Abstract
Lymphatic malformations (LMs) are congenital vascular anomalies characterized by dilated and cystic lymphatic channels. They are subdivided into macrocystic and microcystic lesions based upon the predominant size of the cysts involved. However, significant differences in clinical characteristics, treatment outcomes, and prognosis between macrocytic and microcytic disease suggest variation in underlying biologic and genetic influences. Indirect differential expression analysis revealed that 426 genes are significantly different (p < 0.01) in a small sample of LM subtypes. Functional analyses on the differentially expressed gene sets showed that microcystic LM gene expression favors a prooncogenic profile with upregulation of MYC target genes and cell cycle proteins, whereas macrocystic expression demonstrates hypoxic events that lead to angiogenesis and cell proliferation. Therefore, microcystic and macrocystic LMs, although histologically and physiologically similar, may occur under the influence of vastly different biological pathways and mechanisms of action.
Keywords: : lymphatic malformations, macrocystic lymphatic malformation, microcystic lymphatic malformation, gene microarray, pediatric lymphatic malformations
Introduction
Lymphatic malformations (LMs) are low-flow vascular anomalies characterized by the presence of abnormal lymphatic channels with progressive cystic dilation.1,2 Woven within normal soft tissue, these congenital lesions are present in roughly 1 of 250 live births, creating a mass effect and distortion of involved structures. Most patients with LM present before the age of 2 years, with more complicated disease identified at younger ages.2,3 LMs slowly grow by vascular expansion but may acutely progress under conditions that induce inflammation or increase lymphatic flow, such as infection. Patient symptoms are linked to LM impingement and inflammation of adjacent tissue.4 Surgical extirpation and sclerotherapy, the radiographic injection of caustic agents, are used to control and cure LM. Some lesions have proven more resistant to therapy. Recently, novel pharmacotherapy, mTOR inhibitors, has been employed for massive and complicated disease.2,5,6
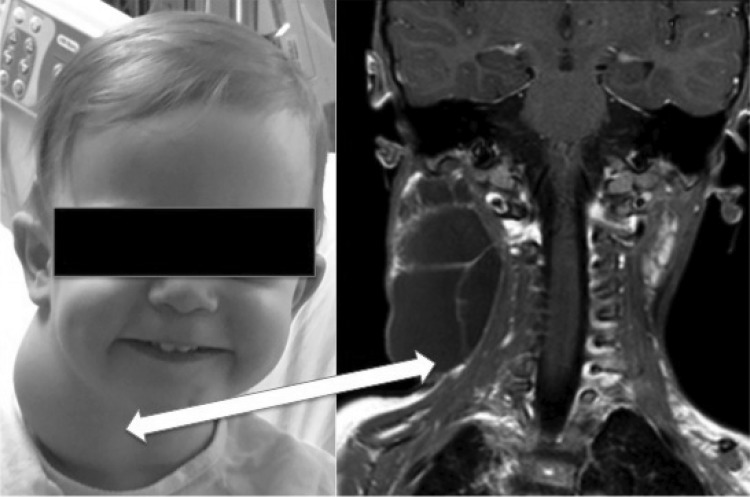
LMs are radiographically classified as either microcystic or macrocystic based on the predominant diameter of the dilated lymphatic lumens present. Cysts <1 cm in diameter are considered microcystic.7,8 Frequently both macrocysts and microcysts are present in the same lesion, which is then labeled as a “mixed” LM. Clinically, LMs are differentiated based on their physical appearance, compressibility, degree of infiltration, and radiographic findings. Purely macrocystic LMs are large fluid-filled compartments lined by septae with distinct vesicular structures that impress upon, and do not infiltrate into, local soft tissue (Fig. 1). Treatment is unlikely to damage adjacent structures, as they can be free mobilized or avoided with surgery and sclerotherapy, respectively.
FIG. 1.
Radiographic and clinical appearance of macrocystic lymphatic malformation in a child. Note the large cystic channels with pressure against local soft tissues.
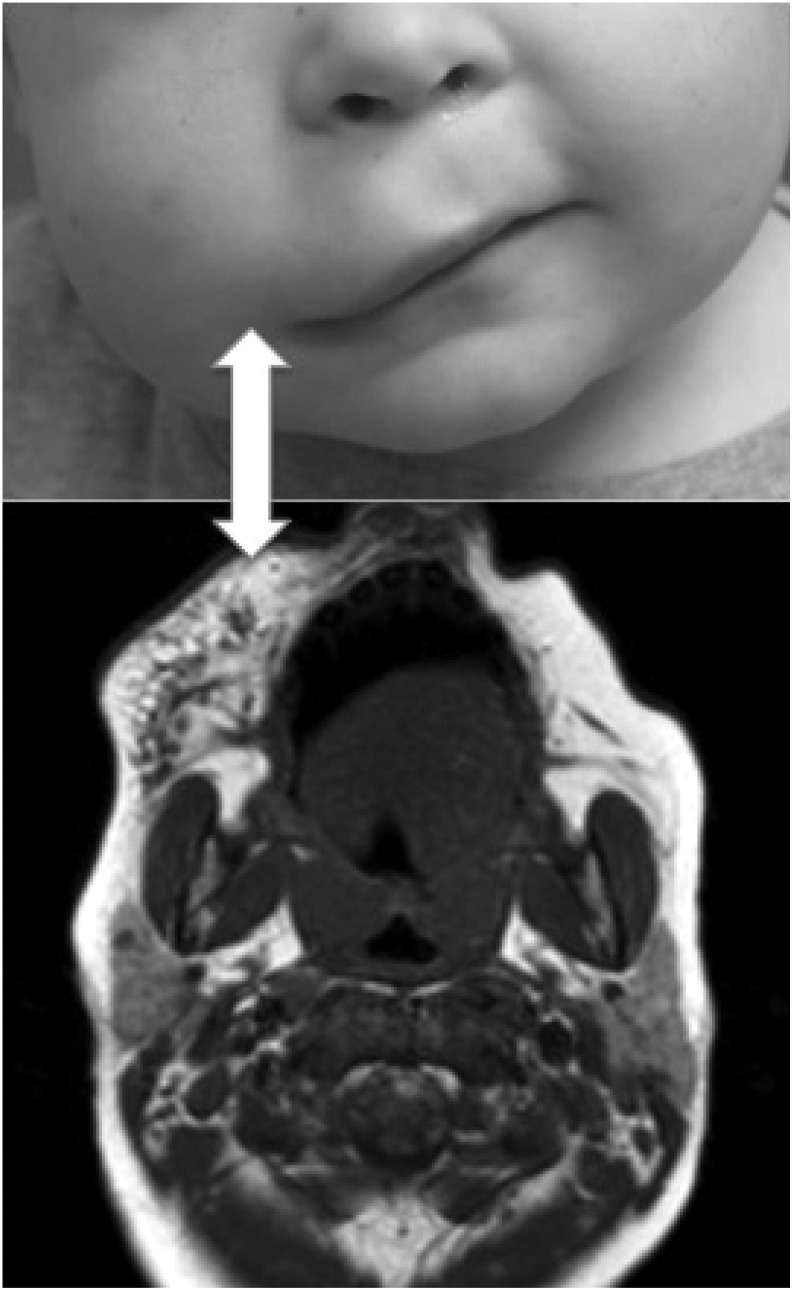
Microcystic disease, in contrast, infiltrates deeply within skin, fat, and muscle that can make selective and effective therapy quite difficult (Fig. 2).3,4 Therefore, very disparate cure rates are reported between macrocystic and microcystic disease, with the former more favorable to treatment.9 Intercalation and reactive fibrosis of microcystic LM responds less readily to sclerotherapy, with one study citing as low as a 14% complete response.2,4,10
FIG. 2.
Radiographic and clinical appearance of microcystic lympathic malformation in a child. Note the infiltrative microcysts making it difficult to delineate disease from normal tissue.
Despite disparate structure, radiographic appearance, and clinical responses, few genetic or autocrine differences between microcystic and macrocystic LMs have been discovered.3 Even histological analysis, showing disordered endothelium lined lymphatic lumens with a complex extracellular matrix, cannot reliably distinguish between LM subtypes.11 Nonetheless, an association between various syndromes (e.g., Klippel–Trenaunay) and LM is an active field of research. Recent genetic studies have reported that LMs are caused by somatic mutations on PIK3CA.12–14
Herein, we analyze and compare the transcriptome of microcystic to macrocystic malformations to better understand fundamental physiologic differences and genetic influences on these lesions. Such characterization might allow for the development of genetic testing during the systematic evaluation and classification of these congenital masses and the conception of new and personalized treatments tailored to their aberrant cellular signaling.
Materials and Methods
Subjects and tissue collection methods
This study was reviewed and approved by the Institutional Review Board of the University of Arkansas for Medical Sciences. Fresh tissue was harvested by surgical removal from 18 children of various ages after acquiring written informed consent from their families. Upon harvesting, tissues were stored at −80°C until RNA extraction was performed. We analyzed six microcystic, seven macrocystic, and five normal samples. The classification of the LMs was based on clinical, radiographic, and histological findings. Description of the samples and location of the LM are described in Table 1. It is worth mentioning that the normal samples were taken from skin and subcutaneous tissue from healthy children.
Table 1.
Description of the Samples and Location of the Lymphatic Malformations
| Sample ID | Type of LM | Location |
|---|---|---|
| 109-LM | Microcystic | Face (left) |
| 070111-LMA | Microcystic | Tongue |
| 052810-LMB | Microcystic | Lip |
| 091710-LMB | Microcystic | Neck (left) |
| 111210-LMB | Microcystic | Tongue |
| 120210-LMB1 | Microcystic | Superficial parotid (left) |
| 070810-LMA-r | Macrocystic | Tongue |
| 59-LM | Macrocystic | Neck (left) |
| 73-LM | Macrocystic | Postauricular (left) |
| 99-LM | Macrocystic | Neck (right) |
| 01111-LMA | Macrocystic | Occiput (left) |
| 041311-LMA | Macrocystic | Face |
| 0708010-LMA | Macrocystic | Tongue |
| 030411-NOR | Normal | Palate/terragoid fossa (left) |
| 262630-NOR | Normal | Cheek/jaw (left) |
| 52100-NOR | Normal | Cheek/upper lip/paranasal (right) |
| 101910-NOR | Normal | Cheek (right) |
| 111910-NOR | Normal | Leg (right) |
LM, lymphatic malformation.
Microarray gene expression profiling
Total RNA was isolated and purified from frozen tissues using the RNeasy Plus Mini Kit (Qiagen, Valencia, CA). The Illumina TotalPrep RNA Amplification Kit (Illumina, San Diego, CA) was used to prepare biotinylated antisense complementary RNA (cRNA) from 500 ng of high-quality total RNA for subsequent global gene expression profiling by the Pharmacogenomics Analysis Laboratory (Central Arkansas Veterans Healthcare System, Little Rock, AR). Quality and quantity of cRNA were determined by Ribogreen fluorescence and Agilent bioanalyzer electropherograms (Agilent Technologies, Santa Clara, CA). A total of 750 ng of cRNA per sample was loaded into the Illumina Human HT-12 v4 Gene Expression BeadChip for hybridization at 58°C for 17 hours.
After blocking, staining, and washing, the microarrays were scanned on the Illumina iScan system. Data on gene-level intensities were extracted and background was corrected using the GenomeStudio software from Illumina. For each array, the expression level, detection p-value, and average number of beads per gene were extracted with the BeadArray package from Bioconductor (www.bioconductor.org). Quantile normalization and logarithmic transformation of the intensities were carried out with the Bioconductor package limma. These data used in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE98742 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE98742).
Gene differential expression analysis
We used the Bioconductor limma package to determine differential gene expression. In brief, for any given gene, we fit a linear model that encodes the difference between two experimental conditions, followed by an empirical Bayes method that moderates standard errors of the estimated log-fold changes.15 Owing to the relatively small sample size, a more stringent cutoff p-value of 0.01 for the differential expression model and a minimum fold change (FC) of 1.5 were used in lieu of a multiple testing methodology. This method yielded differential expression of microcystic and macrocystic LMs with respect to normal controls. However, we are interested in the differences between LMs eliminating possible noise of normal gene expression, thus we used an indirect comparison for estimation of the log ratio log2(Microcystic/Macrocystic) through the so-called double difference, namely, log2(Microcystic/Control)—log2(Macrocystic/Control).
The results of this indirect comparison produced our master list from which all the subsequent gene enrichment and functional analyses are based upon (see Supplementary Data for the complete list of differentially expressed genes; Supplementary Data are available online at www.liebertpub.com/lrb).
Gene enrichment analysis
Gene lists generated from the differential expression were fed into DAVID for mapping into a list of associated biological annotations.16,17 In this case, gene ontology terms described as biological processes were selected. We also conducted a gene set enrichment analysis (GSEA) with the hallmark gene sets to identify phenotype differences in the macrocystic versus microcystic gene sets.18
Results
Gene expression differences between macrocystic and microcystic LMs
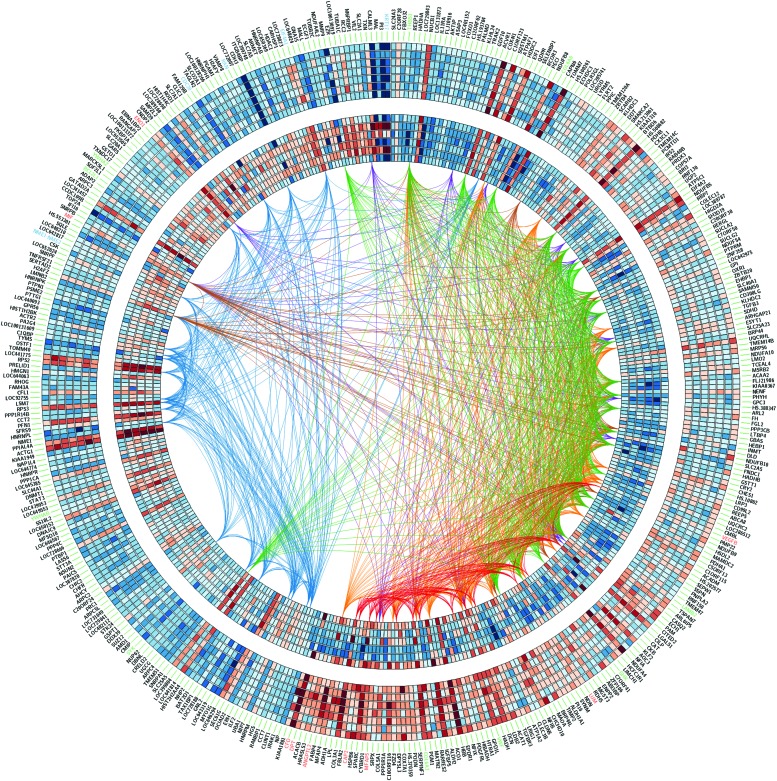
We sought genes that were differentially expressed between each malformation type relative to healthy control samples with statistical level of significance set at 0.01, and a fold change of at least 1.5. For microcystic LM, there were 315 differentially expressed genes with respect to normal controls, whereas 330 differentially expressed genes were discovered for their macrocystic counterpart. About two-thirds of the differentially expressed genes were unique for each of these malformations (67% and 66% for macrocystic and microcystic, respectively), with 108 genes expressed in common to normal controls (Supplementary Data). After indirect differential analysis, 426 genes were found to be differentially expressed at a p-value ≤0.01 for macrocystic and microcystic LMs, as detailed in Table 2 and depicted in Figure 3.
Table 2.
Quantification of Differential Expression Between Microcystic and Macrocystic Lymphatic Malformations
| Comparison | No. of genes (total) | Upregulated in microcystic | Upregulated in macrocystic |
|---|---|---|---|
| Microcystic-(control) vs. Macrocystic-(control) | 426 | 192 | 234 |
FIG. 3.
Heatmap of the 426 differentially expressed genes by indirect difference analysis. The inner circles are microcystic LMs and the outer circles correspond to macrocystic LMs. Gene names discussed in the text are colored in red for macrocystic, blue for microcystic, and green for members of the PI3K/AKT pathway. Links join genes on the same hallmark pathway and they are colored as follows: MYC—blue, epithelial–mesenchymal transition—red, hipoxia—purple, fatty acid methabolism—brown, oxidative phosphorylation—green, adipogenesis—orange. A color version of this figure is available in the online article at www.liebertpub.com/lrb
Gene enrichment and functional analyses
To better understand the collective impact of the indirect comparison, the gene master list was split into two sublists: the first one containing all the genes upregulated in the microcystic malformations with respect to the macrocystic malformations and the second list accounting for all the genes upregulated in macrocystic samples with respect to their microcystic counterpart (Table 2). Both lists were fed into GSEA for determination of hallmark gene sets, and into DAVID for functional analysis of KEGG pathways.
Microcystic
The gene ontology for biological processes showed that terms related to apoptosis or cell death were highly significant (p < 0.001), namely regulation of cell death (GO:0010941), apoptosis (GO:0042981), and programmed cell death (GO:0043067). We further investigate this gene set in GSEA with the Hallmark genes data set, and the MYC targets gene set had a nominal p-value of 0.003. Among the members of the MYC-hallmark data set, the eukaryotic translation initiation factor 4A1 (EIF4A1) was overexpressed (FC = 2.7, p = 0.007). The RNA helicase eIF4A stimulates the translation of messenger RNAs with secondary structures on the untranslated 5′ region, which includes the translation of potent proteins for proliferation, angiogenesis, and survival such as cyclin D1, c-Myc, surviving, vascular endothelial growth factor (VEGF), and Bcl-2.19,20
Other overexpressed members of the MYC-hallmark data set were the Ras-related protein (RAN; FC = 2.42, p = 0.001), which is necessary for survival and mitosis of cancer cells, and the cell-division cycle protein 20 (CDC20; FC = 2.3, p = 0.021). CDC20 is the mitotic coactivator of the anaphase-promoting complex/cyclosome that once activated triggers anaphase entry during mitosis.21 It has been reported that p53 protein inhibits tumor cell growth through an indirect regulation of CDC20 and upregulation of the latter has been observed in a number of tumors.22 The upregulation of the metastatic suppressor (NME1; FC = 1.72, p = 0.02) observed on the microcystic LM suggests a reduction on cell spreading and motility.23 The largest upregulation was observed on the gene type 1 keratin 17 (KRT17; FC = 6.9, p = 0.05), which plays an important role in wound healing and its overexpression is related to tumor progression in epithelial ovarian cancer.24
Macrocystic
The gene ontology for biological processes results showed a highly significant oxidation–reduction (GO:005514) and generation of precursor metabolites and energy (GO:0006091) (p < 1.4 × 10−11). Other relevant terms were related to cell adhesion (GO:007155 and GO:0022610) with p-values <0.02. The GSEA highlighted the following hallmark lists: epithelial–mesenchymal transition, adipogenesis, fatty acid metabolism, oxidative phosphorylation, and hypoxia.
Even though the master regulator to the cellular response to hypoxia-1 was not present in our lists, other upregulated genes such as the alpha-enolase-1 (FC = 2.14, p = 0.003) and the macrophage migration inhibitory factor (FC = 0.02, p = 0.04) suggested the presence of a hypoxic milieu.25 The upregulation of the VEGF-B (FC = 2.36, p = 0.005), and the angiopoietin-like 2 (ANGPTL2; FC = 6.02, p = 0.001) points out angiogenic events and sprout formation.
Other angiogenic processes on the macrocystic LMs can be derived from the microfibril-associated glycoprotein 2 (FC = 4.56, p = 0.02) that promotes cell sprouting.26 Another highly upregulated gene was cyclase-associated protein 2 (FC = 5.06, p = 0.003) that regulates the actin cytoskeleton and—under hypoxic conditions—promotes cell migration, proliferation, and contraction in vascular cells.27 Angiogenesis on endothelial cells seems to be stimulated through the transforming growth factor (TGF-β1) pathway from the extracellular matrix protein dermatopontin (FC = 8, p = 0.002), or from the proteoglycan lumican (FC = 2.83, p = 0.04). It is worth mentioning that one of the largest overexpressed genes on the macrocystic LMs was the complement factor D (FC = 8.11, p = 0.012), but it did not show in any of our functional lists; however, its dramatic upregulation suggests the activation of the alternative pathway of the complement that can damage the vasculature.28
Conclusions
LMs are localized lesions that consist of dilated lymphatic vessels and sacs separated by fibrous septae but disconnected from the normal lymphatic system. Current LMs classification is based on locules dimensions, namely LMs are microcystic if locules are <1 to 2 cm, and macrocystic if locules are >2 cm. Despite the fact that there is abundant literature on genetic mechanisms of angiogenesis and lymphangiogenesis, little is known about their mechanisms of action in LMs. In this study, we sought the transcriptomic differences between a group of patients with micro and macro LMs in comparison with healthy controls through Illumina microarrays.
Indirect differential expression analysis revealed that 426 genes are significantly different between microcystic and macrocystic LMs having a common healthy control, which suggest that both LMs have distinctive mechanisms of development and action. We conducted functional analysis to find putative pathways related to each of the genes overexpressed for each LMs, followed by literature search to find possible roles of those genes for the development of LMs.
In microcystic LMs, antiapoptotic and proliferative mechanisms mimic several oncogenic pathways such as the stimulation for translation of c-Myc, VEGFs, and others growth factors through the upregulation of E1F4A1, and the regulation of mitotic activators (e.g., CDC20 and RAN). Another highly overexpressed gene included KRT17 that has been associated with the development of ovarian cancer.
These findings may elucidate the infiltrative nature, historic recurrence rate, and resistance to sclerotherapy observed on microcystic malformations. Also, the upregulation of NME1, which reduces cell spread in melanoma cell due to the formation of fibronectin deposits, may explain the condensed and fibrotic nature of microcystic lesions compared with the large vesicles and lumen present in the macrocystic counterparts. It is worth mentioning that the matrix metalloproteinase-9 was overexpressed on the microcystic LMs, possibly due to the downregulation of the tissue inhibitor of metalloproteinase-2. The continuous activation of proteases dissolves the extra cellular matrix by releasing proangiogenic factors and influences endothelial cell migration and invasion.
As opposed to the largely oncogenic processes shown in microcystic LMs, analysis of macrocystic malformations showed upregulation of processes related to cellular respiration, epithelial–mesenchymal transition, and adaptation to hypoxia. Specific gene products of interest within this data set are TGF-β3, VEGF-B, and ANGPTL2 that are involved in myriad processes of cell differentiation, embryogenesis, cell adhesion, and antiapoptotic events that adversely impact normal lymphatic development. Abnormal regulation of TGFs is associated with genetic syndromes and may suggest that macrocystic LMs result from a failure in appropriate regulation and pruning of the developing lymphatic networks. Also, the overexpression angiogenic factors (e.g., VEGFs) may represent some form of autocrine stimulation or a response to hypoxic microenvironments created from abnormal and sequestered lymphatic channels.
Hypoxic events highlighted by our analysis may also be seen as a consequence of the adaptation to such microenvironmental milieu or the response to angiogenic autocrine signaling. The local lack of oxygen may contribute to the formation of the large lumen seen with this malformation due to their influence on endothelial growth, migration, and remodeling of the extracellular matrix.
Regardless of the direct impact of the specific factors highlighted in our gene enrichment analysis, we showed that the transcriptomes of microcystic and macrocystic LMs contain large differences. As such, two very similar developmental malformations, characterized histologically, can be considered as unique clinical entities. This may lead to further consideration of genomic markers for identification and, ultimately, treatment of these lesions.
It is important to note that 11 genes from the PI3K/AKT pathway were differentially expressed (gene names are in green in Fig. 3 plus VEGF-B) but no difference in expression of PIK3CA was found. Given the relevance of PIK3CA on LMs,12–14 it would be pertinent to search for mutations on genes from PI3K/AKT pathway that may explain the observed differences on gene expression between macrocystic and microcystic LMs.
Weaknesses of this project involve the small sample size that was used for the bioinformatics analyses and the difficulty in harvesting pure lymphatic channels from surgical specimens. However, we plan to address this limitation in the future by increasing the sample size, isolating lymphatic vessels, and incorporating other sequencing technologies to further investigate the genetic or epigenetic drivers that make these two LMs so different.
Supplementary Material
Acknowledgments
The authors thank the Liam's Land Grant for their generous donation and dedication to finding a cure for LMs. This project was partially supported by the COBRE Center for Translational Pediatric Research P20GM121293.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Defnet AM, Bagrodia N, Hernandez SL, Gwilliam N, Kandel JJ. Pediatric lymphatic malformations: Evolving understanding and therapeutic options. Pediatr Surg Int 2016; 32:425–433 [DOI] [PubMed] [Google Scholar]
- 2.Perkins JA, Manning SC, Tempero RM, et al. . Lymphatic malformations: Review of current treatment. Otolaryngol Head Neck Surg 2010; 142:795–803, 803.e1 [DOI] [PubMed] [Google Scholar]
- 3.Perkins JA, Manning SC, Tempero RM, et al. . Lymphatic malformations: Current cellular and clinical investigations. Otolaryngol Head Neck Surg 2010; 142:789–794 [DOI] [PubMed] [Google Scholar]
- 4.Richter GT, Friedman AB. Hemangiomas and vascular malformations: Current theory and management. Int J Pediatr 2012; 2012:645678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagrodia N, Defnet AM, Kandel JJ. Management of lymphatic malformations in children. Curr Opin Pediatr 2015; 27:356–363 [DOI] [PubMed] [Google Scholar]
- 6.Swetman GL, Berk DR, Vasanawala SS, Feinstein JA, Lane AT, Bruckner AL. Sildenafil for severe lymphatic malformations. N Engl J Med 2012; 366:384–386 [DOI] [PubMed] [Google Scholar]
- 7.Mulliken JB, Glowacki J. Hemangiomas and vascular malformations in infants and children: A classification based on endothelial characteristics. Plast Reconstr Surg 1982; 69:412. [DOI] [PubMed] [Google Scholar]
- 8.Vogel AM, Warner BW. Toward an understanding of lymphatic malformations. Gastroenterol Hepatol (N Y) 2013; 9:195–196 [PMC free article] [PubMed] [Google Scholar]
- 9.Smith RJ. Lymphatic malformations. Lymphat Res Biol 2004; 2:25–31 [DOI] [PubMed] [Google Scholar]
- 10.Alomari AI, Karian VE, Lord DJ, Padua HM, Burrows PE. Percutaneous sclerotherapy for lymphatic malformations: A retrospective analysis of patient-evaluated improvement. J Vasc Interv Radiol 2006; 17:1639–1648 [DOI] [PubMed] [Google Scholar]
- 11.Chen EY, Hostikka SL, Oliaei S, Duke W, Schwartz SM, Perkins JA. Similar histologic features and immunohistochemical staining in microcystic and macrocystic lymphatic malformations. Lymphat Res Biol 2009; 7:75–80 [DOI] [PubMed] [Google Scholar]
- 12.Glaser K, Dickie P, Neilson D, Osborn A, Dickie BH. Linkage of metabolic defects to activated PIK3CA alleles in endothelial cells derived from lymphatic malformation. Lymphat Res Biol 2018; 16:43–55 [DOI] [PubMed] [Google Scholar]
- 13.Luks VL, Kamitaki N, Vivero MP, et al. . Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J Pediatr 2015; 166:1048e1–1054.e1–e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osborn AJ, Dickie P, Neilson DE, et al. . Activating PIK3CA alleles and lymphangiogenic phenotype of lymphatic endothelial cells isolated from lymphatic malformations. Hum Mol Genet 2015; 24:926–938 [DOI] [PubMed] [Google Scholar]
- 15.Hahne F, Huber W, Gentleman R, Falcon S. Bioconductor case studies. New York: Springer; 2008 [Google Scholar]
- 16.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang DW, Sherman BT, Tan Q, et al. . DAVID bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 2007; 35:W169–W175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, et al. . Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res 2008; 68:631–634 [DOI] [PubMed] [Google Scholar]
- 20.Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci U S A 2013; 110:13339–13344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lischetti T, Nilsson J. Regulation of mitotic progression by the spindle assembly checkpoint. Mol Cell Oncol 2015; 2:e970484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kidokoro T, Tanikawa C, Furukawa Y, Katagiri T, Nakamura Y, Matsuda K. CDC20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene 2008; 27:1562–1571 [DOI] [PubMed] [Google Scholar]
- 23.Horak CE, Lee JH, Elkahloun AG, et al. . Nm23-H1 suppresses tumor cell motility by down-regulating the lysophosphatidic acid receptor EDG2. Cancer Res 2007; 67:7238–7246 [DOI] [PubMed] [Google Scholar]
- 24.Wang YF, Lang HY, Yuan J, et al. . Overexpression of keratin 17 is associated with poor prognosis in epithelial ovarian cancer. Tumour Biol 2013; 34:1685–1689 [DOI] [PubMed] [Google Scholar]
- 25.Ji RC. Hypoxia and lymphangiogenesis in tumor microenvironment and metastasis. Cancer Lett 2014; 346:6–16 [DOI] [PubMed] [Google Scholar]
- 26.Albig AR, Roy TG, Becenti DJ, Schiemann WP. Transcriptome analysis of endothelial cell gene expression induced by growth on matrigel matrices: Identification and characterization of MAGP-2 and lumican as novel regulators of angiogenesis. Angiogenesis 2007; 10:197–216 [DOI] [PubMed] [Google Scholar]
- 27.Kosmas K, Eskandarnaz A, Khorsandi AB, et al. . CAP2 is a regulator of the actin cytoskeleton and its absence changes infiltration of inflammatory cells and contraction of wounds. Eur J Cell Biol 2015; 94:32–45 [DOI] [PubMed] [Google Scholar]
- 28.Hertle E, Arts IC, van der Kallen CJ, et al. . The alternative complement pathway is longitudinally associated with adverse cardiovascular outcomes. The CODAM study. Thromb Haemost 2016; 115:446–457 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.