Given our prior observations demonstrating decreased levels of miR-125 family members in PLGG, we selected Res186, Res259 (both lacking BRAF alterations) and BT66 (which carries a BRAF-KIAA1549 fusion) for further functional experiments. Since miR-125b-5p expression levels were very low in these lines, and there is a lack of a protein product to use in measuring knockdown efficiency, only overexpression experiments were performed. miR-125b was overexpressed in these lines using lentiviral based miRNA plasmids (Fig. 5B). MiR-125b overexpression in Res186 (~3 fold), Res259 (~6 fold) and BT66 (~7 fold) uniformly resulted in decreased cell growth (Fig. 5C,D, Supplementary Fig. 3). Differences in miR-125b expression after transfection were likely related to genetic and phenotypic differences between the cells since they are PLGG-derived but with differences in genetic composition.
Figure 5.
Overexpression of miR-125b decreases cell growth. (A) qRT-PCR of miR-125b-5p expression in pediatric cell lines. (B) Overexpression of miR-125b in Res186, Res259 and BT66 (BRAF fusion+) cells. All data were normalized with miR-control infected cells. (**p < 0.01 compare to miR-control). (C) Cell growth curve of Res186, Res259 and BT66 cells infected with miR-control and miR-125b. (*p < 0.05 and **p < 0.01 compare to miR-control). (D) Quantification of BrdU incorporation is at left and BT66 analyzed data of flow cytometer is at right (*p < 0.05 compare to miR-control). Results represent the average of at least three biological replicates and data were analyzed with a two-tailed Student’s t-test.
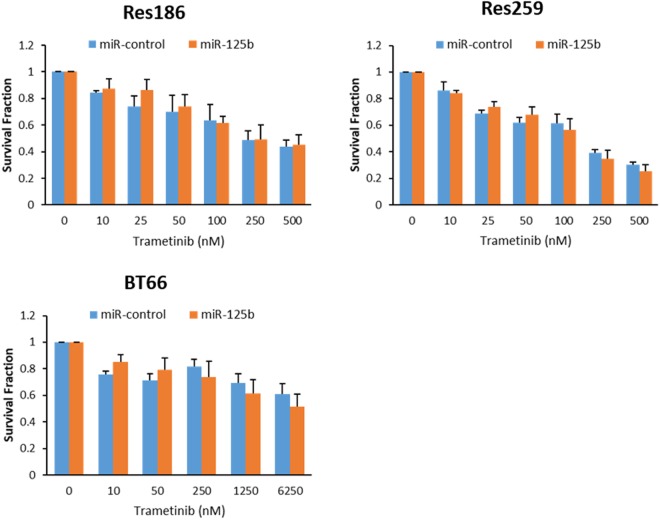
Since MAPK is activated at the genetic level in almost all PLGG6, MEK inhibitors have been used in preclinical and clinical trials for the treatment of this disease. Treatment with the MEK inhibitor trametinib had no significant effect on growth in cells with miR-125b overexpression when compared with control (Fig. 6).
Figure 6.
Treatment with the MEK-inhibitor Trametinib has no effect on cell growth in cells with miR-125b overexpression. Evaluation of cell growth after treatment with the MEK-inhibitor Trametinib in BT66, Res186, and Res259 cells overexpressing miR-125b. All data were normalized with DMSO control. The data represents the average of at least 3 biologic replicates.
miR-125b overexpression leads to senescence in BRAF-KIAA1549 PA cells and apoptosis in pediatric low grade glioma lines (PLGG)
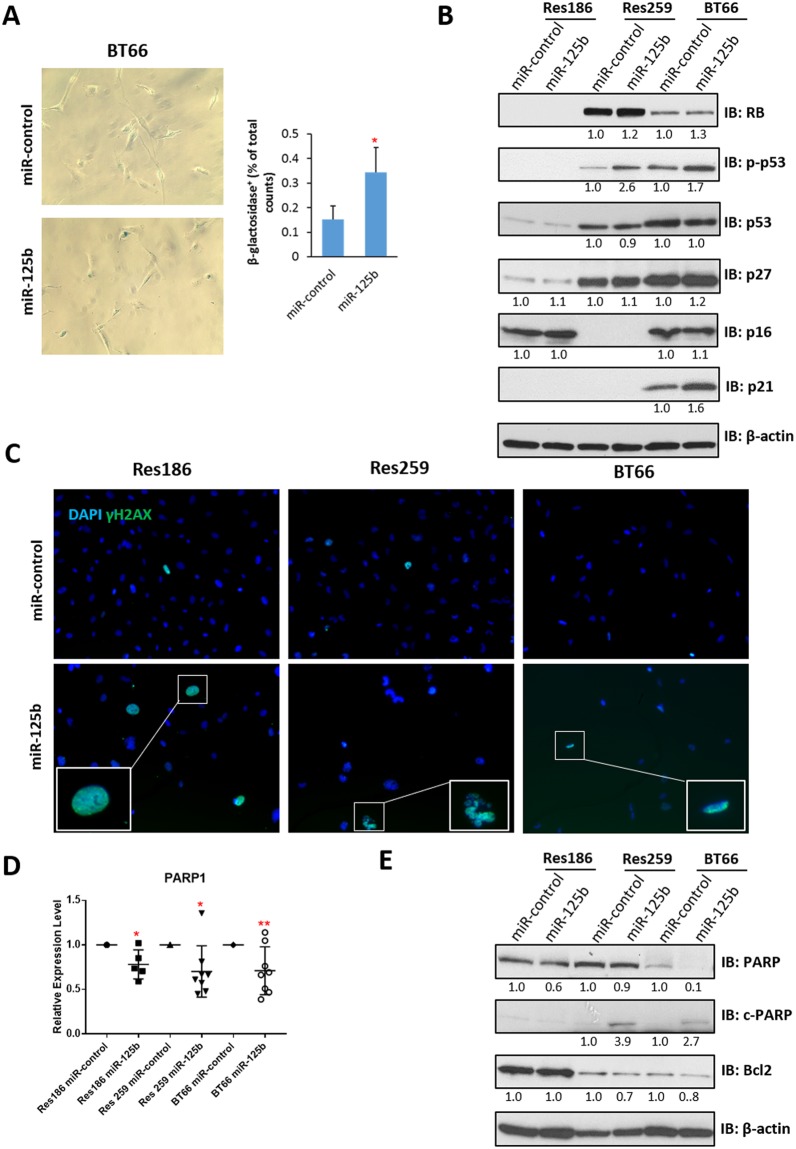
Prior melanoma research has demonstrated that miR-125b induces cell senescence28. Given the high frequency of BRAF mutations in melanoma and PLGG, we hypothesized that senescence may develop in PLGG with BRAF alterations. Indeed, overexpression of miR-125b led to induction of acidic β-galactosidase activity in the BT66 cell line (Fig. 7A), but not in Res186 and Res259 (Supplementary Fig. 4). Next, testing for the levels and activation of proteins associated with senescence pathways was performed. The RB1/p16 senescence pathway is dysfunctional in Res186 (with lack of RB1) and Res 259 (with lack of p16). BT66 demonstrated an increase in the senescence marker p21 while p27 was minimally increased (Fig. 7B, Supplementary Fig. 5). BT66 and Res259 had increased levels of p-p53, consistent with activation of the pathway (Fig. 7B). Absence of p53 in Res186 is expected given that it has a TP53 mutation. The results suggest that senescence after miR-125b overexpression is mediated through TP53/p21 rather than RB1/p16, which is concordant with data published by Nyholm et al. in melanoma cells which lack p1628.
Figure 7.
Overexpression of miR-125b induces senescence and apoptosis. (A) Representative photomicrographs (200x) of β-galactosidase staining on BT66 cells infected with miR-control or miR-125b, quantification of staining is at right. (*p < 0.05 compare to miR-control). (B) Western blot of senescence proteins p16, p21, p27, p53 and RB. β-actin was used as loading control. Protein levels were quantified and normalized to miR-control. (C) Representative photomicrographs (200x) of γ-H2AX shows increasing of γ-H2AX positive cells with miR-12b overexpression in all cell lines tested. (D) qRT-PCR of PARP1 expression in Res186, Res259 and BT66 (BRAF fusion+) cells infected with miR-control or miR-125b. All data were normalized to miR-control for each cell lines. (*p < 0.05, **p < 0.01). (E) Western blot of apoptotic protein PARP and the anti-apoptotic protein Bcl2. β-actin was used as loading control. Protein levels were quantified and normalized to miR-control (uncropped Western blots presented as Supplementary Fig. 4). Results represent the average of at least three biological replicates and data were analyzed with a two-tailed Student’s t-test.
Given the decrease in cell growth noted after miR-125b we next tested whether apoptosis was a factor. Prior reports have found “panstaining” with γ-H2AX identifies apoptotic cells35, so we stained cells that overexpressed miR-125b or miR-125b control using γ-H2AX immunofluorescence. An increased number of γ-H2AX positive cells with a diffuse pattern of staining were present in Res186, Res259 and BT66 after miR-125b overexpression consistent with apoptosis (Fig. 7C). Apoptosis was noted in all three cells lines, including cells with mutant and functional p53. This suggests that apoptosis after miR-125b overexpression in these cells is at least in part p53 independent.
Next, we looked for expression levels of known miR-125b targets, focusing on genes involved in apoptosis. miR-125b overexpression resulted in variable downregulation of these targets at the mRNA level in some but not all the cell lines, including BBC3 (Res259 and BT66) and BMF (Res 259) (Supplementary Fig. 6). The DNA repair enzyme Poly (ADP-ribose) polymerase 1 (PARP1) is also a predicted miR-125b target that was downregulated at the mRNA (20–30%) (Fig. 7D) and protein levels (Fig. 7E, Supplementary Fig. 5) in Res186, Res259 and BT66 overexpressing miR-125b. Static levels of cleaved PARP, a result of apoptosis, were increased in Res259 and BT66 (Fig. 7E). A small increase in cleaved PARP was also identified in Res186 but only when cultured longer (>1 week).
miR-125b overexpression decreases cell invasion
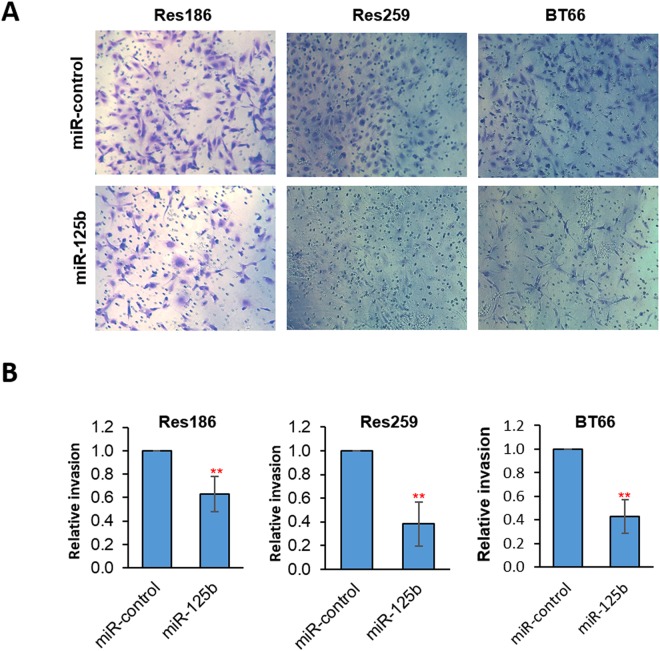
We tested whether miR-125b overexpression affects other glioma cell properties in addition to cell growth. mir-125b overexpression resulted in decreased invasion in Res186, Res259 and BT66 using the transwell assay (Fig. 8).
Figure 8.
Overexpression of miR-125b decreases cell invasion. Transwell invasion assay of Res186, Res259 and BT66 (BRAF fusion+) cells infected with miR-control or miR-125b, migrated cells were photographed (200x) by inverted light microscopy (A) or quantitated with Cell-Titer blue (B). All data were normalized to miR-control. (**p < 0.01). Results represent the average of at least three biological replicates and data were analyzed with a two-tailed Student’s t-test.
Discussion
In the current study, miR-125 members emerged as a microRNA group downregulated in low grade, predominantly pediatric, and glioneuronal tumors compared to diffuse astrocytomas and non-neoplastic brain. The precise reason for this finding is unclear, but our results showed a non-significant trend for lower miR-125b expression in tumors with BRAF alterations, with the caveat that only BRAF p.V600E and BRAF duplications were tested and several other rarer alterations may also affect BRAF and other components of the MAPK pathway in PLGG6. When looking at miR-125b levels in pediatric glioma-derived cell lines, we did find variable expression in all cell lines tested, but in general the levels were lower in glioma lines, low and high grade, than in neural stem cell and astrocyte cultures. This raises the possibility that interactions with the microenvironment in vivo, may also play a role in miR-125b levels, as it has been reported for other microRNAs in cancer36. Overexpression of miR-125b resulted in apoptosis in all three cell lines tested. Senescence, however, was identified only in BT66, a line derived from a PA with BRAF-KIAA1549 fusion, but not in Res186 and Res259. Our data suggests that miR-125b downregulation may play a role in preventing senescence in tumors that depend on BRAF for cell growth. Known and predicted targets of miR-125b reported in prior studies of cancers characterized by BRAF alterations include MLK3, KLF13, CXCL11, and FOXA1. In our study, we found variable levels of miR-125b targets at the mRNA level. The DNA repair enzyme PARP1, however, was a predicted target of miR-125b that was downregulated at the mRNA and protein levels in miR-125b overexpressing cells, particularly BT66. The detection of PARP cleavage products is a reflection of apoptosis, and these were present as well in our studies to various extents in all cell lines tested. Prior studies in ovarian cancer have shown that siRNA PARP knockdowns lead to decreased PARP levels, increased cleaved PARP and apoptosis37. Of interest, combination of PARP and MEK inhibitors have been attempted in RAS activated cancers38 and may represent a therapeutic opportunity for PLGG. Our observations of miR-125b dysregulation have translational implications, since decreased serum miR-125b has been reported in glioma patients, supporting its potential utility as a biomarker39, and efforts to combine miR-125b with chemotherapy and targeted therapies are being explored40. We did not see an additive effect of MEK inhibition with trametinib, which is being increasingly applied to tumors with MAPK pathway activation, on cell growth in cells with miR-125b knockdown. Whether this lack of additional effect was a feature of this specific inhibitor or related to a more broad mechanism of MEK inhibitor remains unclear. However, our data suggests that the study of future combination therapies including anti-miR-125b as a strategy may require approaches with other pathway inhibitors and/or chemotherapeutic agents.
There are some limitations regarding the cell lines used for this study. Res186 and Res259 lack known BRAF mutations. BT66 maintains a BRAF fusion, but was engineered to bypass senescence through the SV40 large T antigen, which is not a feature of PA in general41. These efforts represent important attempts to study the biology of PLGG, given that cell lines and models of this disease are relatively unavailable, given the limitations posed by oncogene-induced senescence as mentioned above29.
In summary, we identified miR-125b as a biomarker that is under-expressed in PLGG subsets. Overexpression resulted in decreased cell growth, as well as increased apoptosis and senescence in cells containing activated BRAF. Further studies should clarify the specific mechanism of action for this key microRNA and its possible applicability in the management of PLGG patients.
Materials and Methods
Patients and Samples
Tumors for in situ hybridization were studied using two tissue microarrays, which are constructed using four cores (0.6 mm diameter). A total of 125 patients (median age 17 years, range 1–75) with low grade glioma and glioneuronal tumors (defined as WHO grade I or II) including pilocytic astrocytomas (n = 43), diffuse astrocytoma (n = 24), low grade glioneuronal tumor NOS (n = 3), low grade astrocytoma NOS (n = 8), pleomorphic xanthoastrocytoma (n = 8), angiocentric glioma (n = 4), ganglioglioma (n = 13), dysembryoplastic neuroepithelial tumor (n = 4), and subependymal giant cell astrocytoma (n = 2). Given the rarity of these tumors and the need to broadly represent all the major categories of low grade glial and glioneuronal neoplasms in these tissue microarrays, in addition to tumors developing in pediatric patients (defined as 18 years of age and younger prior to pathologic diagnosis) (n = 66), tumors with the same pathologic characteristics developing in adults (n = 59) were also included in these TMAs. Whole slides were obtained from 3 non-neoplastic seizure brain cases (ages 32, 4 and 2 years) and autopsy samples from a 51 year-old adult, and two fetuses (35 and 25 weeks).
BRAF duplication was evaluated using FISH42 and BRAF p.V600E was evaluated using sequencing43 or immunohistochemistry. Immunohistochemistry was performed using an antibody specific for BRAF p.V600E mutant protein (VE1, Ventana, prediluted). All experiments were followed under approval of the Institutional Review Board (IRB) at Johns Hopkins (IRB study number NA_00015113). All research was performed in accordance with all relevant ethical and institutional guidelines/regulations, and the data contains no patient identifiers. The analysis of samples was retrospective and a waiver of consent for the use of patient tissue was applied using all IRB guidelines.
microRNA profiling data
MicroRNA expression data was obtained from two separate experiments obtained with different platforms. First, global microRNA profiles from 43 PA and 5 non-neoplastic brain control frozen samples were obtained with the Agilent Human miRNA Microarray V3 platform in a previously published study14. Fold change was calculated as previously reported14, comparing expression in tumor and non-neoplastic brain, after miRNA signal intensities from GeneView files were subjected to quantile normalization using GeneSpring GX, version 11, according to the standard software recommendations (Agilent Technologies). Second, global microRNA profiles from 41 glial/glioneuronal tumors and 4 non-neoplastic brain FFPE samples were obtained using the Nanostring digital counting system in another previously published study13. Demographics for individual cases utilized in these studies are outlined in the prior publications.
Chromogenic in situ hybridization studies
For microRNA in situ hybridization experiments, Ex miRCURY LNA™ Detection probes, 5′-DIG and 3′-DIG labeled (Exiquon, Woburn, MA), were used. The target probes recognized miR-125b and had the following sequence: 5′-TCACAAGTTAGGGTCTCAGGGA-3′. Positive controls to assess the effectiveness of the reaction included probes recognizing U6 and miR-124, a known brain enriched micrRNA. A probe composed of the scrambled sequence of the miR target was used as a negative control. The FFPE optimization kit was used per manufacturer recommendations. In brief, deparaffination was started by placing slides flat on a warm surface for 1 hour at 60 °C, followed by immersions in xylene X3 for 5 minutes each, and serial passage in ethanol at 100%, 95% and 70%, and PBS. This was followed by incubation with Proteinase-K for 20 minutes at 37 °C. Slides were placed PBS and dehydrated using serial ethanol washes (70%, 95%, 100%). Hybridization mix, with separate slides for target and control probes, was applied and incubated for 1 hour at 55 °C in a hybridizer (DAKO, Ft. Collins, CO). Slides were washed in serial SSC buffers at concentrations of 5x, 1x, and 0.2x. Then, slides were incubated with blocking solution for 15 minutes, followed by application of anti-DIG reagent for 60 minutes (with the reagent replenished after 30 minutes). Slides were washed in PBS-T and incubated with AP-substrate for 2 hours at 30 °C, with fresh solution replenished after 1 hour. Slides were incubated at room temperature in KTBT solution for 5 minutes x2, washed with water and counterstained with Nuclear Fast Red™, rinsed, serially dehydrated with Ethanol (70%, 95%, 100%), and coverslipped using mounting media for light microscopy evaluation. Strength of hybridization was scored using a 4 tiered scale semiquantifiable scale, ranging from 3 (strong, similar to brain), 2 (moderate), 1 (weak), 0 (negative), similar to standard evaluations of cytoplasmic immunohistochemical stains as outline in our prior work (REF). Scoring was performed by a neuropathologist (FJR) blinded to demographic characteristics and pathologic diagnosis, using an Olympus BX43 microscope with a 40X objective. Additional figures were taken using 10X and 20X objectives.
Cell culture
Pediatric glioma cell lines Res186, Res259 (PLGG derived lines lacking BRAF alterations) and UW479 were kindly provided by Dr. Chris Jones (Institute of Cancer Research, Sutton, UK)44, and maintained in Dulbecco modified Eagle medium/ F12 Ham medium supplemented with 10% heat-inactivated fetal bovine serum (ThermoFisher Scientific). BT66 originated from a pilocytic astrocytoma containing a BRAF-KIAA1549 fusion. These cells contain doxycycline-inducible expression of Simian Vacuolating Virus 40 Large T Antigen (SV40-TAg) and stable expressed human Telomerase reverse transcriptase (TERT). BT66 cells were kindly provided by Dr. Till Milde (German Cancer Research Center, Heidelberg, Germany) and maintained in astrocyte growth medium (Lonza) with 1% doxycycline (MilliporeSigma)41, cells around passage 38–50 were used for all experiments. TrypLE Express was used for cell dissociation (ThermoFisher Scientific). Pediatric glioblastoma cell line KNS-42 was obtained from the Japan Cancer Research Resources cell bank; CHLA-200 and SJ-GBM2 were obtained from the Children’s Oncology Group (COG) Cell Culture and Xenograft Repository. JHH-CRC1 was derived from a pediatric patient with NF1 associated pilocytic astrocytoma (unpublished data). Human astrocytes were purchased from ScienCell Research Laboratories. Human neural stem cells (hNSC) were kindly provided by Dr. Eric Raabe (Johns Hopkins University, Baltimore, USA)45. All cells were cultured in a humidified 37 °C incubator with 5% CO2. Cell lines were routinely tested for mycoplasma and maintained in the culture for less than 2 months after thawed from liquid nitrogen. Short tandem repeat (STR) identity testing was performed for Res186, Res259 and BT66 on January 2018.
miRNA overexpression
Lentiviral based miRNA plasmids were obtained from Biosettia, including expression plasmids pLV-hsa-miR-125b-1 (miR-125b) and pLV-miR-control (miR-control). To produce lentiviruses, 293T cells were transfected with miRNA plasmid and VSVG packaging plasmids mixture using Lipofectamine 2000 (ThermoFisher Scientific). Lentiviral supernatants were collected at 48 h post transfection, passed through a 0.45 micron filter then kept as frozen aliquots at −80 °C until needed. Res186, Res259 and BT66 cells were infected with virus in the presence of polybrene (5 µg/ml, MilliporeSigma).
Cell growth and cell survival assay
The CellTiter-Blue cell viability assay kit (Promega) was used to count viable cells. 72 hours after virus infection, cells were seeded in 96 well plates at a density of 1000 cells per well in growth media. Twenty microliters of CellTiter-Blue reagent was added to 100 µl medium and incubated for 1 hour at 37 °C incubator. Fluorescence (560 nmEx/590 nmEm) was continuously measured for 5 days using TECAN plate reader. For drug treatment, cells were cultured in the growth media with 2% heat inactived FBS with different doses of trametinib (Skellchem) for 5 days, DMSO was used as vehicle control. Relative cell numbers were converted with standard curve of each cell line.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Primers for miRNA were designed by using miRprimer software46, sequences can be found in Supplementary Table 1, small RNA RNU48 was used as the endogenous control. Primer sequences for mRNA target genes were listed in Supplementary Table 2, HPRT1 was used as the endogenous control. Total RNA was isolated from cultured cells using miRNeasy mini kit (QIAGEN), reverse transcription of miRNA was performed according to Peter K Bush’s protocol46, and cDNA for target genes were produced using QuantiTect reverse transcription kit (QIAGEN). qRT-PCR was performed using PowerUp SYBR Green Master Mix (ThermoFisher Scientific) according to the manufacturer’s protocol. The relative fold changing was calculated using the formula 2ΔCT.
Bromodeoxyuridine (BrdU) incorporation assay
72 hours after virus infection, cells were incubated with 10 µM BrdU (MilloporeSigma) for 2 hours, dissociated and fixed with 70% ethanol overnight at 4 °C. Cells were washed with PBS and incubated with 2 N HC for 30 minutes at 37 °C. After three times of PBS washing, cells were permeabilized with 0.1% Triton and stained with BrdU-PE (eBioscience) for 30 minutes at room temperature, analyzed with Muse flow cytometer (MilloporeSigma).
In vitro invasion assay
Cell invasion assay was analyzed using a 24 well system with growth factor reduced Matrigel coated transwell inserts containing 8 µM pore membrane (Fisher Scientific). 72 hours after virus infection, 1 × 105 cells in 250 µl of DMEM/F12 were added on top of each membrane and fill bottom with 750 µl of complete growth medium. After 22 hours, migrated cells from membrane were fixed and stained with 0.05% crystal violet in 50% methanol or dissociated by using TrypLE Express (ThermoFisher Scientific), relative cell numbers were measured by using Cell-Titer blue reagent (Promega) and converted with standard curve of each cell line.
Western blotting
Cells were lysed in RIPA lysis buffer supplemented with protease inhibitors (MilliporeSigma). Protein extracts were loaded on 4–12% Bis-Tris gel. Antibodies used for western blot were: Bcl2 (#2870, 1:1000, Cell Signaling Technology), p16 (sc-56330,1:200, Santa Cruz), p21 (#2947, 1:1000, Cell Signaling Technology), p27 (sc-1641,1:500, Santa Cruz), p53 (#9282, 1:1000, Cell Signaling Technology), Phospho-p53 (Ser15) (#9286, 1:1000, Cell Signaling Technology), PARP (#9542, 1:1000, Cell Signaling Technology), Cleaved PARP (Asp214) (#5625, 1:1000, Cell Signaling Technology), RB (#9309, 1:2000, Cell Signaling Technology), β-actin (sc-47778, 1:5000, Santa Cruz). The appropriate HRP-conjugated secondary antibodies (1:5000, Cell Signaling Technology) was used and blots were revealed by chemiluminescence (ECL, PerkinElmer). For Phospho-p53 and p53 western blots, membrane was stripped with restore western blot stripping buffer (ThermoFisher Scientific) and re probed. Densitometry analysis was performed using ImageJ software.
Senescence Assay
β-galactosidase staining was performed as per the manufacturer’s instructions (#9680, Cell Signaling Technology). Cells were infected with miR-125b or miR-control virus, 72 hours later 50000 cells were plated on 12-well plates. After 48 hoursculture, cells were fixed and stained for overnight. Cells were photographed by inverted light microscopy with 20x objective.
Immunofluorescence staining for γ-H2AX
For immunofluorescence staining, cells were cultured on the glass coverslips and fixed with 4% paraformaldehyde for 15 min. After three times washing with PBS, cells were incubated for 45 min in blocking buffer containing PBS, 1% bovine serum albumin, 0.1% Triton-X-100 and 5% donkey serum, then incubated overnight at 4 °C with anti-γ-H2AX antibody (#2870, 1:200, Cell Signaling Technology). Cells were washed three times with PBS and incubated for 1 hour with Fluorescein (FITC) conjugated Donkey Anti-rabbit IgG (H + L) (711-095-152, 1:100, Jackson ImmunoResearch Laboratories). DAPI (MilliporeSigma) solution was applied to stain nuclei. Cells were mounted with Prolong Gold anti-fade mounting media (ThermoFisher Scientific) and visualized by Motic AE31 inverted microscopy with a 20x objective.
Statistical Analysis
Means and ranges were used to describe the in situ hybridization scores as appropriate. Statistical tests were performed using Associations with clinicopathologic variables was assessed with contingency tables and the Chi Square test. Continuous variables were compared with the Wilcoxon/Kruskal-Wallis Test. Statistical analyses were performed using JMP v10 software (SAS Institute). Analyses were 2-sided with p < 0.05 considered statistically significant. For cell culture and functional assays, data were presented as mean ± s.d, with p < 0.05 considered statistically significant. All in vitro experiments described in the above sections (including gene expression data, cell proliferation data, H2Ax assays, senescence assays), were performed in at least three biological replicates and data were analyzed with a two-tailed Student’s t-test.
Electronic supplementary material
Acknowledgements
This project was funded in part by the Childhood Brain Tumor Foundation, the Pilocytic/Pilomyxoid Fund, including Lauren’s First and Goal, and the Stick it to Brain Tumors Annual Women’s Ice Hockey Tournament (F.J.R., CGE); the PLGA foundation (F.J.R, CGE), NIH grant P30 CA006973 to the Sidney Kimmel Comprehensive Cancer Center (PI: W. Nelson) and Capes (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – process number: 1371-14-2) a foundation from the Education and Culture Ministry of Brazil (to ACALS).
Author Contributions
Ming Yuan performed and planned experiments, analyzed data, participated in the writing of the manuscript. Ana Cristina A.L. Da Silva performed and planned experiments, analyzed data, participated in the writing of the manuscript. Antje Arnold performed and planned experiments, analyzed data, participated in the writing of the manuscript. Laurence Okeke performed and planned experiments, analyzed data, approved manuscript in final form. Heather Ames performed and planned experiments, analyzed data, approved manuscript in final form. Lina S. Correa-Cerro performed experiments, analyzed data, approved manuscript in final form. M. Adelita Vizcaino performed experiments, analyzed data, approved manuscript in final form. Cheng-Ying Ho performed experiments, analyzed data, approved manuscript in final form. Charles G. Eberhart planned experiments, analyzed data, approved manuscript in final form. Fausto J. Rodriguez performed and planned experiments, analyzed data, supervised experiments, participated in the writing of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ming Yuan and Ana Cristina A. L. Da Silva contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-30942-4.
References
- 1.Bar EE, Lin A, Tihan T, Burger PC, Eberhart CG. Frequent gains at chromosome 7q34 involving BRAF in pilocytic astrocytoma. Journal of neuropathology and experimental neurology. 2008;67:878–887. doi: 10.1097/NEN.0b013e3181845622. [DOI] [PubMed] [Google Scholar]
- 2.Forshew T, et al. Activation of the ERK/MAPK pathway: a signature genetic defect in posterior fossa pilocytic astrocytomas. J Pathol. 2009;218:172–181. doi: 10.1002/path.2558. [DOI] [PubMed] [Google Scholar]
- 3.Jones DT, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer research. 2008;68:8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfister S, et al. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest. 2008;118:1739–1749. doi: 10.1172/JCI33656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sievert AJ, et al. Duplication of 7q34 in pediatric low-grade astrocytomas detected by high-density single-nucleotide polymorphism-based genotype arrays results in a novel BRAF fusion gene. Brain Pathol. 2009;19:449–458. doi: 10.1111/j.1750-3639.2008.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones DT, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45:927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramkissoon, L. A. et al. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc Natl Acad Sci USA, 10.1073/pnas.1300252110 (2013). [DOI] [PMC free article] [PubMed]
- 8.Zhang, J. et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet, 10.1038/ng.2611 (2013). [DOI] [PMC free article] [PubMed]
- 9.Ames H, Halushka MK, Rodriguez F. J. miRNA Regulation in Gliomas: Usual Suspects in Glial Tumorigenesis and Evolving Clinical Applications. J Neuropathol Exp Neurol. 2017;76:246–254. doi: 10.1093/jnen/nlx005. [DOI] [PubMed] [Google Scholar]
- 10.White FV, et al. Nonrandom chromosomal gains in pilocytic astrocytomas of childhood. Hum Pathol. 1995;26:979–986. doi: 10.1016/0046-8177(95)90087-X. [DOI] [PubMed] [Google Scholar]
- 11.Birks DK, et al. Survey of MicroRNA expression in pediatric brain tumors. Pediatr Blood Cancer. 2011;56:211–216. doi: 10.1002/pbc.22723. [DOI] [PubMed] [Google Scholar]
- 12.Jones TA, et al. Molecular analysis of pediatric brain tumors identifies microRNAs in pilocytic astrocytomas that target the MAPK and NF-kappaB pathways. Acta Neuropathol Commun. 2015;3:86. doi: 10.1186/s40478-015-0266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ames HM, Yuan M, Vizcaino MA, Yu W, Rodriguez FJ. MicroRNA profiling of low-grade glial and glioneuronal tumors shows an independent role for cluster 14q32.31 member miR-487b. Mod Pathol. 2017;30:204–216. doi: 10.1038/modpathol.2016.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho CY, et al. MicroRNA profiling in pediatric pilocytic astrocytoma reveals biologically relevant targets, including PBX3, NFIB, and METAP2. Neuro Oncol. 2013;15:69–82. doi: 10.1093/neuonc/nos269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sredni ST, Huang CC, Bonaldo Mde F, Tomita T. MicroRNA expression profiling for molecular classification of pediatric brain tumors. Pediatric blood & cancer. 2011;57:183–184. doi: 10.1002/pbc.23105. [DOI] [PubMed] [Google Scholar]
- 16.Iyer A, et al. MicroRNA-146a: a key regulator of astrocyte-mediated inflammatory response. PloS one. 2012;7:e44789. doi: 10.1371/journal.pone.0044789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prabowo AS, et al. Differential expression and clinical significance of three inflammation-related microRNAs in gangliogliomas. Journal of neuroinflammation. 2015;12:97. doi: 10.1186/s12974-015-0315-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 19.Sempere LF, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013;6:6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, Mao W, Zheng S, Ye J. Epidermal growth factor receptor-regulated miR-125a-5p–a metastatic inhibitor of lung cancer. FEBS J. 2009;276:5571–5578. doi: 10.1111/j.1742-4658.2009.07238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott GK, et al. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–1486. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 23.Jia HY, et al. MicroRNA-125b functions as a tumor suppressor in hepatocellular carcinoma cells. Int J Mol Sci. 2012;13:8762–8774. doi: 10.3390/ijms13078762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morelli E, et al. Selective targeting of IRF4 by synthetic microRNA-125b-5p mimics induces anti-multiple myeloma activity in vitro and in vivo. Leukemia. 2015;29:2173–2183. doi: 10.1038/leu.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu N, et al. MiR-125b acts as an oncogene in glioblastoma cells and inhibits cell apoptosis through p53 and p38MAPK-independent pathways. Br J Cancer. 2013;109:2853–2863. doi: 10.1038/bjc.2013.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Z, et al. miR-125b inhibits Connexin43 and promotes glioma growth. Cell Mol Neurobiol. 2013;33:1143–1148. doi: 10.1007/s10571-013-9980-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu N, et al. miR-125b regulates the proliferation of glioblastoma stem cells by targeting E2F2. FEBS Lett. 2012;586:3831–3839. doi: 10.1016/j.febslet.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 28.Nyholm AM, et al. miR-125b induces cellular senescence in malignant melanoma. BMC Dermatol. 2014;14:8. doi: 10.1186/1471-5945-14-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raabe EH, et al. BRAF activation induces transformation and then senescence in human neural stem cells: a pilocytic astrocytoma model. Clin Cancer Res. 2011;17:3590–3599. doi: 10.1158/1078-0432.CCR-10-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacob K, et al. Genetic aberrations leading to MAPK pathway activation mediate oncogene-induced senescence in sporadic pilocytic astrocytomas. Clin Cancer Res. 2011;17:4650–4660. doi: 10.1158/1078-0432.CCR-11-0127. [DOI] [PubMed] [Google Scholar]
- 31.Tacutu R, Budovsky A, Yanai H, Fraifeld VE. Molecular links between cellular senescence, longevity and age-related diseases - a systems biology perspective. Aging (Albany NY) 2011;3:1178–1191. doi: 10.18632/aging.100413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lafferty-Whyte K, Cairney CJ, Jamieson NB, Oien KA, Keith WN. Pathway analysis of senescence-associated miRNA targets reveals common processes to different senescence induction mechanisms. Biochim Biophys Acta. 2009;1792:341–352. doi: 10.1016/j.bbadis.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Glud M, et al. MicroRNA miR-125b induces senescence in human melanoma cells. Melanoma Res. 2011;21:253–256. doi: 10.1097/CMR.0b013e328345333b. [DOI] [PubMed] [Google Scholar]
- 34.McCall MN, et al. Toward the human cellular microRNAome. Genome Res. 2017;27:1769–1781. doi: 10.1101/gr.222067.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivashkevich A, Redon CE, Nakamura AJ, Martin RF, Martin OA. Use of the gamma-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett. 2012;327:123–133. doi: 10.1016/j.canlet.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki HI, Katsura A, Matsuyama H, Miyazono K. MicroRNA regulons in tumor microenvironment. Oncogene. 2015;34:3085–3094. doi: 10.1038/onc.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldberg MS, et al. Nanoparticle-mediated delivery of siRNA targeting Parp1 extends survival of mice bearing tumors derived from Brca1-deficient ovarian cancer cells. Proc Natl Acad Sci USA. 2011;108:745–750. doi: 10.1073/pnas.1016538108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, C. et al. Rational combination therapy with PARP and MEK inhibitors capitalizes on therapeutic liabilities in RAS mutant cancers. Sci Transl Med9, 10.1126/scitranslmed.aal5148 (2017). [DOI] [PMC free article] [PubMed]
- 39.Wei X, Chen D, Lv T, Li G, Qu S. Serum MicroRNA-125b as a Potential Biomarker for Glioma Diagnosis. Mol Neurobiol. 2016;53:163–170. doi: 10.1007/s12035-014-8993-1. [DOI] [PubMed] [Google Scholar]
- 40.Shi L, Fei X, Wang Z, You Y. PI3K inhibitor combined with miR-125b inhibitor sensitize TMZ-induced anti-glioma stem cancer effects through inactivation of Wnt/beta-catenin signaling pathway. In Vitro Cell Dev Biol Anim. 2015;51:1047–1055. doi: 10.1007/s11626-015-9931-x. [DOI] [PubMed] [Google Scholar]
- 41.Selt F, et al. Establishment and application of a novel patient-derived KIAA1549:BRAF-driven pediatric pilocytic astrocytoma model for preclinical drug testing. Oncotarget. 2017;8:11460–11479. doi: 10.18632/oncotarget.14004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez FJ, et al. BRAF duplications and MAPK pathway activation are frequent in gliomas of the optic nerve proper. J Neuropathol Exp Neurol. 2012;71:789–794. doi: 10.1097/NEN.0b013e3182656ef8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin A, et al. BRAF alterations in primary glial and glioneuronal neoplasms of the central nervous system with identification of 2 novel KIAA1549:BRAF fusion variants. J Neuropathol Exp Neurol. 2012;71:66–72. doi: 10.1097/NEN.0b013e31823f2cb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bax DA, et al. Molecular and phenotypic characterisation of paediatric glioma cell lines as models for preclinical drug development. PLoS One. 2009;4:e5209. doi: 10.1371/journal.pone.0005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao XG, et al. LIN28A facilitates the transformation of human neural stem cells and promotes glioblastoma tumorigenesis through a pro-invasive genetic program. Oncotarget. 2013;4:1050–1064. doi: 10.18632/oncotarget.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Busk PK. A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinformatics. 2014;15:29. doi: 10.1186/1471-2105-15-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.