Abstract
Isolation of entomopathogenic nematodes from Mizoram, northeastern part of India includes a Heterorhabditis species never been recorded in India. The morphological and multigene (ITS rRNA, 28S rRNA and COI) sequences analysis had revealed that the isolated Heterorhabditis belongs to Heterorhabditis baujardi, originally described from Vietnam. The phylogenetic tree (both MP and ML) revealed that the H. baujardi belongs to H. indica clade, and further, forms a monophyletic clade (99/98% and 94/92% bootstrap support for ITS and 28S respectively) with H. amazonensis, H. floridensis, H. mexicana and H. taysearae forming H. baujardi sub clade. The multigene characterization revealed that both the ITS and 28S rRNA showed similar result in resolving the phylogenetic relationship of the genus Heterorhabditis, with the ITS rRNA being superior based on the strong bootstrap support, whereas, the cytochrome c oxidase I (M1–M6 partition) can be a good supportive tool for species delimitation. This is the first report of H. baujardi from India envisaging its future use as a biological control agent, and further incorporated into the IPM.
Keywords: Biological control, Cytochrome c oxidase I, Entomopathogenic nematode, ITS rRNA, 28S rRNA
Introduction
Entomopathogenic nematodes (EPNs), under the families Steinernematidae and Heterorhabditidae are natural insect parasites symbiotically associated with the enterobacteria viz. Xenorhabdus in steinernematids and Photorhabdus in heterorhabditids (Boemare 2002). Due to the nematode-bacteria association, which help in rapid killing of various insect pest, EPNs are used as a successful tool in biological control and integrated pest management programs throughout the world (Griffin et al. 2005). The efficient use of EPNs in biological control program relies on the isolation of native species as they are adapted to the prevailing environment and climatic conditions of that particular area (Stock et al. 1999).
At present, there are ninety-five Steinernema and sixteen Heterorhabditis species distributing world-wide (Hunt and Subbotin 2016), with the exception of Antarctica (Hominick 2002). In India, numerous surveys have been conducted to isolate the indigenous EPNs, resulting in the discovery of a new H. indica (Poinar et al. 1992) from Coimbatore, Kerala. There are altogether eleven species of valid entomopathogenic nematodes documented from India (Ganguly and Singh 2000; Hussaini et al. 2001; Ganguly et al. 2002, 2011; Lalramliana and Yadav 2010; Kulkarni et al. 2012; Bhat et al. 2017; Lalramnghaki et al. 2017), and the valid species includes Heterorhabditis bacteriophora, H. indica, S. abbasi (= S. thermophilum), S. bicornutum, S. carpocapsae (= S. meghalayense), S. glaseri, S. hermaphroditum (= S. dharanai), S. riobrave, S. sangi, S. siamkayai and S. surkhetense.
Due to the overgrowing complications and confusion arising with the use of morphological base species identification among the nematodes, integration with molecular approach is being widely used today. The molecular markers, ITS and D2-D3 of 28S rRNA regions, have been used commonly in species delimitation and resolving phylogenetic relationships in EPN taxonomy (Spiridonov and Subbotin 2016). The objective of the study is to isolate and identify the entomopathogenic nematode from Mizoram, northeastern India, and characterize the three molecular markers viz. ITS rRNA, 28S rRNA and COI (M1–M6 partition) for further use in species delimitation and resolving phylogenetic relationship of the genus Heterorhabditis.
Materials and methods
Isolation and identification
The nematode was isolated from soil samples collected near Tamdil Lake (23.741N 92.951E), Mizoram, NE India following Bedding and Akhurst (1975). Ten (10) numbers of last instar larvae of Galleria mellonella were used as an insect baits and observed for a period of 15 days. The nematodes were extracted following Kaya and Stock (1997), and all the extracted nematodes which passed through the reinfection process were stored for further use.
Different stages (30 each) of the nematode, viz. infective juveniles, hermaphroditic females, amphimictic females and males, were randomly selected for morphological studies. The adults were dissected out from the insect cadavers in Ringer’s solution and killed at 60 °C. Tri-ethanolamine-formalin (TAF) fixative was used for fixation, processed to anhydrous glycerine (Seinhorst 1959), and then observed under Olympus CX41 microscope.
Extraction of DNA was done from a single hermaphrodite female nematode. Gene amplification were performed using primers TW81 (5′-GTTTCCGTAGGTGAACCTGC-3′) forward and AB28 (5′-ATATGCTTAAGTTCAGCGGGT-3′) reverse for ITS (Joyce et al. 1994); (5′-CGATAGCGAACAAGTACCGAGAG-3′) forward and (5′-CCTGCTCAGGCATAGTTCACCATC-3′) reverse for 28 S (Qiu et al. 2011); and LCO 1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) forward and HCO 2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) reverse for COI (M1–M6 partition) (Folmer et al. 1994). The conditions for PCR amplification of ITS and 28S region was similar and follows Lalramnghaki et al. (2017); and a condition for COI was: 94 °C for 3 min (1 cycle), followed by 94 °C for 1 min (36 cycles), 50 °C for 30 s and 72 °C for 45 s, followed by a final extension at 72 °C for 7 min. Sequencing of the PCR products were done in both directions at Agrigenome, Kochi, Kerala, India.
Alignment and analysis of the sequences
A software package, FinchTV 1.4.0 (Geospiza, Inc.; Seattle, WA, USA; http://www.geospiza.com), was used for editing the sequences generated, and alignment was performed using Clustal X 1.64 (Thompson et al. 1997). Genetic distance was calculated by pairwise comparisons of sequences using the Kimura 2-Parameter method under Gamma distribution. The phylogenetic relationship was generated using the maximum likelihood (ML) and the maximum parsimony (MP), with all the available representative species of the genus Heterorhabditis retrieved from GenBank. The ML tree was constructed using T92 + G (Tamura-3-parameter + Gamma distribution) and K2 + G (Kimura 2 parameter + Gamma distribution) substitution models with 1000 bootstraps for ITS and 28S rRNA dataset respectively, based on the lowest BIC scores (Bayesian Information Criterion). The MP tree was constructed using the Close-Neighbor-Interchange algorithm. Caenorhabditis elegans (KX572972 and MF192964 for ITS and 28S respectively) obtained from the GenBank were used as the outgroup taxon. All positions containing gaps and missing data were eliminated. The analysis involved 21 and 19 nucleotide sequences and a total of 658 and 587 positions in the final dataset for ITS and 28S respectively. All the analyses were conducted in MEGA7 (Kumar et al. 2016).
Results
Morphological identification
The morphometric and morphological analysis of all stages of the isolated nematode (Table 1) revealed that it belongs to Heterorhabditis baujardi (Phan et al. 2003), originally described from Vietnam. All the measurements, including ratios (E, F, SW), and the morphological characters observed viz. shape of spicules and gubernaculums etc. clearly indicate that the isolated nematode is conspecific to H. baujardi.
Table 1.
Comparative morphometrics of Heterorhabditis baujardi (n = 30)
| Character | Infective juvenile | After Phan et al. (2003) | Hermaphrodite | After Phan et al. (2003) | Amphimictic female | After Phan et al. (2003) | Male | After Phan et al. (2003) |
|---|---|---|---|---|---|---|---|---|
| Body length | 571.3 ± 5.3 (525.0–615.0) |
487–622 | 3548 ± 38.9 (3250–3970) |
3105–5400 | 2185 ± 14.39 (2060–2290) |
1335–2385 | 814.7 ± 14.3 (710–902.5) |
641–1072 |
| Body width | 20.8 ± 0.4 (17.5–25.0) |
18–22 | 219.6 ± 3.9 (190–250) |
180–330 | 135.6 ± 1.5 (120–150) |
90–165 | 44.6 ± 0.6 (40.0–50.0) |
38–53 |
| Oesophagus length | 109.5 ± 1.4 (97.5–120.0) |
107–124 | 198.2 ± 1.19 (180–205) |
147–222 | 135.6 ± 1.4 (122.5–147.5) |
123–185 | 102.9 ± 1.0 (97.5–110) |
91–132 |
| EP | 91.5 ± 0.6 (87.5–95.5) |
90–105 | 164.6 ± 2.6 (150–185) |
119–v197 | 108.2 ± 1.3 (97.5–115) |
98–156 | 88 ± 0.7 (82.5–92.5) |
71–107 |
| Tail length | 101.2 ± 0.7 (95.0–107.5) |
79–105 | 91.4 ± 1.2 (80–105) |
66–126 | 89.8 ± 1.6 (77.5–107.5) |
65–110 | 36.8 ± 0.6 (32.5–40) |
25–33 |
| Anal body width | 15.3 ± 0.4 (12.5–17.5) |
11–14 | 56.6 ± 0.9 (50.0–65.0) |
42–72 | 34.8 ± 0.6 (30–37.5) |
24–47 | 24.7 ± 0.5 (22.5–27.5) |
16–27 |
| NR | 77.8 ± 1.16 (67.5–85.0) |
74–87 | 126.4 ± 1.1 (120–135) |
92–147 | 89.5 ± 1.2 (80–95) |
75–123 | 60.6 ± 1.4 (52.5–67.5) |
54–83 |
| V% | – | – | 44.0 ± 0.3 (41.4–48.6) |
37–64 | 46.7 ± 0.3 (41.3–48.4) |
43–55 | – | – |
| A | 27.7 ± 0.5 (23.9–31.8) |
26–32 | 16.3 ± 0.3 (13.0–19.2) |
11–19 | 16.5 ± 0.13 (15.2–17.4) |
11–19 | 18.3 ± 0.2 (16.3–19.9) |
16–24 |
| B | 5.2 ± 0.1 (4.6–5.9) |
4.4–5.5 | 17.9 ± 0.2 (15.8–19.9) |
14–31 | 16.7 ± 0.1 (16–18) |
10–28 | 7.9 ± 0.2 (6.7–9.3) |
6.4–11 |
| C | 5.7 ± 0.1 (5.2–6.1) |
4.6–7.3 | 39 ± 0.76 (30.9–44.6) |
29–55 | 24 ± –0.4 (19.7–26.9) |
19–28 | 22.2 ± 0.6 (18.4–27.6) |
22–39 |
| D | 0.78 ± 0.01 (0.74–0.86) |
0.77–0.90 | 0.83 ± 0.01 (0.73–0.92) |
– | 0.7 ± 0.01 (0.63–0.78) |
– | 0.8 ± 0.0 (0.8–0.9) |
– |
| E | 0.91 ± 0.01 (0.89–0.92) |
0.91–1.08 | 1.76 ± 0.04 (1.42–2.11) |
– | 1 ± 0.01 (0.9–1.14) |
– | 2.4 ± 0.1 (2.1–2.8) |
– |
| F | 0.21 ± 0.02 (0.18–0.23) |
0.19–0.27 | 3.15 ± 0.07 (2.67–3.88) |
– | 1.9 ± 0.04 (1.6–2.2) |
– | 1.2 ± 0.0 (1.1–1.4) |
– |
| Spicule length | – | – | – | – | – | – | 44.6 ± 0.4 (42.5–47.5) |
31–47 |
| Gubernaculum length | – | – | – | – | – | – | 24.1 ± 0.6 (20–27.5) |
14–26 |
| SW = SPL/ABW | – | – | – | – | – | – | 1.81 ± 0.03 (1.54–2.00) |
1.38–2.60 |
| GS = GL/SPL | – | – | – | – | – | – | 0.54 ± 0.01 (0.47–0.61) |
0.38–0.61 |
Measurements are in μm, and data are expressed in the form of mean ± SD (range)
EP= distance of excretory pore from anterior end; NR = distance of nerve ring position from anterior end; V%= distance of vulva position from anterior end/body length × 100; A= body length/body width; B= body length/oesophagus length; C= body length/tail length; D= EP/oesophagus length; E= EP/tail length; F= body width/tail length
Molecular identification and analysis
The length of the partial sequences obtained were 829 bp (ITS1 = 395 bp, 5.8S = 154 bp, ITS2 = 211 bp and 28S = 69 bp), 904 and 651 bp long for ITS, 28 S rRNA and COI (M1–M6) respectively. The edited sequences were deposited in GenBank (NCBI accession number MF618319–MF618322, MF621012–MF621014 and MF621249–MF621252 for ITS, 28S and COI respectively). The calculated average base composition revealed that all the developed gene sequences of H. baujardi were A + T rich (55.1, 54.9 and 67.4% for ITS, 28S and COI respectively).
The four developed ITS rRNA sequences of the isolated nematode exhibit high similarity (99.0–100%) with that of all the GenBank species identified as H. baujardi, with the exception of H. baujardi (EU363039). Other species showing higher similarity includes H. amazonensis and H. floridensis with 98.0% similarity. The species labelled as H. baujardi (EU363039) may not be the true species since, alongwith H. mexicana, H. sononrensis and H. taysearae, it shows 97.0% similarity with H. baujardi, and therefore excluded from the analysis. Further, it has been observed that all other Heterorhabditis species available in the GenBank, including H. indica and H. noenieputensis, showed ≤ 88.0% similarity. The calculated genetic distance inferred from ITS rRNA further proved that the isolated nematode is conspecific to H. baujardi (0.0 ± 0.0% average intraspecies distance) and distinct (15.5 ± 2.1% average interspecies distance) from all other Heterorhabditis species available in the GenBank (Table 2).
Table 2.
Estimates of genetic divergence between ITS rRNA sequences of Heterorhabditis spp. using the Kimura 2-parameter model
| H. baujardi PUC-HeTD1 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.03 | 0.03 | 0.06 | |
| H. baujardi PUC-HeTD2 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.03 | 0.03 | 0.06 | |
| H. baujardi PUC-HeTD3 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.03 | 0.03 | 0.06 | |
| H._baujardi PUC-HeTD4 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.03 | 0.03 | 0.06 | |
| H. baujardi AF548768 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.03 | 0.03 | 0.06 | |
| H. amazonensis DQ665222 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.03 | 0.03 | 0.06 | |
| H. floridensis DQ372922 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.01 | 0.01 | 0.01 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.03 | 0.02 | 0.03 | 0.03 | 0.06 | |
| H. taysearae EF043443 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.02 | 0.02 | 0.00 | 0.01 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.03 | 0.03 | 0.06 | |
| H. mexicana AY321478 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.02 | 0.01 | 0.01 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.03 | 0.02 | 0.03 | 0.03 | 0.06 | |
| H. noenieputensis JN620538 | 0.11 | 0.11 | 0.11 | 0.11 | 0.11 | 0.11 | 0.12 | 0.12 | 0.12 | 0.00 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.03 | 0.06 | |
| H. indica AY321483 | 0.11 | 0.11 | 0.11 | 0.11 | 0.11 | 0.11 | 0.12 | 0.12 | 0.12 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.03 | 0.06 | |
| H. bacteriophora AY321477 | 0.27 | 0.27 | 0.27 | 0.27 | 0.27 | 0.27 | 0.29 | 0.30 | 0.29 | 0.26 | 0.26 | 0.01 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.06 | |
| H. georgiana EU099032 | 0.27 | 0.27 | 0.27 | 0.27 | 0.27 | 0.28 | 0.29 | 0.29 | 0.30 | 0.26 | 0.26 | 0.02 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.06 | |
| H. beicherriana HQ896630 | 0.27 | 0.27 | 0.27 | 0.27 | 0.27 | 0.28 | 0.29 | 0.29 | 0.30 | 0.27 | 0.27 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.06 | |
| H_atacamensis HM230723 | 0.28 | 0.28 | 0.28 | 0.28 | 0.28 | 0.27 | 0.29 | 0.29 | 0.30 | 0.26 | 0.26 | 0.19 | 0.19 | 0.18 | 0.01 | 0.00 | 0.01 | 0.01 | 0.01 | 0.05 | |
| H. downesi AY321482 | 0.29 | 0.29 | 0.29 | 0.29 | 0.29 | 0.29 | 0.31 | 0.30 | 0.31 | 0.28 | 0.29 | 0.19 | 0.19 | 0.18 | 0.03 | 0.01 | 0.01 | 0.01 | 0.01 | 0.05 | |
| H. safricana EF488006 | 0.30 | 0.30 | 0.30 | 0.30 | 0.30 | 0.29 | 0.31 | 0.30 | 0.32 | 0.28 | 0.28 | 0.19 | 0.20 | 0.19 | 0.02 | 0.04 | 0.01 | 0.01 | 0.01 | 0.06 | |
| H. marelatus AY321479 | 0.30 | 0.30 | 0.30 | 0.30 | 0.30 | 0.29 | 0.31 | 0.31 | 0.32 | 0.29 | 0.29 | 0.19 | 0.19 | 0.19 | 0.03 | 0.04 | 0.03 | 0.01 | 0.01 | 0.05 | |
| H. megidis AY321480 | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 | 0.32 | 0.32 | 0.33 | 0.31 | 0.31 | 0.21 | 0.22 | 0.22 | 0.06 | 0.05 | 0.07 | 0.07 | 0.02 | 0.06 | |
| H. zealandica EF530041 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.32 | 0.34 | 0.34 | 0.35 | 0.32 | 0.33 | 0.24 | 0.25 | 0.24 | 0.10 | 0.11 | 0.11 | 0.10 | 0.14 | 0.06 | |
| Caenorhabditis elegans_KX572972 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | 0.77 | 0.78 | 0.76 | 0.77 | 0.79 | 0.78 | 0.77 | 0.79 | 0.75 | 0.74 | 0.74 | 0.76 | 0.75 | 0.79 | 0.78 |
Standard error estimate(s) are shown above the diagonal
There are no species labelled as H. baujardi for 28S rRNA (D2–D3) available in the GenBank. The isolated nematode showed 99.0% similarity with Heterorhabditis sp. (KY024496 and KY055370), H. amazonensis (EU099036), H. floridensis (EU099034) and H. mexicana (EU100414). The alignment of the sequences revealed that it has 2, 3, 5, 11 and 11 bp difference with Heterorhabditis sp. (KY024496), Heterorhabditis sp. (KY055370), H. amazonensis (EU099036), H. floridensis (EU099034) and H. mexicana (EU100414) respectively. Other species showing higher similarity includes Heterorhabditis indica and H. noenieputensis (97.0% similarity) with 25–26 bp difference, H. bacteriophora, H. beicherriana, H. georgiana, H. marelatus and H. safricana (94.0%) with 50–54 bp, H. megidis and H. zealandica (93.0–94.0%) with 58–62 bp, and H. atacamensis (with 93.0%) with 40 bp difference (only 594 bp sequenced in this species). The analysis of the sequences of 28S showed that the genetic distance (K2P distance) (Table 3) between the three developed sequences is 0.0% with H. amazonensis, 1.0% with H. floridensis and H. mexicana, 3.0% with H. indica and H. noenieputensis, and 6.0–8.0% with all other Heterorhabditis species analysed. Further, the distance between GenBank data of H. noenieputensis and H. indica is also 0.0%.
Table 3.
Estimates of genetic divergence between 28S rRNA sequences of Heterorhabditis spp. using the Kimura 2-parameter model
| H.baujardi PUC-HeTD1 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | |
| H.baujardi PUC-HeTD2 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | |
| H.baujardi PUC-HeTD3 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | |
| Heterorhabditis sp. KY024496 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | |
| Heterorhabditis sp. KY055370 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | |
| H. amazonensis EU099036 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | |
| H. floridensis EU099034 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | |
| H. mexicana EU100414 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | |
| H. indica JQ178379 | 0.03 | 0.03 | 0.03 | 0.04 | 0.04 | 0.03 | 0.04 | 0.03 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | |
| H. noenieputensis JX624110 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.04 | 0.03 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | |
| H. atacamensis HM230724 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.01 | 0.01 | 0.03 | |
| H. bacteriophora EU099037 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.07 | 0.07 | 0.04 | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | |
| H. beicherriana HQ896631 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.07 | 0.07 | 0.04 | 0.01 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | |
| H. georgiana EU099033 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.06 | 0.06 | 0.06 | 0.07 | 0.07 | 0.04 | 0.01 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | |
| H. marelatus EU100412 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.06 | 0.07 | 0.07 | 0.07 | 0.07 | 0.01 | 0.05 | 0.05 | 0.05 | 0.00 | 0.01 | 0.01 | 0.03 | |
| H. safricana EU100416 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.06 | 0.07 | 0.07 | 0.06 | 0.06 | 0.01 | 0.05 | 0.05 | 0.05 | 0.01 | 0.01 | 0.01 | 0.03 | |
| H. zealandica EU099035 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.02 | 0.05 | 0.05 | 0.05 | 0.02 | 0.02 | 0.01 | 0.03 | |
| H. megidis EU100413 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.07 | 0.08 | 0.07 | 0.08 | 0.08 | 0.02 | 0.06 | 0.06 | 0.06 | 0.02 | 0.02 | 0.02 | 0.02 | |
| Caenorhabditis elegans MF192964_ | 0.30 | 0.30 | 0.30 | 0.31 | 0.31 | 0.30 | 0.30 | 0.31 | 0.29 | 0.29 | 0.29 | 0.29 | 0.29 | 0.29 | 0.29 | 0.29 | 0.28 | 0.28 |
Standard error estimate(s) are shown above the diagonal
There are only three sequences of the cytochrome oxidase I (M1–M6) available in the GenBank viz. Heterorhabditis bacteriophora (JN572120), H. bacteriophora (EF043402) and H. indica CH16 (KU306236). The developed sequences of H. baujardi exhibited 88% similarity and a distance of 13.0–14.0% with all the three species.
Molecular phylogeny
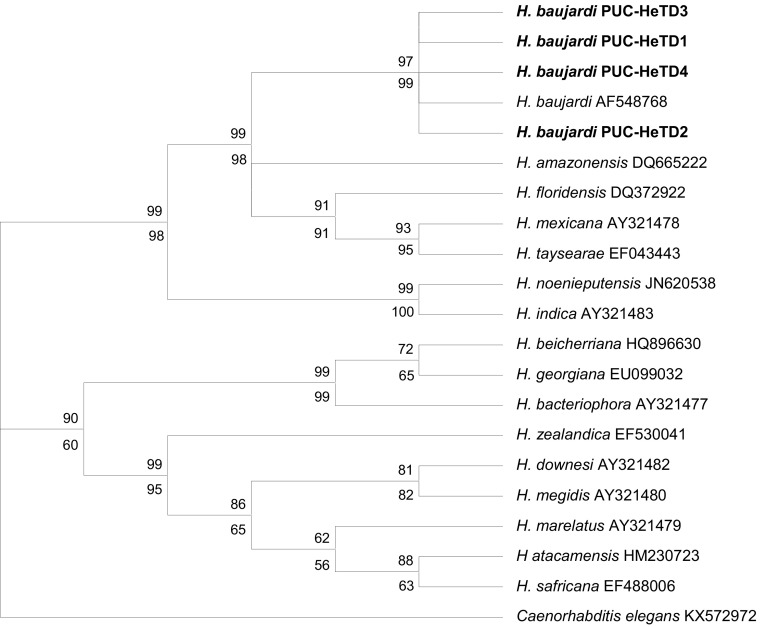
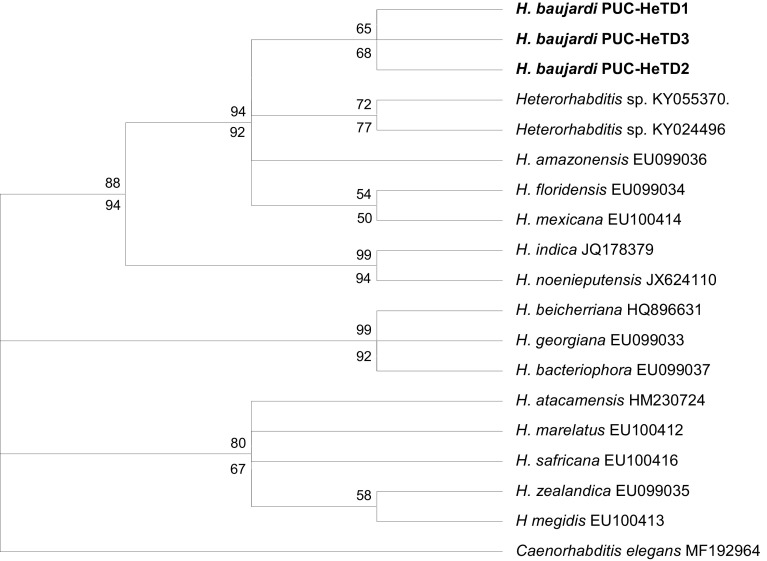
The phylogeny constructed using maximum likelihood (ML) and maximum parsimony (MP) revealed identical trees and both the ITS and 28S data sets exhibited similar topology with a strong bootstrap support. The COI (M1–M6 region) did not yield any useful phylogenetic tree due to lack of comparable sequences. The phylogenetic relationships of the isolated nematode and other sequences of Heterorhabditis species (16 species for ITS and 13 species for 28S) from the GenBank are shown in the Figs. 1 (for ITS) and 2 (for 28S) (tree length = 684, CI = 0.7343, RI = 0.9022 for ITS; and tree length = 225, CI = 0.7217, RI = 0.8827 for 28S). The phylogeny using the two gene data sets revealed that H. baujardi belongs to H. indica clade, and further, forms a monophyletic clade (99/98% and 94/92% bootstrap support for ITS and 28S respectively) with H. amazonensis, H. floridensis, H. mexicana and H. taysearae forming H. baujardi sub clade. The other two species viz. Heterorhabditis indica and H. noenieputensis clustered separately (with 99/100% and 99/94% bootstrap support for ITS and 28S respectively) forming a distinct H. indica sub clade. Apart from Heterorhabditis indica clade, the other two clades viz. H. bacteriophora clade (H. bacteriophora, H, beicherriana and H. georgiana) and H. megidis clade (H. atacamensis, H. downesis, H. marelatus, H. megidis, H. safricana and H. zealandica) clustered cohesively with a strong bootstrap support.
Fig. 1.
Phylogenetic relationship of Heterorhabditis baujardi with other Heterorhabditis spp. based on analysis of the ITS rRNA region. Numbers indicated at the nodes represents bootstrap proportion values (50% or more, 1000 replicates) where numbers on the upper node represent the maximum parsimony and those on the lower node represent the maximum likelihood. Numbers after each species and isolate indicate the GenBank Accession numbers
Fig. 2.
Phylogenetic relationship of Heterorhabditis baujardi with other Heterorhabditis spp. based on analysis of the 28S rRNA region. Numbers indicated at the nodes represents bootstrap proportion values (50% or more, 1000 replicates) where numbers on the upper node represent the maximum parsimony and those on the lower node represent the maximum likelihood. Numbers after each species and isolate indicate the GenBank Accession numbers
Discussion
The type locality of H. baujardi is the forest of Backan, Ninhbinh and Kontum province, Vietnam (Phan et al. 2003), and is further reported from other countries of Southeast Asia, South America and Africa (Kanga et al. 2012; Dolinski et al. 2008). Considering the close proximity of the type locality and wide distribution of the species, there is a high chance that it might have been commonly encountered on Indian soil, however, no occurrence of the species has been reported from India so far.
Accurate identification of the EPNs is very important prior to use as biological control agent since different species exhibited different adaptations and behaviour (Valadas et al. 2014). Simulteneously, identification of Heterorhabditis species, in particular, based on morphology is not an easy task because of limited morphological variation (Dolinski et al. 2008). In contrast to the previous suggestion (Stock and Kaya 1996), where both the body length and tail length of IJ and the body length and testis reflexion of males being the useful diagnostic tools, Phan et al. (2003) suggested that ratios E and F, body diameter of IJ, and spicule length, gubernaculum length and ratio SW of males should be considered for better discrimination of Heterorhabditis spp. Apart from these, the shape and size of the spicule and gubernaculum are another important diagnostic tools. Besides the slight variation observed, the morphometric measurement and their analysis conformed to the original description of H. baujardi. The slight differences may be due to genetic or environmental variations inflicted by different geographical habitats (Rolston et al. 2009; Bolnick et al. 2011).
The ITS rRNA region is useful and widely accepted genetic markers for species delimitation and assessing phylogenetic relationship in Steinernema species, however, the spanning 5.8S region is too conserved to resolve the connection among the species (Nguyen et al. 2001). Similarly, though the tree constructed separately (data not shown here) using each region (ITS1, 5.8S and ITS2) revealed a similar topology, the genetic distance calculated revealed that 5.8S could not discriminate H. baujardi sub group (H. amazonensis, H. floridensis, H. mexicana and H. taysearae) with 0.0% distance, and further, 2.0–5.0% distance with all other species. However, the ITS1 and ITS2 region can distinguish all the species of Heterorhabditis very well with interspecies distance ranging from 1.0–40.0% and 2.0–85.0% respectively.
Reconstruction of molecular phylogeny of the genus Heterorhabditis has been built by several workers. Phylogenetic analysis of H. baujardi inferred from ITS rRNA and D2-D3 region of 28S rRNA showed similar topology, where the species belongs to ‘indica’ group of Nguyen et al. (2008) and Spiridonov and Subbotin (2016). It further forms a monophyletic group with H. amazonensis, H. floridensis, H. mexicana and H. taysearae as ‘baujardi’ subclade of Spiridonov and Subbotin (2016). Comparison of the two markers (ITS and 28S rRNA) resulted that the ITS rRNA gene sequences revealed a better bootstrap support for constructing phylogenetic relationships of Heterorhabditis species. The 28S rRNA region, although commonly used marker for phylogenetic reconstruction, may not be the best tool to distinguish closely related Heterorhabditis species. This is evident from the lack of significant difference between the gene sequences of H. baujardi–H. amazonensis pair and the H. indica–H. noenieputensis pair. The interspecies distance calculated among all the available sequences are also low with 0.0% (H. amazonensis) to 8.0% (H. megidis). Further analysis of the ITS gene revealed that the ITS1 sequence length of H. baujardi is 395 bp long, similar to H. amazonensis, constituting the longest among the species whereas the ITS2 sequence length is 211 bp long similar to H. marelatus constituting the shortest among all the described species. It is interesting to note that the length of the ITS1 region of the present study and Phan et al. (2003) is different (395 bp in this study vs. 406 bp). However, the difference may be attributed to the inclusion of the flanking 18S region by Phan et al. (2003).
The mitochondrial gene, cytochrome c oxidase I (M1–M6 partition), is a reliable tool for Metazoan species delimitation, including helminthes (Hebert et al. 2003). However, Derycke et al. (2007, 2008, 2010) reported the dominance of I3–M11 partition over M1–M6 partition of COI in terms of resolving taxonomic ambiguities and uncovering cryptic diversity among marine nematodes. Analysis of the GenBank data (data not shown here) revealed that the most COI sequences of Heterorhabditis available in the GenBank is I3–M11 partition where it exhibit a different topology from other two genes with a strong bootstrap support, however, a reasonable genetic distance among each other (0.0% intraspecies and 5.0–16.0% interspecies K2P distance). It is difficult to make a concrete conclusion regarding the reliability and use of the COI sequences (M1–M6 partition) as a tool for EPN species delimitation due to inadequate sequences available in the GenBank. Nevertheless, analysis with the available sequence of the region revealed that species labelled as H. bacteriophora (JN572120) may be a case of misidentification because of the wide genetic distance (14.0%) with H. bacteriophora (EF043402), and further, the high similarity (99.0%) and narrow gap of genetic distance (1.0%) with H. indica CH16 (KU306236) ascertained their conspecificity. Furthermore, the developed COI sequences of H. baujardi showed a wide genetic distance (13.0–14.0%) with both H. indica and H. bacteriophora proving the COI (M1–M6 partition) applicable to distinguish the Heterorhabditis species. We therefore agreed that the COI (both M1–M6 and I3–M11 partition) may not be compatible to ITS and 28S rRNA in construction of phylogeny, however, can be a strong supporting tool for species identification of the genus Heterorhabditis.
In conclusion, characterization of both the ITS and 28S rRNA revealed similar topology in resolving the phylogenetic relationship of the genus Heterorhabditis, with the ITS rRNA being superior based on the strong bootstrap support. The cytochrome c oxidase I (M1–M6 partition), although incompatible to other genes like ITS and 28S rRNA in resolving phylogeny, can be a good supportive tool for species delimitation. The present study constitutes the first report of H. baujardi from India and, with the confirmation of this species, twelve valid EPN species are presently known from India. Moreover, the isolation of this species envisages its future use as a biological control agent, and further incorporated into the IPM.
Acknowledgements
We are grateful to DST-SERB, Government of India for the financial support (No. SB/EMEQ-071/2014); and Dr Tawnenga, Principal, Pachhunga University College for providing laboratory facilities. VRL acknowledge the Department of Biotechnology, Government of India for Institutional Biotech Hub (Advance Level).
Author’s contribution
VL—Collect soil samples, nematode extraction, morphological analysis and edited the manuscript; LR—Data analysis and interpretation, drafted the manuscript; HCL—Collect soil samples, molecular analysis and edited the manuscript; VR—Bioinformatics analysis and interpretation, edited the manuscript.
Compliance with ethical standards
Conflict of interest
We wish to confirm that there are no known conflicts of interest associated with this publication.
References
- Bedding RA, Akhurst RJ. A simple technique for the determination of insect parasitic rhabditid nematodes in soil. Nematologica. 1975;21:109–110. doi: 10.1163/187529275X00419. [DOI] [Google Scholar]
- Bhat AH, Istkhar AKC, Půža V, San-Blas E. First report and comparative study of Steinernema surkhetense (Rhabditida: Steinernematidae) and its symbiont bacteria from subcontinental India. J Nematol. 2017;49:92–102. doi: 10.21307/jofnem-2017-049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boemare NE. Biology, taxonomy and systematics of Photorhabdus and Xenorhabdus. In: Gaugler R, editor. Entomopathogenic nematology. Wallingford: CABI Publishing; 2002. pp. 35–36. [Google Scholar]
- Bolnick DI, Amarasekare P, Araújo MS, Bürger R, Levine JM, Novak M, Rudolf VHW, Schreiber SJ, Urban MC, Vasseur D. Why intraspecific trait variation matters in community ecology. Trends Ecol Evolut. 2011;26:183–192. doi: 10.1016/j.tree.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derycke S, Backejau T, Vlaeminck C, Vierstraete A, Vanflecteren J, Vinex M, Moens T. Spatiotemporal analysis of population genetic structure in Geomonhystera disjuncta (Nematoda, Monhysteridae) reveals high levels of molecular diversity. Mar Biol. 2007;151:1799–1812. doi: 10.1007/s00227-007-0609-0. [DOI] [Google Scholar]
- Derycke S, Fonseca G, Vierstraete A, Vinex M, Moens T. Disentangling taxonomy within the Rhabditis (Pellioditis) marina (Nematode, Rhabditidae) species complex using molecular and morphological tools. Zool J Linn Soc. 2008;152:1–15. doi: 10.1111/j.1096-3642.2007.00365.x. [DOI] [Google Scholar]
- Derycke S, Vanaverbeke J, Rigaux A, Backeljau T, Moens T. Exploring the use of cytochrome oxidase c subunit 1 (COI) for DNA barcoding of free-living marine nematodes. PLos One. 2010;5:e13716. doi: 10.1371/journal.pone.0013716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinski C, Kamitani FL, Machado IR, Winter CE. Molecular and morphological characterization of heterorhabditid entomopathogenic nematodes from the tropical rainforest in Brazil. Mem Inst Oswaldo Cruz. 2008;103:150–159. doi: 10.1590/S0074-02762008000200005. [DOI] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Ganguly S, Singh LK. Steinernema thermophilum sp. n. (Rhabditida: Steinernematidae) from India. Int J Nematol. 2000;10:183–191. [Google Scholar]
- Ganguly S, Singh M, Lal M, Singh LK, Vyas RV, Patel DJ. New record of an entomopathogenic nematode, Steinernema riobrave Cabanillas, Poinar and Raulston, 1994 from Gujarat, India. Indian J Nematol. 2002;32:223. [Google Scholar]
- Ganguly S, Rathore KS, Sushil K, Singh M. Steinernema meghalayensis sp. n. (Rhabditida: Steinernematidae) from north-eastern hilly region of India. Indian J Nematol. 2011;41:83–97. [Google Scholar]
- Griffin CT, Boemare NE, Lewis EE. Biology and behaviour. In: Grewal PS, Ehlers RU, Shapiro-Ilan DI, editors. Nematodes as biocontrol agents. Oxon: CABI Publishing; 2005. pp. 47–64. [Google Scholar]
- Hebert PDN, Ratnasingham NS, deWaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B Biol Sci. 2003;270:S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hominick WM. Biogeography. In: Gaugler R, editor. Entomopathogenic nematology. Wallingford: CABI Publishing; 2002. pp. 115–143. [Google Scholar]
- Hunt DJ, Subbotin SA. Taxonomy and systematics. In: Nguyen KB, editor. Hunt DJ. Leiden: Advances in entomopathogenic nematode taxonomy and phylogeny. Nematology monographs and perspectives. Brill Publishing; 2016. pp. 13–58. [Google Scholar]
- Hussaini SS, Ansari MA, Ahmad W, Subbotin SA. Identification of some Indian populations of Steinernema species (Nematoda) by RFLP analysis of ITS region of rDNA. Int J Nematol. 2001;11:73–76. [Google Scholar]
- Joyce SA, Reid A, Driver F, Curran J (1994) Application of polymerase chain reaction (PCR) methods to the identification of entomopathogenic nematodes. In: Burnell AM, Ehlers RU, Masson JP (eds) Proceeding of symposium & workshop, St. Patrick’s College, Maynooth, Co. Kildare, Ireland. European Commission, DGXII, Luxembourg, pp 178–187
- Kanga FN, Waeyenberge L, Hauser S, Moens M. Distribution of entomopathogenic nematodes in Southern Cameroon. J Invertebr Pathol. 2012;109:41–51. doi: 10.1016/j.jip.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Kaya HK, Stock SP. Techniques in insect nematology. In: Lacey LA, editor. Manual of techniques in insect pathology. San Diego: Academic Press; 1997. pp. 281–324. [Google Scholar]
- Kulkarni N, Rizvi AN, Kumar V, Paunikar S, Mishra VK. Morphological and molecular characterization of Steinernema dharanaii sp. n., a new entomopathogenic nematode from India. Indian J Trop Biodiv. 2012;20:107–116. [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalramliana, Yadav AK. Occurrence of entomopathogenic nematodes (Rhabditida: Steinernematidae and Heterorhabditidae) in Meghalaya, NE India. Sci Vis. 2010;10:89–100. [Google Scholar]
- Lalramnghaki HC, Vanlalhlimpuia, Vanramliana, Lalramliana Characterization of a new isolate of entomopathogenic nematode, Steinernema sangi (Rhabditida, Steinernematidae), and its symbiotic bacteria Xenorhabdus vietnamensis (γ-Proteobacteria) from Mizoram, north-eastern India. J Parasit Dis. 2017;41:1123–1131. doi: 10.1007/s12639-017-0945-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KB, Maruniak J, Adams BJ. Diagnostic and phylogenetic utility of the rDNA internal transcribed spacer sequences of Steinernema. J Nematol. 2001;33:73–82. [PMC free article] [PubMed] [Google Scholar]
- Nguyen KB, Půža V, Mracek Z. Steinernema cholashanense n. sp. (Rhabditida, Steinernematidae) a new species of entomopathogenic nematode from the province of Sichuan, Chola Shan Mountains, China. J Invertebr Pathol. 2008;97:251–264. doi: 10.1016/j.jip.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Phan KL, Subbotin SA, Nguyen NC, Moens M. Heterorhabditis baujardi sp. n. (Rhabditida: Heterorhabditidae) from Vietnam and morphometric data for H. indica populations. Nematology. 2003;5:367–382. doi: 10.1163/156854103769224368. [DOI] [Google Scholar]
- Poinar GO, Jr, Karunakar GK, David H. Heterorhabditis indicus n. sp. (Rhabditida: Nematoda) from India: separation of Heterorhabditis spp. by infective juveniles. Fundam Appl Nematol. 1992;15:467–472. [Google Scholar]
- Qiu L, Zhao J, Wu Z, Lv Z, Pang Y. Steinernema pui sp. n. (Rhabditida, Steinernematidae), a new entomopathogenic nematode from Yunnan, China. Zootaxa. 2011;2767:1–13. [Google Scholar]
- Rolston A, Meade C, Boyle S, Kakouli-Duarte T, Downes M. Intraspecific variation among isolates of the entomopathogenic nematode Steinernema feltiae from Bull Island, Ireland. Nematology. 2009;11:439–451. doi: 10.1163/156854109X447015. [DOI] [Google Scholar]
- Seinhorst JW. A rapid method for transfer of nematodes from fixative to anhydrous glycerine. Nematologica. 1959;4:67–69. doi: 10.1163/187529259X00381. [DOI] [Google Scholar]
- Spiridonov SE, Subbotin SA. Phylogeny and phylogeography of Heterorhabditis and Steinernema. In: Nguyen KB, editor. Advances in entomopathogenic nematode taxonomy and phylogeny. Nematology monographs and perspectives. Leiden: Brill Publishing; 2016. pp. 413–427. [Google Scholar]
- Stock SP, Kaya HK. A multivariate analysis of morphometric characters of Heterorhabditis species (Nemata: Heterorhabditidae) and the role of morphometrics in the taxonomy of species of the genus. J Parasitol. 1996;82:806–813. doi: 10.2307/3283895. [DOI] [PubMed] [Google Scholar]
- Stock SP, Pryor BM, Kaya HK. Distribution of entomopathogenic nematodes (Steinernematidae and Heterorhabditidae) in natural habitats in California. Biodiv Conserv. 1999;8:535–549. doi: 10.1023/A:1008827422372. [DOI] [Google Scholar]
- Thompson JD, Toby J, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadas V, Laranjo M, Mota M, Oliveira S. A survey of entomopathogenic nematode species in continental Portugal. J Helminthol. 2014;88:327–341. doi: 10.1017/S0022149X13000217. [DOI] [PubMed] [Google Scholar]